Abstract
Adaptive laboratory evolution (ALE) has emerged as a powerful tool in basic microbial research and strain development. In the context of metabolic science and engineering, it has been applied to study gene knockout responses, expand substrate ranges, improve tolerance to process conditions, and to improve productivity via designed growth coupling. In recent years, advancements in ALE methods and systems biology measurement technologies, particularly genome sequencing and 13C metabolic flux analysis (13C-MFA), have enabled detailed study of the mechanisms and dynamics of evolving metabolism. In this review, we discuss a range of studies that have applied flux analysis to adaptively evolved strains, as well as modeling frameworks developed to predict and interpret evolved fluxes. These efforts link mutations to fitness-enhanced phenotypes, identify bottlenecks and approaches to resolve them, and address systems concepts such as optimality.
Keywords: Adaptive evolution, metabolism, flux analysis, fast growth, genotype-phenotype relationship
INTRODUCTION
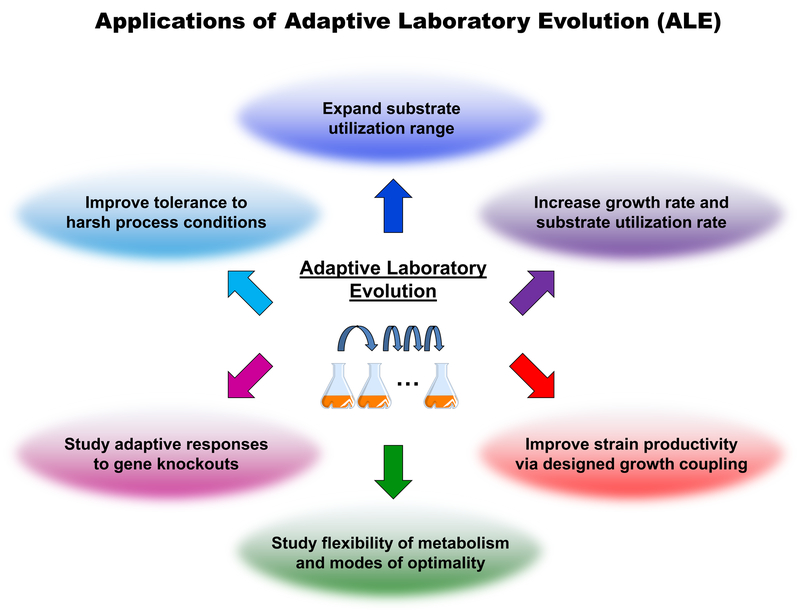
Evolution is a defining characteristic of biological systems, and along with codability and molecular precision is a key attribute in the engineering of biology for applications like biochemical conversion [1]. Harnessing the mechanisms of evolution, adaptation, and selection for developing complex desired phenotypes is particularly valuable when, as is often the case, limited understanding precludes fully rational approaches. In such efforts, an organism is cultured in a condition of interest for many generations, and fitness (typically growth rate) is often improved as beneficial mutations are selected for and accumulate. This approach, called adaptive laboratory evolution (ALE) has been applied to enhance fitness in non-standard environments, including resistance to toxic solvents and other (by)products that inhibit industrial fermentation [2–6], and elevated temperature [7–10], which may be desirable to reduce downstream separations cost or reduce contamination risk (Figure 1).
Figure 1.
Applications of adaptive laboratory evolution (ALE) in strain development for biomanufacturing and in basic science research.
Metabolism is of central interest to biochemical engineering applications such as biocatalysis and metabolic engineering. In the context of metabolism, adaptive evolution is often employed to improve fitness in new environmental (e.g., substrate) or genetic contexts (e.g., knockout of a previously utilized pathway). Significant changes are typically observed, as most organisms are not fully optimized for any one condition. For example, even on its preferred substrate (glucose) E. coli has been shown to evolve to increase its growth rate 1.6-fold [11,12]. ALE has generated large fitness improvements for many other substrates, and even established growth on non-native substrates like 1,2-propanediol and citrate [13,14]. Such efforts are a powerful way to expand the substrate repertoire and efficiency of chassis strains for bioprocessing [15,16]. ALE has also been applied to strains in conjunction with (typically following) genetic engineering, such as gene knockouts. These studies can provide fundamental insight into the flexibility of metabolism and various modes of optimality, or can be involved in targeted strain design efforts. For example, the design algorithm OptKnock [17] can be used to couple growth and production such that fitness improvements via ALE necessarily lead to increased production rates.
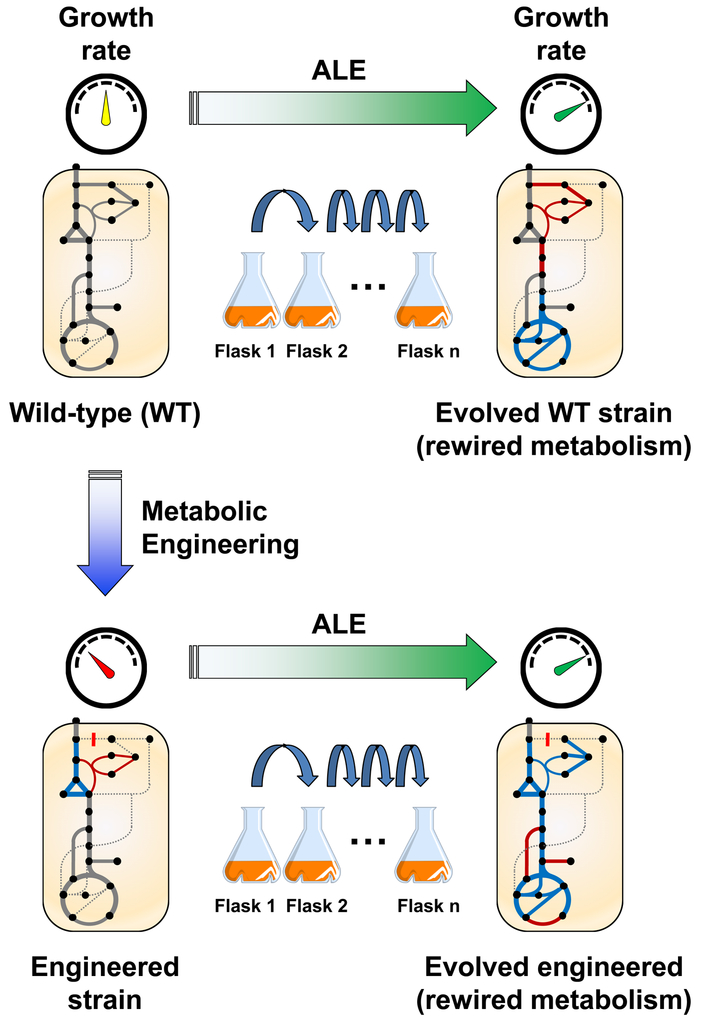
The results from ALE efforts are most informative when the mechanisms of fitness improvement are elucidated by combined genetic and phenotypic analysis. Sequencing of the end-point strains identifies the mutations that have accumulated, and then the mechanisms of improvement can be studied via the relevant systems measurements. Causality can be demonstrated by recapitulating the mutations in the starting strain, or inferred by frequency across a larger number of replicates (i.e. distinguished from random “hitch-hiker” mutations) [11]. In the context of metabolism, the phenotypic outcome of interest is often the changes in metabolic rates or fluxes (Figure 2). The simplest of these to measure are external rates, including substrate uptake, product secretion, and growth rates. These provide limited visibility into changes in intracellular pathway usage, which are resolved using isotopic (e.g. 13C) tracers and metabolic flux analysis (13C-MFA) [18,19]. There is a myriad of in silico approaches for predicting evolved fluxes, mostly around the idea of constrained optimization. In this review we will summarize the state of metabolic flux analysis in evolved systems and highlight promising areas for future study.
Figure 2.
In adaptive laboratory evolution (ALE) an organism is cultured for many generations under specific conditions of interest. In the process, the fitness (typically growth rate) is improved as beneficial mutations are selected for and accumulate, and metabolism is rewired to facilitate the enhanced phenotype.
MODELING FRAMEWORKS
The basis of the predominant family of metabolic models, constraint-based reconstruction and analysis (COBRA), is tightly associated with evolutionary principles. An annotated genome is used to identify the available metabolic reaction network, and thus the set of possible flux solutions consistent with this stoichiometry. Within this solution space, specific predictions can be made by optimization of an ‘objective function’, a method known as flux balance analysis (FBA) [20]. The biological relevance and accuracy of various objective functions have been widely debated [21,22]. The most common is the biomass objective function, which states that the cell will maximize its growth subject to the stoichiometric and substrate uptake limitations. This is justified as reflecting the result of natural evolution, which would select for optimal fitness, i.e. growth [23]. However, the natural fitness landscape can be different and significantly more complex than that in controlled laboratory conditions. For example, in its natural life cycle E. coli contends with wildly changing temperature, pH, and nutrient conditions as it traverses the mammalian gut, moves between hosts, and spends part of its life cycle in open environments. Biomass optimization is expected to be a more robust assumption following ALE in controlled conditions, and has indeed been usefully applied to assess the outcome of ALE on a wide range of diverse substrates including glucose, glycerol, acetate, succinate, malate, α-ketoglutarate, lactate, and pyruvate [24,25]. When assessing fitness trajectories from ALE with FBA, alternative optima must be considered [26], as in many cases different metabolic strategies can achieve similar levels of fitness.
The evolutionary justifications of FBA have long been recognized to carry less weight when predicting fluxes immediately following genetic perturbations [27], as these can introduce significant unnatural constraints on metabolism. Predicting these responses, and those after subsequent adaptive evolution, has been an area of major research interest. Such models provide conceptual frameworks for interpreting experimental measurements with respect to metabolic bottlenecks, regulatory interactions, and flexibility. In engineering practice, they are used in strain design [17]. In addition to FBA, several alternative models have been proposed for predicting fluxes following genetic perturbation and ALE. Minimization of metabolic adjustment (MOMA) [27] minimizes the sum of squares difference (Euclidean distance) between wild-type and perturbed (e.g., gene knockout) strain. This was hypothesized to reflect the initial, unevolved response. Regulatory on/off minimization (ROOM) [28] instead uses the Hamming distance, which minimizes the number of significant flux changes (favoring fewer, larger changes rather than a large number of small adjustments). This was hypothesized to reflect the regulatory adjustments made during ALE, and along with FBA was expected to provide more useful predictions for the evolved phenotype. Another approach, termed RELATCH for relative change [29], implements different assumptions to make flux predictions prior to and after ALE. For the unevolved responses, RELATCH limits latent pathway activation and large increases in enzyme usage. For post-ALE predictions, these restrictions are relaxed, allowing for significant regulatory adjustment and flux rewiring.
MEASURING EVOLVED METABOLISM VIA EXTERNAL FLUXES
The most accessible measures of metabolic phenotype are the external fluxes, including substrate uptake rates and product secretion rates. These provide information on the system performance, but limited visibility into mechanisms of flux change and fitness improvement. Several notable early studies compared external flux measurements to FBA predictions in evolved systems. One performed ALE with E. coli on 5 different substrates (glucose, acetate, succinate, malate, and glycerol) [25], while a second used three substrates (α-ketoglutarate, pyruvate, and lactate) at both 30 °C and 37 °C [24]. Both used FBA approaches to identify “lines of optimality” (LO), where the optimal respective rates of carbon substrate uptake and O2 uptake are predicted. In most cases, evolved strains “converged” to the LO as predicted. Discrepancies in the predicted and measured performance during growth on pyruvate were suggested to have resulted from missing reactions in the model. In another study, six different E. coli knockout strains (of major metabolic genes ackA, frd, pck, ppc, tpi, and zwf) were evolved on 4 different substrates [30]. FBA accurately predicted the final growth phenotype within 10% for 78% (39/50) of the endpoint strains tested. These results collectively gave support to the ideas of constrained optimality in evolved systems. For more detailed understanding of the mechanisms of evolution in metabolism, including whether the optimal solutions are convergent or multiple, observation of intracellular fluxes was needed.
MEASURING INTRACELLULAR FLUXES VIA 13C-MFA
A landmark study of metabolic fluxes of evolved E. coli knockout strains was published in 2006 [31]. Stable-isotope 13C glucose tracers were used to estimate the intracellular fluxes of the unevolved parent and two evolved descendants each of Δpgi, Δppc, Δpta, and Δtpi. Three of these four knockouts caused dramatic reductions in growth rates (from 0.63 h−1 in the wild-type to less than 0.23 h−1), but growth was recovered to near-wild-type rates (0.5–0.6 h−1) in all cases. The flux results revealed several notable effects, for example the expression-driven increase in the normally latent methylglyoxal pathway in Δtpi. In Δpgi, the replicate evolved strains achieved similar growth rates with significant differences in pathway usage. All of these results also informed an interpretation of transient and evolved perturbation responses that focused on latent pathway activation and subsequent re-repression [32]. For example, an active PCK (gluconeogenic reaction from OAC to PEP) reaction was observed in unevolved Δpgi and Δtpi, which was reduced on a relative basis in the evolved strains. As this reaction is not optimal for growth on glucose (it causes a futile cycle), it makes sense that its activity would be repressed by evolution. Similar observations were made in some cases for the Entner-Doudoroff (ED) pathway and glyoxylate shunt.
More recently, additional 13C-MFA studies of ALE strains have called this interpretation into question and led to new insights. A set of 10 evolved Δpgi strains, originally described with respect to external rates and mutations in 2010 [33], was reassessed for intracellular fluxes [34]. The number of replicate strains in this study was valuable for identifying frequently mutated genes to guide investigations into mechanism. The previously reported increase and subsequent decrease in relative flux through the ED, PCK, and glyoxylate shunt pathways was observed again. However, these were shown to be artifacts of the dramatic changes in absolute glucose uptake rate; the absolute fluxes (mmol/gDW/h) were not significantly different in the wild-type, unevolved, or evolved strains. Rather than regulatory responses impacting expression and flux capacity of these pathways, this result indicated that such latent pathways are likely constitutively active at low levels to provide flexibility in changing environments. Another observation was frequent (8/10 strains) mutations to the pyridine cofactor transhydrogenase genes, which corresponded to large changes to the corresponding flux. In the wild-type, excess NADH is converted to NADPH by PntAB, but by forcing flux through the NADPH-producing oxidative pentose phosphate pathway (oxPPP), the pgi knockout necessitates a reversal of this flux (NADPH to NADH by SthA). Frequent mutations in the PTS (glucose transport complex) subunit crr, which is also involved in gene regulation via the global regulator Crp that controls the transcription of over 100 genes, suggested new questions for further study.
While fluxes may be expected to shift to facilitate recovery of severely perturbed strains, what about wild-type strains evolved for faster growth? Even on its preferred substrate, glucose, E. coli increases its growth rate by up to 1.6-fold following ALE [11]. A follow-up study showed that, in fact, uptake-normalized fluxes (i.e., pathway usage) did not appreciably change in six evolved wild-type E. coli MG1655 strains. The differences between them was much less than that between WT MG1655 and other E. coli WT strains, particularly BL21 [35]. This result is consistent with a broad and proportional increase in the activity of metabolic enzymes, and reflects that wild-type metabolism already allocates flux through its pathways in an efficient manner. For both wild-type and knockout strains, the interplay of global regulators and metabolism is a promising area of ongoing investigation. For example, a ubiquitous mutation in the RNAP subunit rpoB was shown to cause a broad change in gene expression partially responsible for the faster metabolism in WT ALE strains [36]. In other cases, evolved phenotypes of interest such as co-utilization of glucose and xylose in Thermus thermophilus have been understood by mapping fluxes via 13C-MFA [37].
FUTURE DIRECTIONS AND OPPORTUNITIES
Adaptive evolution coupled with metabolic analysis is clearly a powerful approach for basic investigation into metabolism, including for hypothesis generation, as well as for gleaning specific and useful knowledge for biotechnology. For example, a recent ALE study of an E. coli Δzwf strain with a blocked oxPPP, mutations were observed that resulted in increased transhydrogenase activity opposite to those observed in the Δpgi-ALE study (NADPH-generating) [38]. These mutations observed across both studies could provide additional tools for metabolic engineering where cofactor balancing is often a crucial consideration. In these examples, transhydrogenase activity apparently was significantly rate-limiting to the unevolved mutants. By using this approach across other mutants or in other conditions, nature’s solutions to other types of rate limitations could be elucidated, providing a wealth of new targets for the metabolic engineering toolbox.
For these reasons, it would be useful to undertake a more complete and systematic approach to characterizing metabolic responses to ALE. These could include more knockouts in central carbon metabolism [39], global regulators, or secondary pathways of interest [40]. Recent studies of gene knockout phenotypes provide a framework for selecting targets [41], such as the slowest growing which would be expected to undergo the most dramatic changes. In addition to providing insights into rate limitations, these efforts could reveal the nature and limits of other unnatural modes of metabolism such as if whole pathways are blocked or restricted. Generating high-performance strains with unique pathway usage, e.g. “no TCA cycle” or “ED pathway-exclusive”, would be interesting for both testing the limits of in silico prediction and as novel chassis strains.
Further study of evolved wild-type strains will also continue to provide valuable insights into biological limitations and engineering efficient chassis organisms. Comparing the fluxes, mutations, and other ‘omics’ of other evolved E. coli strains, such as the aforementioned BL21, will reveal whether all strains ‘converge’ to a single optima or evolve toward distinct high-performance states. This can be extended to other substrates and conditions of industrial interest as well. For example, efforts to engineer E. coli to consume alternative substrates like methanol or CO2 will likely involve evolution, and 13C-MFA is likely to be an important part of the design-build-test cycle [42–46]. Comparison to other organisms with distinct environmental niches and evolutionary histories will also be quite interesting. For example, are the marine strain Vibrio natriegens [47,48] or the thermophile Geobacillus LC300 [49,50] able to further improve upon their exceptional rates of growth, or are they already ‘optimal’ for growth on glucose? Are these microbes less adaptable than E. coli to alternative substrates or harsher culture conditions?
Lastly, creative variations on the ALE experimental process itself may reveal additional nuances. More complicated fitness landscapes can be explored, for example, by alternating the conditions such as substrate [51] or other variables such as temperature [8]. In the case of alternating between glucose and a second substrate (xylose, glycerol, or acetate), different strategies emerged: in some cases a single “generalist” strain emerged, while in others two “specialist” strains persisted [51]. Such experimental designs have rich potential for exploring ecologically relevant scenarios. Another possibility for manipulating the experimental design is to change the order of ALE and genetic perturbation. If a wild-type is evolved first for fast growth on glucose, will the nature of its knockout responses change [52–56]? How does that strain compare to the reverse, original case (knockout then ALE), and what if a second round of ALE was performed subsequently to the knockout? The path and context-dependency of evolution would be explored in these types of studies, with potential to add a significant layer of understanding.
CONCLUSIONS
Measuring, modeling, and understanding evolved metabolic systems is an important and fruitful area of research. The continued application of 13C-MFA and whole genome sequencing to understand the mechanisms of adaptive evolution will generate biological hypotheses, identify engineering targets, and provide fundamental insights into how evolution occurs and how metabolism functions. The ability to explain causal mutations is a useful test of biological knowledge, and offers clear guidance for follow-up investigation to close knowledge gaps. Diverse types of insights are possible depending on the purpose and design of evolution experiments. We propose that further study of fluxes in adaptively evolved strains, particularly following gene knockouts, would be a worthwhile effort for generating understanding of metabolic bottlenecks and potential engineering strategies.
ACKNOWLEDGEMENTS
This work was supported by NSF MCB-1616332 grant.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
Papers of special interest (*)
Papers of outstanding interest (**)
- 1.Woolston BM, Edgar S, Stephanopoulos G: Metabolic engineering: past and future. Annu Rev Chem Biomol Eng 2013, 4:259–288. [DOI] [PubMed] [Google Scholar]
- 2.Atsumi S, Wu TY, Machado IM, Huang WC, Chen PY, Pellegrini M, Liao JC: Evolution, genomic analysis, and reconstruction of isobutanol tolerance in Escherichia coli. Mol Syst Biol 2010, 6:449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horinouchi T, Tamaoka K, Furusawa C, Ono N, Suzuki S, Hirasawa T, Yomo T, Shimizu H: Transcriptome analysis of parallel-evolved Escherichia coli strains under ethanol stress. BMC Genomics 2010, 11:579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lam FH, Ghaderi A, Fink GR, Stephanopoulos G: Biofuels. Engineering alcohol tolerance in yeast. Science 2014, 346:71–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mundhada H, Seoane JM, Schneider K, Koza A, Christensen HB, Klein T, Phaneuf PV, Herrgard M, Feist AM, Nielsen AT: Increased production of L-serine in Escherichia coli through Adaptive Laboratory Evolution. Metab Eng 2017, 39:141–150. [DOI] [PubMed] [Google Scholar]
- 6.Reyes LH, Almario MP, Winkler J, Orozco MM, Kao KC: Visualizing evolution in real time to determine the molecular mechanisms of n-butanol tolerance in Escherichia coli. Metab Eng 2012, 14:579–590. [DOI] [PubMed] [Google Scholar]
- 7.Caspeta L, Chen Y, Ghiaci P, Feizi A, Buskov S, Hallstrom BM, Petranovic D, Nielsen J: Biofuels. Altered sterol composition renders yeast thermotolerant. Science 2014, 346:75–78. [DOI] [PubMed] [Google Scholar]
- 8.Deatherage DE, Kepner JL, Bennett AF, Lenski RE, Barrick JE: Specificity of genome evolution in experimental populations of Escherichia coli evolved at different temperatures. Proc Natl Acad Sci U S A 2017, 114:E1904–E1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandberg TE, Pedersen M, LaCroix RA, Ebrahim A, Bonde M, Herrgard MJ, Palsson BO, Sommer M, Feist AM: Evolution of Escherichia coli to 42 degrees C and subsequent genetic engineering reveals adaptive mechanisms and novel mutations. Mol Biol Evol 2014, 31:2647–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tenaillon O, Rodriguez-Verdugo A, Gaut RL, McDonald P, Bennett AF, Long AD, Gaut BS: The molecular diversity of adaptive convergence. Science 2012, 335:457–461. [DOI] [PubMed] [Google Scholar]
- 11.LaCroix RA, Sandberg TE, O’Brien EJ, Utrilla J, Ebrahim A, Guzman GI, Szubin R, Palsson BO, Feist AM: Use of adaptive laboratory evolution to discover key mutations enabling rapid growth of Escherichia coli K-12 MG1655 on glucose minimal medium. Appl Environ Microbiol 2015, 81:17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sandberg TE, Long CP, Gonzalez JE, Feist AM, Antoniewicz MR, Palsson BO: Evolution of E. coli on [U-13C]Glucose Reveals a Negligible Isotopic Influence on Metabolism and Physiology. PLoS One 2016, 11:e0151130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blount ZD, Borland CZ, Lenski RE: Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc Natl Acad Sci U S A 2008, 105:7899–7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee DH, Palsson BO: Adaptive evolution of Escherichia coli K-12 MG1655 during growth on a Nonnative carbon source, L-1,2-propanediol. Appl Environ Microbiol 2010, 76:4158–4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papapetridis I, Verhoeven MD, Wiersma SJ, Goudriaan M, van Maris AJA, Pronk JT: Laboratory evolution for forced glucose-xylose co-consumption enables identification of mutations that improve mixed-sugar fermentation by xylose-fermenting Saccharomyces cerevisiae. FEMS Yeast Res 2018, 18.* Rational strain angineering, adapative laboratory evolution and genome sequencing were used to improve the simultanoeus co-utilization of glucose and xylose by Saccharomyces cerevisiae.
- 16.LaCroix RA, Palsson BO, Feist AM: A Model for Designing Adaptive Laboratory Evolution Experiments. Appl Environ Microbiol 2017, 83.* This paper describes the computational platform ALEsim that can be used to better design adaptive laboratory evolution experiments. Passage size was identified as a key parameter be optimized to balance the outcomes of experiments with available resources.
- 17.Burgard AP, Pharkya P, Maranas CD: Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol Bioeng 2003, 84:647–657. [DOI] [PubMed] [Google Scholar]
- 18.Antoniewicz MR: Parallel labeling experiments for pathway elucidation and 13C metabolic flux analysis. Curr Opin Biotechnol 2015, 36:91–97. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez JE, Antoniewicz MR: Tracing metabolism from lignocellulosic biomass and gaseous substrates to products with stable-isotopes. Curr Opin Biotechnol 2017, 43:86–95. [DOI] [PubMed] [Google Scholar]
- 20.Edwards JS, Palsson BO: The Escherichia coli MG1655 in silico metabolic genotype: its definition, characteristics, and capabilities. Proc Natl Acad Sci U S A 2000, 97:5528–5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia Sanchez CE, Torres Saez RG: Comparison and analysis of objective functions in flux balance analysis. Biotechnol Prog 2014, 30:985–991. [DOI] [PubMed] [Google Scholar]
- 22.Schuetz R, Kuepfer L, Sauer U: Systematic evaluation of objective functions for predicting intracellular fluxes in Escherichia coli. Mol Syst Biol 2007, 3:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edwards JS, Covert M, Palsson B: Metabolic modelling of microbes: the flux-balance approach. Environ Microbiol 2002, 4:133–140. [DOI] [PubMed] [Google Scholar]
- 24.Fong SS, Marciniak JY, Palsson BO: Description and interpretation of adaptive evolution of Escherichia coli K-12 MG1655 by using a genome-scale in silico metabolic model. J Bacteriol 2003, 185:6400–6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ibarra RU, Edwards JS, Palsson BO: Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature 2002, 420:186–189. [DOI] [PubMed] [Google Scholar]
- 26.Mahadevan R, Schilling CH: The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab Eng 2003, 5:264–276. [DOI] [PubMed] [Google Scholar]
- 27.Segre D, Vitkup D, Church GM: Analysis of optimality in natural and perturbed metabolic networks. Proc Natl Acad Sci U S A 2002, 99:15112–15117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shlomi T, Berkman O, Ruppin E: Regulatory on/off minimization of metabolic flux changes after genetic perturbations. Proc Natl Acad Sci U S A 2005, 102:7695–7700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J, Reed JL: RELATCH: relative optimality in metabolic networks explains robust metabolic and regulatory responses to perturbations. Genome Biol 2012, 13:R78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fong SS, Palsson BO: Metabolic gene-deletion strains of Escherichia coli evolve to computationally predicted growth phenotypes. Nat Genet 2004, 36:1056–1058. [DOI] [PubMed] [Google Scholar]
- 31.Fong SS, Nanchen A, Palsson BO, Sauer U: Latent pathway activation and increased pathway capacity enable Escherichia coli adaptation to loss of key metabolic enzymes. J Biol Chem 2006, 281:8024–8033. [DOI] [PubMed] [Google Scholar]
- 32.Cornelius SP, Lee JS, Motter AE: Dispensability of Escherichia coli’s latent pathways. Proc Natl Acad Sci U S A 2011, 108:3124–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charusanti P, Conrad TM, Knight EM, Venkataraman K, Fong NL, Xie B, Gao Y, Palsson BO: Genetic basis of growth adaptation of Escherichia coli after deletion of pgi, a major metabolic gene. PLoS Genet 2010, 6:e1001186.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Long CP, Gonzalez JE, Feist AM, Palsson BO, Antoniewicz MR: Dissecting the genetic and metabolic mechanisms of adaptation to the knockout of a major metabolic enzyme in Escherichia coli. Proc Natl Acad Sci U S A 2018, 115:222–227.** In this comprehensive study, whole-genome sequencing and 13C metabolic flux analysis were applied to ten adaptively evolved E. coli Δpgi strains to elucidate the genetic and metabolic mechanisms of adaptation to the deletion of a key glycolytic enzyme.
- 35.Long CP, Gonzalez JE, Feist AM, Palsson BO, Antoniewicz MR: Fast growth phenotype of E. coli K-12 from adaptive laboratory evolution does not require intracellular flux rewiring. Metab Eng 2017, 44:100–107.** Intracellular metabolic fluxes of six adaptively evolved E. coli strains and three different wild-type E. coli strains were quantified by 13C metabolic flux analysis. Interestingly, it was found that the relative metabolic pathway usage changed little following adaptive laboratory evolution.
- 36.Utrilla J, O’Brien EJ, Chen K, McCloskey D, Cheung J, Wang H, Armenta-Medina D, Feist AM, Palsson BO: Global Rebalancing of Cellular Resources by Pleiotropic Point Mutations Illustrates a Multi-scale Mechanism of Adaptive Evolution. Cell Syst 2016, 2:260–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cordova LT, Lu J, Cipolla RM, Sandoval NR, Long CP, Antoniewicz MR: Co-utilization of glucose and xylose by evolved Thermus thermophilus LC113 strain elucidated by C metabolic flux analysis and whole genome sequencing. Metab Eng 2016, 37:63–71.* The thermophilic bacterium Thermus thermophilus was evolved to simultaneously co-utilize glucose and xylose. Whole-genome sequencing identified causal mutations and 13C metabolic flux analysis was used to quantify the metabolism of the evolved strain.
- 38.Chou HH, Marx CJ, Sauer U: Transhydrogenase promotes the robustness and evolvability of E. coli deficient in NADPH production. PLoS Genet 2015, 11:e1005007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Long CP, Antoniewicz MR: Metabolic flux analysis of Escherichia coli knockouts: lessons from the Keio collection and future outlook. Curr Opin Biotechnol 2014, 28:127–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wytock TP, Fiebig A, Willett JW, Herrou J, Fergin A, Motter AE, Crosson S: Experimental evolution of diverse Escherichia coli metabolic mutants identifies genetic loci for convergent adaptation of growth rate. PLoS Genet 2018, 14:e1007284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Long CP, Gonzalez JE, Sandoval NR, Antoniewicz MR: Characterization of physiological responses to 22 gene knockouts in Escherichia coli central carbon metabolism. Metab Eng 2016, 37:102–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Antonovsky N, Gleizer S, Noor E, Zohar Y, Herz E, Barenholz U, Zelcbuch L, Amram S, Wides A, Tepper N, et al. : Sugar Synthesis from CO2 in Escherichia coli. Cell 2016, 166:115–125.** The authors expressed the non-native Calvin-Benson-Bassham cycle in E. coli and then adaptively evolved the strain. The resulting strain produced all carbohydrates and related metabolites in upper metabolism solely from CO2.
- 43.Gonzalez JE, Bennett RK, Papoutsakis ET, Antoniewicz MR: Methanol assimilation in Escherichia coli is improved by co-utilization of threonine and deletion of leucine-responsive regulatory protein. Metab Eng 2018, 45:67–74. [DOI] [PubMed] [Google Scholar]
- 44.Meyer F, Keller P, Hartl J, Groninger OG, Kiefer P, Vorholt JA: Methanol-essential growth of Escherichia coli. Nat Commun 2018, 9:1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whitaker WB, Jones JA, Bennett RK, Gonzalez JE, Vernacchio VR, Collins SM, Palmer MA, Schmidt S, Antoniewicz MR, Koffas MA, et al. : Engineering the biological conversion of methanol to specialty chemicals in Escherichia coli. Metab Eng 2017, 39:49–59. [DOI] [PubMed] [Google Scholar]
- 46.Woolston BM, King JR, Reiter M, Van Hove B, Stephanopoulos G: Improving formaldehyde consumption drives methanol assimilation in engineered E. coli. Nat Commun 2018, 9:2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Long CP, Gonzalez JE, Cipolla RM, Antoniewicz MR: Metabolism of the fast-growing bacterium Vibrio natriegens elucidated by 13C metabolic flux analysis. Metab Eng 2017, 44:191–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weinstock MT, Hesek ED, Wilson CM, Gibson DG: Vibrio natriegens as a fast-growing host for molecular biology. Nat Methods 2016, 13:849–851. [DOI] [PubMed] [Google Scholar]
- 49.Cordova LT, Antoniewicz MR: (13)C metabolic flux analysis of the extremely thermophilic, fast growing, xylose-utilizing Geobacillus strain LC300. Metab Eng 2016, 33:148–157. [DOI] [PubMed] [Google Scholar]
- 50.Cordova LT, Long CP, Venkataramanan KP, Antoniewicz MR: Complete genome sequence, metabolic model construction and phenotypic characterization of Geobacillus LC300, an extremely thermophilic, fast growing, xylose-utilizing bacterium. Metab Eng 2015, 32:74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sandberg TE, Lloyd CJ, Palsson BO, Feist AM: Laboratory Evolution to Alternating Substrate Environments Yields Distinct Phenotypic and Genetic Adaptive Strategies. Appl Environ Microbiol 2017, 83:e00410–00417.* In this study, E. coli was evolved under static and fluctuating environmental conditions by switching between glucose and other carbon sources (e.g. xylose). In some cases, a generalist strain emerged, while in another cases specialist subpopulations arose. The diauxic lag phenotypes varied widely across the generalists and specialists.
- 52.McCloskey D, Xu S, Sandberg TE, Brunk E, Hefner Y, Szubin R, Feist AM, Palsson BO: Multiple optimal phenotypes overcome redox and glycolytic intermediate metabolite imbalances in Escherichia coli pgi knockout evolutions. Appl Environ Microbiol 2018:AEM.00823–00818.** In a series of papers, the authors investigated the mechanisms of adaptation to the knockout of a several core metabolic enzymes in E. coli using a range of systems biology tools that included whole-genome sequencing and 13C metabolic flux analysis.
- 53.McCloskey D, Xu S, Sandberg TE, Brunk E, Hefner Y, Szubin R, Feist AM, Palsson BO: Adaptation to the coupling of glycolysis to toxic methylglyoxal production in tpiA deletion strains of Escherichia coli requires synchronized and counterintuitive genetic changes. Metab Eng 2018, 48:82–93. [DOI] [PubMed] [Google Scholar]
- 54.McCloskey D, Xu S, Sandberg TE, Brunk E, Hefner Y, Szubin R, Feist AM, Palsson BO: Adaptive laboratory evolution resolves energy depletion to maintain high aromatic metabolite phenotypes in Escherichia coli strains lacking the Phosphotransferase System. Metab Eng 2018, 48:233–242. [DOI] [PubMed] [Google Scholar]
- 55.McCloskey D, Xu S, Sandberg TE, Brunk E, Hefner Y, Szubin R, Feist AM, Palsson BO: Growth Adaptation of gnd and sdhCB Escherichia coli Deletion Strains Diverges From a Similar Initial Perturbation of the Transcriptome. Front Microbiol 2018, 9:1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McCloskey D, Xu S, Sandberg TE, Brunk E, Hefner Y, Szubin R, Feist AM, Palsson BO: Evolution of gene knockout strains of E. coli reveal regulatory architectures governed by metabolism. Nat Commun 2018, 9:3796. [DOI] [PMC free article] [PubMed] [Google Scholar]