Abstract
Many proteoforms—arising from alternative splicing, post-translational modifications (PTM), or paralogous genes—have distinct biological functions, such as histone PTM proteoforms. However, their quantification by existing bottom-up mass-spectrometry (MS) methods is undermined by peptide-specific biases. To avoid these biases, we developed and implemented a first-principles model (HIquant) for quantifying proteoform stoichiometries. We characterized when MS data allow inferring proteoform stoichiometries by HIquant and derived an algorithm for optimal inference. We applied this algorithm to infer proteoform stoichiometries in two experimental systems that supported rigorous bench-marking: alkylated proteoforms spiked-in at known ratios and endogenous histone 3 PTM proteoforms quantified relative to internal heavy standards. When compared with the benchmarks, the proteoform stoichiometries interfered by HIquant without using external standards had relative error of 5–15% for simple proteoforms and 20–30% for complex proteoforms. A HIquant server is implemented at: https://web.northeastern.edu/slavov/2014_HIquant/
Keywords: Algorithms, Bioinformatics, Bioinformatics Software, Mathematical Modeling, Mass Spectrometry
Alternative mRNAs splicing and post-translational modifications (PTM)1 produce multiple protein isoforms per gene, termed proteoforms by Smith et al. (1). Furthermore, protein isoforms can be produced by distinct but highly homologous open reading frames, i.e. paralogous genes. Despite having similar sequence, proteoforms and protein isoforms often have distinct, even opposite biological functions (2, 3). For examples: (1) some Bcl-x isoforms promote apoptosis whereas other Bcl-x isoforms inhibit apoptosis (4); (2) the methylation of histone 3 can cause either transcriptional activation (lysine 4) or repression (lysine 9) depending on the modified lysine (5); (3) pyruvate kinase isoforms have different metabolic regulation, activities, and roles in aerobic glycolysis (6–8); and (4) minor amino acid differences between actin and tropomyosin isoforms alter the tropomyosin position on actin (9).
Understanding such systems demands accurate methods to distinguish and quantify proteoform abundances (3, 10). However, the high sequence homology among proteoforms makes their quantification by bottom-up LC-MS/MC very challenging because protease digestion (which is an essential part of high-throughput bottom-up proteomics) is likely to produce mostly peptides that are shared by multiples proteoforms (11–13). One approach to overcoming the challenge of shared peptides and distinguishing among proteoforms is to analyze the intact proteoforms without protease digestion, which is known as top-down proteomics (10, 14–16). Combining advances in chromatographic separation of proteins with top-down proteomics has enabled quantifying functionally distinct isoforms of skeletal and cardiac muscle proteins (9, 17), combinatorial histone modifications (18, 19) and discovery analysis of thousands of human proteoforms (10, 20). Distinguishing proteoforms has been powered by efficient fragmentation methods, such as electron capture dissociation (ECD) (21) and electron transfer dissociation (ETD) (22). Combining these methods with infrared photoactivation (23) and with clever algorithms has made it possible to localize PTMs and identify the sequences and modified sites even for peptides with multiple covalent modifications (24, 25). Such methods allow quantifying the ratios among modifications occurring on the same peptide.
Although advances in protein separation and top-down methods are becoming increasingly powerful (10, 16, 20), bottom-up methods remain more widely used and still afford higher throughput (26, 27). When analyzed by bottom-up methods, many proteoform-specific modifications occur on distant parts of the corresponding proteins and upon protease digestion will be found on different peptides. Furthermore, most of the peptides may be shared among proteoforms (11–13). Here we focus on quantifying the stoichiometry among such proteoforms from peptides quantified by bottom-up LC-MS/MS across multiple samples.
One validated approach to quantifying the stoichiometry among such proteoforms relies on external standards (28, 29). This method can afford high accuracy even for complex proteoforms (29). However, their wider use has been limited by expense and applied only to special cases that allow chemical modification of cell lysates, e.g. phosphorylation (30) and acetylation (31, 32). In the absence of external standards, the quantification of complex proteoform stoichiometries remains very challenging because the ratios between proteoform-specific peptides do not necessarily reflect the ratios between the corresponding proteoforms (33); precursor ion areas corresponding to the same phospho-site in the same sample can differ over 100-fold depending on the choice of protease (34). This discrepancy arises because a measured peptide level (precursor ion area) depends not only on the abundance of the corresponding protein(s) but also on extraneous factors including protein digestion, peptide ionization efficiency, the presence of other co-eluting peptides, and chromatographic aberrations (34–36). These extraneous factors break the equivalence between the abundance of a peptide and its precursor ion area and thus make protein quantification much more challenging than DNA quantification by sequencing. This problem is compounded when PTM peptides have been enriched, and thus their intensities scaled by unknown enrichment-dependent factors.
EXPERIMENTAL PROCEDURES
We used two experimental approaches to derive bench-mark estimates of proteoform stoichiometries: (1) We mixed alkylated proteoforms of the dynamic universal proteomics standard (UPS2) into known ratios and (2) We quantified stoichiometries among histone PTMs by parallel reaction monitoring (PRM) relative to peptide standards with known absolute abundances. Each of these methods is described in the subsections below, and the resulting bench-mark estimates of proteoform stoichiometries were used to evaluate HIquant inferences.
Quantifying Alkylated proteoforms of UPS2
Our first test of HIquant on experimental data with bench-marked proteoforms was based on alkylated proteoforms of UPS2 that we mixed into predetermined ratios. To achieve such mixtures, we started by splitting a UPS standard into two equal parts, part A and part B. Then:
Part A was reduced with TCEP, and Cys were alkylated with iodoacetamide.
Part B was reduced with TCEP, and Cys were alkylated with vinylpyridine.
The alkylated proteoforms from both part A and part B were diluted to a total volume of 320 μl and spiked into yeast samples according to Table I.
Table I.
| TMT-126 | TMT-127 | TMT-128 | TMT-129 | TMT-130 | TMT-131 | |
|---|---|---|---|---|---|---|
| Part A (iodoacetamide) | 10 | 10 | 50 | 50 | 100 | 80 |
| Part B (vinylpyridine) | 50 | 100 | 40 | 50 | 10 | 50 |
This range of mixing ratios, spanning an order of magnitude, was chosen as it includes the range of relative changes commonly measured reliably by mass-spectrometry. The fractional site occupancy between the proteoforms varied within 10-fold in each direction, the range over which HIquant is likely to give the most accurate results. As discussed below, the ratios among proteoforms whose levels differ by many orders of magnitude may be better inferred from the precursor ion areas of their unique peptides.
After the spike in of UPS alkylated proteoforms, each sample was digested by trypsin overnight and labeled with the corresponding TMT tag according to manufacturer's protocol. The labeled samples were mixed into a set and processed as described previously (8). Briefly, the set-sample was injected from an auto-sampler into the trapping column (75 μm column ID, 5 cm length, and packed with 5 μm beads with 20 nm pores; from Michrom Bioresources, Inc.) and washed for 15 min; the sample was eluted to analytic column (Waters columns with 75 μm ID, 15 cm length, and packed with HSS T3 1.8 μm beads) with a gradient from 2 to 32% of buffer B (0.1% formic acid in ACN) over 180 min gradient and fed into LTQ Orbitrap Elite (Thermo Fisher, San Jose, CA). The instrument was set to run in TOP 20 MS/MS mode method with dynamic exclusion. After MS1 scan in Orbitrap with 60K resolving power, each ion was submitted to an HCD MS/MS with 15K or 30K resolving power and to CID MS/MS scan subsequently. All quantification data were derived from HCD spectra.
In the particular case of UPS2, trypsin digestion resulted in enough peptides to constrain the HIquant model (Fig. 1) and to support accurate inference (Fig. 2B, 2C). More complex proteoforms may require more peptides than those quantified from a trypsin digestion. In such cases, to better constrain the inference, one might increase the number of peptides used by HIquant by compiling peptides quantified from several digestions, each of which using a different protease (34, 36). Because HIquant uses only the relative quantification and is insensitive to systematic biases, including proteoform-specific digestion efficiency, it can easily accommodate peptides quantified from digestions with different proteases or from different enrichment methods.
Fig. 1.

Model for inferring stoichiometries among proteoforms and paralogous proteins independently from peptide-specific biases. A, One shared (X2) and three unique (X1, X3, and X4) peptides of H3 proteoforms illustrate a very simple case of HIquant. HIquant models the peptide levels measured across conditions (x) as a supposition of the protein levels (p), scaled by unknown peptide-specific biases/nuisances (z). These coupled equations can be written in a matrix form whose solution infers the methylation stoichiometry independently from the nuisances (z). B, The general form of the model for K proteoforms (or homologous proteins) with M peptides quantified across N conditions can be formulated and solved. In many, albeit not all, cases an optimal and unique solution can be found, even in the absence of unique peptides; see supplemental Fig. S1 and Supplemental Information.
Fig. 2.
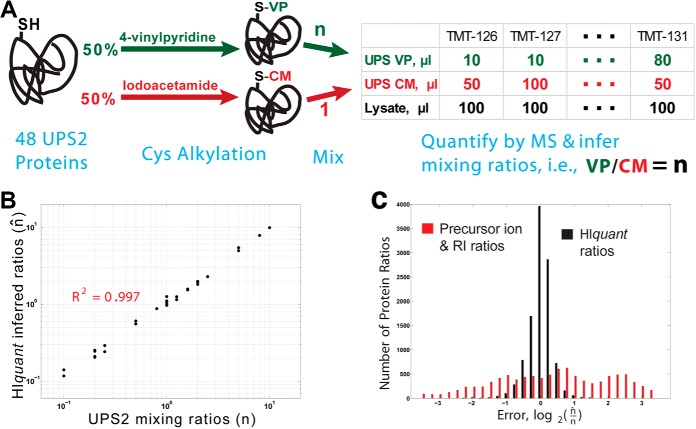
HIquant accurately quantifies ratios across alkylated proteoforms of a spiked-in standard. A, Schematic diagram of a validation experiment. We prepared a gold standard of proteoforms from the dynamic universal proteomics standard (UPS2) whose cysteines were covalently modified either with iodoacetamide or with vinylpyridine. Upon digestion, these modified UPS proteins generate many shared peptides (peptides not containing cysteine) and a few unique peptides (peptides containing cysteine). The modified UPS2 proteins were mixed with one another at known ratios (n), mixed with yeast lysate, digested and quantified by MS. The proteoform ratios that HIquant inferred from the MS data (n̂) were compared with the mixing ratios. B, The ratios across the alkylated isoforms of UPS2 inferred by HIquant (n̂, y axis) accurately reflect the mixing ratios (n, x axis). C, The mixing and inferred ratios in panel B span 2-orders of magnitude, which is much larger than the dynamic range of relative error. To zoom in on the relative errors, we plotted a distribution of log2(n/n̂) for 1, 500 HIquant problems generated by sampling with replacement peptides from all UPS2 proteins. For HIquant, this distribution indicates small error, with median error below 11%. However the ratios estimated just from the precursor intensities of the unique peptides for each proteoform show significantly higher relative error, mostly likely because of peptide-specific variability in digestion and ionization.
Analysis of Mass-Spec Spectra
Mass/charge spectra were analyzed by MaxQuant (37) (version 1.4.1.2) or SEQUEST HT run via the Proteome Discoverer (64 bit version 1.4.0.288, Thermo). All searches were run on a Windows server 2008 64 bit operating system with 64 CPU blades and 256 GB of RAM with the following general parameters. Parent ion mass tolerance was set to 20 ppm, mass tolerance for MS/MS ions was set to 0.02 Da for HCD and to 0.6 Da for CID spectra. For all searches, minimal peptide length was specified as 6 amino acids and maximal peptide length as 50 amino acids. The peptide charge state was limited to +7 for searches with MaxQuant. Searches were performed against the yeast uniprot database downloaded from www.yeastgenome.org in October 2014 containing 6,750 entries, the fasta sequences of the UPS standard, and common contaminants. Searches had trypsin specificity, allowing 2 missed cleavages. Asn and Gln deamidation, Cys alkylation by either iodoacetamide or vinylpyridine, and Met oxidation were included as variable modifications in the search parameters. There were no fixed modifications.
The search results from all search engines were filtered at 1% false discovery rate (FDR) on both protein and on peptide levels using the Percolator (Version 2.05 Build Date May 6 2013). The results exported for further analysis included all peptide spectrum matches (PSM) that were assigned to one or more proteins and passed the statistical significance filter. These results were outputted in the “evidence.txt” file for MaxQuant and in a peptide-level-results text file for Proteome Discoverer.
PRM of Histone PTMs Relative to Heavy Peptide Standards
To evaluate HIquant inferences, we also used PTM stoichiometries estimated from a parallel reaction monitoring (PRM) assay quantifying each peptide relative to an internal heavy standard. Creech et al. have published a detailed description of the assay (29), and here we will only briefly summarize it. Drug-treated cells were collected by centrifugation. After cell lysis, histones were extracted with sulfuric acid and were precipitated with trichloroacetic acid. Samples comprised of 10 μg, a 5-fold reduction compared with Creech et al. (29), were propionylated, desalted, and digested overnight with trypsin. After a second round of propionylation, the samples were desalted using C18 Sep-Pak Cartridge (Waters). A mix of isotopically labeled synthetic peptides was spiked-in to each sample prior to MS analysis. Peptides were separated on a C18 column (EASY-nLC 1000, Thermo Scientific) and analyzed by MS in a PRM mode (Q Exactive-plus, Thermo Scientific) as described previously (29). Detailed SOPs for P100 and GCP assays, including synthetic peptide master mixture formulation, can be found at https://panoramaweb.org/labkey/wiki/LINCS/Overview%20Information/page.view?name=sops.
Inference Model for HIquant
To infer proteoform stoichiometry, we use a simple model that is illustrated in Fig. 1A with proteoforms of histone H3 and in supplemental Fig. S1 with paralogous ribosomal proteins and phosphoproteoforms of pyruvate dehydrogenase. HIquant explicitly models peptide levels measured across conditions as a superposition of the levels of the proteins from which the peptides originate, Fig. 1A. In this model, shared peptides serve as indispensable internal standards; they couple the equations for different peptides and thus make possible estimating stoichiometries between homologous proteins and proteoforms. The simple example in Fig. 1A generalizes to any number of proteins/proteoforms (M) and any number of conditions greater than 1 (N > 1) as the system in Fig. 1B shows. HIquant solves this system and infers the protein levels (P) independently from the extraneous noise (Z; coming from protein-digestion, peptide-ionization differences, sample loss during enrichment, and even coisolation interference); Z is also inferred as part of the solution and discarded. A related superposition model has been used before with peptides quantified at one condition (13). However, for a single condition, the model cannot quantify the proteins independently from the nuisance Z because all problems described by system 1 in Fig. 1 are under-determined, i.e. have infinite number of solutions (Proof 1; Supplemental Information). Thus, for a single condition, the model cannot take advantage of the robust corresponding-ion pairs, i.e. ratios between ions with the same chemical composition. In contrast, HIquant infers ratios across proteins and their PTMs solely from the corresponding-ion ratios. This is possible because when N > 1, the system in Fig. 1B often has a unique solution up to a single scaling constant, even when all peptides are shared, e.g. the problem defined by the design matrix in supplemental Fig. S1C. We characterize the conditions under which HIquant has a unique solution for the abundances of individual proteoforms and derive algorithms that use convex-optimization to find the optimal solution given the data; see Malioutov and Slavov (38) and Supplemental Information.
Validating Inference of Proteoform Stoichiometry
Our model (Fig. 1B) aims to make proteoform quantification insensitive to many systematic biases. For example, incomplete cleavage of a peptide, e.g. only 5% of the peptide is released during enzyme digestion, is fully absorbed into the corresponding nuisance and does not affect inferred protein levels if the cleavage is 5% for all conditions/samples. Analogously, if coisolation interference compresses the fold-changes of a peptide, the systematic component of the compression is fully absorbed by the nuisances. Unlike systematic biases, random noise in the data is not absorbed by the nuisances; it can degrade the quality of the inference. To assess the reliability of the inferred proteoform abundances, HIquant carefully evaluates the inference and assigns confidence levels. The evaluation uses inference features, such as fraction of explained variance, eigenvalue spectrum spacing and noise sensitivity; see Supplemental Information.
We sought to experimentally evaluate HIquant's ability to infer the proteoform stoichiometry in samples for which proteoform stoichiometries are accurately determined by other methods. The first method included creating and mixing alkylated proteoforms. The second method included quantifying histone H3 proteoforms relative to heavy peptide standards with known abundances.
Validation Based on Spiked-in Alkylated Proteoforms
We aimed to create proteoform mixtures with known stoichiometries so that they can be used to assess the accuracy of stoichiometries inferred by HIquant. To this end, the dynamic universal proteomics standard (UPS2) was split into two equal parts, A and B. In part A, cysteines were covalently modified with iodoacetamide, and in part B with vinylpyridine as described in the methods and shown in Fig. 2A. We mixed part A and B in predefined ratios (n) and spiked each mixing ratio into an yeast sample. All samples were labeled with TMT, and the relative peptide levels quantified from the reporter ions at the MS2 level.
These alkylated UPS proteoforms have mostly shared peptides (peptides not containing cysteine) and only one or a few unique peptides per proteoform (peptides containing cysteine). HIquant modeled the relative levels of these peptides as shown in Fig. 1 and solved the model to infer the stoichiometries of the alkylated proteoforms (n̂), which should correspond to the mixing ratios. Indeed, we find that the actual mixing ratios (n) for all quantified proteoforms correlate strongly to the inferred ratios (n̂) as shown in Fig. 2B. To examine the accuracy of the inferred ratios n̂ more closely, we sought to generate a distribution of errors between the inferred and the expected mixing ratios (n̂/n) for many HIquant problems, more than the number of UPS2 proteoforms. To do so, we took advantage of the fact that the A/B ratios of the alkylated proteoforms for different UPS2 proteins should be the same because the proteins were mixed simultaneously and therefore in equal proportions. Thus, we created 1500 HIquant problems by sampling with replacement shared peptides (not containing cysteine) and unique peptides (containing cysteine), and then inferred the n̂ ratios between the iodoacetamide and vinylpyridine modified proteoforms, one ratios for each TMT channel and 6 per HIquant problem. The corresponding distribution of relative errors is shown in Fig. 2C along with the errors for ratios estimated from the abundances of the precursor ions. The median error is below 11% for ratios inferred by HIquant and a substantially larger error for the ratios between precursor ion areas of unique peptides (Fig. 2C).
Validation Based on Histone 3 Proteoforms Quantified by PRM
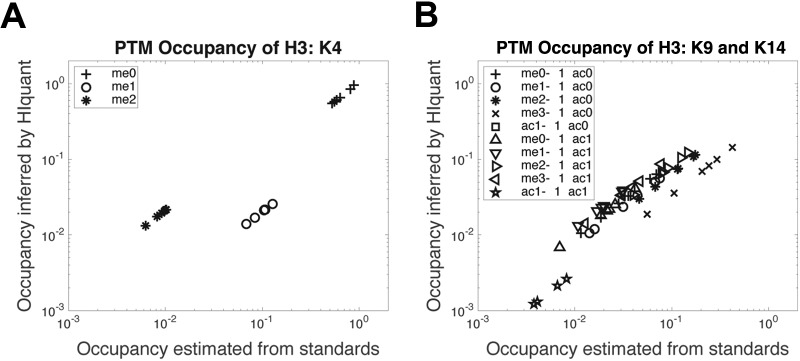
Next, we sought to evaluate the ability of HIquant to infer stoichiometries of more complex PTM proteoforms, those of histone H3. We rigorously quantified endogenous proteoform stoichiometries by a previously developed assay based on external standards (MasterMix) with known concentrations (29). For the test, we used peptides quantified by parallel reaction monitoring (PRM) across 7 drug perturbations. Fractional site occupancies were estimated based on the external standards as described before by Creech et al. (29). Independently, the same stoichiometries were inferred by HIquant only from the relative levels of the indigenous peptides, without using the MasterMix concentrations. The comparison of these estimates indicates good agreement (Fig. 3), supporting the ability of HIquant to infer fractional site occupancy even when the same site may be modified by different PTMs. The estimates from the external standards and from HIquant are very close but also show some systematic deviations. Those deviations may arise because of incomplete protein digestion that is hard to control for with peptide standards, measurement noise corrupting the solution inferred by HIquant or proteforms not explicitly included in the model. The abundances of some proteoforms with quantified peptides is over 1000-fold lower than the abundance of the main proteoforms. They and their corresponding peptides were omitted from the HIquant inference because their quantification requires unrealistically high accuracy of relative quantification; see Supplemental Information and Discussion.
Fig. 3.
HIquant accurately infers stoichiometries and confidence intervals across PTM site occupancies of histone 3. A, Histone 3 peptides were quantified by SRM across 7 perturbations, and the fractional site occupancies for K4 methylation estimated by two methods: Estimates inferred by HIquant without using external standards are plotted against the corresponding estimates based on MasterMix external standards with known concentrations (29). Each marker shape corresponds to the PTM site(s) shown in the legend; methylation is denoted with “me” and acetylation with “ac” followed by the number of methyl/acetyl groups. B, The validation method from (a) was extended to another set of more complex fractional site occupancies on K9 methylation and K14 acetylation.
DISCUSSION
The idea of using ratios between chemically identical ions is a cornerstone of quantitative proteomics (28, 39). It has been used for two decades in the context of relative quantification of proteins based on unique peptides (40) and even applied to the special case of inferring phosphorylation site occupancy (11, 33). Our work expands and generalizes this idea to all peptides, to stoichiometries of complex proteoforms, and to unlimited number of conditions. Crucially, HIquant allows accurate, efficient, and numerically stable inference resulting in reliability estimates.
HIquant requires and depends upon accurate relative quantification. This limitation is largely and increasingly mitigated by technological developments allowing accurate estimates of corresponding ion ratios. Such improvements include instrumentation advances (41, 42), interference free tandem mass tags (43), and enhanced peptide quantification from mass-spectra (27) However, these technological developments on their own do not allow accurate estimates of PTM site occupancy from bottom-up LC-MS/MS (34). HIquant's dependence on the accuracy of relative quantification increases with increasing difference in the abundance of proteoforms. If the levels of two proteins differ by more than 3–6 orders of magnitude, this difference is likely better inferred from the precursor ion areas of the unique peptides. The associated noise (because of variability in protein digestion and ionization) is generally below 100-fold (36) and thus smaller than the signal. HIquant's utility is particularly relevant when proteins and proteoforms have comparable abundances (within 10–100 fold difference) but distinct functions (44, 45) and thus accurate quantification is essential for quantifying relatively small differences in abundance. Quantifying such proteoforms is an exciting frontier essential for understanding post-transcriptional regulation (46, 47) and defining cell-types from single cell proteomes (48).
The general form of HIquant described in Fig. 1C indicates that HIquant is not limited to proteoforms, even broadly defined. Rather, HIquant can be applied to any set of proteins sharing a peptide. Here we emphasize the application to proteoforms because existing bottom-up methods are better suited for quantifying the stoichiometry between proteins with low homology that generate many unique peptides. For proteins with multiple unique peptides, some of the peptide-specific bias (from variation in protein-digestion and peptide-ionization efficiency) is likely to be averaged out and reduced. However, this bias is a more serious problem for proteoforms with only one or only a few unique peptides (34). For such proteoforms, HIquant can allow estimating stoichiometries accurately using only ratios between chemically identical ions.
Supplemental Information
Supplemental information includes Extended Experimental Procedures, Mathematical Proofs, and Supplemental Figs. can be found in the Supplemental Information. The supplemental website for interactive data analysis can be found at: https://web.northeastern.edu/slavov/2014_HIquant/ Python code implementing HIquant is available at: https://github.com/nslavov/HIquant.
DATA AVAILABILITY
The data are available at ProteomeXchange (accession ID: PXD008557, url: http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD008557) via MassIVE (accession ID: MSV000081857, url: https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=38f67398ac7a4547b68f343ca1aff905).
Supplementary Material
Acknowledgments
We thank S. Carr, A. Ivanov, S. MacNamara, Y. Katz, and MA. Blanco for help, critical discussions and feedback. N.S. started this work while a postdoc with Alexander van Oudenaarden and thanks him for generous support.
Footnotes
* This work was supported by grants to N.S. from NIGMS of the NIH under Award Number DP2GM123497, a SPARC grant from the Broad Institute to N.S and S.C, and a NEU Tier 1 grant to N.S.
 This article contains supplemental Figures.
This article contains supplemental Figures.
1 The abbreviations used are:
- PTM
- post-translational modifications
- PSM
- peptide spectrum matches
- FDR
- false discovery rate
- PRM
- parallel reaction monitoring.
REFERENCES
- 1. Smith L. M., Kelleher N. L., et al. (2013) Proteoform: a single term describing protein complexity. Nat. Methods 10, 186–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Soria P. S., McGary K. L., and Rokas A. (2014) Functional divergence for every paralog. Mol. Biol. Evolution 31, 984–992 [DOI] [PubMed] [Google Scholar]
- 3. Aebersold R., Agar J. N., Amster I. J., Baker M. S., Bertozzi C. R., et al. (2018) How many human proteoforms are there? Nat. Chem. Biol. 14, 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schwerk C., and Schulze-Osthoff K. (2005) Regulation of apoptosis by alternative pre-mRNA splicing. Mol. Cell 19, 1–13 [DOI] [PubMed] [Google Scholar]
- 5. Berger S. L. (2007) The complex language of chromatin regulation during transcription. Nature 447, 407–412 [DOI] [PubMed] [Google Scholar]
- 6. Tanaka T., Harano Y., Sue F., and Morimura H. (1967) Crystallization, characterization and metabolic regulation of two types of pyruvate kinase isolated from rat tissues. J. Biochem. 62, 71–91 [DOI] [PubMed] [Google Scholar]
- 7. Christofk H. R., Vander Heiden M. G., Harris M. H., Ramanathan A., Gerszten R. E., Wei R., Fleming M. D., Schreiber S. L., and Cantley L. C. (2008) The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452, 230–233 [DOI] [PubMed] [Google Scholar]
- 8. Slavov N., Budnik B., Schwab D., Airoldi E., and van Oudenaarden A. (2014) Constant growth rate can be supported by decreasing energy flux and increasing aerobic glycolysis. Cell Reports 7, 705–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lehman W., Hatch V., Korman V., Rosol M., Thomas L., Maytum R., Geeves M.A., Van Eyk J.E., Tobacman L.S., and Craig R. (2000) Tropomyosin and actin isoforms modulate the localization of tropomyosin strands on actin filaments. J. Mol. Biol. 302, 593–606 [DOI] [PubMed] [Google Scholar]
- 10. Toby T. K., Fornelli L., and Kelleher N. L. (2016) Progress in top-down proteomics and the analysis of proteoforms. Ann. Rev. Anal. Chem. 9, 499–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dost B., Bandeira N., Li X., Shen Z., Briggs S. P., and Bafna V. (2012) Accurate mass spectrometry based protein quantification via shared peptides. J. Computational Biol. 19, 337–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stastna M., and Van Eyk J. E. (2012) Analysis of protein isoforms: can we do it better? Proteomics 12, 2937–2948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gerster S., Kwon T., Ludwig C., Matondo M., Vogel C., et al. (2014) Statistical approach to protein quantification. Mol. Cell. Proteomics 13, 666–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kelleher N. L. (2004) Top-down proteomics. Anal. Chem. 76, 197A–203A [PubMed] [Google Scholar]
- 15. Ge Y., Rybakova I. N., Xu Q., and Moss R. L. (2009) Top-down high-resolution mass spectrometry of cardiac myosin binding protein C revealed that truncation alters protein phosphorylation state. Proc. Natl. Acad. Sci. U.S.A. 106, 12658–12663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Siuti N., and Kelleher N. L. (2007) Decoding protein modifications using top-down mass spectrometry. Nat. Methods 4, 817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sheng S., Chen D., and Van Eyk J. E. (2006) Multidimensional liquid chromatography separation of intact proteins by chromatographic focusing and reversed phase of the human serum proteome optimization and protein database. Mol. Cell. Proteomics 5, 26–34 [DOI] [PubMed] [Google Scholar]
- 18. Young N. L., DiMaggio P. A., Plazas-Mayorca M. D., Baliban R. C., Floudas C. A., and Garcia B. A. (2009) High throughput characterization of combinatorial histone codes. Mol. Cell. Proteomics 8, 2266–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tian Z., Tolić N., Zhao R., Moore R. J., Hengel S. M., Robinson E. W., Stenoien D. L., Wu S., Smith R. D., and Paša-Toliæ L. (2012) Enhanced top-down characterization of histone post-translational modifications. Genome Biol. 13, R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tran J. C., Zamdborg L., Ahlf D. R., Lee J. E., Catherman A. D., Durbin K. R., Tipton J. D., Vellaichamy A., Kellie J. F., Li M., Wu C., Sweet S. M., Early B. P., Siuti N., LeDuc R. D., Compton P. D., Thomas P. M., and Kelleher N. L. (2011) Mapping intact protein isoforms in discovery mode using top-down proteomics. Nature 480, 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zubarev R. A., Kelleher N. L., and McLafferty F. W. (1998) Electron capture dissociation of multiply charged protein cations. A nonergodic process. J. Am. Chem. Soc. 120, 3265–3266 [DOI] [PubMed] [Google Scholar]
- 22. Syka J. E., Coon J. J., Schroeder M. J., Shabanowitz J., and Hunt D. F. (2004) Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 101, 9528–9533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Riley N. M., Sikora J. W., Seckler H. S., Greer J. B., Fellers R. T., LeDuc R. D., Westphall M. S., Thomas P. M., Kelleher N. L., and Coon J. J. (2018) The value of activated ion electron transfer dissociation for high-throughput top-down characterization of intact proteins. Anal. Chem. 90, 8553–8560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. DiMaggio P. A., Young N. L., Baliban R. C., Garcia B. A., and Floudas C. A. (2009) A mixed integer linear optimization framework for the identification and quantification of targeted post-translational modifications of highly modified proteins using multiplexed electron transfer dissociation tandem mass spectrometry. Mol. Cell. Proteomics 8, 2527–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guan S., and Burlingame A. L. (2010) Data processing algorithms for analysis of high resolution MSMS spectra of peptides with complex patterns of posttranslational modifications. Mol. Cell. Proteomics 9, 804–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gillet L. C., Leitner A., and Aebersold R. (2016) Mass spectrometry applied to bottom-up pro- teomics: entering the high-throughput era for hypothesis testing. Ann. Rev. Anal. Chem. 9, 449–472 [DOI] [PubMed] [Google Scholar]
- 27. Sinitcyn P., Rudolph J. D., and Cox J. (2018) Computational methods for understanding mass spectrometry–based shotgun proteomics data. Annu. Rev. Biomed. Data Sci. 1, 207–234 [Google Scholar]
- 28. Ong S. E., and Mann M. (2005) Mass spectrometry–based proteomics turns quantitative. Nat. Chem. Biol. 1, 252. [DOI] [PubMed] [Google Scholar]
- 29. Creech A. L., Taylor J. E., Maier V. K., Wu X., Feeney C. M., Udeshi N. D., Peach S. E., Boehm J. S., Lee J. T., Carr S. A., and Jaffe J. D. (2015) Building the connectivity map of epigenetics: Chromatin profiling by quantitative targeted mass spectrometry. Methods 72, 57–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu R., Haas W., Dephoure N., Huttlin E. L., Zhai B., Sowa M. E., and Gygi S. P. (2011) A large-scale method to measure absolute protein phosphorylation stoichiometries. Nat. Methods 8, 677–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Weinert B. T., Iesmantavicius V., Moustafa T., Scholz C., Wagner S. A., Magnes C., Zechner R., and Choudhary C. (2014) Acetylation dynamics and stoichiometry in saccharomyces cerevisiae. Mol. Syst. Biol. 10, 716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Baeza J., Dowell J. A., Smallegan M. J., Fan J., Amador-Noguez D., Khan Z., and Denu J. M. (2014) Stoichiometry of site-specific lysine acetylation in an entire proteome. J. Biol. Chem. 289, 21326–21338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Olsen J. V., Vermeulen M., Santamaria A., Kumar C., Miller M. L., Jensen L. J., Gnad F., Cox J., Jensen T. S., Nigg E. A., and Brunak S., Mann M. (2010) Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signaling 3, ra3–ra3 [DOI] [PubMed] [Google Scholar]
- 34. Giansanti P., Aye T. T., van den Toorn H., Peng M., van Breukelen B., and Heck A. J. (2015) An augmented multiple-protease-based human phosphopeptide atlas. Cell Reports 11, 1834–1843 [DOI] [PubMed] [Google Scholar]
- 35. Lu P., Vogel C., Wang R., Yao X., and Marcotte E. M. (2006) Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 25, 117–124 [DOI] [PubMed] [Google Scholar]
- 36. Peng M., Taouatas N., Cappadona S., van Breukelen B., Mohammed S., Scholten A., and Heck A. J. (2012) Protease bias in absolute protein quantitation. Nat. Methods 9, 524–525 [DOI] [PubMed] [Google Scholar]
- 37. Cox J., and Mann M. (2008) Maxquant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 38. Malioutov D., and Slavov N. (2014) Convex total least squares. J. Machine Learning Res. 32, 109–117 [Google Scholar]
- 39. Blagoev B., Ong S. E., Kratchmarova I., and Mann M. (2004) Temporal analysis of phosphotyrosine-dependent signaling networks by quantitative proteomics. Nature Biotechnol. 22, 1139–1145 [DOI] [PubMed] [Google Scholar]
- 40. Altelaar A., Frese C. K., Preisinger C., Hennrich M. L., Schram A. W., Timmers H. T., Heck A. J., and Mohammed S. (2013) Benchmarking stable isotope labeling based quantitative proteomics. J. Proteomics 88, 14–26 [DOI] [PubMed] [Google Scholar]
- 41. Zubarev R. A., and Makarov A. (2013) Orbitrap mass spectrometry. Anal. Chem. 85, 5288–5296 [DOI] [PubMed] [Google Scholar]
- 42. Nagornov K. O., Kozhinov A. N., and Tsybin Y. O. (2017) Fourier transform ion cyclotron resonance mass spectrometry at the cyclotron frequency. J. Am. Soc. Mass Spectrom. 28, 768–780 [DOI] [PubMed] [Google Scholar]
- 43. Winter S. V., Meier F., Wichmann C., Cox J., Mann M., and Meissner F. (2018) EASI-tag enables accurate multiplexed and interference-free MS2-based proteome quantification. Nat. Methods 15, 527–530 [DOI] [PubMed] [Google Scholar]
- 44. Slavov N., Semrau S., Airoldi E., Budnik B., and van Oudenaarden A. (2015) Differential stoichiometry among core ribosomal proteins. Cell Reports 13, 865–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Emmott E. P., Jovanovic M., and Slavov N. (2018) Ribosome stoichiometry: from form to function. Trends in Biochemical Sciences (in press) Peer J. Preprints 6, e26991v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. van den Berg P. R., Budnik B., Slavov N., and Semrau S. (2017) Dynamic post-transcriptional regulation during embryonic stem cell differentiation. bioRxiv doi: 10.1101/123497 [DOI] [Google Scholar]
- 47. Franks A., Airoldi E., and Slavov N. (2017) Post-transcriptional regulation across human tissues. PLoS Computational Biol. 13, e1005535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Budnik B., Levy E., Harmange G., and Slavov N. (2018) SCoPE-MS: mass-spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. Genome Biol. 19, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data are available at ProteomeXchange (accession ID: PXD008557, url: http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD008557) via MassIVE (accession ID: MSV000081857, url: https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=38f67398ac7a4547b68f343ca1aff905).