Abstract
Preprocessing of functional MRI (fMRI) involves numerous steps to clean and standardize data before statistical analysis. Generally, researchers create ad-hoc preprocessing workflows for each new dataset, building upon a large inventory of tools available. The complexity of these workflows has snowballed with rapid advances in acquisition and processing. We introduce fMRIPrep, an analysis-agnostic tool that addresses the challenge of robust and reproducible preprocessing for fMRI data. FMRIPrep automatically adapts a best-in-breed workflow to the idiosyncrasies of virtually any dataset, ensuring high-quality preprocessing with no manual intervention. By introducing visual assessment checkpoints into an iterative integration framework for software-testing, we show that fMRIPrep robustly produces high-quality results on a diverse fMRI data collection. Additionally, fMRIPrep introduces less uncontrolled spatial smoothness than commonly used preprocessing tools. FMRIPrep equips neuroscientists with a high-quality, robust, easy-to-use and transparent preprocessing workflow, which can help ensure the validity of inference and the interpretability of their results.
INTRODUCTION
Functional magnetic resonance imaging (fMRI) is a commonly used technique to map human brain activity1. However, the blood-oxygen-level dependent (BOLD) signal measured by fMRI is typically mixed with non-neural sources of variability2. Preprocessing identifies the nuisance sources and reduces their effect on the data3,4, and further addresses particular imaging artifacts and the anatomical localization of signals5. For instance, slice-timing6 correction (STC), head-motion correction (HMC), and susceptibility distortion correction (SDC) address particular artifacts, while co-registration, and spatial normalization are concerned with signal localization (Supplementary Note 1). Extracting a signal that is most faithful to the underlying neural activity is crucial to ensure the validity of inference and interpretability of results7. Thus, a primary goal of preprocessing is to reduce sources of false positive errors without inducing excessive false negative errors. An illustration of false positive errors familiar to most researchers is finding activation outside of the brain due to faulty spatial normalization. As a more practical example, Power et al. demonstrated that unaccounted-for head-motion in resting-state fMRI generated systematic correlations that could be misinterpreted as functional connectivity8. Conversely, false negatives can result from a number of preprocessing failures, such as anatomical misregistration across individuals which reduces statistical power.
Workflows for preprocessing fMRI produce two broad classes of outputs. First, preprocessed time-series derive from the original data after the application of retrospective signal corrections, temporal/spatial filtering, and the resampling onto a target space appropriate for analysis (e.g. a standardized anatomical reference). Second, experimental confounds are additional time-series such as physiological recordings and estimated noise sources that are useful for analysis (e.g. to be modeled as nuisance regressors). Some commonly used confounds include: motion parameters, framewise displacement9 (FD), spatial standard deviation of the data after temporal differencing (DVARS8), global signals, etc. Preprocessing may include further steps for denoising and estimation of confounds. For instance, dimensionality reduction methods based on principal components analysis (PCA) or independent components analysis (ICA), such as component-based noise correction (CompCor10) or automatic removal of motion artifacts (ICA-AROMA11).
The neuroimaging community is well equipped with tools that implement the majority of the individual steps of preprocessing described so far (Table 1). These tools are readily available within software packages including AFNI12, ANTs13, FreeSurfer14, FSL15, Nilearn16, or SPM17. Despite the wealth of accessible software and multiple attempts to outline best practices for preprocessing2,5,7,18, the large variety of data acquisition protocols have led to the use of ad-hoc pipelines customized for nearly every study19. In practice, the neuroimaging community lacks a preprocessing workflow that reliably provides high-quality and consistent results on arbitrary datasets.
Table 1.
State-of-the-art neuroimaging offers a large catalog of readily available software tools. FMRIPrep integrates best-in-breed tools for each of the preprocessing tasks that its workflow covers, except for steps implemented as part of the development of fMRIPrep (in-house implementations). Tasks listed on the first column are described in detail in Supplementary Note 1.
| Preprocessing task | fMRIPrep includes | Alternatives (not included within fMRIPrep) |
|---|---|---|
| Anatomical T1w brain-extraction | antsBrainExtraction.sh (ANTs) | bet (FSL), 3dSkullstrip (AFNI), MRTOOL (SPM Plug-in) |
| Anatomical surface reconstruction | recon-all (FreeSurfer) | CIVET, BrainSuite, Computational Anatomy (SPM Plug-in) |
| Head-motion estimation (and correction) | mcflirt (FSL) | 3dvolreg (AFNI), spm_realign (SPM), cross_realign_4dfp (4dfp), antsBrainRegistration (ANTs) |
| Susceptibility-derived distortion estimation (and unwarping) | 3dqwarp (AFNI) | fugue and topup (FSL), FieldMap and HySCO (SPM Plug-ins) |
| Slice-timing correction | 3dTshift (AFNI) | slicetimer (FSL), spm_slice_timing (SPM), interp_4dfp (4dfp) |
| Intra-subject registration | bbregister (FreeSurfer), flirt (FSL) | 3dvolreg (AFNI), antsRegistration (ANTs), Coregister (SPM GUI) |
| Spatial normalization (inter-subject co-registration) | antsRegistration (ANTs) | @auto_tlrc (AFNI), fnirt (FSL), Normalize (SPM GUI) |
| Surface sampling | mri_vol2surf (FreeSurfer) | SUMA (AFNI), MNE, Nilearn |
| Subspace projection denoising (ICA, PCA, etc) | melodic (FSL), ICA-AROMA | Nilearn, LMGS (SPM Plug-in) |
| Confounds | in-house implementation | fsl_motion_outliers (FSL), TAPAS PhysIO (SPM Plug-in) |
| Detection of nonsteady-states | in-house implementation | Ad-hoc implementations, manual setting |
RESULTS
FMRIPrep is a robust and convenient tool for researchers and clinicians to prepare both task-based and resting-state fMRI data for analysis. Its outputs enable a broad range of applications, including within-subject analysis using functional localizers, voxel-based analysis, surface-based analysis, task-based group analysis, resting-state connectivity analysis, and many others.
A modular design alongside BIDS allow for a flexible, adaptive workflow
FMRIPrep is composed of sub-workflows that are dynamically assembled into different configurations depending on the input data. These building blocks combine tools from widely-used, open-source neuroimaging packages (Table 1). The workflow engine Nipype20 is used to stage the workflows and to deal with execution details (such as resource management). As presented in Figure 1, the workflow comprises two major blocks, separated into anatomical and functional MRI processing streams. The Brain Imaging Data Structure21 (BIDS, Supplementary Note 2) allows fMRIPrep to precisely identify the structure of the input data and gather all the available metadata (e.g. imaging parameters) with no manual intervention. FMRIPrep reliably self-adapts to dataset irregularities such as missing acquisitions or runs through a set of heuristics.
Figure 1. FMRIPrep is an fMRI preprocessing tool that adapts to the input dataset.
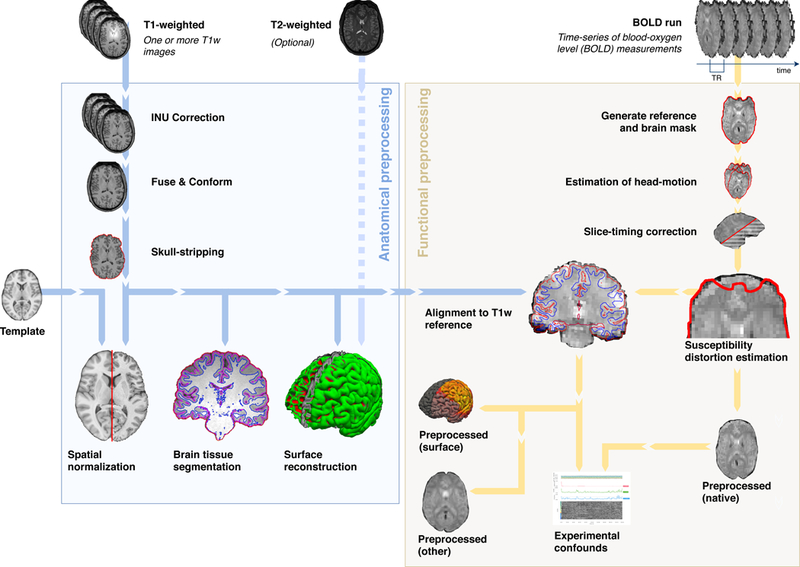
Leveraging the Brain Imaging Data Structure (BIDS21), the software self-adjusts automatically, configuring the optimal workflow for the given input dataset. Thus, no manual intervention is required to locate the required inputs (one T1-weighted image and one BOLD series), read acquisition parameters (such as the repetition time –TR– and the slice acquisition-times) or find additional acquisitions intended for specific preprocessing steps (like field maps and other alternatives for the estimation of the susceptibility distortion).
Visual reports ease quality control and maximize transparency
Users can assess the quality of preprocessing with an individual report generated per participant (see Supplementary Figure 1). Reports contain dynamic and static mosaic views of images at different quality control points along the preprocessing pipeline. Written in hypertext markup language (HTML), reports can be opened with any web browser, are amenable to integration within online science services (e.g. OpenNeuro, or CodeOcean22), and maximize shareability between peers. These reports effectively minimize the amount of time required for assessing the quality of the results. As an additional transparency enhancement, reports include a citation boilerplate that follows the guidelines by Poldrack et al.23, and gives due credit to all authors of all of the individual pieces of software used within fMRIPrep.
Highlights of fMRIPrep within the neuroimaging context
FMRIPrep is analysis-agnostic to currently-available analysis choices, as it supports a wide range of higher-level analysis and modeling options. Alternative workflows such as afni_proc.py (AFNI12), feat (FSL15), C-PAC24 (configurable pipeline for the analysis of connectomes), Human Connectome Project (HCP25) Pipelines26, or the Batch Editor of SPM, are not agnostic because they prescribe particular methodologies to analyze the preprocessed data. Important limitations to compatibility with downstream analysis derive from the coordinates space of the outputs and the regular (volume) vs. irregular (surface) sampling of the BOLD signal. For example, HCP Pipelines supports surface-based analyses on subject or template space. Conversely, C-PAC and feat are volume-based only. Although afni_proc.py is volume-based by default, pre-reconstructed surfaces can be manually set for sampling the BOLD signal prior to analysis. FMRIPrep allows a multiplicity of output spaces including subject-space and atlases for both volume-based and surface-based analyses. While fMRIPrep avoids including processing steps that may limit further analysis (e.g. spatial smoothing), other tools are designed to perform preprocessing that supports specific analysis pipelines. For instance, C-PAC performs several processing steps towards the connectivity analysis of resting-state fMRI. Further advantages of fMRIPrep are described in Online Methods, and include the “fieldmap-less” susceptibility distortion correction (SDC), the community-driven development and high-standards of software engineering, and the focus on reproducibility.
FMRIPrep yields high-quality results on diverse data
We iteratively maximized the robustness and overall quality of the results generated by fMRIPrep using the two-stage validation framework shown in Supplementary Figure 2. In a Phase I for fault-discovery, we tested fMRIPrep on a set of 30 datasets from OpenfMRI (see Table 2). Since data showing substandard quality are known to likely degrade the outcomes of image processing, we used MRIQC27 to select the set of test images. Phase I concluded with the release of fMRIPrep version 1.0 on December 6, 2017. We addressed the quality assurance and reliability validation in Phase II. Figure 2 illustrates how the quality of results improved during Phase II. After Phase II, 50 datasets out of the total 54 were rated above the “acceptable” average quality level. The remaining 4 datasets were all above the “poor” level and in or nearby the “acceptable” rating. Correspondingly, Supplementary Figure 3 shows the individual evolution of every dataset at each of the seven quality control points. Phase II concluded with the release of fMRIPrep version 1.0.8 on February 22, 2018. Supplementary Results 1 presents some examples of issues resolved during validation.
Table 2.
Data from OpenfMRI used in evaluation. S: number of sessions; T: number of tasks; R: number of BOLD runs; Modalities: number of runs for each modality, per subject (FM indicates acquisitions for susceptibility distortion correction); Part. IDs (phase): participant identifiers included in testing phase; N: total of unique participants; TR: repetition time (s); #TR: length of time-series (volumes); Resolution: voxel size of BOLD series (mm).
| DS000X XX |
Scanner | S | T | R | Modalities | Part. IDs (Phase I) | Part. IDs (Phase II) | N | TR | #TR | Resolution |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 00154 | SIEMENS | 1 | 1 | 21 | 1 T1w, 3 BOLD | 02, 03, 09, 15 | 01, 02, 07, 08 | 7 | 2.0 | 6300 | 3.124.00 |
| 00255 | SIEMENS | 1 | 3 | 48 | 1 T1w, 6 BOLD | 01, 11, 14, 15 | 02, 03, 04, 10 | 8 | 2.0 | 9510 | 3.125.00 |
| 00356 | SIEMENS | 1 | 1 | 6 | 1 T1w, 1 BOLD | 03, 07, 09, 11 | 02, 09, 10, 11 | 6 | 2.0 | 956 | 3.124.00 |
| 00557 | SIEMENS | 1 | 1 | 21 | 1 T1w, 3 BOLD | 01, 03, 06, 14 | 01, 04, 05, 15 | 7 | 2.0 | 5040 | 3.124.00 |
| 00758 | SIEMENS | 1 | 3 | 46 | 1 T1w, 5 BOLD | 09, 11, 18, 20 | 03, 04, 08, 12 | 8 | 2.0 | 8205 | 3.124.00 |
| 00859 | SIEMENS | 1 | 2 | 38 | 1 T1w, 5 BOLD | 04, 09, 12, 14 | 10, 12, 13, 15 | 7 | 2.0 | 6808 | 3.124.39 |
| 009 | SIEMENS | 1 | 4 | 48 | 1 T1w, 6 BOLD | 01, 03, 09, 10 | 17, 18, 21, 23 | 8 | 2.0 | 10528 | 3.004.00 |
| 01160 | SIEMENS | 1 | 4 | 41 | 1 T1w, 5 BOLD | 01, 03, 06, 08 | 03, 09, 11, 14 | 7 | 2.0 | 8041 | 3.125.00 |
| 017 | SIEMENS | 2 | 2 | 48 | 4 T1w, 9 BOLD | 2, 4, 7, 8 | 2, 5, 7, 8 | 5 | 2.0 | 8736 | 3.124.00 |
| 03034,61 | SIEMENS | 1 | 8 | 30 | 1 T1w, 7 BOLD | 10[440,638,668,855] | 4 | 2.2 | 6254 | 3.004.00 | |
| 03162 | SIEMENS | 107 | 9 | 191 | 29 T1w, 18 T2w, 46 FM, 191 BOLD | 01 | 1 | 1.2 | 79017 | 2.552.54 | |
| 05163 | SIEMENS | 1 | 1 | 54 | 2 T1w, 7 BOLD | 03, 04, 05, 13 | 02, 04, 06, 09 | 7 | 2.0 | 10800 | 3.126.00 |
| 05264 | SIEMENS | 1 | 2 | 28 | 2 T1w, 4 BOLD | 06, 08, 12, 14 | 05, 10, 12, 13 | 7 | 2.0 | 6300 | 3.126.00 |
| 053 | SIEMENS | 1 | 3 | 32 | 1 T1w, 8 BOLD | 002, 003, 005, 006 | 4 | 1.2 | 10712 | 2.402.40 | |
| 101 | SIEMENS | 1 | 1 | 16 | 1 T1w, 2 BOLD | 06, 08, 16, 19 | 05, 11, 17, 20 | 8 | 2.0 | 2416 | 3.004.00 |
| 10265–67 | SIEMENS | 1 | 1 | 16 | 1 T1w, 2 BOLD | 05, 19, 22, 23 | 08, 10, 16, 20 | 8 | 2.0 | 2336 | 3.004.00 |
| 10568,69 | GE | 1 | 1 | 71 | 1 T1w, 11 BOLD | 1, 2, 3, 6 | 1, 4, 5, 6 | 6 | 2.5 | 8591 | 3.503.75 |
| 10770 | SIEMENS | 1 | 1 | 14 | 1 T1w, 2 BOLD | 02, 05, 20, 29 | 05, 36, 39, 47 | 7 | 3.0 | 2315 | 3.003.00 |
| 10871 | GE | 1 | 1 | 41 | 1 T1w, 5 BOLD | 01, 03, 07, 17 | 03, 10, 24, 26 | 7 | 2.0 | 7860 | 3.444.50 |
| 10972 | SIEMENS | 1 | 1 | 12 | 1 T1w, 2 BOLD | 02, 10, 39, 47 | 02, 11, 15, 39 | 6 | 2.0 | 2148 | 3.003.54 |
| 11073 | GE | 1 | 1 | 80 | 1 T1w, 10 BOLD | 07, 09, 17, 18 | 01, 02, 03, 06 | 8 | 2.0 | 14880 | 3.444.01 |
| 11474 | GE | 2 | 5 | 70 | 2 T1w, 10 BOLD | 01, 05, 07, 08 | 02, 03, 04, 07 | 7 | 5.0 | 10626 | 4.004.00 |
| 11575,76 | SIEMENS | 1 | 3 | 24 | 1 T1w, 3 BOLD | 31, 68, 77, 78 | 04, 33, 67, 79 | 8 | 2.5 | 3288 | 4.004.00 |
| 11677–80 | PHILIPS | 1 | 2 | 36 | 1 T1w, 6 BOLD | 02, 08, 10, 15 | 08, 12, 15, 17 | 6 | 2.0 | 6120 | 3.004.00 |
| 11981 | SIEMENS | 1 | 1 | 31 | 1 T1w, 3 BOLD | 10, 51, 59, 74 | 11, 26, 56, 58 | 8 | 1.5 | 7564 | 3.124.00 |
| 12082 | SIEMENS | 1 | 1 | 11 | 1 T1w, 2 BOLD | 04, 05, 08, 24 | 4 | 1.5 | 2376 | 3.124.00 | |
| 12183 | SIEMENS | 1 | 1 | 28 | 1 T1w, 4 BOLD | 01, 04, 05, 20 | 01, 18, 22, 26 | 7 | 1.5 | 5656 | 3.124.00 |
| 13384 | PHILIPS | 2 | 1 | 24 | 2 T1w, 6 BOLD | 06, 21, 22, 23 | 4 | 1.7 | 3480 | 4.004.00 | |
| 14085 | PHILIPS | 1 | 1 | 36 | 1 T1w, 9 BOLD | 05, 27, 32, 33 | 4 | 2.0 | 7380 | 2.803.00 | |
| 148 | GE | 1 | 1 | 12 | 1 T1w, 1 T2w, 3 BOLD | 09, 26, 28, 33 | 4 | 1.8 | 3162 | 3.003.00 | |
| 15786 | PHILIPS | 1 | 1 | 4 | 1 T1w, 1 BOLD | 04, 21, 23, 28 | 4 | 1.6 | 1485 | 4.003.99 | |
| 15887 | SIEMENS | 1 | 1 | 4 | 1 T1w, 1 BOLD | 064, 081, 122, 149 | 4 | 2.0 | 1240 | 3.003.30 | |
| 16488 | SIEMENS | 1 | 1 | 4 | 1 T1w, 1 BOLD | 006, 012, 019, 027 | 4 | 1.5 | 1480 | 3.503.50 | |
| 16889 | SIEMENS | 1 | 1 | 4 | 1 T1w, 1 BOLD | 08, 27, 30, 49 | 4 | 2.5 | 2112 | 3.003.00 | |
| 17090–92 | GE | 1 | 4 | 48 | 1 T1w, 12 BOLD | 1700, 1708, 1710, 1713 | 4 | 3.0 | 2160 | 3.443.40 | |
| 17193 | SIEMENS | 1 | 2 | 20 | 1 T1w, 5 BOLD | control0[4,8,14], mdd03 | 4 | 3.0 | 2066 | 2.903.00 | |
| 17794 | SIEMENS | 1 | 1 | 4 | 1 T1w, 1 BOLD | 04, 07, 10, 11 | 4 | 3.0 | 920 | 3.003.00 | |
| 20095 | SIEMENS | 1 | 1 | 4 | 1 T1w, 1 BOLD | 2004, 2011, 2012, 2014 | 4 | 2.5 | 480 | 3.284.29 | |
| 20596 | SIEMENS | 1 | 2 | 12 | 1 T1w, 3 BOLD | 01, 05, 06, 07 | 4 | 2.2 | 4103 | 3.003.00 | |
| 20897 | SIEMENS | 1 | 1 | 4 | 1 T1w, 1 BOLD | 27, 45, 56, 69 | 4 | 2.5 | 1200 | 3.443.00 | |
| 21298,99 | SIEMENS | 1 | 2 | 40 | 1 T1w, 10 BOLD | 07, 13, 20, 29 | 4 | 3.0 | 5808 | 3.124.00 | |
| 213100 | SIEMENS | 1 | 1 | 4 | 1 T1w, 1 BOLD | 06, 10, 12, 13 | 4 | 2.0 | 1120 | 3.003.99 | |
| 214101 | SIEMENS | 1 | 1 | 4 | 1 T1w, 1 BOLD | EESS0[06,31,33,34] | 4 | 1.6 | 1364 | 3.445.00 | |
| 216102 | GE | 1 | 1 | 16 | 1 T1w, 4 BOLD (ME) | 01, 02, 03, 04 | 4 | 3.5 | 2688 | 3.003.00 | |
| 218103 | PHILIPS | 1 | 1 | 12 | 1 T1w, 3 BOLD | 02, 07, 12, 17 | 4 | 1.5 | 6709 | 2.882.88 | |
| 219103 | PHILIPS | 1 | 1 | 14 | 1 T1w, 3 BOLD | 04, 09, 10, 12 | 4 | 1.5 | 7807 | 2.882.88 | |
| 220104 | PHILIPS, SIEMENS | 3 | 1 | 12 | 3 T1w, 3 BOLD | tbi[03,05,06,10] | 4 | 2.0 | 1728 | 3.004.00 | |
| 221 | SIEMENS | 2 | 1 | 15 | 1 MP2RAGE, 9 FM, 3 BOLD | 010[016,064,125,251] | 4 | 2.5 | 9855 | 2.302.30 | |
| 224105 | SIEMENS | 12 | 6 | 399 | 4 T1w, 4 T2w, 10 FM, 79 BOLD | MSC[05,06,08,09] | MSC[05,08,09,10] | 5 | 2.2 | 88528 | 4.004.00 |
| 228 | SIEMENS | 1 | 1 | 4 | 1 T1w, 1 BOLD | pixar[001,017,103,132] | 4 | 2.0 | 672 | 3.063.29 | |
| 229106 | SIEMENS | 1 | 1 | 12 | 1 T1w, 3 BOLD | 02, 05, 07, 10 | 4 | 2.0 | 4680 | 3.443.00 | |
| 231107 | SIEMENS | 1 | 1 | 12 | 1 T1w, 3 BOLD | 01, 02, 03, 09 | 4 | 2.0 | 4548 | 2.022.00 | |
| 233108 | PHILIPS | 1 | 2 | 80 | 2 T1w, 10 BOLD | rid0000[12,24,36,41] | rid0000[01,17,31,32] | 8 | 2.0 | 15680 | 3.003.00 |
| 237109 | SIEMENS | 1 | 1 | 41 | 1 T1w, 5 BOLD | 03, 08, 11, 12 | 01, 03, 04, 06 | 7 | 1.0 | 19844 | 3.003.00 |
| 2439 | SIEMENS | 1 | 1 | 13 | 1 T1w, 1 BOLD | 012, 032, 042, 071 | 023, 066, 089, 094 | 8 | 2.5 | 2884 | 4.004.004.00 |
| Total | 2176 | 120 | 325 | 304 | 551769 | ||||||
Figure 2. Integrating visual assessment into the software testing framework effectively increases the quality of results.
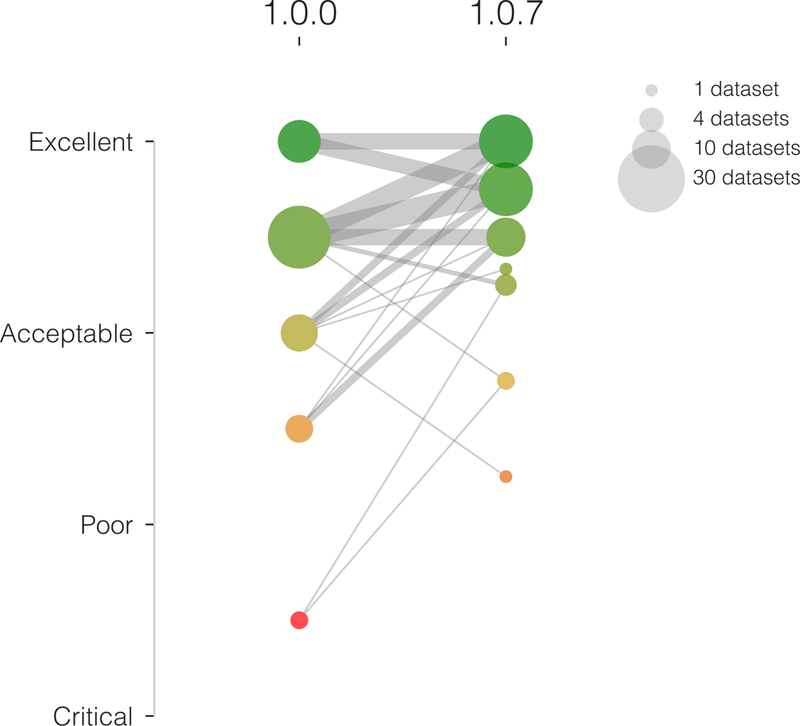
In an early assessment of quality using fMRIPrep version 1.0.0, the overall rating of two datasets was below the “poor” category and four below the “acceptable” level (left column of colored circles). After addressing some outstanding issues detected by the early assessment, the overall quality of processing is substantially improved (right column of circles), and no datasets are below the “poor” quality level. Only two datasets are rated below the “acceptable” level in the second assessment (using fMRIPrep version 1.0.7).
FMRIPrep prevents loss of spatial accuracy via smoothing
We demonstrate that the focus on robustness against data irregularity does not come at a cost in quality of the preprocessing outputs. Moreover, as shown in Figure 3A, the preprocessing outcomes of FSL feat are smoother than those of fMRIPrep. Although preprocessed data were resampled to an isotropic voxel size of 2.0×2.0×2.0 [mm], the smoothness estimation (before the prescribed smoothing step) for fMRIPrep was below 4.0mm, very close to the original resolution of 3.0×3.0×4.0 [mm] of these data. We calculated standard deviation maps in MNI space28 for the temporal average map derived from preprocessing with both alternatives. Visual inspection of these variability maps (Figure 3B) reveals a higher anatomical accuracy of fMRIPrep over feat, likely reflecting the combined effects of a more precise spatial normalization scheme and the application of “fieldmap-less” SDC. FMRIPrep outcomes are particularly better aligned with the underlying anatomy in regions typically warped by susceptibility distortions such as the orbitofrontal lobe, as demonstrated by close-ups in Supplementary Figure 4. We also compared preprocessing done with fMRIPrep and FSL’s feat in two common fMRI analyses. First, we performed within subject statistical analysis using feat –the same tool provides preprocessing and first-level analysis– on both sets of preprocessed data. Second, we perform a group statistical analysis using ordinary least-squares (OLS) mixed modeling (flame29, FSL). In both experiments, we applied identical analysis workflows and settings to both preprocessing alternatives. The first-level analysis showed that the thresholded activation count maps for the go vs. successful stop contrast in the “stopsignal” task were very similar (Figure 4). It can be seen that the results from both pipelines identified activation in the same regions. However, since data preprocessed with feat are smoother, the results from fMRIPrep are more local and better aligned with the cortical sheet. The overlap of statistical maps, as well as Pearson’s correlation, were tightly related to the smoothing of the input data. In the group analysis, fMRIPrep and feat perform equivalently (see Supplementary Results 2).
Figure 3. FMRIPrep affords the researcher finer control over the smoothness of their analysis.
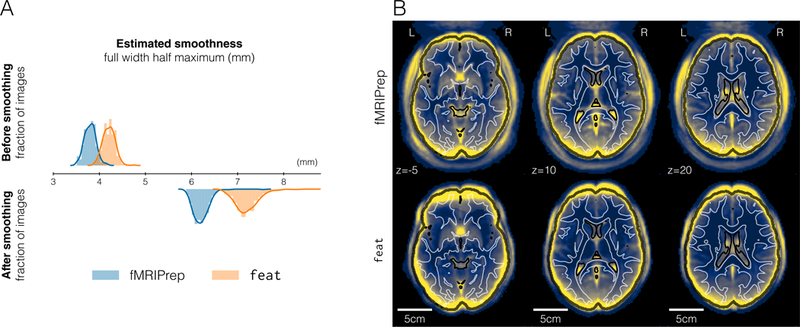
A | Estimating the spatial smoothness of data before and after the initial smoothing step of the analysis workflow confirmed that results of preprocessing with feat are intrinsically smoother. B | Mapping the standard deviation of averaged BOLD time-series displayed greater variability around the brain outline (represented with a black contour) for data preprocessed with feat. This effect is generally associated with a lower performance of spatial normalization28. Reference contours correspond to the brain tissue segmentation of the MNI atlas.
Figure 4. The activation count maps from fMRIPrep are better aligned with the underlying anatomy.
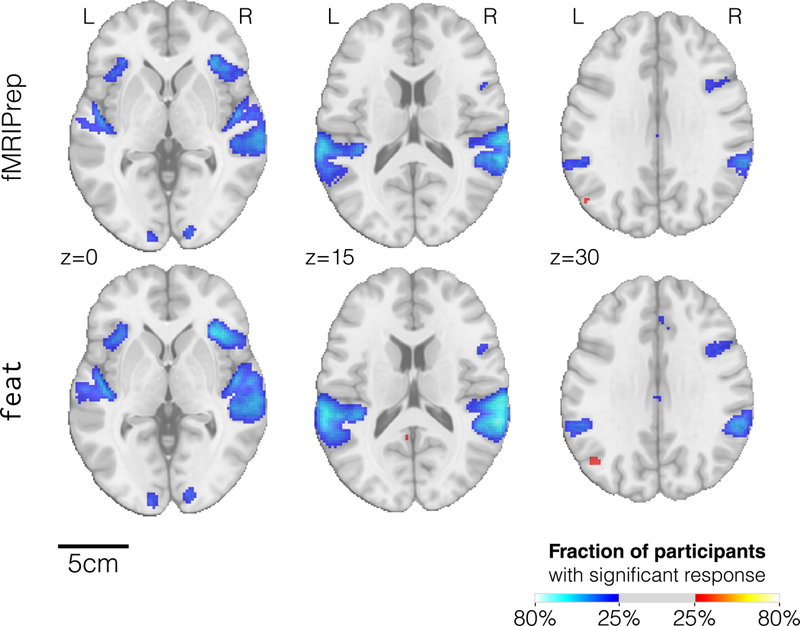
The mosaics show thresholded activation count maps for the go vs. successful stop contrast in the “stopsignal” task after preprocessing using either fMRIPrep (top row) or FSL’s feat (bottom row), with identical single subject statistical modeling. Both tools obtained similar activation maps, with fMRIPrep results being slightly better aligned with the underlying anatomy.
DISCUSSION
FMRIPrep is an fMRI preprocessing workflow developed to excel at four aspects of scientific software: robustness to data idiosyncrasies, high quality and consistency of results, maximal transparency, and ease-of-use. We describe how using the Brain Imaging Data Structure (BIDS21) along with a flexible design allows the workflow to self-adapt to the idiosyncrasy of inputs (sec. A modular design alongside BIDS allow for a flexible, adaptive workflow). The workflow (briefly summarized in Figure 1) integrates state-of-art tools from widely used neuroimaging software packages at each preprocessing step (see Table 1).
Some other relevant facets of fMRIPrep and how they relate to existing alternative pipelines are presented in sec. Highlights of fMRIPrep within the neuroimaging context. We stress that fMRIPrep is developed with the best software engineering principles, which are fundamental to ensure software reliability. The pipeline is easy to use for researchers and clinicians without extensive computer engineering experience, and produces comprehensive visual reports (Supplementary Figure 1).
In sec. FMRIPrep yields high-quality results on diverse data, we demonstrate the robustness of fMRIPrep on a representative collection of data from datasets associated with different studies (Table 2). We then interrogate the quality of those results with the individual inspection of the corresponding visual reports by experts (sec. Visual reports ease quality control and maximize transparency and the corresponding summary in Figure 2). A comparison to FSL’s feat (sec. FMRIPrep prevents loss of spatial accuracy via smoothing) demonstrates that fMRIPrep achieves higher spatial accuracy and introduces less uncontrolled smoothness (Figure 3, 4). Group 𝑝-statistical maps only differed on their smoothness (sharper for the case of fMRIPrep). The fact that first-level and second-level analyses resulted in small differences between fMRIPrep and our ad-hoc implementation of a feat-based workflow indicates that the individual preprocessing steps perform similarly when they are fine-tuned to the input data. That justifies the need for fMRIPrep, which autonomously adapts the workflow to the data without error-prone manual intervention. To a limited extent, that also mitigates some concerns and theoretical risks that arise from analytical degrees-of freedom19 available to researchers. FMRIPrep stands out amongst pipelines because it automates the adaptation to the input dataset without compromising the quality of results.
One limitation of this work is the use of visual (the reports) and semi-visual (e.g. Figure 3, 4) assessments for the quality of preprocessing outcomes. Although some frameworks have been proposed for the quantitative evaluation of preprocessing on task-based (such as NPAIRS30) and resting-state31 fMRI, they impose a set of assumptions on the test data and the workflow being assessed that severely limit their suitability in general. The modular design of fMRIPrep defines an interface to each processing step, which will permit the programmatic evaluation of the many possible combinations of software tools and processing steps. That will also enable the use of quantitative testing frameworks to pursue the minimization of Type I errors without the cost of increasing Type II errors. The range of possible applications for fMRIPrep also presents some boundaries. For instance, very narrow field-of-view (FoV) images often do not contain enough information for standard image registration methods to work correctly. Reduced FoV datasets from OpenfMRI were excluded from the evaluation since they are not yet fully supported by fMRIPrep. Extending fMRIPrep’s support for these particular images is already a future line of the development road-map. FMRIPrep may also under-perform for particular populations (e.g. infants) or when brains show nonstandard structures, such as tumors, resected regions or lesions. Despite these challenges, fMRIPrep performed robustly on data from a simultaneous MRI/electrocorticography study, which is extremely challenging to analyze due to the massive BOLD signal drop-out near the implanted cortical electrodes (see Supplementary Figure 5). In addition, fMRIPrep’s modular architecture makes it straightforward to extend the tool to support specific populations or new species by providing appropriate atlases of those brains. This future line of work would be particularly interesting in order to adapt the workflow to data collected from rodents and nonhuman primates.
Approximately 80% of the analysis pipelines investigated by Carp19 were implemented using either AFNI12, FSL15, or SPM17. Ad-hoc pipelines adapt the basic workflows provided by these tools to the particular dataset at hand. Although workflow frameworks like Nipype20 ease the integration of tools from different packages, these pipelines are typically restricted to just one of these alternatives (AFNI, FSL or SPM). Otherwise, scientists can adopt the acquisition protocols and associated preprocessing software of large consortia26,32 like the Human Connectome Project (HCP) or the UK Biobank33. The off-the-shelf applicability of these workflows is contravened by important limitations on the experimental design. Therefore, researchers typically opt to recode their custom preprocessing workflows with nearly every new study19. That practice entails a “pipeline debt”, which requires the investment on proper software engineering to ensure an acceptable correctness and stability of the results (e.g. continuous integration testing) and reproducibility (e.g. versioning, packaging, containerization, etc.). A trivial example of this risk would be the leakage of magic numbers that are hard-coded in the source (i.e. a crucial imaging parameter that inadvertently changed from one study to the next one). Until fMRIPrep, an analysis-agnostic approach that builds upon existing software instruments and optimizes preprocessing for robustness to data idiosyncrasies, quality of outcomes, ease-of-use, and transparency, was lacking.
The rapid increase in volume and diversity of data, as well as the evolution of available techniques for processing and analysis, presents an opportunity for significantly advancing research in neuroscience. The drawback resides in the need for progressively complex analysis workflows that rely on decreasingly interpretable models of the data. Such context encourages “black-box” solutions that efficiently perform a valuable service but do not provide insights into how the tool has transformed the data into the expected outputs. Black-boxes obscure important steps in the inductive process mediating between experimental measurements and reported findings. This way of moving forward risks producing a future generation of cognitive neuroscientists who have become experts in using sophisticated computational methods, but have little to no working knowledge of how data were transformed through processing. Transparency is often identified as a treatment for these problems. FMRIPrep ascribes to “glass-box” principles, which are defined in opposition to the many different facets or levels at which black-box solutions are opaque. The visual reports that fMRIPrep generates are a crucial aspect of the glass-box approach. Their quality control checkpoints represent the logical flow of preprocessing, allowing scientists to critically inspect and better understand the underlying mechanisms of the workflow. A second transparency element is the citation boilerplate that formalizes all details of the workflow and provides the versions of all involved tools along with references to corresponding scientific literature. A third asset for transparency is the thorough documentation which delivers additional details on each of the building blocks that are represented in the visual reports and described in the boilerplate. Further, fMRIPrep is open-source since its inception: users have access to all the incremental additions to the tool through the history of the version-control system. The use of GitHub (https://github.com/poldracklab/fmriprep) grants access to the discussions held during development, allowing the retrieval of how and why the main design decisions were made. GitHub also provides an excellent platform to foster the community with useful tools such as source browsing, code review, bug tracking and reporting, submission of new features and bug fixes through pull requests, etc. The modular design of fMRIPrep enhances its flexibility and improves transparency, as the main features of the software are more easily accessible to potential collaborators. In combination to some coding style and contribution guidelines, this modularity has enabled multiple contributions by peers and the creation of a rapidly growing community that would be difficult to nurture behind closed doors. A number of existing tools have implemented elements of “glass-box” philosophy (for example visual reports in feat, documentation in C-PAC, open source community of Nilearn), but the complete package (visual reports, educational documentation, reporting templates, collaborative open source community) is still rare among scientific software. FMRIPrep’s transparent and accessible development and reporting aims to better equip fMRI practitioners to perform reliable, reproducible, statistical analyses with a high-standard, consistent, and adaptive preprocessing instrument.
DATA
Data used in the validation of fMRIPrep
Participants were drawn from a multiplicity of studies available in OpenfMRI, accessed on September 30, 2017. Studies were sampled uniformly (four participants each), except for DS000031 that consists of only one participant. Data selection criteria are described below. Magnetic resonance imaging (MRI) data were acquired at multiple scanning centers, with the following frequencies of vendors: ∼70% SIEMENS, ∼14% PHILIPS, ∼14% GE. Data were acquired by 1.5T and 3T systems running varying software versions. Acquisition protocols, as well as the particular acquisition parameters (including relevant BOLD settings such as the repetition time −TR−, the echo time −TE−, the number of TRs and the resolution) also varied with each study. However, only datasets including at least one T1-weighted (T1w) and one BOLD per subject run were included. Datasets containing BIDS errors (DS000210), and degenerate data (many T1w images of DS000223 are skull-stripped) at the time of access were discarded. Similarly, very-narrow FoV BOLD datasets (DS000172, DS000217, and DS000232) were also excluded. In total, 54 datasets (46 single-session datasets, 8 multi-session) were included in this assessment. Table 2 overviews the particular properties of each dataset, summarizing the large heterogeneity of the resource.
This evaluation covered0i 54 studies out of a total of 58 studies in OpenfMRI that included the two required imaging modalities (T1w and BOLD). Therefore, by covering 93% of the studies in OpenfMRI, we ensured a large heterogeneity in terms of acquisition protocols, settings, instruments and parameters that is necessary to demonstrate the robustness of fMRIPrep against the variability in input data features.
Data used in the comparison to FSL feat
We reuse the UCLA Consortium for Neuropsychiatric Phenomics LA5c Study34, a dataset that is publicly available on OpenfMRI under data accession DS000030. During the experiment, subjects performed six tasks, a block of rest, and two anatomical scans. The study includes imaging data of a large group of healthy individuals from the community, as well as samples of individuals diagnosed with schizophrenia, bipolar disorder, and attention-deficit/hyperactivity disorder. As described in their data descriptor34, MRI data were acquired on one of two 3T Siemens Trio scanners, located at the Ahmanson-Lovelace Brain Mapping Center (syngo MR B15) and the Staglin Center for Cognitive Neuroscience (syngo MR B17). FMRI data were collected using an echo-planar imaging (EPI) sequence (slice thickness=4mm, 34 slices, TR=2s, TE=30ms, flip angle=90deg, matrix 64×64, FoV=192mm, oblique slice orientation). Additionally, a T1w image is available per participant (MPRAGE, TR=1.9s, TE=2.26ms, FoV=250mm, matrix=256×256, sagittal plane, slice thickness=1mm, 176 slices). For this experiment, only images including both the T1w and the functional scans corresponding the Stop Signal task (referred to as “stopsignal”) were included (totaling N=257 participants).
Stop Signal task.
Participants were instructed to respond quickly to a “go” stimulus. During some of the trials, at unpredictable times, a stop signal would appear after the stimulus is presented. During those trials, the subject has to inhibit any planned response. In this experiment, we specifically look into the difference between the brain activation during a successful stop trial and a go trial (contrast: Go - StopSuccess). Thus, we expect to see brain regions responsible for response inhibition (negative) and motor response (positive). Further details on the task are available with the dataset descriptor34.
THE FMRIPREP WORKFLOW
Preprocessing anatomical images
The T1w image is corrected for intensity non-uniformity using N4BiasFieldCorrection35 (ANTs), and skull-stripped using antsBrainExtraction.sh (ANTs). Skull-stripping is performed through coregistration to a template, with two options available: the OASIS template36 (default) or the NKI template37. Using visual inspection, we have found that this approach outperforms other common approaches, which is consistent with previous reports26. When several T1w volumes are found, the intensity non-uniformity-corrected versions are first fused into a reference T1w map of the subject with mri_robust_template38 (FreeSurfer). Brain surfaces are reconstructed from the subject’s T1w reference (and T2w images if available) using recon-all39 (FreeSurfer). The brain mask estimated previously is refined with a custom variation of a method (originally introduced in Mindboggle40) to reconcile ANTs-derived and FreeSurfer-derived segmentations of the cortical gray matter (GM). Both surface reconstruction and subsequent mask refinement are optional and can be disabled to save run time when surface-based analysis is not needed. Spatial normalization to the ICBM 152 Nonlinear Asymmetrical template41 (version 2009c) is performed through nonlinear registration with antsRegistration42 (ANTs), using brain-extracted versions of both the T1w reference and the standard template. ANTs was selected due to its superior performance in terms of volumetric group level overlap43. Brain tissues –cerebrospinal fluid (CSF), white matter (WM) and GM– are segmented from the reference, brain-extracted T1w using fast44 (FSL).
Preprocessing functional runs
For every BOLD run found in the dataset, a reference volume and its skull-stripped version are generated using an in-house methodology (described in Supplementary Note 3). Then, head-motion parameters (volume-to-reference transform matrices, and corresponding rotation and translation parameters) are estimated using mcflirt45 (FSL). Among several alternatives (see Table 1), mcflirt is used because its results are comparable to other tools46 and it stores the estimated parameters in a format that facilitates the composition of spatial transforms to achieve one-step interpolation (see below). If slice timing information is available, BOLD runs are (optionally) slice time corrected using 3dTshift (AFNI12). When field map information is available, or the experimental “fieldmap-less” correction is requested (see “Fieldmap-less” susceptibility distortion correction), SDC is performed using the appropriate methods (see Supplementary Figure 6). This is followed by co-registration to the corresponding T1w reference using boundary-based registration47 with nine degrees of freedom (to minimize remaining distortions). If surface reconstruction is selected, fMRIPrep uses bbregister (FreeSurfer). Otherwise, the boundary based coregistration implemented in flirt (FSL) is applied. In our experience, bbregister yields the better results47 due to the high resolution and the topological correctness of the GM/WM surfaces driving registration. To support a large variety of output spaces for the results (e.g. the native space of BOLD runs, the corresponding T1w, FreeSurfer’s fsaverage spaces, the template used as target in the spatial normalization step, etc.), the transformations between spaces can be combined. For example, to generate preprocessed BOLD runs in template space (e.g. MNI), the following transforms are concatenated: head-motion parameters, the warping to reverse susceptibility-distortions (if calculated), BOLD-to-T1w, and T1w-to-template mappings. The BOLD signal is also sampled onto the corresponding participant’s surfaces using mri_vol2surf (FreeSurfer), when surface reconstruction is being performed. Thus, these sampled surfaces can easily be transformed onto different output spaces available by concatenating transforms calculated throughout fMRIPrep and internal mappings between spaces calculated with recon-all. The composition of transforms allows for a single-interpolation resampling of volumes using antsApplyTransforms (ANTs). Lanczos interpolation is applied to minimize the smoothing effects of linear or Gaussian kernels48. Optionally, ICA-AROMA can be performed and corresponding “non-aggressively” denoised runs are then produced. When ICA-AROMA is enabled, the time-series are first smoothed and then denoised, following the description of the original method11.
Extraction of nuisance time-series
To avoid restricting fMRIPrep’s outputs to particular analysis types, the tool does not perform any temporal denoising by default. Nonetheless, it provides researchers with a diverse set of confound estimates that could be used for explicit nuisance regression or as part of higher-level models. This lends itself to decoupling preprocessing and behavioral modeling as well as evaluating robustness of final results across different denoising schemes. A set of physiological noise regressors are extracted for the purpose of performing component-based noise correction (CompCor10). Principal components are estimated after high-pass filtering the BOLD time-series (using a discrete cosine filter with 128s cut-off) for the two CompCor variants: temporal (tCompCor) and anatomical (aCompCor). Six tCompCor components are then calculated from the top 5% variable voxels within a mask covering the subcortical regions. This subcortical mask is obtained by heavily eroding the brain mask, which ensures it does not include cortical GM regions. For aCompCor, six components are calculated within the intersection of the aforementioned mask and the union of CSF and WM masks calculated in T1w space, after their projection to the native space of each functional run (using the inverse BOLD-to-T1w transformation). FD and DVARS are calculated for each functional run, both using their implementations in Nipype (following the definitions by Power et al.8). Three global signals are extracted within the CSF, the WM, and the whole-brain masks using Nilearn16. If ICA-AROMA11 is requested, the “aggressive” noise-regressors are collected and placed within the corresponding confounds files. Since the non-aggressive cleaning with ICA-AROMA is performed after extraction of other nuisance signals, the “aggressive” regressors can be used to orthogonalize those other nuisance signals to avoid the risk of re-introducing nuisance signal within regression. In addition, a “non-aggressive” version of preprocessed data is also provided since this variant of ICA-AROMA denoising cannot be performed using only nuisance regressors.
“Fieldmap-less” susceptibility distortion correction
Many legacy and current human fMRI protocols lack the MR field maps necessary to perform standard methods for SDC. As described in Supplementary Figure 6, the BIDS dataset is queried to discover whether extra acquisitions containing field map information are available. When no fieldmap information is found, fMRIPrep adapts the “fieldmap-less” correction for diffusion EPI images introduced by Wang et al.49. They propose using the same-subject T1w reference as the undistorted target in a nonlinear registration scheme. To maximize the similarity between the T2★ contrast of the EPI scan and the reference T1w, the intensities of the latter are inverted. To regularize the optimization of the deformation field, only displacements along the phase-encoding direction are allowed, and the magnitude of the displacements is modulated using priors. To our knowledge, no other existing pipeline applies “fieldmap-less” SDC to the BOLD images. Further details on the integration of the different SDC techniques and particularly this “fieldmap-less” option are found in Supplementary Note 3.
FMRIPrep is thoroughly documented, community-driven, and developed with high-standards of software engineering
Preprocessing pipelines are generally well documented, however the extreme flexibility of fMRIPrep makes its proper documentation substantially more challenging. As for other large scientific software communities, fMRIPrep contributors pledge to keep the documentation thorough and updated along coding iterations. Packages also differ on the involvement of the community: while fMRIPrep includes researchers in the decision making process and invites their suggestions and contributions, other packages have a more closed model where the feedback from users is more limited (e.g. a mailing list). In contrast to other pipelines, fMRIPrep is community-driven. This paradigm allows the fast adoption of cutting-edge advances on fMRI preprocessing, which tend to render existing workflows (including fMRIPrep) obsolete. For example, while fMRIPrep initially performed STC before HMC, we adapted the tool to the recent recommendations of Power et al.18 upon a user’s requestii. This model has allowed the user base to grow rapidly and enabled substantial third-party contributions to be included in the software, such as the support for processing multi-echo datasets. The open-source nature of fMRIPrep has permitted frequent code reviews that are effective in enhancing the software’s quality and reliability50. Supplementary Note 4 describes how the community interacts, discusses the code review process, and underscores how the modular design of fMRIPrep successfully facilitates contributions from peers. Finally, fMRIPrep undergoes continuous integration testing (see Supplementary Fig. SN4.1), a technique that has recently been proposed as a means to ensure reproducibility of analyses in computational sciences51,52. Additional comparison points, such as the graphical user interface of several preprocessing workflows, are given in Supplementary Note 5.
Ensuring reproducibility with strict versioning and containers
For enhanced reproducibility, fMRIPrep fully supports execution via the Docker (https://docker.com) and Singularity53 container platforms. Container images are generated and uploaded to a public repository for each new version of fMRIPrep. These containers are released with a fixed set of software versions for fMRIPrep and all its dependencies, maximizing run-to-run reproducibility in an easy way. This helps address the widespread lack of reporting of specific software versions and the large variability of software versions, which threaten the reproducibility of fMRI analyses19. Except for C-PAC, alternative pipelines do not provide official support for containers. The adoption of the BIDS-Apps51 container model makes fMRIPrep amenable to a multiplicity of infrastructures and platforms: PC, high-performance computing, Cloud, etc.
VALIDATION OF FMRIPREP ON DIVERSE DATA
The general validation framework presented in Supplementary Figure 2 implements a testing plan elaborated prior the release of version 1.0 of the software. The plan is divided into two validation phases in which different data samples and validation procedures are applied. Table 2 describes the data samples used on each phase. In Phase I, we ran fMRIPrep on a manually selected sample of participants that are potentially challenging to the tool’s robustness, exercising the adaptiveness to the input data. Phase II focused on the visual assessment of the quality of preprocessing results on a large and heterogeneous sample.
Methodology and test plan
To ensure that fMRIPrep fulfills the specifications on reliability and scientific-software standards, the tool undergoes a thorough acceptance testing plan. The plan is structured in three phases: the first was aimed at the discovery of faults, the second at the evaluation of the robustness, and the final phase at the full coverage of OpenfMRI. To note, an early test Phase 0 was conducted as a proof of concept for the tool.
Validation Phase I – Fault-discovery testing.
During Phase I, a total of 120 subjects from 30 different datasets (see Table 2) were manually identified as low-quality using MRIQC27. Data showing substandard quality are known to likely degrade the outcomes of image processing, and therefore they are helpful to test software reliability. This sub-sample of OpenfMRI underwent preprocessing in the Stampede2 supercomputer of the Texas Advanced Computer Center (TACC), Austin, TX. Results were visually inspected and failures reported in the GitHub repository. Once software faults were fixed, fMRIPrep 1.0.0 of was released and the Phase II of validation was launched.
Validation Phase II – Quality assurance and reliability testing.
In this second phase, the coverage of OpenfMRI was extended to 54 available datasets (Table 2), randomly selecting four participants per dataset (with replacement of participants covered in Phase I). A total of 325 participantsiii were preprocessed in the Sherlock cluster of Stanford University, Stanford, CA. Validation Phase II integrated a protocol for the screening of results into the software testing (Supplementary Figure 2). Three raters evaluated each participant’s report following the protocol described below. Their ratings are made available with the corresponding reports for scrutiny.
Protocol for manual assessment.
Each visual report generated in Phase II was inspected by one expert (selected randomly between authors CJM, KJG and OE) at seven quality checkpoints: i) overall performance; ii) surface reconstruction from anatomical MRI; iii) T1w brain mask and tissue segmentation; iv) spatial normalization; v) brain mask and regions-of-interest (ROIs) for CompCor application in native BOLD space (“BOLD ROIs”); vi) intra-subject BOLD-to-T1w co-registration; and vii) SDC. Experts were instructed to assign a score on a scale from 1 (poor) to 3 (excellent) at each quality control point. A special rating score of 0 (unusable) was assigned to tasks that failed in a critical way hampering further preprocessing. Poor (1) was assigned when fMRIPrep did not critically failed at the task, but the outcome would likely affect negatively downstream analysis. For example, when “fieldmap-less” correction unwarped in the expected direction, although some distorted areas remained (or were overcorrected), then the acceptable (2) rating was assigned. Finally, excellent (3) was assigned when the expert did not notice any substantial defect that would indicate a lower rating. Supplementary Figure 3 shows the evolution of the quality ratings at the seven checkpoints at the beginning and completion of Phase II (indicated by versions 1.0.0 and 1.0.7, respectively).
COMPARISON TO AN ALTERNATIVE PREPROCESSING TOOL
For comparison, data were preprocessed with two alternate pipelines: fMRIPrep 1.0.8 and FSL’s feat 5.0.10. We then performed identical analyses on each dataset preprocessed with either pipeline. On the first level analysis, we calculate a 𝑡-statistic map per participant for the task under analysis (N=257). Second level analyses were performed in a specific resampling scheme to allow a statistical comparison between the pipelines: two random (non-overlapping) subsets of 𝑛 participants are repeatedly entered into a group level analysis. The first step is the experimental manipulation resulting in two conditions: (1) the data are preprocessed with fMRIPrep, and (2) the data are preprocessed using feat. The next two steps are identical for both conditions.
Preprocessing
Preprocessing with fMRIPrep is described using the corresponding citation boilerplate (Supplementary Box SN3.1). We configured feat using its graphical user interface (GUI) and generated a template.fsf file, which can be found in GitHubiv. We manually extended execution to all participants in our sample creating the script fsl_feat_wrapper.py that accompanies the template.fsf file in GitHub. As it can be seen on the template.fsf file, we disabled band-pass filtering and spatial smoothing to make results of preprocessing comparable. Both processing steps (temporal filtering and spatial smoothing) were implemented in a common, subsequent analysis workflow described below. Additionally, we manually configured the ICBM 152 Nonlinear Asymmetrical template41 version 2009c as target for spatial normalization. Finally, we manually resampled the preprocessed BOLD files into template space using FSL’s flirt.
Mapping the BOLD variability on standard space.
To investigate the spatial consistency of the average BOLD across participants, we calculated standard deviation maps in MNI space for the temporal average map28 derived from preprocessing with both alternatives.
Smoothness.
We used AFNI’s 3dFWHMx to estimate the (average) smoothness of the data at two check- points: i) before the first-level analysis workflow, and ii) after applying a 5.0mm full-width half-maximum (FWHM) spatial smoothing, which was the first step of the analysis workflow described in the following.
First-level statistical analysis
We analyzed the “stopsignal” task data using FSL and AFNI tools, integrated in a workflow using Nipype. Spatial smoothing was applied using AFNI’s 3dBlurInMask with a Gaussian kernel of FWHM=5mm. Activity was estimated using a general linear model (GLM) with FSL’s feat. For the one condition under comparison (go - successful), one task regressor was included with a fixed duration of 1.5s. An extra regressor was added with equal amplitude, but the duration equal to the reaction time. These regressors were orthogonalized with respect to the fixed duration regressor of the same condition. Predictors were convolved with a double-gamma canonical hemodynamic response function. Temporal derivatives were added to all task regressors to compensate for variability on the hemodynamic response function. Furthermore, the six rigid-motion parameters (translation in three directions, rotation in three directions) were added as regressors to avoid confounding effects of head-motion. We included a high-pass filter (100Hz) in FSL’s feat.
Activation-count maps.
The statistical map for each participant was binarized at 𝑧=±1.65 (which corresponds to a two-sided test of 𝑝<0.1). Then, the average of these maps is computed across participants. The average negative map (percentage of subjects showing a negative effect with 𝑧 < −1.65) is subtracted from the average positive map to indicate the direction of effects. High values in certain regions and low values in other regions show a good overlap of activation between subjects.
Second-level statistical analysis
Subsequent to the single subject analyses, two random, non-overlapping subsamples of 𝑛 subjects were taken and entered into a second level analysis. We vary the sample size 𝑛 of groups between 10 and 120. We ran the group level analyses based on two variants of the first level: with a prescribed smoothing of 5.0mm FWHM, and without such smoothing step. The resampling process was repeated 200 times per group sample-size and smoothing condition. To investigate the implications of either pipeline on the group analysis use-case, we ran the same OLS mixed modeling using FSL’s flame on each two disjoint subsets of randomly selected subjects and resampling repetition. We calculated several metrics of spatial agreement on the resulting maps of (uncorrected) 𝑝-statistical values. We also calculated the spatial agreement of the thresholded statistical maps, binarized with a threshold chosen to control for the false-discovery rate (FDR) at 5% (using FSL’s fdr command).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Laura and John Arnold Foundation, NIH grants NBIB R01EB020740, NIMH R24MH114705, R24MH117179, and NINDS U01NS103780. JD has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement No 706561. The authors thank S. Nastase and T. van Mourik for their thoughtful open reviews of a pre-print version of this paper.
LIST OF ACRONYMS AND ABBREVIATIONS
- BIDS
Brain Imaging Data Structure
- BOLD
blood-oxygen-level dependent
- C-PAC
configurable pipeline for the analysis of connectomes
- CompCor
component-based noise correction
- CSF
cerebrospinal fluid
- DVARS
spatial standard deviation of the data after temporal differencing
- EPI
echo-planar imaging
- FD
framewise displacement
- FDR
false-discovery rate
- fMRI
functional MRI
- FoV
field-of-view
- FWHM
full-width half-maximum
- GM
gray matter
- GUI
graphical user interface
- HCP
Human Connectome Project
- HMC
head-motion correction
- HTML
hypertext markup language
- ICA
independent components analysis
- ICA-AROMA
automatic removal of motion artifacts
- MRI
magnetic resonance imaging
- OLS
ordinary least-squares
- PCA
principal components analysis
- ROI
region-of-interest
- SDC
susceptibility distortion correction
- STC
slice-timing correction
- T1w
T1-weighted
- TR
repetition time
- WM
white matter
Footnotes
COMPETING FINANCIAL INTERESTS STATEMENT
The authors declare no competing interests.
ETHICAL COMPLIANCE
We complied with all relevant ethical regulations. This study reuses publicly available data acquired at many different institutions. Protocols for all the original studies have been approved by corresponding ethical boards.
REPORTING SUMMARY
Further information on research design is available in the Life Sciences Reporting Summary linked to this article.
DATA AVAILABILITY
All original data used in this work are publicly available through the OpenNeuro platform (formerly, OpenfMRI). Derivatives generated with fMRIPrep in this work are available at https://s3.amazonaws.com/fmriprep/index.html. The expert ratings collected after visual assessment of all reports are available through FigShare (doi:10.6084/m9.figshare.6196994.v3).
SOFTWARE AVAILABILITY
FMRIPrep’s source code is available at GitHub (https://github.com/poldracklab/fmriprep). We use Zenodo to generate new digital object identifiers for each new release of fMRIPrep, version 1.1.4 (doi:10.5281/zenodo.1340696) being the latest one. FMRIPrep is licensed under the BSD 3-Clause “New” or “Revised” License. Software is distributed as a Python package (https://pypi.org/project/fmriprep/), as a Docker container (https://hub.docker.com/r/poldracklab/fmriprep/), and as a CodeOcean capsule22.
i Data coverage is a metric used in software engineering that measures the area that a given test or test-set covers with respect to the full domain of possible input data.
iii DS000031 contains one participant only.
REFERENCES
- 1.Poldrack RA & Farah MJ Progress and challenges in probing the human brain. Nature 526, 371–379 (2015). doi: 10.1038/nature15692. [DOI] [PubMed] [Google Scholar]
- 2.Power JD, Plitt M, Laumann TO & Martin A Sources and implications of whole-brain fMRI signals in humans. NeuroImage 146, 609–625 (2017). doi: 10.1016/j.neuroimage.2016.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindquist MA The Statistical Analysis of fMRI Data. Statistical Science 23, 439–464 (2008). doi: 10.1214/09-STS282. [DOI] [Google Scholar]
- 4.Caballero-Gaudes C & Reynolds RC Methods for cleaning the BOLD fMRI signal. NeuroImage 154, 128–149 (2017). doi: 10.1016/j.neuroimage.2016.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strother SC Evaluating fMRI preprocessing pipelines. IEEE Engineering in Medicine and Biology Magazine 25, 27–41 (2006). doi: 10.1109/MEMB.2006.1607667. [DOI] [PubMed] [Google Scholar]
- 6.Sladky R et al. Slice-timing effects and their correction in functional MRI. NeuroImage 58, 588–594 (2011). doi: 10.1016/j.neuroimage.2011.06.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashburner J Preparing fMRI Data for Statistical Analysis In Filippi M (ed.) fMRI Techniques and Protocols, no. 41 in Neuromethods, 151–178 (Humana Press, 2009). doi: 10.1007/978-1-60327-919-2_6. [DOI] [Google Scholar]
- 8.Power JD, Barnes KA, Snyder AZ, Schlaggar BL & Petersen SE Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. NeuroImage 59, 2142–2154 (2012). doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Power JD et al. Methods to detect, characterize, and remove motion artifact in resting state fMRI. NeuroImage 84, 320–341 (2014). doi: 10.1016/j.neuroimage.2013.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Behzadi Y, Restom K, Liau J & Liu TT A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. NeuroImage 37, 90–101 (2007). doi: 10.1016/j.neuroimage.2007.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pruim RHR et al. ICA-AROMA: A robust ICA-based strategy for removing motion artifacts from fMRI data. NeuroImage 112, 267–277 (2015). doi: 10.1016/j.neuroimage.2015.02.064. [DOI] [PubMed] [Google Scholar]
- 12.Cox RW & Hyde JS Software tools for analysis and visualization of fMRI data. NMR in Biomedicine 10, 171–178 (1997). doi:. [DOI] [PubMed] [Google Scholar]
- 13.Avants BB et al. A reproducible evaluation of ANTs similarity metric performance in brain image registration. NeuroImage 54, 2033–44 (2011). doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischl B FreeSurfer. NeuroImage 62, 774–781 (2012). doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW & Smith SM FSL. NeuroImage 62, 782–790 (2012). doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 16.Abraham A et al. Machine learning for neuroimaging with scikit-learn. Frontiers in Neuroinformatics 8 (2014). doi: 10.3389/fninf.2014.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friston KJ, Ashburner J, Kiebel SJ, Nichols TE & Penny WD Statistical parametric mapping : the analysis of functional brain images (Academic Press, London, 2006). [Google Scholar]
- 18.Power JD, Plitt M, Kundu P, Bandettini PA & Martin A Temporal interpolation alters motion in fMRI scans: Magnitudes and consequences for artifact detection. PLOS ONE 12, e0182939 (2017). doi: 10.1371/journal.pone.0182939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carp J The secret lives of experiments: Methods reporting in the fMRI literature. NeuroImage 63, 289–300 (2012). doi: 10.1016/j.neuroimage.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 20.Gorgolewski K et al. Nipype: a flexible, lightweight and extensible neuroimaging data processing framework in Python. Frontiers in Neuroinformatics 5, 13 (2011). doi: 10.3389/fninf.2011.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorgolewski KJ et al. The Brain Imaging Data Structure, a format for organizing and describing outputs of neuroimaging experiments. Scientific Data 3, 160044 (2016). doi: 10.1038/sdata.2016.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esteban O et al. FMRIPrep Software Capsule. Code Ocean Capsule: Nature Methods (2018). doi: 10.24433/CO.ed5ddfef-76a3-4996-b298-e3200f69141b. [DOI] [Google Scholar]
- 23.Poldrack RA et al. Guidelines for reporting an fMRI study. NeuroImage 40, 409–414 (2008). doi: 10.1016/j.neuroimage.2007.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sikka S et al. Towards automated analysis of connectomes: The configurable pipeline for the analysis of connectomes (C-PAC). In 5th INCF Congress of Neuroinformatics, vol. 117 (Munich, Germany, 2014). doi: 10.3389/conf.fninf.2014.08.00117. [DOI] [Google Scholar]
- 25.Van Essen D et al. The Human Connectome Project: A data acquisition perspective. NeuroImage 62, 2222–2231 (2012). doi: 10.1016/j.neuroimage.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glasser MF et al. The minimal preprocessing pipelines for the Human Connectome Project. NeuroImage 80, 105–124 (2013). doi: 10.1016/j.neuroimage.2013.04.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Esteban O et al. MRIQC: Advancing the automatic prediction of image quality in MRI from unseen sites. PLOS ONE 12, e0184661 (2017). doi: 10.1371/journal.pone.0184661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calhoun VD et al. The impact of T1 versus EPI spatial normalization templates for fMRI data analyses. Human Brain Mapping 38, 5331–5342 (2017). doi: 10.1002/hbm.23737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beckmann CF, Jenkinson M & Smith SM General multilevel linear modeling for group analysis in FMRI. NeuroImage 20, 1052–1063 (2003). doi: 10.1016/S1053-8119(03)00435-X. [DOI] [PubMed] [Google Scholar]
- 30.Strother SC et al. The Quantitative Evaluation of Functional Neuroimaging Experiments: The NPAIRS Data Analysis Framework. NeuroImage 15, 747–771 (2002). doi: 10.1006/nimg.2001.1034. [DOI] [PubMed] [Google Scholar]
- 31.Karaman M, Nencka AS, Bruce IP & Rowe DB Quantification of the Statistical Effects of Spatiotemporal Processing of Nontask fMRI Data. Brain Connectivity 4, 649–661 (2014). doi: 10.1089/brain.2014.0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alfaro-Almagro F et al. Image processing and Quality Control for the first 10,000 brain imaging datasets from UK Biobank. NeuroImage (2017). doi: 10.1016/j.neuroimage.2017.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller KL et al. Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nature Neuroscience 19, 1523–1536 (2016). doi: 10.1038/nn.4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS-ONLY REFERENCES
- 34.Poldrack RA et al. A phenome-wide examination of neural and cognitive function. Scientific Data 3, 160110 (2016). doi: 10.1038/sdata.2016.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tustison NJ et al. N4ITK: Improved N3 Bias Correction. IEEE Transactions on Medical Imaging 29, 1310–1320 (2010). doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marcus DS et al. Open Access Series of Imaging Studies (OASIS): Cross-sectional MRI Data in Young, Middle Aged, Nondemented, and Demented Older Adults. Journal of Cognitive Neuroscience 19, 1498–1507 (2007). doi: 10.1162/jocn.2007.19.9.1498. [DOI] [PubMed] [Google Scholar]
- 37.Nooner KB et al. The NKI-Rockland Sample: A Model for Accelerating the Pace of Discovery Science in Psychiatry. Frontiers in Neuroscience 6 (2012). doi: 10.3389/fnins.2012.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reuter M, Rosas HD & Fischl B Highly accurate inverse consistent registration: A robust approach. NeuroImage 53, 1181–1196 (2010). doi: 10.1016/j.neuroimage.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dale AM, Fischl B & Sereno MI Cortical Surface-Based Analysis: I. Segmentation and Surface Reconstruction. NeuroImage 9, 179–194 (1999). doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- 40.Klein A et al. Mindboggling morphometry of human brains. PLOS Computational Biology 13, e1005350 (2017). doi: 10.1371/journal.pcbi.1005350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fonov V, Evans A, McKinstry R, Almli C & Collins D Unbiased nonlinear average age-appropriate brain templates from birth to adulthood. NeuroImage 47, Supplement 1, S102 (2009). doi: 10.1016/S1053-8119(09)70884-5. [DOI] [Google Scholar]
- 42.Avants B, Epstein C, Grossman M & Gee J Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Medical Image Analysis 12, 26–41 (2008). doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klein A et al. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. NeuroImage 46, 786–802 (2009). doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Brady M & Smith S Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Transactions on Medical Imaging 20, 45–57 (2001). doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- 45.Jenkinson M, Bannister P, Brady M & Smith S Improved Optimization for the Robust and Accurate Linear Registration and Motion Correction of Brain Images. NeuroImage 17, 825–841 (2002). doi: 10.1006/nimg.2002.1132. [DOI] [PubMed] [Google Scholar]
- 46.Oakes TR et al. Comparison of fMRI motion correction software tools. NeuroImage 28, 529–543 (2005). doi: 10.1016/j.neuroimage.2005.05.058. [DOI] [PubMed] [Google Scholar]
- 47.Greve DN & Fischl B Accurate and robust brain image alignment using boundary-based registration. NeuroImage 48, 63–72 (2009). doi: 10.1016/j.neuroimage.2009.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lanczos C Evaluation of Noisy Data. Journal of the Society for Industrial and Applied Mathematics Series B Numerical Analysis 1, 76–85 (1964). doi: 10.1137/0701007. [DOI] [Google Scholar]
- 49.Wang S et al. Evaluation of Field Map and Nonlinear Registration Methods for Correction of Susceptibility Artifacts in Diffusion MRI. Frontiers in Neuroinformatics 11 (2017). doi: 10.3389/fninf.2017.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McIntosh S, Kamei Y, Adams B & Hassan AE The Impact of Code Review Coverage and Code Review Participation on Software Quality: A Case Study of the Qt, VTK, and ITK Projects. In Proceedings of the 11th Working Conference on Mining Software Repositories, MSR 2014, 192–201 (ACM, New York, NY, USA, 2014). doi: 10.1145/2597073.2597076. [DOI] [Google Scholar]
- 51.Gorgolewski KJ et al. BIDS Apps: Improving ease of use, accessibility, and reproducibility of neuroimaging data analysis methods. PLOS Computational Biology 13, e1005209 (2017). doi: 10.1371/journal.pcbi.1005209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beaulieu-Jones BK & Greene CS Reproducibility of computational workflows is automated using continuous analysis. Nature Biotechnology 35, 342 (2017). doi: 10.1038/nbt.3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kurtzer GM, Sochat V & Bauer MW Singularity: Scientific containers for mobility of compute. PLOS ONE 12, e0177459 (2017). doi: 10.1371/journal.pone.0177459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schonberg T et al. Decreasing Ventromedial Prefrontal Cortex Activity During Sequential Risk-Taking: An fMRI Investigation of the Balloon Analog Risk Task. Frontiers in Neuroscience 6 (2012). doi: 10.3389/fnins.2012.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aron AR, Gluck MA & Poldrack RA Long-term test-retest reliability of functional MRI in a classification learning task. NeuroImage 29, 1000–1006 (2006). doi: 10.1016/j.neuroimage.2005.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xue G & Poldrack RA The Neural Substrates of Visual Perceptual Learning of Words: Implications for the Visual Word Form Area Hypothesis. Journal of Cognitive Neuroscience 19, 1643–1655 (2007). doi: 10.1162/jocn.2007.19.10.1643. [DOI] [PubMed] [Google Scholar]
- 57.Tom SM, Fox CR, Trepel C & Poldrack RA The Neural Basis of Loss Aversion in Decision-Making Under Risk. Science 315, 515–518 (2007). doi: 10.1126/science.1134239. [DOI] [PubMed] [Google Scholar]
- 58.Xue G, Aron AR & Poldrack RA Common Neural Substrates for Inhibition of Spoken and Manual Responses. Cerebral Cortex 18, 1923–1932 (2008). doi: 10.1093/cercor/bhm220. [DOI] [PubMed] [Google Scholar]
- 59.Aron AR, Behrens TE, Smith S, Frank MJ & Poldrack RA Triangulating a Cognitive Control Network Using Diffusion-Weighted Magnetic Resonance Imaging (MRI) and Functional MRI. Journal of Neuroscience 27, 3743–3752 (2007). doi: 10.1523/JNEUROSCI.051907.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Foerde K, Knowlton BJ & Poldrack RA Modulation of competing memory systems by distraction. Proceedings of the National Academy of Sciences 103, 11778–11783 (2006). doi: 10.1073/pnas.0602659103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gorgolewski KJ, Durnez J & Poldrack RA Preprocessed Consortium for Neuropsychiatric Phenomics dataset. F1000Research 6, 1262 (2017). doi: 10.12688/f1000research.11964.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Laumann TO et al. Functional System and Areal Organization of a Highly Sampled Individual Human Brain. Neuron 87, 657–670 (2015). doi: 10.1016/j.neuron.2015.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alvarez R, Jasdzewski G & Poldrack RA Building memories in two languages: an fMRI study of episodic encoding in bilinguals. In SfN Neuroscience (Orlando, FL, US, 2002). [Google Scholar]
- 64.Poldrack RA et al. Interactive memory systems in the human brain. Nature 414, 546–550 (2001). doi: 10.1038/35107080. [DOI] [PubMed] [Google Scholar]
- 65.Kelly AMC, Uddin LQ, Biswal BB, Castellanos FX & Milham MP Competition between functional brain networks mediates behavioral variability. NeuroImage 39, 527–537 (2008). doi: 10.1016/j.neuroimage.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 66.Mennes M et al. Inter-individual differences in resting-state functional connectivity predict task-induced BOLD activity. NeuroImage 50, 1690–1701 (2010). doi: 10.1016/j.neuroimage.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mennes M et al. Linking inter-individual differences in neural activation and behavior to intrinsic brain dynamics. NeuroImage 54, 2950–2959 (2011). doi: 10.1016/j.neuroimage.2010.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Haxby JV et al. Distributed and Overlapping Representations of Faces and Objects in Ventral Temporal Cortex. Science 293, 2425–2430 (2001). doi: 10.1126/science.1063736. [DOI] [PubMed] [Google Scholar]
- 69.Hanson SJ, Matsuka T & Haxby JV Combinatorial codes in ventral temporal lobe for object recognition: Haxby (2001) revisited: is there a face area? NeuroImage 23, 156–166 (2004). doi: 10.1016/j.neuroimage.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 70.Duncan KJ, Pattamadilok C, Knierim I & Devlin JT Consistency and variability in functional localisers. NeuroImage 46, 1018–1026 (2009). doi: 10.1016/j.neuroimage.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wager TD, Davidson ML, Hughes BL, Lindquist MA & Ochsner KN Prefrontal-Subcortical Pathways Mediating Successful Emotion Regulation. Neuron 59, 1037–1050 (2008). doi: 10.1016/j.neuron.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moran JM, Jolly E & Mitchell JP Social-Cognitive Deficits in Normal Aging. Journal of Neuroscience 32, 5553–5561 (2012). doi: 10.1523/JNEUROSCI.5511-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Uncapher MR, Hutchinson JB & Wagner AD Dissociable Effects of Top-Down and Bottom-Up Attention during Episodic Encoding. Journal of Neuroscience 31, 12613–12628 (2011). doi: 10.1523/JNEUROSCI.0152-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gorgolewski KJ et al. A test-retest fMRI dataset for motor, language and spatial attention functions. GigaScience 2, 1–4 (2013). doi: 10.1186/2047-217X-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Repovs G & Barch DM Working memory related brain network connectivity in individuals with schizophrenia and their siblings. Frontiers in Human Neuroscience 6 (2012). doi: 10.3389/fnhum.2012.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Repovs G, Csernansky JG & Barch DM Brain Network Connectivity in Individuals with Schizophrenia and Their Siblings. Biological Psychiatry 69, 967–973 (2011). doi: 10.1016/j.biopsych.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Walz JM et al. Simultaneous EEG-fMRI Reveals Temporal Evolution of Coupling between Supramodal Cortical Attention Networks and the Brainstem. Journal of Neuroscience 33, 19212–19222 (2013). doi: 10.1523/JNEUROSCI.2649-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Walz JM et al. Simultaneous EEG-fMRI reveals a temporal cascade of task-related and default-mode activations during a simple target detection task. NeuroImage 102, 229–239 (2014). doi: 10.1016/j.neuroimage.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Conroy BR, Walz JM & Sajda P Fast Bootstrapping and Permutation Testing for Assessing Reproducibility and Interpretability of Multivariate fMRI Decoding Models. PLOS ONE 8, e79271 (2013). doi: 10.1371/journal.pone.0079271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Walz JM et al. Prestimulus EEG alpha oscillations modulate task-related fMRI BOLD responses to auditory stimuli. NeuroImage 113, 153 163 (2015). doi: 10.1016/j.neuroimage.2015.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Velanova K, Wheeler ME & Luna B Maturational Changes in Anterior Cingulate and Frontoparietal Recruitment Support the Development of Error Processing and Inhibitory Control. Cerebral Cortex 18, 2505–2522 (2008). doi: 10.1093/cercor/bhn012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Padmanabhan A, Geier CF, Ordaz SJ, Teslovich T & Luna B Developmental changes in brain function underlying the influence of reward processing on inhibitory control. Developmental Cognitive Neuroscience 1, 517–529 (2011). doi: 10.1016/j.dcn.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Geier CF, Terwilliger R, Teslovich T, Velanova K & Luna B Immaturities in Reward Processing and Its Influence on Inhibitory Control in Adolescence. Cerebral Cortex 20, 1613–1629 (2010). doi: 10.1093/cercor/bhp225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cera N, Tartaro A & Sensi SL Modafinil Alters Intrinsic Functional Connectivity of the Right Posterior Insula: A Pharmacological Resting State fMRI Study. PLOS ONE 9, e107145 (2014). doi: 10.1371/journal.pone.0107145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Woo C-W, Roy M, Buhle JT & Wager TD Distinct Brain Systems Mediate the Effects of Nociceptive Input and Self-Regulation on Pain. PLOS Biology 13, e1002036 (2015). doi: 10.1371/journal.pbio.1002036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Smeets PAM, Kroese FM, Evers C & de Ridder DTD Allured or alarmed: Counteractive control responses to food temptations in the brain. Behavioural Brain Research 248, 41–45 (2013). doi: 10.1016/j.bbr.2013.03.041. [DOI] [PubMed] [Google Scholar]
- 87.Pernet CR et al. The human voice areas: Spatial organization and inter-individual variability in temporal and extra-temporal cortices. NeuroImage 119, 164–174 (2015). doi: 10.1016/j.neuroimage.2015.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Verstynen TD The organization and dynamics of corticostriatal pathways link the medial orbitofrontal cortex to future behavioral responses. Journal of Neurophysiology 112, 2457–2469 (2014). doi: 10.1152/jn.00221.2014. [DOI] [PubMed] [Google Scholar]
- 89.Bursley JK, Nestor A, Tarr MJ & Creswell JD Awake, Offline Processing during Associative Learning. PLOS ONE 11, e0127522 (2016). doi: 10.1371/journal.pone.0127522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ella G, David M & Avi K Learning from the other limb’s experience: sharing the trained M1 representation of the motor sequence knowledge. The Journal of Physiology 594, 169–188 (2015). doi: 10.1113/JP270184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gabitov E, Manor D & Karni A Patterns of Modulation in the Activity and Connectivity of Motor Cortex during the Repeated Generation of Movement Sequences. Journal of Cognitive Neuroscience 27, 736–751 (2014). doi: 10.1162/jocn_a_00751. [DOI] [PubMed] [Google Scholar]
- 92.Gabitov E, Manor D & Karni A Done That: Short-term Repetition Related Modulations of Motor Cortex Activity as a Stable Signature for Overnight Motor Memory Consolidation. Journal of Cognitive Neuroscience 26, 2716–2734 (2014). doi: 10.1162/jocn_a_00675. [DOI] [PubMed] [Google Scholar]
- 93.Lepping RJ, Atchley RA & Savage CR Development of a validated emotionally provocative musical stimulus set for research. Psychology of Music 44, 1012–1028 (2016). doi: 10.1177/0305735615604509. [DOI] [Google Scholar]
- 94.Park C-A & Kang C-K Sensing the effects of mouth breathing by using 3-tesla MRI. Journal of the Korean Physical Society 70, 1070–1076 (2017). doi: 10.3938/jkps.70.1070. [DOI] [Google Scholar]
- 95.Iannilli E et al. Effects of Manganese Exposure on Olfactory Functions in Teenagers: A Pilot Study. PLOS ONE 11, e0144783 (2016). doi: 10.1371/journal.pone.0144783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim J, Wang J, Wedell DH & Shinkareva SV Identifying Core Affect in Individuals from fMRI Responses to Dynamic Naturalistic Audiovisual Stimuli. PLOS ONE 11, e0161589 (2016). doi: 10.1371/journal.pone.0161589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tétreault P et al. Brain Connectivity Predicts Placebo Response across Chronic Pain Clinical Trials. PLOS Biology 14, e1002570 (2016). doi: 10.1371/journal.pbio.1002570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chakroff A et al. When minds matter for moral judgment: intent information is neurally encoded for harmful but not impure acts. Social Cognitive and Affective Neuroscience 11, 476–484 (2016). doi: 10.1093/scan/nsv131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Koster-Hale J, Saxe R, Dungan J & Young LL Decoding moral judgments from neural representations of intentions. Proceedings of the National Academy of Sciences 110, 5648–5653 (2013). doi: 10.1073/pnas.1207992110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gao X et al. My Body Looks Like That Girls: Body Mass Index Modulates Brain Activity during Body Image Self-Reflection among Young Women. PLOS ONE 11, e0164450 (2016). doi: 10.1371/journal.pone.0164450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Romaniuk L, Pope M, Nicol K, Steele D & Hall J Neural correlates of fears of abandonment and rejection in borderline personality disorder. Wellcome Open Research 1, 33 (2016). doi: 10.12688/wellcomeopenres.10331.1. [DOI] [Google Scholar]
- 102.Cohen AD, Nencka AS, Lebel RM & Wang Y Multiband multi-echo imaging of simultaneous oxygenation and flow timeseries for resting state connectivity. PLOS ONE 12, e0169253 (2017). doi: 10.1371/journal.pone.0169253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dalenberg JR, Weitkamp L, Renken RJ, Nanetti L & Horst G. J. t. Flavor pleasantness processing in the ventral emotion network. PLOS ONE 12, e0170310 (2017). doi: 10.1371/journal.pone.0170310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Roy A et al. The evolution of cost-efficiency in neural networks during recovery from traumatic brain injury. PLOS ONE 12, e0170541 (2017). doi: 10.1371/journal.pone.0170541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gordon EM et al. Precision Functional Mapping of Individual Human Brains. Neuron 95, 791–807.e7 (2017). doi: 10.1016/j.neuron.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Veldhuizen MG et al. Integration of Sweet Taste and Metabolism Determines Carbohydrate Reward. Current Biology 27, 2476–2485.e6 (2017). doi: 10.1016/j.cub.2017.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Greene DJ et al. Behavioral interventions for reducing head motion during MRI scans in children. NeuroImage 171, 234–245 (2018). doi: 10.1016/j.neuroimage.2018.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nastase SA et al. Attention Selectively Reshapes the Geometry of Distributed Semantic Representation. Cerebral Cortex 27, 4277–4291 (2017). doi: 10.1093/cercor/bhx138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kanazawa Y et al. Phonological memory in sign language relies on the visuomotor neural system outside the left hemisphere language network. PLOS ONE 12, e0177599 (2017). doi: 10.1371/journal.pone.0177599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


