Abstract
Purpose:
Effective targeted therapies are lacking for refractory and relapsed T-cell Acute Lymphoblastic Leukemia (T-ALL). Suppression of the NOTCH pathway using gamma-secretase inhibitors (GSIs) is toxic and clinically not effective. The goal of this study was to identify alternative therapeutic strategies for T-ALL.
Experimental Design:
We performed a comprehensive analysis of our high throughput drug screen across hundreds of human cell lines including fifteen T-ALL models. We validated and further studied the top hit, navitoclax (ABT-263). We used multiple human T-ALL cell lines as well as primary patient samples, and performed both, in vitro experiments and in vivo studies on patient-derived xenograft models.
Results:
We found that T-ALL are hypersensitive to navitoclax, an inhibitor of BCL2 family of anti-apoptotic proteins. Importantly, GSI-resistant T-ALL are also susceptible to navitoclax. Sensitivity to navitoclax is due to low levels of MCL-1 in T-ALL. We identify an unsuspected regulation of mTORC1 by the NOTCH pathway, resulting in increased MCL-1 upon GSI treatment. Finally, we show that pharmacological inhibition of mTORC1 lowers MCL-1 levels and further sensitizes cells to navitoclax in vitro and leads to tumor regressions in vivo.
Conclusions:
Our results support the development of navitoclax, as single agent and in combination with mTOR inhibitors, as a new therapeutic strategy for T-ALL, including in the setting of GSI resistance.
Keywords: leukemia, MCL-1, NOTCH1, GSI, ABT-263, targeted therapy, mTOR
INTRODUCTION
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive blood cancer that accounts for 15% of pediatric and 25% of adult ALL cases 1. Although significant advances in chemotherapy regimens have led to high rates of complete remission, close to 50% of adults and 25% of pediatric cases relapse within the first two years 2. Unfortunately, for patients with relapsed or refractory leukemia, the prognosis remains poor, emphasizing the need to develop more effective anti-leukemic targeted therapies.
Although the detailed molecular mechanisms that account for the aggressive nature and poor therapeutic response of T-ALL remain to be fully elucidated, studies have shown that mutations activating the NOTCH pathway are common in this disease. In particular, mutations in the gene encoding NOTCH1, are present in almost one half of patients 3. Upon ligand binding, NOTCH1 is sequentially cleaved by ADAM and γ-secretase proteases. The cleaved NOTCH intracellular domain (NICD) translocates to the nucleus, where it associates with Recombinant signal Binding Protein (RBP-J), and members of the mastermind-like (MAML 1–4) family, to turn on a transcription module vital for T-cell development. NOTCH1 mutations in T-ALL patients activate downstream signaling in the absence of ligand (HD domain mutations), or extend the half life of NICD (PEST domain mutations), leading to constitutive up-regulation of the pathway 4. In rare cases, similar pathway-activating mutations have also been discovered in NOTCH3 5. In addition, mutations in FBXW7, the ubiquitin ligase that binds and degrades NICD, are found in 10–15% of T-ALL cases 6; they increase the stability of NICD, thus allowing the NOTCH signal to persist. Due to the high prevalence of NOTCH pathway activating mutations in this disease, translational efforts have been focused on inhibiting this pathway.
Gamma-secretase inhibitors (GSIs) block NOTCH translocation to the nucleus and all downstream signals emanating thereof. They have been shown to induce cell-cycle arrest and, in some cases, apoptosis, and have demonstrated efficacy in mouse xenograft models of T-ALL 7. However, so far, GSIs have had limited success in patients. An initial clinical trial undertaken in the context of T-ALL demonstrated severe on target gastrointestinal toxicities 8–10. In addition, a number of T-ALL cell lines as well as primary T-ALL cells are resistant to gamma-secretase inhibition, in some instances due to loss of the tumor suppressor PTEN 11,12 13. Currently, patients depend solely on chemotherapy regimens and the prognosis of patients with relapsed or resistant T-ALL remains bleak.
In search of alternatives to conventional chemotherapy, we conducted a high-throughput drug screen across hundreds of human cancer cell lines 14. The screen revealed that both, GSI-sensitive and resistant T-ALL cell lines were highly sensitive to treatment with the BH3-mimetic, ABT-263. This sensitivity was due, at least in part, to low levels of MCL-1, an anti-apoptotic protein that is known to counter the activity of ABT-263 15,16. Interestingly, our work revealed that blocking the NOTCH pathway in GSI-resistant models results in de-repression of mTORC1, increased MCL-1 expression, and mitigated responses to ABT-263. Importantly, MCL-1 levels can be further lowered by pharmaceutical inhibition of mTORC1, enhancing ABT-263 activity in vitro and in vivo.
MATERIALS AND METHODS
Information about cell lines, reagents, and immunoblotting experiments are detailed under supplementary information.
Cell lines were obtained from commercial sources (ATCC, DSMZ and equivalent) and stocks created. Authentication of cell lines was further performed using STR. All lines were tested for mycoplasma routinely and only mycoplasma free cells were used. Cells were not kept in culture for extended periods of time. Typically, from frozen stocks derived from the commercial repository, cells were not kept in culture for more than 2 months.
High-Throughput Drug Screen
Data for the 888-cell line screen were obtained through our collaborative HTS efforts: Genomics of Drug Sensitivity in Cancer - http://www.cancerrxgene.org.
Cell Viability assays
HTS screening validation and further studies were performed as follows: 180 μl of a 400,000 cells/ml cell suspension was used to seed a 96-well plate. The next day, drugs were added to the cells, to a final volume of 200 μl/well. All drugs were serially diluted over a 9-point, 256-fold concentration range. 72h after drug treatment, 50 μl/well of CellTiter-Glo (Promega) solution was added and the plate was read on a SpectraMax M5 (Molecular Devices) luminometer. For 7d assays, we followed the same protocol as above, except, a suspension of 100,000 cells/ml was used to seed the wells.
FACS Apoptosis
Cells were seeded at 0.5 × 106 cells/well in 6-well plates. The next day triplicate wells were treated with respective drugs. After 48h of treatment, cells were harvested, washed with PBS, resuspended in Annexin binding buffer (BD Biosciences). Cells were stained with propidium iodide (BD Biosciences) and Annexin V Cy5 (Biovision) and analyzed on a LSRII flow cytometer (BD Biosciences).
Xenograft Mouse Studies
A suspension of 10 ×106 MOLT4 cells was inoculated subcutaneously into the left flanks of 6- to 8-week-old female athymic nude mice. Tumors were monitored until they reached approximately 500–800 mm3. At this time, mice were randomized to control and treatment groups (n = 4–5/group). All drugs were administered once a day by oral gavage. ABT-263 was dissolved in a mixture of 60% Phosal 50 PG, 30% PEG 400 and 10% EtOH and administered at 80 mg/kg. AZD8055 was dissolved in Captisol, and administered at 16 mg/kg. AZD8055 was administered 1.5h prior to ABT-263 for combination treatments. Tumors were measured twice-weekly using calipers. For pharmacodynamic analyses, tumor-bearing mice were administered with drugs or vehicle for 3d. Three hours after the last treatment, tumors were excised and snap-frozen in liquid nitrogen for immunoblotting. All experiments were approved by the Massachusetts General Hospital Animal Care and Use Committee.
In vivo studies with primary T-ALL patient-derived xenografts
NOD-scid Il2rγ−/− (NSG) mice were intravenously injected with 1 × 10^6 primary human T-ALL blasts. Once the leukemic burden reached 55% (TALL-X-7) or 65% (TALL-X-2) human leukemic blasts (as determined by hCD45 staining) in the peripheral blood, mice were randomized to one of four treatment groups. Vehicle (Captisol and 60% Phosal 50 PG, 30% PEG 400 and 10% EtOH), AZD8055 (16mg/kg, diluted in Captisol), ABT-263 (80mg/kg, diluted in 60% Phosal 50 PG, 30% PEG 400 and 10% EtOH) or both AZD8055 and ABT-263 were administered by oral gavage for 3 weeks using a 6-day on, 1-day off regimen. Mice were monitored daily and sacrificed when moribund. To assess leukemic burden, animals were sacrificed following 2 weeks of treatment and the percentage of human CD45+ leukemic cells in mouse spleen, bone marrow and peripheral blood were determined by flow cytometry. All mouse procedures used in these PDX studies are approved by the University of Massachusetts Medical School Institutional Animal Care and Use Committee. Primary T-ALL sample collection and in vitro studies are described under supplementary information.
RESULTS
T-ALL cell lines are sensitive to ABT-263
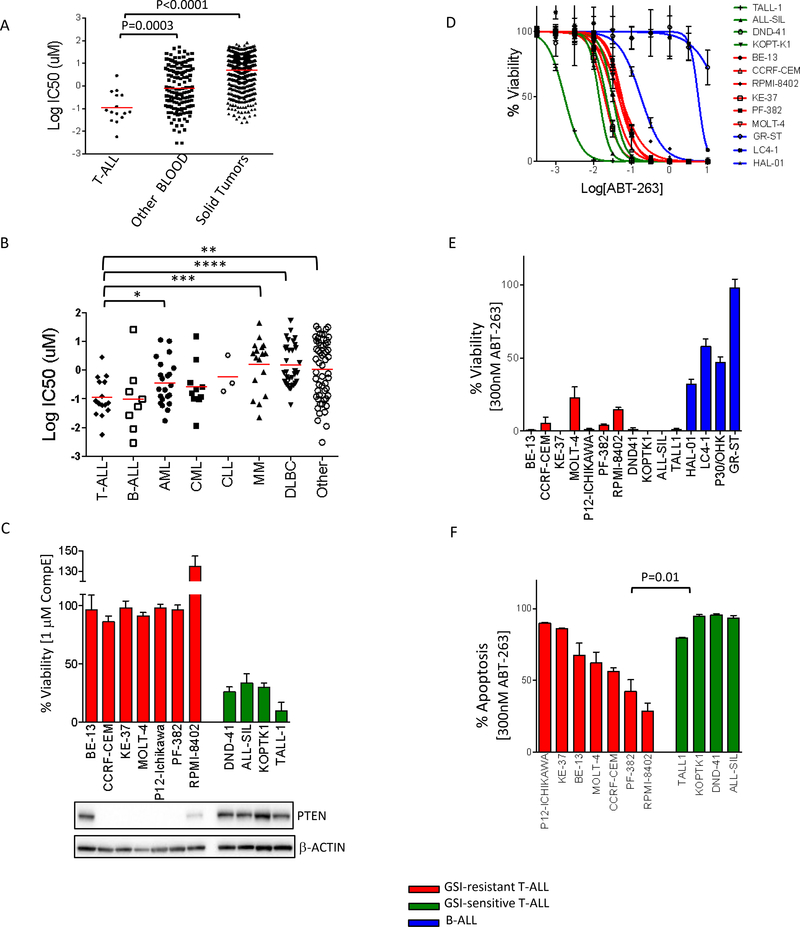
Data from a high throughput screen that we performed 14, wherein we tested the efficacy of 130 drugs on a panel of 888 human cancer cell lines, uncovered that T-ALL lines are highly sensitive to the BH-3 mimetic, ABT-263 (Mean IC50 = 0.1 uM for T-ALL lines, 0.8 uM for other blood cell lines, 5.0 uM for solid tumor cell lines) (Fig 1A, B), with 60% of the T-ALL lines in the screen having IC50s less than 100nM (Sup Fig S1A). To validate the results of the high-throughput screen we used a group of 11 T-ALL cell lines consisting of seven GSI-resistant lines, and four GSI-sensitive lines (Fig 1C). All eleven T-ALL cell lines, except TALL-1, had mutations in NOTCH1 (Sup Table S1), and expressed NICD (Sup Fig S1B), demonstrating that the NOTCH pathway is broadly active in these models of the disease. TALL-1 had a mutation in NOTCH3, leading to ligand-free activation of NOTCH3 5. As shown before, there was no correlation between NICD levels and sensitivity to GSI 17. Also, in agreement with previously published results, most GSI-resistant lines did not express wild type PTEN 13 (Fig 1C). BE-13 expressed PTEN, but had a nonsense mutation (W164*) in the TSC1 locus (TSC1 is a negative regulator of mTORC1); RPMI-8402 showed weak PTEN expression but both alleles of PTEN are mutated 18. We performed dose response experiments across three days of treatment with ABT-263 and included 3 B-ALL lines for comparison. We found that, indeed, the IC50s for most of the T-ALL cell lines were at least an order of magnitude lower than for the B-ALL cell lines in our panel (Fig 1D, Sup Fig S1C). In a 3-day assay, 300 nM ABT-263 was very toxic to the T-ALL cell lines as compared to B-ALL lines (Fig 1E). Further, ABT-263 treatment resulted in more than 80% of the cells undergoing apoptosis in multiple models (Fig 1F). Interestingly, the GSI-sensitive lines exhibited higher levels of apoptosis than the GSI-resistant lines (p-value = 0.01), although the IC50 values of the two groups were not significantly different (Sup Fig S1D). Overall, these data indicate that, a major component of the response to ABT-263 is, as expected, induction of apoptosis and that T-ALL lines are particularly sensitive to this compound.
Figure 1. GSI-resistant cell lines are sensitive to ABT-263.
(A) Sensitivity values (log10 IC50 μM) of 888 human cell lines treated with ABT-263, comparing T-ALLs, other leukemic and lymphatic lines (blood) and solid tumors. Each dot represents one cell line and each red bar is the geometric mean for that group. An unpaired t-test was used to assess the differences in mean IC50 between the T-ALLs and the other two groups. Data obtained from our high throughput screen collaboration - Genomics of Drug Sensitivity in Cancer - http://www.cancerrxgene.org (B) Sensitivity values (log10 IC50 μM) of 163 hematopoietic and lymphoid cell lines treated with ABT-263. These cell lines, which in Fig (A) were clustered together under T-ALL and Other Blood groups, are further classified based on cancer type. Each dot represents one cell line and each red bar is the geometric mean for that group. Data was analyzed using student t test (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001). B-ALL, CML and CLL are not significantly different when compared to T-ALL (C) Top: A panel of GSI-resistant (red) and GSI-sensitive (green) T-ALL lines were treated either with DMSO or with 1 μM CompE for 7 days. Percentage viability compared to DMSO treatment was measured using CellTiter-Glo. The values shown are an average of two biological replicates, ± SD. Bottom: For each cell line, 15 μg of cell lysates were subjected to western blotting and probed with the indicated antibodies. (D) Dose response curves of GSI-resistant (red) and GSI-sensitive (green) T-ALL lines and B-ALL lines (blue) to ABT-263. Cell lines were treated with ABT-263 for 3 days, and viability was measured with CellTiter-Glo (E) 14 human ALL lines, including GSI-resistant T-ALLs (red), GSI-sensitive T-ALLs (green) or B-ALLs (blue), were treated with 300 nM ABT-263 for 3 days. Viability was measured using CellTiter-Glo and plotted as a percentage (drug treated samples / DMSO-treated controls). The values shown are an average of two biological replicates, each in triplicate ± SD. (F) GSI-resistant (red) and GSI-sensitive (green) T-ALL cell lines were treated with 300 nM ABT-263 for 48h, after which cells were stained with propidium iodide and Annexin-V. Apoptosis was measured by FACS analysis as percentage annexin-positive cells, and normalized against DMSO-treated controls. The experiment was done in triplicate and the values shown are mean ± SD.
We next tested whether ABT-199 (venetoclax), a BCL-2 specific inhibitor with a more favorable toxicity profile than ABT-26319, was also effective against T-ALL cell lines. Two AML lines that were previously shown to be sensitive to ABT-199 20 were used as positive controls. The T-ALL lines were not sensitive to ABT-199 (Sup Fig S2A, B). Since ABT-199 targets BCL-2 but not the anti-apoptotic protein BCL-xL (encoded by the BCL2L1 gene), high levels of BCL-xL can cause resistance to this drug 21,22. Consistent with this, T-ALL lines had higher levels of BCL-xL protein as compared to the AML lines (Sup Fig S2C). Analysis of cell lines from an expression dataset (CCLE) 18 confirmed that BCL-xL mRNA levels were higher in T-ALL samples compared to AML samples (Sup Fig S2D). These results indicate that ABT-199 is unlikely to be active against T-ALL.
Low MCL-1 levels are responsible for hypersensitivity to ABT-263
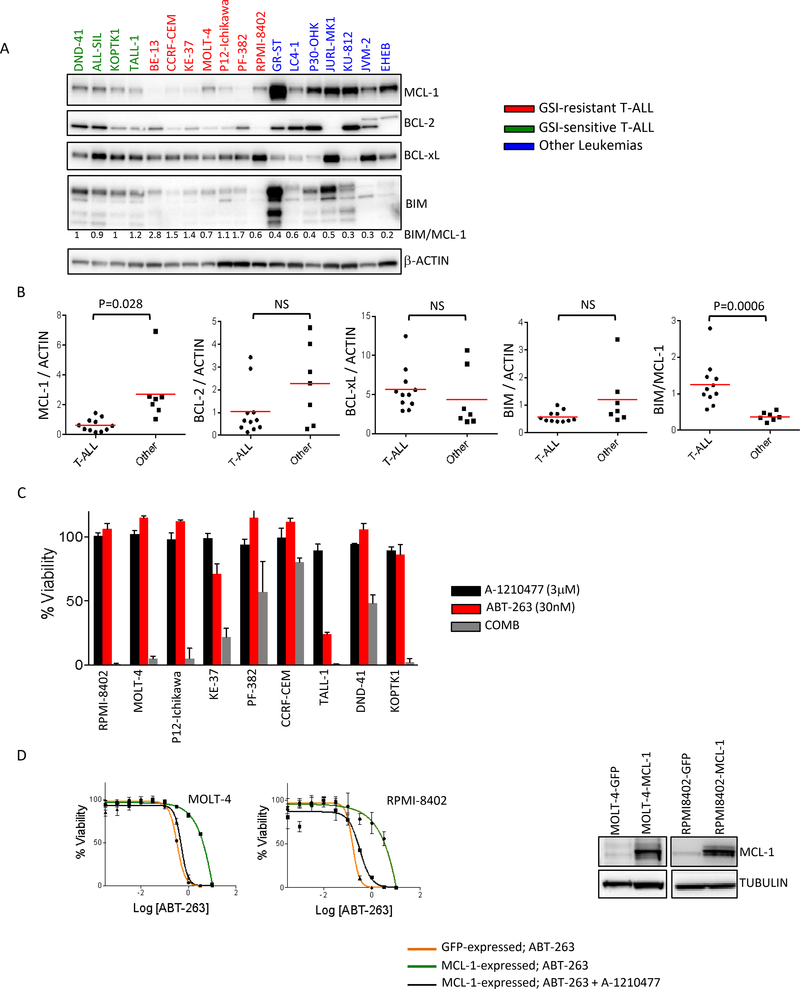
ABT-263 targets several anti-apoptotic BCL2 family members such as BCL-2, BCL-xL and BCL-w, but not others such as MCL-1 and BCL2A1 and high expression of MCL-1 is known to confer resistance to ABT-263 15,16,23,24. Previous data have shown that upon inhibition of BCL-2/BCL-xL/BCL-w with ABT-263, MCL-1 can sequester the newly released BIM from BCL-2/BIM and BCL-xL/BIM complexes, preventing free BIM-initiated apoptosis 25. Consistent with these findings, multivariate analysis of expression and mutational data of our large cell line collection, identified MCL-1 as the top resistance genomic feature explaining response to ABT-263 in our high-throughput screen 14,26. To investigate the relevance of BCL-2 family members in regulating response to ABT-263 in T-ALL, we analyzed the expression of BCL-2 family proteins (BCL-2, BCL-xL, MCL-1 and BIM) across a group of blood cancer cell lines including the 11 T-ALL models (Fig 2A). Quantitation of the western blots indicated that the levels of MCL-1 protein were considerably lower in the T-ALL cell lines examined compared to samples from other leukemic cell lines (Fig 2B). In addition, analysis of our mRNA expression dataset across cell lines (http://www.cancerrxgene.org) revealed that levels of MCL-1 mRNA were significantly lower in the T-ALL lines compared to 145 lines from other blood cancers (Sup Fig S3A). These results suggested that low MCL-1 levels underlie ABT-263 sensitivity in T-ALL. Intriguingly, while the GSI-sensitive lines are somewhat more sensitive to ABT-263 than the GSI-resistant ones, they have higher levels of MCL-1 than the GSI-resistant T-ALLs. We and others had previously shown that the ratio of BIM/MCL-1 is an important determinant of response to ABT-263 27,28. We thus examined levels of the pro-apoptotic protein BIM in these lines. The GSI-sensitive lines have higher levels of BIM, than the GSI-resistant lines, which could counterbalance for the higher levels of MCL-1 in these lines. Indeed, the BIM/MCL-1 ratio is significantly higher for the T-ALL lines as compared to the other leukemic lines (Fig 2A, B). Further, the BIM/MCL-1 ratios of the GSI-sensitive T-ALLs are similar to those of the GSI-resistant T-ALLs (Fig 2A, B). Thus, differences in MCL-1 levels are often counterbalanced by differences in BIM levels and the BIM/MCL1 ratio best explains ABT-263 sensitivity profile of the T-ALL lines.
Figure 2. Low MCL-1 levels are responsible for hypersensitivity to ABT-263.
(A) 15 μg of cell extract from each of the GSI-sensitive (green) and GSI-resistant (red) T-ALL lines and 7 other leukemic cell lines (blue) was separated by SDS-PAGE and probed with the indicated antibodies. The cell lines in blue represent B-ALL (GR-ST, LC4–1, P30-OHK), CML (JURL-MK1, KU-812) and CLL (JVM-2, EHEB). Independent experiments were performed at least three times and a representative result is shown. (B) Scatter plots representing quantitated amounts of MCL-1/BCL2/BCL-xL/BIM protein normalized to β-Actin, for the 18 cell lines in (A). The proteins were quantitated using GeneTools from SynGene. An unpaired t-test was used to assess the significance of differences in mean between the two groups (C) T-ALL cell lines were treated for 72h with 3μM of the MCL-1 inhibitor A-1210477 (black), 30nM ABT-263 (red) or a combination of 3μM of A-1210477 and 30nM ABT-263 (grey). Viability was measured using CellTiter-Glo and normalized to DMSO-treated samples. (D) Two human T-ALL cell lines, MOLT-4 and RPMI-8402, were infected with lentiviral particles expressing GFP alone (GFP) or GFP-IRES-MCL-1 (MCL-1). GFP expressing cells were sorted and tested for sensitivity to ABT-263 alone or in combination with A-1210477 (the MCL-1 inhibitor). Dose response curves of the MCL-1 overexpressing lines treated alone (green) or in combination (grey) and the controls (orange) are shown. The experiments were done two times and the values shown are mean ± SD. Equal amounts of lysates from these lines were separated by SDS-PAGE and probed with MCL-1 and α-tubulin antibodies.
To confirm that MCL-1 protein levels influence ABT-263 sensitivity in T-ALL, we co-treated the cell lines with ABT-263 and an MCL-1-specific inhibitor, A-1210477. Cells were further sensitized to ABT-263 in the presence of the MCL-1 inhibitor, with several cell lines showing high synergy (Fig 2C, Sup Fig S3B). It was not immediately obvious as to why cells with very low levels of MCL-1 were responding robustly to the combination of ABT-263 and the MCL-1 inhibitor. In order to gain insights into the mechanisms leading to synergy, we treated the T-ALL lines with ABT-263 and examined MCL-1. As noted previously 25,27,29, MCL-1 levels increase in almost all the lines treated with ABT-263 (Sup Fig S4). The MCL-1 inhibitor by itself, was not effective in these lines (Fig 2C). On the other hand, low dose ABT-263 was potentiated by MCL1 inhibition. The effect of MCL-1 inhibition is thus likely through compensation of the MCL-1 increase induced by ABT-263. In fact, in cell lines such as PF-382 and CCRF-CEM, which show little increases in MCL-1 levels, the combination is not very effective (57% and 80% viability, respectively) and does not provide benefit over single agent ABT-263.
Further, we overexpressed MCL-1 in two cell lines and examined sensitivity to ABT-263. Cell lines overexpressing MCL-1 were more resistant to ABT-263 than the parental and control lines overexpressing GFP (Fig 3D). The MCL-1 overexpressing lines were re-sensitized to ABT-263 in the presence of the MCL-1 inhibitor. These results show that MCL-1 expression levels are a major determinant of the sensitivity of T-ALL cells to ABT-263. In addition, targeting MCL-1 can further sensitize T-ALL cells to ABT-263.
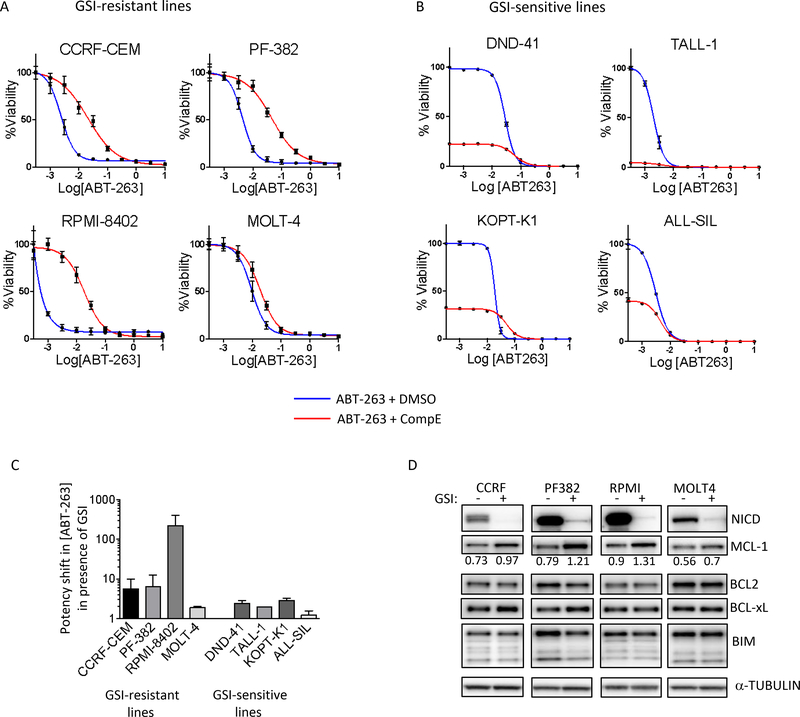
Figure 3. NOTCH inhibition desensitizes human T-ALL cell lines to ABT-263.
GSI-resistant (A) and GSI-sensitive (B) human T-ALL cell lines were pre-treated with DMSO or 1 μM CompE for 72h, followed by treatment with varying concentrations of ABT-263, in the continued presence of DMSO or 1 μM CompE for another 72h. At the end of the six-day period, cell viability was measured with CellTiter-Glo and expressed as a percentage of drug-treated cells to DMSO-treated cells. The experiments were done twice, in triplicate each and the values shown are mean ± SD. (C) Bar graph shows the shift in the potency of ABT-263 upon GSI treatment. Values are calculated using 50% viability loss over DMSO and GSI alone, for single agent and GSI combination (GSI alone = 100%) respectively. (D) GSI-resistant T-ALL lines were treated with DMSO or 1 μM CompE for 5 days, harvested, lysed and subjected to western blot analysis using the indicated antibodies. Independent experiments were done three times and a representative result is shown. MCL-1 levels, quantitated using GeneTools from SynGene, normalized to α-tubulin, are shown under MCL-1 blot.
NOTCH inhibition desensitizes T-ALL to ABT-263
Since GSIs are used to limit the growth of T-ALLs, we combined ABT-263 with GSIs to see if there was a further effect on viability. We treated human T-ALL cell lines with a well-studied GSI, compound E (CompE), for 3 days before exposing the cells to ABT-263. Interestingly, rather than sensitizing the cells, GSI treatment rendered the GSI-resistant lines 10–100 fold more resistant to ABT-263 (Fig 3A, C). Furthermore, while in the case of GSI-sensitive lines the effect on ABT-263 was limited, GSI treatment still made these cells less sensitive to low doses of ABT-263 (10–100 nM) (Fig 3B, C). These findings were recapitulated with two additional GSIs, MRK-003 and RO4929097 (Sup Fig S5A, B). Since blocking NOTCH processing and activity with GSI made the lines more resistant to ABT-263, we investigated whether the NOTCH pathway was mechanistically linked to the sensitivity of human T-ALL cells to ABT-263. GSI-resistant cell lines (where we observed large ABT-263 dose potency shifts, Fig 3C) were treated with CompE, and probed for MCL-1. Indeed, we saw a consistent increase in MCL-1 protein levels across the models, with the level of change in MCL-1 matching the changes in ABT-263 potency in the viability experiments (Fig 3D and compare with Fig 3A). Similar changes were not seen amongst other BCL2 family members upon GSI treatment. Thus, taken together, our results suggest that increase in MCL-1 is likely to be an important part of the effect of GSI on ABT-263 sensitivity.
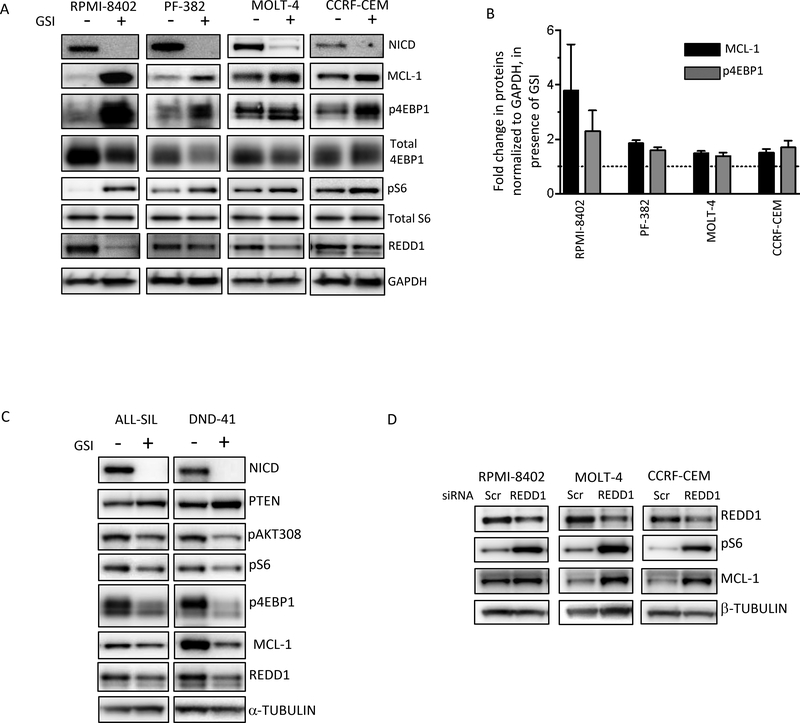
GSI treatment affects mTORC1 activity and MCL-1 levels
To investigate whether the increase in MCL-1 protein upon GSI treatment (Fig 3D) was due to changes at the transcript level, we treated the GSI-resistant cell lines with CompE and measured MCL-1 mRNA levels. MCL-1 mRNA levels did not change upon treatment with GSI (Sup Fig S6A), suggesting that differences in MCL-1 protein expression reflect post-transcriptional effects either through synthesis or stability. We turned to translational regulation of MCL-1 because previous studies had demonstrated important control of MCL-1 levels through cap-dependent translation regulated by mTORC1 30–33. We reasoned that this could also offer an opportunity to improve the effects of ABT-263 by using drug combinations. Indeed, treatment with the mTOR inhibitor, AZD8055, which decreased phosphorylation of 4EBP1 and S6, also lowered MCL-1 expression in T-ALL cell lines (Sup Fig S6B). To determine whether the regulation of MCL-1 protein levels by the NOTCH pathway (Fig 3D) is through mTORC1, we treated GSI-resistant cell lines with compE and measured mTORC1 activity. Indeed, treatment with a GSI led to decreases in intra-cellular NOTCH1, and concomitant increase in mTORC1 activity, as measured by phosphorylation 4EBP1 and ribosomal protein S6 (Fig 4A). The cell line that showed larger changes in MCL-1 levels (RPMI-8402, Fig 4A, B), also exhibited the highest increase in phospho-4EBP1. To confirm that NOTCH1 does indeed suppress mTORC1, we knocked down NOTCH1 in one of the cell lines and examined whether we recapitulate the results seen with GSI inhibitors. Indeed, knockdown of NOTCH1 led to increased mTORC1 activity and increased MCL-1 levels (Sup Fig S6C) and was accompanied by loss in sensitivity to ABT-263 (Sup Fig S6D, E).
Figure 4. NOTCH1 represses mTORC1 activity and MCL-1 levels in GSI-resistant cell lines.
(A) GSI-resistant lines were treated with DMSO or 1 μM CompE for 5 days, harvested, lysed and subjected to western blot analysis using the indicated antibodies. Independent experiments were done three times and a representative result is shown. (B) Changes in levels of MCL-1 and p4EBP1 upon GSI treatment, normalized to GAPDH, were quantitated using GeneTools from SynGene, and presented on a graph. The experiments were done at least 3 times and values shown are mean ± SD (C) GSI-sensitive cell lines were mock-treated or treated with 1 μM CompE for 5 days, harvested, lysed and subjected to Western blot analysis using the indicated antibodies. (D) GSI-resistant lines were transfected with Scramble siRNAs or siRNAs against REDD1 (DDIT4). After 72 hrs cells were harvested, lysed and subjected to western blot analysis using the indicated antibodies.
These results were intriguing, since previous studies had shown that NOTCH1 activates the PI3K-AKT pathway, leading to mTOR activation 11,13. Notably, those studies were performed in PTEN-competent cells with NOTCH1 acting via PTEN to regulate changes in AKT activity 13. By contrast, the cell lines of interest here are GSI resistant lines that are functionally PTEN null 13. In agreement with previous results showing positive regulation of AKT by NOTCH1, when NOTCH1 was inhibited in GSI-sensitive and PTEN wild-type cell lines, we observed an up-regulation of PTEN and suppression of mTORC1 and MCL-1 (Fig 4C). It is unclear at this point why ABT-263 sensitivity is marginally lowered by GSI-treatment in the GSI-sensitive cells. It is possible that post GSI treatment these GSI-sensitive cells are less prone to apoptosis.
To gain insights into how the NOTCH pathway regulates mTORC1, we examined the levels and activation status of regulators of mTORC1. Interestingly, based on phosphorylation of AKT itself and two of its known substrates ATP-citrate lyase and GSK3α/β (Sup Fig S6F), AKT activity was not consistently modified by GSI treatment across the models. In one cell line (PF-382), AKT was activated upon GSI treatment, while in others (MOLT-4 and RPMI-8402) it was inhibited, and in yet another cell line (CCRF-CEM) it appeared unchanged. Thus, AKT regulation did not seem to explain the consistent activation effect seen on mTORC1 in the presence of GSI. Further, we did not see consistent changes in the phosphorylation of TSC2 or AMPK, major upstream regulators of mTORC1 (Sup Fig 6F). Interestingly, a less well-characterized regulator of mTORC1, REDD1, was seen as the best candidate to explain changes in mTORC1 activity upon GSI treatment. REDD1 was previously shown, in other contexts, to be a negative regulator of mTORC1 and exert its effect on mTORC1 by activating TSC1/TSC2 (without change in TSC expression) 34. Indeed, in T-ALL cell lines too, knocking down REDD1 led to increased mTORC1 activity as well as higher levels of MCL-1 (Fig 4D). Furthermore, previous studies have shown that the NOTCH pathway regulates REDD1 (encoded by the DDIT4 gene) 35,36. In three of the four cell lines, we observed that REDD1 levels decreased upon GSI treatment (Fig 4A). However, one cell line (CCRF) showed increased mTORC1 activity without changes in REDD1 levels, suggesting that other factors regulate mTORC1 activity in those cells. Interestingly, REDD1 levels were also lowered in two PTEN competent models (Fig 4C). However, this did not translate into higher mTORC1 activity in those models. This might be due to the PTEN-AKT regulation of mTORC1 overcoming the effect of REDD1 in those models (see pAKT308 in Fig 4C). Overall, our data suggest that NICD suppression by GSI treatment and mTORC1 activation is due, at least in part, to a decrease in REDD1 levels. Consistent with previous findings 31,32 mTORC1 suppression is accompanied by lowering of MCL-1 protein levels. In the context of already low MCL-1 protein levels due to low MCL-1 transcript levels, suppression of mTORC1 renders T-ALL cells highly sensitive to ABT-263.
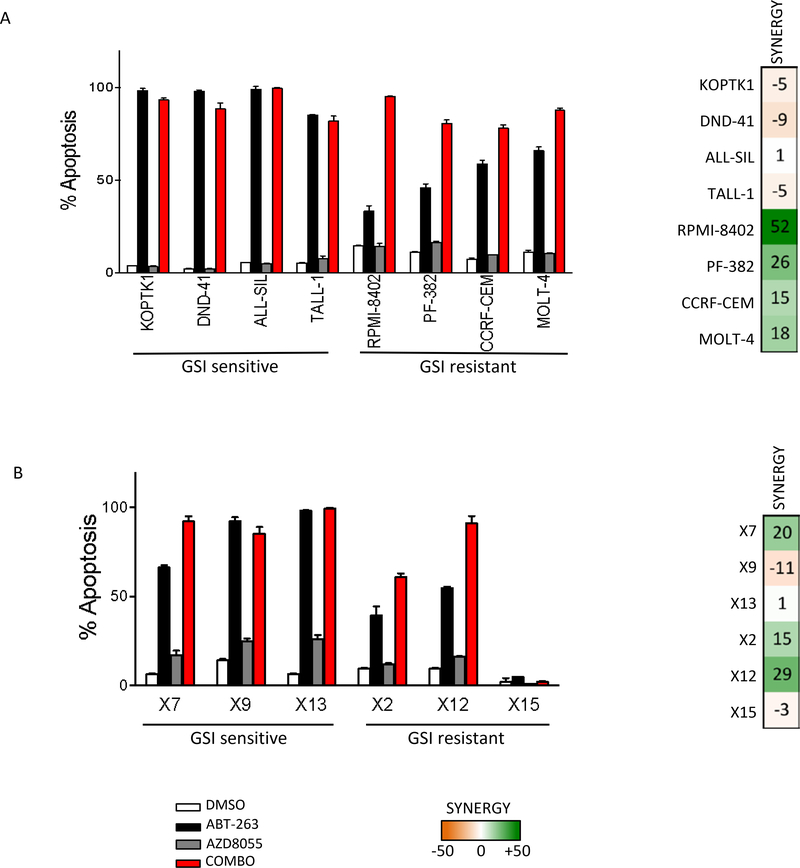
Combination of AZD8055 and ABT-263 triggers high apoptosis in T-ALL cell lines and primary T-ALL samples in vitro
Based on our results so far, including the effect of mTORC1 inhibition on MCL-1 (Sup Fig S6B), we tested whether we could further sensitize T-ALL to ABT-263 using an mTORC1 inhibitor. Indeed, the combination treatment of ABT-263 with the mTOR inhibitor AZD8055, resulted in decreased cell viability in both GSI-sensitive and resistant T-ALL cell lines (Sup Fig S7A). In several lines the combination synergistically reduced viability (Sup Fig S7B). Consistent with apoptosis playing a major role in the combination effect, the combined treatment of ABT-263 and AZD8055 led to massive apoptosis (90%) in most T-ALL models tested (Fig 5A, Sup Fig S8).
Fig 5. Combination of AZD8055 and ABT-263 triggers high apoptosis in T-ALL cell lines and primary T-ALL models in vitro.
(A) Human T-ALL cell lines were treated with DMSO, 300 nM ABT-263, 500 nM AZD8055 or a combination of 300 nM ABT-263 and 500 nM AZD8055 for 48h, after which cells were stained with propidium iodide and Annexin-V. Apoptosis was measured by FACS analysis as percentage of cells that are Annexin-V positive. The experiment was done in triplicate and the percentage apoptosis values are shown for each treatment with mean ± SD. (B) Primary T-ALL cells from six patients were treated with DMSO, 1 μM ABT-263, 500 nM AZD8055 or a combination of 1 μM ABT-263 and 500 nM AZD8055 for 3days, after which cells were stained with propidium iodide and Annexin-V. Apoptosis was measured by FACS analysis as percentage of Annexin-V positive cells. The experiment was done in triplicate and the percentage apoptosis values are shown for each treatment with mean ± SD.
We next examined primary T-ALL samples obtained from patients at the time of diagnosis or relapse (Sup Fig S9A). As shown previously, of the 6 primary T-ALL samples, 3 were GSI-sensitive and 3 were GSI-resistant (Sup Fig S9B) 37. We note here that three of the models (TALL-X-12, TALL-X-13 and TALL-X-15) had wild type NOTCH1, with low but detectable NICD (Sup Fig S9A, C), and, as with established cell lines, there was no correlation between NICD levels and sensitivity to GSI (Sup Fig S9B, C). We treated all six primary T-ALL samples with vehicle, ABT-263, AZD8055 or a combination of the two and measured viability after 3 days of treatment. In all but one case, the combination treatment inhibited leukemic growth better than either single agent alone (Sup Fig S10A). Annexin V/PI staining revealed that the combined therapy induced more apoptosis than either drug alone (Fig 5B, Sup Figs S11, S12). Interestingly, one of the models (TALL-X-15) was impervious to ABT-263 either as single agent or in combination with AZD0855 when measuring apoptosis (Fig 5B) (More in discussion).
Based on this panel of cell lines and primary cultures, the combination effect over single-agent ABT-263 appeared more pronounced in the GSI-resistant than in the GSI-sensitive models (Fig 5A, B). Indeed, ABT-263 treatment alone was very effective at killing the GSI-sensitive models tested (over 90% in several models), and the combination treatment could thus not increase apoptosis much further. This translates into higher level of synergy in the GSI-resistant models as compared to the GSI-sensitive ones (Fig 5A, B, Sup Fig S10A).
We also analyzed lysates from one GSI-sensitive (TALL-X-7) and one GSI-resistant (TALL-X-2) sample after 6h of drug treatment. As with established cell lines, ABT-263 treatment increases MCL-1 levels in the primary T-ALLs too (Sup Fig S10B). Further, mTOR inhibition yields decreased phospho-S6 and phospho-4EBP1, and MCL-1 levels. In the combined treatment, mTOR inhibition likely acts synergistically with ABT-263 by preventing a surge in MCL-1, in some cases (TALL-X-2) yielding MCL-1 levels even lower than in untreated conditions (Sup Fig S10B).
Efficacy of the Combination AZD8055 and ABT-263 in subcutaneous xenograft and primary patient-derived models
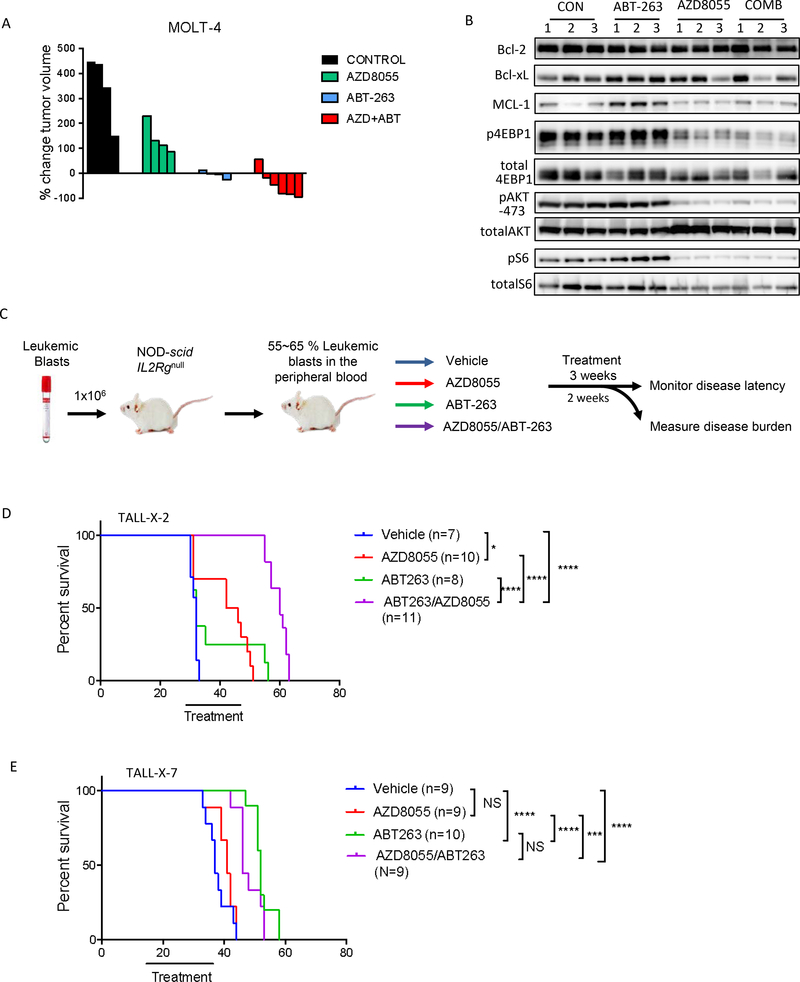
We first examined efficacy of the combination treatment using a conventional subcutaneous xenograft model of the MOLT-4 cell line. Consistent with the in vitro findings, the growth of MOLT-4 leukemic cells was significantly impaired in vivo by treatment with ABT-263. Importantly, the combination of ABT-263 and AZD8055 was more efficacious than single agents alone and, in fact, led to tumor regressions (Fig 6A). Pharmacodynamic studies of the tumor extracts indicate that AZD8055 effectively reduced mTORC1 activity and lowered MCL-1 levels (Fig 6B).
Fig 6. The combination of AZD8055 and ABT-263 causes tumor regression in vivo:
(A) GSI-resistant MOLT-4 cells were grown as xenograft tumors in Nu/Nu mice. Mice were randomized into 4 treatment cohorts: control (no drug), 16 mg/kg AZD8055, 80 mg/kg ABT-263, or their combination. Waterfall plot showing percentage change in tumor volume (relative to initial volume) for individual tumors in tumor-bearing mice, treated for 21 days. (B) For pharmaco-dynamic studies, tumor-bearing mice were treated as in (A), for 3 days. On the third day, tumors were harvested 2h after drug treatment and snap frozen. Proteins were extracted, 15 μg protein from each sample was run on an SDS-PAGE and subjected to western blotting with the indicated antibodies. (C) Schematic of the in vivo experiment with primary T-ALL cells. Briefly, NOD-scid IL2rg−/− (NSG) mice were intravenously injected with primary human T-ALL blasts. When human leukemic blasts reached 55–65% mouse peripheral blood, mice were randomized to one of four treatment groups indicated. 2–6 mice from each group were sacrificed after 2 weeks of treatment to assess leukemic burden (data shown in Fig S10, S11). The remaining mice were treated for a total of 3 weeks, after which survival was monitored. (D, E) Kaplan-Meier survival curves are shown for mice engrafted with TALL-X-2 (D) or TALL-X-7 (E) patient samples. The difference in overall survival between the treatment groups was assessed by log-rank test (*p<0.05, ***p<0.001, ****p<0.0001).
We then tested the efficacy of ABT-263 and AZD8055 dual therapy in models of primary human T-ALL. We engrafted two primary T-ALL samples, one GSI-sensitive (TALL-X-7) and one GSI-resistant (TALL-X-2), into NOD-scid Il2rγ−/− (NSG) mice. When leukemic burden reached 50–65% CD45-positive human leukemic blasts in the peripheral blood, the mice were randomized to one of 4 treatment groups and treated for 21 days. 2–6 mice from each treatment group were sacrificed after 14 days and effects on leukemic burden were determined (Fig 6C). Mice were weighed regularly to evaluate whether the combination treatment was deleterious to their health; all treatments were well tolerated (Sup Fig S13). Our results indicate that in the GSI-resistant model (TALL-X-2), the combination treatment significantly reduced leukemia burden and prolonged survival more effectively than either single agent alone (Fig 6D, Sup Fig S14). Consistent with the trend of GSI-sensitive models in-vitro, treatment with the single agent ABT-263 proved highly efficacious for TALL-X-7 in vivo (Fig 6E, Sup Fig S15). Absolute numbers, as well as percentages, of human CD45+ cells were significantly reduced in the blood, bone marrow and spleen upon ABT-263 treatment and there was very little additional benefit with the combination, likely because of profound response to single agent ABT-263 (Sup Fig S15). This was also reflected in the survival curves for this model; while ABT-263 treatment significantly improved the life-span of these leukemic mice, there was no added advantage with combination therapy compared to monotherapy (Fig 6E).
Taken together, our results suggest that ABT-263 is a promising therapeutic option as a single agent in some T-ALL, and that the addition of mTOR inhibitors could further improve efficacy, in particular, in GSI resistant T-ALLs.
DISCUSSION
ABT-263 has shown promising results in phase I clinical trials of relapsed or refractory CLL 38,39, but has only been examined to a limited extent in the context of T-ALL 40,41. Our studies show that low MCL-1 levels make T-ALL highly susceptible to ABT-263, whereas they are unsuitable for ABT-199 treatment, probably due to high levels of BCL-xL. While low MCL-1 mRNA levels likely lead to low protein levels, our work clearly shows that in GSI-resistant models, NOTCH represses mTORC1 and suppresses MCL-1. This crosstalk between two important growth pathways adds an interesting layer of regulation, where active NOTCH signaling leads to a further decrease in MCL-1 protein levels in T-ALL, making them vulnerable to ABT-263 therapy.
While the activation of AKT/mTOR by NOTCH1 has been described previously in GSI-sensitive, PTEN wildtype T-ALL models11,13, the repression of mTORC1 by NOTCH1 in GSI-resistant human T-ALLs is less well studied. Inhibiting NOTCH1 was previously shown to increase AKT-308 phosphorylation in Jurkat, CCRF-CEM and MOLT3 – all PTEN-depleted, GSI-resistant lines 42. While this is in general agreement with our results showing an increase in mTORC1 activity upon GSI treatment, in the series of models we studied we did not consistently observe AKT activation, leading us to study other possible influencers of mTOR activation that NOTCH was affecting (REDD1). In another study, NOTCH inhibition was shown to activate the PI3K pathway in wild-type and KRASG12D murine models of T-ALL 43.
It is somewhat surprising that two seemingly contradictory crosstalk events exist between NOTCH and the PI3K/mTOR pathway. On one hand, NOTCH activates AKT via PTEN suppression in some lines; on the other hand NOTCH represses mTORC1 downstream of AKT and at least in part through REDD1. Notably, in addition to activating AKT/mTORC1, the product of PI3K, PIP3, plays many other roles 44–46. This network wiring might allow decoupling in part the PIP3 production from downstream mTORC1 activation.
Our work, as well as other reports, shows that the NOTCH pathway regulates REDD1 levels 35,36. In our studies the effect on REDD1 upon manipulation of the NOTCH pathway was very consistent and substantial across experiments in some models but less pronounced in others. REDD1 is a stress-induced protein, that is up-regulated in response to many stimuli and is regulated by a number of transcription factors, including HIF1α, p63, p53, and ATF434,47–49. Because REDD1 is regulated by so many inputs it is possible that the effect of NOTCH inhibition is not readily observed in some of the cell lines over other inputs. It is also possible that NOTCH regulates mTORC1 through other effectors in addition to REDD1 in these models. Regardless, our data support that mTORC1 is inhibited by NOTCH in GSI-resistant T-ALL lines, with REDD1 playing an important role connecting NOTCH and mTORC1 in many of these lines.
Our studies identify BCL-2/BCL-xL targeting as a candidate therapeutic strategy for the treatment of T-ALLs. In fact, TALL-X-15 primary patient model is the only one that, in our hands, is not sensitive to ABT-263. This clone harbors a TP53 loss of function mutation (p.H168P) and TP53 mutations in T-ALL are associated with high risk of relapse and very poor survival 50. This model was derived from a patient who relapsed after multiple failed treatments and it might have acquired additional mutations or epigenetic characteristics making it impervious to ABT-263.
We show that ABT-263 based therapies are effective in GSI-sensitive as well as GSI-resistant models. This is especially significant since preclinical studies and initial clinical indications strongly suggest that a number of tumors with active NOTCH pathways might be impervious to NOTCH inhibition 11. Further, leukemic cells with mutations in FBXW7 are not responsive to GSIs 17. The three cell lines and one primary T-All model (X-7, Sup Table1) with FBXW7 mutations in our study, respond well to ABT-263 and the combination treatment.
Our work demonstrates that ABT-263 alone, or in combination with mTOR inhibitors, should be explored as a potential targeted therapy for T-ALL patients. There is some indication from our studies that GSI-resistant tumors might benefit more from the combination therapy, whereas GSI-sensitive tumors respond well to ABT-263 alone. One possible approach to clinical deployment would be to test leukemic cells from patients ex-vivo for sensitivity to GSI, ABT-263 and ABT-263 plus mTOR inhibitor and depending on the outcome, proceed with appropriate therapeutic strategy.
Supplementary Material
TRANSLATIONAL RELEVANCE.
Our work identifies the BH3-mimetic navitoclax (ABT-263), either as a single agent or in combination with mTOR inhibitors, as a potential therapeutic strategy for T-ALL patients. Importantly, our results show that tumors that are unresponsive to gamma-secretase inhibitors (GSIs) or to chemotherapy, could also benefit. In addition, no benefit or even antagonism was seen, in combining GSIs with navitoclax.
Acknowledgments
Financial Support: This study was supported by grants from the Wellcome Trust 086357 and 102696 (to C.H.B., U.M. and M.J.G.) and from the NIH R01CA167708 (to S.G.). A.H.C. and M.K. are supported by NIH grant R01 CA096899 and an Innovator Award from the Alex’s Lemonade Stand Foundation. A.C.F. is supported by a National Cancer Institute Career Development Award (NCI K22 CA175276–02), NIH R01CA215610 and by the George and Lavinia Blick Research Fund. J.A.E. was supported by a grant from NIH R01CA140594.
Footnotes
Conflict-of-Interest Disclosure: C.H.B. receives sponsored research from Novartis and Amgen. J.A.E. and C.C are employees of Novartis. The other authors have nothing to disclose.
REFERENCES
- 1.Ferrando AA, Neuberg DS, Staunton J, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1(1):75–87. [DOI] [PubMed] [Google Scholar]
- 2.Pui CH, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet. 2008;371(9617):1030–1043. [DOI] [PubMed] [Google Scholar]
- 3.Weng AP, Ferrando AA, Lee W, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269–271. [DOI] [PubMed] [Google Scholar]
- 4.Grabher C, von Boehmer H, Look AT. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat Rev Cancer. 2006;6(5):347–359. [DOI] [PubMed] [Google Scholar]
- 5.Bernasconi-Elias P, Hu T, Jenkins D, et al. Characterization of activating mutations of NOTCH3 in T-cell acute lymphoblastic leukemia and anti-leukemic activity of NOTCH3 inhibitory antibodies. Oncogene. 2016;35(47):6077–6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Girardi T, Vicente C, Cools J, De Keersmaecker K. The genetics and molecular biology of T-ALL. Blood. 2017;129(9):1113–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tammam J, Ware C, Efferson C, et al. Down-regulation of the Notch pathway mediated by a gamma-secretase inhibitor induces anti-tumour effects in mouse models of T-cell leukaemia. Br J Pharmacol. 2009;158(5):1183–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Es JH, van Gijn ME, Riccio O, et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435(7044):959–963. [DOI] [PubMed] [Google Scholar]
- 9.Deangelo RMS DJ, Silverman LB, Stock W, Attar EC, Fearen I, Dallob A, Matthews C, Stone J, Freedman SJ and Aster J. A phase I clinical trial of the notch inhibitor MK-0752 in patients with T-cell acute lymphoblastic leukemia/lymphoma (T-ALL) and other leukemias. Journal of Clinical Oncology, 2006 ASCO Annual Meeting Proceedings. 2006;24(18S). [Google Scholar]
- 10.Gounder MM, Schwartz GK. Moving forward one Notch at a time. J Clin Oncol. 2012;30(19):2291–2293. [DOI] [PubMed] [Google Scholar]
- 11.Chan SM, Weng AP, Tibshirani R, Aster JC, Utz PJ. Notch signals positively regulate activity of the mTOR pathway in T-cell acute lymphoblastic leukemia. Blood. 2007;110(1):278–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samon JB, Castillo-Martin M, Hadler M, et al. Preclinical analysis of the gamma-secretase inhibitor PF-03084014 in combination with glucocorticoids in T-cell acute lymphoblastic leukemia. Mol Cancer Ther. 2012;11(7):1565–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palomero T, Sulis ML, Cortina M, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13(10):1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garnett MJ, Edelman EJ, Heidorn SJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483(7391):570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konopleva M, Contractor R, Tsao T, et al. Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer Cell. 2006;10(5):375–388. [DOI] [PubMed] [Google Scholar]
- 16.van Delft MF, Wei AH, Mason KD, et al. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell. 2006;10(5):389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Neil J, Grim J, Strack P, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med. 2007;204(8):1813–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Souers AJ, Leverson JD, Boghaert ER, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med. 2013;19(2):202–208. [DOI] [PubMed] [Google Scholar]
- 20.Pan R, Hogdal LJ, Benito JM, et al. Selective BCL-2 inhibition by ABT-199 causes on-target cell death in acute myeloid leukemia. Cancer Discov. 2014;4(3):362–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin KH, Winter PS, Xie A, et al. Targeting MCL-1/BCL-XL Forestalls the Acquisition of Resistance to ABT-199 in Acute Myeloid Leukemia. Sci Rep. 2016;6:27696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tahir SK, Smith ML, Hessler P, et al. Potential mechanisms of resistance to venetoclax and strategies to circumvent it. BMC Cancer. 2017;17(1):399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin X, Morgan-Lappe S, Huang X, et al. ‘Seed’ analysis of off-target siRNAs reveals an essential role of Mcl-1 in resistance to the small-molecule Bcl-2/Bcl-XL inhibitor ABT-737. Oncogene. 2007;26(27):3972–3979. [DOI] [PubMed] [Google Scholar]
- 24.Yecies D, Carlson NE, Deng J, Letai A. Acquired resistance to ABT-737 in lymphoma cells that up-regulate MCL-1 and BFL-1. Blood. 2010;115(16):3304–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faber AC, Coffee EM, Costa C, et al. mTOR inhibition specifically sensitizes colorectal cancers with KRAS or BRAF mutations to BCL-2/BCL-XL inhibition by suppressing MCL-1. Cancer Discov. 2014;4(1):42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iorio F, Knijnenburg TA, Vis DJ, et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell. 2016;166(3):740–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faber AC, Farago AF, Costa C, et al. Assessment of ABT-263 activity across a cancer cell line collection leads to a potent combination therapy for small-cell lung cancer. Proc Natl Acad Sci U S A. 2015;112(11):E1288–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spender LC, Inman GJ. Phosphoinositide 3-kinase/AKT/mTORC1/2 signaling determines sensitivity of Burkitt’s lymphoma cells to BH3 mimetics. Mol Cancer Res. 2012;10(3):347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams MM, Lee L, Hicks DJ, et al. Key Survival Factor, Mcl-1, Correlates with Sensitivity to Combined Bcl-2/Bcl-xL Blockade. Mol Cancer Res. 2017;15(3):259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faber AC, Li D, Song Y, et al. Differential induction of apoptosis in HER2 and EGFR addicted cancers following PI3K inhibition. Proc Natl Acad Sci U S A. 2009;106(46):19503–19508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsieh AC, Costa M, Zollo O, et al. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer Cell. 2010;17(3):249–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mills JR, Hippo Y, Robert F, et al. mTORC1 promotes survival through translational control of Mcl-1. Proc Natl Acad Sci U S A. 2008;105(31):10853–10858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wendel HG, Silva RL, Malina A, et al. Dissecting eIF4E action in tumorigenesis. Genes Dev. 2007;21(24):3232–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brugarolas J, Lei K, Hurley RL, et al. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18(23):2893–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Helbig C, Gentek R, Backer RA, et al. Notch controls the magnitude of T helper cell responses by promoting cellular longevity. Proc Natl Acad Sci U S A. 2012;109(23):9041–9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moellering RE, Cornejo M, Davis TN, et al. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462(7270):182–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roderick JE, Tesell J, Shultz LD, et al. c-Myc inhibition prevents leukemia initiation in mice and impairs the growth of relapsed and induction failure pediatric T-ALL cells. Blood. 2014;123(7):1040–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balakrishnan K, Gandhi V. Bcl-2 antagonists: a proof of concept for CLL therapy. Invest New Drugs. 2013;31(5):1384–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts AW, Seymour JF, Brown JR, et al. Substantial susceptibility of chronic lymphocytic leukemia to BCL2 inhibition: results of a phase I study of navitoclax in patients with relapsed or refractory disease. J Clin Oncol. 2012;30(5):488–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chonghaile TN, Roderick JE, Glenfield C, et al. Maturation stage of T-cell acute lymphoblastic leukemia determines BCL-2 versus BCL-XL dependence and sensitivity to ABT-199. Cancer Discov. 2014;4(9):1074–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suryani S, Carol H, Chonghaile TN, et al. Cell and molecular determinants of in vivo efficacy of the BH3 mimetic ABT-263 against pediatric acute lymphoblastic leukemia xenografts. Clin Cancer Res. 2014;20(17):4520–4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hales EC, Orr SM, Larson Gedman A, Taub JW, Matherly LH. Notch1 receptor regulates AKT protein activation loop (Thr308) dephosphorylation through modulation of the PP2A phosphatase in phosphatase and tensin homolog (PTEN)-null T-cell acute lymphoblastic leukemia cells. J Biol Chem. 2013;288(31):22836–22848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dail M, Wong J, Lawrence J, et al. Loss of oncogenic Notch1 with resistance to a PI3K inhibitor in T-cell leukaemia. Nature. 2014;513(7519):512–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ebi H, Costa C, Faber AC, et al. PI3K regulates MEK/ERK signaling in breast cancer via the Rac-GEF, P-Rex1. Proc Natl Acad Sci U S A. 2013;110(52):21124–21129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vasudevan KM, Barbie DA, Davies MA, et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell. 2009;16(1):21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Costa C, Ebi H, Martini M, et al. Measurement of PIP3 levels reveals an unexpected role for p110beta in early adaptive responses to p110alpha-specific inhibitors in luminal breast cancer. Cancer Cell. 2015;27(1):97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ellisen LW, Ramsayer KD, Johannessen CM, et al. REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Mol Cell. 2002;10(5):995–1005. [DOI] [PubMed] [Google Scholar]
- 48.Sofer A, Lei K, Johannessen CM, Ellisen LW. Regulation of mTOR and cell growth in response to energy stress by REDD1. Mol Cell Biol. 2005;25(14):5834–5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang H, Kubica N, Ellisen LW, Jefferson LS, Kimball SR. Dexamethasone represses signaling through the mammalian target of rapamycin in muscle cells by enhancing expression of REDD1. J Biol Chem. 2006;281(51):39128–39134. [DOI] [PubMed] [Google Scholar]
- 50.Salmoiraghi S, Montalvo ML, Ubiali G, et al. Mutations of TP53 gene in adult acute lymphoblastic leukemia at diagnosis do not affect the achievement of hematologic response but correlate with early relapse and very poor survival. Haematologica. 2016;101(6):e245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.