Abstract
Steroid hormone receptors regulate gene transcription in a highly tissue-specific manner. The local chromatin structure underlying promoters and hormone response elements is a major component involved in controlling these highly restricted expression patterns. Chromatin remodeling complexes, as well as histone and DNA modifying enzymes, are directed to gene-specific regions and create permissive or repressive chromatin environments. These structures further enable proper communication between transcription factors, co-regulators and basic transcription machinery. The regulatory elements active at target genes can be either constitutively accessible to receptors or subject to rapid receptor-dependent modification. The chromatin states responsible for these processes are in turn determined during development and differentiation. Thus access of regulatory factors to elements in chromatin provides a major level of cell selective regulation.
Keywords: chromatin remodeling, DNA methylation, DNase I hypersensitivity, enhancer, histone modifications, nuclear receptors, nucleosome positioning, promoter
Introduction
Steroid hormone receptors (SHRs) are transcription factors (TFs) that become activated after binding to steroid hormones. Upon activation, SHRs regulate specific target genes in order to accomplish an appropriate physiological response. The transcriptional response is highly cell-specific and can be achieved on multiple levels with chromatin structure and accessibility implicated as a key step. Although many advances have been made in recent years, the role that chromatin structure plays in the regulation of genes by nuclear receptors (NRs) is only beginning to be understood.
Stimulation with ligand leads to a series of rapidly occurring steps. First, hormone binding to the receptor takes place either in the cytoplasm or in the nucleus and is followed by ligand-specific changes in receptor conformation. These changes are accompanied by dissociation of the receptor from heat shock factors (e.g. heat shock protein 90, Hsp90). If initial localization of the receptor is cytoplasmic, translocation to the nucleus follows. While in the nucleus, hormone–receptor complexes are recruited, usually as dimers, to defined DNA sequences termed hormone response elements (HREs) [1]. HREs either are located in close proximity to transcription start sites (TSSs) of target genes or function as enhancers and control transcription from distal loci. Sequence specificity of an HRE serves as a precise docking element for an appropriate NR to bind. However, it is chromatin, not naked DNA, that makes up an environment for SHRs and other TFs to regulate gene transcription. Herein, we discuss the mechanisms by which DNA sequence and local chromatin structure control the NR response in a cell-specific and promoter-specific manner. The main emphasis will be put on the formation and detection of chromatin structures due to the nucleosome reorganization and the role of chromatin remodeling complexes in this process (see also accompanying review [2,3]). Additionally, the spatial organization of the genome in the nucleus and the role it plays in directing the physical association of enhancers and promoters are also important components of hormone signaling. An increasing effort is being placed on explaining how distant regulatory elements are brought together in a functional manner. This subject has been addressed elsewhere, however, and will not be brought up in the current review [4].
The basic building block of chromatin architecture in eukaryotic cells is a nucleosome, which consists of 147 bp of DNA wrapped 1.65 times around a histone octamer (two molecules each of H2A, H2B, H3 and H4) [5,6]. This complex is stabilized by strong interactions between the DNA phosphate backbone and lysine and arginine residues on the surface of the octamer, while the unstructured N-terminal histone tails protrude outside the nucleosome core and are the subjects of numerous modifications [7]. Histones are known to be the most evolutionary conserved proteins since histone equivalents and a simplified chromatin structure have been observed in Archaeabacteria [8]. It has been suggested that the primary/ancestral function of these prototype proteins is regulatory rather than structural and the function of DNA compaction evolved much later either as a result of or as a necessary precondition for increasing sizes of the genomes [8].
Eukaryotic chromatin can be divided into two extreme groups: an active (inducible) form called euchromatin and an inactive (silent) form known as heterochromatin [9]. Although a gene resides in a euchromatin compartment it does not mean that it is actively transcribed. In fact euchromatin can have a highly repressive effect on gene transcription and plays an important role in buffering the transcriptional noise. In inducible gene expression (i.e. by hormone), chromatin provides an environment for suppression of the gene before the stimulus and fast activation of the same after the stimulus.
SHRs and their model systems
All SHRs are modular proteins composed of six domains (A–F) [1,10]. The divergent A/B region contains the transcription activation function domain 1 (AF1) and is followed by two domains with high degree of sequence conservation: the DNA binding domain (DBD, region C) and the ligand binding domain (LBD, region E). DBD and LBD are separated by a flexible hinge region (region D) encompassing a nuclear localization signal (NLS). The multifunctional carboxyl terminus domain is a less conserved region which takes part in ligand-dependent activation (AF2). Both AFs act cooperatively to link receptor with basal TFs and co-regulators.
Approximately 50 NRs have been identified in mammals; however, most of them still lack a designated ligand. Glucocorticoid (GR), androgen (AR), progesterone (PR) and mineralocorticoid receptors (MR) form a subgroup with high homology within the DBD. As a result, all four receptors bind to similar sequence motifs, originally described as glucocorticoid response elements (GREs) [11]. GREs are composed of palindromic repeats of a hexanucleotide sequence separated by three non-conserved base pairs with each HRE half-site being bound by one receptor monomer [12,13].
Out of the multitude of potential binding sites, the receptor occupies only a small subset of them in a given cell type. Similarly, the observed overlap between glucocorticoid-mediated expression profiles between cell lines is modest [14–16]. Since the DNA sequence is identical in every cell, the mechanism of tissue-specific regulation must lie beyond the genetic composition of regulatory elements. Possible mechanisms by which tissue-specific regulation is dictated include differential expression of receptors (and receptors’ isoforms) and other co-factors, metabolism of ligands, and expression of selective modulators [17,18]. In addition, chromatin structure can play a role in the tissue-specific regulation of genes [10,17,19,20]. Specific structural alterations to the chromatin permit the binding of the receptor and Pol II transcriptional machinery. The process involves a variety of chromatin remodeling activities, all of them dependent on energy stored in ATP [19,21,22]. Once remodeled, these sites become ‘open’ and can be measured by their accessibility to DNase I digestion [23]. Two other activities, histone modifications and DNA methylation, have also been described as participating in the generation of open or closed chromatin structures.
It is logical to assume that hormone-dependent genes are placed in less permissive chromatin. This would allow for fine-tuned regulation and would prevent constitutive activation. In fact, studies have shown that transcription from chromatin templates, but not from transiently transfected DNA, is properly regulated by hormone in both GR- and ER-mediated response [24–26]. Furthermore, localization of the otherwise inducible pS2 promoter in a highly active chromatin compartment causes its constitutive and hormone-independent activation [27]. This proves that permissive chromatin is, at least in some cases, paramount over the TF requirements.
Current understanding of GR-regulated gene expression is based on extensive analysis of two gene model systems: the long terminal repeat of the mouse mammary tumor virus (MMTV-LTR) [28–30] and the glucocorticoid responsive unit of the rat tyrosine aminotransferase gene (Tat-GRU) [31]. The MMTV-LTR serves as a proximal promoter GRE whereas the GRU of the Tat gene is an enhancer located − 2.5 kb from the TSS. Nevertheless, both of them show a similar reliance on ATP-dependent remodeling activity upon hormone activation which results in increased accessibility to DNase I and other nucleases and leads to the recruitment of several TFs [31,32]. The MMTV-LTR, when assembled into chromatin, forms a well described nucleosomal structure with six (A–F) positioned nucleosomes and binding sites for GR, nuclear factor 1 (NF1), octamer transcription factor (OTF) and TATA binding protein [28,33,34]. Activation of MMTV by hormone results in the receptor binding to GREs within the nucleosomes B–C followed by a chromatin transition within this region [35–37].
However, in order to examine the role of chromatin and chromatin remodeling in hormone-regulated gene expression it remains crucial to sample the biological processes as they happen within a higher order chromatin organization. To accomplish this, a tandem array of MMTV-LTR repeats has been integrated near the centromere of chromosome 4 in 3134 (murine mammary epithelial adenocarcinoma) cells forming a system with well defined nucleosome positioning and localization of TF binding sites. The array consists of 2 Mb of 200 MMTV-LTR copies encompassing 800–1200 GR binding sites and can be visualized in living cells by the binding of green fluorescent protein (GFP) tagged versions of steroid receptors or associated factors [38]. This system is an excellent model for studying NR binding in vivo [38–40].
These described model systems are indispensable for examining chromatin dynamics and the results obtained by using them are cited throughout the review. However, they represent only a small subset of possible regulatory processes. Thus genome-wide studies are necessary in order to research the complexity of DNA sequences and protein components of chromatin.
DNA sequence as a factor in nucleosomal positioning and tissue-specific recognition by NRs
Contrary to previous assumptions, most NR binding events are not proximal to TSSs but are found at considerable distances from the promoter, and are distributed almost evenly between upstream and downstream sequences [41]. Sixty-three percent of GREs are found further than 10 kb from the TSS and only 9% of GREs [41], 4% of estrogen response elements (EREs) [42] and a similar number of androgen response elements (AREs) [43,44] have been mapped within −800 to +200 bp from TSSs of known genes.
NRs recognize short specific motifs but their binding certainly takes much more than simple sequence recognition. In the genome there are numerous sequences which could potentially be recognizable by each of the receptors. For example, in the murine genome we estimate the number of potential binding sites for the GR to be approximately 4 × 106. The vast majority of these sites are never occupied by a receptor, some are recognized only in a tissue-specific manner and a small number seem to be bound and activated ubiquitously across different cell lines (Fig. 1). Similarly, only 14% of computationally predicted EREs show genuine ER binding [45] and only a fraction of AREs are observed to be functional [43]. One factor in determining the occupancy of a specific site by a receptor might be the neighboring sequence. It has been proposed that the native GREs as well as AREs are in fact composite elements composed of multiple factor binding sites (i.e. GR and AP-1, ETS, SP1, C/EBP, HNF4) [41,46]. The individual loci that feature the GRE binding site and GRE composite architecture (up to 1 kb) remain evolutionarily conserved even if the sequences of GRE motifs themselves have been shown to be quite diverse. This allows the conservation of loci to serve as a good predictor of occupancy by the receptor in vivo [47]. Furthermore, the variety of GRE sequences provides another level of selective regulation. It has been suggested that the core GR binding sequence might, similarly to the effect of different ligands, impose unique allosteric restrictions on the receptor itself. This in turn could alter the types of co-regulators associated with NRs uniquely based on DNA sequence [48–50].
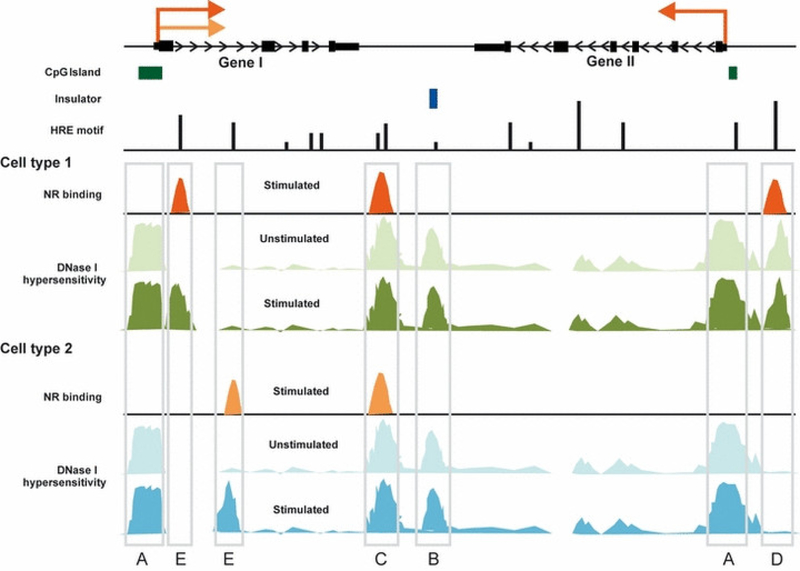
Fig. 1.
Tissue-specific chromatin architecture revealed in localization of DHSs. A schematic representation of DHSs before and after hormone stimulation in two cell types. The majority of hormone-responsive genes have a TSS that is embedded within a localized region of DNase I hypersensitivity. These promoter regions are generally hypersensitive across multiple cell types, and usually correlate with CpG islands (A). Common and preprogrammed DHSs present at distal regulatory elements often overlap with insulators (B). Hormone receptors recognize short DNA motifs (HREs), but only a small percentage of them are occupied by a receptor in a given cell type. NR binding occurs usually at distal enhancers and is highly correlated with the presence of accessible chromatin regions (C, D, E). Only a small fraction of enhancer-related DHSs are universally utilized in multiple cell lines and they usually represent hormone-independent chromatin structures (pre-programmed DHSs) (C). Most distal DHSs are tissue-specific and can be either hormone-independent (D) or appear only after hormone stimulation (inducible DHSs) (E). Thus the presence of a DHS and subsequent receptor/transcription factor binding results in a hormone-dependent and tissue-specific transcriptional regulation of a particular gene (gene II). A gene can be activated by the same hormone receptor in different tissues, although through different regulatory elements (gene I; elements C and E).
Within the chromatin architecture TF binding sites tend to cluster in linker DNA. However, there is still a large fraction of the regulatory elements that are buried inside the nucleosome [51]. Some factors are able to recognize and interact with their cognate elements even if they are placed within a nucleosome, but for many of them the affinity decreases by 10- to 100-fold [52–58]. Therefore, nucleosome positioning can play an important role in regulating TF access to specific DNA sequences.
The first observation that nucleosome position can be determined by sequence-dependent modulations of DNA structure was made more than 30 years ago [59]. Thanks to the most recent genome-wide analyses of nucleosome positioning an increasing number of reports followed suggesting strongly that the information about the nucleosome positioning might be embedded within the DNA sequence itself [60–64]. Evolution may have selected for specific arrangements of nucleosomes and indeed it is observed that a large fraction of nucleosomes are well positioned in vivo [51,65,66]. Nucleosomes within promoter regions often show reproducible, non-random organization which could potentially serve as another level of regulation for TF binding. Six nucleosomes within the earlier described MMTV-LTR promoter tend to occupy exactly the same positions in vivo as they do after they are assembled in vitro. Similar observations are made when the osteocalcin promoter is reconstituted in vitro using SWI/SNF complexes as remodelers [67].
The published yeast-based models predict that nucleosome occupancy at promoters and functional TF binding sites is low (termed nucleosome-free regions or nucleosome-depleted regions) and that there are more stable nucleosomes at nonfunctional sites [57,60]. One can imagine that sequences evolved to encode unstable nucleosomes and thus facilitate their accessibility for TFs and transcription machinery. Indeed DNase I hypersensitive sites (DHSs) are found to be enriched in nucleosome-excluding sequences, including short repeats of adenine (A16), long CCG triplet repeats and TGGA repeats [61]. In contrast to yeast, the analysis of human regulatory sequences predicts that there is a higher nucleosome occupancy in chromatin in vivo [68]. Thus, the preference for high nucleosome occupancy at the regulatory elements can be amenable for the restricted and tissue-specific regulation observed in higher eukaryotes but not in yeast.
In agreement with that, the inducible genes in yeast are also characterized by promoters that have been described as ‘covered’ with nucleosomes that are able to compete efficiently with TFs’ binding [69]. These kinds of promoters tend to contain a TATA box and numerous binding sites for different TFs and are highly dependent on chromatin remodeling (Fig. 2). They also display higher plasticity and noisy expression and are more sensitive to genetic perturbations, and thus are more prone to change their expression under evolutionary pressure [70,71]. In contrast, the chromatin architecture for yeast genes which are constitutively active is characterized by an open promoter structure, the presence of a nucleosome-depleted region with well positioned nucleosomes further upstream, and H2A.Z histone variants at the +1 and −1 nucleosomes [69].
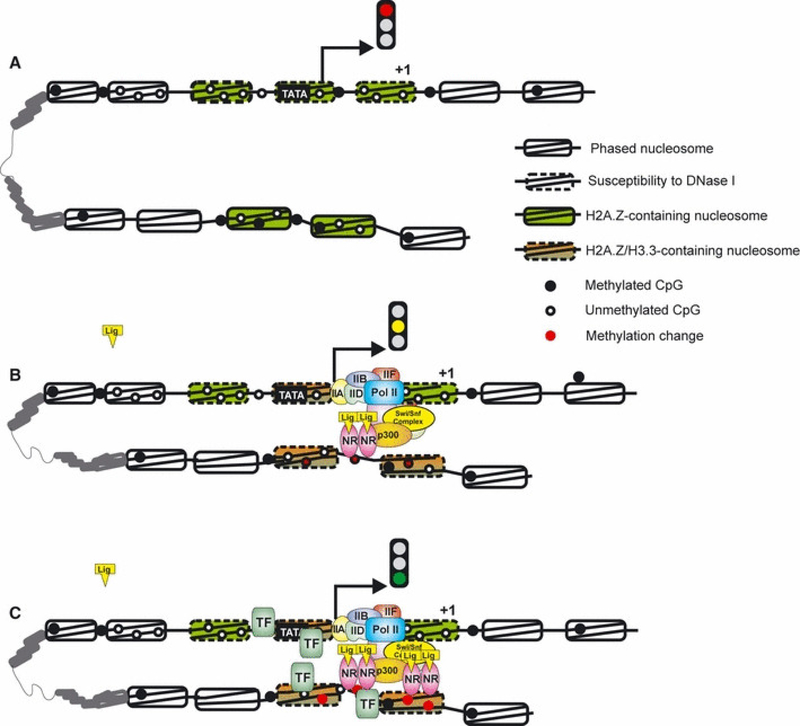
Fig. 2.
Dynamics of chromatin structures at inducible genes. (A) Inducible genes are regulated by a ‘covered’ class of promoters characterized by the presence of a TATA box and nucleosomes competing efficiently with TFs for access to DNA. Both promoters and enhancers are marked as chromatin structures staged for remodeling by the H2A.Z histone variant. In addition, enhancers available for subsequent receptor binding have a decreased level of DNA methylation. (B) Induction (i.e. hormone stimulation) leads to localized incorporation of H3.3 and formation of very labile H2A.Z/H3.3 nucleosomes at both the promoter and enhancer. These nucleosomes are very dynamic and can be easily ejected thus enabling TF binding. At enhancers, the receptor binding leads to nucleosome reorganization where two stable nucleosomes flank the receptor binding sites. Additionally, the +1 nucleosome at the promoter has been reported to move 30 bp downstream leaving space for RNA Pol II and the basic transcriptional machinery to dock at the TSS. Mediator complexes hold the promoter and enhancer together and changes in DNA methylation (red dots) are observed in at least a subset of enhancers. (C) Full transcriptional response is achieved due to synchronized binding of hormone receptor and other TFs, as well as to additional receptor binding events at neighboring HREs.
The differences in nucleosome positioning between active and silenced genes in human cells have also been examined recently [66]. The promoters of expressed genes are characterized by several well positioned nucleosomes, whereas only one nucleosome downstream from the TSS (+1) is phased when silenced genes are considered. The position of the first nucleosome upstream from the TSS (−1) in inactive promoters is replaced in active genes by Pol II binding and this results in a shift of the +1 nucleosome 30 bp towards the 3’ end. Also, within the functional enhancers, nucleosomes become more localized after activation in a way such that potential binding sites are moved to more accessible positions within the linker regions [66]. Specifically, androgen treatment dismisses a central nucleosome present at AREs allowing for ARs to bind. After remodeling the AR binding site is also found to be flanked by a pair of well positioned nucleosomes marked with H3-K4me2 or H3-K9,14ac [72,73].
As mentioned before, the studies based on yeast models suggest that intrinsic DNA sequence features have a dominant role in nucleosome organization in vivo [60,64]. However, discrepancies exist between nucleosome positions observed in vivo and computational predictions based on thermodynamic properties of DNA–histone interactions. One would expect these differences to be an integral part of inducible or cell-type-specific gene regulation with nucleosome location further modulated by the presence of specific features such as histone variants, DNA methylation and, to a lesser extent, histone modifications. In the last two cases, however, it is more difficult to differentiate between the direct effect of a modification on his-tone–DNA interactions and the indirect influence it has on another factor’s binding, which could consequently affect nucleosome positioning. Nevertheless, a strong link between CpG methylation and nucleosome positioning has been suggested based on observations that the presence of a methyl group can directly influence DNA bendability (dependent on the specific DNA sequence and extent of DNA methylation) [60,74]. On the other hand, nucleosome positioning has been observed to influence genome-wide methylation patterns by preferentially targeting DNA methyltransferases to nucleosome-bound DNA than to linker regions [75]. Further discrepancies between predicted versus observed nucleosome locations are believed to come from the competition between nucleosomes and TFs for access to DNA and the activity of chromatin remodelers. It has recently been reported that in yeast cells the depletion of the remodeling complex RISC caused the nucleosome-free regions to shrink and in vivo nucleosome occupancy to obtain positions reflecting the theoretical predictions more closely [76].
Overall, the results show that genomes encode and preserve both the sequences recognized by NRs and the positioning and stability of nucleosomes in regions that are critical for gene regulation. Those regions can be further rearranged which is accompanied by changes in DNA sensitivity to nucleases such as DNase I and restriction enzymes. Sites within the DNA which are accessible to DNase I are termed hypersensitive (DNase I hypersensitive site, DHS).
DNase I hypersensitivity as a marker of many different regulatory elements
Mapping DHSs is believed to be an effective method for determining the localization of the functional regulatory elements including promoters, enhancers, silencers, insulators and locus control regions [77]. DHSs have been identified in six cell lines within 1% of the genome as a part of an ENCODE project [78] and across the whole genome for CD4+ T cells[79]. Only 16–22% of sites are consistently present in all cell lines proving that the majority of gene regulatory elements are cell-type-specific. These shared sites have been further characterized by close (< 2 kb) proximity to TSSs, high CpG content, and binding of basal transcription machinery or CTCF (Fig. 1). Overall, the results indicate that the common DHSs belong to housekeeping promoters or, when distal, to insulators but not to enhancers. Cell-type-specific DHSs, on the other hand, are more distal, and are found to be enriched for binding sites of proteins known for their enhancer function (p300), sequence motifs for TFs, and cell-type-specific histone modifications [78,79]. Recent studies have suggested that proximal DHSs which do not overlap with promoters are associated with activating histone marks (H3-K4me3, AcH3) usually found at promoters [78,79]. Distal sites, however, are more enriched in H3-K27me3, H3-K9me2 and H3-K9me3 marks, while those found in the transcribed regions have higher levels of H3-K27me1 and H3-K9me1 (Fig. 3)[79]. Chromatin immunoprecipitation (ChIP) analyses have shown that the hypersensitive sites, both proximal and distant, are enriched for the H2A.Z histone variant, which has been reported to be a subject of exchange upon hormone treatment [80]. Therefore, it is argued that H2A.Z is associated with chromatin sites that are staged for remodeling and TF binding (Fig. 2A).
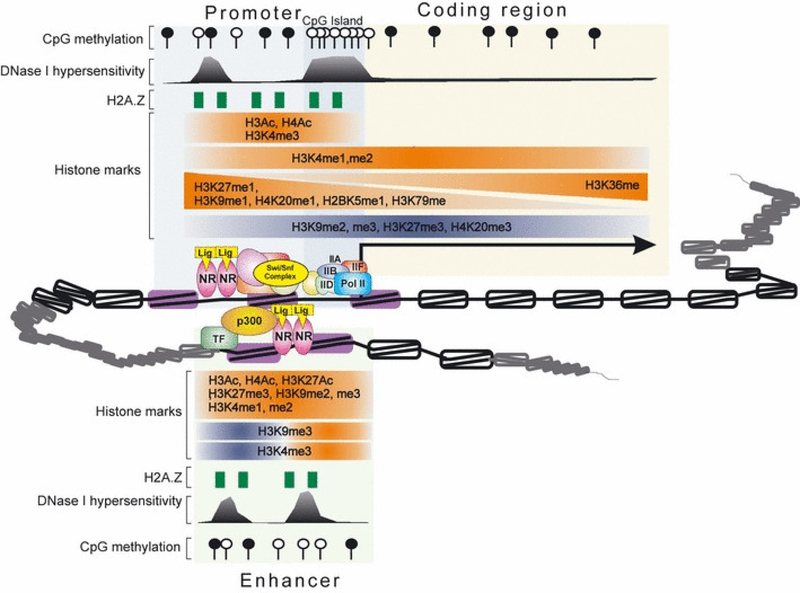
Fig. 3.
Characteristics of local chromatin structures within promoters, enhancers and coding regions. The non-random positioning of a nucleosome is dictated by DNA sequence, activity of remodeling complexes (like SWI/SNF) and competition of the nucleosome with TFs for access to specific DNA sequences. The regulatory regions are characterized by high turnover of histone proteins (depicted by purple nucleosomes). The histone marks identified at the promoters and enhancers of active (red) and silent (blue) genes are indicated. The gradients reflect changes of histone marks across the coding region. Contradictory observations about the presence of H3-K9me3 and H3-K4me3 within enhancer regions have been reported. Both promoters and enhancers are marked by DHSs and H2A.Z histone variants. Most promoters are characterized by increased density of CpG dinucleotides (CpG islands) which are usually unmethylated (open circles). Enhancers also show highly localized CpG enrichment with DNA methylation status correlating with their activity. The CpG dinucleotides are under-represented within coding regions and contain high methylation levels (filled circles) in order to prevent spurious transcription.
Correlation of DHSs with gene expression has shown that all expressed genes are marked by a DHS at the TSS [79]. However, although the presence of a DHS might be necessary for gene expression, it is clearly not sufficient. Inactive genes that are characterized by the presence of a DHS may be in a transcriptionally poised state. This is supported by an observation that activating histone marks and Pol II binding are also present at these genes. In contrast, promoter regions near silenced genes with no DHSs showed no evidence of these marks [79].
We have found that GR binding invariably occurs at nuclease-accessible sites [80,81] (Fig. 1). When profiles are compared between two cell lines, the lack of response to GR regulation is consistently correlated with the lack of GR binding and the absence of chromatin transition at the corresponding sites. Interestingly, the hypersensitive sites either pre-exist in chromatin (pre-programmed), or appear only after stimulation with hormone (de novo) [80,81]. The finding that GR interacts with the pre-existing DHSs is surprising as GR has classically been considered to be a pioneer factor which triggers the initiation of chromatin remodeling processes.
Steroid receptors have been frequently shown to induce DNase I hypersensitivity within the region of their binding sites [82–84]. Although there is strong evidence for histone loss after hormone induction, the remodeled site is not completely nucleosome-free and the question about the nature of DNA hypersensitivity to nucleolytic attack stays open. Two possibilities are taken into account: the nucleosome can either be repositioned to the neighboring regions or be temporarily unfolded from the template. The meticulous study performed at the Tat-GRU speaks in favor of the latter possibility [85]. No modification of the distribution of nucleosome frames has been observed while H1 and H3 interaction is clearly lost upon remodeling. The significance of H1 loss for transcription activation has also been shown using MMTV as a model [86,87]. Once a nucleosome’s binding becomes weaker and DNA becomes accessible, synergistic binding between receptors and other TFs is observed (Fig. 2C). On the MMTV promoter PR binding to the exposed element enables NF1 access to DNA. This in turn facilitates more PR binding to the remaining elements resulting in a full transcriptional response. The transcription is significantly compromised by the NF1 depletion or mutations in NF1 binding site [88]. Importantly, the synergistic binding between receptor and NF1 to MMTV is strongly dependent on the nucleosomal structure and is not observed for naked DNA [33].
Furthermore, we suggest that the vast majority of localized reorganization events are not stable but in fact represent a highly dynamic process. We have proposed that the rapid exchange observed for TFs and response elements in chromatin [38,39,89,90] has a direct correlation to chromatin remodeling [37,39,91,92]. The nucleosomes at promoter regions are also characterized by a high turnover rate independent of whether they are in active or repressed state [71,93]. This constant movement, assembly and disassembly of nucleosomes is a product of ATP-dependent remodeling activity.
Chromatin remodeling activity and achieving an open/accessible chromatin structure
Chromatin remodeling appears to be the first step in an ordered sequence of events required for hormone-regulated transcription. During the remodeling reaction DNA can be transiently unwound from a nucleosome or a nucleosome can be moved to a neighboring position (sliding) [94]. These reactions are energy-dependent and are executed by protein complexes that were first identified in yeast-based screens as mutations that control gene transcription triggered by extracellular signals [95–97]. The ATP-dependent remodeling engines can exist in multiple forms, usually as large (~ 2 MDa) multiprotein complexes with a core catalytic ATPase subunit and a team of auxiliary factors. The nature of an ATPase subunit underlies the current classification of remodeling complexes into four major classes: SWI/SNF, ISWI, Mi-2/NuRD and INO80 [94,98]. They also differ in the mechanism by which chromatin remodeling is executed (sliding, looping etc.). Both SWI/SNF and ISWI can slide nucleosomes along DNA; however, SWI/SNF may additionally be able to create stable DNA loops within nucleosome structures and remove/exchange histone dimers or octamers [94,99,100]. At the level of promoter activity regulation it translates into an ability to generate nucleosome-free regions (when coupled to his-tone chaperones), exchange canonical histone dimers for histone variants or, if there is not enough space for repositioning, expose specific DNA sequences as loops. Therefore, the SWI/SNF complexes are perceived as the most potent in rearranging promoter structures during transcriptional activation and as such are the best exploited in the studies of SHR-regulated transcription [101–105]. The ATPase subunit in human SWI/SNF complexes is either BRG1 or BRM, and is associated with up to a dozen additional factors including BRG1-associated factors (BAFs) [106–108]. It is worth mentioning that both the ATPase core and a composition of BAFs can be responsible for promoter-specific and tissue-specific regulation [107, 109,110].
Extensive studies based on the MMTV model have shown that SWI/SNF-dependent chromatin remodeling is a necessary prerequisite for optimal hormone-dependent transcription and in this case GR can utilize both BRG1- and BRM- containing complexes [36,105]. GR does not contact BRG1 directly but rather through the associated factors BAF57 and BAF60a which are common for both BRG1 and BRM complexes [111]. Transfection experiments with dominant negative forms of either BRG1 or BRM have resulted in an inhibition of transcription, lack of both Pol II loading and chromatin transition, as well as compromised decondensation of the MMTV array [105]. Furthermore, using a UV laser crosslinking approach it has been possible to establish highly transient and periodic interactions of GR with the MMTV template during the remodeling reaction. This is further reflected by periodic binding of SWI/SNF, H2A and H2B [112]. The suggested model requires that receptor binding is aided during the early phase of the nucleosome remodeling reaction, but when the remodeling reaction is completed and nucleosomes return to the basal state, receptors are actively removed from the promoter. In human cells lacking BRG1 and BRM (i.e. SW-13) transactivation by GR is weak and can be selectively enhanced by the ectopic expression of either BRG1 or BRM [102]. However, it cannot be substituted by the activity of ISWI or Mi-2 complexes, both present in SW-13 [19]. BRG1 remodeling action is also specifically required for PR- and AR-dependent activation of MMTV-LTR chromatin [113,114] as well as for ER-regulated genes [103,104,115]. These results suggest that SWI/SNF complexes are commonly utilized by NRs for creating chromatin transition states during hormone induction. On the other hand, the transcription profile obtained after overexpression of a dominant negative form of BRG1 shows significant reduction in only 40% of glucocorticoid-activated genes [80]. An even smaller effect (11%) is observed when glucocorticoid-repressed genes are analyzed. Consistent with that, only a subset of DHSs, both preexisting and hormone-inducible, are dependent on BRG1 action in these cells. Hence, the contribution of other remodeling complexes seems to be an obvious possibility.
The picture that emerges is that receptor-based gene regulation is always dependent on the presence of remodeled chromatin. However, this feature can either be formed during cell development, and continuously present, or be triggered by the receptor itself only after hormone stimulation (Fig. 1). In addition, chromatin remodeling alone is not sufficient for transcriptional activation and depends on the context of the DNA and histone modifications. In some cases opening the chromatin structure by chromatin remodeling enzymes is necessary for subsequent acetylation of histones [116–118]. In other cases, the recruitment of chromatin remodeling complexes must be preceded by RNA Pol II binding and histone acetylation which in turn creates binding sites for bromodomain-containing proteins, i.e. BAFs [85,119,120].
Thus the action of remodeling complexes should not be separated from the action of histone and DNA modifying enzymes, as they operate simultaneously on the same sequences and influence each other. In fact, large multifunctional complexes have been found in vivo where the chromatin remodelers are associated with histone-modifying enzymes including histone deacetylases (HDACs, NCoR complex), histone methyltransferases, such as CARM1 (nucleosomal methylation activation complex, NUMAC), as well as other proteins with co-regulatory functions (mSin3a, BRCA1, TOPO II, actin). Furthermore, Mi2/NURD, part of the NCoR complex, can repress NR-dependent transcription [121,122] and is targeted to specific areas of chromatin through recruitment by transcription repressors or by factors that recognize methylated DNA.
Histone modifications and histone variants as a part of gene architecture and transcription regulation
Over 60 different residues within histone tails have been identified as targets for post-translational modifications (reviewed in [7,55]). The most common histone modifications are acetylation or ubiquination of lysine residues, methylation of arginine and lysine residues, and phosphorylation of serine and threonine residues. Acetylation usually occurs cumulatively on multiple lysine residues and utilizes different histone acetyltransferases (HATs) in a seemingly non-specific manner. In contrast, other histone marks are deposited by a specific enzyme on a defined residue. Furthermore, methylation can exist as monomethylation, dimethylation or trimethylation with different methyltransferases being active at each step. All modifications can affect one another and many of them are positively or negatively correlated [7,55,123].
The mechanisms by which histone modifications exert their function include alterations in DNA–nucleosome and nucleosome–nucleosome interactions as well as in the recruitment of non-histone regulatory proteins (reviewed in [7,55]). The internucleosomal and intranucleosomal interactions can become relaxed simply due to the change in the net charge of nucleosomes caused by most (methylation is an exception) modifications. Among them, lysine acetylation is believed to be the most potent due to both its ability to neutralize the basic charge and its abundance. This idea is supported by the experimental observation that acetylated histones are easier to displace from DNA both in vivo [124] and in vitro [120,125]. Recently, acetylation of H3-K14 has been shown to be essential for nucleosome eviction [126]. The effect of histone modifications can also go beyond local contacts and directly influence higher-order chromatin structure. For example, acetylation of H4-K16 is known to inhibit the formation of 30 nm fibers [127] as discussed in detail in an accompanying review [2]. The alternative function of histone modifications is to create a ‘code’ which can be recognized and read by other proteins. Proteins with chromo-like domains can bind to methylated his-tone residues, whereas acetylation is recognized by bromodomains. These proteins, in turn, provide enzymatic activities which further influence chromatin dynamics and function.
Globally, active euchromatin and inactive heterochromatin are marked by different histone modifications. Acetylation of H3 and H4 and methylation of H3-K4, H3-K36 and H3-K79 are characteristic of active chromatin whereas low levels of acetylation and high levels of H3-K9me3, H3-K27me3 and H4-K20 methylation are associated with inactive chromatin [7]. These modifications frequently spread along extended chromosomal regions and are sharply separated from each other by boundary elements associated with the insulator binding protein CTCF [128,129].
Within euchromatin, actively transcribed genes are further characterized by a set of features that show a more complex and localized pattern within enhancers, the core promoter, coding regions and the 3’ end of the gene [7,128,130] (Fig. 3). Multiple studies have proved that H3 and H4 histones within the TSSs are generally acetylated [128,131–134]. As far as other modifications are concerned, high levels of all three states of H3-K4 methylation and H2A.Z form a peak within the promoter and TSS regions whereas H3-K27me1, H3-K79me, H2B-K5me1, H4-K20me1 and H3-K9me1 are associated with the entire transcribed region. Unlike other marks, H3-K36me1 tends to accumulate towards the 3’ end of the gene. Interestingly, the signatures of both promoters and insulators appear to be invariant across different cell lines [129,135] and several studies have shown that both active and inactive promoters are associated with histone acetylation and H3-K4me3 [128,136–138]. Chromatin modification patterns at inducible genes have also proved to be relatively stable during activation of resting T cells with active modifications being already in place [139]. In contrast to that, enhancers are believed to be the most variable elements and display a highly cell-type-specific pattern of histone modifications.
Identification of enhancer elements has not been an easy undertaking because of their distant localization from regulated genes and the lack of specific sequence elements. Attempts made thus far to identify enhancer regions have been based on sequence conservation, the position of DHSs [78,79,140] or p300 binding to DNA outside the promoter regions [135,136,141]. Based on the latter, over 55 000 enhancers have been identified in only two cell lines (K562 and HeLa), thus leading to the prediction of 105–106 enhancers existing in total [135].
Once enhancer elements have been recognized their chromatin characteristics can be described (Fig. 3). Similar to promoters, enhancer elements are marked by H3 acetylation, H3-K4 monomethylation, H2A.Z and H3-K9me1, but lack other promoter-specific modifications [128,132,134,135,142,143]. Surprisingly, H3-K27me3, previously ascribed to the repressive chromatin, has also been identified within enhancer elements. The combination of H3-K4me1 and H3-K4me3 has been proposed as the strongest discriminator between enhancers and promoters with enhancers being deprived of trimethylation [140,142,144]. However, this might not be the universal feature since it has recently been shown that H3-K4me3 is also present at the enhancers when a DHS-based approach is applied to identify these regions [140]. Furthermore, each of the modifications including H3-K4me1, 2 and 3, H3-K9me1 and H2A.Z have been detected at only 20–40% of putative enhancers suggesting that they are found only in unique subgroups [128,140]. No significant correlation between specific modification patterns at the enhancer regions and gene expression has been observed [140].
Even if current literature lacks the global overview of histone marks specifically in terms of regulation by steroid receptors there is no reason to assume that their common pattern would be different from that mentioned above. Arginine methylation of both H3R2 and H4R3 has been previously suggested to play a role in NR-mediated transcription activation [145]; however, none of these marks showed any characteristic patterns in a genome-wide analysis [128]. It is still unknown how many enhancers can be identified based on their characteristics before hormone induction and to what extent the chromatin marks within enhancer elements can change after induction and receptor binding. When HeLa cells are treated with interferon-c only 25% of STAT1 binding is observed within predicted enhancers [135]. Analysis of GR binding sites, however, shows that most fall into DHS regions existing before hormone stimulation and only 15% of binding events are followed by a chromatin transition and possibly by changes in at least some of the chromatin signatures [80,81]. Dynamic changes of histone modifications after hormone induction have not been globally analyzed yet but have been observed on selected steroid-hormone-regulated promoters. The changes are shown to be limited to a few or even a single nucleosome. At the MMTV promoter, GRE-containing nucleosomes B and C, but not F, undergo rapid (30 min after stimulation) deacetylation at H3 and H4, both of which show a high level of acetylation in the 3134 cell line before induction [146]. Similar experiments in T47D breast cancer cells have shown that H4 deacetylation is preceded by initial acetylation which precisely coincides with the time of recruitment of progesterone receptor, Pol II and p300 [147].
However, the most detailed data concerning dynamic recruitment of co-regulators and histone modification changes have been collected for the ER-regulated pS2 gene. In the initial experiments the increase of acetylation of histone H3 and H4 at ER-regulated cathepsin D and pS2 promoters was observed within 10 min of estrogen treatment, peaking at 1 h post-treatment and decreasing to near-basal levels within 6 h. The transient increase in histone acetylation coincided with a transient increase in the association of SRC-3, p300, CBP and RNA Pol II, as well as a transient increase in the transcription level [148,149]. Further experiments that included a more detailed time course and using α-amanitin-synchronized cells demonstrated in fact cyclic behavior of histone acetylation with peaks every 40–60 min corresponding to productive transcription cycles [150,151].
Core histones not only undergo covalent post-translational modifications but can also be exchanged with histone variants (reviewed in [55,152]). Differences between variants and canonical histones can be as small as a few amino acids, as in the case of H3.3, or can apply to larger domains within the histone tails (MacroH2A) or in the histone fold domains (H2ABdb). In contrast to canonical histones, histone variants are expressed mainly outside of S phase and are thought to be deposited into nucleosomes in a replication-independent manner by means of specific protein complexes. H2A.Z can be incorporated into a nucleosome either by Swr1 through ATP-dependent histone exchange reactions [153] or via the help of replication-independent chaperones like Nap1 [154], and H3.3 is assembled by histone regulator A (HIRA). Incorporation of histone variants into chromatin impacts its structure in various ways [155]. For example, histone H2A.Z replaces histone H2A at the promoter sites, insulators and enhancers and when co-assembled with H3.3 forms a very unstable structure that can be ejected following gene activation [156,157] (Fig. 2B). This observation sheds a new light on the previously observed nucleosome-free regions at active promoters and enhancers. These nucleosome-free regions may now be explained by the presence of highly labile H2A.Z/H3.3-containing nucleosomes that are easily displaced by TFs. This fact has been overlooked before because of H2A.Z/H3.3 disruption in the moderate salt concentrations usually used for chromatin purification [156]. It has been suggested that transcription of as many as 30% of genes can be regulated through incorporation of H2A.Z [158]. Taken together, histone modifications and histone variants play a role in defining the chromatin architecture of active or potentially active gene transcription units and undergo dynamic changes during ligand stimulation.
DNA methylation and MeCP2 – link to chromatin architecture and remodeling
DNA methylation of cytosine is the most abundant covalent DNA modification. In differentiated cells DNA methylation appears almost exclusively in the CpG context and is deposited there by one of the DNA methyltransferases (Dnmt1, Dnmt3a or Dnmt3b) [159]. In mammalian genomes the distribution of a methyl mark is described as a global methylation pattern [160]. Approximately 98% of CpG dinucleotides are located within the CpG-poor regions and 80% of them are methylated. The observed high level of DNA methylation within gene bodies and non-coding regions is believed to serve as a suppressor of transcriptional noise by preventing the spurious transcription initiation from cryptic promoters [160,161]. The remaining 2% of genomic CpGs are densely grouped in short stretches located mostly at the 5’ end of the genes [160] (Fig. 3). These patches are referred to as CpG islands and typically stay unmethylated independent of the gene expression [162,163].
High CpG density promoters are associated with two classes of genes, commonly expressed housekeeping genes and highly regulated key developmental genes, whereas low CpG density promoters are generally linked to tissue-specific genes [164]. In contrast to CpG-rich sequences, CpG-poor regulatory elements are more prone to active and de novo methylation and demethylation, which might provide yet another level of gene regulation. As mentioned before, cis regulatory elements active in a particular cell type are often associated with marks of open chromatin such as H3-K4me2 or H3-K4me1 [131,142]. It has been shown that CpGs found at H3-K4me2-enriched sites (outside of promoters and CpG islands) have significantly lower DNA methylation levels than those at H3-K4me2-depleted sites, and this relationship is particularly strong for CpGs located within highly conserved non-coding elements [164]. We have observed a high correlation between CpG methylation status and activity of enhancer elements at GR binding sites with demethylation strongly linked to chromatin accessibility and GR binding (Wiench M, John S, Baek S, Johnson TA, Sung M-H, Escobar T, Simmons CA, Pearce KH, Biddie SC, Sabo PJ, Thurman RE, Stamatoyannopoulos JA & Hager GL, unpublished results). The lack of DNA methylation, often restricted to a single CpG site, has been shown to be the remnant of interactions with pioneer TFs (such as PU.1 or C/EBPβ) in early embryonic development [165]. These interactions might be necessary to prevent the enhancers from assembling into repressive chromatin structures during development and to make them amenable for subsequent activation of tissue-specific genes. Thus activation of these genes may be possible only after specific interactions and changes in chromatin structure have occurred at earlier stages in development.
DNA methylation leads to transcriptional repression through several mechanisms. It can directly affect the TF’s binding to its DNA recognition element as well as the positioning and stability of a nucleosome. However, it primarily acts indirectly through the action of proteins like MeCP2 and MBD1–4 that selectively bind methylated CpGs through their methyl-CpG binding domain (MBD) [166]. Various members of the MBD family display different DNA binding specificities. They can recognize and bind to more complex sequences than a single methylated CpG. For example, high affinity binding by MeCP2 requires four or more A/T base pairs adjacent to the methylated CpG [167].
Even if methylation marks a DNA molecule directly, the silencing effect is observed only after the DNA template is assembled into chromatin [168,169]. Moreover, MeCP2 has been shown to bind methylated DNA only in a nucleosome context [170]. Hence, the mechanism through which DNA methylation and MBDs accomplish the silencing effect employs both chromatin modification and remodeling activities. Different sets of proteins have been identified to bind a nucleosome when DNA methylation is combined with different histone modifications [170]. Furthermore, MeCP2 has been found to interact with histone deacetylases (HDAC2, Sin3A) [171,172], histone methyltransferases (SUV39H1) [173] and remodeling complexes [174]. Both MeCP2 and BRM were shown to be associated with each other on the same sequences within hypermethylated promoters, and treatment with inhibitors of DNA methylation (5-aza-2’-deoxycytidine, 5Aza-dC) results in a loss of methylation, loss of BRM and MeCP2 binding and reactivation of transcription [174]. In cancer cells, changes in DNA methylation promoted by SWI/SNF complexes induce transcriptional activation and rescue the transcription of CD44 and E-cadherin [175]. Although MeCP2 as well as MBD2 are likely to be responsible for initial recruitment of chromatin remodelers, studies in vitro and in vivo suggest that chromatin remodeling activities further facilitate binding of MBD proteins to those methylated sites that are not initially accessible on nucleosomal templates and by doing so further stimulate MBD-mediated gene repression [174].
In many vertebrates site-specific demethylation affects tissue-specific genes [176]. Unlike plants, methylation of DNA in mammals has long been considered to be a stable mark that is removed only during a passive process which involves replication. Recently, evidence showing that the demethylation process might be active and dynamic is accumulating. Demethylation is often restricted to regulatory elements outside the core promoter where transcription factors bind and where chromatin is hypersensitive to DNase I [164]. It has been reported that the Bdnf regulatory region is demethylated by 25–45% and MeCP2 is redistributed 2–3 days after depolarization of neuron cells which is accompanied by increase in Bdnf synthesis [177]. In another study, prolonged glucocorticoid treatment caused demethylation of all four CpGs located within the remodeled area of a Tat-GRU while neighboring CpGs located outside the remodeled area remained methylated. In this case the demethylation is completed in 3 days and allows for the recruitment of additional transcription factors [178].
A kinetics profile of 2–3 days does not point to an active and efficient process. However, very fast and cyclical changes (with a periodicity of about 100 min) of DNA methylation and demethylation have also been observed within the promoters of pS2 and several other ER-regulated genes [179]. It is not clear yet what kind of mechanism is primarily involved in the demethylation but it has been suggested previously that it most probably involves the creation of nicks in DNA 3’ to the methylcytidine [176,180]. A recent report confirms this observation and suggests that deamination paired with glycosylation enzymatic activities (AID/MBD4) and a base excision repair process are involved [181]. Rapid demethylation after activation seems to be a common event at hormone-inducible elements since it has been observed during ER [179,182], vitamin D receptor [183] and GR (Wiench et al., submitted) regulation even though the mechanism is poorly understood (Fig. 2).
In addition to its role in gene silencing, MeCP2 has been described as a chromosomal architecture element. MeCP2 has been shown to mediate the formation of complex chromatin structure by promoting chromatin looping at the Dlx5 gene with the loop itself marked by an H3-K9me2 repressive mark [184]. A very recent report describes MeCP2 in neuronal cells as a highly abundant nuclear protein that might replace H1 binding and globally alter chromatin state [185]. The described action of MeCP2 is dependent on cytosine methylation. However, another study reported that the MeCP2 protein can organize chromatin independently of DNA methylation and in the absence of a functional MBD domain. The addition of MeCP2 to unmethylated nucleosomal arrays leads to significant chromatin compaction, greater than that achieved by histone H1[87]. At the ratio of one MeCP2 molecule per nucleo-some, electron microscopy revealed formation of a novel 60S ellipsoidal structure. Also, DNA when methylated is able to adopt a distinctive chromatin structure after being assembled into chromatin which results in a loss of DNase I hypersensitivity [186,187]. This is in agreement with the fact that demethylated CpGs are more often found at regions where chromatin remodeling creates DHSs (Figs 2 and 3). Regions of DNA methylation have also been found to be deficient in H2A.Z, a marker of inducible chromatin, and in fact these two chromatin features seem to be mutually exclusive in both plants [188] and mammals [158].
To summarize, DNA methylation appears to be a more dynamic feature than previously thought especially within distal regulatory elements where it teams up with other chromatin signatures defined by the presence of DHSs and characteristic histone modifications. The specificity can be achieved (a) by utilizing different DNA methyltransferases to deposit the methyl mark and (b) by recognition of the mark by different DNA methyl binding proteins. Methyl-CpG binding proteins have been shown recently to provide a functional link between DNA methylation, chromatin remodeling and histone modifications, as well as to serve as structural proteins of chromatin.
Conclusions
During the last two decades model systems have been developed and well exploited to study the mechanisms governing steroid hormone gene regulation in the chromatin environment. Those systems have made it possible to explore the effects of nucleosome positioning, the changes in the nucleosome structure in response to ligand stimulation, and the role chromatin remodeling complexes and histone modifying enzymes have during promoter progression. However, it is apparent that they represent unique examples rather than a universal type of regulated promoter. Thus, to overcome this limitation a new set of methods has been developed to study the epigenome on a high throughput basis. The necessary precondition is to reliably identify promoters, enhancers and other regulatory elements within the genome. Recent reports discussed in this review show that this step can be achieved. The next step is to prove their functionality and to characterize them by the presence of specific chromatin signatures. The results of the first attempts have been published during the last several months proving the complexity of the system and revealing even more questions. Thanks to ChIP-chip, ChIP-seq, DNase I-seq, MeDIP-seq and even genome-wide bisulfite sequencing becoming more available and affordable, a big leap forward can be made in understanding how local chromatin architecture affects tissue-specific gene regulation and regulation by NRs.
Acknowledgements
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. T.B. Miranda is funded in part by the NIGMS Pharmacology Research Associate Fellowship.
Abbreviations
- AF
activation function
- 5-Aza-dC
5-aza-2’-deoxycytidine
- AR
androgen receptor
- ARE
androgen response element
- BAF
BRG1-associated factor
- ChIP
chromatin immunoprecipitation
- DBD
DNA binding domain
- DHS
DNase I hypersensitive site
- ER
estrogen receptor
- ERE
estrogen response element
- GFP
green fluorescent protein
- GR
glucocorticoid receptor
- GRE
glucocorticoid response element
- GRU
glucocorticoid responsive unit
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- HRE
hormone response element
- LBD
ligand binding domain
- LTR
long terminal repeat
- MBD
methyl-CpG binding domain
- MMTV
mouse mammary tumor virus
- NF1
nuclear factor 1
- NLS
nuclear localization signal
- NR
nuclear receptor
- PR
progesterone receptor
- SHR
steroid hormone receptor
- TF
transcription factor
- TSS
transcription start site
References
- 1.Beato M & Klug J (2000) Steroid hormone receptors: an update. Hum Reprod Update 6, 225–236. [DOI] [PubMed] [Google Scholar]
- 2.Cockerill PN (2011) Structure and function of active chromatin and DNase I hypersensitive sites. FEBS J 278, 2182–2210. [DOI] [PubMed] [Google Scholar]
- 3.Bednar J & Dimitrov S (2011) Chromatin under mechanical stress: from single 30 nm fibers to single nucleosomes. FEBS J 278, 2231–2243. [DOI] [PubMed] [Google Scholar]
- 4.Hakim O, Sung MH & Hager GL (2010) 3D shortcuts to gene regulation. Curr Opin Cell Biol 22, 305–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richmond TJ, Finch JT, Rushton B, Rhodes D & Klug A (1984) Structure of the nucleosome core particle at 7Å resolution. Nature 311, 532–537. [DOI] [PubMed] [Google Scholar]
- 6.Luger K, Mader AW, Richmond RK, Sargent DF & Richmond TJ (1997) Crystal structure of the nucleo-some core particle at 2.8Å resolution. Nature 389, 251–260. [DOI] [PubMed] [Google Scholar]
- 7.Kouzarides T (2007) Chromatin modifications and their function. Cell 128, 693–705. [DOI] [PubMed] [Google Scholar]
- 8.Felsenfeld G & Groudine M (2003) Controlling the double helix. Nature 421, 448–453. [DOI] [PubMed] [Google Scholar]
- 9.Horn PJ & Peterson CL (2006) Heterochromatin assembly: a new twist on an old model. Chromosome Res 14, 83–94. [DOI] [PubMed] [Google Scholar]
- 10.Kinyamu HK & Archer TK (2004) Modifying chromatin to permit steroid hormone receptor-dependent transcription. Biochim Biophys Acta 1677, 30–45. [DOI] [PubMed] [Google Scholar]
- 11.Scheidereit C, Geisse S, Westphal HM & Beato M (1983) The glucocorticoid receptor binds to defined nucleotide sequences near the promoter of mouse mammary tumour virus. Nature 304, 749–752. [DOI] [PubMed] [Google Scholar]
- 12.Beato M (1989) Gene regulation by steroid hormones. Cell 56, 335–344. [DOI] [PubMed] [Google Scholar]
- 13.Luisi BF, Xu WX, Otwinowski Z, Freedman LP, Yamamoto KR & Sigler PB (1991) Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature 352, 497–505. [DOI] [PubMed] [Google Scholar]
- 14.Rogatsky I, Wang JC, Derynck MK, Nonaka DF, Khodabakhsh DB, Haqq CM, Darimont BD, Garabedian MJ & Yamamoto KR (2003) Target-specific utilization of transcriptional regulatory surfaces by the glucocorticoid receptor. Proc Natl Acad Sci USA 100, 13845–13850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.John S, Johnson TA, Sung MH, Koch-Paiz CA, Davis SR, Walker R, Meltzer P & Hager GL (2009) Kinetic complexity of the global response to glucocorticoid receptor action. Endocrinology 150, 1766–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang JC, Derynck MK, Nonaka DF, Khodabakhsh DB, Haqq C & Yamamoto KR (2004) Chromatin immunoprecipitation (ChIP) scanning identifies primary glucocorticoid receptor target genes. Proc Natl Acad Sci USA 101, 15603–15608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collingwood TN, Urnov FD & Wolffe AP (1999) Nuclear receptors: coactivators, corepressors and chromatin remodeling in the control of transcription. J Mol Endocrinol 23, 255–275. [DOI] [PubMed] [Google Scholar]
- 18.Hsiao PW, Deroo BJ & Archer TK (2002) Chromatin remodeling and tissue-selective responses of nuclear hormone receptors. Biochem Cell Biol 80, 343–351. [DOI] [PubMed] [Google Scholar]
- 19.Trotter KW & Archer TK (2007) Nuclear receptors and chromatin remodeling machinery. Mol Cell Endocrinol 265–266, 162–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aoyagi S, Trotter KW & Archer TK (2005) ATP-dependent chromatin remodeling complexes and their role in nuclear receptor-dependent transcription in vivo. Vitam Horm 70, 281–307. [DOI] [PubMed] [Google Scholar]
- 21.Elbi C, Walker DA, Romero G, Sullivan WP, Toft DO, Hager GL & DeFranco DB (2004) Molecular chaperones function as steroid receptor nuclear mobility factors. Proc Natl Acad Sci USA 101, 2876–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agresti A, Scaffidi P, Riva A, Caiolfa VR & Bianchi ME (2005) GR and HMGB1 interact only within chromatin and influence each other’s residence time. Mol Cell 18, 109–121. [DOI] [PubMed] [Google Scholar]
- 23.Sabo PJ, Humbert R, Hawrylycz M, Wallace JC, Dorschner MO, McArthur M & Stamatoyannopoulos JA (2004) Genome-wide identification ofDNaseI hypersensitive sites using active chromatin sequence libraries. Proc Natl Acad Sci USA 101, 4537–4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bresnick EH, Rories C & Hager GL (1992) Evidence that nucleosomes on the mouse mammary tumor virus promoter adopt specific translational positions. Nucleic Acids Res 20, 865–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee H-L & Archer TK (1994) Nucleosome mediated disruption of transcription factor:chromatin initiation complexes at the mouse mammary tumour virus long terminal repeat in vivo. Mol Cell Biol 14, 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kraus WL & Kadonaga JT (1998) p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev 12, 331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oduro AK, Fritsch MK & Murdoch FE (2008) Chromatin context dominates estrogen regulation of pS2 gene expression. Exp Cell Res 314, 2796–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richard-Foy H & Hager GL (1987) Sequence specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J 6, 2321–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Archer TK, Cordingley MG, Wolford RG & Hager GL (1991) Transcription factor access is mediated by accurately positioned nucleosomes on the mouse mammary tumor virus promoter. Mol Cell Biol 11, 688–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Truss M, Bartsch J, Hache RJ & Beato M (1993) Chromatin structure modulates transcription factor binding to the mouse mammary tumor virus (MMTV) promoter. J Steroid Biochem Mol Biol 47, 1–10. [DOI] [PubMed] [Google Scholar]
- 31.Grange T, Roux J, Rigaud G & Pictet R (1991) Cell-type specific activity of two glucocorticoid responsive units of rat tyrosine aminotransferase gene is associated with multiple binding sites for C/EBP and a novel liver-specific nuclear factor. Nucleic Acids Res 19, 131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trotter KW & Archer TK (2004) Reconstitution of glucocorticoid receptor-dependent transcription in vivo. Mol Cell Biol 24, 3347–3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vicent GP, Zaurin R, Ballare C, Nacht AS & Beato M (2009) Erk signaling and chromatin remodeling in MMTV promoter activation by progestins. Nucl Recept Signal 7, e008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bresnick EH, John S, Berard DS, Lefebvre P & Hager GL (1990) Glucocorticoid receptor-dependent disruption of a specific nucleosome on the mouse mammary tumor virus promoter is prevented by sodium butyrate. Proc Natl Acad Sci USA 87, 3977–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fragoso G, Pennie WD, John S & Hager GL (1998) The position and length of the steroid-dependent hypersensitive region in the mouse mammary tumor virus long terminal repeat are invariant despite multiple nucleosome B frames. Mol Cell Biol 18, 3633–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fryer CJ & Archer TK (1998) Chromatin remodeling by the glucocorticoid receptor requires the BRG1 complex. Nature 393, 88–91. [DOI] [PubMed] [Google Scholar]
- 37.Fletcher TM, Xiao N, Mautino G, Baumann CT, Wolford RG, Warren BS & Hager GL (2002) ATP-dependent mobilization of the glucocorticoid receptor during chromatin remodeling. Mol Cell Biol 22, 3255–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McNally JG, Mueller WG, Walker D, Wolford RG & Hager GL (2000) The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science 287, 1262–1265. [DOI] [PubMed] [Google Scholar]
- 39.Rayasam GV, Elbi C, Walker DA, Wolford RG, Fletcher TM, Edwards DP & Hager GL (2005) Ligand specific dynamics of the progesterone receptor in living cells and during chromatin remodeling in vitro. Mol Cell Biol 25, 2406–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mueller WG, Walker D, Hager GL & McNally JG (2001) Large scale chromatin decondensation and recondensation in living cells and the role of transcription. J Cell Biol 154, 33–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.So AY, Chaivorapol C, Bolton EC, Li H & Yamamoto KR (2007) Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor. PLoS Genet 3, e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF et al. (2006) Genome-wide analysis of estrogen receptor binding sites. Nat Genet 38, 1289–1297. [DOI] [PubMed] [Google Scholar]
- 43.Jia L, Berman BP, Jariwala U, Yan X, Cogan JP, Walters A, Chen T, Buchanan G, Frenkel B & Coetzee GA (2008) Genomic androgen receptor-occupied regions with different functions, defined by histone acetylation, coregulators and transcriptional capacity. PLoS ONE 3, e3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Q, Li W, Liu XS, Carroll JS, Janne OA, Keeton EK, Chinnaiyan AM, Pienta KJ & Brown M (2007) A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell 27, 380–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vega VB, Lin CY, Lai KS, Kong SL, Xie M, Su X, Teh HF, Thomsen JS, Yeo AL, Sung WK et al. (2006) Multiplatform genome-wide identification and modeling of functional human estrogen receptor binding sites. Genome Biol 7, R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bolton EC, So AY, Chaivorapol C, Haqq CM, Li H & Yamamoto KR (2007) Cell- and gene-specific regulation of primary target genes by the androgen receptor. Genes Dev 21, 2005–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.So AY, Cooper SB, Feldman BJ, Manuchehri M & Yamamoto KR (2008) Conservation analysis predicts in vivo occupancy of glucocorticoid receptor-binding sequences at glucocorticoid-induced genes. Proc Natl Acad Sci USA 105, 5745–5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meijsing SH, Pufall MA, So AY, Bates DL, Chen L & Yamamoto KR (2009) DNA binding site sequence directs glucocorticoid receptor structure and activity. Science 324, 407–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lefstin JA & Yamamoto KR (1998) Allosteric effects of DNA on transcriptional regulators. Nature 392, 885–888. [DOI] [PubMed] [Google Scholar]
- 50.Lefstin JA, Thomas JR & Yamamoto KR (1994) Influence of a steroid receptor DNA-binding domain on transcriptional regulatory functions. Genes Dev 8, 2842–2856. [DOI] [PubMed] [Google Scholar]
- 51.Schnitzler GR (2008) Control of nucleosome positions by DNA sequence and remodeling machines. Cell Biochem Biophys 51, 67–80. [DOI] [PubMed] [Google Scholar]
- 52.Beato M & Eisfeld K (1997) Transcription factor access to chromatin. Nucleic Acids Res 25, 3559–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pina B, Brüggemeier U & Beato M (1990) Nucleosome positioning modulates accessibility of regulatory proteins to the mouse mammary tumor virus promoter. Cell 60, 719–731. [DOI] [PubMed] [Google Scholar]
- 54.Pina B, Barettino D, Truss M & Beato M (1990) Structural features of a regulatory nucleosome. J Mol Biol 216, 975–990. [DOI] [PubMed] [Google Scholar]
- 55.Li B, Carey M & Workman JL (2007) The role of chromatin during transcription. Cell 128, 707–719. [DOI] [PubMed] [Google Scholar]
- 56.Bernstein BE, Liu CL, Humphrey EL, Perlstein EO & Schreiber SL (2004) Global nucleosome occupancy in yeast. Genome Biol 5, R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee CK, Shibata Y, Rao B, Strahl BD & Lieb JD (2004) Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet 36, 900–905. [DOI] [PubMed] [Google Scholar]
- 58.Sekinger EA, Moqtaderi Z & Struhl K (2005) Intrinsic histone–DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol Cell 18, 735–748. [DOI] [PubMed] [Google Scholar]
- 59.Satchwell SC, Drew HR & Travers AA (1986) Sequence periodicities in chicken nucleosome core DNA. J Mol Biol 191, 659–675. [DOI] [PubMed] [Google Scholar]
- 60.Segal E & Widom J (2009) What controls nucleosome positions? Trends Genet 25, 335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Montecino M, Stein JL, Stein GS, Lian JB, Van Wijnen AJ, Cruzat F, Gutierrez S, Olate J, Marcellini S & Gutierrez JL (2007) Nucleosome organization and targeting of SWI/SNF chromatin-remodeling complexes: contributions of the DNA sequence. Biochem Cell Biol 85, 419–425. [DOI] [PubMed] [Google Scholar]
- 62.Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP & Widom J (2006) A genomic code for nucleosome positioning. Nature 442, 772–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Richmond TJ (2006) Genomics: predictable packaging. Nature 442, 750–752. [DOI] [PubMed] [Google Scholar]
- 64.Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, Leproust EM, Hughes TR, Lieb JD, Widom J et al. (2009) The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458, 362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ozsolak F, Song JS, Liu XS & Fisher DE (2007) High-throughput mapping of the chromatin structure of human promoters. Nat Biotechnol 25, 244–248. [DOI] [PubMed] [Google Scholar]
- 66.Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G & Zhao K (2008) Dynamic regulation of nucleosome positioning in the human genome. Cell 132, 887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gutierrez J, Paredes R, Cruzat F, Hill DA, Van Wijnen AJ, Lian JB, Stein GS, Stein JL, Imbalzano AN & Montecino M (2007) Chromatin remodeling bySWI/SNF results in nucleosome mobilization to preferential positions in the rat osteocalcin gene promoter. J Biol Chem 282, 9445–9457. [DOI] [PubMed] [Google Scholar]
- 68.Tillo D, Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Field Y, Lieb JD, Widom J, Segal E & Hughes TR (2010) High nucleosome occupancy is encoded at human regulatory sequences. PLoS ONE 5, e9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cairns BR (2009) The logic of chromatin architecture and remodelling at promoters. Nature 461, 193–198. [DOI] [PubMed] [Google Scholar]
- 70.Tirosh I, Barkai N & Verstrepen KJ (2009) Promoter architecture and the evolvability of gene expression. J Biol 8, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tirosh I & Barkai N (2008) Two strategies for gene regulation by promoter nucleosomes. Genome Res 18, 1084–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Berman BP, Frenkel B, Coetzee GA & Jia L (2010) Androgen receptor responsive enhancers are flanked by consistently-positioned H3-acetylated nucleosomes. Cell Cycle 9, 2249–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M et al. (2010) Nucleosome dynamics define transcriptional enhancers. Nat Genet 42, 343–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pennings S, Allan J & Davey CS (2005) DNA methylation, nucleosome formation and positioning. Brief Funct Genomic Proteomic 3, 351–361. [DOI] [PubMed] [Google Scholar]
- 75.Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ et al. (2010) Relationship between nucleosome positioning and DNA methylation. Nature 466, 388–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hartley PD & Madhani HD (2009) Mechanisms that specify promoter nucleosome location and identity. Cell 137, 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Heintzman ND & Ren B (2009) Finding distal regulatory elements in the human genome. Curr Opin Genet Dev 19, 541–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xi H, Shulha HP, Lin JM, Vales TR, Fu Y, Bodine DM, McKay RD, Chenoweth JG, Tesar PJ, Furey TS et al. (2007) Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet 3, e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS & Crawford GE (2008) High-resolution mapping and characterization of open chromatin across the genome. Cell 132, 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.John S, Sabo PJ, Johnson TA, Sung MH, Biddie SC, Lightman SL, Voss TC, Davis SR, Meltzer PS, Stamatoyannopoulos JA et al. (2008) Interaction of the glucocorticoid receptor with the global chromatin landscape. Mol Cell 29, 611–624. [DOI] [PubMed] [Google Scholar]
- 81.John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL & Stamatoyannopoulos JA (2011) Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet 43, 264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kaye JS, Pratt-Kaye S, Bellard M, Dretzen G, Bellard F & Chambon P (1986) Steroid hormone dependence of four DNase I-hypersensitive regions located within the 7000-bp 5¢-flanking segment of the ovalbumin gene. EMBO J 5, 277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jantzen K, Fritton HP, Igo-Kemenes T, Espel E, Janich S, Cato AC, Mugele K & Beato M (1987) Partial overlapping of binding sequences for steroid hormone receptors and DNaseI hypersensitive sites in the rabbit uteroglobin gene region. Nucleic Acids Res 15, 4535–4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Becker PB, Renkawitz R & Schutz G (1984) Tissue-specific DNaseI hypersensitive sites in the 5¢-flanking sequences of the tryptophan oxygenase and the tyro-sine aminotransferase genes. EMBO J 3, 2015–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Flavin M, Cappabianca L, Kress C, Thomassin H & Grange T (2004) Nature of the accessible chromatin at a glucocorticoid-responsive enhancer. Mol Cell Biol 24, 7891–7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bresnick EH, Bustin M, Marsaud V, Richard-Foy H& Hager GL (1992) The transcriptionally-active MMTV promoter is depleted of histone H1. Nucleic Acids Res 20, 273–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Georgel PT, Fletcher TM, Hager GL & Hansen JC (2003) Formation of higher-order secondary and tertiary chromatin structures by genomic mouse mammary tumor virus promoters. Genes Dev 17, 1617–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vicent GP, Zaurin R, Nacht AS, Font-Mateu J, Le Dily F & Beato M (2010) Nuclear factor 1 synergizes with progesterone receptor on the mouse mammary tumor virus promoter wrapped around a histoneH3/H4 tetramer by facilitating access to the central hormone-responsive elements. J Biol Chem 285, 2622–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Klokk TI, Kurys P, Elbi C, Nagaich AK, Hendarwanto A, Slagsvold T, Chang CY, Hager GL & Saatcioglu F (2007) Ligand-specific dynamics of the androgen receptor at its response element in living cells. Mol Cell Biol 27, 1823–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sharp ZD, Mancini MG, Hinojos CA, Dai F, Berno V, Szafran AT, Smith KP, Lele TT, Ingber DE & Mancini MA (2006) Estrogen-receptor-alpha exchange and chromatin dynamics are ligand- and domain-dependent. J Cell Sci 119, 4101–4116. [DOI] [PubMed] [Google Scholar]
- 91.Nagaich AK, Walker DA, Wolford RG & Hager GL (2004) Rapid periodic binding and displacement of the glucocorticoid receptor during chromatin remodeling. Mol Cell 14, 163–174. [DOI] [PubMed] [Google Scholar]
- 92.Fletcher TM, Ryu B-W, Baumann CT, Warren BS, Fragoso G, John S & Hager GL (2000) Structure and dynamic properties of the glucocorticoid receptor-induced chromatin transition at the MMTV promoter. Mol Cell Biol 20, 6466–6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N & Rando OJ (2007) Dynamics of replication-independent histone turnover in budding yeast. Science 315, 1405–1408. [DOI] [PubMed] [Google Scholar]
- 94.Narlikar GJ, Fan HY & Kingston RE (2002) Cooperation between complexes that regulate chromatin structure and transcription. Cell 108, 475–487. [DOI] [PubMed] [Google Scholar]
- 95.Winston F & Carlson M (1992) Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet 8, 387–391. [DOI] [PubMed] [Google Scholar]
- 96.Roberts SM & Winston F (1997) Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with theSnf/Swi and Srb/mediator complexes. Genetics 147, 451–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sudarsanam P & Winston F (2000) The Swi/Snf family nucleosome-remodeling complexes and transcriptional control. Trends Genet 16, 345–351. [DOI] [PubMed] [Google Scholar]
- 98.Eberharter A & Becker PB (2004) ATP-dependent nucleosome remodelling: factors and functions. J Cell Sci 117, 3707–3711. [DOI] [PubMed] [Google Scholar]
- 99.Racki LR & Narlikar GJ (2008) ATP-dependent chromatin remodeling enzymes: two heads are not better, just different. Curr Opin Genet Dev 18, 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bouazoune K, Miranda TB, Jones PA & Kingston RE (2009) Analysis of individual remodeled nucleosomes reveals decreased histone-DNA contacts created by hSWI/SNF. Nucleic Acids Res 37, 5279–5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yoshinaga SK, Peterson CL, Herskowitz I & Yamamoto KR (1992) Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science 258, 1598–1604. [DOI] [PubMed] [Google Scholar]
- 102.Muchardt C & Yaniv M (1993) A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J 12, 4279–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chiba H, Muramatsu M, Nomoto A & Kato H (1994) Two human homologues of Saccharomyces cerevisiae SWI2/SNF2 and Drosophila brahma are transcriptional coactivators cooperating with the estrogen receptor and the retinoic acid receptor. Nucleic Acids Res 22, 1815–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ichinose H, Garnier JM, Chambon P & Losson R (1997) Ligand-dependent interaction between the estro gen receptor and the human homologues of SWI2/SNF2. Gene 188, 95–100. [DOI] [PubMed] [Google Scholar]
- 105.Johnson TA, Elbi C, Parekh BS, Hager GL & John S (2008) Chromatin remodeling complexes interact dynamically with a glucocorticoid receptor regulated promoter. Mol Biol Cell 19, 3308–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang W, Xue Y, Zhou S, Kuo A, Cairns BR & Crab-tree GR (1996) Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev 10, 2117–2130. [DOI] [PubMed] [Google Scholar]
- 107.Wang W, Cote J, Xue Y, Zhou S, Khavari PA, Biggar SR, Muchardt C, Kalpana GV, Goff SP, Yaniv M et al. (1996) Purification and biochemical heterogeneity of the mammalian SWI–SNF complex. EMBO J 15, 5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 108.Yan Z, Cui K, Murray DM, Ling C, Xue Y, Gerstein A, Parsons R, Zhao K & Wang W (2005) PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev 19, 1662–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lemon B, Inouye C, King DS & Tjian R (2001) Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature 414, 924–928. [DOI] [PubMed] [Google Scholar]
- 110.Decristofaro MF, Betz BL, Rorie CJ, Reisman DN, Wang W & Weissman BE (2001) Characterization of SWI/SNF protein expression in human breast cancer cell lines and other malignancies. J Cell Physiol 186, 136–145. [DOI] [PubMed] [Google Scholar]
- 111.Hsiao PW, Fryer CJ, Trotter KW, Wang W & Archer TK (2003) BAF60a mediates critical interactions between nuclear receptors and the BRG1 chromatin-remodeling complex for transactivation. Mol Cell Biol 23, 6210–6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nagaich AK & Hager GL (2004) UV laser cross-linking: a real-time assay to study dynamic protein/DNA interactions during chromatin remodeling. Sci STKE 256, PL13. [DOI] [PubMed] [Google Scholar]
- 113.Vicent GP, Zaurin R, Nacht AS, Li A, Font-Mateu J, Le DilyF, Vermeulen M, Mann M & Beato M (2009) Two chromatin remodeling activities cooperate during activation of hormone responsive promoters. PLoS Genet 5, e1000567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fryer CJ, Nordeen SK & Archer TK (1998) Antiprogestins mediate differential effects on glucocorticoid receptor remodeling of chromatin structure. J Biol Chem 273, 1175–1183. [DOI] [PubMed] [Google Scholar]
- 115.DiRenzo J, Shang Y, Phelan M, Sif S, Myers M, Kingston RE & Brown M (2000) BRG-1 is recruited to estrogen-responsive promoters and cooperates with factors involved in histone acetylation. Mol Cell Biol 20, 7541–7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li Q, Imhof A, Collingwood TN, Urnov FD & Wolffe AP (1999) p300 stimulates transcription instigated by ligand-bound thyroid hormone receptor at a step sub sequent to chromatin disruption. EMBO J 18, 5634–5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dilworth FJ, Fromental-Ramain C, Yamamoto K & Chambon P (2000) ATP-driven chromatin remodeling activity and histone acetyltransferases act sequentially during transactivation by RAR/RXR in vitro. Mol Cell 6, 1049–1058. [DOI] [PubMed] [Google Scholar]
- 118.Ito T, Ikehara T, Nakagawa T, Kraus WL & Muramatsu M (2000) p300-mediated acetylation facilitates the transfer of histone H2A-H2B dimers from nucleosomes to a histone chaperone. Genes Dev 14, 1899–1907. [PMC free article] [PubMed] [Google Scholar]
- 119.Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T & Thanos D (2000) Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103, 667–678. [DOI] [PubMed] [Google Scholar]
- 120.Hassan AH, Neely KE & Workman JL (2001) Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell 104, 817–827. [DOI] [PubMed] [Google Scholar]
- 121.Underhill C, Qutob MS, Yee SP & Torchia J (2000) A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J Biol Chem 275, 40463–40470. [DOI] [PubMed] [Google Scholar]
- 122.Kehle J, Beuchle D, Treuheit S, Christen B, Kennison JA, Bienz M & Muller J (1998) dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science 282, 1897–1900. [DOI] [PubMed] [Google Scholar]
- 123.Suganuma T & Workman JL (2008) Crosstalk among histone modifications. Cell 135, 604–607. [DOI] [PubMed] [Google Scholar]
- 124.Reinke H & Horz W (2003) Histones are first hyper-acetylated and then lose contact with the activated PHO5 promoter. Mol Cell 11, 1599–1607. [DOI] [PubMed] [Google Scholar]
- 125.Chandy M, Gutierrez JL, Prochasson P & Workman JL (2006) SWI/SNF displaces SAGA-acetylated nucleosomes. Eukaryot Cell 5, 1738–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Luebben WR, Sharma N & Nyborg JK (2010) Nucleo-some eviction and activated transcription require p300 acetylation. Proc Natl Acad Sci USA 107, 19254–19259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR & Peterson CL (2006) Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311, 844–847. [DOI] [PubMed] [Google Scholar]
- 128.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I & Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837. [DOI] [PubMed] [Google Scholar]
- 129.Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K & Zhao K (2009) Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res 19, 24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bernstein BE, Meissner A & Lander ES (2007) The mammalian epigenome. Cell 128, 669–681. [DOI] [PubMed] [Google Scholar]
- 131.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ III, Gingeras TR et al. (2005) Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120, 169–181. [DOI] [PubMed] [Google Scholar]
- 132.Roh TY, Cuddapah S & Zhao K (2005) Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev 19, 542–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Roh TY, Ngau WC, Cui K, Landsman D & Zhao K (2004) High-resolution genome-wide mapping of his-tone modifications. Nat Biotechnol 22, 1013–1016. [DOI] [PubMed] [Google Scholar]
- 134.Koch CM, Andrews RM, Flicek P, Dillon SC, Karaoz U, Clelland GK, Wilcox S, Beare DM, Fowler JC, Couttet P et al. (2007) The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res 17, 691–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW et al. (2009) Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA et al. (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39, 311–318. [DOI] [PubMed] [Google Scholar]
- 137.Guenther MG, Levine SS, Boyer LA, Jaenisch R & Young RA (2007) A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130, 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ng HH, Ciccone DN, Morshead KB, Oettinger MA & Struhl K (2003) Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc Natl Acad Sci USA 100, 1820–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Barski A, Jothi R, Cuddapah S, Cui K, Roh TY, Schones DE & Zhao K (2009) Chromatin poises miRNA- and protein-coding genes for expression. Genome Res 19, 1742–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ et al. (2008) Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40, 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F et al. (2009) ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457, 854–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE et al. (2007) Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Roh TY, Wei G, Farrell CM & Zhao K (2007) Genome-wide prediction of conserved and nonconserved enhancers by histone acetylation patterns. Genome Res 17, 74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Won KJ, Chepelev I, Ren B & Wang W (2008) Prediction of regulatory elements in mammalian genomes using chromatin signatures. BMC Bioinformatics 9, 547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lee J & Bedford MT (2002) PABP1 identified as an arginine methyltransferase substrate using high-density protein arrays. EMBO Rep 3, 268–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sheldon LA, Becker M & Smith CL (2001) Steroid hormone receptor-mediated histone deacetylation and transcription at the mouse mammary tumor virus promoter. J Biol Chem 276, 32423–32426. [DOI] [PubMed] [Google Scholar]
- 147.Aoyagi S & Archer TK (2007) Dynamic histone acetylation/deacetylation with progesterone receptor-mediated transcription. Mol Endocrinol 21, 843–856. [DOI] [PubMed] [Google Scholar]
- 148.Chen H, Lin RJ, Xie W, Wilpitz D & Evans RM (1999) Regulation of hormone-induced histone hyper-acetylation and gene activation via acetylation of an acetylase. Cell 98, 675–686. [DOI] [PubMed] [Google Scholar]
- 149.Shang Y, Hu X, DiRenzo J, Lazar MA & Brown M (2000) Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103, 843–852. [DOI] [PubMed] [Google Scholar]
- 150.Reid G, Hubner MR, Metivier R, Brand H, Denger S, Manu D, Beaudouin J, Ellenberg J & Gannon F (2003) Cyclic, proteasome-mediated turnover of unliganded and liganded ERalpha on responsive promoters is an integral feature of estrogen signaling. Mol Cell 11, 695–707. [DOI] [PubMed] [Google Scholar]
- 151.Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M & Gannon F (2003) Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115, 751–763. [DOI] [PubMed] [Google Scholar]
- 152.Talbert PB & Henikoff S (2010) Histone variants – ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol 11, 264–275. [DOI] [PubMed] [Google Scholar]
- 153.Mizuguchi G, Shen X, Landry J, Wu WH, Sen S & Wu C (2004) ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303, 343–348. [DOI] [PubMed] [Google Scholar]
- 154.Park YJ & Luger K (2006) Structure and function of nucleosome assembly proteins. Biochem Cell Biol 84, 549–558. [DOI] [PubMed] [Google Scholar]
- 155.Altaf M, Auger A, Covic M & Cote J (2009) Connection between histone H2A variants and chromatin remodeling complexes. Biochem Cell Biol 87, 35–50. [DOI] [PubMed] [Google Scholar]
- 156.Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K & Felsenfeld G (2009) H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet 41, 941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Jin C & Felsenfeld G (2007) Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev 21, 1519–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Conerly ML, Teves SS, Diolaiti D, Ulrich M, Eisenman RN & Henikoff S (2010) Changes in H2A.Z occupancy and DNA methylation during B-cell lymphomagenesis. Genome Res 20, 1383–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Goll MG & Bestor TH (2005) Eukaryotic cytosine methyltransferases. Annu Rev Biochem 74, 481–514. [DOI] [PubMed] [Google Scholar]
- 160.Suzuki MM & Bird A (2008) DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet 9, 465–476. [DOI] [PubMed] [Google Scholar]
- 161.Suzuki MM, Kerr AR, de Sousa D & Bird A (2007) CpG methylation is targeted to transcription units in an invertebrate genome. Genome Res 17, 625–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M & Schubeler D (2007) Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 39, 457–466. [DOI] [PubMed] [Google Scholar]
- 163.Illingworth R, Kerr A, Desousa D, Jorgensen H, Ellis P, Stalker J, Jackson D, Clee C, Plumb R, Rogers J et al. (2008) A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol 6, e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB et al. (2008) Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454, 766–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Smale ST (2010) Pioneer factors in embryonic stem cells and differentiation. Curr Opin Genet Dev 20, 519–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Clouaire T & Stancheva I (2008) Methyl-CpG binding proteins: specialized transcriptional repressors or structural components of chromatin? Cell Mol Life Sci 65, 1509–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Klose RJ, Sarraf SA, Schmiedeberg L, McDermott SM, Stancheva I & Bird AP (2005) DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol Cell 19, 667–678. [DOI] [PubMed] [Google Scholar]
- 168.Buschhausen G, Wittig B, Graessmann M & Graess-mann A (1987) Chromatin structure is required to block transcription of the methylated herpes simplex virus thymidine kinase gene. Proc Natl Acad Sci USA 84, 1177–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kass SU, Landsberger N & Wolffe AP (1997) DNA methylation directs a time-dependent repression of transcription initiation. Curr Biol 7, 157–165. [DOI] [PubMed] [Google Scholar]
- 170.Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M & Kouzarides T (2010) Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell 143, 470–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J & Wolffe AP (1998) Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet 19, 187–191. [DOI] [PubMed] [Google Scholar]
- 172.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN & Bird A (1998) Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393, 386–389. [DOI] [PubMed] [Google Scholar]
- 173.Fuks F, Hurd PJ, Deplus R & Kouzarides T (2003) The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res 31, 2305–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Harikrishnan KN, Chow MZ, Baker EK, Pal S, Bassal S, Brasacchio D, Wang L, Craig JM, Jones PL, Sif S et al. (2005) Brahma links the SWI/SNF chromatin-remodeling complex with MeCP2-dependent transcriptional silencing. Nat Genet 37, 254–264. [DOI] [PubMed] [Google Scholar]
- 175.Banine F, Bartlett C, Gunawardena R, Muchardt C, Yaniv M, Knudsen ES, Weissman BE & Sherman LS (2005) SWI/SNF chromatin-remodeling factors induce changes in DNA methylation to promote transcriptional activation. Cancer Res 65, 3542–3547. [DOI] [PubMed] [Google Scholar]
- 176.Kress C, Thomassin H & Grange T (2001) Local DNA demethylation in vertebrates: how couldit be performed and targeted? FEBS Lett 494, 135–140. [DOI] [PubMed] [Google Scholar]
- 177.Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G & Sun YE (2003) DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science 302, 890–893. [DOI] [PubMed] [Google Scholar]
- 178.Thomassin H, Flavin M, Espinas ML & Grange T (2001) Glucocorticoid-induced DNA demethylation and gene memory during development. EMBO J 20, 1974–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kangaspeska S, Stride B, Metivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, Benes V, Gannon F & Reid G (2008) Transient cyclical methylation of promoter DNA. Nature 452, 112–115. [DOI] [PubMed] [Google Scholar]
- 180.Kress C, Thomassin H & Grange T (2006) Active cytosine demethylation triggered by a nuclear receptor involves DNA strand breaks. Proc Natl Acad Sci USA 103, 11112–11117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Rai K, Huggins IJ, James SR, Karpf AR, Jones DA & Cairns BR (2008) DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell 135, 1201–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Metivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G et al. (2008) Cyclical DNA methylation of a transcriptionally active promoter. Nature 452, 45–50. [DOI] [PubMed] [Google Scholar]
- 183.Kim MS, Kondo T, Takada I, Youn MY, Yamamoto Y, Takahashi S, Matsumoto T, Fujiyama S, Shirode Y, Yamaoka I et al. (2009) DNA demethylation in hormone-induced transcriptional derepression. Nature 461, 1007–1012. [DOI] [PubMed] [Google Scholar]
- 184.Horike S, Cai S, Miyano M, Cheng JF & Kohwi-Shigematsu T (2005) Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat Genet 37, 31–40. [DOI] [PubMed] [Google Scholar]
- 185.Skene PJ, Illingworth RS, Webb S, Kerr AR, James KD, Turner DJ, Andrews R & Bird AP (2010) Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol Cell 37, 457–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Nguyen CT, Gonzales FA & Jones PA (2001) Altered chromatin structure associated with methylation-induced gene silencing in cancer cells: correlation of accessibility, methylation, MeCP2 binding and acetylation. Nucleic Acids Res 29, 4598–4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Keshet I, Lieman-Hurwitz J & Cedar H (1986) DNA methylation affects the formation of active chromatin. Cell 44, 535–543. [DOI] [PubMed] [Google Scholar]
- 188.Zilberman D, Coleman-Derr D, Ballinger T & Henikoff S (2008) Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature 456, 125–129. [DOI] [PMC free article] [PubMed] [Google Scholar]