Summary
Diabetes mellitus is caused by either loss of pancreatic islets β-cells (Type 1 Diabetes, T1D), insufficient insulin release in the islet β-cells coupled with insulin resistance in target tissues (Type 2 Diabetes, T2D), or impaired insulin release (genetic forms of diabetes and, possibly, T1D subtypes). The investigation of the islet proteome could elucidate facets of the pathogenesis of diabetes. Enzymatically isolated and cultured (EIC) islets are frequently used to investigate biochemical signaling pathways that could trigger β-cell changes and death in diabetes. However, they cannot fully reflect the natural protein composition and disease process of in vivo islets due to the stress from isolation procedures and in vitro culture. The laser capture microdissection method employs a high-energy laser source to separate the desired cells from the remaining tissue section in an environment which is well conserved and close to the natural condition. Here, we describe a label-free proteomic workflow of laser capture microdissected (LCM) human islets from fresh-frozen pancreas sections of cadaveric donors to obtain an accurate and unbiased profile of the pancreatic islet proteome. The workflow includes preparation of frozen tissue section, staining and dehydration, LCM islets collection, islet protein digestion, label free Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS), database search, and statistical analysis.
Keywords: LCM human pancreatic islets, Label-free proteomics, LC-MS/MS, MaxQuant, Perseus
1. Introduction
Diabetes mellitus is caused by either loss of pancreatic islets β-cells (Type 1 Diabetes, T1D), insufficient insulin release in the islet β-cells coupled with insulin resistance in target tissues (Type 2 Diabetes, T2D), or impaired insulin release (genetic forms of diabetes and, possibly, T1D subtypes). The investigation of the islet proteome could elucidate facets of the pathogenesis of diabetes. Enzymatically isolated and cultured (EIC) islets (1-3) have been frequently used to investigate biochemical signaling pathways that could trigger β-cell changes and death. However, such in vitro models have some limitations: they do not fully reflect what happens in vivo due to a lack of the natural environment where islets exist and due to the changes in cell physiology induced by isolation and culture. The procedure of enzymatic isolation of pancreatic islets causes major structural changes and induces up-regulation of stress-related genes in islets (4). Furthermore, EIC islets frequently contain a significant percentage of contaminating acinar cells and duct cells (5). Alternatively, human pancreatic tissue can be collected from cadaveric individuals and preserved frozen for further laser-capture microdissected (LCM) isolation. LCM employs a high-energy laser source to separate the desired cells from the remaining tissue section (6), a strategy that can minimize the contamination of surrounding tissue. LCM isolation also enables the extraction of samples from an environment which is well conserved and close to the natural condition, to better investigate cell physiology (7), cell biology (8), cell transcriptome (4), and proteome (9). The exploration of the proteome signature of LCM islets with an unbiased method may provide information on the changes of protein composition occurring in dysfunctional islets, even with limited sample amounts, which may facilitate understanding of the pathogenesis of diabetes.
Here we describe a workflow for label-free proteomic analysis of LCM islets obtained from sections of fresh-frozen human pancreas. This method enables accurate and unbiased profiling of the pancreatic islet proteome. The strategy avoids enzymatic treatment for cell dissociation and in vitro culture, and is designed to maintain protein composition close to that of the original tissue. The method can be easily adapted to other tissues, organs, and species. The workflow covers preparation of frozen tissue sections, immunohistochemical staining of reference sections, staining and dehydration for LCM, LCM of pancreatic islets and acinar tissue, preparation of samples for proteomic analysis, label free Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) data acquisition, database search for protein identification, quantification, and statistical analysis for the determination of the proteins differentially expressed between LCM islets and LCM acinar tissue.
2. Materials
Common solvents and reagents [acetic acid, dithiothreitol (DTT), iodoacetamide (IAA), Ammonium Bicarbonate (NH4HCO3), Hydrochloric acid (HCl), Formic Acid (FA), Acetonitrile (CH3CN)] were purchased from Sigma-Aldrich (St. Louis, MO). Tissue-Tek O.C.T. compound and 100% ethanol were purchased from VWR. Toluidine blue O and Anti-insulin antibody clone K36aC10 were purchased from Sigma-Aldrich. Leica polyethylene naphthalate (PEN) Membrane slides were purchased from Leica. PBS was purchased from Gibco Life Technologies. Kimwipes and Drierite were purchased from VWR. Peroxo-Block™ was purchased from Thermofisher Scientific. Histostain Plus Broadspectrum AEC kit was purchased from Invitrogen. Elite Mini PAP Pen was purchased from Diagnostic BioSystems (Pleasanton, CA). PPS Silent Surfactant was purchased from Expedeon (San Diego, CA). The BCA protein assay was obtained from ThermoFisher Scientific (Rockford, IL), the sequencing-grade Trypsin was purchased from Promega (Madison WI). All solvents used are HPLC-grade.
Instrumentation: cryotome (Leica Cryotome CM3050 S, Leica), histology slide scanner (PathScan Enabler IV, with workstation and PathScan Enabler software, Meyer Instruments), laser microdissection system (Leica Microscope LS LMD, with workstation and Leica LMD software, Leica), liquid chromatography and mass spectrometry system (UltiMate 3000 RSLCnano system and a Q Exactive HF mass spectrometer coupled with an EASY-Spray ion source, ThermoFisher Scientific). Details of reagents and materials used in each step are listed below.
2.1. For frozen tissue sections
Tissue-Tek Cryomold Standard.
Tissue-Tek O.C.T. compound.
100% ethanol.
Leica PEN Membrane slides.
2.2. Immunohistochemical staining of reference sections
Elite Mini PAP Pen.
10 mM phosphate-buffered saline (PBS) pH 7.4.
Anti-insulin antibody clone K36aC10 1:1000 dilution (see Note 1).
Ready-to-use biotinylated secondary antibody (from Histostain Plus Broadspectrum AEC kit).
HRP Substrate/Chromogen reagents: AEC Single Solution.
2.3. For LCM staining and dehydration
100% ethanol.
70% ethanol: combine 70mL ethanol and 30mL H2O.
90% ethanol: combine 90mL ethanol and 10mL H2O.
0.5% w/v Toluidine blue O staining solution was prepared in 70% ethanol (see Note 2).
Drierite desiccant.
2.4. For LCM of pancreatic islets and acinar tissue
50 mM NH4HCO3 pH 8: add 0.40 g of NH4HCO3 to 100 mL of H2O.
2.5. For protein digestion
Prepare 1% PPS by adding 100 μL 50 mM NH4HCO3 to 1mg PPS bottle (see Note 3).
50 mM dithiothreitol (DTT): weigh 0.77 mg of DTT in a microcentrifuge tube, and add 100 μL DI water (see Note 4).
50 mM iodoacetamide (IAA): weigh 0.925 mg of IAA in a microcentrifuge tube, and add 100 μL DI water (see Note 5).
Trypsin stock solution: prepare 1 μg/μL in 50 mM acetic acid, and store at −20°C before use.
2 M HCl: 16.52mL 37% HCl and add H2O to 100 mL.
2.6. For LC-MS/MS
Buffer A: 0.1% FA; Buffer B: 0.1% FA in CH3CN.
3. Methods
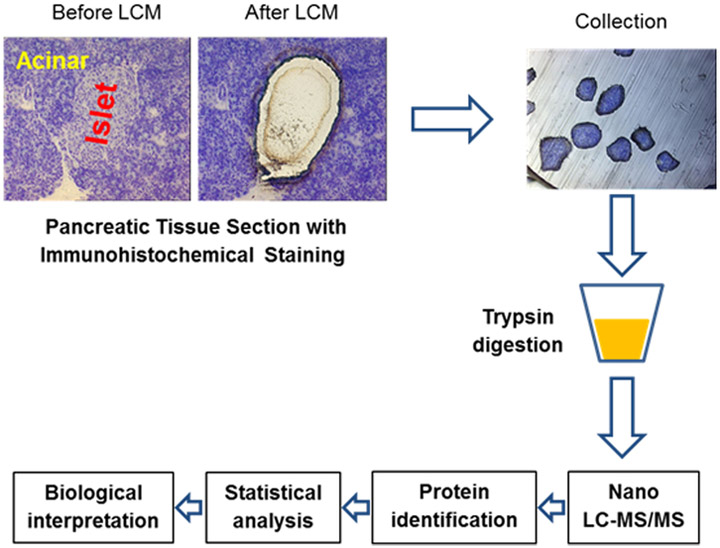
The major procedures involved in analysis of LCM human islets proteome is shown in Figure 1. The human pancreas tissue from three donors was used in this study. Three technical replicates of LCM islets were collected from each donor and 6 islet equivalent human pancreatic islets were collected from each replication. Meanwhile, the same equivalent acinar tissue was collected from surrounding of the same islets. Two technique replicates were collected for each donor. LCM acinar tissue was used to confirm no contamination in LCM islets.
Figure 1.
Schematic representation of the experimental work flow.
3.1. Preparation of frozen tissue sections
Exercise care while processing pancreatic tissue: avoid squeezing or stretching the tissue, use a scalpel to obtain blunt cuts.
Resect pancreatic tissue blocks from the neck region of cadaveric pancreata from organ donors.
Collect tissue fragments of approximately 1cm × 0.5cm and position fragments in the center of cryomolds.
Embed tissue in Tissue-Tek O.C.T. compound and immediately freeze at −80 °C by placing the mold holding the tissue on top of dry ice (see Note 6).
Cut the blocks into sections of 10 μm thickness with a cryotome, with temperature set at −20 °C (see Note 7).
Transfer three pancreas sections onto each of 10 Leica PEN Membrane slides.
Prepare extra reference sections on a regular glass slide for insulin immunohistochemistry and mapping (see Note 8).
3.2. Immunohistochemical staining of reference sections
Fix the reference sections in 10% formalin for 15min (see Note 9).
Wash 4 times in PBS.
Leave a drop of PBS on each tissue section and draw a circle with a PAP pen to surround each section.
Remove the PBS.
Add Peroxo-Block™ for 45 sec. Wash immediately.
Add 100 μL of anti-insulin antibody clone K36aC10 solution (dilution 1:300) to each section to completely cover tissue.
Incubate in a humidified chamber at room temperature for 60 min.
Rinse with PBS for 5 min, 3 times.
Add 100 μL of secondary antibody to each section to completely cover tissue and incubate for 10 min.
Rinse with PBS for 5 min, 3 times.
Add enough enzyme conjugate solution (from the Histostain Plus Broadspectrum AEC kit) to each section to completely cover tissue and incubate for 10 min.
Rinse with PBS for 2 min, 3 times.
Add chromogen AEC Single Solution and incubate 5-10min.
Scan reference sections with a PathScan Enabler IV instrument to obtain maps of the entire sections and identify stained insulin-containing islets.
3.3. Staining and dehydration for LCM
The staining and dehydration protocol is performed with 8 clean Coplin jars, prefilled with 50 mL of 70% (jar # 1-5), 90% (jar #6), and 100% (jar #7, 8) ethanol.
Jars # 1 to 5 are maintained chilled on ice during the staining, jars # 6 to 8 are maintained at room temperature to avoid condensation after dehydration.
The PEN membrane slides with tissue sections are dipped for 30 seconds in each jar from #1 to #3 of the ethanol series.
After jar #3, each slide is drained by gently placing the side edge of the glass on a Kimwipe, then placed horizontally, with the tissue sections on top.
The sections are stained for 90 seconds by adding 200 μL of Toluidine blue O staining solution, then drained and transferred to jar # 4, to continue dehydration (see Note 10).
The slides are dipped for 30 seconds in each jar following the numerical order (jar # 4-8), to obtain dehydration.
The stained and dehydrated slide is drained with a Kimwipe and placed under a laminar flow hood for 4 minutes to enable ethanol evaporation.
The slide is placed in a slide box containing the desiccant Drierite wrapped in Kimwipes and closed with tape (see Note 11).
3.4. LCM of pancreatic islets and acinar tissue (see Note 12)
The stage of the Leica Microscope LS LMD system is positioned in a clear acrylic box (microdissection chamber) where the atmosphere can be controlled.
1 hour before microdissection, the workplace is cleaned and the microdissection chamber is dehydrated using 2 kg of fresh Drierite to minimize the humidity and enable membrane microdissection.
The Leica LMD software is used to set up the laser (see Table 1), to initialize the instrument and to control the movements of the laser on the tissue section.
The PEN membrane slide with the stained tissue is positioned on the stage, with the tissue facing down.
Empty sterile collection tubes are placed under the cutting area (RNAse-free Eppendorf tubes, 500 μL volume, flat cap).
The scans of the reference sections are used to map the insulin-containing islets.
Pancreatic islets are identified in the toluidine-stained tissue by visualizing in bright field and in phase-contrast with 10 × magnification. In bright field, pancreatic islets appear as clusters of cells with lightly colored cytoplasm, whereas the surrounding acinar tissue is composed by cells with darker cytoplasm (see Figure 1). In phase-contrast, islet cells appear finely granulated. Visualize islet borders at 20 × magnification (see Note13).
Pancreatic islets and pancreatic exocrine tissue are collected in separate tubes. The area of each microdissection is annotated.
The volume of microdissected tissue is calculated by multiplying the total area collected by 10μm (thickness of the section).
Microdissected islets are collected into the cap of 500 μL sterile tubes (see Note 14, Figure 1).
Acinar tissue is microdissected from neighboring areas and collected into separate tubes.
Additional tissue is microdissected until the total volume for each sample corresponds to 1.06 × 107 μm3, 6 Islet Equivalents (see next paragraph).
Microdissection session should not last more than 60 minutes (stain additional tissue sections every 60 minutes).
Carefully remove the collection tubes from the microdissection chamber.
Resuspend the microdissected tissue with 50 μL of 50 mM NH4HCO3.
Close the tube, centrifuge the resuspended tissue for 2 minutes at 13,000 rpm (see Note 15)
Freeze by placing on dry ice, maintain frozen at −80 °C.
Table 1.
System configuration for laser capture microdissection with the Leica LMD instrument
| Parameters | 10x magnification | 20x magnification |
|---|---|---|
| Aperture | 10 | 13 |
| Intensity | 40 | 35 |
| Speed | 4 | 7 |
| Offset | 26 | 34 |
| Ap Diff | 8 | 8 |
| Option | Med | Med |
3.5. Conversion of LCM areas to volumes and to IEQ.
The total volume of isolated islets can be expressed as number of islet equivalents (IEQ) (10). An IEQ corresponds to the volume of a ‘standard’ islet, a sphere with a diameter d = 150 μm and a volume of VIEQ = 1.77 × 106 μm3. 1 IEQ contains approximately 1560 islet cells (11). The area of laser-captured tissue is recorded, and the volume is calculated by multiplying the total area collected for the thickness of the tissue section (10 μm). The target total volume of each microdissected sample is 1.06 × 107 μm3, corresponding to 6 IEQ (6 × 1.77 × 106 μm3). At any time during the collection, the total volume of laser-captured tissue can be divided by the standard volume of 1 IEQ (VIEQ = 1.77 × 106 μm3) to obtain the corresponding number of laser-captured IEQ.
3.6. Protein digestion
Add 6 μL of 1% pps silent surfactant (PPS) to extract and solubilize hydrophobic proteins,
Add 1.5 μL of 50 mM DTT and incubate at 95°C for 6 min.
Sonicate sample for 3 min
Alkylate with 7.5 μL 50 mM iodoacetamide for 25 min at 45 °C in the dark.
Add 1μg stock trypsin at 37 °C overnight.
Hydrolyze PPS by adding 12 μL 2 M HCl at room temperature for 2 h.
Centrifuge samples at 16,000 g for 12 min and separate the supernatant for LC-MS/MS analysis (see Note 16).
3.7. LC-MS/MS analysis
Protocols for LC-MS/MS analysis can vary because of diversity of LC systems (manufacturer, column, solvent composition, gradient, flow rate, etc.) and MS instruments (manufacturer, electrospray condition, fragmentation, MS parameters, analyzer, etc.). The following is the practice routinely used in our laboratory.
The LC-MS/MS platform consists of an UltiMate 3000 RSLCnano system and a Q Exactive HF mass spectrometer coupled with an EASY-Spray ion source (ThermoFisher Scientific).
Peptide separation is performed on a PepMap C18 analytical column (2 μm particle, 50 cm × 75 μm, ThermoFisher Scientific). Injection volume is 2.5 μL (0.5 μg peptide amount loaded into column) per sample (see Note 17).
A binary solvent system consisting of 0.1% FA in water (solvent A) and 0.1% FA in CH3CN (solvent B) is used at a flow rate of 250 nL min−1 (see Note 18).
LC separation is performed using the following gradient setting: hold at 4% B for 3 min (for desalting), from 4% to 8% B in 0.1 min, 8% to 40% B in 90 min (effective gradient), 40% to 90% B in 0.1% min, hold at 90% B for 10 min (for washing column), 90% to 4% B in 0.1 min, and hold at 4% B for 17 min for re-equilibrating column (see Note 19).
MS data are acquired in profile mode and resolution for full scan (400 to 2000 m/z) is set to 120,000 (at m/z 200) with maximum ion injection time of 50 ms, and automatic gain control (AGC) target of 1e6.
MS/MS data are acquired with data-dependent method of top 15. An isolation window of 1.4 m/z is used to isolate precursor ions for fragmentation by higher-energy collisional dissociation (HCD) at normalized collision energy of 28. Resolution for MS/MS spectrum is set to 15,000 (at m/z 200) with maximum ion injection time of 100 ms. AGC target for MS/MS scans is 1e5.
Precursor ions with single, seven and higher charge states are excluded from fragmentation, and dynamic exclusion time is set to 20 sec.
3.8. Database search for protein identification and quantification
Many database search software packages are available for this purpose. MaxQuant is demonstrated here (12).
The acquired datasets (.raw files) are analyzed using MaxQuant and the built-in Andromeda search engine against a UniProt human database (see Note 20).
Variable modifications include protein N-terminal acetylation and methionine oxidation.
Fixed modifications contain cysteine carbamidomethylation.
A maximum of 2 missed cleavages are allowed for the search.
Trypsin/P is selected as the specific proteolytic enzyme (see Note 21).
For label free quantification, “match between runs” is selected (see Note 22).
The false discovery rate (FDR) cut off used for both peptides and proteins is 0.01 (1%) using decoy database.
Only the razor/unique peptides are used for quantitative calculations.
The other parameters are the default settings in MaxQuant software for processing orbitrap-type data.
3.9. Statistical analysis
The search results in ProteinGroups.txt generated by MaxQuant are directly processed by Perseus software.(13) The differentially expressed proteins are identified by statistical analysis tools built in Perseus.
Import the quantitative data from ProteinGroups.txt into Perseus.
The potential contaminants, reverse hits and proteins only identified by modification site are excluded.
Filter out the protein with unique peptides less than 1.
The protein intensities are log2–transformed.
Categorize the samples into two groups: LCM islets and LCM acinar tissue.
Filter out the proteins not quantified in all the samples
Two-samples tests coupled with Benjamini-Hochberg (FDR cut off of 0.05) correction are performed to identify the differentially expressed proteins.(14)
3.10. Additional resources for data analysis and biological interpretation
http://string-db.org : a database of known and predicted protein interactions. The interactions include direct (physical) and indirect (functional) associations; they are derived from four sources: Genomic context, High-throughput experiments, Coexpression, Previous knowledge. This tool can be used to interpolate proteins in functional and interaction networks. The participation of proteins in networks was established by references in literature.
http://www.proteinatlas.org : a database of Antibody-based Proteomics. This tool enables the analysis of gene and protein expression in various human tissues. The database presents data related to the binding specificity of commercially-available antibodies.
http://compartments.jensenlab.org : a subcellular localization database. The database integrates evidence on protein subcellular localization from manually curated literature, high-throughput analyses, automatic text mining, and sequence-based prediction methods.
4. Notes
Note 1: The dilution of the anti-insulin antibody should be prepared freshly.
Note 2: The 0.5% w/v Toluidine blue O staining solution in 70% ethanol should be prepared freshly.
Note 3: PPS solution should be prepared freshly. Once the package is opened to air, the contents should be immediately reconstituted in aqueous buffer (pH 7-8), protected from elevated temperatures, and used within 12 hours.
Note 4: Stock DTT solution should be freshly prepared before use.
Note 5: Keep the IAA solution in the dark.
Note 6: Wipe the inner chamber and the stage of the cryotome with 100% ethanol.
Note 7: Change the blade, wipe the inner chamber and the stage of the cryotome with 100% ethanol after each sample in order to avoid contamination.
Note 8: Sections are maintained frozen and stored at −80 °C.
Note 9: Immunohistochemical staining of reference slides with insulin antibodies enables the identification and mapping of islets with β-cells.
Note 10: Toluidine blue O staining of frozen pancreas sections enables good discrimination of islets and acinar tissue: islets appear lightly colored compared to the surrounding acinar tissue; moreover, endocrine cells have a characteristic granulated or ‘rugged’ aspect in phase contrast illumination. If the humidity in the microdissection chamber is too high (as indicated by a pink drierite), the tissue section may re-hydrate in one hour or less: this determines visible tissue degradation and hampers further laser-capture microdissection.
Note 11: Insulin-containing islets were mapped via conventional immunohistochemical staining of reference sections, and Toluidine Blue O staining was used to guide the laser-capture microdissection in ethanol-dehydrated sections.
Note 12: Each LCM session lasted a total of 60 minutes, to avoid tissue rehydration and degradation.
Note 13: The setup for Laser capture microdissection is optimized by the operator and adjusted to the nature of the samples.
Note 14: The collection tubes should be sterile and RNase/DNase/Protease free.
Note 15: Centrifugation of the tissue at this stage enabled us to avoid loss of tissue.
Note 16: A rough estimation of solvent volume used for reconstitution of peptides in each fraction can be determined by the amount of peptides loaded onto column for fractionation and the number of final fractions. For instance, 100 μg divided by 24 fractions yields 4.2 μg per fraction, and the preparation of samples at 0.2 μg/μL requires the addition of 21.0 μL of solvent for reconstitution.
Note 17: The injection volume depends on the sample loop of autosampler, column loading capacity, and MS detector, therefore injection volume need to be adjusted based on the actual set-up.
Note 18: A flow rate of 250 nL/min for C18 50 cm × 75 μm i.d. column results in around 550 to 600 bar column pressure when heating column at 35°C.
Note 19: The gradient used for peptide separation can be modified depending on separation performance. However, all samples must be run under the same condition to limit variations between samples.
Note 20: The database information needs to include the type, sequence entry number, and releasing date of database.
Note 21: The selection of enzyme used for search is based on the enzyme that is chosen for protein digestion in section 3.6.
Note 22: “Match between runs” should be selected because it can improve the search results for less missing values.
Acknowledgements
This work was supported by the National Institutes of Health (R01 DK114345) and by the Diabetes Research Institute Foundation.
References
- 1.Schrimpe-Rutledge AC; Fontès G; Gritsenko MA; Norbeck Angela D.; Anderson DJ; Waters M; Adkins ,JN; Smith RD; Poitout V; Metz TO, Discovery of Novel Glucose-Regulated Proteins in Isolated Human Pancreatic Islets Using LC–MS/MS-Based Proteomics. J Proteome Res 2012, 11, (7), 3520–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waanders LF; Chwalek K; Monetti M; Kumar C; Lammert E; Mann M, Quantitative proteomic analysis of single pancreatic islets. Proc Natl Acad Sci U S A 2009, 106, (45), 18902–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eizirik DL; Sammeth M; Bouckenooghe T; Bottu G; Sisino G; Igoillo-Esteve M; Ortis F; Santin I; Colli ML; Barthson J; Bouwens L; Hughes L; Gregory L; Lunter G; Marselli L; Marchetti P; McCarthy MI; Cnop M, The human pancreatic islet transcriptome: expression of candidate genes for type 1 diabetes and the impact of pro-inflammatory cytokines. PLoS Genet 2012, 8, (3), e1002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marselli L; Thorne J; Ahn YB; Omer A; Sgroi DC; Libermann T; Otu HH; Sharma A; Bonner-Weir S; Weir GC, Gene expression of purified beta-cell tissue obtained from human pancreas with laser capture microdissection. J Clin Endocrinol Metab 2008, 93, (3), 1046–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marselli L; Thorne J; Dahiya S; Sgroi DC; Sharma A; Bonner-Weir S; Marchetti P; Weir GC, Gene expression profiles of Beta-cell enriched tissue obtained by laser capture microdissection from subjects with type 2 diabetes. PLoS One 2010, 5, (7), e11499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonner RF; Emmert-Buck M; Cole K; Pohida T; Chuaqui R; Goldstein S; Liotta LA, Laser capture microdissection: molecular analysis of tissue. Science 1997, 278, (5342), 1481,1483. [DOI] [PubMed] [Google Scholar]
- 7.Sturm D; Marselli L; Ehehalt F; Richter D; Distler M; Kersting S; Grutzmann R; Bokvist K; Froguel P; Liechti R; Jorns A; Meda P; Baretton GB; Saeger HD; Schulte AM; Marchetti P; Solimena M, Improved protocol for laser microdissection of human pancreatic islets from surgical specimens. Journal of visualized experiments 2013, (71), 50231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marciniak A; Cohrs CM; Tsata V; Chouinard JA; Selck C; Stertmann J; Reichelt S; Rose T; Ehehalt F; Weitz J; Solimena M; Slak Rupnik M; Speier S, Using pancreas tissue slices for in situ studies of islet of Langerhans and acinar cell biology. Nat Protoc 2014, 9, (12), 2809–22. [DOI] [PubMed] [Google Scholar]
- 9.Nishida Y; Aida K; Kihara M; Kobayashi T, Antibody-validated proteins in inflamed islets of fulminant type 1 diabetes profiled by laser-capture microdissection followed by mass spectrometry. PLoS One 2014, 9, (10), e107664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ricordi C; Gray DW; Hering BJ; Kaufman DB; Warnock GL; Kneteman NM; Lake SP; London NJ; Socci C; Alejandro R; et al. , Islet isolation assessment in man and large animals. Acta Diabetol Lat 1990, 27, (3), 185–95. [DOI] [PubMed] [Google Scholar]
- 11.Pisania A; Weir GC; O’Neil JJ; Omer A; Tchipashvili V; Lei J; Colton CK; Bonner-Weir S, Quantitative analysis of cell composition and purity of human pancreatic islet preparations. Lab Invest 2010, 90, (11), 1661–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tyanova S; Temu T; Cox J, The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc 2016, 11, (12), 2301–2319. [DOI] [PubMed] [Google Scholar]
- 13.Tyanova S; Temu T; Sinitcyn P; Carlson A; Hein MY; Geiger T; Mann M; Cox J, The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 2016, 13, (9), 731–40. [DOI] [PubMed] [Google Scholar]
- 14.Tusher VG; Tibshirani R; Chu G, Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 2001, 98, (9), 5116–21. [DOI] [PMC free article] [PubMed] [Google Scholar]