Hepatitis E virus genotype 1 (HEV-1) is associated with large epidemics. Notably, HEV subtype 1e (HEV-1e) has caused HEV outbreaks in sub-Saharan Africa.
ABSTRACT
Hepatitis E virus genotype 1 (HEV-1) is associated with large epidemics. Notably, HEV subtype 1e (HEV-1e) has caused HEV outbreaks in sub-Saharan Africa. We report here the second full-length genome sequence of an HEV-1e strain (NG/17-0503) from a recent outbreak in Nigeria in 2017. It shares 94.2% identity with an HEV-1e strain from Chad.
ANNOUNCEMENT
Hepatitis E virus (HEV) is the most common causative agent of acute viral hepatitis. Eight HEV genotypes (HEV-1 to HEV-8) have been described, of which five are well recognized as human pathogens (1). HEV-1 and HEV-2 are waterborne, transmitted through fecal-oral routes, and are responsible for large HEV outbreaks in resource-limited countries. HEV-3 and HEV-4 are linked to zoonotic transmission and cause sporadic infections in industrialized countries. It has been reported that HEV subtype 1e (HEV-1e) was responsible for a large outbreak in sub-Saharan Africa (Chad). The T3 strain (GenBank accession no. AY204877) was obtained from an infected person from France during an outbreak in Chad in 1983 and represents the only complete genome sequence of HEV-1e available (2). We report the second full-length genome sequence of an HEV-1e strain (NG/17-0503) from a recent outbreak in Nigeria in 2017. The virus was isolated from a 35-year-old resident of the Mobbar local government area of Borno State, Nigeria. The patient’s serum tested positive for HEV antibodies using Wantai HEV IgM rapid test and Wantai HEV IgM enzyme-linked immunosorbent assay (ELISA; Sanbio, The Netherlands).
Viral RNA was extracted from serum using the QIAamp viral RNA minikit with the QIAcube BioRobot workstation (Qiagen, Hilden, Germany), followed by cDNA synthesis using SuperScript first-strand synthesis (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions. Real-time reverse transcription-PCR (RT-PCR) targeting the HEV open reading frames (ORF) 2 and 3 showed viremic HEV infection, with a viral load of 2.22 × 10E+6 IU/ml. Sequence analysis of partial ORF1 (307 bp) and ORF2 (401 bp) implemented using the BLASTn search engine (https://blast.ncbi.nlm.nih.gov) indicated that NG/17-0503 had highest nucleotide identity with the T3 strain. Real-time RT-PCR and consensus nested RT-PCR were conducted as previously described (3). The complete viral genome was amplified in fragments using the Kapa HiFi HotStart ReadyMix PCR kit (Roche, Mannheim, Germany) with HEV-1 universal and NG/17-0503 genome-specific primers (Table 1). The 5ʹ and 3ʹ ends were amplified using 5ʹ and 3ʹ rapid amplification of cDNA ends (Roche). All HEV amplicons were sequenced using Sanger sequencing with a BigDye Terminator 3.1 kit (Thermo Fisher, USA) in sense and antisense directions. The whole-genome sequence was assembled and analyzed using Geneious software 10.0.5 (Biomatters Ltd., Auckland, New Zealand) (4). Phylogenetic analyses were performed with the MEGA 7.0.26 software (5).
TABLE 1.
Primers used for NG/17-0503 complete genome sequencing
| Primera | Sequence (5′ to 3′) | Locationb |
|---|---|---|
| HEV-289_f | CTTGGGCCTTGAGTGTGCTA | 4443–4462 |
| HEV-290_f | CCCTATCCAGCGCGTTATACAT | 222–243 |
| HEV-291_r | ACCGACAGTAACCTTGTAGCTG | 1089–1068 |
| HEV-292_r | CATGAGACGGTCCCAGATATGG | 999–978 |
| HEV-293_f | CCTGTGTCGGGTGGAATGAA | 5091–5110 |
| HEV-294_r | GGACTGGTCATACTCGGCAG | 6595–6576 |
| HEV-295_f | ACGAAGGGTCCGATGTTGAC | 1532–1551 |
| HEV-296_r | AATGGCTGGGATCTGGTTCG | 3210–3191 |
| HEV-297_r | GACTCTAGCAGCAGTGTGGG | 3087–3068 |
| HEV-298_f | GATCCCAGCCATTGACTTCGAA | 3198–3219 |
| HEV-299_r | TAGCACACTCAAGGCCCAAG | 4462–4443 |
| HEV-300_r | TGTCGTCAAAAGCATCCCCA | 4351–4332 |
| HEV-302_f | TGCCACTGTAGAACCATGATCC | 1396–1417 |
| HEV-303_r | GGTAGATAAAGCTCATCCCCGG | 2771–2750 |
| HEV-304_r | GCGTCAAAACTAGGACCGATTG | 2729–2708 |
| HEV-311_f | GTGCTATTATGGAGGAGTGCGG | 4457–4478 |
| HEV-312_r | AGCACTATCGAATCATCACCTT | 4694–4673 |
| HEV-321_r | ACAGAGCATAACAAGGCCAGAA | 5983–5962 |
| HEV-322_f | ATGCTGTTGGTGGCTATGCTAT | 5745–5766 |
| HEV-323_r | CCTGGATAACTACACGGGATTCC | 6470–6448 |
| HEV-324_f | CATATCCGGGTCCTATGTGGTAC | 1575–1597 |
| HEV-325_r | GCATCAACYTCCGACCAAGT | 2156–2137 |
| HEV-326_f | GCATGTYTGGGAGTCGGC | 2082–2099 |
Forward primer designations end with _f; reverse primer designations end with _r.
Numbering is according the HEV prototype strain Burma (GenBank accession no. M73218).
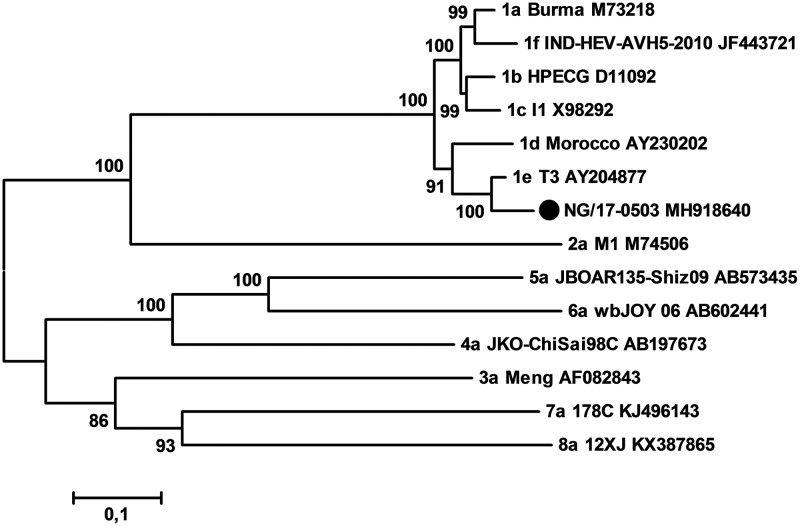
The complete genome of NG/17-0503 is 7,284 nucleotides in length, excluding the poly(A) tail, with a G+C content of 57.6%. The genome contains three ORFs (ORF1, 1,693 amino acids; ORF2, 660 amino acids; ORF3, 123 amino acids) encoding the HEV viral proteins. NG/17-0503 shared the highest identity of 94.2% with the T3 strain from Chad, followed by 88.7% with an HEV-1d Moroccan strain (GenBank accession no. MH918640). Sequences were aligned using the MAFFT algorithm (6). Phylogenetically, NG/17-0503 and T3 formed a separate subcluster within HEV-1 (Fig. 1).
Fig 1.
Phylogenetic tree based on complete genome sequences of representative proposed HEV reference strains. Each branch is labeled with the subtype designation, the strain name, and the GenBank accession number. The Nigerian NG/17-0503 strain in this study is marked with a solid circle. A maximum likelihood method based on the general time-reversible model with gamma distributed with invariant sites was inferred. The values at nodes indicate the bootstrap values (using 1,000 replications).
In conclusion, we identified and characterized the full-length genome of an HEV-1e strain circulating during an outbreak in Nigeria in 2017. Sequence and phylogenetic analyses showed that NG/17-0503 is the second full-length HEV-1e genome available to date. Since the Chad HEV-1e strain from 1983 was from a neighboring country of Nigeria, the detection of NG/17-0503 is strongly suggestive of local transmission of this endemic virus over decades.
Data availability.
The complete genome sequence of NG/17-0503 has been deposited in the GenBank database under the accession no. MH918640.
ACKNOWLEDGMENTS
We are grateful for the excellent technical assistance of Marcel Schulze and Steffen Zander (RKI).
This research was supported by the Global Outbreak Alert and Response Network (GOARN) Program of the World Health Organization (WHO). O.A.A. is funded by the German Academic Exchange Service (DAAD), Bonn, Germany. D.H. is funded by the Claussen-Simon-Stiftung (Claussen-Simon Foundation) “Dissertation Plus” program, Germany. B.W. is funded by the China Scholarship Council (CSC), Beijing, China. F.A.O. and O.A. are funded by a subproject of the Global Health Protection Program supported by the Federal Ministry of Health on the basis of a decision by the German Bundestag, the Partnership in Postgraduate Education (PPE), Robert Koch Institute, Berlin, Germany.
The funders, CSC, DAAD, Claussen-Simon Foundation, and PPE, had no role in the study design, data collection and interpretation, or the decision to submit the work for publication. The content is the responsibility of the authors and does not represent the views of the funders.
REFERENCES
- 1.Nan Y, Wu C, Zhao Q, Zhou EM. 2017. Zoonotic hepatitis E virus: an ignored risk for public health. Front Microbiol 8:2396. doi: 10.3389/fmicb.2017.02396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Cuyck H, Juge F, Roques P. 2003. Phylogenetic analysis of the first complete hepatitis E virus (HEV) genome from Africa. FEMS Immunol Med Microbiol 39:133–139. doi: 10.1016/S0928-8244(03)00241-4. [DOI] [PubMed] [Google Scholar]
- 3.Wang B, Harms D, Papp CP, Niendorf S, Jacobsen S, Lutgehetmann M, Pischke S, Wedermeyer H, Hofmann J, Bock CT. 2018. Comprehensive molecular approach for characterization of hepatitis E virus genotype 3 variants. J Clin Microbiol 56:e01686-17. doi: 10.1128/JCM.01686-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang B, Harms D, Hofmann J, Ciardo D, Kneubuhl A, Bock CT. 2017. Identification of a novel hepatitis E virus genotype 3 strain isolated from a chronic hepatitis E virus infection in a kidney transplant recipient in Switzerland. Genome Announc 5:e00345-17. doi: 10.1128/genomeA.00345-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete genome sequence of NG/17-0503 has been deposited in the GenBank database under the accession no. MH918640.