Significance
Substrate specificity landscape of a protease enzyme is the set of all substrate sequences that are recognized/cut (and, as importantly, not recognized/cut) by the enzyme. Accurate and rapid elucidation of these landscapes for any given protease is key for the design of novel targeted proteases to prevent unwarranted off-target cleavage, and provides insight into the functional robustness of naturally occurring proteases. We developed a structure-guided approach for predicting protease substrate specificity landscapes, in which data from experiments in yeast and molecular simulations are combined using machine learning. Using this approach, we comprehensively map the sequence−energetics−function landscape of the hepatitis C virus NS3/4A protease and its drug-resistant variants.
Keywords: protease, sequence−function mapping, substrate specificity, machine learning, molecular modeling
Abstract
Biophysical interactions between proteins and peptides are key determinants of molecular recognition specificity landscapes. However, an understanding of how molecular structure and residue-level energetics at protein−peptide interfaces shape these landscapes remains elusive. We combine information from yeast-based library screening, next-generation sequencing, and structure-based modeling in a supervised machine learning approach to report the comprehensive sequence−energetics−function mapping of the specificity landscape of the hepatitis C virus (HCV) NS3/4A protease, whose function—site-specific cleavages of the viral polyprotein—is a key determinant of viral fitness. We screened a library of substrates in which five residue positions were randomized and measured cleavability of ∼30,000 substrates (∼1% of the library) using yeast display and fluorescence-activated cell sorting followed by deep sequencing. Structure-based models of a subset of experimentally derived sequences were used in a supervised learning procedure to train a support vector machine to predict the cleavability of 3.2 million substrate variants by the HCV protease. The resulting landscape allows identification of previously unidentified HCV protease substrates, and graph-theoretic analyses reveal extensive clustering of cleavable and uncleavable motifs in sequence space. Specificity landscapes of known drug-resistant variants are similarly clustered. The described approach should enable the elucidation and redesign of specificity landscapes of a wide variety of proteases, including human-origin enzymes. Our results also suggest a possible role for residue-level energetics in shaping plateau-like functional landscapes predicted from viral quasispecies theory.
Predicting the impact of genetic diversity on molecular recognition specificity of enzymes is of fundamental importance in molecular biology and also has implications for the design of novel enzymes with controllable molecular recognition properties. The balance between mutational tolerance and functional plasticity is encapsulated in the notion of mutational landscapes (1), which are high-dimensional maps that relate the function of individual biomolecular variants to their functional and/or evolutionary fitness (2, 3). Recent empirically determined sequence−function mappings of proteins (4–12) have enabled the partial construction of mutational landscapes. Typically, sequence−function mapping of proteins and protein−protein interactions involves partial enumeration of the possible sequence diversity (for example, all single mutations and a subset of double mutations at a large number of protein residue positions) and high-throughput functional evaluation coupled with deep sequencing (13–17). The astronomical size of sequence space, however, limits comprehensive elucidation of sequence−function landscapes with any one experimental approach. Computational biophysical methods may, in principle, assist in creation and analysis of functional and fitness landscapes (18, 19). Indeed, mutational landscapes of simple protein models, such as lattice models, have been extensively investigated using biophysical evolutionary theory and computational simulations (20–30), and connections with population genetics theories have been discovered (20, 31, 32). While pioneering and crucial insights have been obtained in these studies, chemically realistic atomic resolution structure-based elucidation of functional landscapes has remained elusive.
The genomes of several RNA viruses, e.g., human immunodeficiency virus (HIV) and hepatitis C virus (HCV), encode polyproteins, which are processed posttranslationally by viral proteases during maturation (33). The activity of HCV NS3 protease is key for viral maturation, as it cleaves exclusively at four specific sites in the viral polyprotein (Fig. 1A) to release individual nonstructural proteins (34) and also mediates inactivation of key human immunity proteins (35). The cleavage specificity of the protease is thus a key determinant of viral fitness, and its proper functioning includes negative specificity—the lack of cleavage of noncanonical sites on the viral protein and most host cell proteins—but how and whether these features are encoded in the protease−substrate interactions at a molecular level is currently unknown. Furthermore, while RNA viruses such as HCV are believed to be under strong purifying selection against nonsynonymous mutations (36–38), due to the extremely high error rates of the associated RNA polymerases (39–41), these viruses can exist as a population of variants called quasispecies (42, 43) even within a single host individual (44). Indeed, spontaneous emergence of diverse mutations (including drug-resistant mutations) was demonstrated in continuous evolution studies of the protease (45) and in viral replicon assays coupled to ultradeep sequencing (46). Can molecular interaction fidelity be maintained in the face of a large mutational load, and what, if any, are the limits imposed on the allowed genetic diversity by the underlying molecular interactions? The degeneracy of the genetic code, the thermodynamic and kinetic stabilities of RNA and proteins, and the presence of molecular chaperones may all contribute to the mutational robustness of the structures of individual viral biomolecules under a high mutational load (41). However, the mechanism by which key viral protein-based interactions—for example, protease−substrate interactions critical for viral propagation—may encode “fuzziness” (47) is not well understood.
Fig. 1.
Overview of workflow and results. (A) The HCV viral polyprotein depicting marked biological cleavage sites for the HCV NS3 protease. (B) Overview of the experimental and computational workflow. The construct shown (LY104) is the vector used for testing in the yeast-based assay. The substrate was cloned in the region between the FLAG and HA signaling tags. Extent of cleavage was measured as a ratio of FLAG/HA, with a ratio of 1 indicating that the substrate was uncleaved and a ratio of 0 indicating that the substrate was cleaved. (C) Validation of FACS gates for cleaved (blue), partially cleaved (black), and uncleaved (red) sequences using the yeast surface display assay. (D–G) Heatmaps showing per-position amino acid frequencies for (D) sequences taken from in vivo samples of HCV patients (8,726 sequences) compared with (E) sequences determined by our assay as cleaved (7,472 sequences), (F) sequences determined by our assay as partially cleaved (8,737 sequences), and (G) sequences determined by our assay as uncleaved (14,702 sequences).
Here, we use a combination of experimental (biochemical) and computational techniques to elucidate the specificity landscape of the interaction between HCV NS3/4A protease enzyme and its substrates. Using yeast surface display, next-generation sequencing, and a machine learning approach which combines features from experimental data and atomistic computational simulations (utilizing the Rosetta and Amber force fields) that we recently developed (48, 49), we construct the specificity landscapes (with cleavability assignments made for 3.2 million substrate pentapeptide sequences) of the HCV NS3/4A protease and three of its known drug-resistant variants (50).
Results
We mapped the protease−substrate interaction landscape for the HCV NS3/4A protease by considering all possible pentapeptide sequence combinations at positions P6 through P2 (Schechter and Berger nomenclature) (51) in the substrate. Positions P1 and P1′, between which the scissile bond is present, were maintained as the canonical C and A, respectively. In the remainder of this paper, we refer to individual pentapeptide patterns (e.g., the canonical cleavage sites DEMEE, EDVVC, ECTTP, and ALVTP) and omit the identity of the P1, P1′ residues. By mapping the substrate diversity for HCV NS3/4A wild-type protease as well as three drug-resistant protease mutants, we explore the interaction landscape for both protease and substrate diversity.
Exploration of the (P6-P2) Specificity Landscape of the HCV NS3/4A Protease Reveals a Diverse Specificity Profile.
To mimic the viral intrachain arrangement of substrate libraries and the protease, we utilized a modified version of the assay described by Iverson and coworkers (52) (Fig. 1B and SI Appendix, Fig. S1A). A mutagenic library was created incorporating degenerate codons at P6 to P2 specificity defining substrate positions (53, 54). In this assay, substrates are transported to the surface of yeast cells in a cleavage-dependent manner: The degree of cleavage is estimated by measuring the relative levels of substrate-flanking FLAG and HA tags using fluorescent-labeled antibodies. We have previously used this assay to test known and novel substrates of the HCV protease (48). A first round of yeast surface display assay and Fluorescence Assisted Cell Sorting (FACS) was performed with an inactive protease variant (S139A) to select for high expression of library variants, remove sequences containing stop codons in the substrate region, and deplete substrate sequences that are cleaved by yeast endoplasmic reticulum (ER) proteases (55).
The resulting substrate variants from the preselection were subjected to rounds of yeast surface display assay and FACS with an active-protease−containing construct to select cleaved, partially cleaved, and uncleaved variants using three sorting gates (Fig. 1B), based on the relative levels of anti-HA and anti-FLAG fluorescence values. The FLAG/HA ratio ranges between 0 for completely cleaved substrates and 1 for completely uncleaved substrates. Sorting gates were defined based on the distribution of populations observed for known cleaved and uncleaved sequences (48). This procedure was coupled with rounds of growth and selection to improve the signal-to-noise ratio for variants in each pool. Specificity profiles of the unselected population and isolated functional variants were determined using next-generation sequencing technology (Illumina NextSeq).
We identified a total of ∼379,000 unique sequences in the background pool corresponding to ∼12% of the possible amino acid diversity (3.2 million). Analysis of technical replicates as well as the overlap between the sequence pools was used to determine a count threshold (normalized count of 11) to remove noise from the sequencing data (SI Appendix, Supplementary Methods and Fig. S2). Based on these criteria, we identified 7,472, 8,737, and 14,702 unique pentapeptide sequences in the cleaved, partially cleaved, and uncleaved pools, respectively. In parallel, we performed Rosetta simulations on all 3.2 million sequences in the P6 to P2 region to determine energetic features of the protease−substrate models. We then used a support vector machine (SVM) in a supervised machine learning approach to predict the complete protease−substrate interaction landscape that incorporated sequence information procured from the aforementioned library and Rosetta-generated energetic features (Fig. 1B).
Several novel substrates identified from the three variant populations were tested as clonal populations in the yeast surface display assay system (Fig. 1C and SI Appendix, Fig. S3) to validate that individual sequences fall into the gates used for selection from the library (SI Appendix, Fig. S4 A–C). Some sequences were chosen because they were either different from any of the canonical cleavage sites at three or more positions (e.g., PSTVL) or implicated in epistatic networks (e.g., LSLIP; see SI Appendix, Supplementary Discussion), or were identified using a stricter enrichment threshold as described in Materials and Methods. A subset of these sequences was also tested in vitro to ensure that the cleavage properties observed in the yeast system were reproduced with purified protease and substrates (SI Appendix, Fig. S4D). The process of transforming quantitative cleavage efficiencies to discrete functional pools of cleaved, partially cleaved, and uncleaved substrates by definition involves a loss of information regarding relative binding efficiencies of substrates within each pool; however, clear differences in FLAG/HA ratio between the three categorizations are apparent (Fig. 1C).
We next analyzed the cleaved, partially cleaved, and uncleaved sequence sets obtained from deep sequencing. The cleaved specificity profile has greater diversity than the substrates identified from viral genomes sequenced from patient populations (SI Appendix, Supplementary Methods and Fig. 1D). For example, we observe that a more diverse subset of amino acids is tolerated at substrate positions P6 and P5 in our cleaved and partially cleaved pools (Fig. 1 E and F) whereas the patient-derived genomes display a high enrichment of Asp and Glu specifically at these positions. The relative abundance (compared with all 20 amino acids, normalized to 1) of Asp at P6 is 0.08 in the cleaved population compared with 0.03 in partially cleaved and 0.02 in uncleaved populations; the relative abundance of Glu at position P5 in the cleaved population is 0.06 compared with 0.03 and 0.01 in partially cleaved and uncleaved populations. In contrast, these abundances are P6-Asp-0.47, and P5-Glu-0.35 in the patient-derived genome data. Overall, the viral genome-derived specificity profile has a lower information content (i.e., lower diversity) than that of the cleaved specificity profile obtained using our method (1.92 bits vs. 3.85 bits, out of a maximum 4.32 bits), pointing to the relative flatness of the extended substrate specificity profile (SI Appendix, Table S10) that can be recognized by the protease.
Strikingly, even though the actual sequences in each pool are chosen to be distinct (SI Appendix, Fig. S9B), the overall specificity profiles of the three sequence sets (cleaved, partially cleaved, and uncleaved) are similar (Fig. 1 E–G): The cosine similarities (ranges between 0 and 1, dissimilar to similar) of cleaved to uncleaved, cleaved to partially cleaved, and partially cleaved to uncleaved are 0.61, 0.86, and 0.84, respectively. Thus, cleaved sequences are more similar overall to partially cleaved sequences than to uncleaved ones. Additionally, there are several differences in the enrichment of certain residues at each position in the three pools. For example, we found prolines enriched at position P2 in the cleaved (relative abundance of 0.2) and partially cleaved populations (relative abundance of 0.14) compared with the uncleaved set (relative abundance 0.04), which, in turn, prefers proline at P3 (0.23 relative abundance). These trends correspond well with the fact that two out of four canonical cleaved sequences have proline at P2 (ECTTP and ALVTP). While some of the above trends are also reflected in the sequences we tested during method validation (Fig. 1C), it is evident that overall sequence composition or individual positional enrichments cannot be directly used to predict the pool assignments of individual sequences. For example, His is somewhat enriched (relative abundance of 0.09 in cleaved population) at P6 in the cleaved sequence pool, but the sequence HNTSN is experimentally determined to be in the uncleaved pool (Fig. 1C and SI Appendix, Fig. S3). We conclude that interactions between amino acids at various substrate positions (mediated possibly through interaction networks in the protease) influence the cleavability, thereby motivating the need for an analysis of the specificity landscape using properties of whole pentapeptide sequences and models of their complexes with the protease.
Clustering Among Cleaved, Partially Cleaved, and Uncleaved Substrates.
To visualize the functionally labeled sequence space of the experimentally derived substrates, we generated a force-directed graph (Fig. 2A) (56, 57) in which each node represents a sequence and is colored according to the functional pool to which it belongs. Nodes are connected by an edge if they differ by one amino acid (Hamming distance = 1). Cleaved substrates exhibit significant clustering in the resulting graph (Fig. 2A). To examine the landscape in greater detail around the cleaved sequences, we generated a subgraph of the cleaved sequences (Fig. 2B), identified four clusters in this graph using the Gephi (56) modularity algorithm, and determined corresponding profiles for each cluster. One identified cluster is clearly related to a canonical substrate, DEMEE, the starting point for our library generation protocol.
Fig. 2.
Sequences of a given cleavage status cluster together. (A) Force-directed graph of experimentally investigated substrates in amino acid sequence space. Blue nodes are cleaved, red are uncleaved, and black is partially cleaved. Edges connect nodes that are within one hamming distance of each other. Orphan nodes and dyads are not shown. (B) Force-directed graph of cleaved sequences. Colors denote clusters, which are shown as specificity profiles outlined in the same color as the corresponding cluster. (C) Fraction of neighbors that are cleaved (blue bars), partially cleaved (black bars), and uncleaved (red bars) for cleaved, partially cleaved, and uncleaved sequences. Frequencies are normalized so that the integral of each histogram is equal to 1.
To determine whether the clustering behavior observed in the cleaved sequence pool is also found in the partially cleaved and uncleaved pools, we calculated the fraction of neighbors in the same functional pool for all sequences (Fig. 2C). We find that, similar to cleaved sequences, uncleaved sequences are most frequently surrounded by uncleaved neighbors, indicating clustering behavior for this functional pool as well. On average, cleaved sequence neighbors are 66.4% cleaved, and uncleaved sequence neighbors are 83.3% uncleaved. Partially cleaved sequences are the least clustered among the three pools, having, on average, 53% neighbors belonging to the same pool. These distributions indicate that, in the specificity landscape, clusters of partially cleaved sequences surround clusters of cleaved and uncleaved ones.
To delineate how the three functional populations, which appear to be individually clustered in sequence space, are connected to each other, we used the PageRank metric (58). This metric predicts the likelihood of reaching a node given a random walk on the substrate specificity landscape starting from a chosen sequence. The sequence that was used as the template for library generation, DEMEE, was chosen as the starting point in this analysis. Partially cleaved substrates have, on average, higher pageranks (SI Appendix, Fig. S5A) than either cleaved or uncleaved substrates, indicating that they are more connected to other nodes in the graph. The mean pagerank of the partially cleaved sequences is 0.71 × 10−4, while the mean pageranks of the cleaved and uncleaved sequences are 0.26 × 10−4 and 0.35 × 10−4, respectively. These connectivity patterns imply that, in the experimentally determined landscape, where the sampling is largely limited to the region around the canonical sequence DEMEE, increased mutational distance from DEMEE is correlated with loss of function: Partially cleaved sequences have intermediate distance, and uncleaved sequences are more sequence-distant. The high connectivity of partially cleaved nodes to other clusters, as determined by PageRank and neighbor analysis, suggests that the local topology of the specificity landscape is smooth. The loss of cleavability upon mutation is not abruptly precipitous [as observed for other systems (7)] but occurs, on average, via partially cleaved nodes. However, these average connectivity properties do not indicate the likelihood of individual mutational trajectories on the landscape, for which more quantitative analysis with catalytic efficiencies of cleavage of individual variants will be required.
The graph generated by the experimentally derived sequences is incomplete (∼30,000 nodes out of the 3.2 million possible). To test whether the observed clustering and PageRank distributions are an artifact of the limited sampling of the experiment, we generated 10 random graphs (e.g., SI Appendix, Fig. S6A). The random graphs were generated by choosing new ending node assignments for each edge randomly while preserving the cleavability label of all nodes. In these control random graphs, where functional assignments and mutational distance are decoupled by construction, the pageranks trend is cleaved (mean: 0.17 × 10−4) < partially cleaved (mean: 0.28 × 10−4) << uncleaved (mean: 0.47 × 10−4), as opposed to cleaved < uncleaved << partially cleaved in the experimentally derived graph, further highlighting the relationship between mutational distance and function preservation.
Energetic Features Derived from Rosetta Modeling Enable Reconstruction of the Complete Protease−Pentapeptide Substrate Landscape.
While the experimentally derived populations of the cleaved, partially cleaved, and uncleaved sequences display striking clustering patterns in sequence space, they include a small fraction of the entire sequence diversity in the P6 to P2 region (3.2 million sequences). To predict cleavability of all possible 3.2 million sequences in the interaction landscape, we used an SVM method that we developed previously (48). Briefly, each sequence was threaded onto a bound complex based on a modeled near-attack conformation, and the complex was then relaxed to accommodate the substrate while maintaining favorable catalytic geometry. Energy evaluation of each of the 3.2 million complexes was performed using Rosetta and Amber simulation packages.
A binary classification (cleaved/uncleaved) SVM was trained on two sets of sequences. The first set was a subset of experimentally identified sequences that were identified using more stringent criteria than the original set of experimentally categorized sequences (1,817 cleaved and 3,605 uncleaved sequences; see Materials and Methods for details). The second set included 196 cleaved sequences and 1,943 uncleaved sequences identified by Shiryaev et al. (59), and both sets together yielded 7,338 unique sequences. Training features consisted of structure-based features (energies of interaction) and sequence-based features (see Materials and Methods and SI Appendix, Fig. S7A). We initially cross-validated the SVM on the training set using an 80:20 split with 100 iterations, which yielded an average AUROC (area under receiver operating characteristic) of 0.96 (SI Appendix, Fig. S7D) indicating high recapitulation of training data (perfect performance would lead to an AUROC of 1). We then used the SVM to predict cleaved and uncleaved labels for the remaining 3,192,658 sequences. These predictions have a precision of 0.95 at a recall level of 0.89 for an overall accuracy of 0.95 (SI Appendix, Fig. S7C) for the experimentally derived assignments that were left out of the training set (5,906 cleaved sequences and 11,087 uncleaved sequences). For testing of SVM predictions, we selected six sequences that were not found within the experimentally defined set and had predicted distances greater than 2 from the SVM-calculated separation hyperplane. For all of the tested sequences, we find good agreement with the SVM-based predictions (Fig. 3A): The experimentally observed FLAG/HA ratios are ∼0 and ∼1 for the predicted cleaved and uncleaved test sequences, respectively. To validate that the successful predictions are not localized to specific regions of sequence space, we visualized a subgraph of predicted cleaved sequences, present at a distance of >2 from the hyperplane constructed by the SVM learning procedure (SI Appendix, Fig. S7B). The experimentally identified cleaved sequences are distributed evenly across sequence clusters in the predicted cleaved population graph, further indicating that the machine learning procedure performs evenly across the sequence diversity of cleaved sequences.
Fig. 3.
Structural modeling provides insights into basis for cleavage. (A) Validation assay performed for three predicted cleaved, one partially cleaved, and three uncleaved sequences using a yeast surface display-based technique. (B and C) The volume occupied by (B) TATTA and (C) EDVVC. EDVVC occupies an optimal volume, making good contacts with the protease residue sidechains. TATTA fits in the available space but does not make optimal contacts, thus resulting in suboptimal interaction energetics making TATTA a suboptimal substrate. (D) Structure of two models, ECTIP (cleaved) and RPGPG (uncleaved).
Structural and Energetic Bases for Observed Specificity Patterns.
Having obtained and validated predictions of cleavability by combining experimental and computational data using supervised learning, we turned to structural models of protease−substrate complexes to obtain insight into the underlying structural basis of observed specificity patterns. For example, a comparative analysis of the partially cleaved substrate “TATTA” and canonical substrate “EDVVC” reveals that the former, composed of small residues, does not completely occupy the substrate cavity volume, whereas EDVVC occupies the entire cavity (Fig. 3 B and C). The lack of voids at the interface and several hydrogen bonds formed by the canonical substrate lead to better binding [binding interaction energy = −80.2 Rosetta energy units (Reu), as opposed to −77.5 Reu for TATTA], resulting in better cleavage for this substrate. Apart from sidechain-based interaction patterns, models also capture backbone conformational changes that affect the orientation of the substrate in the active site. For example, in the model corresponding to the sequence RPGPG (uncleaved), the proline present at P3 in RPGPG (Fig. 3D) bends the peptide chain away from the protease, resulting in breaking of the crucial backbone hydrogen bond patterns that are characteristic of protease−substrate interactions (60).
Connectivity Properties of the Experimentally Determined and Computationally Reconstructed Landscapes.
Having computed the entire P6 to P2 specificity landscape, we next examined the connectivity patterns between cleaved and uncleaved sequences in this reconstructed landscape. As with the experimentally determined landscape, the reconstructed landscape also shows evidence of clustering between cleaved and uncleaved nodes (SI Appendix, Fig. S5 H and I): Most neighbors of (un)cleaved nodes are (un)cleaved. While the clustering properties are similar, the lack of a partially cleaved category in the computationally derived dataset and incompleteness of the experimental dataset also lead to some differences in the measured connectivity properties of these graphs (SI Appendix, Fig. S5 F and G).
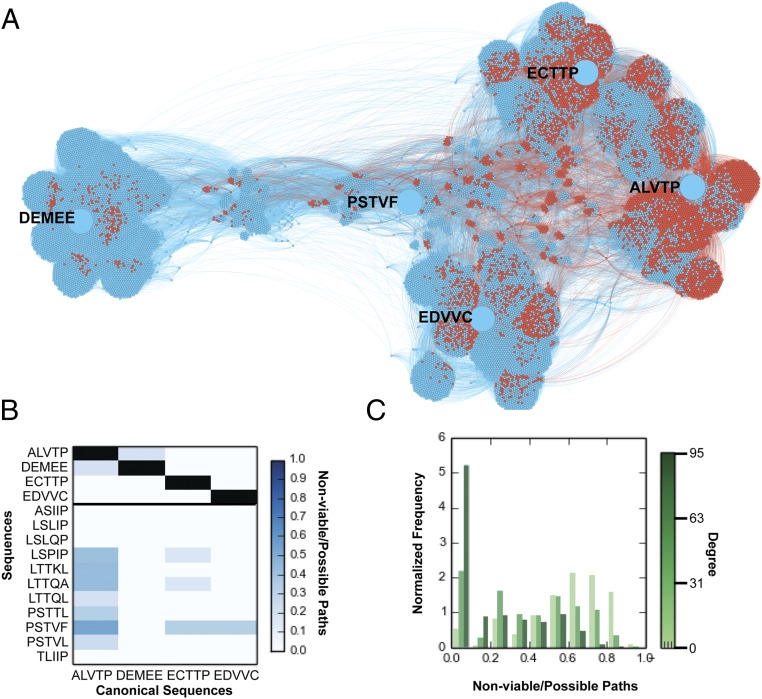
Both the reconstructed and experimentally derived landscapes feature several “novel” cleaved sequence patterns (defined as more than three substitutions away from a canonical recognition motif). To determine the accessibility of novel sequences, we investigated the network connectivity between identified (and individually experimentally validated) novel cleaved and canonical cleaved sequences. As an example, we generated a subgraph of the sequence space connecting the canonical cleaved sequences (DEMEE, EDVVC, ECTTP, and ALVTP) with each other as well as the novel cleaved sequences, e.g., PSTVF (Fig. 4A). Analysis of all internode shortest paths in the predicted graph shows that there exist many paths between canonical and novel sequences that do not include uncleaved nodes (viable paths), while some paths involve traversal of at least one predicted uncleaved node (nonviable paths).
Fig. 4.
Mutational trajectories between novel sequences and canonical cleaved sequences contain nonviable paths. (A) Force-directed graph between the five canonical sequences: DEMEE, ECTTP, EDVVC, ALVTP, and the novel cleaved sequence PSTVF (depicted by large blue nodes). The graph includes the intermediate sequences between PSTVF and all canonical sequences, as well as all of the neighbors of these sequences. The cleaved sequences in the mutational trajectories are denoted by blue nodes, and the uncleaved are denoted by red. Cleavage statuses are predicted by the SVM. (B) Nonviable path fraction from canonical sequences to other canonical sequences and novel sequences. (C) Histogram depicting the nonviable path fraction frequencies between each novel sequence and its closest canonical sequence. Histogram is shown separately for binned degrees.
For every novel cleaved sequence, we identified all shortest paths between that sequence and each canonical cleaved sequence, and then asked whether each of these paths was a viable trajectory (i.e., did not include an uncleaved sequence). For example, for PSTVF, we found that there were 120 possible shortest paths to DEMEE, e.g., PSTVF → PSTVE → PSTEE → PSMEE → PEMEE → DEMEE. None of the shortest paths between PSTVF and DEMEE included uncleaved nodes; thus, 100% of the shortest paths were viable. In contrast, out of the 24 possible shortest paths between PSTVF and ECTTP, only 8 of them did not include any uncleaved nodes; thus, the fraction of viable shortest paths between PSTVF and ECTTP is 33%. The relatively high fraction of nonviable paths (66%) between PSTVF and ECTTP indicates a potentially higher barrier between PSTVF and ECTTP compared with DEMEE. We similarly calculated the fraction of nonviable shortest paths between every canonical sequence and every novel predicted cleaved sequence. Canonical sequences have a lower fraction of nonviable paths between themselves than between canonical sequences and the novel sequences (Fig. 4B); the canonical to canonical nonviable path fractions have a mean of 0.0 and range from 0 to 0.2, while the canonical to novel nonviable path fractions have a mean of 0.1 and range from 0 to 0.5. Finally, the canonical sequence ALVTP has a higher proportion of nonviable paths than the other canonical sequences, and is surrounded to a greater extent by uncleaved neighbors, suggesting that the degree of a node (number of cleaved neighbors) is correlated with its reachability from canonical sequence nodes. Interestingly, it has been found that the ALVTP site is a suboptimal substrate of the protease compared with other canonical sites and, at times, is not processed in vivo (59). Thus, it is likely that the polyprotein context also plays a role in the cleavage of different sites in vivo.
We next investigated whether the nonviable path fraction of novel cleaved nodes to canonical nodes is correlated with the number of cleaved neighbors of a node. We divided the novel sequences into three groups based on their number of cleaved neighbors: 0 to 31 cleaved neighbors, 32 to 63 cleaved neighbors, and 64 to 95 cleaved neighbors. We find that those novel cleaved sequences that have a higher degree have, on average, a higher fraction of viable trajectories to canonical nodes (Fig. 4C). Thus, as expected for a highly clustered graph, the higher single mutational tolerance of a given novel sequence is correlated with its ability to be reachable from/to canonical sequences that are at least three amino acid substitutions away in sequence space.
We note that the nature of the qualitative bins within which substrates are classified in our analyses precludes evaluating the evolutionary likelihood of individual mutational trajectories. In particular, paths that appear to be completely viable (i.e., contain no uncleaved nodes) may contain nodes that are of lower catalytic efficiencies and/or fitness than the starting point, and therefore are less likely to be traversed. Therefore, our analysis considers all paths between nodes and focuses on relative nonviable path fraction, e.g., nonviable path fraction between canonical and other canonical substrates vs. nonviable path fraction between canonical and novel substrates. An uncleaved node in a path is guaranteed to make it nonviable, but demonstrating that two nodes which have 100% viable paths between themselves form a neutral network would require more quantitative characterization of the catalytic efficiencies and molecular fitness of all of the nodes.
Specificity Landscapes of Drug-Resistant Protease Variants.
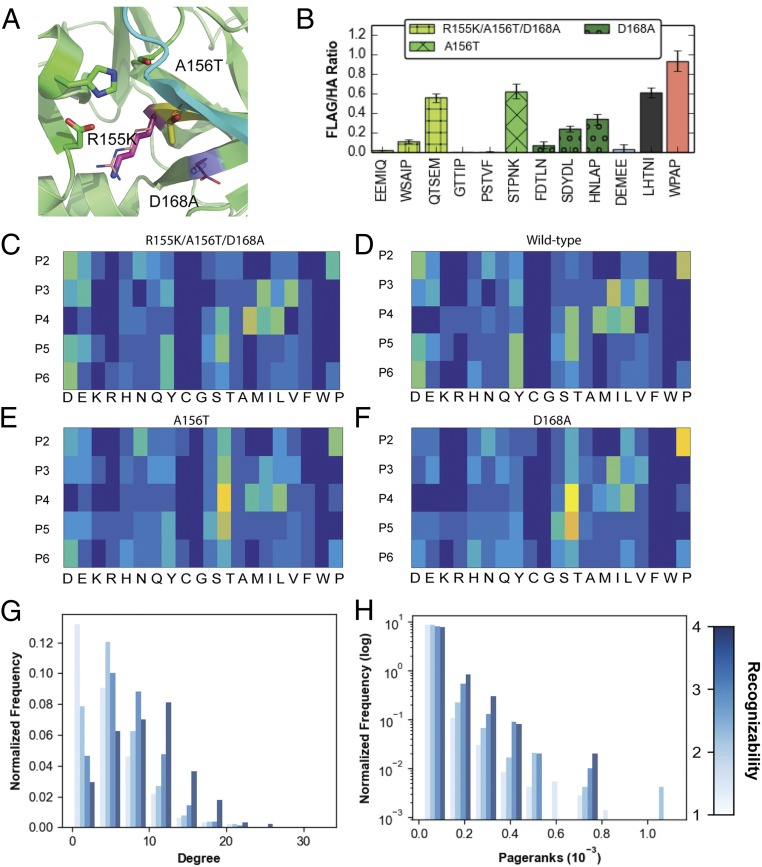
As the NS3/4A protease plays a key role in the viral assembly and maturation process, it is a target for therapeutics that aim at neutralizing viral activity. However, due to prevalence of quasispecies that are lurking at low levels in the population (61), several viral variants get exposed to the drug. Some of these develop resistance, and propagate to form drug-resistant mutants (DRMs). To investigate how drug-resistant variants of the protease affect the mutational robustness, we explored the specificity landscape for three DRMs: A156T, D168A, and R155K/A156T/D168A (Fig. 5). We find that the DRM proteases have similar specificity profiles to those of each other and to that of the wild-type protease, with cosine similarities of DRM specificity profiles to wild-type specificity profiles that range between 0.89 and 0.99 (Fig. 5 C–F). However, the sequences cleaved by the variant and wild-type proteases are distinct, with an 8 to 21% overlap in sequences between the each of the various DRMs and wild-type proteases (SI Appendix, Fig. S9 I and J). Upon comparing the graphical properties of the specificity landscapes of the various protease variants, we observe that substrates that are experimentally detected in the cleaved pools of a greater number of protease variants (more recognizable) are more reachable (higher pageranks) and more connected (higher degree) in each graph (Fig. 5 G and H). These data indicate that more recognizable substrates appear to be more robust to changes in the protease. Thus, functional clustering in sequence space appears to be a robust feature of the molecular recognition between the HCV NS3/4 protease variants and their substrates.
Fig. 5.
DRMs have similar structures and similar specificity profiles. (A) Drug-resistant variant structures. Mutations are outlined in sticks, and WT residues are outlined in lines. Active site residues are represented as green sticks. (B) Validation assay performed using yeast surface display for each of the mutants. (C–F) Mutant cleaved sequence specificity profiles for the (C) triple mutant, (F) D168A, (E) A156T, and (D) wild type, showing that the mutants have very similar specificity profiles with slight variation compared with the WT. (G and H) Substrate sequences that are recognized by a greater number of variants have higher (G) degrees and (H) pageranks.
Discussion
We combined information gleaned from library screening in yeast, deep sequencing, and structure-based modeling, using a machine learning framework to delineate the protease−substrate interaction landscape of HCV NS3/4A protease. These results provide atomic-resolution insight into the bases for both positive and negative specificity. We used a yeast surface display-based assay that relies on the cleavage of the substrate region in the ER of yeast followed by cell sorting into gates and deep sequencing. We note that our assay is qualitative, and does not permit association of the detected signal from deep sequencing with quantitative cleavability of substrates. Indeed, while we have validated that assignments to the three different pools are accurate with at least ∼20 individual sequences, the identified cleaved and partially cleaved substrates may represent a range of catalytic efficiencies within their pools. On the other hand, the assay construct with the protease and substrate on the same chain is a good representation of the situation in the virus, where the substrates of the protease are part of the same polyprotein (although both cis and trans cleavages occur), leading to high effective concentrations of substrates ([S] >> KM) in vivo. Under these saturating conditions in the virus and in our assay, we argue that selectivity and catalytic efficiency are both determined to a great extent by the goodness of fit of various substrates in the protease active site (i.e., by the relative binding between the different substrates). Similarly, our SVM-based machine learning approach to combine experimental and computational data also is not without errors, showing a false-positive rate of ∼5 to 10% on the experimental data. While we have validated several predictions on individual sequences (Figs. 1, 3, and 5), it is possible that some individual sequences may be mispredicted. However, the overall trends regarding the connectivity patterns observed for the entire landscape should be robust to the misprediction noise. Further ongoing development of the computational and experimental methods that we utilized is expected to help make the approach outlined here more quantitatively accurate.
We note that our graph theoretic analyses (Fig. 4), which draw from similar previous analyses (10, 62), report qualitatively on the local topology of the recognition specificity landscape, which is related to, but distinct from, a molecular fitness landscape. This difference arises as functional bins, whose boundaries are derived from experiments and within which substrates are classified (cleaved, partially cleaved, uncleaved), serve as a qualitative proxy for the catalytic efficiency and associated molecular fitness; binning necessarily flattens functional differences within a pool, and there is loss of information. Overall clustering and connectivity properties thus qualitatively describe the specificity landscape. On the molecular fitness landscape, the likelihood/fitness of individual mutational trajectories may depend quantitatively on the relative catalytic efficiency of traversed nodes. Definitive demonstration of specific network topologies (e.g., a neutral network) would require a more quantitative approach for molecular fitness characterization.
Our results provide a biophysical baseline for understanding how HCV protease substrates may sample genetic diversity while maintaining function. For example, our analysis suggests that viral evolution occurring at the substrate sites in the polyprotein could also contribute to drug resistance. Due to the high interconnectedness of partially cleaved and fully cleaved clusters, novel sequences that are better substrates of drug-resistant variants may easily arise. This mode of substrate coevolution-based drug resistance has been observed in HIV-1 (63). At the same time, our analysis of the dominant HCV sequences obtained from patients suggests that the protease−substrate interactions may also contribute to negative selection (SI Appendix, Supplementary Discussion) and help limit the acquisition of heterogeneity. The molecular interaction between the protease and substrates, while key for viral survival, does not directly determine evolutionary fitness and is one of the many evolutionary forces at play, especially in the “wild” (64). Other factors such as the impact of genetic diversity on intrahost population size, stability, size and structure of the viral RNA genome, and interactions between the host and viral machineries and other environment-dependent factors are also important to consider while considering the evolution of HCV (46). Thus, apart from the connectivity properties of the specificity landscape (Fig. 4), a confluence of various factors, including the lack of sampling of genetic diversity by the virus in the wild and a relatively small number of genomes sequenced, may contribute to the lack of genetic diversity observed in patient-derived sequences of HCV protease substrates.
In summary, our exploration of a viral molecular specificity landscape uncovers novel specificities for the HCV NS3/4A protease. The developed specificity landscape enumeration approach is general, and combining experimental deep sequencing and structural modeling at a matching high throughput, followed by supervised machine learning, may be useful for elucidating a significantly larger space of sequence−function relationships for a variety of other natural or designed protease enzyme systems.
Materials and Methods
See SI Appendix for detailed descriptions of experimental procedures and computational methods and for additional data: detailed overview (SI Appendix, Fig. S1), threshold determination (SI Appendix, Fig. S2), flow cytometry (SI Appendix, Figs. S3 and S4), graph metrics (SI Appendix, Fig. S5), experimentally derived graphs (SI Appendix, Fig. S6), SVM classification (SI Appendix, Figs. S7, S16, and S17), negative selection (SI Appendix, Fig. S8), population overlap (SI Appendix, Fig. S9), positive and negative epistasis (SI Appendix, Fig. S10), specificity profile comparisons (SI Appendix, Fig. S11), sorting replicates (SI Appendix, Fig. S12), sorting gates for all enriched populations (SI Appendix, Fig. S13), molecular cloning experimental overview (SI Appendix, Fig. S14), and sequence subsampling with more stringent criteria (SI Appendix, Fig. S15).
Supplementary Material
Acknowledgments
We thank D. Zorine, L. Cuypers, H. Khiabanian, D. Kumar, T. Choi, E. Sontag, and S. Annavarappu for technical assistance, and J. Marcotrigiano, T. Whitehead, A. Keating, M. Harms, and D. Tawfik for helpful suggestions. We also thank Y. Li, B. Iverson, and G. Georgiou for sharing the LY104 plasmid used in the yeast ER sequestration screening assay experiments. This work was supported by NSF Grant MCB1716623 (to S.D.K.) and NSF Graduate Research Fellowship Grant DGE-1433187 (to A.B.R.). This work used resources from the Rutgers Discovery Informatics Institute, which is supported by Rutgers and the State of New Jersey.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805256116/-/DCSupplemental.
References
- 1.Smith JM. Natural selection and the concept of a protein space. Nature. 1970;225:563–564. doi: 10.1038/225563a0. [DOI] [PubMed] [Google Scholar]
- 2.Wright S. Evolution in Mendelian populations. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Visser JA, Krug J. Empirical fitness landscapes and the predictability of evolution. Nat Rev Genet. 2014;15:480–490. doi: 10.1038/nrg3744. [DOI] [PubMed] [Google Scholar]
- 4.Fowler DM, et al. High-resolution mapping of protein sequence-function relationships. Nat Methods. 2010;7:741–746. doi: 10.1038/nmeth.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hietpas RT, Jensen JD, Bolon DN. Experimental illumination of a fitness landscape. Proc Natl Acad Sci USA. 2011;108:7896–7901. doi: 10.1073/pnas.1016024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim I, Miller CR, Young DL, Fields S. High-throughput analysis of in vivo protein stability. Mol Cell Proteomics. 2013;12:3370–3378. doi: 10.1074/mcp.O113.031708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sarkisyan KS, et al. Local fitness landscape of the green fluorescent protein. Nature. 2016;533:397–401. doi: 10.1038/nature17995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wrenbeck EE, Azouz LR, Whitehead TA. Single-mutation fitness landscapes for an enzyme on multiple substrates reveal specificity is globally encoded. Nat Commun. 2017;8:15695. doi: 10.1038/ncomms15695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Firnberg E, Labonte JW, Gray JJ, Ostermeier M. A comprehensive, high-resolution map of a gene’s fitness landscape. Mol Biol Evol. 2014;31:1581–1592. doi: 10.1093/molbev/msu081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Podgornaia AI, Laub MT. Protein evolution. Pervasive degeneracy and epistasis in a protein-protein interface. Science. 2015;347:673–677. doi: 10.1126/science.1257360. [DOI] [PubMed] [Google Scholar]
- 11.Bandaru P, et al. Deconstruction of the Ras switching cycle through saturation mutagenesis. eLife. 2017;6:e27810. doi: 10.7554/eLife.27810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McLaughlin RN, et al. The spatial architecture of protein function and adaptation. Nature. 2012;491:138–142. doi: 10.1038/nature11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fowler DM, Fields S. Deep mutational scanning: A new style of protein science. Nat Methods. 2014;11:801–807. doi: 10.1038/nmeth.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reich LL, Dutta S, Keating AE. SORTCERY-A high-throughput method to affinity rank peptide ligands. J Mol Biol. 2015;427:2135–2150. doi: 10.1016/j.jmb.2014.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klesmith JR, Bacik JP, Wrenbeck EE, Michalczyk R, Whitehead TA. Trade-offs between enzyme fitness and solubility illuminated by deep mutational scanning. Proc Natl Acad Sci USA. 2017;114:2265–2270. doi: 10.1073/pnas.1614437114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenson JM, Ryan JA, Grant RA, Letai A, Keating AE. Epistatic mutations in PUMA BH3 drive an alternate binding mode to potently and selectively inhibit anti-apoptotic Bfl-1. eLife. 2017;6:e25541. doi: 10.7554/eLife.25541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Louie RHY, Kaczorowski KJ, Barton JP, Chakraborty AK, McKay MR. Fitness landscape of the human immunodeficiency virus envelope protein that is targeted by antibodies. Proc Natl Acad Sci USA. 2018;115:E564–E573. doi: 10.1073/pnas.1717765115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodrigues JV, et al. Biophysical principles predict fitness landscapes of drug resistance. Proc Natl Acad Sci USA. 2016;113:E1470–E1478. doi: 10.1073/pnas.1601441113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butler TC, Barton JP, Kardar M, Chakraborty AK. Identification of drug resistance mutations in HIV from constraints on natural evolution. Phys Rev E. 2016;93:022412. doi: 10.1103/PhysRevE.93.022412. [DOI] [PubMed] [Google Scholar]
- 20.Echave J, Wilke CO. Biophysical models of protein evolution: Understanding the patterns of evolutionary sequence divergence. Annu Rev Biophys. 2017;46:85–103. doi: 10.1146/annurev-biophys-070816-033819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sikosek T, Chan HS. Biophysics of protein evolution and evolutionary protein biophysics. J R Soc Interface. 2014;11:20140419. doi: 10.1098/rsif.2014.0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding F, Dokholyan NV. Emergence of protein fold families through rational design. PLoS Comput Biol. 2006;2:e85. doi: 10.1371/journal.pcbi.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DePristo MA, Weinreich DM, Hartl DL. Missense meanderings in sequence space: A biophysical view of protein evolution. Nat Rev Genet. 2005;6:678–687. doi: 10.1038/nrg1672. [DOI] [PubMed] [Google Scholar]
- 24.Yang JR, Liao BY, Zhuang SM, Zhang J. Protein misinteraction avoidance causes highly expressed proteins to evolve slowly. Proc Natl Acad Sci USA. 2012;109:E831–E840. doi: 10.1073/pnas.1117408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bornberg-Bauer E, Chan HS. Modeling evolutionary landscapes: Mutational stability, topology, and superfunnels in sequence space. Proc Natl Acad Sci USA. 1999;96:10689–10694. doi: 10.1073/pnas.96.19.10689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bloom JD, Wilke CO, Arnold FH, Adami C. Stability and the evolvability of function in a model protein. Biophys J. 2004;86:2758–2764. doi: 10.1016/S0006-3495(04)74329-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Nimwegen E, Crutchfield JP, Huynen M. Neutral evolution of mutational robustness. Proc Natl Acad Sci USA. 1999;96:9716–9720. doi: 10.1073/pnas.96.17.9716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manhart M, Morozov AV. Protein folding and binding can emerge as evolutionary spandrels through structural coupling. Proc Natl Acad Sci USA. 2015;112:1797–1802. doi: 10.1073/pnas.1415895112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sailer ZR, Harms MJ. High-order epistasis shapes evolutionary trajectories. PLoS Comput Biol. 2017;13:e1005541. doi: 10.1371/journal.pcbi.1005541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Serohijos AW, Shakhnovich EI. Merging molecular mechanism and evolution: Theory and computation at the interface of biophysics and evolutionary population genetics. Curr Opin Struct Biol. 2014;26:84–91. doi: 10.1016/j.sbi.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bershtein S, Serohijos AW, Shakhnovich EI. Bridging the physical scales in evolutionary biology: From protein sequence space to fitness of organisms and populations. Curr Opin Struct Biol. 2017;42:31–40. doi: 10.1016/j.sbi.2016.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yost S, Marcotrigiano J. Viral precursor polyproteins: Keys of regulation from replication to maturation. Curr Opin Virol. 2013;3:137–142. doi: 10.1016/j.coviro.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scheel TK, Rice CM. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med. 2013;19:837–849. doi: 10.1038/nm.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 36.Sanjuán R, Moya A, Elena SF. The distribution of fitness effects caused by single-nucleotide substitutions in an RNA virus. Proc Natl Acad Sci USA. 2004;101:8396–8401. doi: 10.1073/pnas.0400146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Acevedo A, Brodsky L, Andino R. Mutational and fitness landscapes of an RNA virus revealed through population sequencing. Nature. 2014;505:686–690. doi: 10.1038/nature12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Visher E, Whitefield SE, McCrone JT, Fitzsimmons W, Lauring AS. The mutational robustness of influenza a virus. PLoS Pathog. 2016;12:e1005856. doi: 10.1371/journal.ppat.1005856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Domingo E, Holland JJ. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 40.Holland J, et al. Rapid evolution of RNA genomes. Science. 1982;215:1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- 41.Lauring AS, Frydman J, Andino R. The role of mutational robustness in RNA virus evolution. Nat Rev Microbiol. 2013;11:327–336. doi: 10.1038/nrmicro3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andino R, Domingo E. Viral quasispecies. Virology. 2015;479-480:46–51. doi: 10.1016/j.virol.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eigen M. Viral quasispecies. Sci Am. 1993;269:42–49. doi: 10.1038/scientificamerican0793-42. [DOI] [PubMed] [Google Scholar]
- 44.Cristina J, del Pilar Moreno M, Moratorio G. Hepatitis C virus genetic variability in patients undergoing antiviral therapy. Virus Res. 2007;127:185–194. doi: 10.1016/j.virusres.2007.02.023. [DOI] [PubMed] [Google Scholar]
- 45.Dickinson BC, Packer MS, Badran AH, Liu DR. A system for the continuous directed evolution of proteases rapidly reveals drug-resistance mutations. Nat Commun. 2014;5:5352. doi: 10.1038/ncomms6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geller R, et al. Highly heterogeneous mutation rates in the hepatitis C virus genome. Nat Microbiol. 2016;1:16045. doi: 10.1038/nmicrobiol.2016.45. [DOI] [PubMed] [Google Scholar]
- 47.Tokuriki N, Oldfield CJ, Uversky VN, Berezovsky IN, Tawfik DS. Do viral proteins possess unique biophysical features? Trends Biochem Sci. 2009;34:53–59. doi: 10.1016/j.tibs.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 48.Pethe MA, Rubenstein AB, Khare SD. Large-scale structure-based prediction and identification of novel protease substrates using computational protein design. J Mol Biol. 2017;429:220–236. doi: 10.1016/j.jmb.2016.11.031. [DOI] [PubMed] [Google Scholar]
- 49.Rubenstein AB, Pethe MA, Khare SD. MFPred: Rapid and accurate prediction of protein-peptide recognition multispecificity using self-consistent mean field theory. PLoS Comput Biol. 2017;13:e1005614. doi: 10.1371/journal.pcbi.1005614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Romano KP, et al. The molecular basis of drug resistance against hepatitis C virus NS3/4A protease inhibitors. PLoS Pathog. 2012;8:e1002832. doi: 10.1371/journal.ppat.1002832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 52.Yi L, et al. Engineering of TEV protease variants by yeast ER sequestration screening (YESS) of combinatorial libraries. Proc Natl Acad Sci USA. 2013;110:7229–7234. doi: 10.1073/pnas.1215994110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benatuil L, Perez JM, Belk J, Hsieh CM. An improved yeast transformation method for the generation of very large human antibody libraries. Protein Eng Des Sel. 2010;23:155–159. doi: 10.1093/protein/gzq002. [DOI] [PubMed] [Google Scholar]
- 54.Kowalsky CA, et al. High-resolution sequence-function mapping of full-length proteins. PLoS One. 2015;10:e0118193. doi: 10.1371/journal.pone.0118193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Q, et al. Profiling protease specificity: Combining yeast ER sequestration screening (YESS) with next generation sequencing. ACS Chem Biol. 2017;12:510–518. doi: 10.1021/acschembio.6b00547. [DOI] [PubMed] [Google Scholar]
- 56.Amat CB. Gephi Cookbook, Revista Española Documentación Científica. 2016;39:e124. [Google Scholar]
- 57.Jacomy M, Venturini T, Heymann S, Bastian M. ForceAtlas2, a continuous graph layout algorithm for handy network visualization designed for the Gephi software. PLoS One. 2014;9:e98679. doi: 10.1371/journal.pone.0098679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brin S, Page L. The anatomy of a large-scale hypertextual web search engine. Comput Networks Isdn Syst. 1998;30:107–117. [Google Scholar]
- 59.Shiryaev SA, et al. New details of HCV NS3/4A proteinase functionality revealed by a high-throughput cleavage assay. PLoS One. 2012;7:e35759. doi: 10.1371/journal.pone.0035759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tyndall JD, Nall T, Fairlie DP. Proteases universally recognize beta strands in their active sites. Chem Rev. 2005;105:973–999. doi: 10.1021/cr040669e. [DOI] [PubMed] [Google Scholar]
- 61.Farci P, et al. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science. 2000;288:339–344. doi: 10.1126/science.288.5464.339. [DOI] [PubMed] [Google Scholar]
- 62.Steinberg B, Ostermeier M. Environmental changes bridge evolutionary valleys. Sci Adv. 2016;2:e1500921. doi: 10.1126/sciadv.1500921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dam E, et al. ANRS 109 Study Group Gag mutations strongly contribute to HIV-1 resistance to protease inhibitors in highly drug-experienced patients besides compensating for fitness loss. PLoS Pathog. 2009;5:e1000345. doi: 10.1371/journal.ppat.1000345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boucher JI, Bolon DN, Tawfik DS. Quantifying and understanding the fitness effects of protein mutations: Laboratory versus nature. Protein Sci. 2016;25:1219–1226. doi: 10.1002/pro.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.