Abstract
Mammals diversified by colonizing drastically different environments, with each transition yielding numerous molecular changes including losses of protein function. While not initially deleterious, these losses could subsequently carry deleterious pleiotropic consequences. Here we use phylogenetic methods to identify convergent functional losses across independent marine mammal lineages. In one extreme case, Paraoxonase 1 (PON1) accrued lesions in all marine lineages, while remaining intact in all terrestrial mammals. These lesions coincide with PON1 enzymatic activity loss in marine species’ blood plasma. This convergent loss is likely explained by parallel shifts in marine ancestors’ lipid metabolism and/or bloodstream oxidative environment affecting PON1’s role in fatty acid oxidation. PON1 loss also eliminates marine mammals’ main defense against neurotoxicity from specific man-made organophosphorus compounds, implying potential risks in modern environments.
One Sentence Summary:
Organophosphate toxicity may threaten modern marine mammals due to their ancestors’ repeated loss of PON1 for oxidative or metabolic reasons.
As the ancestors of aquatic marine mammals adopted obligate aquatic lifestyles, they evolved many adaptive changes, such as those that improved locomotion in and perception of their new environment (1, 2). Many of these morphological and physiological changes occurred in parallel in distinct lineages of marine mammals, including cetaceans, pinnipeds, and sirenians. Although convergent trait changes are frequently adaptive, environmental transitions can also result in non-adaptive convergent trait loss due to release from functional constraint. Examples of convergently reduced or lost traits include olfaction in marine mammals (3–5), bitter taste receptors in carnivorous tetrapods (6), and eyes in subterranean species (7–9). Any convergent evolutionary change in the context of a given environment can carry negative consequences in a different environment as a result of pleiotropy (one genetic locus influencing multiple phenotypes).
To characterize how mammals responded to selective pressures imposed by the marine environment, we identified genes that convergently lost function in marine mammals. We identified candidate pseudogenes with observed early stop codons and/or frameshifts (genetic lesions) in 58 eutherian mammals’ genomes in a 100-way vertebrate alignment (http://genome.ucsc.edu/). Using our predicted pseudogene calls, we then tested, for each gene, whether its pattern of functional loss was better explained by a model with one loss rate throughout the mammalian phylogeny or by a model in which the loss rate was dependent upon the terrestrial/marine state of a given branch, using a likelihood ratio test (LRT) (10). To ensure that our results were not strongly influenced by errors in pseudogene calling, we performed manual checks of lesion calls against reference genomes for our top genes, along with comparisons of pseudogene calls at highly conserved genes for marine and terrestrial species (11). We used simulations to estimate empirical gene-specific P-values and study-wide (multiple-test-corrected) false discovery rates (FDR) for all genes (11) (Tables 1 and S1). The set of genes with the strongest evidence for a higher loss rate on marine lineages was strongly enriched for functions related to chemosensation, driven by many olfactory and taste receptors (Tables S2–S5). These results are consistent with previous behavioral, anatomical, and genetic studies indicating a reduction of smell and taste in marine mammals (4, 12, 13).
Table 1.
Top 10 manually validated genes with evidence for marine-specific loss
| Gene | Loss rate (independent) | Marine loss rate (dependent) | Terrestrial loss rate (dependent) | LRT statistic | Empirical P-value | FDR | Description |
|---|---|---|---|---|---|---|---|
| PON1 | 0.672 | 49.7 | 0 | 22.24 | 3.08 × 10−6 | 0.0154 | paraoxonase 1 |
| ORIOZI | 1.15 | 100 | 0.467 | 19.99 | 7.25 × 10−6 | 0.0201 | olfactory receptor |
| OR8D4 | 1.25 | 100 | 0.510 | 19.21 | 1.60 × 10−5 | 0.0201 | olfactory receptor |
| TAS2R1 | 1.32 | 100 | 0.535 | 19.20 | 1.60 × 10−5 | 0.0201 | taste receptor |
| OR1F2P | 2.03 | 100 | 1.18 | 15.86 | 5.40 × 10−5 | 0.0831 | olfactory receptor |
| GSTM1 | 1.48 | 100 | 0.762 | 15.82 | 3.90 × 10−5 | 0.0831 | glutathione S-transferase mu 1 |
| OR6K2 | 2.02 | 100 | 1.22 | 15.79 | 4.50 × 10−5 | 0.0831 | olfactory receptor |
| OR51D1 | 1.13 | 49.3 | 0.466 | 15.59 | 8.60 × 10−5 | 0.0831 | olfactory receptor |
| TAAR5 | 1.17 | 48.2 | 0.484 | 15.16 | 9.90 × 10−5 | 0.0936 | trace amine associated receptor 5 |
| OR4C13 | 1.77 | 100 | 0.915 | 14.88 | 7.00 × 10−5 | 0.0972 | olfactory receptor |
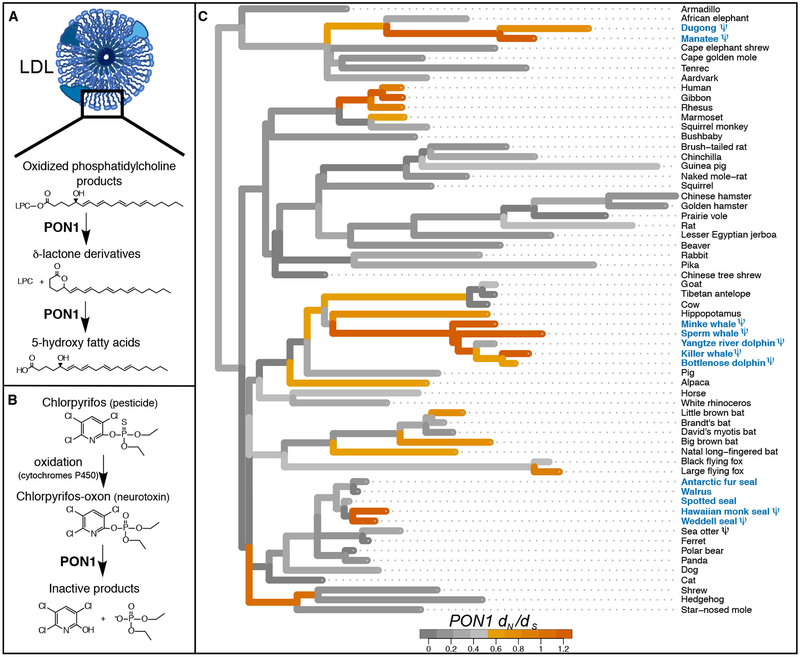
We also observed a striking pattern of convergent loss in the marine environment at Paraoxonase 1 (PON1) (Table 1) (11). PON1 encodes a bloodstream enzyme that reduces oxidative damage to lipids in low- and high-density lipoprotein particles (LDL and HDL, respectively), potentially preventing atherosclerotic plaque formation (14, 15) (Fig. 1A). PON1 also hydrolyzes the oxon forms of specific organophosphate compounds, such that it is the main line of defense against some man-made pesticide byproducts, including chlorpyrifos oxon and diazoxon (Fig. 1B) (16). The PON1 coding sequence contains genetic lesions in the cetacean, pinniped, and sirenian lineages but is intact in all 53 terrestrial mammal genomes surveyed (Fig. 1C; Table S1).
Fig. 1. PON1 functions and evolutionary history.
Illustration of PON1’s proposed roles in (A) preventing oxidative damage to low- and high-density lipoproteins (14, 15) and (B) detoxifying the oxon byproduct/metabolite of a common organophosphorus pesticide, chlorpyrifos (25). (C) Evolutionary rate of PON1 coding sequence across the phylogeny of 62 eutherian mammals. Branch lengths represent dN, and colors represent dN/dS (see color legend). dN/dS values greater than 1.2 were set to 1.2. Blue: marine species. ψ: genetic lesion(s) present.
To estimate when PON1 function was lost in the three marine mammal clades, we obtained PON1 sequences for 14 additional species including three cetaceans, the dugong, and two pinnipeds, and we estimated evolutionary rates across the mammalian phylogeny (11) (Figs. 1C and S1). We observed shared genetic lesions among all sequenced cetaceans and a different shared lesion in sirenians (Fig. S2), and the inferred ratio of non-synonymous-to-synonymous substitutions (dN/dS) was not significantly different from one on the ancestral branches of both clades (cetacean ancestor dN/dS = 1.09, P = 0.79; sirenian ancestor dN/dS = 1.20, P = 0.57). This suggests that PON1 lost functional constraint in the ancestral cetacean lineage soon after its split with the ancestral hippopotamid lineage, approximately 53 MYA (95% confidence interval lower bound: 34.5 MYA) (11, 17). In sirenians, functional loss occurred soon after their split with the ancestral elephantid lineage, approximately 64 MYA (lower bound 41.7 MYA) (17).
In pinnipeds, we observed clear evidence of PON1 functional loss only among a subset of species within family Phocidae, wherein Weddell seal and Hawaiian monk seal PON1 sequences contained non-shared genetic lesions (Fig. S2). Because these branches are short, it is difficult to estimate precisely when functional loss occurred in pinnipeds; however, there was likely at least one loss since the Phocidae:Otarioidea split approximately 21 MYA (95% CI: 0 – 21 MYA). This incomplete loss could reflect either a difference between the selective environments experienced by pinnipeds and those experienced by other marine mammals, or it could reflect pinnipeds’ more recent colonization of the marine environment (pinnipeds: 24 MYA, cetaceans: 44.7 – 37.3 MYA, sirenians: 47.1 – 43.9 MYA) (18).
PON1’s functional loss in marine mammals may be related to its role in lipid metabolism via fatty acid beta-oxidation (19) (Tables S6 and S7). Compared to their terrestrial relatives, the diets of both herbivorous and carnivorous aquatic mammals contain a higher proportion of w-3 relative to ω−6 polyunsaturated fatty acids (PUFAs) (20), and these PUFAs differ in their capacity to sustain oxidative damage (21). Marine and terrestrial mammals also have vastly different antioxidant profiles (22, 23), presumably due to the extreme oxidative stress experienced during diving, with repeated cycles of hypoxia and reperfusion. Rewiring of either lipid metabolism or antioxidant networks in ancient marine mammals could have obviated the function of PON1. Supporting the antioxidant hypothesis, the Weddell seal, which carries PON1 lesions, is one of the longest diving pinnipeds known, in contrast to the shorter diving walrus and Antarctic fur seal, which lack lesions but share an aquatic diet (24). However, two semi-aquatic mammals, the sea otter and the beaver, which are more moderate divers (24), also have either lesions or substitutions at sites predicted to be necessary for PON1 function (Fig. S2; Table S8).
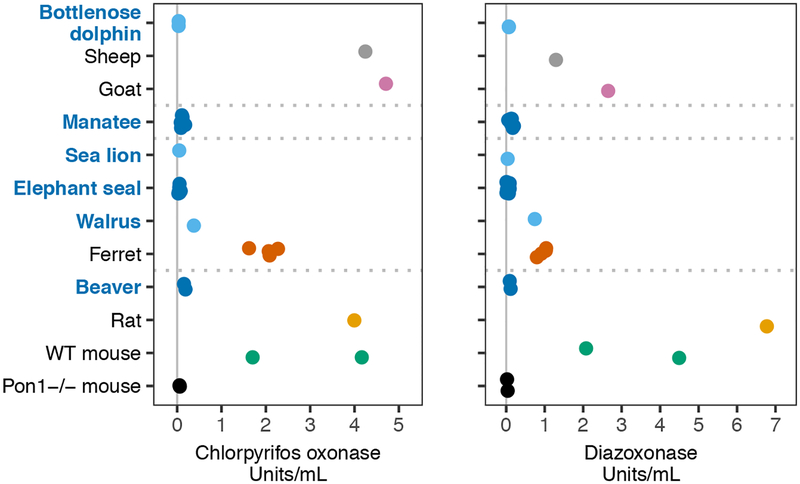
Whatever the cause, loss of PON1 function could carry negative pleiotropic consequences for the health of marine mammals repeatedly exposed to man-made organophosphate compounds. PON1 alone is protective against the highly toxic oxon forms of the heavily used pesticides chlorpyrifos and diazinon; these oxons are formed from the parent compounds in the environment and in vivo by cytochromes P450 (25) (Fig. 1B). We tested blood plasma from six marine and semi-aquatic species for capacity to hydrolyze these and other PON1 substrates (Figs. 2 and S3). The plasma from all but one of the assayed marine and semi-aquatic species showed activity levels against the PON1 substrates that more closely resembled those of the Pon1 knockout (Pon1−/−) mouse than those of terrestrial outgroups. Thus, the genetic deterioration of PON1 has left these species without a mechanism to break down specific neurotoxic compounds.
Fig. 2. Blood plasma enzymatic activity against two organophosphate-derived substrates.
Points represent rates of hydrolysis of chlorpyrifos oxon (left) or diazoxon (right) in μmol/min/mL for plasma from marine and semi-aquatic species (in blue) and terrestrial outgroups. Values for sheep, goat, and rat are from Furlong et al. (33), who performed assays under the same experimental conditions as in this study. Control assays of alkaline phosphatase activity show samples were not degraded (Fig. S3).
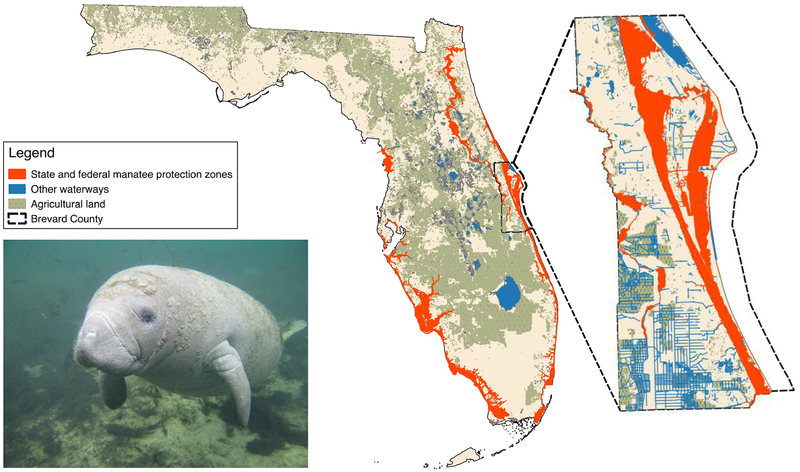
Given the sensitivity of Pon1−/− mice to organophosphate exposure (26), the inability of most marine mammal plasma to detoxify organophosphates suggests the potential for neurotoxicity if sufficient levels of these compounds accumulate in these animals’ habitats or food sources. In Florida, agricultural use of organophosphate pesticides is common, and runoff can drain into manatee habitats. In Brevard County, where an estimated 70% of Atlantic Coast manatees migrate or seasonally reside (27, 28), agricultural lands frequently abut manatee protection zones and waterways (Fig. 3). Limited sampling upstream of Manatee Bay has measured levels of chlorpyrifos as high as 0.023 μg/L (29), and levels could be much higher directly after pesticide applications (30). Dugongs may be at risk of exposure to organophosphorus pesticides that are used in the sugar cane industry along the Queensland coast and have been detected at 5 – 270 pg/L in coastal river systems (31). Carnivorous marine mammals may also ingest these compounds through their diets of invertebrates and fish, which have shown evidence of bioaccumulation of organophosphates in Arctic populations (32). In order to improve our understanding of the extent of exposure and attendant risk marine mammals face, we recommend increased monitoring of marine mammal habitats, as well as the testing of tissues from deceased animals for biomarkers of organophosphate exposure.
Fig. 3. Manatee and adjacency of its habitat to agricultural land use.
Left: Florida manatee (photo by Robert K. Bonde, 2006). Center: Manatee protection zones and agricultural land in Florida. Right: Manatee protection zones, waterways, and agricultural land in Brevard County.
The presence of these potential risks to many marine mammals due to their loss of PON1 function provides a clear example of the tradeoffs possible in evolution: although PON1 functional loss was not deleterious and may even have been beneficial in ancestral marine environments, it may carry detrimental fitness consequences in modern environments.
Supplementary Material
Acknowledgments:
We would like to thank Brandon Small for assistance with molecular work; Kelly Goulet and Vanessa Fravel for supplying marine mammal samples; Seema Lakdawala and the Lakdawala lab for supplying ferret samples; and Aldons Lusis, Diana Shih, and Aaron Tward for supplying Pon1 knockout mice; as well as all members of the Clark and Chikina labs and Kyle Dolan for feedback.
Funding: This study was funded by NIH grants R01HG009299 and U54 HG008540 to NLC and MC. AK was supported by NIH T32 training grant T32 EB009403 as part of the HHMI-NIBIB Interfaces Initiative. Dugong samples were collected with funds from the Winifred Violet Scott Foundation and the Sea World Research and Rescue Foundation. The collection of manatee samples was funded by the U.S. Geological Survey.
Footnotes
Competing interests: The authors declare no competing financial interests.
Data and materials availability: The data reported in this paper are tabulated in the Supplementary Materials. Resequencing data for PON1 coding sequence in dugong is available in GenBank (accession MF197755). Scripts used in analyses are available at https://github.com/nclark-lab/MarineFxLoss. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
References and Notes:
- 1.Fish FE, Howle LE, Murray MM, Hydrodynamic flow control in marine mammals. Integr. Comp. Biol 48, 788–800 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Wartzok D, Ketten DR, in Biology of Marine Mammals, Reynolds J, Rommel S, Eds. (Smithsonian Institution Press, Washington DC, 1999), pp. 117–175. [Google Scholar]
- 3.McGowen M, Clark C, Gatesy J, The Vestigial Olfactory Receptor Subgenome of Odontocete Whales: Phylogenetic Congruence between Gene-Tree Reconciliation and Supermatrix Methods. Syst. Biol 57, 574–590 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Bills ML, thesis, University of Florida (2011). [Google Scholar]
- 5.Chikina M, Robinson JD, Clark NL, Hundreds of Genes Experienced Convergent Shifts in Selective Pressure in Marine Mammals. Mol. Biol. Evol 33, 2182–92 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li D, Zhang J, Diet shapes the evolution of the vertebrate bitter taste receptor gene repertoire. Mol. Biol. Evol 31, 303–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Protas M, Conrad M, Gross JB, Tabin C, Borowsky R, Regressive evolution in the Mexican cave tetra, Astyanax mexicanus. Curr. Biol 17, 452–454 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeffery WR, Regressive evolution in Astyanax cavefish. Annu. Rev. Genet 43, 25–47 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Partha R et al. , Subterranean mammals show convergent regression in ocular genes and enhancers, along with adaptation to tunneling. Elife 6, e25884 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pagel M, Meade A, BayesTraits (2013). [Google Scholar]
- 11.Materials and methods are available as supplementary materials at the Science website.
- 12.Marino L, Cetacean brains: how aquatic are they? Anat. Rec. (Hoboken) 290, 694–700 (2007). [DOI] [PubMed] [Google Scholar]
- 13.McGowen MR, Gatesy J, Wildman DE, Molecular evolution tracks macroevolutionary transitions in Cetacea. Trends Ecol. Evol 29, 336–346 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Rosenblat M, Aviram M, Paraoxonases role in the prevention of cardiovascular diseases. BioFactors 35, 98–104 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Mackness MI, Arrol S, Durrington PN, Paraoxonase prevents accumulation of lipoperoxides in low-density lipoprotein. FEBS Lett 286, 152–4 (1991). [DOI] [PubMed] [Google Scholar]
- 16.Li WF et al. , Catalytic efficiency determines the in-vivo efficacy of PON1 for detoxifying organophosphorus compounds. Pharmacogenetics 10, 767–79 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Meredith RW et al. , Impacts of the cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science 334, 521–524 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Thewissen J, Nummela S, in Sensory Evolution on the Threshold: Adaptations in Secondarily Aquatic Vertebrates, Thewissen J, Nummela S, Eds. (University of California Press, Berkeley and Los Angeles, California, 2008), pp. 1–28. [Google Scholar]
- 19.Furlong CE, Marsillach J, Jarvik GP, Costa LG, Paraoxonases-1, −2 and −3: What are their functions? Chem. Biol. Interact 259, 51–62 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koussoroplis A-M et al. , From Aquatic to Terrestrial Food Webs: Decrease of the Docosahexaenoic Acid/Linoleic Acid Ratio. Lipids 43, 461–466 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Miyashita K, Nara E, Ota T, Oxidative Stability of Polyunsaturated Fatty Acids in an Aqueous Solution. Biosci. Biotechnol. Biochem 57, 1638–1640 (1993). [Google Scholar]
- 22.Vázquez-Medina JP, Zenteno-Savín T, Elsner R, Ortiz RM, Coping with physiological oxidative stress: a review of antioxidant strategies in seals. J. Comp. Physiol. B 182, 741–750 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cantú-Medellín N, Byrd B, Hohn A, Vázquez-Medina JP, Zenteno-Savín T, Differential antioxidant protection in tissues from marine mammals with distinct diving capacities. Shallow/short vs. deep/long divers. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol 158, 438–443 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Mirceta S et al. , Evolution of Mammalian Diving Capacity Traced by Myoglobin Net Surface Charge. Science 340, 1234192–1234192 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Furlong CE, Genetic variability in the cytochrome P450-paraoxonase 1 (PON1) pathway for detoxication of organophosphorus compounds. J. Biochem. Mol. Toxicol 21, 197–205 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Shih DM et al. , Mice lacking serum paraoxonase are susceptible to organophosphate toxicity and atherosclerosis. Nature 394, 284–287 (1998). [DOI] [PubMed] [Google Scholar]
- 27.Deutsch CJ et al. , Seasonal movements, migratory behavior, and site fidelity of West Indian manatees along the Atlantic coast of the United States. Wildl. Monogr, 1–77 (2003). [Google Scholar]
- 28.Martin J et al. , Combining information for monitoring at large spatial scales: first statewide abundance estimate of the Florida manatee. Biol. Conserv 186, 44–51 (2015). [Google Scholar]
- 29.Carriger JF, Rand GM, Aquatic risk assessment of pesticides in surface waters in and adjacent to the Everglades and Biscayne National Parks: I. Hazard assessment and problem formulation. Ecotoxicology 17, 660–679 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Aguirre-Rubí JR et al. , Chemical contamination assessment in mangrove-lined Caribbean coastal systems using the oyster Crassostrea rhizophorae as biomonitor species. Environ. Sci. Pollut. Res. Int (2017), doi: 10.1007/s11356-017-9159-2. [DOI] [PubMed] [Google Scholar]
- 31.Shaw M et al. , Monitoring pesticides in the Great Barrier Reef. Mar. Pollut. Bull 60, 113–22 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Morris AD et al. , Current-use pesticides in seawater and their bioaccumulation in polar bear-ringed seal food chains of the Canadian Arctic. Environ. Toxicol. Chem 35, 1695–1707 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Furlong CE et al. , Genetic and temporal determinants of pesticide sensitivity: role of paraoxonase (PON1). Neurotoxicology 21, 91–100. [PubMed] [Google Scholar]
- 34.Felsenstein J, PHYLIP (Phylogeny Inference Package) version 3.6 (2005).
- 35.Gouy M, Guindon S, Gascuel O, SeaView Version 4: A Multiplatform Graphical User Interface for Sequence Alignment and Phylogenetic Tree Building. Mol. Biol. Evol 27, 221–224 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ, Basic local alignment search tool. J. Mol. Biol 215, 403–10 (1990). [DOI] [PubMed] [Google Scholar]
- 37.Kent WJ et al. , The human genome browser at UCSC. Genome Res 12, 996–1006 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parra G, Bradnam K, Ning Z, Keane T, Korf I, Assessing the gene space in draft genomes. Nucleic Acids Res 37, 289–97 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parra G, Bradnam K, Korf I, CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23, 1061–7 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Mudunuri U, Che A, Yi M, Stephens RM, bioDBnet: the biological database network. Bioinformatics 25, 555–6 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Casper J et al. , The UCSC Genome Browser database: 2018 update. Nucleic Acids Res 46, D762–D769 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pagel M, Meade A, Bayesian Analysis of Correlated Evolution of Discrete Characters by Reversible-Jump Markov Chain Monte Carlo. Am. Nat 167, 808–825 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Harmon LJ, Weir JT, Brock CD, Glor RE, Challenger W, GEIGER: investigating evolutionary radiations. Bioinformatics 24, 129–31 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Tusher VG, Tibshirani R, Chu G, Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci 98, 5116–5121 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benjamini Y, Hochberg Y, Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 57 (1995), pp. 289–300. [Google Scholar]
- 46.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z, GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10, 48 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liberzon A et al. , Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith CL, Eppig JT, The mammalian phenotype ontology: enabling robust annotation and comparative analysis. Wiley Interdiscip. Rev. Syst. Biol. Med 1, 390–9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bininda-Emonds ORP et al. , The delayed rise of present-day mammals. Nature 446, 507–512 (2007). [DOI] [PubMed] [Google Scholar]
- 50.He K et al. , Talpid Mole Phylogeny Unites Shrew Moles and Illuminates Overlooked Cryptic Species Diversity. Mol. Biol. Evol 34, 78–87 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Douady CJ, Douzery EJP, Molecular estimation of eulipotyphlan divergence times and the evolution of “Insectivora”. Mol. Phylogenet. Evol 28, 285–96 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Bibi F, A multi-calibrated mitochondrial phylogeny of extant Bovidae (Artiodactyla, Ruminantia) and the importance of the fossil record to systematics. BMC Evol. Biol 13, 166 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cantalapiedra JL et al. , Dietary innovations spurred the diversification of ruminants during the Caenozoic. Proc. R. Soc. B Biol. Sci 281, 20132746–20132746 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fulton TL, Strobeck C, Molecular phylogeny of the Arctoidea (Carnivora): effect of missing data on supertree and supermatrix analyses of multiple gene data sets. Mol. Phylogenet. Evol 41, 165–81 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Flynn JJ, Finarelli JA, Zehr S, Hsu J, Nedbal MA, Molecular phylogeny of the Carnivora (Mammalia): assessing the impact of increased sampling on resolving enigmatic relationships. Syst. Biol 54, 317–37 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Yang Z, PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Paradis E, Claude J, Strimmer K, APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Guindon S et al. , New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol 59, 307–321 (2010). [DOI] [PubMed] [Google Scholar]
- 59.Seim I et al. , Genome analysis reveals insights into physiology and longevity of the Brandt’s bat Myotis brandtii. Nat. Commun 4, 2078–2079 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lok S et al. , De Novo Genome and Transcriptome Assembly of the Canadian Beaver (Castor canadensis). G3 Genes, Genomes, Genet 7 (2017) (available at http://www.g3journal.org/content/7/2/755.full). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arnason U et al. , Pinniped phylogeny and a new hypothesis for their origin and dispersal. Mol. Phylogenet. Evol 41, 345–354 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Yim H-S et al. , Minke whale genome and aquatic adaptation in cetaceans. Nat. Genet 46, 88–92 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eckalbar WL et al. , Transcriptomic and epigenomic characterization of the developing bat wing. Nat. Genet 48, 528–536 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miller W et al. , Polar and brown bear genomes reveal ancient admixture and demographic footprints of past climate change. Proc. Natl. Acad. Sci. U. S. A 109, E2382–90 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jones S et al. , The Genome of the Northern Sea Otter (Enhydra lutris kenyoni). Genes (Basel) 8, 379 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou X et al. , Baiji genomes reveal low genetic variability and new insights into secondary aquatic adaptations. Nat. Commun 4, 2708 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Z, Schwartz S, Wagner L, Miller W, A Greedy Algorithm for Aligning DNA Sequences. J. Comput. Biol 7, 203–214 (2000). [DOI] [PubMed] [Google Scholar]
- 68.Humble E et al. , A draft fur seal genome provides insights into factors affecting SNP validation and how to mitigate them. Mol. Ecol. Resour 16, 909–921 (2016). [DOI] [PubMed] [Google Scholar]
- 69.Kent WJ, BLAT--the BLAST-like alignment tool. Genome Res 12, 656–64 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gao X, Han J, Lu Z, Li Y, He C, De novo assembly and characterization of spotted seal Phoca largha transcriptome using Illumina paired-end sequencing. Comp. Biochem. Physiol. Part D Genomics Proteomics 8, 103–110 (2013). [DOI] [PubMed] [Google Scholar]
- 71.Sedlazeck FJ, Rescheneder P, von Haeseler A, NextGenMap: fast and accurate read mapping in highly polymorphic genomes. Bioinformatics 29, 2790–2791 (2013). [DOI] [PubMed] [Google Scholar]
- 72.Li H et al. , The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Higdon JW et al. , Phylogeny and divergence of the pinnipeds (Carnivora: Mammalia) assessed using a multigene dataset. BMC Evol. Biol 7, 216 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fabre P-H et al. , A glimpse on the pattern of rodent diversification: a phylogenetic approach. BMC Evol. Biol 12, 88 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Agnarsson I, Zambrana-Torrelio CM, Flores-Saldana NP, May-Collado LJ, A time-calibrated species-level phylogeny of bats (Chiroptera, Mammalia). PLoS Curr (2011), doi: 10.1371/currents.RRN1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McGowen MR, Spaulding M, Gatesy J, Divergence date estimation and a comprehensive molecular tree of extant cetaceans. Mol. Phylogenet. Evol 53, 891–906 (2009). [DOI] [PubMed] [Google Scholar]
- 77.Yang Z, PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci 13, 555–6 (1997). [DOI] [PubMed] [Google Scholar]
- 78.Meredith RW, Gatesy J, Murphy WJ, Ryder OA, Springer MS, Molecular Decay of the Tooth Gene Enamelin (ENAM) Mirrors the Loss of Enamel in the Fossil Record of Placental Mammals. PLoS Genet 5, e1000634 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bustamante CD, Nielsen R, Hartl DL, A Maximum Likelihood Method for Analyzing Pseudogene Evolution: Implications for Silent Site Evolution in Humans and Rodents. Mol. Biol. Evol 19, 110–117 (2002). [DOI] [PubMed] [Google Scholar]
- 80.Stamper MA, Bonde RK, in Sirenian Conservation: Issues and Strategies in Developing Countries, Hines EM, Reynolds JEI, Aragones LV, Mignucci-Giannoni AA, Marmontel M, Eds. (University Press of Florida, Gainesville, FL, 2012), pp. 139–147. [Google Scholar]
- 81.Bonde RK et al. , Biomedical health assessments of the Florida manatee in Crystal River - providing opportunities for training during the capture, handling, and processing of this endangered aquatic mammal. J. Mar. Anim. Their Ecol 5, 17–28 (2012). [Google Scholar]
- 82.Lanyon JM, Sneath HL, Long T, Bonde RK, Physiological Response of Wild Dugongs to Out-of-Water Sampling for Health Assessment. Aquat. Mamm 36, 46–58 (2010). [Google Scholar]
- 83.Lanyon JM et al. , A Method for Capturing Dugongs in Open Water. Aquat. Mamm 32, 196–201 (2006). [Google Scholar]
- 84.Se Fum Wong S et al. , A Simple Method for DNA Isolation from Clotted Blood Extricated Rapidly from Serum Separator Tubes. Clin. Chem 53, 522–524 (2007). [DOI] [PubMed] [Google Scholar]
- 85.Miller SA, Dykes DD, Polesky HF, A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16, 1215 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Clark NL, Aquadro CF, A novel method to detect proteins evolving at correlated rates: identifying new functional relationships between coevolving proteins. Mol. Biol. Evol 27, 1152–1161 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Richter RJ, Jarvik GP, Furlong CE, Paraoxonase 1 (PON1) status and substrate hydrolysis. Toxicol. Appl. Pharmacol 235, 1–9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Walter K, Schütt C, in Methods of Enzymatic Analysis, Bergmeyer H-U, Ed. (Academic Press Inc., New York, Vol. 2, 1974), pp. 860–4. [Google Scholar]
- 89.E. C. O. S. U.S. Fish and Wildlife Service, Manatee, West Indian (Trichechus manatus) Final Critical Habitat FCH_Trichechus_manatus_19770922 [Data set] (2010).
- 90.F. and W. R. I. Florida Fish and Wildlife Conservation Commission, State Manatee Protection Zones in Florida [Data set] (2016), (available at http://geodata.myfwc.com/datasets/a95041ab2f034955a693b7b69f8e2ee9_14).
- 91.T. products U.S. Census Bureau, 2016 TIGER/Line Shapefiles: Water: Linear Hydrography: Brevard County [Data set] (2016).
- 92.T. products U.S. Census Bureau, Cartographic Boundary Shapefiles - Counties 500k [Data set] (2017).
- 93.T. products U.S. Census Bureau, Cartographic Boundary Shapefiles – States 500k (2017).
- 94.F. D. of E. Protection, Lakes (areas) [Data set] (2002), (available at http://geodata.dep.state.fl.us/datasets/97b765ff2b70400d8bcab23fbe2a5e88_0).
- 95.D. of E. Protection, Statewide Land Use Land Cover [Data set] (2017), (available at http://geodata.dep.state.fl.us/datasets/2f0e5f9a180a412fbd77dc5628f28de3_3).
- 96.Subramanian A et al. , Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A 102, 15545–50 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Blake JA et al. , Mouse Genome Database (MGD)-2017: community knowledge resource for the laboratory mouse. Nucleic Acids Res 45, D723–D729 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Harel M et al. , Structure and evolution of the serum paraoxonase family of detoxifying and anti-atherosclerotic enzymes. Nat. Struct. Mol. Biol 11, 412–9 (2004). [DOI] [PubMed] [Google Scholar]
- 99.Ben-David M et al. , Catalytic Versatility and Backups in Enzyme Active Sites: The Case of Serum Paraoxonase 1. J. Mol. Biol 418, 181–196 (2012). [DOI] [PubMed] [Google Scholar]
- 100.Yeung DT, Lenz DE, Cerasoli DM, in The Paraoxonases: Their Role in Disease Development and Xenobiotic Metabolism, Mackness B, Mackness M, Aviram M, Paragh G, Eds. (Springer; Netherlands, 2008), pp. 151–170. [Google Scholar]
- 101.Josse D et al. , Identification of residues essential for human paraoxonase (PON1) arylesterase/organophosphatase activities. Biochemistry 38, 2816–25 (1999). [DOI] [PubMed] [Google Scholar]
- 102.Josse D, Xie W, Masson P, Lockridge O, Human serum paraoxonase (PON1): identification of essential amino acid residues by group-selective labelling and site-directed mutagenesis. Chem. Biol. Interact 119–120, 71–8 (1999). [DOI] [PubMed] [Google Scholar]
- 103.Yeung DT et al. , Structure/function analyses of human serum paraoxonase (HuPON1) mutants designed from a DFPase-like homology model. Biochim. Biophys. Acta 1702, 67–77 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.