Abstract
Objective:
Elite controllers (EC), defined as persons maintaining undetectable levels of HIV-1 replication in the absence of antiretroviral therapy, represent living evidence that sustained, natural control of HIV-1 is possible, at least in relatively rare instances. Understanding the complex immunologic and virologic characteristics of these specific patients holds promise for inducing drug-free control of HIV-1 in broader populations of HIV-1 infected patients.
Design:
We used an unbiased transcriptional profiling approach to characterize CD8+ T cells, the strongest correlate of HIV-1 immune control identified thus far, in a large cohort of EC (n=51); HAART-treated patients (n=32) and HIV-1 negative (n=10) served as reference cohorts.
Methods:
We isolated mRNA from total CD8+ T cells isolated from PBMC of each subject followed by microarray analysis of the transcriptional signatures.
Results:
We observed profound transcriptional differences (590 transcripts, FDR-adjusted p<0.05) between EC and HAART-treated patients. Interestingly, metabolic and signaling pathways governed by mTOR and eIF2, known for their key roles in regulating cellular growth, proliferation, and metabolism, were among the top functions enriched in the differentially expressed genes, suggesting a therapeutically actionable target as a distinguishing feature of spontaneous HIV-1 immune control. A subsequent bootstrapping approach distinguished five different subgroups of EC, each characterized by distinct transcriptional signatures. However, despite this marked heterogeneity, differential regulation of mTOR and eIF2 signaling remained the dominant functional pathway in three of these EC subgroups.
Conclusions:
These studies suggest that mTOR and eIF2 signaling may play a remarkably universal role for regulating CD8+ T cell function from EC.
Keywords: Elite controllers, CD8+ T cells, transcriptional analysis, mTOR, eIF2
Introduction
Elite controllers (EC), a rare group of HIV-1-infected individuals (typically <1%) who maintain extremely low or undetectable levels of HIV-1 replication in the absence of antiretroviral therapy, may represent the closest possible approximation to a long-term remission or cure of HIV-1 infection [1]. Despite extremely low plasma HIV-1 RNA levels, these patients harbor clearly detectable populations of virally-infected cells, and can readily transmit virus to other individuals [2–4]. Prior studies have investigated a wide spectrum of host factors that may contribute to this remarkable phenotype, but failed to identify a unifying immune mechanism that is universally responsible for viral control in these patients. In part, this may be due to a constitutive heterogeneity among EC, who may include multiple distinct subgroups characterized by discrete mechanisms of viral control [5,6]. Nevertheless, empiric evidence from genome-wide association studies and other immunogenetic association studies, along with functional immunologic investigations and experimental animal data clearly suggest that cytotoxic T cells can have a profound impact on natural HIV-1 immune control, and arguably represent the best correlate of immune protection against HIV-1 disease progression [6–14]. CD8+ T cells can recognize viral peptide/MHC class I complexes and execute a variety of antiviral effector functions to restrict viral replication. Notably, a number of prior studies described significant qualitative differences between HIV-1-specific T cell responses from controllers and progressors, and suggested that continuously-ongoing viral replication in progressors drives T cells into an exhausted state, characterized by upregulation of inhibitory cell surface markers, defective proliferative activities, shortened telomeres, and reduced expression of perforin, granzyme B and the Th1 signature transcription factor T-bet [15–23]. Yet, most of these prior studies were biased towards specific pre-defined molecules or hypotheses, and did not allow for analysis of holistic and global characteristics distinguishing CD8+ T cells from individuals with natural control of HIV-1 infection. In the present study, we conducted unbiased microarray-supported transcriptional profiling studies of cytotoxic CD8+ T cells in a large cohort of HIV-1 EC, in comparison to HIV-1 negative persons and individuals with pharmacological viral suppression.
Methods
Patients
HIV-1-infected EC study participants (n=51) and HIV-1 negative control persons (n=10) were recruited from the Massachusetts General Hospital and the Brigham and Women’s Hospital (both in Boston, MA). The definition of EC varies widely in categorizing HIV-1 infected patient cohorts [24]. However, the definition used for inclusion of EC patients in this study is as follows: undetectable viral load using commercially-available HIV-1 plasma PCR assays (usually <50 or <75 copies/mL) on at least two consecutive occasions, in the absence of treatment with antiretroviral agents for at least two years. Occasional detection above the undetectable threshold (always <200 copies/mL) was allowed so long as subsequently measured plasma HIV-1 levels were below detection thresholds on at least two consecutive occasions. Peripheral blood mononuclear cell (PBMC) samples were used according to protocols approved by the local Institutional Review Board. Study subjects gave written informed consent to participate in accordance with the Declaration of Helsinki. Additionally, previously described HIV-1 infected study subjects treated with suppressive HAART (n=32) were used for this analysis [25]. Demographical characteristics of all the study subjects are summarized in Table S1 and S2.
Isolation of CD8+ T cells from PBMC
CD8+ T cells were isolated from PBMC of patient samples using an automated magnetic cell enrichment device (autoMACS, Miltenyi Biotec). Isolated CD8+ T cells routinely reached >95% purity. Samples from all cohorts were processed simultaneously for microarray analysis.
Transcriptional profiling (mRNA) using microarray analysis
Following mRNA extraction from the sorted cells (mirVana™ miRNA Isolation Kit, Ambion), whole-genome transcriptional profiling was performed using Illumina HumanHT-12 V4 microarrays according to standard protocols. Data retrieved from the Illumina software were background corrected (Illumina beadstudio software) and quantile normalized using the Arraystar normalization function. Subsequent data analysis was restricted to genes with significant expression (p<0.05) in at least 95% of samples. The microarray dataset was deposited to GEO (accession number GSE87620).
Data Analysis
Differentially expressed genes were identified using the R/Bioconductor package, LIMMA, with a cut-off False Discovery Rate (FDR)-adjusted p-value of <0.05 and fold change >1.5 fold [26]. First and second components from principal component analysis (PCA) were used to demonstrate the similarity/difference of expression patterns among all samples. Differentially-expressed genes were subjected to unsupervised hierarchical clustering analysis and visualized using heatmaps and volcano plots, using R [27]. Identification of canonical pathways, diseases and functions, as well as roles of upstream regulators of differentially-expressed genes were determined by Ingenuity Pathway Analysis (IPA, QIAGEN Inc.) and subsequently visualized using GraphPad Prism for Mac (GraphPad Software, La Jolla, CA, USA). Functional annotations of the differentially expressed genes were also determined using the Database for Annotation, Visualization and Integrated Discovery or DAVID [28,29]. Gene ontology of the biological processes were subsequently visualized using REVIGO [30]. To identify subgroups of EC, we performed a bootstrapping analysis with 1000 hierarchical clustering permutations, each consisting of 500 transcripts randomly selected out of to the total pool of 4921 transcripts that were detectable in at least 49 (96%) EC. Patients clustering with each other in >60% of these permutations were assigned to identical subgroups.
Results
Distinct transcriptional signatures of CD8+ T cells from HIV EC
Cytotoxic T cells have repeatedly been identified as a predominant correlate of HIV-1 immune control, but distinct molecular signatures most closely associated with effective antiviral activity of CD8+ T cells remain an area of remarkable uncertainty [5–7,31,32]. Individuals with natural control of HIV-1 replication to low or undetectable levels arguably represent the most informative patient cohort for characterizing protective CD8+ T cells, but relatively small sample sizes, combined with a narrow and exclusive focus on antigen-specific cells in prior studies may have limited investigative power to detect discrete transcriptional profiles of CD8+ T cells in this specific patient population [16,17,33–36]. To address this, we conducted a detailed analysis of genome-wide transcriptional programs of CD8+ T cells from a large cohort of EC (n=51), using unbiased microarray-based transcriptional profiling. HIV-1 patients treated with suppressive HAART (n=32), as well as a cohort of HIV-1 uninfected, healthy individuals (n=10) were recruited for comparative purposes.
This global comparison of gene expression patterns in CD8+ T cells demonstrated profound differences between HAART-treated patients and EC, with a total of 590 genes that were differentially expressed between these two cohorts using robust statistical criteria (FDR-adjusted p<0.05, fold-change difference in gene expression >1.5) (Figure 1A). Gene expression profiles between EC and HIV-1 negative individuals were remarkably similar, and displayed not a single transcript meeting the definition for differential gene expression, highlighting that CD8+ T cell transcriptional profiles from these two study cohorts were essentially indistinguishable. In contrast, a total of 47 transcripts were differentially expressed between HIV-1 negative and HAART treated patients (Figure 1A and Figure S1). Principal component analysis (PCA) and a hierarchical clustering analysis confirmed considerable differences in global gene expression patterns between EC and HAART-treated persons, while transcriptional profiles of HIV-1 negative subjects appeared to intermingle between these two study cohorts (Figure 1B and C). Notably, transcripts that were differentially-expressed between HIV-1 negative subjects and HAART-treated persons almost entirely overlapped with genes that differed between EC and HAART-treated patients, and the directional expression profiles of these 40 transcripts (encoding for 34 mapped genes functionally involved in cell death, proliferation, differentiation and cytokine response) were almost identical in both comparisons; this suggests that these 40 transcripts represent a molecular signature unique to HAART-treated persons (Figure 1D and E). Together, these data demonstrate distinct global gene expression patterns of CD8+ T cells in HIV-1 patients with immunological control of HIV-1 infection, compared to those with pharmacological control of HIV-1 infection.
Figure 1: Distinct transcriptional signatures of CD8+ T cells from HIV-1 EC.
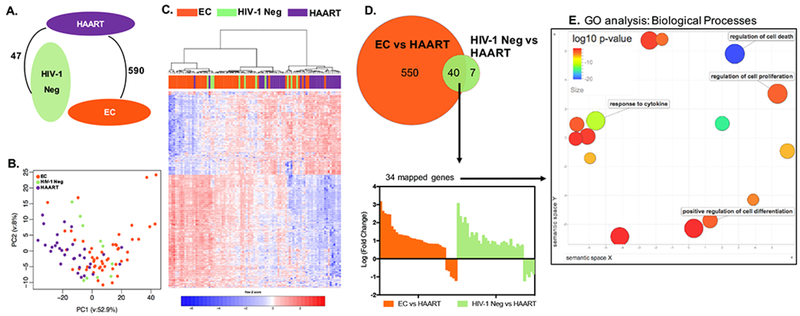
(A) Number of differentially expressed genes between EC, HIV-1 negative (labeled as HIV-1 Neg in all figures), and HAART-treated patients (FDR-adjusted p<0.05 and fold change >1.5). There was no significant transcriptional difference between CD8+ T cells from EC and HIV-1 negative persons in this analysis. (B) Principal Component Analysis (PCA) of the transcriptional profiles of CD8+ T cells from EC, HIV-1 negative, and HAART-treated patients. (C) Heat map reflecting 597 differentially expressed transcripts between CD8+ T cells from EC, HIV-1 negative, and HAART treated patients. The differentially expressed transcripts were chosen from 5377 transcripts that were significantly expressed in at least 95% of the samples. (D) Upper panel: Venn diagram depicting the differentially expressed transcripts between EC vs HAART-treated patients (orange circle) and HIV-1 negative vs HAART-treated individuals (green circle). Lower panel: Bar diagram reflecting directional transcriptional differences of 34 mapped genes distinguishing HAART-treated persons from EC and HIV-1 negative persons. (E) Gene ontology (GO) analysis of the 34 differentially expressed genes selected in (D). The plot generated using REVIGO reflects clustering of semantic similarities between GO terms of Biological Processes as determined by DAVID. Color coding corresponds to log10 (p-value) of GO terms in differentially expressed genes, also determined using DAVID. Terms with high functional relevance for CD8+ T cell biology are highlighted.
Metabolic pathways distinguish transcriptional signatures of EC
We subsequently focused on a closer analysis of the transcripts distinguishing EC from HAART-treated patients. Out of the 590 transcripts with different expression between EC and HAART-treated patients, a total of 342 were up- and 248 were down-regulated in EC (Figure 2A). A downstream effect analysis using IPA predicted that transcripts related to protein synthesis and metabolism, and genes involved in viral RNA replication and infection were enriched in EC, while transcripts related to cell death and necrosis were repressed in these patients (Figure 2B). Upstream regulators computationally inferred to govern the distinct transcriptional profile of CD8+ T cells from EC included the common gamma-chain cytokines IL-2, IL-7 and IL-15, as well as the inflammatory cytokines TNF-α and IL-1β (Figure 2C). We subsequently entered differentially expressed transcripts from EC into a weighed IPA analysis able to computationally translate altered gene expression patterns into directional changes of canonical biological pathways. These investigations demonstrated that CD8+ T cells from EC exhibited activation of closely-related metabolic pathways centered around the eukaryotic initiation factor 2 (eIF2), the mammalian target of rapamycin (mTOR), and Phosphoinositide-3-Kinases (PI3K). mTOR, a downstream effector of PI3Ks, acts as a nutrient and oxygen sensor and can exert critical influence on cellular gene translation, in part through modulation of eIF2 (Figure 2D). Notably, transcripts involved in oxidative stress and hypoxia, regulated at least partially by mTOR, were also increased in EC. In line with these findings, metabolic activation, protein synthesis, and TCR signaling were inferred as differentially regulated pathways in EC by alternative computational gene ontology prediction tools (Figure 2E and Figure S2). Previous reports also suggest similar types of coordinated signaling and metabolic control of CD8+ T cell functions [37–39]
Figure 2: Differential metabolic programs characterize transcriptional profiles of CD8+ T cells from EC.
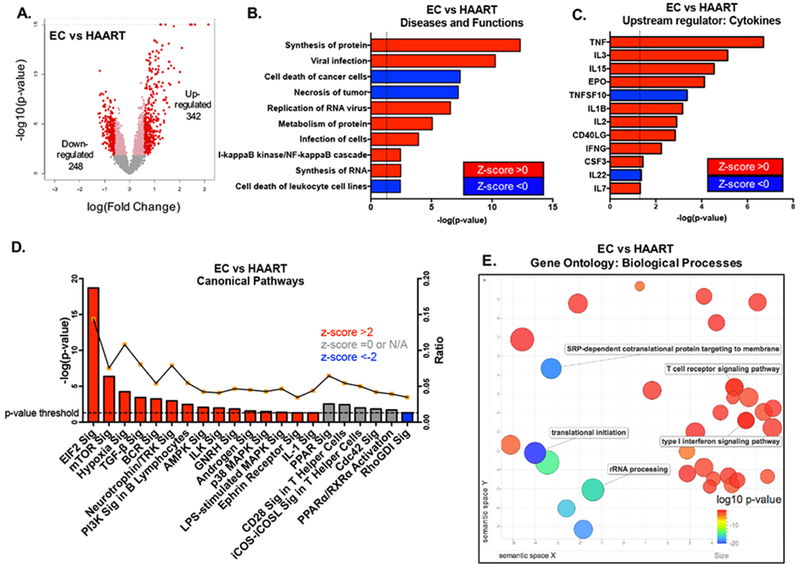
(A) Volcano plot of differentially expressed genes between CD8+ T cells from EC and HAART-treated patients (FDR-adjusted p<0.05, fold change >1.5). (B) Predicted biological functions of genes differentially expressed between EC and HAART-treated patients, as determined using Ingenuity Pathway Analysis (IPA). Dotted line depicts the threshold -log(p-value) of 1.3 or p-value≤0.05. Z-scores reflect predicted directional changes of genes function, positive z-score corresponds to functional activation while negative z-score reflects functional de-activation. (C) Upstream cytokines predicted to account for transcriptional differences between EC vs HAART-treated patients. Dotted line depicts the threshold -log(p-value) of 1.3 or p-value≤0.05. (D) Canonical pathways enriched in differentially regulated genes between EC and HAART, as determined using IPA. The right y-axis reflects the ratio between the number of differentially expressed genes that map to a particular pathway divided by the total number of genes curated to that pathway. Dotted line depicts the threshold -log(p-value) of 1.3 or p-value≤0.05. (E) Gene ontology (GO) analysis of the 590 differentially expressed transcripts between EC and HAART samples. The REVIGO plot reflects clustering of semantic similarities between the biological GO terms and is color coded according to log10(p-value) of the GO term as determined using DAVID analysis of the identified genes within the list of 590 transcripts.
Unbiased transcriptional profiling identifies distinct subgroups of EC
EC are operationally defined by low or undetectable viral loads, but numerous studies suggest that they may represent a heterogeneous group of patients with distinct underlying mechanisms of immune control [40]. To identify possible transcriptional differences between subgroups of EC, a bootstrapping approach was chosen, based on 1000 hierarchical clustering permutations of 500 randomly selected transcripts. Patients clustering with each other in >60% of these permutations were assigned to identical subgroups. Using this analytic strategy, we were able to categorize 49 members of our EC cohort to one of 5 different subgroups, termed EC1- EC5 in this manuscript, whose gene expression signatures were clearly distinguishable using a hierarchical clustering approach (Figure 3A) and by principal component analysis (Figure 3B). The numbers of transcripts that were differentially-expressed between these subgroups of EC ranged from 564 to 3327, and were highest for inter-subgroup comparisons with EC5, the smallest of all EC subgroups identified (Figure 3C). EC5 showed also the most striking transcriptional differences in comparison to HAART-treated individuals and HIV-1 negative persons, followed by EC3 and EC1; EC2 and EC4 showed comparatively minor differences relative to HAART-treated patients (Figure 3C and D). A similar pattern was noted in comparison to HIV-1 negative individuals, with EC5, EC3 and EC1 displaying the most profound transcriptional differences to this reference group (Figure 2C and E). Notably, despite the marked gene expression changes between these EC subgroups, we noted that patients’ age, the duration of HIV-1 infection, the ratio of CD4:CD8 cells or total CD4 counts, the neutralizing breadth of HIV-1-specific antibodies or the breadth and magnitude of HIV-1-specific CTL responses determined by interferon-gamma ELISpot assays were not significantly different between the different subgroups (Figure S3). Additionally, the expression intensity of the immune checkpoint molecules Tim-3, LAG3, and PD-1, did not markedly differ among total CD8+ T cells from the detected subgroups of EC (Figure S4B), despite the distinct expression profile of such markers previously observed on HIV-1-specific CD8+ T cells from controllers [41,42]. However, CTLA4 expression was slightly, albeit significantly, increased in HIV-1 negative individuals when compared to CD8+ T cells from total EC and the EC4 subgroup (Figure S4A, S4B).
Figure 3: Transcriptional profiling of CD8+ T cells identifies distinct subgroups of EC.
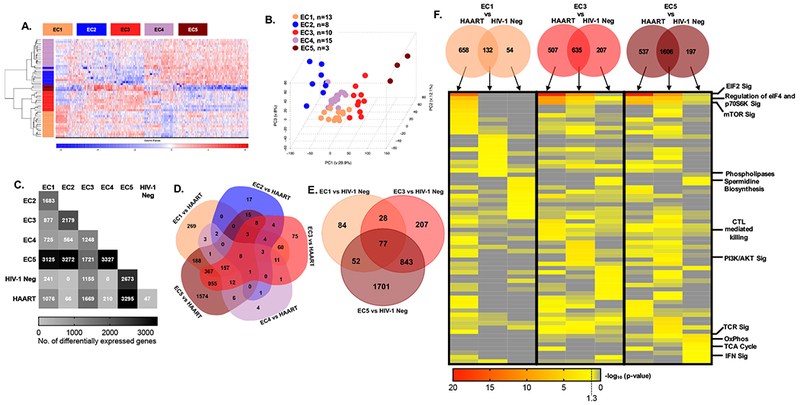
(A) Heat map showing differentially expressed genes among five EC subgroups identified using a bootstrapping approach; 49 out of 51 EC patients who met the analysis criteria described in the methods section were included. (B) PCA analysis of EC subgroups. (C) Matrix diagram indicating the frequency of differentially expressed genes between EC subgroups as well as HIV-1 negative and HAART treated patients. (D-E) Venn diagrams depicting the number of differentially-expressed genes distinguishing EC subgroups from HAART-treated patients (D) or HIV-1 negative study persons (E). No statistically significant transcriptional differences were detected between EC2 vs HIV-1 negative persons or EC4 vs HIV-1 negative study subjects. (F) Transcripts that distinguish EC1, 3, and 5 from HAART (C, D) and HIV-1 negative (C, E) CD8+ T cells were used to generate the Venn diagrams demonstrating genes differentially expressed between indicated EC subgroups and either HAART-treated individuals, HIV-1 negative study subjects or both. The columns of the heat map correspond to the individual segments of the Venn diagram as indicated by the arrows. Heat map depicts unbiased analysis of top ten canonical pathways enriched in each indicated gene set as determined using IPA. Selected pathway annotations are indicated on the right of the heat map.
Transcriptional signatures of mTOR and eIF2 in EC subgroups
To more closely analyze the functional differences between these EC subgroups, we focused on transcripts that distinguished EC subgroups from either HIV-1 negative study subjects, HAART-treated control patients or both reference cohorts, and selectively entered these transcripts into a predictive canonical pathway analysis supported by IPA. This analysis demonstrated that transcripts involved in translational control by eIF2, eIF4, p70S6K and mTOR signaling pathways, previously identified as key functional regulators distinguishing the EC cohort as a whole from the HAART-treated samples (Figure 2D), were most clearly detectable in EC1 (Figure 3F). However, relative to the control cohorts, mTOR and eIF signaling pathways were also predicted to be significantly involved in all other EC subgroups, except for EC2 (Figure 4A and B). Yet, individual gene members of the eIF2 and mTOR signaling pathways frequently showed strikingly different, and sometimes opposing gene expression patterns between the EC subgroups, consistent with discrete activation and de-activation of eIF2/eIF4/mTOR pathway members in EC subgroups (Figure 4B-E). These observations support the role of eIF2 and mTOR-governed metabolic pathways as distinguishing features of CD8+ T cells from EC, but highlight considerable variations in metabolic pathway regulation between the EC subgroups, potentially involving receptor downstream intermediates such as PI3K/AKT or p38 signaling (Figure 4E). Moreover, each of the analyzed EC subgroups were associated with distinct, although partially overlapping functional annotations (Figure 4F-G). For instance, signatures of EC3 and EC5 seemed to be enriched for transcripts related to T cell maturation, activation, proliferation, as well as CTL cytotoxicity, as opposed to EC2 in whom these functions were less obvious (Figure 4F-H). Additionally, EC2 showed predicted signatures of increased cell death (Figure 4H), while EC1, EC3, and EC5 were enriched for transcripts predicted to induce cell survival and viability (Figure 4G-H). These data suggest that discrete transcriptional profiles of CD8+ T cells in EC subgroups may contribute differentially to HIV-1 immune control in EC subgroups.
Figure 4: Functional variations in transcriptional signatures among the EC subgroups.
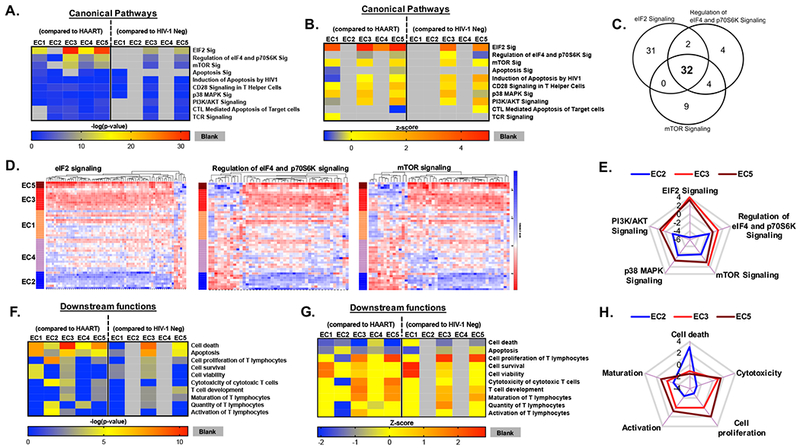
(A-B) Functional annotations of differentially expressed genes between indicated EC subgroups and HAART-treated patients or HIV-1 negative persons. Data show p-values (A) and z-scores (B) of selected Canonical Pathways determined by IPA. Functional annotations with p-values <0.05 for EC1, EC3 and EC5 are included. Grey segments represent a lack of value as determined using IPA. (C) Venn diagram depicting overlaps in the genes involved in the top three canonical pathway hits of genes distinguishing EC subgroups and control cohorts. (D) Heat maps reflecting expression profile of transcripts involved in the top three canonical pathways highlighted in (C). (E) Comparative analysis of the top five interconnected canonical pathways in EC2, EC3, and EC5. The radar plot depicts predicted activation (positive z-score) or de-activation (negative z-score) of the canonical pathways in indicated EC subgroups, as determined using IPA analysis. EC3 did not have any z-score associated with p38 MAPK signaling pathway. (F-G) Downstream CD8+ T cell functions were predicted using differentially expressed genes between indicated EC subgroups and HAART-treated patients or HIV-1 negative donors. Heat maps show p-values (F) and Z-scores (G) of selected CD8+ T cell functions determined using IPA. Grey segments represent a lack of value as determined using IPA or a lack of significant differentially expressed transcripts in input for IPA analysis. (H) Comparative analysis of the relevant functions in EC2, EC3, and EC5. The radar plot depicts predicted up-regulation (positive z-score) or down-regulation (negative z-score) of the selected functions in indicated EC subgroups, as determined using IPA analysis.
Discussion
HIV-1 infection can lead to drastically different clinical outcomes, ranging from EC with a functional cure of HIV-1 infection to persons with rapid progression to clinical immune deficiency and death. How natural, drug-free control of HIV-1 replication can be achieved seems to vary considerably among different individuals, but CD8+ T cells represent the immune compartment most frequently invoked in HIV-1 immune control in humans, and most definitively validated as correlates of SIV immune protection in SIV-infected animals. Antiretroviral effects of CD8+ T cells have been mostly attributed to the small subset of HIV-1-specific CD8+ T cells, which can be detected and visualized by intracellular cytokine staining or MHC class I tetramers [32,43–45]. These cells can recognize cognate HIV-1 peptides presented by MHC class I complexes and execute antiviral effector functions upon antigen stimulation. However, increasing evidence suggest that antiviral effects of CD8+ T cells may not be restricted to the HIV-1-specific CD8+ T cells, but more broadly involve the entire CD8+ T cell compartment. For instance, elegant studies by Cartwright et al. demonstrate that depletion of total CD8+ T cells in ART-treated SIV-infected rhesus macaques leads to blips of viremia, arguably resulting from re-ignition of viral replication that is otherwise suppressed by CD8+ T cells [46]. Moreover, work by Nishimura et al showed that treatment of SIV-infected rhesus macaques with broadly-neutralizing antibodies can induce a CD8+ T cell-mediated form of viral immune control [47]. In the present work, we have interrogated the entire CD8+ T cell compartment of a large cohort of EC, using a transcriptional profiling approach. Surprisingly, these studies failed to reflect a specific antiviral effector profile of CD8+ T cells in EC, but instead identified a distinct program of signaling networks governed by PI3K/AKT, mTOR, and eIF2, an interconnected family of cellular signal transduction molecules with critical roles for regulating cellular growth, proliferation, and metabolism, as distinguishing aspects of CD8+ T cells from EC. Notably, mTOR has also been shown to play a pivotal role in controlling HIV transcription and latency in CD4+ T cells, and pharmaceutical inhibitors of the mTORC1 sub-complex such as rapamycin/sirolimus protect against a variety of age-related pathologies, leading to extended lifespan in model organisms [48–51]. Remarkably, sirolimus is currently evaluated as a modulator of antiviral immune responses and viral reservoir sizes in HIV-1 infected patients in a multicenter study, and such an interventional study may allow to more specifically identify mechanisms by which mTOR modulation may contribute to natural control of HIV-1 (ClinicalTrials.gov Identifier: NCT02440789). Although we found that mTOR-associated transcripts were altered in the vast majority of EC relative to the control cohorts, we noted that directional gene expression patterns of such transcripts varied considerably among EC subgroups, suggesting distinct effects of the mTOR signaling pathway in subgroups of such patients. Future comparisons of mTOR-associated transcriptional signatures in EC and HAART-treated HIV-1 patients undergoing treatment with sirolimus may allow to delineate possible therapeutic benefits of mTORC1 inhibition during HIV-1 infection, and help to identify optimized biomarkers of metabolic and immunologic changes associated with sirolimus therapy.
An important finding of this study is the transcriptional heterogeneity in CD8+ T cells from EC. This observation corresponds well to a series of prior studies highlighting marked immunological differences among EC, despite a seemingly identical clinical phenotype defined by undetectable levels of viral replication in the absence of antiretroviral therapy [5,6]. In fact, previous studies have identified diverse groups of EC subgroups based on the presence or absence of protective HLA class I alleles, the magnitude and functionality of HIV-1 CD8+ T cells, the susceptibility of CD4+ T cells to HIV-1 in ex vivo infection assays, and the transcriptional signatures of CD4+ T cells [5,6,52]. Together with the findings reported here, these results strongly suggest that natural control of HIV-1 in the absence of treatment is likely a result of combinations of different immune mechanisms that may directly or indirectly contribute to restriction of HIV-1 replication. In the future, a systematic parallel assessment of transcriptional gene expression patterns, virological reservoir parameters, immune phenotypes and functions and epigenetic profiles in large numbers of EC may be necessary to detect more comprehensive, multi-faceted characteristics that distinguish EC, and mechanistically contribute to antiviral immune control. Moreover, use of improved techniques such as single-cell RNA-seq [53] may allow for a more personalized, “precision medicine”-oriented approach for identifying individual components of immune protection in EC. Given the considerable morbidity that HIV-1 continues to exert worldwide, despite improved availability and access to potent antiretroviral therapy, such a large-scale effort to characterize EC may represent a promising approach to identify and, ultimately, induce mechanisms of natural HIV-1 control in broader populations of patients.
Supplementary Material
Figure S1: Heat map of 47 differentially expressed genes (FDR-adjusted p<0.05, fold change >1.5) between CD8+ T cells from HIV-1 negative and HAART treated patients.
Figure S2: (A) Enrichment of KEGG Pathways using DAVID Analysis within genes differentially expressed between EC and HAART. (B) Differentially expressed genes between EC and HAART that are involved in significantly enriched biological processes and pathways in Annotation Cluster 1 using DAVID analysis.
Figure S3: Box and Whisker plots reflecting (A) patients’ age at the time of sampling, (B) duration of HIV infection, (C) ratio of CD4:CD8 at the time of sampling, and (D) CD4 count at the time of sampling are reported. Plots reflect minimum, maximum, and 25%, 50% and 75% percentiles; outliers are shown individually using open circles. (E) Viral load measured by single copy viral RNA assay and (F) breadth of neutralizing antibody as measured in virus neutralization assay where the result was considered positive if > 3× negative control, are also reported in indicated EC subgroups. Breadth (G) or magnitude (H) of HIV-1-specific CD8+ T cells against optimal epitopes in indicated EC subgroups are also shown. (E-H) Not all parameters were measured in every patient due to sample availability and each dot represents an individual in the subgroup.
Figure S4: EC bulk CD8+ T cells do not have high expression of exhaustion markers when compared with HIV-1 negative and HAART treated patients. (A) Normalized expression values of T cell exhaustion markers CTLA4, Tim-3, LAG3, and PD1 are compared between EC, HIV-1 negative, and HAART treated patients. (B) Normalized expression values of the same exhaustion markers are compared between EC subgroups, HIV-, and HAART treated patient CD8+ T cells. Kruskal-Wallis test was performed for each plot with Dunn’s multiple comparisons test to determine statistical significance. * denotes p-value<0.05. The detection threshold of expression value is set at 3.17716 or log2(8), reflecting significant expression (p-value <0.05) and indicated with the dashed line.
Acknowledgments
We would like to thank the Flow Cytometry and Imaging Core Facility at the Ragon Institute of MGH, MIT and Harvard for their help with cell isolation. We also want to thank the clinical coordinators at the Ragon Institute or MGH, MIT and Harvard for their help with patient recruitment, sample collection, and careful recordkeeping.
Funding Sources: This work was supported for the US National Institutes of Health (grants HL134539, AI078799, AI116228, AI098484, and HL126554 to XGY; 2T32AI007387 to FZC). PBMC sample collection was supported by the Bill and Melinda Gates Foundation (OPP 1066973), the Mark and Lisa Swartz Foundation, the Ragon Institute of MGH, MIT and Harvard, and the International HIV Controller Consortium.
Footnotes
Conflicts of Interest: The authors declare no conflicts of interest.
References
- 1.Walker BD, Yu XG. Unravelling the mechanisms of durable control of HIV-1. Nat Rev Immunol 2013; 13:487–498. [DOI] [PubMed] [Google Scholar]
- 2.Julg B, Pereyra F, Buzón MJ, Piechocka-Trocha A, Clark MJ, Baker BM, et al. Infrequent recovery of HIV from but robust exogenous infection of activated CD4(+) T cells in HIV elite controllers. CLIN INFECT DIS 2010; 51:233–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blankson JN, Bailey JR, Thayil S, Yang H-C, Lassen K, Lai J, et al. Isolation and characterization of replication-competent human immunodeficiency virus type 1 from a subset of elite suppressors. Journal of Virology 2007; 81:2508–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckheit RW, Allen TG, Alme A, Salgado M, O’Connell KA, Huculak S, et al. Host factors dictate control of viral replication in two HIV-1 controller/chronic progressor transmission pairs. Nat Commun 2012; 3:716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sáez-Cirión A, Sinet M, Shin SY, Urrutia A, Versmisse P, Lacabaratz C, et al. Heterogeneity in HIV Suppression by CD8 T Cells from HIV Controllers: Association with Gag-Specific CD8 T Cell Responses. The Journal of Immunology 2009; 182:7828–7837. [DOI] [PubMed] [Google Scholar]
- 6.Pereyra F, Addo MM, Kaufmann DE, Liu Y, Miura T, Rathod A, et al. Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. J Infect Dis 2008; 197:563–571. [DOI] [PubMed] [Google Scholar]
- 7.International HIV Controllers Study, Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PIW, et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 2010; 330:1551–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pereyra F, Heckerman D, Carlson JM, Kadie C, Soghoian DZ, Karel D, et al. HIV control is mediated in part by CD8+ T-cell targeting of specific epitopes. Journal of Virology 2014; 88:12937–12948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Migueles SA, Sabbaghian MS, Shupert WL, Bettinotti MP, Marincola FM, Martino L, et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proceedings of the 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Migueles SA, Mendoza D, Zimmerman MG, Martins KM, Toulmin SA, Kelly EP, et al. CD8(+) T-cell Cytotoxic Capacity Associated with Human Immunodeficiency Virus-1 Control Can Be Mediated through Various Epitopes and Human Leukocyte Antigen Types. EBioMedicine 2015; 2:46–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, et al. A Whole-Genome Association Study of Major Determinants for Host Control of HIV-1. Science 2007; 317:944–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 2006; 107:4781–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brennan CA, Ibarrondo FJ, Sugar CA, Hausner MA, Shih R, Ng HL, et al. Early HLA-B*57-Restricted CD8+ T Lymphocyte Responses Predict HIV-1 Disease Progression. Journal of Virology 2012; 86:10505–10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferre AL, Hunt PW, Critchfield JW, Young DH, Morris MM, Garcia JC, et al. Mucosal immune responses to HIV-1 in elite controllers: a potential correlate of immune control. Blood 2009; 113:3978–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmann M, Pantazis N, Martin GE, Hickling S, Hurst J, Meyerowitz J, et al. Exhaustion of Activated CD8 T Cells Predicts Disease Progression in Primary HIV-1 Infection. PLoS Pathog 2016; 12:e1005661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaiha GD, McKim KJ, Woods M, Pertel T, Rohrbach J, Barteneva N, et al. Dysfunctional HIV-specific CD8+ T cell proliferation is associated with increased caspase-8 activity and mediated by necroptosis. Immunity 2014; 41:1001–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quigley M, Pereyra F, Nilsson B, Porichis F, Fonseca C, Eichbaum Q, et al. Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nat Med 2010; 16:1147–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elahi S, Dinges WL, Lejarcegui N, Laing KJ, Collier AC, Koelle DM, et al. Protective HIV-specific CD8+ T cells evade Treg cell suppression. Nat Med 2011; 17:989–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams K, Seiss K, Beamon J, Pereyra F, Rosenberg ES, Walker BD, et al. Epigenetic regulation of telomerase expression in HIV-1-specific CD8+ T cells. AIDS 2010; 24:1964–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hersperger AR, Martin JN, Shin LY, Sheth PM, Kovacs CM, Cosma GL, et al. Increased HIV-specific CD8+ T-cell cytotoxic potential in HIV elite controllers is associated with T-bet expression. Blood 2011; 117:3799–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hersperger AR, Pereyra F, Nason M, Demers K, Sheth P, Shin LY, et al. Perforin expression directly ex vivo by HIV-specific CD8 T-cells is a correlate of HIV elite control. PLoS Pathog 2010; 6:e1000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Migueles SA, Osborne CM, Royce C, Compton AA, Joshi RP, Weeks KA, et al. Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity 2008; 29:1009–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Migueles SA, Laborico AC, Shupert WL, Sabbaghian MS, Rabin R, Hallahan CW, et al. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat Immunol 2002; 3:1061–1068. [DOI] [PubMed] [Google Scholar]
- 24.Gurdasani D, Iles L, Dillon DG, Young EH, Olson AD, Naranbhai V, et al. A systematic review of definitions of extreme phenotypes of HIV control and progression. AIDS 2014; 28:149–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ouyang Z, Buzon MJ, Zheng L, Sun H, Yu XG, Bosch RJ, et al. Transcriptional Changes in CD8(+) T Cells During Antiretroviral Therapy Intensified With Raltegravir. Open Forum Infect Dis 2015; 2:ofv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.TEAM RRC. R: A language and environment for statistical computing. 2008. [Google Scholar]
- 28.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 29.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009; 37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011; 6:e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sáez-Cirión A, Lacabaratz C. HIV controllers exhibit potent CD8 T cell capacity to suppress HIV infection ex vivo and peculiar cytotoxic T lymphocyte activation phenotype. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shasha D, Karel D, Angiuli O, Greenblatt A, Ghebremichael M, Yu X, et al. Elite controller CD8+ T cells exhibit comparable viral inhibition capacity, but better sustained effector properties compared to chronic progressors. J Leukoc Biol Published Online First: July 2016. doi: 10.1189/jlb.4A0915-422R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu JQ, Dwyer DE, Dyer WB, Yang YH, Wang B, Saksena NK. Genome-wide analysis of primary CD4+ and CD8+ T cell transcriptomes shows evidence for a network of enriched pathways associated with HIV disease. Retrovirology 2011; 8:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu JQ, Dwyer DE, Dyer WB, Yang YH, Wang B, Saksena NK. Transcriptional profiles in CD8+ T cells from HIV+ progressors on HAART are characterized by coordinated up-regulation of oxidative phosphorylation enzymes and interferon responses. Virology 2008; 380:124–135. [DOI] [PubMed] [Google Scholar]
- 35.Martinez-Mariño B, Foster H, Hao Y, Levy JA. Differential gene expression in CD8(+) cells from HIV-1-infected subjects showing suppression of HIV replication. Virology 2007; 362:217–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Payne TL, Blackinton J, Frisbee A, Pickeral J, Sawant S, Vandergrift NA, et al. Transcriptional and posttranscriptional regulation of cytokine gene expression in HIV-1 antigen-specific CD8+ T cells that mediate virus inhibition. Journal of Virology 2014; 88:9514–9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trautmann L, Mbitikon-Kobo F-M, Goulet J-P, Peretz Y, Shi Y, Van Grevenynghe J, et al. Profound metabolic, functional, and cytolytic differences characterize HIV-specific CD8 T cells in primary and chronic HIV infection. Blood 2012; 120:3466–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu JQ, Dwyer DE, Dyer WB, Yang YH, Wang B, Saksena NK. Transcriptional profiles in CD8+ T cells from HIV+ progressors on HAART are characterized by coordinated up-regulation of oxidative phosphorylation enzymes and interferon responses. Virology 2008; 380:124–135. [DOI] [PubMed] [Google Scholar]
- 39.Hukelmann JL, Anderson KE, Sinclair LV, Grzes KM, Murillo AB, Hawkins PT, et al. The cytotoxic T cell proteome and its shaping by the kinase mTOR. Nat Immunol 2015; 17:104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vigneault F, Woods M, Buzon MJ, Li C, Pereyra F, Crosby SD, et al. Transcriptional profiling of CD4 T cells identifies distinct subgroups of HIV-1 elite controllers. Journal of Virology 2011; 85:3015–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sabins NC, Harman BC, Barone LR, Shen S, Santulli-Marotto S. Differential Expression of Immune Checkpoint Modulators on In Vitro Primed CD4(+) and CD8(+) T Cells. Front Immunol 2016; 7:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marra A, Scognamiglio G, Peluso I, Botti G, Fusciello C, Filippelli A, et al. Immune Checkpoint Inhibitors in Melanoma and HIV Infection. Open AIDS J 2017; 11:91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ndhlovu ZM, Proudfoot J, Cesa K, Alvino DM, McMullen A, Vine S, et al. Elite controllers with low to absent effector CD8+ T cell responses maintain highly functional, broadly directed central memory responses. Journal of Virology 2012; 86:6959–6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ndhlovu ZM, Kamya P, Mewalal N, Kloverpris HN, Nkosi T, Pretorius K, et al. Magnitude and Kinetics of CD8+ T Cell Activation during Hyperacute HIV Infection Impact Viral Set Point. Immunity 2015; 43:591–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ndhlovu ZM, Stampouloglou E, Cesa K, Mavrothalassitis O, Alvino DM, Li JZ, et al. The breadth of expandable memory CD8+ T cells inversely correlates with residual viral loads in HIV elite controllers. Journal of Virology Published Online First: 12 August 2015. doi: 10.1128/JVI.01527-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cartwright EK, Spicer L, Smith SA, Lee D, Fast R, Paganini S, et al. CD8(+) Lymphocytes Are Required for Maintaining Viral Suppression in SIV-Infected Macaques Treated with Short-Term Antiretroviral Therapy. Immunity 2016; 45:656–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishimura Y, Gautam R, Chun T-W, Sadjadpour R, Foulds KE, Shingai M, et al. Early antibody therapy can induce long-lasting immunity to SHIV. Nature 2017; 543:559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Besnard E, Hakre S, Kampmann M, Lim HW, Hosmane NN, Martin A, et al. The mTOR Complex Controls HIV Latency. Cell Host Microbe 2016; 20:785–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin AR, Pollack RA, Capoferri A, Ambinder RF, Durand CM, Siliciano RF. Rapamycin-mediated mTOR inhibition uncouples HIV-1 latency reversal from cytokine-associated toxicity. J Clin Invest 2017; 127:651–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rai P, Plagov A, Lan X, Chandel N, Singh T, Lederman R, et al. mTOR plays a critical role in p53-induced oxidative kidney cell injury in HIVAN. Am J Physiol Renal Physiol 2013; 305:F343–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pan H, Finkel T. Key proteins and pathways that regulate lifespan. J Biol Chem 2017; 292:6452–6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vigneault F, Woods M, Buzon MJ, Li C, Pereyra F, Crosby SD, et al. Transcriptional profiling of CD4 T cells identifies distinct subgroups of HIV-1 elite controllers. Journal of Virology 2011; 85:3015–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martin-Gayo E, Cole MB, Kolb KE, Ouyang Z, Cronin J, Kazer SW, et al. A Reproducibility-Based Computational Framework Identifies an Inducible, Enhanced Antiviral State in Dendritic Cells from HIV-1 Elite Controllers. Genome Biol 2018; 19:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Heat map of 47 differentially expressed genes (FDR-adjusted p<0.05, fold change >1.5) between CD8+ T cells from HIV-1 negative and HAART treated patients.
Figure S2: (A) Enrichment of KEGG Pathways using DAVID Analysis within genes differentially expressed between EC and HAART. (B) Differentially expressed genes between EC and HAART that are involved in significantly enriched biological processes and pathways in Annotation Cluster 1 using DAVID analysis.
Figure S3: Box and Whisker plots reflecting (A) patients’ age at the time of sampling, (B) duration of HIV infection, (C) ratio of CD4:CD8 at the time of sampling, and (D) CD4 count at the time of sampling are reported. Plots reflect minimum, maximum, and 25%, 50% and 75% percentiles; outliers are shown individually using open circles. (E) Viral load measured by single copy viral RNA assay and (F) breadth of neutralizing antibody as measured in virus neutralization assay where the result was considered positive if > 3× negative control, are also reported in indicated EC subgroups. Breadth (G) or magnitude (H) of HIV-1-specific CD8+ T cells against optimal epitopes in indicated EC subgroups are also shown. (E-H) Not all parameters were measured in every patient due to sample availability and each dot represents an individual in the subgroup.
Figure S4: EC bulk CD8+ T cells do not have high expression of exhaustion markers when compared with HIV-1 negative and HAART treated patients. (A) Normalized expression values of T cell exhaustion markers CTLA4, Tim-3, LAG3, and PD1 are compared between EC, HIV-1 negative, and HAART treated patients. (B) Normalized expression values of the same exhaustion markers are compared between EC subgroups, HIV-, and HAART treated patient CD8+ T cells. Kruskal-Wallis test was performed for each plot with Dunn’s multiple comparisons test to determine statistical significance. * denotes p-value<0.05. The detection threshold of expression value is set at 3.17716 or log2(8), reflecting significant expression (p-value <0.05) and indicated with the dashed line.


