Abstract
P-glycoprotein (Pgp) is a multidrug resistance transporter that limits the penetration of a wide range of neurotherapeutics into the brain including opioids. The diphenylpropylamine opioids methadone and loperamide are structurally similar, but loperamide has about a 4-fold higher Pgp-mediated transport rate. In addition to these differences, they showed significant differences in their effects on Pgp-mediated ATP hydrolysis. The activation of Pgp-mediated ATP hydrolysis by methadone was monophasic, while loperamide activation of ATP hydrolysis was biphasic implying methadone has a single binding site and loperamide has two binding sites on Pgp. Quenching of tryptophan fluorescence with these drugs and digoxin showed competition between the opioids and that loperamide does not compete for the digoxin binding site. Acrylamide quenching of tryptophan fluorescence to probe Pgp conformational changes revealed that methadone-and loperamide-induced conformational changes were distinct. These results were used to develop a model for Pgp-mediated transport of methadone and loperamide where opioid binding and conformational changes are used to explain the differences in the opioid transport rates between methadone and loperamide.
Keywords: ABC transporters, P-glycoprotein, fluorescence, nuclear magnetic resonance (NMR), opioids
INTRODUCTION
The blood-brain barrier (BBB) is an intricate network of highly selective cells that protect the brain from toxic insults.1 The ATP binding cassette (ABC) transporter P-glycoprotein (Pgp) is highly expressed on the apical surface of the BBB.1–3 Pgp is a highly promiscuous protein that effluxes a structurally and chemically diverse range of compounds away from the brain into the bloodstream.3–5 While this function is critical for neurohomeostasis and for preventing the penetration of toxic insults, Pgp’s role at the BBB prevents the entry of many neurotherapeutics and represents a formidable hurdle to drug development.6–8 Over the last three decades, considerable effort has been made to improve penetration of drugs by inhibiting Pgp, but this has been unsuccessful in the clinic because of unforeseen toxicity.9–11 As a result, there has been considerable interest in the pharmaceutical industry to identify the molecular features of drugs that drive Pgp-mediated transport.
Most of our structural understanding of Pgp comes from X-ray crystallography and a cryo-electron microscopy study.5,12–17 Previously determined X-ray crystal structures of mouse Pgp revealed a 140 kDa monomer consisting of twelve transmembrane helices and two nucleotide binding domains (NBDs).5,13–16 The NBDs lie in the interior of the cell and bind ATP, resulting in large conformational changes that bring the NBDs together, as seen in the analogous bacterial transporters.5,13,18,19 Twelve transmembrane helices create an enormous 6000 Å3 binding cavity capable of binding chemically diverse molecules and multiple substrates simultaneously.5,13,20,21
The diphenylpropylamine opioid neurotherapeutic drug class has been used to manage pain and is designed to target the μ-opioid receptors in the central nervous system (CNS).22,23 Unfortunately, penetration of some diphenylpropylamine opioids into the CNS is stymied by Pgp-mediated efflux.24–27 On the extremes are the diphenylpropylamine opioids loperamide and methadone. Loperamide has extremely poor penetration into the CNS as a result of Pgp efflux.25 An in vitro study in MDR1-transfected cells found that loperamide had relatively high Pgp-mediated transport with efflux ratios ranging from 6.5 to 9.9.4,28,29 Several in vivo studies have demonstrated Pgp-mediated transport of loperamide. Pgp-knockout mice had more than a 10-fold higher level of loperamide in their brains than wild-type.30,31 Both rats and humans have shown elevated levels of loperamide in the presence of the Pgp-specific inhibitor cyclosporine A (CsA).32 In contrast, the diphenylpropylamine derivative methadone shows very good penetration into the CNS and the brain and is used for treatment of opioid addiction despite its structural similarities to loperamide (Fig. 1).23,33 Methadone was found to inhibit Pgp-mediated efflux of rhodamine-123 and calcein-AM in Caco-2 and HEK293 cells.34,35 Methadone has also been shown to be transported by Pgp in Pgp-transfected cells and had efflux ratios around 2 that decreased in the presence of a Pgp specific inhibitor.28 Transport has also been demonstrated in vivo with Pgp knockout mice having greater than a 3-fold methadone accumulation in the brain versus wild type.23,30,31
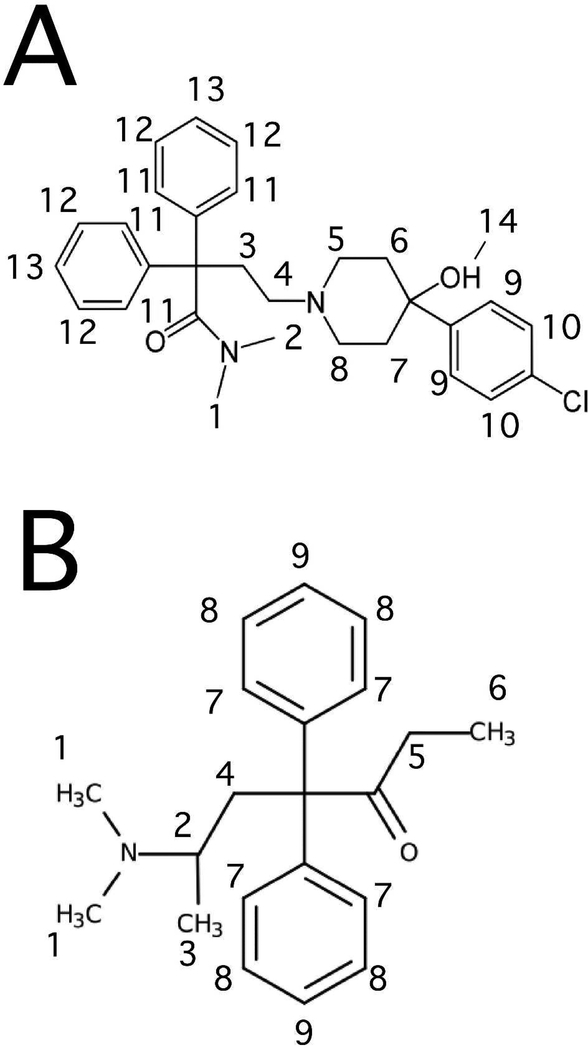
Figure 1.
The molecular structures of (A) loperamide and (B) methadone with protons labeled.
Because of large differences in the transport properties of methadone and loperamide, this study represents an opportunity to bridge gaps in the understanding of neurotherapeutic drug transport by Pgp. In this study, we examined the effect of these drugs on Pgp-mediated ATP hydrolysis. Their interactions with Pgp were probed by fluorescence spectroscopy and an NMR technique known as saturation transfer double difference (STDD) NMR. Finally, drug-induced Pgp conformational changes were investigated through acrylamide quenching of tryptophan fluorescence. These results and previous transport studies23,28,30,31 were used to create conformationally-driven model of diphenylpropylamine opioid transport by Pgp.
EXPERIMENTAL
Materials
Loperamide was purchased from MP Biomedicals (Santa Ana, CA). Digoxin, ethylene glycol tetraacetic acid (EGTA), and imidazole were purchased from Alfa Aesar (Tewksbury, MA). The detergent n-dodecyl-β-D-maltoside (DDM) was purchased from EMD Millipore Corporation (San Diego, CA). Cholesterol, disodium ATP, and Tris-HCl were purchased from Amresco (Solon, OH). Escherichia (E.) coli total lipid extract powder was purchased from Avanti Polar Lipids Inc (Alabaster, AL). Dithiothreitol (DTT) was purchased from Gold Biotechnology (Olivette, MO). Deuterated dithiothreitol was purchased from CDN Isotopes (Quebec, Canada). HEPES and acrylamide were purchased from Calbiochem (San Diego, CA). Deuterium oxide was purchased from Cambridge Isotope Laboratories, Inc (Tewksbury, MA). Sodium orthovanadate (Na3VO4) was purchased from Enzo Life Sciences (Farmingdale, NY). Ammonium chloride (NH4Cl), deuterated dimethyl sulfoxide, and methadone were purchased from Sigma-Aldrich (Milwaukee, WI). All remaining chemicals used in this study were purchased from Thermo Fischer Scientific (Waltham, MA).
Methods
Purification and Reconstitution of Pgp
The His-tagged wild type mouse Pgp (Abcb1a, MDR3) was overexpressed in Pichia pastoris and purified as previously described in two steps with nickel-nitrilotriacetic acid (Ni-NTA) (Thermo-Fischer Scientific) and diethylaminoethyl cellulose (DEAE) resin (Thermo-Fischer Scientific).36,37 Pgp solubilized in n-dodecyl-β-D-maltoside (DDM) was reconstituted into 400 nm liposomes using a previously described procedure.38–40 Liposomes consisted of 80% wt/vol Avanti Total E. Coli Lipid Extract (Avanti Polar Lipids, Alabaster, AL) and 20% wt/v cholesterol with a lipid-to-protein ratio of 0.16 mg ml−1.38–41 The Avanti Total E. Coli Lipid Extract was chosen because Pgp reconstituted with the lipid has high ATP hydrolysis activity and has been used in many investigations of human and mouse Pgp e.g.37,42,43 The 20% wt/v cholesterol was added to the lipid mixture to improve Pgp’s ATPase activity.41,44 To create the liposomes, lipids and cholesterol were dissolved in 10 ml of chloroform to a concentration of 10 mg ml−1. The solution was evaporated using a Buchi Rotovap Model R-114 (Buchi) to remove the chloroform. The resulting dried lipid film was resuspended in 10 mL of 0.1 mM EGTA and 50 mM Tris-HCl (pH 7.4). The solution was freeze-thawed in liquid nitrogen at least 10 times. Liposomes were extruded using a LIPEX extruder with a 400 nm cutoff filter 11 times (Northern Lipids). DDMsolubilized protein (~100 μM) was dialyzed against HEPES buffer (20 mM HEPES, 100 mM NaCl, 5 mM MgCl2, 2 mM DTT, pH 7.4) for 2 h to remove excess free detergent. The DDM-solubilized Pgp and liposomes were then combined to a final concentration of ~50 μM and 4 mg ml−1, respectively, and incubated for one hour at room temperature. This was followed by a second dialysis for two hours against HEPES buffer to promote integration of Pgp into liposomes. Aliquots of prepared proteoliposomes were stored at −80°C in HEPES buffer. Protein concentration was determined using an extinction coefficient of 1.28 ml mg−1 cm−1.36 The purity of Pgp determined through SDS-PAGE analysis was greater than 95%.37,38 Experimental trials for these studies were performed on proteoliposomes from both the same and different batches. No significant batch-dependent differences were observed between the experimental trials (data not shown).
ATPase Activity Assay
The ATPase activity assay developed by Chifflet, et. al. was employed to determine the Pgp-mediated ATP hydrolysis in the presence of methadone and loperamide as we did previously.38–40 Experiments were performed on a FlexStation 3 spectrometer (Molecular Devices, Sunnyvale, CA), as previously described.38–40,45 The free inorganic phosphate (Pi) was measured through the formation of the Pi-molybdenum complex, which has a strong absorbance signal at 850 nm. The ATPase activity of methadone and loperamide was measured in the presence of 50 nM Pgp reconstituted into liposomes in Chifflet buffer (150 mM NH4Cl, 5 mM MgSO4, 0.02% wt/vol NaN3, 50 mM Tris HCl, pH 7.4).38–40,45
Nonlinear regression was used to fit the ATPase kinetics curves, as this type of analysis is more accurate than traditional linear regression models such as Lineweaver-Burke, Hans-Woolf, and Eadie-Hofstee plots.46–50 The Pgp-coupled ATP hydrolysis curves were fit in Igor Pro 6.2 software (Wavemetrics, Tigard, OK). For monophasic Pgp-coupled ATP hydrolysis, the modified Michaelis-Menten equation (Eq. 1) was used50,51:
| (1) |
where v is the ATP hydrolysis rate, Vmax is the maximum ATP hydrolysis, [L] is the ligand concentration, Km is the Michaelis-Menten constant, and vbasal is the basal ATPase activity. For curves showing biphasic ATP hydrolysis kinetics, the substrate inhibition equation (Eq. 2) was used50,51:
| (2) |
where KI is the inhibitory constant.
Fluorescence Quenching
Intrinsic protein fluorescence quenching can be used to measure dissociation constants of ligands to Pgp.38,39,52,53 An Olis DM 45 spectrofluorimeter (Olis Corp., Bogart, GA) was used to measure the drug-induced quenching of protein fluorescence. A 10 nm bandpass filter was put on the excitation and emission paths of the spectrofluorimeter to remove Rayleigh bands from the fluorescence emission spectra. Samples contained 1 μM Pgp reconstituted into liposomes in 100 mM potassium phosphate buffer (pH 7.4) with 2 mM DTT to prevent protein aggregation.38–40 Protein fluorescence emission was measured between 300 to 500 nm with an emission maximum around 333 nm after excitation at 280 or 295 nm.54 The wavelengths 280 and 295 nm were chosen for excitation of digoxin and methadone, respectively, because Pgp showed the greatest quenching response to the drugs at these wavelengths. Drug-induced fluorescence quenching was corrected (Fcorrected) for background fluorescence, dilution, and inner filter effects using Equation 338,54:
| (3) |
where F is the measured protein fluorescence at 333 nm, B is the background, ε is the extinction coefficient for excitation (εex) and emission (εem), b is the pathlength for excitation (bex) and emission (bem), and [Q] is the quenching ligand concentration. Extinction coefficients for methadone, loperamide and digoxin are listed in Table S1. The fluorescence emission spectra that were used as background in calculations using Eq. 3 were produced from samples containing the solvent and the drug. Subtracting this spectra removed potentially interfering weak spectral contributions from Raman scattering emanating from the solvent.55
Protein fluorescence quenching can occur by two mechanisms: static or dynamic. Static quenching occurs when a ligand complexes with the protein and is directly related to ligand affinity to the protein.54 Conversely, quenching caused by random collisions between ligand and the protein is known as dynamic quenching.54 To distinguish between the two mechanisms, protein fluorescence quenching titrations were performed at different temperatures as described previously.38,39,54 The following version of the Stern-Volmer equation (Eq. 4) was used to fit the corrected fluorescence (Fcorrected) of monophasic fluorescence quenching curves38,54:
| (4) |
where Fcorrected,0 is fluorescence without a quenching ligand, KSV is the Stern-Volmer constant, [Q] is the concentration of quenching ligand, and Funquenched is an offset related to the unquenched fluorescence. Biphasic fluorescence quenching curves were fit with Equation 5:
| (5) |
where FL,0 and FH,0 are the fluorescence amplitudes at low and high quenching ligand concentration, respectively. KSV,L and KSV,H are the Stern-Volmer constants at low and high ligand concentration, respectively.
Acrylamide quenching is a technique that is used to probe changes in the solvent accessibility of tryptophan residues as a result of the protein undergoing conformational changes.39,52,54,56,57 Acrylamide is a polar molecule that cannot penetrate into lipid bilayers or the hydrophobic interior of proteins such as Pgp.58 Fluorescence emission of Pgp reconstituted into liposomes was monitored at 333 nm after excitation at 295 nm. Samples contained 1 μM Pgp reconstituted into liposomes in potassium phosphate buffer (pH 7.4) with 2 mM DTT. Fluorescence intensities were corrected with Equation 3 for inner filter effects and background. Stern-Volmer plots were produced by plotting Fcorrected,0/Fcorrected versus acrylamide concentration. The slope of these curves was used to estimate the degree of acrylamide quenching with the slope equal to KSV by Equation 638,54:
| (6) |
To determine if there were significant deviations between the individual trials, analysis of covariance approach (ANCOVA) was performed on each set of Stern-Volmer plots.59 The p-values for the slopes of each group were >>0.05 indicating that they were not significantly different (data not shown). To analyze the differences between the mean and standard deviations of the KSV values in Fig. 6, a two-tailed Student’s unpaired t-test was performed as described.59 Statistical p-values <<0.05 indicate that the KSV values are significantly different, while p-values >>0.05 indicate that the KSV values are similar. The statistical analysis on the Stern-Volmer plots was performed using GraphPad Prism 7 (GraphPad software, San Diego, CA).
Figure 6.
Methadone- and loperamide-induced conformational changes of Pgp. (A) The Stern-Volmer plot of Pgp in the absence of drug (solid squares, solid line) and NATA (dashed line). Stern-Volmer plots of Pgp in the presence of (B) 7.8 μM loperamide and (C) 500 μM loperamide, respectively. (D) Stern-Volmer plot of Pgp with 500 μM methadone. For convenience, the slope of the Stern-Volmer plot without changes (Panel A) is presented as a dashed line in the other panels. These data are the average of three experiments (black squares) and the standard deviation is shown as error bars.
Saturation Transfer Double Difference NMR
All NMR experiments were performed at 25°C on a Var ian INOVA 600 MHz NMR spectrometer with a 5 mm z-gradient 1H{13C/15N} cryoprobe. The 1H NMR peaks for methadone and loperamide were assigned using standard 1H 1D NMR techniques in iNMR (http://www.inmr.net) and Igor Pro 6.2 as previously described.38–40 The 1H NMR peak assignments for methadone and loperamide are shown in Figure S1.
Saturation transfer difference NMR is a technique used to identify ligand functional groups that interact with a protein receptor.38,40,60 This is done by selectively saturating the protein and measuring the degree of saturation transfer from protein to ligand.60–62 In a reconstituted liposome system such as the one used in these studies, saturation transfer between lipid and ligand can contribute to the STD 1H NMR spectrum.60,62 By subtracting the STD NMR spectrum of drug in the presence of liposomes from the STD NMR spectrum of a drug in the presence of protein reconstituted liposomes, this contribution can be removed, yielding the saturation transfer double difference (STDD) spectrum which reflects the specific saturation between the protein receptor and the ligand.39,61,63,64 This NMR technique was used to identify functional groups of loperamide and methadone that are involved in molecular recognition of Pgp.
NMR samples contained 1 μM Pgp reconstituted liposomes in 100 mM potassium phosphate buffer (80% D2O), and 1 mM drug. Control samples were prepared identically with liposomes instead of proteoliposomes.63 A 2s train of 50 ms gaussian saturation pulses was used to selectively excite the protein.60 To suppress water background signals, the water signal was suppressed by the water suppression by gradient tailored excitation (WATERGATE) pulse sequence.65 Difference spectra were acquired by irradiating Pgp on resonance at −1.5 ppm and off resonance at 40 ppm for a total of 512 scans.38,39 The STDD 1H NMR spectrum was created by subtracting the STD spectrum of the control liposome sample with drug from the STD spectrum of the reconstituted Pgp sample with the drug. This difference (ΔI) directly relates to the interactions between drug functional groups and the protein. The STDD amplification factor was calculated from Equation 738:
| (7) |
where [L] is the ligand concentration, [P] is the protein concentration, and I0 is the amplitude of the 1H NMR peaks in the absence of radio frequency saturation.
RESULTS
ATPase activation of Pgp by methadone and loperamide
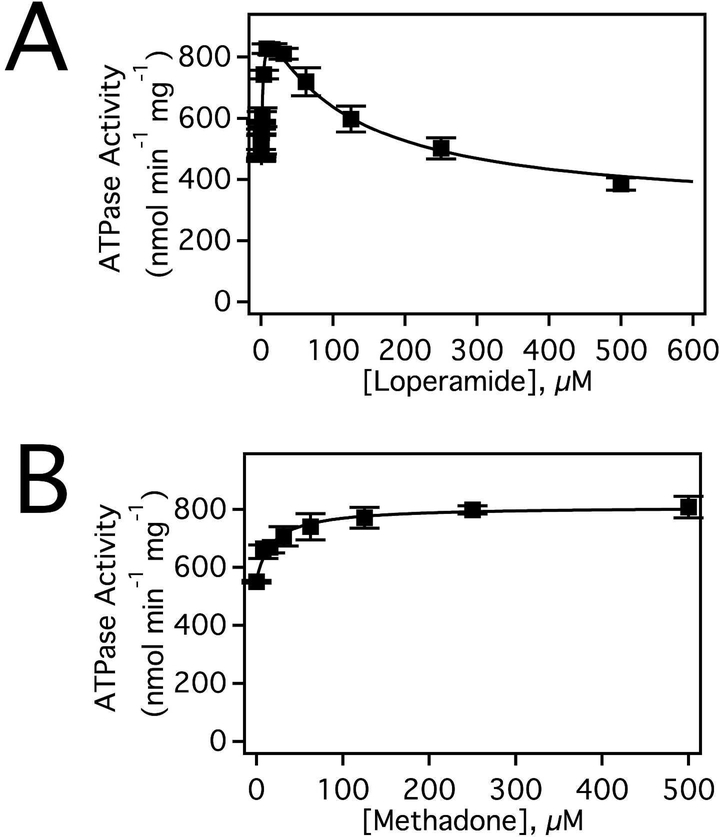
Methadone and loperamide were expected to have significantly different effects on Pgp-mediated ATPase activity because of differences in their transport rates.23,28,30,31 These effects on Pgp-mediated ATP hydrolysis are shown in Figure 2. In the absence of drug, the basal ATPase activity was 536 ± 78 nmol min−1 mg−1, which correlates with previous observations.38,40 Figure 2A shows the ATPase activation of Pgp in the presence of loperamide. The loperamide-induced ATPase activation kinetics of Pgp was biphasic implying that there were at least two loperamide binding sites on Pgp as suggested for other drugs.38,66,67 The kinetics exhibited an apparent maximum around 800 nmol min−1 mg−1 at 7.8 μM loperamide that decreased to ~400 nmol min−1 mg−1 at saturating (500 μM) loperamide. Biphasic ATP hydrolysis kinetics is a common drug-induced effect on Pgp-mediated ATP hydrolysis and has been observed previously with several drugs including verapamil, lidocaine, and quinidine.39,66,68,69 Fitting this curve with Equation 2 yielded VMAX, Km, and Ki values of 963 ± 44 nmol min−1 mg−1, 1.6 ± 0.4 μM and 108 ± 27 μM respectively. In contrast, the kinetics of the methadone-induced activation of ATP hydrolysis were monophasic and reached a maximum velocity ~800 nmol min−1 mg−1 at saturating (500 μM) methadone (Fig. 2B). From Equation 1, a VMAX and a Km for methadone was determined of 810 ± 19.3 nmol min−1 mg−1 and 19.1 ± 4.0 μM, respectively. The monophasic ATP activation kinetics of Pgp in the presence of methadone implies that it has a single binding site on Pgp. Multiple drug binding by loperamide versus a single binding site for methadone could partially explain the higher transport rate of loperamide.
Figure 2.
Loperamide and methadone induced ATPase activation of Pgp. Pgp activation in response to varying concentrations of (A) loperamide and (B) methadone. The fits are shown as a solid line with the data average of three experiments and standard deviation as solid dots and bars, respectively.
Competition studies of methadone and loperamide investigated by intrinsic protein fluorescence quenching by Pgp
Due to differences in their transport28 and their effects on ATP hydrolysis kinetics (Fig. 2), methadone and loperamide were hypothesized to occupy distinct sites on the transporter. To test the hypothesis, the interactions of these drugs were probed by drug-induced fluorescence quenching of intrinsic Pgp fluorescence.52 Consistent with this hypothesis, loperamide did not quench intrinsic Pgp fluorescence, while methadone did (data not shown). These differences in quenching allowed us to examine the effects of loperamide on binding of methadone to Pgp in Figure 3.
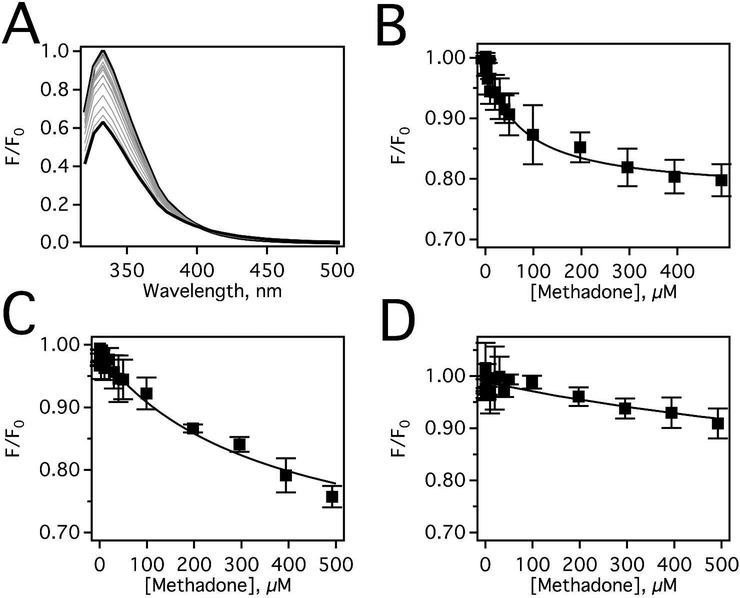
Figure 3.
Methadone-induced fluorescence quenching of Pgp in the presence of loperamide. (A) Pgp fluorescence spectra in the presence a range of methadone concentrations after excitation at 295 nm at 25°C. Pgp emission data at 333 nm as a fu nction of methadone concentration after correction for inner filter effects and background (Eq. 3) in the presence of (B) 0 μM, (C) 7.8 μM, and (D) 500 μM loperamide. The data points represent the average of three experiments and the standard deviation is shown with error bars.
The methadone-induced Pgp fluorescence quenching in the absence of loperamide is shown in Figure 3A. The protein fluorescence reached a maximum at 333 nm, which decreases 40% at saturating concentrations of methadone. The peak amplitudes at 333 nm corrected for inner filter effects were plotted as a function of methadone concentration in Figure 3B. After correcting for inner filter effects, the decrease in quenching is reduced to 20% at saturating methadone. Quenching of Pgp protein fluorescence by methadone was monophasic (Fig. 3B), and fitting to Equation 4 yielded a KSV value of 0.0137 ± 0.0022 μM−1, which corresponds to a KD of 72.7 ± 11.7 μM (KD=1/KSV). Figure 3C shows the effect of 7.8 μM loperamide on methadone binding to Pgp. Fitting of the monophasic fluorescence quenching curve in Figure 3C revealed that the KD increased to 356.3 ± 10.8 μM, implying that loperamide competitively displaces methadone from Pgp. The ~5-fold increase of methadone’s KD at low loperamide concentration implies that the loperamide binding associated with the lower Km was displaced by methadone. In the presence of 500 μM loperamide (Fig. 3D), the KD increased further to 1424.6 ± 317.5 μM. The fact that the KD increases approximately twenty-fold in the presence of loperamide is consistent with overlap between methadone and loperamide binding sites on Pgp. However, having the same binding site seems contradictory to their differences in transport rates, their effects on ATPase activity, and their respective quenching effects of Pgp. An alternative hypothesis is that loperamide shifts Pgp into a conformation that is not favorable for methadone binding, causing displacement of methadone. In other words, loperamide shifts Pgp into a conformation that is distinct from the Pgp conformation in the presence of methadone.
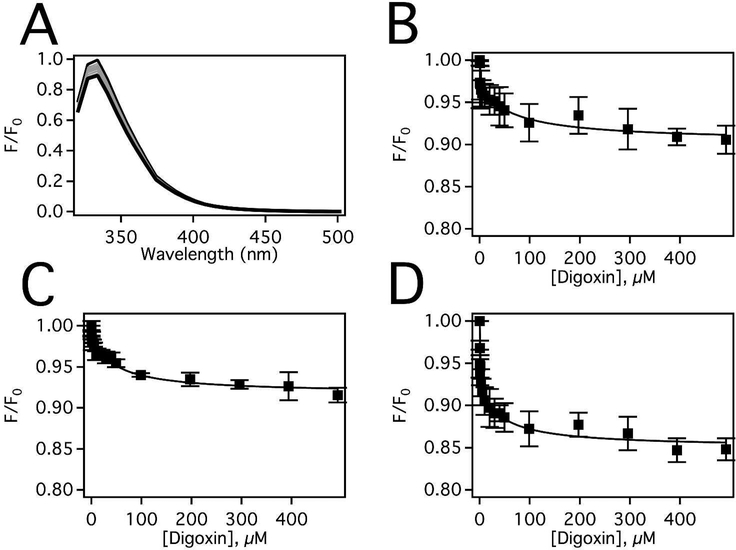
The location of the loperamide and methadone binding sites on Pgp are unknown. Using the fact that loperamide does not quench Pgp fluorescence and that digoxin is hypothesized to bind near G181 (G185 in human Pgp) on Pgp38,70, the effects of loperamide on digoxin binding were examined in Figure 4. Figure 4A shows the Pgp fluorescence emission spectra in the presence of a range of digoxin concentrations, which decreases 20% at saturating digoxin concentration. The inner filter effect corrected protein fluorescence amplitudes at 333 nm in panel A were plotted against digoxin concentration in Figure 4B. As previously observed38, the digoxininduced quenching of Pgp fluorescence curve was monophasic. Fitting the protein fluorescence quenching curve to Equation 4 yielded a KD of 32.0 ± 13.4 μM. Upon the addition of 7.8 μM loperamide (Fig. 4C), fitting to Equation 4 produced a KD of 44.0 ± 11.8 μM that was changed little by the presence of loperamide. At saturating (500 μM) loperamide, the digoxin Pgp fluorescence quenching curve becomes biphasic in Figure 4D. The fluorescence quenching curve was fit to Equation 5, which produced a KSV,L value of 4.59 ± 2.10 μM−1 and KSV,H of 0.0241 ± 0.0108 μM−1. To determine the quenching mechanism of the two phases, the titration was repeated at higher temperature as we did previously.38 At the higher temperature, KSV,L value increased while the KSV,H value decreased, suggesting that digoxin quenches the low concentration phase by a dynamic quenching mechanism and quenches the high concentration phase by a static quenching mechanism (data not shown). In this case, the KSV,H value associated with the high concentration phase correlated to the association constant (KA) and the KD (i.e. 1/KA) of digoxin to Pgp. Fitting the curve to Equation 5 resulted in a KD value of 41.5 ± 18.5 μM, which was essentially unchanged from the KD at other loperamide concentrations. The fact that the KD was unchanged and the quenching of Pgp fluorescence by digoxin becomes biphasic in the presence of saturating loperamide suggests that loperamide and digoxin are bound to the transporter simultaneously and implies that they occupy distinct binding sites on the transporter.
Figure 4.
Digoxin-induced fluorescence quenching of Pgp in the presence of loperamide. (A) Pgp fluorescence spectra in the presence of a range of digoxin concentrations after excitation at 280 nm at 25°C. Pgp emission data at 333 nm as a functi on of digoxin concentration after correction for inner filter effects and background (Eq. 3) in the presence of (B) 0 μM, (C) 7.8 μM, and (D) 500 μM loperamide. The data points represent the average of three experiments and the standard deviation is shown with error bars.
Identification of functional groups on methadone and loperamide that interact with Pgp
Because of their effects on Pgp-mediated ATP hydrolysis and large differences in their transport rates23,28,30,31, we wondered how the interactions of methadone and loperamide differed with Pgp. Therefore, these interactions were investigated using the saturation transfer double difference (STDD) NMR technique, which has been successfully used to examine drug-Pgp interactions and can identify functional groups of the ligand that interact with the transporter.38–40,60,63
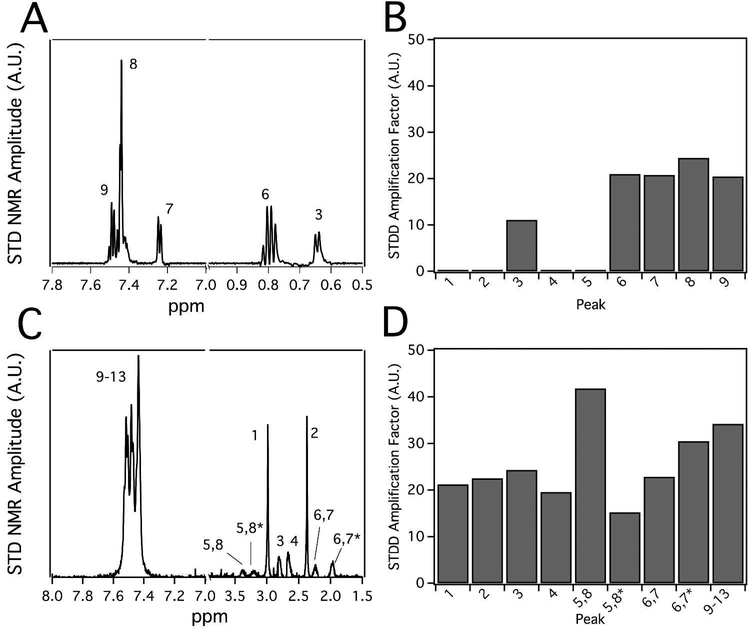
Figure 5 shows the STDD NMR spectra of methadone and loperamide. The 1H NMR peaks of the drugs were labeled according to the molecular structures shown in Figure 1. Figure 5A shows the 1H STDD NMR spectrum of 1 mM methadone in the presence of 1 μM Pgp reconstituted into liposomes. The STDD amplification factors in Figure 5B were calculated using the STDD amplitudes in Figure 5A and Equation 7. No 1H STDD NMR peaks were observed for protons labeled 1, 2, 4 and 5, suggesting that there were no significant interactions between these nuclei and the transporter (see Fig. 1 for labeling scheme). The strongest STDD amplification factors (~20) were observed for the aromatic protons labeled 7–9 and the methyl group labeled 6. The methyl protons labeled 3 had an STDD amplification factor that was half the methyl group labeled 6, implying that its interactions are weaker. Overall, the average STDD amplification factor for methadone was ~10. The methyl group labeled 6 and the protons from the aromatic rings of methadone seem to have the strongest interaction with the transporter.
Figure 5.
The interactions of methadone and loperamide with Pgp investigated by STDD NMR. Representative STDD NMR spectrum of (A) methadone and (C) loperamide. Peak numbers correspond to labels of protons in Figure 1. Data from panels (A, C) were used to calculate the STDD amplification factors (Eq. 7) in panels (B, D).
Figures 5C shows the STDD NMR spectrum of 1 mM loperamide in the presence of 1 μM Pgp reconstituted into liposomes. Using Equation 7, the STDD amplitudes from Figure 5C were used to calculate the STDD amplification factors in Figure 5D. The average STDD amplification factor of loperamide is ~30, which is three times larger than methadone. This increased saturation transfer may be the result of multiple loperamide binding to Pgp. Like methadone, the aromatic protons labeled 9–13 had relatively large STDD amplification factors around 30. Exchanging equatorial and axial protons labeled 5,8 and 5,8* of the piperidine ring had the largest and smallest STDD amplification factors, respectively. In contrast, the differences in the STDD amplification factors for the exchanging equatorial and axial protons labeled 6,7 and 6,7*, respectively, on the opposite end of the piperidine ring were considerably smaller. These large and small differences in the STDD amplification factors imply that Pgp selectively interacts with specific conformations of the piperidine functional group. Unlike methadone, the STDD amplification factors for methyl protons labeled 1 and 2 were very similar. The STDD amplification factors were also similar for the alkyl protons labeled 3 and 4. Although the phenyl groups of loperamide and methadone play a common role in their recognition of Pgp, their overall interactions of methadone and loperamide with Pgp are quite distinct.
Methadone- and loperamide-induced conformational changes of Pgp
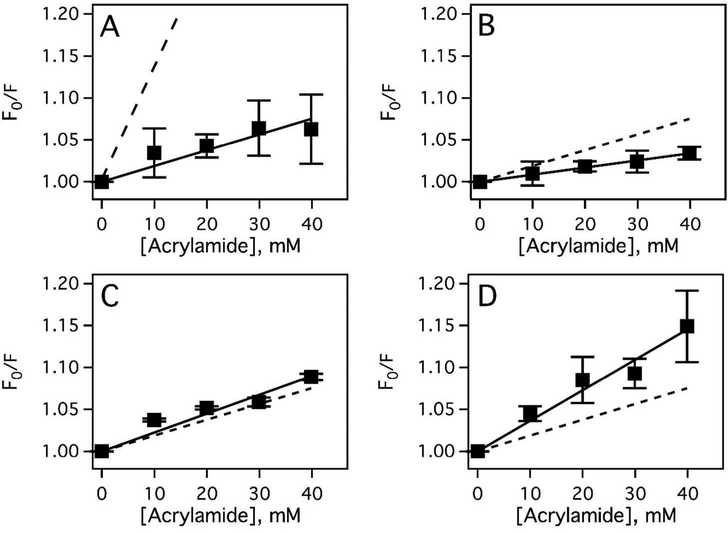
To explain apparent competition between loperamide and methadone, an alternative hypothesis was proposed that the apparent competition was the result of loperamide shifting Pgp into a different conformation than methadone. To test the hypothesis, drug-induced Pgp conformational changes were probed by changes in tryptophan accessibility through acrylamide quenching.38,40 Quantitatively, the changes in Pgp conformation and tryptophan accessibility were estimated with a KSV value (Eq. 6), which is determined from the slope of a Stern-Volmer plot.38–40,54 The Stern-Volmer plots of Pgp in the presence of loperamide and methadone are shown in Figure 6. In the absence of drug, the slope of the Stern-Volmer plot had a KSV value of 1.88 ± 0.20 M−1 (Fig. 6A, closed squares), which is similar to previous results.38–40 The tryptophan analog N-acetyl-L-tryptophanamide (NATA) was used as a positive control representative of complete tryptophan accessibility and it produced a KSV value of 14.2 ± 0.32 M−1 (Fig. 6A, dashed line) in the presence of a range of acrylamide concentrations. The large difference in KSV values between Pgp and NATA are consistent with the fact that only 3 of the 11 tryptophan residues on the transporter are accessible and exposed to bulk solvent in the X-ray crystal structure of Pgp.71,72
In the presence of 7.8 μM loperamide (Fig. 6B), the slope decreased to a KSV value of 0.851 ± 0.002 M−1 suggesting that the bound loperamide reduces solvent accessibility of tryptophan, which could occur by occlusion of the binding site cavity to bulk solvent. The effect of saturating 500 μM loperamide concentration on acrylamide quenching of Pgp is shown in Figure 6C. The KSV value determined from the slope of the Stern-Volmer plot increased about 1.4 M−1 to 2.25 ± 0.17 M−1 (Fig. 6C) (p-value<<0.001, Fig. 6C vs. Fig. 6B), which is consistent with an increase in solvent accessibility. This KSV value was similar to the KSV value in the absence of drugs (p-value =0.0711, Fig. 6C vs. Fig. 6A) implying that they have similar conformations. In contrast, in the presence of saturating 500 μM methadone, the KSV value from the Stern-Volmer plot was significantly higher at 3.65 ± 0.21 M−1 (Fig. 6D). This KSV value was significantly larger than the other KSV values (p-values << 0.001) suggesting that the Pgp conformation with saturating methadone was indeed distinct from the Pgp conformation with loperamide (Figs. 6B and 6C) or without ligands present (Fig. 6A).
DISCUSSION
A number of Pgp-mediated transport models have been proposed to explain the observed drug efflux.39,40,73–76 Partitioning and alternating site models have been proposed where drugs occupy different sites on Pgp during efflux.73,74 Models were suggested where the ligand is cooperative with other ligands and/or ATP during transport.39,75 In addition, there have been conformationally-gated models of Pgp-mediated transport proposed.40,76
The process of transport can be inhibited by several mechanisms as well including competitive77,78, non-competitive77,78 and uncompetitive79 or a combination.38 Inhibition has been proposed to fix Pgp into discrete conformations.80 Pgp transport inhibition has also been shown to be cooperative78,81 or accompanied by activation of ATP hydrolysis.80 Moreover, Pgp inhibitors can potentially undergo transport themselves.82,83
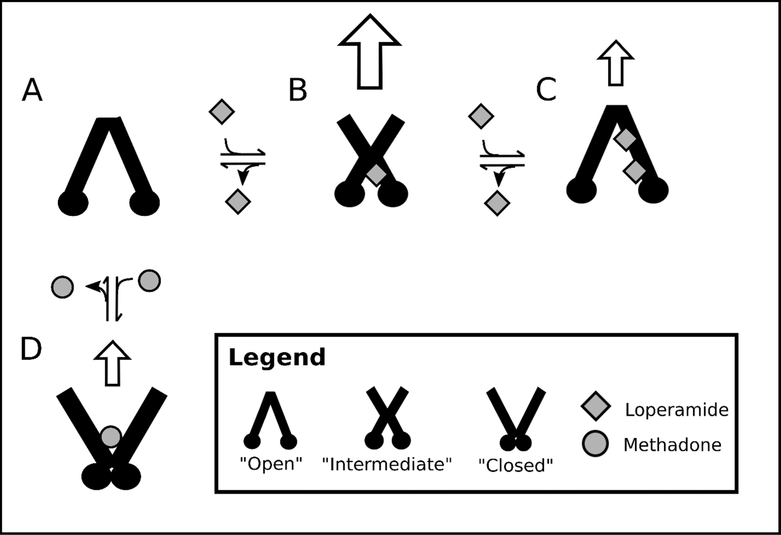
A model of Pgp-mediated transport for methadone and loperamide is presented in Figure 7 that is conformationally-gated and based on competition between these drugs. In our previous studies38–40, we found it useful to model Pgp in three conformations: “closed”, “intermediate”, and “open”. In the “open” conformation, Pgp is in a similar conformation as the Pgp X-ray crystal structures (e.g. 5) with the substrate cavity exposed to the cytosolic (C) side. In the “closed” conformation, the NBDs are in contact and the transporter’s substrate cavity is exposed to the extracellular (EC) space in a conformation that is similar to the analogous bacterial transporter with a non-hydrolyzable ATP analog.18,19 In the “intermediate” conformation, Pgp is between the “open” and “closed” conformations, with the binding cavity exposed to both the EC and C sides of the transporter.
Figure 7.
Conformationally-gated model of methadone and loperamide transport by Pgp. Pgp conformations are represented as “open”, “intermediate”, and “closed”. Methadone and loperamide are represented by grey circles and grey squares, respectively. Panels show (A) Pgp in the absence of drug, Pgp with (B) 1 loperamide and (C) 2 loperamide molecules bound, and (D) Pgp with 1 methadone molecule bound. White arrows indicate the degree of transport by Pgp.
Acrylamide quenching of Pgp fluorescence indirectly probes conformational changes through estimates of tryptophan accessibility.38,52,57 Differences in acrylamide quenching of Pgp in the presence of drugs can suggest distinct drug-bound Pgp-conformations. Higher KSV values suggest Pgp conformations that are more exposed to bulk solvent (i.e. “open”), while lower KSV values suggest more occluded Pgp conformations. The dominant drug-bound Pgp conformation will be dependent on the drug’s affinity to the transporter and its concentration. To convincingly assign Pgp conformations, the relative interaction between the NBDs needs to be known. Luckily, activation of Pgp-mediated ATPase activity is related to the interaction of NBDs, since NBD interaction is a requirement for ATP hydrolysis.84–88 The combined information helped us assign these conformations as we did previously.38–40
X-ray crystal structures of Pgp in the absence of drugs show the transporter in an “open” conformation with NBDs separated.5 Therefore, Pgp is shown in this conformation in Figure 7A. Loperamide is proposed to have two binding sites on Pgp in Figure 7C because it induces biphasic ATPase activation of the transporter (Fig. 2). At low concentrations of loperamide, the KSV value decreased (cf. Fig. 6A and 6B), suggesting a shift toward a more occluded conformation. The KSV value was almost identical to values that were determined for Pgp in the presence of non-hydrolyzable nucleotide analogs.39,57 With non-hydrolyzable nucleotides, Pgp was hypothesized to be in an “intermediate” Pgp conformation.39 Therefore, in the presence of 7.8 μM loperamide, Pgp is shown in an “intermediate” conformation in Figure 7B, where the NBDs are relatively close together and the activation barrier for Pgp-mediated ATP hydrolysis is reduced. Because loperamide causes significant activation of Pgp-mediated hydrolysis (Fig. 2), the drug binding site was posited near the NBDs but not overlapping with the digoxin binding site, since Fig. 4 shows that loperamide and digoxin do not compete with each other. The loperamideinduced activation of ATPase activity and the KSV value of Pgp with saturating loperamide was similar to the ATPase activity and KSV value of Pgp in the absence of ligands (Fig. 2). Therefore, Pgp is shown in an “open” conformation in Figure 7C with two loperamide molecules bound to the transporter. Because there is a reduction in Pgp-mediated ATP hydrolysis in the presence of saturating loperamide, one might expect the transport rate to also decrease. An in vitro study in Caco-2 cells in fact showed that transport was highest at 5 μM loperamide and was inhibited at 100 μM loperamide.89 Inhibition of Pgp at high loperamide concentration is consistent with the clinical observation that elevated loperamide doses cause opiate effects on the CNS.90 Furthermore, both in vitro and in vivo studies have shown that verapamil competitively inhibits loperamide transport by Pgp, which implies that they occupy overlapping binding sites on the transporter.89,91,92 In a previous study from our group, verapamil was proposed to occupy a low affinity site on Pgp near the digoxin binding site and a high affinity site on the EC side of the transporter.38 In our model, we propose that loperamide occupies similar positions on Pgp as verapamil but does not overlap with the digoxin binding site. However, since loperamide already occupies a binding site near the NBDs at low concentration in our model, we hypothesize that the second loperamide occupies a site near the EC side of the transporter in Figure 7C like inhibitors in the Pgp X-ray crystal structures.5,93 In this configuration, the low affinity loperamide could conceptually function as a “wedge” to separate the NBDs as is implied by the location of inhibitors in the Pgp X-ray crystal structures.5,93
Conversely, the KSV value of Pgp in the presence of saturating methadone was about double the KSV value in the absence of ligands (cf. Fig. 6D and Fig. 6A). This suggested that the binding site cavity was more exposed to the bulk solvent with methadone than without methadone. If Pgp was in an “open” conformation with the binding cavity exposed to the C side and the NBDs separated, one would expect Pgp-mediated ATPase activity to be inhibited. On the other hand, if Pgp was in a “closed” conformation with the binding cavity exposed to the EC side and the NBDs together, one would expect Pgp-mediated ATPase activity to be activated. Since there was significant activation of ATP hydrolysis activity by Pgp in the presence of saturating methadone, we propose that Pgp is in the “closed” conformation with methadone. Because of differences in transport and ATPase activity, we also propose that methadone binds near the NBDs, but not overlapping with the loperamide binding sites. In our model, the apparent competition between loperamide and methadone is the result of loperamide shifting Pgp into a conformation that is unfavorable to methadone binding. Because methadone exhibited relatively modest Pgp-mediated transport or inhibited transport,28,30,35 we hypothesize that methadone locks Pgp into a “closed” conformation similar to the model proposed for Pgp inhibition by tariquidar.80
Since this investigation was performed on mouse Pgp, we wondered if the proposed transport model in Fig. 7 applied to human Pgp. There are significant differences in the expression and substrate specificity between mouse and human Pgp.94,95 For isolated brain microvessels, the expression level of mouse Pgp was more than two-fold higher than human Pgp.94 Also, in a study with 640 Pgp substrates with human and mouse Pgp, the efflux ratios differed more than 3-fold for a third of the them.95 However, a study focused on a diverse range of CNS drugs including loperamide found that their efflux ratios differed by less than 2-fold.96 In addition, methadone had an efflux ratio for human Pgp expressing mammalian cells of ~2,28 which was very close to the brain uptake clearance ratio of 2.6 for mice.97 At least for Pgp-mediated transport of loperamide and methadone, we anticipate that the proposed transport model in Fig. 7 will be similar for human Pgp.
Despite their molecular similarity, the interactions of methadone and loperamide with Pgp are quite distinct. Pgp-mediated ATP hydrolysis kinetics in the presence of the drugs suggest that loperamide has two binding sites and methadone has a single binding site. These drugs also shifted Pgp into distinct conformations and provide a mechanism to explain the apparent competition between loperamide and methadone. This information combined with previous transport studies23,28,30,31 lead to a conformationally-gated model for diphenylpropylamine opioid transport by Pgp (Fig. 7). These studies are the first to describe a detailed molecular mechanism of opioid transport by Pgp. We anticipate that these results will aide in identifying neurotherapeutics that are Pgp substrates and will be useful for neurotherapeutic drug development.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr. Ina L. Urbatsch of Texas Tech University Health Sciences Center for her generous gift of Pichia (P.) pastoris with the wild-type mouse Pgp transporter gene and providing protocols for genetically manipulating and purifying Pgp from P. pastoris for our laboratory. Without her contribution and generosity, this research would not be possible.
FUNDING
This work was supported by the National Institute of Health R15 Area Grant (1R15GM1079301A1) and the National Institute of Health R01 Area Grant (R01-1021RR571424).
ABBREVIATIONS
- ABC
ATP-binding cassette
- BBB
blood-brain barrier
- C
cytosolic
- CNS
central nervous system
- DDM
n-dodecyl-β-D-maltoside
- DEAE
diethylaminoethyl cellulose
- DTT
dithiothreitol
- EC
extracellular
- EGTA
ethylene glycol tetraacetic acid
- KA
association constant
- KD
dissociation constant
- KSV
Stern-Volmer constant
- NATA
N-acetyl-L-tryptophanamide
- NBD
nucleotide-binding domain
- NMR
nuclear magnetic resonance
- Pgp
P-glycoprotein
- STD
saturation transfer difference
- STDD
saturation transfer double difference
- WATERGATE
water suppression by gradient tailored excitation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Bendayan R, Lee G, Bendayan M 2002. Functional expression and localization of P-glycoprotein at the blood brain barrier. Microsc Res Tech 57(5):365–380. [DOI] [PubMed] [Google Scholar]
- 2.Lee G, Bendayan R 2004. Functional Expression and Localization of P-glycoprotein in the Central Nervous System: Relevance to the Pathogenesis and Treatment of Neurological Disorders. Pharm Res 21(8):1313–1330. [DOI] [PubMed] [Google Scholar]
- 3.Ramakrishnan P 2003. The role of P-glycoprotein in the blood-brain barrier. Einstein QJ Biol Med 19(1):160–165. [Google Scholar]
- 4.Doan KMM, Humphreys JE, Webster LO, Wring SA, Shampine LJ, Serabjit-Singh CJ, Adkison KK, Polli JW 2002. Passive Permeability and P-Glycoprotein-Mediated Efflux Differentiate Central Nervous System (CNS) and Non-CNS Marketed Drugs. J Pharmacol Exp Ther 303(3):1029–1037. [DOI] [PubMed] [Google Scholar]
- 5.Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, Harrell PM, Trinh YT, Zhang Q, Urbatsch IL, Chang G 2009. Structure of P-glycoprotein reveals a molecular basis for polyspecific drug binding. Science 323(5922):1718–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cutler NR, Sramek JJ, Murphy MF, Riordan H, Bieck P, Carta A 2010. The impending crisis in cns drug development. Critical Pathways to Success in CNS Drug Development:1–13. [Google Scholar]
- 7.Jose M, Thomas SV 2009. Role of multidrug transporters in neurotherapeutics. Annals of Indian Academy of Neurology 12(2):89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaitin KI, Milne CP 2011. A dearth of new meds. Sci Am 305(1):16–16. [DOI] [PubMed] [Google Scholar]
- 9.Binkhathlan Z, Lavasanifar A 2013. P-glycoprotein inhibition as a therapeutic approach for overcoming multidrug resistance in cancer: current status and future perspectives. Curr Cancer Drug Targets 13(3):326–346. [DOI] [PubMed] [Google Scholar]
- 10.Callaghan R, Luk F, Bebawy M 2014. Inhibition of the Multidrug Resistance PGlycoprotein: Time for a Change of Strategy? Drug Metab Disposition 42(4):623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller DS, Bauer B, Hartz AMS 2008. Modulation of P-Glycoprotein at the Blood-Brain Barrier: Opportunities to Improve Central Nervous System Pharmacotherapy. Pharmacol Rev 60(2):196–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frank GA, Shukla S, Rao P, Borgnia MJ, Bartesaghi A, Merk A, Mobin A, Esser L, Earl LA, Gottesman MM, Xia D, Ambudkar SV, Subramaniam S 2016. Cryo-EM Analysis of the Conformational Landscape of Human P-glycoprotein (ABCB1) During its Catalytic Cycle. Mol Pharmacol 90(1):35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, Jaimes KF, Aller SG 2014. Refined structures of mouse P‐glycoprotein. Protein Sci 23(1):34–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ward AB, Szewczyk P, Grimard V, Lee C-W, Martinez L, Doshi R, Caya A, Villaluz M, Pardon E, Cregger C, Swartz DJ, Falson PG, Urbatsch IL, Govaerts C, Steyaert J, Chang G 2013. Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain. Proceedings of the National Academy of Sciences 110(33):1338613391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin MS, Oldham ML, Zhang Q, Chen J 2012. Crystal structure of the multidrug transporter P-glycoprotein from Caenorhabditis elegans. Nature 490(7421):566–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moeller A, Lee SC, Tao H, Speir JA, Chang G, Urbatsch IL, Potter CS, Carragher B, Zhang Q 2015. Distinct conformational spectrum of homologous multidrug ABC transporters. Structure 23(3):450–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenberg MF, Callaghan R, Ford RC, Higgins CF 1997. Structure of the multidrug resistance P-glycoprotein to 2.5 nm resolution determined by electron microscopy and image analysis. J Biol Chem 272(16):10685–10694. [DOI] [PubMed] [Google Scholar]
- 18.Dawson RJP, Hollenstein K, Locher KP 2007. Uptake or extrusion: crystal structures of full ABC transporters suggest a common mechanism. Mol Microbiol 65(2):250–257. [DOI] [PubMed] [Google Scholar]
- 19.Ward A, Reyes CL, Yu J, Roth CB, Chang G 2007. Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proceedings of the National Academy of Sciences 104(48):19005–19010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahmad RS 2004. Identification and Characterization of the Binding Sites of PGlycoprotein for Multidrug Resistance-Related Drugs and Modulators. Current Medicinal Chemistry - Anti-Cancer Agents 4(1):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin C, Berridge G, Higgins CF, Mistry P, Charlton P, Callaghan R 2000. Communication between multiple drug binding sites on P-glycoprotein. Mol Pharmacol 58(3):624–632. [DOI] [PubMed] [Google Scholar]
- 22.McQuay H 1999. Opioids in pain management. The Lancet 353(9171):2229–2232. [DOI] [PubMed] [Google Scholar]
- 23.Wang J-S, Ruan Y, Taylor RM, Donovan JL, Markowitz JS, DeVane CL 2004. Brain penetration of methadone (R)- and (S)-enantiomers is greatly increased by P-glycoprotein deficiency in the blood–brain barrier of Abcb1a gene knockout mice. Psychopharmacology 173(1):132–138. [DOI] [PubMed] [Google Scholar]
- 24.Kharasch ED, Hoffer C, Whittington D 2004. The effect of quinidine, used as a probe for the involvement of P‐glycoprotein, on the intestinal absorption and pharmacodynamics of methadone. Br J Clin Pharmacol 57(5):600–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schinkel AH, Wagenaar E, Mol CA, van Deemter L 1996. P-glycoprotein in the bloodbrain barrier of mice influences the brain penetration and pharmacological activity of many drugs. J Clin Invest 97(11):2517–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Argüelles CF, Torres-López JE, Granados-Soto V 2002. Peripheral antinociceptive action of morphine and the synergistic interaction with lamotrigine. Anesthesiology: The Journal of the American Society of Anesthesiologists 96(4):921–925. [DOI] [PubMed] [Google Scholar]
- 27.Hassan HE, Myers AL, Coop A, Eddington ND 2009. Differential involvement of P-glycoprotein (ABCB1) in permeability, tissue distribution, and antinociceptive activity of methadone, buprenorphine, and diprenorphine: In vitro and in vivo evaluation. J Pharm Sci 98(12):4928–4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tournier N, Declèves X, Saubaméa B, Scherrmann J-M, Cisternino S 2011. Opioid transport by ATP-binding cassette transporters at the blood-brain barrier: implications for neuropsychopharmacology. Curr Pharm Des 17(26):2829–2842. [DOI] [PubMed] [Google Scholar]
- 29.Wandel MDC, Kim MDR, Wood MBCBFRCAM, Wood MBCBFRCPA 2002. Interaction of Morphine, Fentanyl, Sufentanil, Alfentanil, and Loperamide with the Efflux Drug Transporter Pglycoprotein. Anesthesiology 96(4):913–920. [DOI] [PubMed] [Google Scholar]
- 30.Dagenais C, Graff CL, Pollack GM 2004. Variable modulation of opioid brain uptake by Pglycoprotein in mice. Biochem Pharmacol 67(2):269–276. [DOI] [PubMed] [Google Scholar]
- 31.Kalvass JC, Graff CL, Pollack GM 2004. Use of Loperamide as a Phenotypic Probe of mdr1a Status in CF-1 Mice. Pharm Res 21(10):1867–1870. [DOI] [PubMed] [Google Scholar]
- 32.Hsiao P, Unadkat JD 2012. P-glycoprotein-based loperamide-cyclosporine drug interaction at the rat blood-brain barrier: prediction from in vitro studies and extrapolation to humans. Mol Pharm 9(3):629–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hagen NA, Wasylenko E 1999. Methadone: Outpatient Titration and Monitoring Strategies in Cancer Patients. J Pain Symptom Manage 18(5):369–375. [DOI] [PubMed] [Google Scholar]
- 34.Störmer E, Perloff MD, von Moltke LL, Greenblatt DJ 2001. Methadone Inhibits Rhodamine123 Transport in Caco-2 Cells. Drug Metab Disposition 29(7):954–956. [PubMed] [Google Scholar]
- 35.Tournier N, Chevillard L, Megarbane B, Pirnay S, Scherrmann J-M, Declèves X 2010. Interaction of drugs of abuse and maintenance treatments with human P-glycoprotein (ABCB1) and breast cancer resistance protein (ABCG2). Int J Neuropsychopharmacol 13(7):905–915. [DOI] [PubMed] [Google Scholar]
- 36.Bai J, Swartz DJ, Protasevich II, Brouillette CG, Harrell PM, Hildebrandt E, Gasser B, Mattanovich D, Ward A, Chang G, Urbatsch IL 2011. A Gene Optimization Strategy that Enhances Production of Fully Functional P-Glycoprotein in Pichia pastoris. PLoS One 6(8):e22577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lerner-Marmarosh N, Gimi K, Urbatsch IL, Gros P, Senior AE 1999. Large scale purification of detergent-soluble P-glycoprotein from Pichia pastoris cells and characterization of nucleotide binding properties of Wild-type, Walker A, and Walker B mutant proteins. J Biol Chem 274(49):34711–34718. [DOI] [PubMed] [Google Scholar]
- 38.Ledwitch KV, Barnes RW, Roberts AG 2016. Unravelling the complex drug–drug interactions of the cardiovascular drugs, verapamil and digoxin, with P-glycoprotein. Biosci Rep 36(2):e00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ledwitch KV, Gibbs ME, Barnes RW, Roberts AG 2016. Cooperativity between verapamil and ATP bound to the efflux transporter P-glycoprotein. Biochem Pharmacol 118:96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilt LA, Nguyen D, Roberts AG 2017. Insights into the Molecular Mechanism of Triptan Transport by P-glycoprotein. J Pharm Sci 106(6):1670–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eckford PD, Sharom FJ 2008. Interaction of the P-glycoprotein multidrug efflux pump with cholesterol: effects on ATPase activity, drug binding and transport. Biochemistry 47(51):1368613698. [DOI] [PubMed] [Google Scholar]
- 42.Loo TW, Clarke DM 2016. P-glycoprotein ATPase activity requires lipids to activate a switch at the first transmission interface. Biochem Biophys Res Commun 472:379–383. [DOI] [PubMed] [Google Scholar]
- 43.Skrzypek R, Iqbal S, Callaghan R Methods of reconstitution to investigate membrane protein function. Methods In Press. [DOI] [PubMed]
- 44.Rothnie A, Theron D, Soceneantu L, Martin C, Traikia M, Berridge G, Higgins CF, Devaux PF, Callaghan R 2001. The importance of cholesterol in maintenance of P-glycoprotein activity and its membrane perturbing influence. Eur Biophys J 30(6):430–442. [DOI] [PubMed] [Google Scholar]
- 45.Chifflet S, Torriglia A, Chiesa R, Tolosa S 1988. A method for the determination of inorganic phosphate in the presence of labile organic phosphate and high concentrations of protein: Application to lens ATPases. Anal Biochem 168(1):1–4. [DOI] [PubMed] [Google Scholar]
- 46.Martin RB 1997. Disadvantages of double reciprocal plots. J Chem Educ 74(10):1238. [Google Scholar]
- 47.Cook P, Cleland WW. 2007. Enzyme Kinetics and Mechanism. ed.: Garland Science. [Google Scholar]
- 48.Leatherbarrow RJ 1990. Using linear and non-linear regression to fit biochemical data. Trends Biochem Sci 15(12):455–458. [DOI] [PubMed] [Google Scholar]
- 49.Ranaldi F, Vanni P, Giachetti E 1999. What students must know about the determination of enzyme kinetic parameters. Biochemical Education 27(2):87–91. [Google Scholar]
- 50.Segel IH. 1975. Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady State Enzyme Systems. ed.: Wiley, New York. [Google Scholar]
- 51.Roberts AG, Yang J, Halpert JR, Nelson SD, Thummel KT, Atkins WM 2011. The Structural Basis for Homotropic and Heterotropic Cooperativity of Midazolam Metabolism by Human Cytochrome P450 3A4. Biochemistry 50(50):10804–10818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu R, Siemiarczuk A, Sharom FJ 2000. Intrinsic Fluorescence of the P-glycoprotein Multidrug Transporter: Sensitivity of Tryptophan Residues to Binding of Drugs and Nucleotides. Biochemistry 39(48):14927–14938. [DOI] [PubMed] [Google Scholar]
- 53.Sharom FJ, Russell PL, Qu Q, Lu P. 2003. Fluorescence Techniques for Studying Membrane Transport Proteins In Yan Q, editor Membrane Transporters: Methods and Protocols, ed., Totowa, NJ: Humana Press; p 109–128. [DOI] [PubMed] [Google Scholar]
- 54.Lakowicz JR. 2011. Principles of Fluorescence Spectroscopy. 3rd edition ed., New York, NY: Springer; p 954. [Google Scholar]
- 55.Lawaetz AJ, Stedmon CA 2009. Fluorescence intensity calibration using the Raman scatter peak of water. Appl Spectrosc 63:936–940. [DOI] [PubMed] [Google Scholar]
- 56.Russell Paula L, Sharom Frances J 2006. Conformational and functional characterization of trapped complexes of the P-glycoprotein multidrug transporter. Biochem J 399(2):315–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sonveaux N, Vigano C, Shapiro AB, Ling V, Ruysschaert J-M 1999. Ligand-mediated Tertiary Structure Changes of Reconstituted P-glycoprotein: A tryptophan fluorescence quenching analysis. J Biol Chem 274(25):17649–17654. [DOI] [PubMed] [Google Scholar]
- 58.Eckford PDW, Sharom FJ 2008. Functional Characterization of Escherichia coli MsbA: INTERACTION WITH NUCLEOTIDES AND SUBSTRATES. J Biol Chem 283(19):12840–12850. [DOI] [PubMed] [Google Scholar]
- 59.Zar JH. 2009. Biostatistical Analysis. 5 edition ed., Upper Saddle River, NJ: Pearson; p 960. [Google Scholar]
- 60.Mayer M, Meyer B 2001. Group Epitope Mapping by Saturation Transfer Difference NMR To Identify Segments of a Ligand in Direct Contact with a Protein Receptor. J Am Chem Soc 123(25):6108–6117. [DOI] [PubMed] [Google Scholar]
- 61.Venkitakrishnan RP, Benard O, Max M, Markley JL, Assadi-Porter FM. 2012. Use of NMR Saturation Transfer Difference Spectroscopy to Study Ligand Binding to Membrane Proteins In Vaidehi N, Klein-Seetharaman J, editors. Membrane Protein Structure and Dynamics: Methods and Protocols, ed., Totowa, NJ: Humana Press; p 47–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Viegas A, Manso J, Nobrega FL, Cabrita EJ 2011. Saturation-Transfer Difference (STD) NMR: A Simple and Fast Method for Ligand Screening and Characterization of Protein Binding. J Chem Educ 88(7):990–994. [Google Scholar]
- 63.Claasen B, Axmann M, Meinecke R, Meyer B 2005. Direct observation of ligand binding to membrane proteins in living cells by a saturation transfer double difference (STDD) NMR spectroscopy method shows a significantly higher affinity of integrin αIIbβ3 in native platelets than in liposomes. J Am Chem Soc 127(3):916–919. [DOI] [PubMed] [Google Scholar]
- 64.Haselhorst T, Münster-Kühnel AK, Oschlies M, Tiralongo J, Gerardy-Schahn R, Itzstein Mv 2007. Direct detection of ligand binding to Sepharose-immobilised protein using saturation transfer double difference (STDD) NMR spectroscopy. Biochem Biophys Res Commun 359(4):866–870. [DOI] [PubMed] [Google Scholar]
- 65.Piotto M, Saudek V, Sklenář V 1992. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR 2(6):661–665. [DOI] [PubMed] [Google Scholar]
- 66.Litman T, Zeuthen T, Skovsgaard T, Stein WD 1997. Structure-activity relationships of Pglycoprotein interacting drugs: kinetic characterization of their effects on ATPase activity. Biochim Biophys Acta 1361(2):159–168. [DOI] [PubMed] [Google Scholar]
- 67.Aanismaa P, Seelig A 2007. P-Glycoprotein kinetics measured in plasma membrane vesicles and living cells. Biochemistry 46(11):3394–3404. [DOI] [PubMed] [Google Scholar]
- 68.Äänismaa P, Seelig A 2007. P-Glycoprotein Kinetics Measured in Plasma Membrane Vesicles and Living Cells. Biochemistry 46(11):3394–3404. [DOI] [PubMed] [Google Scholar]
- 69.Calabrese EJ 2008. P-Glycoprotein Efflux Transporter Activity Often Displays Biphasic Dose-Response Relationships. Crit Rev Toxicol 38(5):473–487. [DOI] [PubMed] [Google Scholar]
- 70.Gozalpour E, Wilmer MJ, Bilos A, Masereeuw R, Russel FGM, Koenderink JB 2016. Heterogeneous transport of digitalis-like compounds by P-glycoprotein in vesicular and cellular assays. Toxicol In Vitro 32(Supplement C):138–145. [DOI] [PubMed] [Google Scholar]
- 71.Esser L, Zhou F, Pluchino KM, Shiloach J, Ma J, Tang W-k, Gutierrez C, Zhang A, Shukla S, Madigan JP 2017. Structures of the multidrug transporter P-glycoprotein reveal asymmetric ATP binding and the mechanism of polyspecificity. J Biol Chem 292(2):446–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Swartz DJ, Weber J, Urbatsch IL 2013. P-glycoprotein is fully active after multiple tryptophan substitutions. Biochimica et Biophysica Acta (BBA)-Biomembranes 1828(3):11591168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Al-Shawi MK, Omote H 2005. The remarkable transport mechanism of P-Glycoprotein: A multidrug transporter. J Bioenerg Biomembr 37(6):489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Verhalen B, Ernst S, Börsch M, Wilkens S 2012. Dynamic ligand-induced conformational rearrangements in P-glycoprotein as probed by fluorescence resonance energy transfer spectroscopy. J Biol Chem 287(2):1112–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shapiro AB, Ling V 1997. Positively cooperative sites for drug transport by P-glycoprotein with distinct drug specificities. Eur J Biochem 250(1):130–137. [DOI] [PubMed] [Google Scholar]
- 76.Ritchie TK, Kwon H, Atkins WM 2011. Conformational analysis of human ATP-binding cassette transporter ABCB1 in lipid nanodiscs and inhibition by the antibodies MRK16 and UIC2. J Biol Chem 286(45):39489–39496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Garrigos M, Mir LM, Orlowski S 1997. Competitive and non‐competitive inhibition of the multidrug‐resistance‐associated P‐glycoprotein ATPase. Eur J Biochem 244(2):664–673. [DOI] [PubMed] [Google Scholar]
- 78.Litman T, Zeuthen T, Skovsgaard T, Stein WD 1997. Competitive, non-competitive and cooperative interactions between substrates of P-glycoprotein as measured by its ATPase activity. Biochim Biophys Acta 1361(2):169–176. [DOI] [PubMed] [Google Scholar]
- 79.Teng Y-N, Hsieh Y-W, Hung C-C, Lin H-Y 2015. Demethoxycurcumin modulates human P-Glycoprotein function via uncompetitive inhibition of ATPase hydrolysis activity. J Agric Food Chem 63(3):847–855. [DOI] [PubMed] [Google Scholar]
- 80.Loo TW, Clarke DM 2014. Tariquidar inhibits P-glycoprotein drug efflux but activates ATPase activity by blocking transition to an open conformation. Biochem Pharmacol 92(4):558566. [DOI] [PubMed] [Google Scholar]
- 81.Wang E-j, Casciano CN, Clement RP, Johnson WW 2000. Cooperativity in the Inhibition of P-Glycoprotein-Mediated Daunorubicin Transport: Evidence for Half-of-the-Sites Reactivity. Arch Biochem Biophys 383(1):91–98. [DOI] [PubMed] [Google Scholar]
- 82.Anglicheau D, Pallet N, Rabant M, Marquet P, Cassinat B, Meria P, Beaune P, Legendre C, Thervet E 2006. Role of P-glycoprotein in cyclosporine cytotoxicity in the cyclosporinesirolimus interaction. Kidney Int 70(6):1019–1025. [DOI] [PubMed] [Google Scholar]
- 83.Pauli-Magnus C, von Richter O, Burk O, Ziegler A, Mettang T, Eichelbaum M, Fromm MF 2000. Characterization of the major metabolites of verapamil as substrates and inhibitors of Pglycoprotein. J Pharmacol Exp Ther 293(2):376–382. [PubMed] [Google Scholar]
- 84.Beaudet L, Gros P 1995. Functional Dissection of P-glycoprotein Nucleotide-binding Domains in Chimeric and Mutant Proteins Modulation of Drug Resistance Profiles. J Biol Chem 270(29):17159–17170. [DOI] [PubMed] [Google Scholar]
- 85.Hrycyna CA, Ramachandra M, Germann UA, Cheng PW, Pastan I, Gottesman MM 1999. Both ATP Sites of Human P-Glycoprotein Are Essential but Not Symmetric. Biochemistry 38(42):13887–13899. [DOI] [PubMed] [Google Scholar]
- 86.Lawson J, O’Mara ML, Kerr ID 2008. Structure-based interpretation of the mutagenesis database for the nucleotide binding domains of P-glycoprotein. Biochim Biophys Acta 1778(2):376–391. [DOI] [PubMed] [Google Scholar]
- 87.Loo TW, Bartlett MC, Detty MR, Clarke DM 2012. The ATPase activity of the Pglycoprotein drug pump is highly activated when the N-terminal and central regions of the nucleotide-binding domains are linked closely together. J Biol Chem 287(32):26806–26816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Urbatsch IL, Sankaran B, Bhagat S, Senior AE 1995. Both P-glycoprotein nucleotidebinding sites are catalytically active. J Biol Chem 270(45):26956–26961. [DOI] [PubMed] [Google Scholar]
- 89.Crowe A, Wong P 2003. Potential roles of P-gp and calcium channels in loperamide and diphenoxylate transport. Toxicol Appl Pharmacol 193(1):127–137. [DOI] [PubMed] [Google Scholar]
- 90.Dierksen J, Gonsoulin M, Walterscheid JP 2015. Poor Man’s Methadone A Case Report of Loperamide Toxicity. Am J Forensic Med Pathol 36(4):268–270. [DOI] [PubMed] [Google Scholar]
- 91.Himanshu R, Jakir P, Pradnya H, Suneel P, Rahul S 2013. The impact of permeability enhancers on assessment for monolayer of colon adenocarcinoma cell line (CACO-2) used in in vitro permeability assay. Journal of Drug Delivery & Therapeutics 3(3):20–29. [Google Scholar]
- 92.Elkiweri IA, Zhang YL, Christians U, Ng KY, Tissot van Patot MC, Henthorn TK 2009. Competitive substrates for P-glycoprotein and organic anion protein transporters differentially reduce blood organ transport of fentanyl and loperamide: pharmacokinetics and pharmacodynamics in Sprague-Dawley rats. Anesth Analg 108(1):149–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nicklisch SC, Rees SD, McGrath AP, Gokirmak T, Bonito LT, Vermeer LM, Cregger C, Loewen G, Sandin S, Chang G, Hamdoun A 2016. Global marine pollutants inhibit Pglycoprotein: Environmental levels, inhibitory effects, and cocrystal structure. Sci Adv 2(4):e1600001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Uchida Y, Ohtsuki S, Katsukura Y, Ikeda C, Suzuki T, Kamiie J, Terasaki T 2011. Quantitative targeted absolute proteomics of human blood-brain barrier transporters and receptors. J Neurochem 117(2):333–345. [DOI] [PubMed] [Google Scholar]
- 95.Lin JH, Yamazaki M 2003. Role of P-Glycoprotein in pharmacokinetics. Clin Pharmacokinet 42(1):59–98. [DOI] [PubMed] [Google Scholar]
- 96.Feng B, Mills JB, Davidson RE, Mireles RJ, Janiszewski JS, Troutman MD, de Morais SM 2008. In vitro P-glycoprotein assays to predict the in vivo interactions of P-glycoprotein with drugs in the central nervous system. Drug Metab Dispos 36(2):268–275. [DOI] [PubMed] [Google Scholar]
- 97.Mercer SL, Coop A 2011. Opioid analgesics and P-glycoprotein efflux transporters: a potential systems-level contribution to analgesic tolerance. Curr Top Med Chem 11(9):1157–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.