Abstract
The conversion of the normal prion protein (PrPC) into its misfolded, aggregation-prone and infectious (prion) isoform is central to the progression of transmissible spongiform encephalopathies (TSEs) or prion diseases. Since the initial development of a cell free PrP conversion reaction, striking progress has been made in the development of much more continuous prion-induced conversion and amplification reactions. These studies have provided major insights into the molecular underpinnings of prion propagation and enabled the development of ultra-sensitive tests for prions and prion disease diagnosis. This chapter will provide an overview of such reactions and the practical and fundamental consequences of their development.
Introduction
A major issue in coping with any infectious disease is the ability to detect the responsible pathogen. In the case of the transmissible spongiform encephalopathies (TSEs) or prion diseases of mammals, it is increasingly apparent that the pathogen is a misfolded multimeric form of the host’s prion protein (PrP) [15]. This infectious protein, PrPSc, can instigate its own propagation by binding to its normal counterpart, PrPC or PrPsen, and inducing its conversion to a form that tends to be higher in beta sheet content, polymeric, and more protease-resistant. The lack of an agent-specific nucleic acid genome negates the possibility of ultrasensitive detection of prions by nucleic acid amplification methods such as PCR. The fact that the infectious agent is mainly comprised of a host protein also restricts the use of antibody-based detection methods to those based on conformational epitopes or epitope exposure. However, the apparent seeded/templated conformational conversion mechanism of prion propagation can be exploited to detect the presence of prions. Here we summarize recent developments in the characterization and detection of prions using assays based on seeded conversion of PrPC.
Cell-free conversion assays
The ability of PrPres to induce the conversion of PrPC to PrPres was initially demonstrated in cell-free reactions in which brain-derived PrPres was incubated with radioactively labeled PrPC, which, under suitable conditions, bound to the PrPres and became similarly partially protease-resistant [34]. These first generation cell-free conversion (CFC) reactions were shown to be highly specific in ways that correlated with prion transmission barriers [9,30,35,47,48] and strains [6]. However, the newly generated PrPres was usually substoichiometric relative to the initial PrPres seed, and was not demonstrably associated with new infectivity [29]. As a result, these CFC reactions [reviewed in [14]] were not suitable for sensitive detection of PrPres or prions.
Protein-Misfolding Cyclic Amplification (PMCA) and recombinant Protein-Misfolding Cyclic Amplification (rPMCA)
In 2001 Soto and colleagues described a new type of cell-free prion conversion reaction called protein-misfolding cyclic amplification (PMCA) which has greatly improved efficiency, continuity and sensitivity compared to the initial CFC reactions [52]. In the typical PMCA reaction, crude brain extracts are used as a source of the PrPC which is induced to convert by prions or PrPres in the test sample. Under these conditions, PrPres can be amplified to levels that are detectable by immunoblotting. Prion amplification by PMCA involves repeated cycles of incubation and sonication during which growing multimers of PrPres are fragmented by sonication to increase the effective seed concentration. Sensitivity can also be increased by performing serial rounds of PMCA by transferring a small proportion of one reaction into fresh PrPC substrate for the subsequent round. Originally developed using the hamster adapted 263K scrapie strain, the PMCA has now been adapted to many other species [53] such as sheep [56], deer [36], mice [41] and humans [31,32]. Also, Pastrana and colleagues showed the ability of the PMCA to detect relatively PK sensitive forms of PrPSc (i.e. sPrPSc) [45].
PMCA is capable of extremely sensitive detection of PrPres in tissues, including hamster and mouse blood [13,25,50,54] or environmental samples such as water [42]. To improve the assay’s practicality an automated microplate horn system dubbed serial automated PMCA (saPMCA) was developed [50,51]. This system allows the detection of as little as 1.2 ag or ~ 26 molecules of PrPres after seven reaction rounds. The ability of the PMCA to detect miniscule amounts of PrPres and its applicability to human CJD, sheep scrapie and deer CWD support its use as a TSE pre-clinical diagnostic test. Recently, Chen et al. have reported a quantitative PMCA (qPMCA) approach that allows the determination of the concentration of small amounts of prions in biological samples [16]. This quantitation strategy is based on the direct correlation between the amount of PrPSc in a given sample and the number of PMCA rounds necessary to detect it. However, limitations of the assay for routine clinical applications include the need for 1) brain extracts as a source of PrPC substrate, 2) multiple reaction rounds over several days to 3 weeks to achieve the best sensitivity, and 3) PK digestion and immunoblotting of PMCA products which would impede high-throughput applications.
To improve the speed and practicality of the PMCA Atarashi et al used recombinant PrPC (rPrPC) as a substrate instead of brain derived PrPC [2]. rPrPC has the advantage of being easily manipulated genetically, purified in large quantities, and added to PMCA reactions at concentrations sufficient to accelerate conversion. The resulting reaction, designated rPrP-PMCA or, more briefly rPMCA, was shown initially to be able to detect as little as 50 ag of PrPres and to differentiate between scrapie-infected and uninfected hamsters using 2 µl of cerebral spinal fluid (CSF).
A ground-breaking consequence of being able to propagate PrPres in various cell-free reactions was the opportunity to directly evaluate the protein-only hypothesis for TSE prions. Initial indications that synthetic recombinant PrP amyloid fibrils alone can be infectious was reported by Legname and colleagues, who showed that such fibrils could induce or accelerate transmissible neurodegenerative disease in transgenic mice (Tg9944) that overexpressed the same truncated PrP mutant that was used to make the fibrils [37]. However the rPrP fibrils were non-infectious for wild-type mice, indicating that infectivity titers were extremely low. Moreover the initial report left open the possibility that prions were being generated spontaneously in the Tg9944 mice, but evidence to the contrary has recently been published [17].
Much more robust TSE infectivity for wild-type rodents has since been propagated in brain homogenate-based PMCA reactions by Castilla, Soto and colleagues [12]. Their study showed evidence that the biochemical, structural and biological characteristics of PMCA-propagated PrPres was almost indistinguishable from PrPres produced in vivo, except for the observation of substantially longer incubation periods obtained upon inoculation of the former into animals.
Weber and colleagues then focused on the cause of the prolonged incubation periods obtained with sPMCA-generated PrPres [59–61]. They described a sonication-dependent reduction in PMCA-generated PrPres aggregate size and suggested that enhanced clearance of such aggregates might explain the longer incubation periods. This interpretation is consistent with their ability to shorten the incubation periods by adsorbing the sPMCA PrPres products to nitrocellulose particles prior to inoculation.
Green and colleagues demonstrated PMCA amplification of naturally occurring CWD infectivity [27]. They reported an equal level of infectivity being present in both the CWD PMCA conversion product and the original 04–22412 CWD cervid brain homogenate inoculum.
Studies by Deleault, Supattapone and colleagues demonstrated that infectious PrPres propagation can be achieved in a greatly simplified system containing largely purified PrPC from brain and co-purified lipids [21]. Moreover, as previously reported by the same group using another in vitro PrPSc amplification assay [20], the PMCA’s amplification efficiency was improved when carried out in the presence of accessory polyanions, i.e. single stranded synthetic Poly A RNA. In the presence of such RNA molecules, infectious PrPres was even generated spontaneously, i.e., without seeding with brain-derived PrPSc. The PMCA-propagated material had a titer of ≈ 5 × 103 LD50 per ml when intracerebrally (i.c.) inoculated into wild type hamsters. Mechanistic studies have described the selective integration of poly A RNA molecules into stable complexes with PrP molecules in the process of prion formation in vitro, suggesting that even in the absence of PrPSc, polyanionic molecules can induce a molecular reorganization of purified PrPC resulting in a conformation similar to that of PrPSc [26].
More recently, Wang and colleagues [58] described the generation of a recombinant prion with features typical of in vivo-generated prions using three components: rPrPC, POPG (1-palmitoyl-2-oleoylphosphatidylglycerol) and RNA. Following intracerebral inoculation in wild type mice clinical stage of disease was reached at ~150 days. Biochemical characterizations of the PrPres generated, as well as the clinical symptoms, histopathology and second passage behavior induced by its inoculation strongly supported the conclusion they had generated TSE infectivity using these three molecular components.
A concurrent study by Kim and colleagues reported that prions able to induce disease in wild type hamsters can be generated from purified bacterial rPrPC in the absence of any mammalian co-factors using prion-seeded rPMCA [33]. This rPMCA product showed variable attack rates upon inoculation into hamsters and therefore contained low levels of infectivity (incubation time from 119 – 401 days) on the first passage. However, upon second passage all animals became ill with an average incubation period of about 80 days. Lesion profiling indicated that rPMCA had altered the strain characteristics of prions (263K strain) that were initially used to seed the serial rPMCA reactions.
The modulation of conversion of PrPC into PrPres by cofactors was also studied by Abid et al. by using a heterologous PMCA reaction [1]. Their results suggest that the conversion factor involved in prion replication is present in several tissues (e.g. brain, liver, kidney, heart) from different mammalian species and absent in total extracts from other evolutionary lower species such as bacteria and drosophila. This cofactor was found within lipid rafts and most likely was neither a protein nor other molecule that can be denatured by heat. Furthermore, they present evidence that when nucleic acids were depleted from brain homogenate, some other factor promoted the PrP conversion suggesting that more than one type of molecule can act as a cofactor. In a similar study, using both hamster and mouse PMCA, Deleault and colleagues described species specific difference in the use of cofactors for PrPSc propagation [22]. They reported that in the case of mouse PMCA only brain and liver homogenates appeared to contain the conversion cofactor, which also appeared to be protease-resistant and heat stable.
To investigate the role of PrPC glycosylation in modulating conversion efficiency Nishina and colleagues tested the ability of un-, mono- or di-glycosylated PrPC to support prion amplification using both hamster and mouse the PMCA reactions [43]. Their data shows that whereas unglycosylated mouse PrPC is required to propagate homologous RML prions, diglycosylated PrPC is necessary to propagate hamster Sc237 prions, suggesting that the stoichiometry of the PrPC glycoforms influences the efficiency of PrPres formation in vitro. However, more recently, the Supattapone lab also used PMCA to show that PrP glycosylation is not necessary for strain-specific neurotropism [46].
Prion strains and species barrier studies using PMCA
Prion strains are characterized by distinct incubation periods, clinical symptoms and brain lesion profiles, as well as differences in biochemical features of PrPres (e.g. electrophoretic mobility, glycoform pattern, infrared spectrum, and conformational stability). Prion strains (or mixtures of strains) can usually be serially passaged stably in a hosts of a given species and genotype. However, under some circumstances, new strains or mixtures of strains can arise, especially after passage from one host genotype to another. A wealth of evidence suggests that the properties of prion strains are usually maintained by the faithful propagation of different conformers and/or aggregation states of PrPSc [6,7,19,40,55]. However, the occasional biological instability of prion strains implies that propagation of such conformational states can be subject to permutation, most notably when the prion seed has to act on heterologous PrPC molecules.
Several PMCA studies support this concept of prion strain propagation. Castilla et al showed that PMCA generated PrPres seeded with five different murine and four human prion strains retained their specific biochemical properties and, upon injection into wild type animals, the PMCA generated PrPres caused disease with features comparable to the parental strain [11]. Green and colleagues reported that features of the 04–22412 CWD prion strain were kept after PMCA reaction [27]. Collectively, these data are consistent with the idea that prion strain features are encoded, at least to a large extent, by the PrPres conformation. Furthermore, Green and colleagues describe the adaptation of the RML mouse prion strain to Tg(CerPrP) mice, overcoming the mouse-cervid species barrier and creating a new prion strain using PMCA [27]. In a similar study, Castilla and colleagues describe the generation of new prion strains by hamster-mouse interspecies PMCA amplification [10]. In particular, hamster PrPC substrate and mouse brain-derived PrPSc or vice versa, produced new prion strains which caused diseases with pathological and biochemical features that were unlike those of other known prion strains. Barria and colleagues developed mouse and hamster PMCA reaction conditions that allowed spontaneous generation of PrPres in the absence of initial seeding with PrPSc [5]. The spontaneous PrPres was infectious in wild type animals but caused a new disease phenotype, suggesting the creation of a novel prion strain. Finally, the 263K scrapie-seeded recombinant PrP prions propagated in rPMCA produced distinct lesion profiles through two passages in vivo, providing evidence that rPMCA with rPrPC substrate alone with no mammalian cofactors lead to stable changes in strain characteristics [33]. These studies indicate that new prion strains can be generated with interspecies PMCA, unseeded PMCA, or PMCA using solely rPrPC as substrate. Usually, when amplifying PrPSc the PMCA maintains the strain features of the initial seed, probably through precise templating of the PrPC misfolding process towards the formation of an exact replica of itself. When the PMCA is carried out in the absence of seed, with a heterologous or recombinant PrPC substrate, additional conformational options presumably become available which enhance the likelihood of forming a new prion conformer or strain.
Collectively, and remarkably, PMCA-based prion propagation mimics prion propagation in vivo to the extent that one can observe not only the stable propagation of prion strains within a given host, but also the permutation of strains and the spontaneous generation of new strains. However, as prion propagation seems largely to be a protein folding problem, we would not expect to PMCA reactions to recapitulate all aspects of prion strain propagation and transmission barriers that are seen in vivo. In intact cells or tissues, interactions between PrPres and PrPC are highly constrained in three dimensions by GPI anchoring to membranes and by localized interactions between the PrP isoforms and other molecules in their physiologically controlled microenvironments. In contrast, PMCA reactions occur in detergent lysates or extracts in which most such constraints on intermolecular interactions are removed.
Amyloid Seeding Assay (ASA)
The Amyloid Seeding Assay described by Colby and colleagues is a multi-well plate prion amplification assay that uses Thioflavin T (ThT) to detect amplification products [18]. ThT is an amyloid dye that undergoes an enhancement of fluorescence yield when bound to protein amyloid fibrils and is used in this and many other amyloidogenesis assays [38]. The ASA utilizes phosphotungstic acid (PTA) precipitated PrPSc as a seed and recombinant PrP (rPrPC) stored in 6 M guanidine hydrochloride as a substrate. The final guanidine hydrochloride concentration in the reaction (0.4 M) is such that the substrate is likely in a partially unfolded/destabilized state. Other reaction parameters include incubation at 37⁰C, the presence of a 3 mm glass bead in each well to enhance agitation, and continuous shaking of the plate. Notably, the ASA can detect protease-sensitive PrPSc from transgenic mice over expressing the PrP (101L) mutation [17]. ASA applicability to various rodent scrapie experimental models and capability to distinguish between brain samples from sporadic CJD (sCJD) patients and negative control normal brains were described. Furthermore, a 98 % correlation of prion detection by ASA and neuropathological lesions in transgenic mice was described. Nevertheless, as noted by the authors, one weakness of the assay is the need to analyze a high number of replicates per sample because of the variability of the kinetics of ThT positive fibril formation. This problem is exacerbated by the fact that under the ASA conditions, spontaneous (unseeded) rPrPC fibril formation also occurs, but usually with a longer lag phase than those seen with prion-seeded reactions. As detailed below for the real time QuIC assay, spontaneous fibrillization can be largely avoided under other reaction conditions.
Quaking-Induced conversion (QuIC) reactions
To avoid technical complexities associated with sonication in PMCA reactions new assays were developed by Atarashi and colleagues in which sonication was substituted by intermittent shaking as a means to break up prion protein aggregates and produce new PrP seeds in reaction tubes [3,4,44,62]. Such shaken conversion reactions have been dubbed Quaking-Induced Conversion (QuIC) reactions. The first-generation QuIC reactions, herein abbreviated Standard QuIC or SQ, were developed as individual microtube-based reactions that contained detergents and used hamster-adapted 263K scrapie as a seed and hamster rPrPC as substrate [4]. As with the rPMCA [2], scrapie seeds induced the conversion of rPrPC to a specific set of proteinase K-resistant bands (rPrP-res(Sc)) that were visualized on immunoblots. The ability to detect as little as 100 ag PrPSc was demonstrated. Through careful selection of reaction parameters such as shaking regimen, detergent concentrations, incubation time and reaction temperature, virtual elimination of spontaneous (unseeded) conversion of the substrate to proteinase K-resistant product (rPrP-res(spon)) seed can be achieved.
SQ has been used successfully to discriminate between scrapie affected and control hamsters using CSF [4] or nasal lavages [8]. The assay was also applied to the detection of prion seeding activity in brain samples from scrapie affected sheep and a human vCJD patient [44]. Furthermore, good discrimination between cerebral spinal fluid samples from scrapie positive and normal sheep was observed.
To address limitations of SQ and ASA, Atarashi, Wilham and colleagues developed a new prion-seeded rPrP conversion assay that combines features of the ASA (i.e. multiwell plate format and ThT detection of conversion products) and the SQ (e.g. intermittent shaking, rPrPC preparation, and lack of chaotropic salts) [3,62]. This new assay was called Real-Time (RT) QuIC, or herein RTQ, because of its ability to almost continuously monitor the progress of the QuIC reaction in a shaking, temperature-controlled fluorescence plate reader. As with the ASA [18], the multiwell plate format gives the RTQ is more amenable to high-throughput testing of samples. However, in contrast to the ASA, the RTQ conditions can, depending on the rPrPC substrate, virtually eliminate the problem of unseeded, prion-independent amyloid formation. The prion-seeded RTQ conversion products were similar to the ones previously described with SQ [4,44] and showed distinct PK-resistant bands of ~20, 18, 14 and 13 kDa, while control reactions seeded with normal tissue had virtually no PK-resistant products. Circular dichroism (CD) and Fourier transform infrared (FTIR) studies of the RTQ substrate (hamster rPrPC 90–231) and conversion product indicate that the prion-induced structural changes in rPrPC shared some similarities with those occurring in vivo upon conversion of PrPC to PrPSc. Thus, RTQ has promise not only as a prion detection assay, but also as a tool to study the mechanism of prion-induced PrP conversion.
Wilham and colleagues also describe the use of RTQ to quantitate prion seeding activity in biological samples. Serial dilutions of a given sample are used as seeds and the seeding dose (SD) giving 50% ThT-positive replicate reactions (SD50), i.e., the 50% endpoint dilution, is estimated. The SD50 is analogous to the 50% lethal dose (LD50) determined in an endpoint dilution animal bioassay. As is commonly done in determining LD50 values, the estimation of SD50 values can be aided by using Spearman-Kärber [23] or Reed-Muench [49] analyses. This end-point dilution approach to prion quantification is potentially applicable to any prion-seeded amplification assay (e.i. PMCA, rPMCA, ASA). With the RTQ, SD50 concentrations obtained for four hamster scrapie brain homogenate stocks were comparable to LD50 concentrations obtained with hamster end-point dilution bioassays, indicating similar sensitivities for these two types of assays. However, RTQ has several major advantages over animal bioassays, including practicality, high-throughput potential, rapidity and reduced cost.
Quantitation of prions in CSF samples from scrapie positive hamsters by RTQ gave SD50 values of 105.6 and 104.7 per ml, respectively. Detection of prions in brain samples from TSE positive sheep and deer was also described. One important version of RTQ has been shown to have 81% sensitivity and 100% specificity in discriminating sporadic-CJD and non-CJD patients based on CSF samples [3,3].
Of particular interest are prion amplification assays that are capable of detecting prions in blood components such as plasma. However, blood typically has extremely low prion concentrations [i.e., ~13 LD50 per ml, [28]] and contains inhibitors of some of the most sensitive tests such as PMCA [13] and another assay [57]. Recently, a prion specific immunoprecipitation has been integrated with both SQ and RTQ to increase sensitivity and isolate prions from inhibitors such as those present in plasma. When coupled with the immunoprecipitation step the RTQ reaction allowed more sensitive detection of variant CJD brain homogenate diluted into human plasma and also rapid discrimination of plasma and serum samples from scrapie-infected and uninfected hamsters (unpublished observations). These developments should improve prospects for the practical detection of minimal levels of prions in tissues, fluids or environmental samples.
Conclusions
Since the development of the first PrP in vitro conversion reaction [34] much more efficient, continuous and sensitive prion-seeded conversion assays have been developed. These techniques have been used to investigate prion composition and propagation mechanisms as well as prion strain and transmission barrier phenomena. Moreover, these reactions serve as bases for ultra-sensitive prion detection that should facilitate TSE diagnostic tests and screening assays for medical, agricultural and environmental prion contamination.
A pre-clinical TSE diagnostic test should be sensitive enough to detect minimally infectious or even subinfectious quantities of prions and allow amplification/detection of multiple prion strains in a wide variety of biological tissues. As reported by Wilham and colleagues, inhibitory matrix interference can be overcome by diluting the sample until the reaction is no longer affect by the inhibitors present [62]. Another strategy is to capture and concentrate prions from complex mixtures in a manner that is compatible with amplification/detection as we have recently accomplished using immunoprecipitation and others have reported using steel [24] or magnetic particles [39]. Our studies indicate that when used in combination with an improved RTQ reaction, immunoprecipitation provides for sensitivities that are several orders of magnitude greater than those obtained using the metallic particles.
The fact that so far the infectivity of PrP in in vitro conversion products as been shown to be lower than that of bona fide PrPSc suggests that we are still missing important information about the conversion process and how to create an infectious prion The prion-seeded conversion reactions described in this chapter provide valuable tools to investigate these issues.
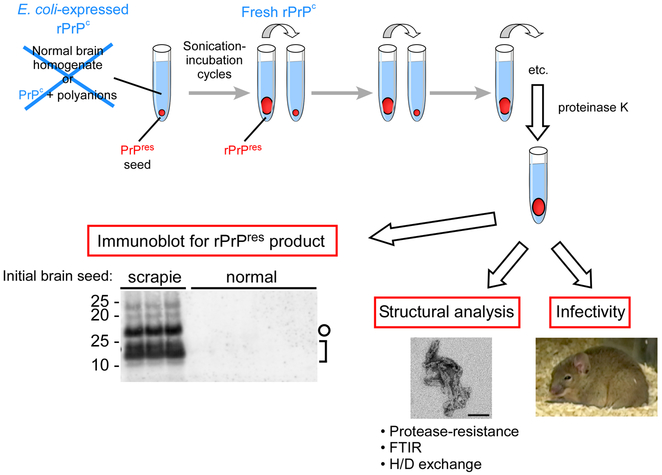
Figure 1.
Summary of the biochemical, structural and biological studies on recombinant-PMCA (r-PMCA) products. Immunoblot and electron microscopy image adapted from Ref. [2].
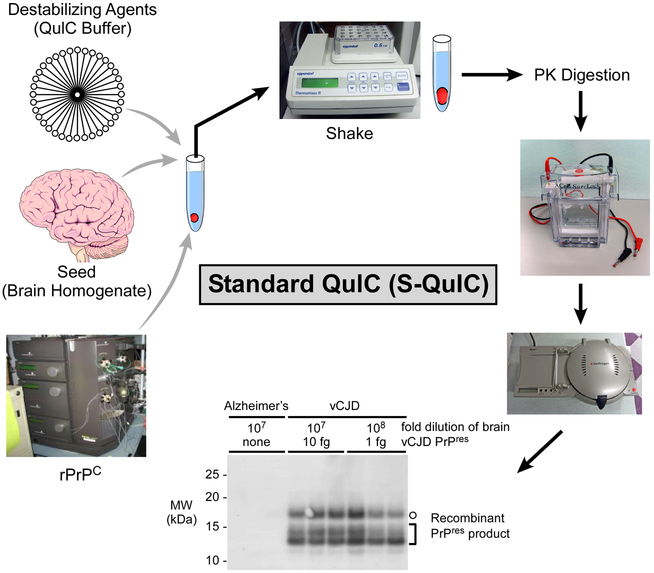
Figure 2. Standard Quaking Induce Conversion Reaction (SQ).
The components of the SQ (rPrPC substrate, brain homogenate seed and conversion buffer) are represented. The final conversion products from reactions seeded with vCJD brain homogenate dilutions containing the indicated amounts of PrPres or Alzheimer’s disease (AD) negative controls are shown. Hamster rPrPC (residues 23–231) was used as substrate and single amplification round reaction proteinase K (PK)-digested products were analyzed using antiserum R20 (C-terminal epitope). Open circles mark 17-kDa fragments and brackets indicate the lower molecular weight bands (10–13 kDa). Immunoblot adapted from Ref. [44].
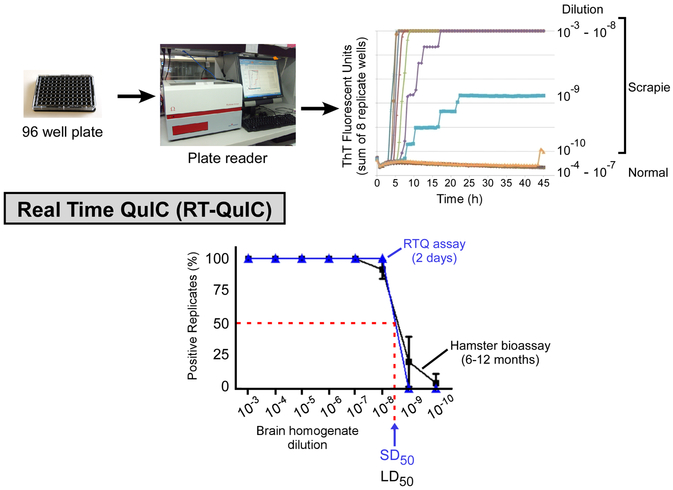
Figure 3. Diagram of Real Time QuIC (RTQ) and comparison of end-point dilution titrations of scrapie brain homogenate by RTQ compared to animal bioassay.
(Top panel) RTQ analysis of normal and scrapie brain homogenate (BH) dilutions using hamster (90–231) rPrPC as a substrate. (Bottom panel) Comparison of hamster brain homogenate end point dilution titrations by RTQ and animal bioassay. The Spearman-Kärber estimate of the SD50 (i.e. seeding dose giving sufficient Thioflavin T fluorescence in half of the replicate wells) per 2 µl of neat brain tissue is indicated. Graphs adapted from Ref. [62].
Acknowledgements
We thank Jason M Wilham for designing the figures and Anita Mora for graphics assistance.
References
- 1.Abid K, Morales R, Soto C (2010) FEBS Lett 584:2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atarashi R, Moore RA, Sim VL, Hughson AG, Dorward DW, Onwubiko HA, Priola SA, Caughey B (2007) Nat Methods 4:645. [DOI] [PubMed] [Google Scholar]
- 3.Atarashi R, Satoh K, Sano K, Fuse T, Yamaguchi N, Ishibashi D, Matsubara T, Nakagaki T, Yamanaka H, Shirabe S, Yamada M, Mizusawa H, Kitamoto T, Klug G, McGlade A, Collins SJ, Nishida N (2011) Nat Med 17:175. [DOI] [PubMed] [Google Scholar]
- 4.Atarashi R, Wilham JM, Christensen L, Hughson AG, Moore RA, Johnson LM, Onwubiko HA, Priola SA, Caughey B (2008) Nat Methods 5:211. [DOI] [PubMed] [Google Scholar]
- 5.Barria MA, Mukherjee A, Gonzalez-Romero D, Morales R, Soto C (2009) PLoS Pathog 5:e1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bessen RA, Kocisko DA, Raymond GJ, Nandan S, Lansbury PT Jr, Caughey B (1995) Nature 375:698. [DOI] [PubMed] [Google Scholar]
- 7.Bessen RA, Marsh RF (1994) J Virol 68:7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bessen RA, Shearin H, Martinka S, Boharski R, Lowe D, Wilham JM, Caughey B, Wiley JA (2010) PLoS Pathogens 6:e1000837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bossers A, PBGM Belt, Raymond GJ, Caughey B, de Vries R, Smits MA (1997) Proc Natl Acad Sci USA 94:4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castilla J, Gonzalez-Romero D, Saa P, Morales R, de CJ, Soto C (2008) Cell 134:757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castilla J, Morales R, Saa P, Barria M, Gambetti P, Soto C (2008) EMBO J 27:2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castilla J, Saa P, Hetz C, Soto C (2005) Cell 121:195. [DOI] [PubMed] [Google Scholar]
- 13.Castilla J, Saa P, Morales R, Abid K, Maundrell K, Soto C (2006) Methods Enzymol 412:3. [DOI] [PubMed] [Google Scholar]
- 14.Caughey B (2003) Br Med Bull 66:109. [DOI] [PubMed] [Google Scholar]
- 15.Caughey B, Baron GS, Chesebro B, Jeffrey M (2009) Annu Rev Biochem 78:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen B, Morales R, Barria MA, Soto C (2010) Nat Methods 7:519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colby DW, Wain R, Baskakov IV, Legname G, Palmer CG, Nguyen HO, Lemus A, Cohen FE, DeArmond SJ, Prusiner SB (2010) PLoS Pathog 6:e1000736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colby DW, Zhang Q, Wang S, Groth D, Legname G, Riesner D, Prusiner SB (2007) Proc Natl Acad Sci U S A 104:20914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collinge J, Clarke AR (2007) Science 318:930. [DOI] [PubMed] [Google Scholar]
- 20.Deleault NR, Geoghegan JC, Nishina K, Kascsak R, Williamson RA, Supattapone S (2005) J Biol Chem 280:26873. [DOI] [PubMed] [Google Scholar]
- 21.Deleault NR, Harris BT, Rees JR, Supattapone S (2007) Proc Natl Acad Sci U S A 104:9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deleault NR, Kascsak R, Geoghegan JC, Supattapone S (2010) Biochemistry 49:3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dougherty RM (1964) Animal virus titration techniques. In: Harris RJC (ed) Techniques in experimental virology. Academic Press, Inc., New York p 183 [Google Scholar]
- 24.Edgeworth JA, Farmer M, Sicilia A, Tavares P, Beck J, Campbell T, Lowe J, Mead S, Rudge P, Collinge J, Jackson GS (2011) Lancet 377:487. [DOI] [PubMed] [Google Scholar]
- 25.Fujihara A, Atarashi R, Fuse T, Ubagai K, Nakagaki T, Yamaguchi N, Ishibashi D, Katamine S, Nishida N (2009) FEBS J 276:2841. [DOI] [PubMed] [Google Scholar]
- 26.Geoghegan JC, Valdes PA, Orem NR, Deleault NR, Williamson RA, Harris BT, Supattapone S (2007) J Biol Chem 282:36341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Green KM, Castilla J, Seward TS, Napier DL, Jewell JE, Soto C, Telling GC (2008) PLoS Pathog 4:e1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gregori L, McCombie N, Palmer D, Birch P, Sowemimo-Coker SO, Giulivi A, Rohwer RG (2004) Lancet 364:529. [DOI] [PubMed] [Google Scholar]
- 29.Hill AF, Antoniou M, Collinge J (1999) J Gen Virol 80:11. [DOI] [PubMed] [Google Scholar]
- 30.Horiuchi M, Priola SA, Chabry J, Caughey B (2000) Proc Natl Acad Sci U S A 97:5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones M, Peden AH, Prowse CV, Groner A, Manson JC, Turner ML, Ironside JW, Macgregor IR, Head MW (2007) J Pathol 213:21. [DOI] [PubMed] [Google Scholar]
- 32.Jones M, Peden AH, Yull H, Wight D, Bishop MT, Prowse CV, Turner ML, Ironside JW, Macgregor IR, Head MW (2009) Transfusion 49:376. [DOI] [PubMed] [Google Scholar]
- 33.Kim JI, Cali I, Surewicz K, Kong Q, Raymond GJ, Atarashi R, Race B, Qing L, Gambetti P, Caughey B, Surewicz WK (2010) J Biol Chem 285:14083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kocisko DA, Come JH, Priola SA, Chesebro B, Raymond GJ, Lansbury PT, Caughey B (1994) Nature 370:471. [DOI] [PubMed] [Google Scholar]
- 35.Kocisko DA, Priola SA, Raymond GJ, Chesebro B, Lansbury PT, Jr., Caughey B (1995) Proc Natl Acad Sci USA 92:3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurt TD, Perrott MR, Wilusz CJ, Wilusz J, Supattapone S, Telling GC, Zabel MD, Hoover EA (2007) J Virol 81:9605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Legname G, Baskakov IV, Nguyen HO, Riesner D, Cohen FE, DeArmond SJ, Prusiner SB (2004) Science 305:673. [DOI] [PubMed] [Google Scholar]
- 38.LeVine H III (1999) Methods Enzymol 309:274. [DOI] [PubMed] [Google Scholar]
- 39.Miller MB, Supattapone S (2011) J Virol 85:2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morales R, Abid K, Soto C (2007) Biochim Biophys Acta 1772:681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murayama Y, Yoshioka M, Okada H, Takata M, Yokoyama T, Mohri S (2007) J Gen Virol 88:2890. [DOI] [PubMed] [Google Scholar]
- 42.Nichols TA, Pulford B, Wyckoff AC, Meyerett C, Michel B, Gertig K, Hoover EA, Jewell JE, Telling GC, Zabel MD (2009) Prion 3:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishina KA, Deleault NR, Mahal SP, Baskakov I, Luhrs T, Riek R, Supattapone S (2006) Biochemistry 45:14129. [DOI] [PubMed] [Google Scholar]
- 44.Orrú CD, Wilham JM, Hughson AG, Raymond LD, McNally KL, Bossers A, Ligios C, Caughey B (2009) Protein Eng Des Sel 22:515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pastrana MA, Sajnani G, Onisko B, Castilla J, Morales R, Soto C, Requena JR (2006) Biochemistry 45:15710. [DOI] [PubMed] [Google Scholar]
- 46.Piro JR, Harris BT, Nishina K, Soto C, Morales R, Rees JR, Supattapone S (2009) J Virol 83:5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raymond GJ, Bossers A, Raymond LD, O’Rourke KI, McHolland LE, Bryant PK III, Miller MW, Williams ES, Smits M, Caughey B (2000) EMBO J 19:4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raymond GJ, Hope J, Kocisko DA, Priola SA, Raymond LD, Bossers A, Ironside J, Will RG, Chen SG, Petersen RB, Gambetti P, Rubenstein R, Smits MA, Lansbury PT Jr., Caughey B (1997) Nature 388:285. [DOI] [PubMed] [Google Scholar]
- 49.Reed LJ, Muench H (1938) Am J Hyg 27:493 [Google Scholar]
- 50.Saa P, Castilla J, Soto C (2006) Science 313:92. [DOI] [PubMed] [Google Scholar]
- 51.Saa P, Castilla J, Soto C (2006) J Biol Chem 281:35245. [DOI] [PubMed] [Google Scholar]
- 52.Saborio GP, Permanne B, Soto C (2001) Nature 411:810. [DOI] [PubMed] [Google Scholar]
- 53.Soto C, Anderes L, Suardi S, Cardone F, Castilla J, Frossard MJ, Peano S, Saa P, Limido L, Carbonatto M, Ironside J, Torres JM, Pocchiari M, Tagliavini F (2005) FEBS Lett 579:638. [DOI] [PubMed] [Google Scholar]
- 54.Tattum MH, Jones S, Pal S, Collinge J, Jackson GS (2010) Transfusion 50:996. [DOI] [PubMed] [Google Scholar]
- 55.Telling GC, Parchi P, DeArmond SJ, Cortelli P, Montagna P, Gabizon R, Mastrianni J, Lugaresi E, Gambetti P, Prusiner SB (1996) Science 274:2079. [DOI] [PubMed] [Google Scholar]
- 56.Thorne L, Terry LA (2008) J Gen Virol 89:3177. [DOI] [PubMed] [Google Scholar]
- 57.Trieschmann L, Navarrete SA, Kaschig K, Torkler S, Maas E, Schatzl H, Bohm G (2005) BMC Biotechnol 5:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang F, Wang X, Yuan CG, Ma J (2010) Science 327:1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weber P, Giese A, Piening N, Mitteregger G, Thomzig A, Beekes M, Kretzschmar HA (2006) Proc Natl Acad Sci U S A 103:15818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weber P, Giese A, Piening N, Mitteregger G, Thomzig A, Beekes M, Kretzschmar HA (2007) Vet Microbiol 123:346. [DOI] [PubMed] [Google Scholar]
- 61.Weber P, Reznicek L, Mitteregger G, Kretzschmar H, Giese A (2008) Biochem Biophys Res Commun 369:924. [DOI] [PubMed] [Google Scholar]
- 62.Wilham JM, Orrú CD, Bessen RA, Atarashi R, Sano K, Race B, Meade-White KD, Taubner LM, Timmes A, Caughey B (2010) PLoS Pathog 6:e1001217. [DOI] [PMC free article] [PubMed] [Google Scholar]