Abstract
Although DNA replication is a fundamental aspect of biology, it is not known what determines where DNA replication starts and stops in the human genome. Here we directly identify and quantitatively compare sites of replication initiation and termination in untransformed human cells. We report that replication preferentially initiates at the transcription start site of genes occupied by high levels of RNA polymerase II, and terminates at their polyadenylation sites, thus ensuring global co-directionality of transcription and replication, particularly at gene 5’ ends. During replication stress, replication initiation is stimulated downstream of genes and termination is redistributed to gene bodies; this globally re-orients replication relative to transcription around gene 3’ ends. These data suggest that replication initiation and termination are coupled to transcription in human cells, and propose a model for the impact of replication stress on genome integrity.
Keywords: DNA replication initiation, DNA replication termination, R-loops, Replication-transcription conflicts, Replication stress, dormant origin firing, Fanconi Anemia
INTRODUCTION
Since the identification of cis-acting sequences responsible for the definition of replication origins in S. cerevisiae1, significant effort has been applied to identify analogous determinants of replication initiation in human cells2,3. Although origins have been observed via several independent techniques to be enriched close to transcribed genes and a range of other chromatin features4–7, a coherent model that encompasses both origin specification and activation has not emerged. Furthermore, throughout eukaryotes many more replication origins are licensed by MCM2–7 loading in G1 than are required to complete S-phase8,9. The pool of excess MCMs is required for survival when dNTPs are depleted by the ribonucleotide reductase inhibitor hydroxyurea (HU)10, and has been proposed to allow the firing of ‘dormant’ replication origins that rescue genome replication after replication fork stalling. The identities of these dormant origins, and how they differ from constitutive origins, have not been defined.
DNA is a one-dimensional template that can be simultaneously acted upon by the replication and transcription machineries. The orientation of essential genes in prokaryotes is biased to avoid head-on collisions11; among eukaryotes, budding yeast and C. elegans show statistical orientation bias of the most highly transcribed genes to the co-directional orientation12,13, and significant co-orientation of transcribed genes with the direction of replication has been noted in human cells7,14. Head-on replication-transcription conflicts are deleterious in eukaryotes, leading to increased DNA damage14 and genomic rearrangements15. How origin location is specified to co-orient replication with transcription in diverse human cell types, and the impact of co-directional transcription on replication-fork progression through genes, remain speculative.
Here, using Okazaki fragment sequencing (Ok-seq) to infer the direction of replication-fork movement, we define transcription initiation efficiency and gene length as independent determinants of replication origin location and firing efficiency. We show that origin firing occurs close to the transcription start site (TSS) of long, transcribed genes, ensuring co-oriented replication of the most highly transcribed regions of the genome. We also show that, under conditions that stimulate dormant replication origin firing, activation of the most efficient constitutive origins is further stimulated. Additionally, we demonstrate widespread localized replication termination at the transcription termination site (TTS) of transcribed genes under unperturbed conditions. Replication termination redistributes to gene bodies during replication stress, resulting in increased replication of gene 3’ ends in the head-on orientation.
RESULTS
Using Ok-seq, we and others have reported on replication initiation7,13,16,17, replication elongation12 and lagging-strand processing12,18. To investigate replication initiation and termination in untransformed human cells, we performed Ok-seq7 on hTERT-immortalized RPE-1 cells. Previous genome-wide studies of mammalian DNA replication show limited agreement in origin calls2,3, with the exception that all identify significant enrichment of origins close to transcribed genes. Therefore, instead of aiming to identify all potential sites at which replication can possibly initiate in the human genome, we focused our analysis on the efficiency of replication initiation and termination in transcribed regions.
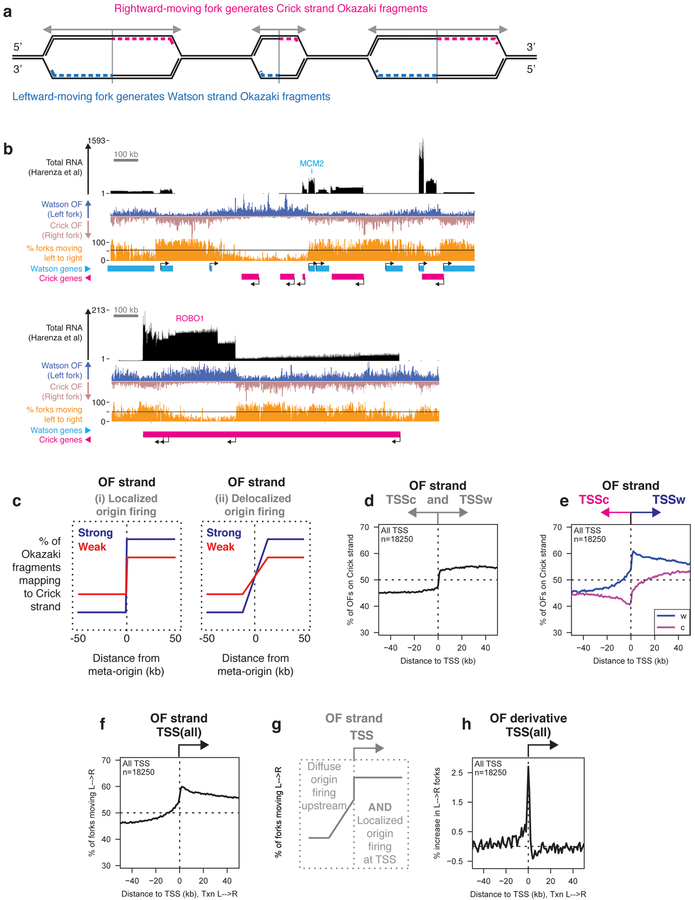
Replication initiates in the immediate vicinity of transcription start sites.
Rightward-moving replication forks generate Okazaki fragments (OFs) that map to the Crick strand, while leftward-moving forks generate Watson-strand fragments (Fig. 1a). Therefore, a replication origin will manifest as an increase in the proportion of OFs mapping to the Crick strand. Okazaki fragment distributions from asynchronously dividing RPE-1 cells showed regions of largely unidirectional replication. Not all genes were associated with replication origins, but OF strand transitions often occurred upstream of transcribed genes (two arbitrary but representative 1.5 Mb regions are shown in Fig. 1b). In addition to initiation upstream of TSS, we occasionally noticed OF strand bias transitions consistent with replication initiation immediately downstream of genes (e.g. ROBO1 in Fig. 1b). Total RNA-seq data used in this work are from a previously published dataset19.
Figure 1. DNA Replication initiates preferentially at transcription start sites.
(A) The relationship between replication direction and Okazaki fragment (OF) strand.
(B) Distribution of OFs and RNA-seq reads from total cellular RNA19 in RPE-1 cells over 1.5 MB regions around the MCM2 and ROBO1 loci. Transcription start sites (TSS) are indicated, and the percentage of replication forks moving from left to right is shown in orange. Unless otherwise indicated, all data in figures 1–7 are from asynchronously dividing hTERT-immortalized RPE-1 cells, and are displayed using 1 kb bins.
(C) Schematic depiction of expected OF distributions around replication origins.
(D) Percent of RPE-1 OFs mapping to the Crick strand (indicating rightward-moving replication forks) across a ±50 kb window around annotated TSS in the human genome. All meta-analyses shown in figures 1–6 were carried out using one replicate: data from a second biological replicate are shown in the supplement.
(E) Percentage of OFs mapping to the Crick strand across a ±50 kb window around the TSS of Watson (W) or Crick (C) genes.
(F) Percentage of replication forks moving from left to right around the TSS of all genes. Here and in subsequent figure panels, data are oriented such that transcription runs from left to right (indicated by ‘Txn L→R’).
(G) Schematic representation of upstream and TSS-proximal replication initiation inferred from data in Figure 1.
(H) Replication initiation frequency, calculated as the first derivative of Okazaki fragment strand bias as a function of position, across a ±50 kb window around TSS. Data are smoothed across two 1 kb bins.
We calculated OF distributions by meta-analysis around specific classes of genomic locus. The efficiency and spatial localization of origin firing will impact the magnitude and gradient of the OF strand bias transition, respectively (schematic in Fig. 1c). As expected, OFs showed no strand bias around random genomic loci (Supplementary Fig. 1a). Consistent with global origin activity at TSS, meta-analysis of OF Crick strand bias over a 50 kb window surrounding all 18,037 annotated TSS showed a symmetrical transition from predominantly leftward- to predominantly rightward-moving forks (Fig. 1d). However, separate analysis of Watson- and Crick-strand genes revealed profound asymmetry based on gene orientation (Fig. 1e). Therefore, we considered OF strand bias relative to gene orientation, such that transcription occurs from left to right (Fig. 1f, schematic in Fig. 1g). All data for TSS analysis were highly reproducible across two biological replicates (Supplementary Fig. 1b–d).
By analyzing the first derivative of replication direction calculated from OF distributions, we can directly infer the change in replication polarity – and therefore the extent of replication initiation – at each position relative to the meta-TSS. A positive value for the increase in the proportion of replication forks moving from left to right indicates origin firing. We observed a strong peak of replication initiation within 2–3 kb of the meta-TSS, preceded by a gradual increase over the ~20kb upstream (Fig. 1h). Replication initiation frequency was consistently negative or zero downstream of TSS, indicating a paucity of replication initiation. Thus, while individually low but cumulatively high levels of initiation occur over a wide region upstream of TSS, initiation is strongly biased to a 2–3 kb region immediately adjacent to the TSS itself and is under-represented or absent in gene bodies. These data are consistent with previous reports that origin firing is predominantly intergenic7,20,21, but extend these observations by localizing the preferred site of initiation immediately upstream of the TSS.
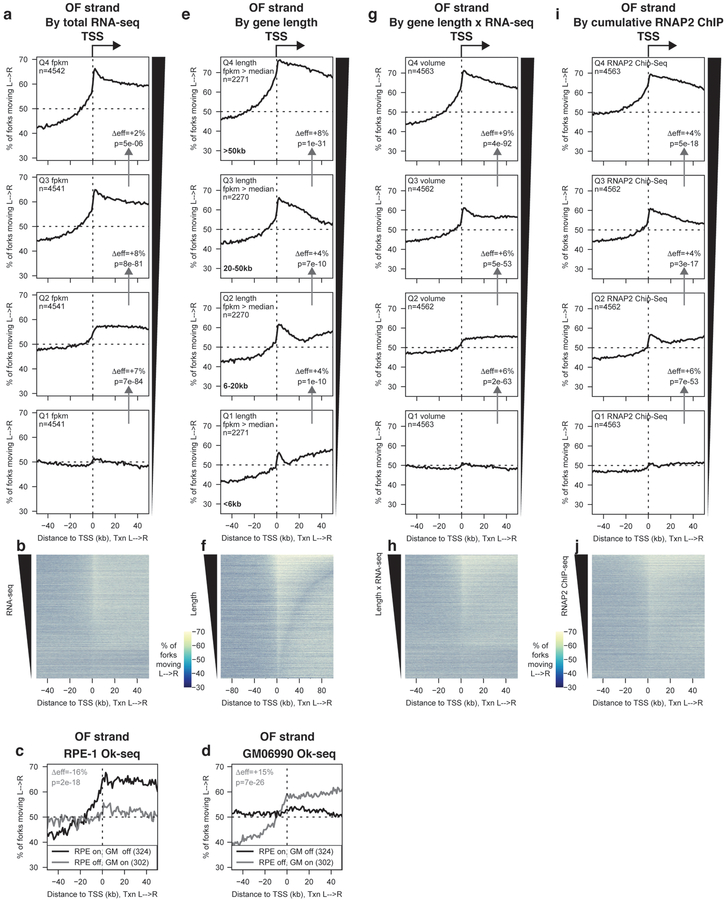
Determinants of replication origin firing efficiency at TSS.
We estimated origin efficiencies by calculating the difference in OF strand bias between the region from 50 to 30 kb upstream and the region 1 to10 kb downstream of the TSS. We present changes in origin efficiency (Δeff), representing the absolute change in the percentage of cells in which a given set of origins will fire. A Δeff of +5% means that origin firing occurs in an additional 5% of cells in the population.
We separated genes into quartiles based on RNA-seq read density from total RNA (FPKM, Fragments Per Kilobase of transcript per Million mapped reads) in RPE-119. TSS of genes with higher RNA-seq read density showed significantly greater change in strand bias than TSS of weakly- or non-transcribed genes (Fig. 2a,b). We verified the specificity of this transcriptional effect by comparing our Ok-seq data from RPE cells with previously published data from GM06990 cells7 around TSS of genes that are uniquely active in only one of these cell types (Fig. 2c,d). Genes transcribed above median levels in RPE-1 but below median levels in GM06990 (RPE on, GM off) showed robust origin activity at their TSS only in RPE-1, and vice versa.
Figure 2. Total RNA polymerase occupancy of a gene predicts replication origin firing efficiency at its TSS.
(A) Percentage of replication forks moving left to right around TSS binned by total RNA-seq read depth quartile (FPKM) from19. p values were calculated using Kruskal-Wallis test, using the regions from 50–30 kb upstream and 1–10 kb downstream of the TSS. Effect sizes, shown as Δeff, were calculated using the same regions. Δeff and p values between adjacent quartiles are indicated on the relevant panels in figure 2a,e,g and h. All statistics for figures 2–6 are presented in the methods section.
(B) Heat map representation of data in (A).
(C&D) Percentage of replication forks moving left to right in (C) RPE-1 or (D) GM06990 cells7 around the TSS of genes whose transcription is off in one cell type (FPKM in first quartile) and on in the other (FPKM > median) as indicated.
(E) Percentage of replication forks moving left to right around TSS of actively transcribed genes (FPKM > median,19) binned by length according to quartiles for transcribed genes.
(F) Heat map representation of data in (E). Note that the x-axis scale differs from other panels to encompass a region ±100 kb from the TSS.
(G) Percentage of replication forks moving left to right around TSS binned by transcriptional volume (FPKM from19 × gene length).
(H) Heat map representation of data in (G).
(I) Percentage of replication forks moving left to right around TSS binned by cumulative RNAP2 ChIP-seq signal within the gene22.
(J) Heat map representation of data in (I).
We separated transcribed genes (FPKM > median) into quartiles by length, and observed that gene length showed a strong positive correlation with origin activity (Fig. 2e,f). Gene length and transcription level are not correlated in RPE-1 cells: therefore, gene length and transcript number independently correlate with origin firing efficiency. Distance to the nearest downstream TSS or transcription termination site (TTS) – both of which are intrinsically dependent on gene length – are substantially stronger determinants of origin efficiency than the distance to the nearest upstream TSS or TTS, which are independent of gene length (Supplementary Fig. 2). Therefore, the effect of gene length on origin efficiency cannot solely be explained by decreased passive replication from nearby ‘competitor’ origins. Transcript levels and gene length both correlate with origin activity in HeLa and GM06990 cells using previously published Ok-seq data7 (Supplementary Fig. 3).
While (assuming equal RNA decay rates) RNA-seq reports the number of RNA molecules synthesized, the number of RNA polymerases occupying a gene for a given FPKM is linearly related to the length of the gene. We therefore analyzed OF strand bias around TSS separated by transcriptional volume (FPKM × gene length). As expected activity was strongly correlated with high transcriptional volume (Fig. 2g,h). We confirmed this by analyzing origin activity as a function of cumulative RNAP2 ChIP-seq signal within the gene body in asynchronous RPE-1 cells22 (Fig. 2i,j).
We conclude that, in unperturbed human cells, the initiation of DNA replication is strongly biased towards the immediate vicinity of TSS that drive transcription of genes with high RNA polymerase II (RNAP2) occupancy. While transcription creates conflicts with replication, the coupling of origin firing to RNAP2 occupancy would bias conflicts towards the co-directional orientation.
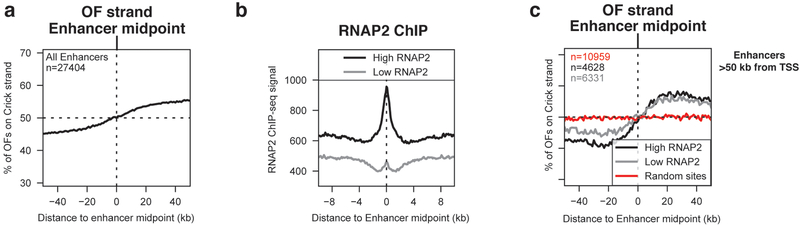
Delocalized replication initiation at enhancers.
We reasoned that, in addition to TSS, other accessible chromatin regions such as enhancers might similarly act as localized replication origins. OF bias around 27,404 enhancers previously identified in RPE-1 cells22 was consistent with origin activity (Fig. 3a). We separated enhancers into high vs low transcription based on RNAP2 ChIP-seq22 (Fig. 3b). Enhancers <50 kb from the nearest TSS showed OF transitions expected for replication origins, but a similar transition was apparent for random sites within the same range (Supplementary Fig. 4), consistent with a ‘bystander effect’ due to origin activity of nearby TSS. Therefore, we considered only enhancers for which the closest TSS is over 50 kb away. We compared OF distributions around enhancer midpoints to the OF distribution around an identical number of random sites, also >50 kb from TSS (Fig. 3c). A transition in OF strand bias was absent for random sites but persisted for enhancers, consistent with bona fide origin activity centered on enhancer midpoints and independent of nearby protein-coding TSS (Fig. 3c). Due to the symmetrical OF signal around enhancers, we calculated relative origin efficiencies using data from ±20–40 kb from the enhancer midpoint. While transcribed enhancers were slightly more active origins than non-transcribed enhancers, both displayed significant origin activity relative to random sites. Origin signal extended over a ~15 kb region on either side of the enhancer. Thus, replication specifically initiates in proximity to enhancers: this effect is slightly stimulated by – but not dependent on – enhancer transcription.
Figure 3. Replication initiates at enhancers, largely independent of enhancer transcription.
(A) Percent of RPE-1 OFs mapping to the Crick strand (indicating rightward-moving replication forks) across a ±50 kb window around the midpoints of enhancers identified in RPE-122
(B) RNAP2 ChIP-seq signal from22 across a ±10kb window around enhancers from (A). Enhancers are separated into high (above median) and low (below median) RNAP2.
(C) Percent of RPE-1 OFs mapping to the Crick strand around enhancers from (A), binned by transcription level and separated by distance from the enhancer to the nearest TSS. For each analysis, an equivalent number of random sites within the same distance of a TSS was used as a control. Δeff and p-values were calculated using the regions ±20–40 kb from the enhancer midpoint. Low RNAP2 vs. random sites (Kruskal-Wallis, p=1.50E-54); High RNAP2 vs. Low RNAP2 (p=4.78E-07). Groups: High RNAP2 (black, n=4628); Low RNAP2 (gray, n=6331); Random sites (red, n=10959).
Modulation of constitutive origin efficiency during replication stress.
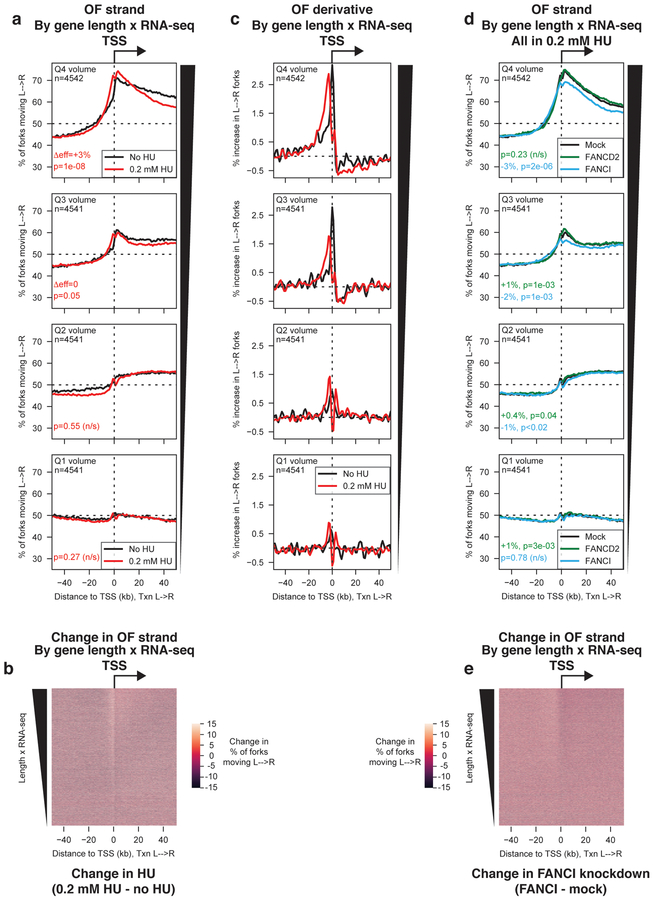
In response to replication stress, origin firing increases23. Origin firing events during growth in hydroxyurea (HU) have previously been reported to occur in the vicinity of transcribed genes24. We treated cells with 0.2 mM HU for 4h prior to OF collection, and additionally depleted either of the Fanconi Anemia (FA) effector proteins FANCD2 or FANCI25,26, by RNAi. This HU treatment decreases average inter-origin distance without activating the checkpoint10: knockdown of FANCI increases inter-origin distance in HU, reflecting reduced origin firing, while knockdown of FANCD2 weakly stimulates origin firing under the same conditions27.
We analyzed the effect of HU treatment on origin efficiency at TSS separated by transcriptional volume (Fig. 4a,b). We observed a significant increase in origin efficiency for the most highly transcribed genes, i.e. the most efficient pre-existing origins. We observed no evidence for a widespread increase in intragenic origin firing upon HU treatment. Indeed, while the overall pattern of origin use was globally similar in the presence and absence of HU, the preferred site of replication initiation moved upstream of the TSS upon HU treatment, as opposed to downstream (Fig. 4c).
Figure 4. Global modulation of origin activity under conditions that increase or suppress dormant origin firing.
(A) Percentage of replication forks moving left to right around TSS binned by transcriptional volume, for cells grown in the absence (black) or presence (red) of 0.2 mM hydroxyurea (HU) for 4h before OF collection. p-values and effect sizes were calculated using Kruskal-Wallis relative to the no HU dataset for each quartile. Δeff and p values between the no HU and low HU sample for each transcriptional quartile are indicated in the relevant panels.
(B) Heat map representation of the change in replication direction around TSS upon treatment with 0.2 mM HU. Genes are sorted by transcriptional volume (length × FPKM). Values were calculated as %L→R0.2 mM HU - %L→ RNo HU
(C) Replication initiation frequency, calculated as the first derivative of Okazaki fragment strand bias as in Fig. 1h, around TSS in the absence or presence of 0.2 mM HU. Data are smoothed over 2 adjacent bins.
(D) Percentage of replication forks moving left to right around TSS binned by transcriptional volume, for cells treated with siRNAs against FANCD2 (green), FANCI (blue), or mock-treated (black), grown in 0.2 mM hydroxyurea for 4h before OF collection. p-values and effect sizes for both RNAi treatments were calculated using Kruskal-Wallis relative to the mock RNAi (low HU) dataset for each quartile. Δeff and p values between the low HU (mock RNAi) and either the FANCD2 (green) or FANCI (blue) knockdown sample for each transcriptional quartile are indicated in the relevant panels and in the methods section.
(E) Heat map representation of the change in replication direction around TSS in 0.2 mM HU upon FANCI knockdown. Genes are sorted by transcriptional volume (length × FPKM). Values were calculated as %L→RFANCI knockdown - %L→ RNo knockdown
We observed that FANCI knockdown significantly reduced origin firing around high volume TSS in HU-treated cells: (Fig. 4d,e and Supplementary Fig. 1d). We did not observe a significant increase in firing of the most efficient pre-existing origins upon FANCD2 knockdown, although firing efficiency was very modestly increased for less efficient origins (Fig. 4d). The effect of FANCI knockdown was independent of gene length, arguing against a global defect in replication-fork mobility through transcribed regions due to reduced FANCI levels (Supplementary Fig. 5). Thus, under conditions of globally increased or decreased origin firing, we find that the firing efficiency of the most active pre-existing origins is the most strongly affected in transcribed regions of the genome. Our data suggest that intragenic replication origins do not contribute significantly to dormant origin firing in RPE-1 cells.
Replication termination is locally enriched at polyadenylation sites of transcribed genes.
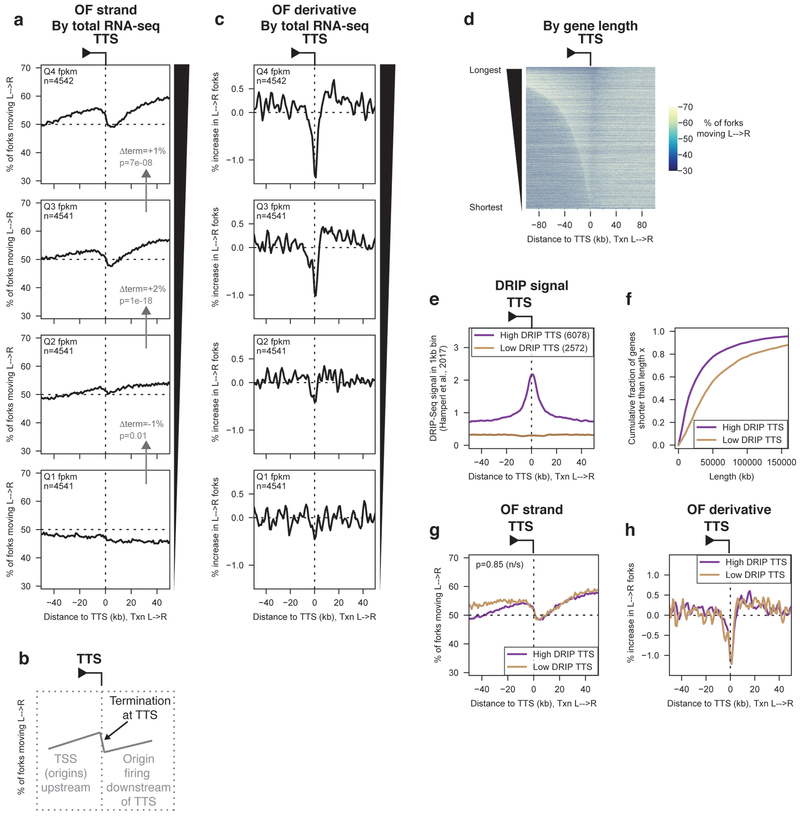
Sites of replication termination generate a transition in OF strand bias opposite to that observed at replication origins – i.e. a decrease in the proportion of replication forks moving from left to right. We queried OF distributions around the annotated polyadenylation site (hereafter referred to as the TTS). The TTS corresponds to the site of pre-mRNA cleavage and polyadenylation, and does not necessarily reflect the site of RNAP2 dissociation28. We quantified replication termination by calculating OF strand bias over the region from −10 kb to +10 kb relative to the TTS. Differences in termination frequency are presented as Δterm. All data for TTS were reproducible across replicate datasets (Supplementary Fig. 6).
Consistent with an over-representation of replication termination events at the TTS of transcribed genes, we observed a significant, transcription-dependent reduction in replication forks moving in the direction of transcription through the gene body, occurring precisely at the TTS (Fig. 5a). This termination signal was surrounded by an increase in left-to-right fork movement, consistent with diffuse origin firing occurring at TSS a variable distance away (Fig. 5b,d). Termination was significantly higher for more highly transcribed genes. Analogously to replication initiation (Fig. 1h), localized replication termination can be inferred from analysis of the change in replication polarity around a meta-locus. The change in Okazaki fragment strand bias in the vicinity of TTS showed a sharp trough immediately adjacent to the TTS itself, indicating that the peak of replication termination is localized, on average, precisely at the TTS for transcribed genes (Fig. 5c).
Figure 5. Widespread, R-loop-independent replication-fork termination occurs at the 3’ ends of transcribed genes under unperturbed conditions.
(A) Percentage of replication forks moving left to right around transcription termination sites (TTS) binned by RNA-seq read depth quartile from19. p-values and effect sizes were calculated using Kruskal-Wallis as for TSS data in previous figures, but using data ±10 kb from the TTS as described in the results and methods sections. Δterm and p values between adjacent quartiles are indicated on the relevant panels and in the methods section.
(B) Schematic representation of Okazaki fragment distributions arising from replication termination at TTS.
(C) Replication initiation frequency, calculated as the first derivative of Okazaki fragment strand bias as in Fig. 1h, around TTS binned by total RNA-seq read density in the gene body as in (A). Negative initiation frequencies correspond to termination. Data are smoothed over 2 adjacent bins.
(E) DRIP-seq signal from (14) around TTS of actively transcribed genes (FPKM from (18) > median) separated into high (purple) and low (brown) DRIP bins based on DRIP-seq signal14 ±10kb from the TTS.
(F) Length distribution of actively transcribed high-DRIP vs low-DRIP genes. The cumulative fraction of high-DRIP vs low-DRIP genes shorter than the length on the x-axis is plotted.
(G) Percentage of replication forks moving left to right around TTS of actively transcribed (FPKM from19 > median) high-DRIP vs low-DRIP genes. p = 0.85 by Kruskal-Wallis.
(H) Replication initiation (termination) frequency around high-DRIP vs low-DRIP TTS.
In addition to the replication termination signal at the TTS, we observed an origin signal – i.e. sustained positive values for the increase in rightward-moving replication– in the region immediately downstream of the gene (Fig. 5b,c). This origin firing signature was apparent only downstream of genes transcribed above the median level (cf. top vs bottom two panels in Fig. 5b,c).
Replication termination at gene 3’ ends is not affected by R-loops.
We analyzed replication termination around genes separated based on their likely levels of R-loop formation at TTS in RPE cells. Most differences in R-loop formation between cell types and even species can be attributed to differences in transcription29: therefore, we separated genes with FPKM > median in RPE into two bins based on the DRIP-seq signal observed ±10 kb from their TTS in HeLa cells14, to obtain high- and low-DRIP TTS gene sets (Fig. 5e). Genes in our high-DRIP set are significantly shorter than those in our low-DRIP set (Fig. 5f).
Analysis of OF strand bias around the TTS of high- and low-DRIP genes indicated equivalent replication termination frequency and localization regardless of the propensity of the region to form R-loops (Fig. 5g,h). The upward gradient observed upstream of the TTS for high-DRIP genes is due to origin firing at TSS, which are closer to TTS due to the length difference between high- and low-DRIP genes (Fig. 5f,g). Results from Fig. 5 were reproducible using previously published Ok-seq data from HeLa and GM06990 cells7 (Supplementary Fig. 7). Therefore, although replication termination can occur throughout the genome, termination events are enriched at the 3’ end of transcribed genes; localized termination occurs independently of the gene’s propensity to form R-loops.
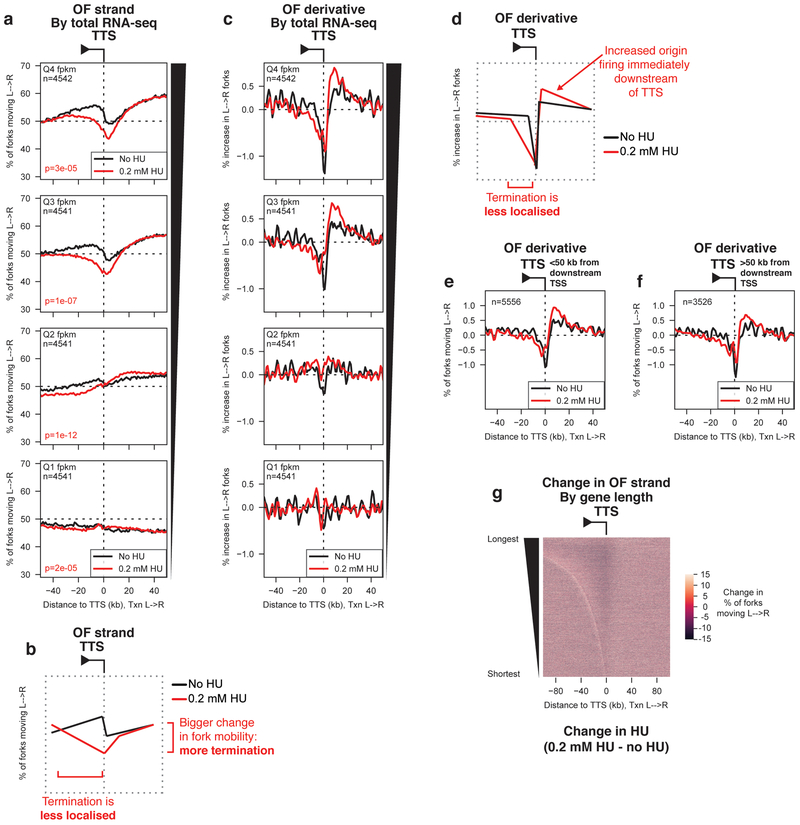
Global re-orientation of replication at gene 3’ ends under replication stress.
For highly transcribed genes, upon HU treatment we observed both an increase in the overall level of gene-associated replication termination, and an increase in the fraction of this termination that is intragenic as opposed to localized at the TTS (Fig. 6a,b). Analysis of the change in replication direction (Fig. 6c,d) – indicated that replication termination is still elevated around TTS, but additionally showed a marked increase in replication initiation within the region ~20 kb downstream of the TTS relative to untreated cells. To test whether this effect was entirely due to increased origin firing at proximal TSS in this downstream region, we separately analyzed the change in OF strand bias around TTS of transcribed genes (FPMK > median) with or without a TSS for a protein-coding gene in the 50 kb immediately downstream (Fig. 6e,f). We observed substantial replication initiation downstream of the TTS even in genes with no nearby TSS. Analogous to efficient origins near TSS (Fig. 4a,c), downstream initiation was more pronounced in HU-treated cells than untreated cells (Fig. 6f).
Figure 6. Redistribution of replication termination to gene bodies under replication stress.
(A) Percentage of replication forks moving left to right around (TTS) binned by RNA-seq read depth quartile from19, for cells grown in the absence (black) or presence (red) of 0.2 mM hydroxyurea (HU) for 4h before OF collection. p-values are calculated by Kruskal-Wallis relative to the no HU dataset for each quartile, and are indicated in red on the appropriate figure panel and in the methods section.
(B) Schematic representation of the change in replication termination observed under replication stress.
(C) Replication initiation (termination) frequency around TTS binned as in (A) in the absence or presence of 0.2 mM HU.
(D) Schematic representation of the change in replication initiation observed downstream of transcribed genes under replication stress.
(E-F) Replication initiation frequency, calculated as the first derivative of Okazaki fragment strand bias, around the TTS of transcribed genes binned by distance of the TTS to the nearest downstream TSS. Positive values correspond to initiation while negative values correspond to termination.
(G) Heat map representation of the change in replication direction relative to transcription upon treatment with 0.2 mM HU. Genes are ordered by length, and data are centered on the TTS. Lighter colors indicate an increase in the proportion of replication forks moving from left to right in the presence of HU. Values were calculated as %L→R0.2 mM HU - %L→ RNo HU. Note that the x-axis scale is different from other figure panels to encompass a range ±100 kb from the TTS.
Taken together, our data demonstrate that replication origin efficiency increases near active TSS in the presence of HU (Fig. 4a): this increases co-orientation of replication and transcription at gene 5’ ends (Figs. 4a and 6g). However, HU treatment also increases the efficiency of origins downstream of genes, reducing the proportion of replication forks co-oriented with transcription at gene 3’ ends. This effect is most prominent for long genes (Fig. 6g).
DISCUSSION
Co-orientation of replication and transcription in multicellular eukaryotes
Our data suggest that DNA replication initiation is coupled to transcription initiation, such that origin firing preferentially initiates immediately adjacent to the TSS of genes with high RNAP2 occupancy (Fig. 2, model in Fig. 7). Therefore, the most highly transcribed genes will have the strongest bias towards co-directional replication in any proliferative cell type, regardless of its transcriptional profile. Many unicellular organisms define replication origins via the use of cis-acting sequences: in these organisms, the orientation of conflict-prone genes has been biased by evolution12,30,31, presumably as a result of damage induced by head-on collisions followed by re-orientation32. However, the use of such cis-acting sequences as the sole means of origin specification in an organism with many transcriptionally distinct proliferative cell types would enforce cell-type-specific head-on replication-transcription conflicts. Thus, while unicellular organisms can co-orient transcription with replication by genome evolution, the alternative strategy – to co-orient replication with transcription – is more robust for organisms with many distinct transcriptomes.
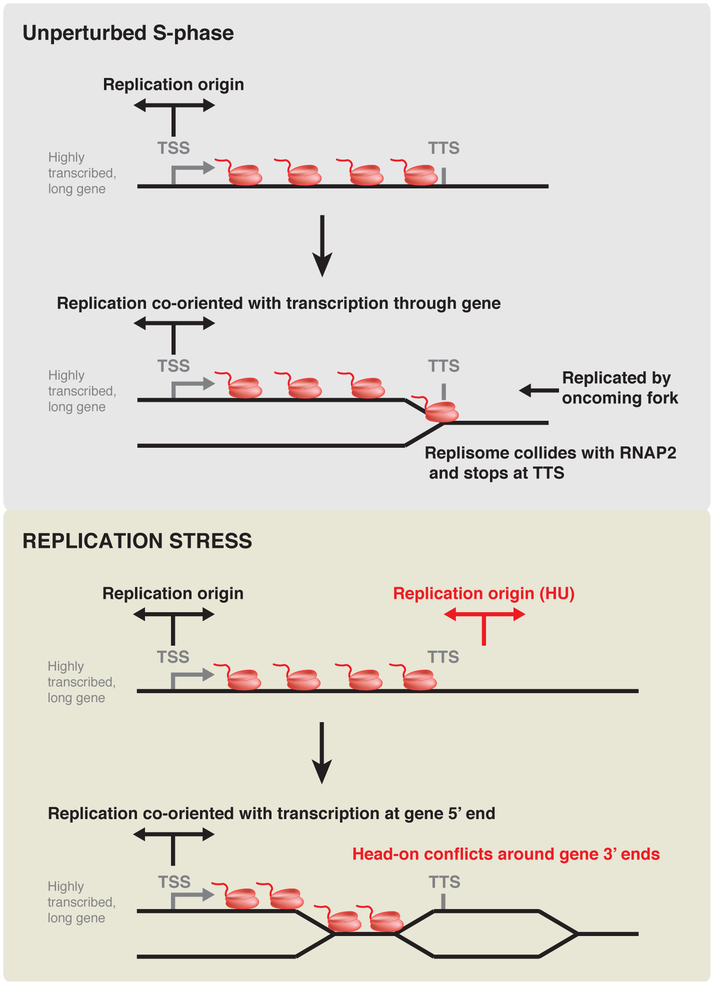
Figure 7. Model for the coupling of replication initiation and termination to transcription.
In addition to replication origin activity immediately upstream of highly transcribed genes at TSS, we observe significant initiation immediately downstream of TTS. If downstream initiation were as efficient as upstream initiation then genes would, on average, be replicated co-directionally with transcription at their 5’ ends and head-on at their 3’ ends. We do not observe such a pattern in unperturbed cells (Figs. 2, 5, Supplementary Figs. 3,7). Firing efficiency of TSS-proximal origins must therefore be, on average, higher than that of TTS-proximal ones.
A simple mechanism to explain the increased replication origin activity of the genes occupied by the most RNAP2 is that chromatin accessibility determines both MCM2–7 loading and the recruitment of replisome components to licensed replication origins. This mechanism would also provide a rationale for the delocalized origin efficiency observed at enhancers (Fig. 3). Chromatin accessibility has previously been reported as the best correlate of replication timing33. Our data do not preclude preferential initiation at specific sequences within regions of open chromatin defined by a cell’s transcriptional profile – for example a recent report that replication initiation in mouse cells correlates with poly(dA:dT) tracts17. Indeed, since poly(dA:dT) sequences disfavor stable nucleosome occupancy34, and human ORC displays increased affinity for A-T rich DNA35 these sites should experience increased ORC recruitment and MCM loading.
Replication initiation outside transcribed regions
Gene distribution is not uniform, and there are clearly fewer actively transcribed genes than the number of origins required to replicate the entire human genome. Inactive regions of the genome lack both spatially localized regions of highly accessible chromatin and a requirement to co-orient replication with transcription: therefore, specifically (as opposed to randomly) localized replication origins would serve little biological purpose in these regions. Transient chromatin opening, likely driven by low-levels of pervasive transcription factor binding or transcription, could ensure a sufficient density of MCM2–7 loading and activation to support genome duplication in these regions through a distribution of origin firing events that largely disfavors initiation at specific individual sites.
Replication termination at gene 3’ ends
Although replication termination can occur throughout the genome, termination events are highly enriched at gene 3’ ends in both unperturbed transformed and non-transformed human cell lines (Fig. 5a–c and Supplementary Fig. 7a,b,e,f). Because replication origins are located at variable distances both upstream and downstream of TTS (Fig. 5d), it is improbable that such localized replication termination would arise passively. The precise termination of replication at TTS can more plausibly be explained by replication-fork stalling or arrest at these sites. RNAP2 and the replication fork normally move at approximately the same speed in eukaryotes. Replication forks co-oriented with transcription should generally not collide with RNAP2 except when the latter is paused. Gene 3’ ends represent prominent RNAP2 pause sites36. We propose that co-directional collisions with paused RNAP2 are prevalent at TTS, and result in localized replication termination at these sites.
Replication-fork stalling due to co-oriented collisions with RNA polymerase at gene 3’ ends has been observed in prokaryotes37 but not, to our knowledge, in eukaryotes. Unlike in prokaryotes38, but similarly to replication-fork arrest at tRNA genes in yeast12, R-loops do not appear to impede replisome mobility despite their impact on the ultimate outcome of collisions14,15,39,40. Screens in HeLa cells39 and S. cerevisiae40 identified RNA splicing and 3’ end formation factors as preventative against DNA damage. It will be interesting to determine the relative contributions of R-loop modulation and replisome mobility to DNA damage when mRNA processing is impaired.
The frequency of replication termination at TTS is lower than the cumulative initiation frequency at TSS (Fig. 1h), consistent with only a fraction of replisomes stopping at TTS. Regardless, replication fork initiation at TSS followed by stalling or arrest at TTS could impair the replication of intergenic regions flanked by convergent, highly transcribed genes. However, the ability of both RNA polymerases41,42 and the replicative helicase43 to push loaded MCM2–7 double hexamers would lead to a redistribution of licensed origins towards these intergenic regions. We predict that a transcriptionally redistributed pool of MCMs, together with MCM loading at these sites due to pervasive transcription at gene 3’ ends, generates the TSS-independent pool of replication origins that we observe downstream of active genes (Figs. 1b, 5c, 6c,f, and Supplementary 7b,f). In support of this hypothesis, TTS-proximal origin activity is observed downstream of genes that are themselves actively transcribed (Figs. 5c, 6c).
Dormant replication origin firing
Under conditions of global origin activation or repression, a proportionally similar activation/repression of all origins would lead to the greatest absolute change in origin efficiency occurring at origins with intrinsically high firing efficiencies. We observe such behavior at TSS upon HU treatment (activation), and upon depletion of FANCI (repression) (Fig. 4a,b,d). This should globally maintain co-orientation of replication and transcription, especially for short genes and at the 5’ ends of long genes. In HU-treated cells, we also observe increased origin firing downstream of genes even without a proximal protein-coding TSS (Fig. 6f). TTS-proximal origins are active even in the absence of HU but become more active upon HU treatment (Figs. 5c and 6c). Average replication fork speed for RPE-1 cells in 0.2 mM HU is approximately 0.67 kb/min as opposed to 1.37 kb/min in untreated cells27. Decreased passive replication by oncoming replisomes (due to reduced replisome speed) would increase the firing efficiency of all licensed origins – particularly those far from origins that normally fire early and with high efficiency. We propose that at least a significant fraction of dormant origin activation during conditions of replication stress is due to the increased firing efficiency of preexisting origins.
Re-orientation of replication-transcription conflicts during replication stress
Upon treatment with low-dose HU, we observed a global re-orientation of replication relative to gene 3’ ends, most prominently for long genes (Fig. 6g). Reorientation of replication is associated with increased origin firing downstream of TTS (Fig. 6c–f). This increased TTS-proximal origin firing could arise from decreased passive replication of these sites due to decreased replication fork speed. Another possibility, not mutually exclusive with decreased passive replication, is that origin firing downstream of genes is actively stimulated by global up-regulation of origin firing. The time required for RNAP2 or the replisome to progress from the 5’ to the 3’ end of a gene is similar due to the matched speeds of these two complexes. Assuming RNAP2 initiating on replicated DNA cannot co-directionally bypass a replisome, the suppression of downstream origin firing by passive replication would predict that all transcribing RNAP2 has time to clear the gene before downstream origin firing normally occurs. Thus, if origin firing downstream of genes is normally suppressed mostly by passive replication, decreased replication fork speed or increased intragenic replisome stalling is unlikely to lead to increased head-on replication-transcription conflicts. By contrast, the activation of downstream origin firing, independently of decreased passive replication, would drive replication into genes still occupied by RNAP2, increasing head-on conflicts and thereby DNA damage and ATR signaling, both of which are strongly orientation-dependent14. HU is genotoxic, and long genes show transcription-dependent instability44. Furthermore, a recent preprint suggests that replication timing is stochastic rather than absolute in human cells45, so higher downstream origin firing efficiency would also result in the early firing of more downstream origins. We propose that such early firing of origins downstream of highly transcribed genes under replication stress increases head-on conflicts at their 3’ ends, contributing to genome instability.
METHODS
Cell Culture/EdU labeling
hTERT immortalized RPE-1 cells (ATCC) were grown in Dulbecco’s modified Eagle’s medium: Nutrient Mixture F-12 (DMEM/F-12) media (Life Technologies) supplemented with 10% fetal bovine serum (FBS), 3% sodium bicarbonate and 1% Pen-Strep. The cell line was authenticated by short tandem repeat (STR) profiling and was tested for mycoplasma using Roche MycoTOOL detection kit. In brief, exponentially growing cells of 50–60% confluency were plated in 150mm dishes (approximately 15–20 plates per sample/replicate). siRNA transfections were done according to27. EdU labeling was done for 2 min (4 min for low-dose HU) at 20 μM final concentration. Cells were either untreated or treated with low-dose HU (200 μM final concentration).
Ok-seq
Okazaki fragments were purified and libraries generated essentially as described7, with the most major modification being the use of gel-purified adaptor duplexes from18. In brief, frozen cell pellet was lysed and gently mixed in DNA lysis buffer (10mM Tris-pH 8), 25mM EDTA and 100 mM NaCl) with 0.5% (vol/vol) SDS and 0.1 mg/ml Proteinase K for overnight at 50°C. DNA was extracted with equal volume of phenol-chloroform and precipitated by ammonium acetate and ethanol. After successive washes, DNA was isolated and resuspended in TE at 4°C. DNA was denatured for 10 min at 95°C, then chilled on ice for 10 min and sized fractionated on a neutral sucrose gradient with an Ultracentrifuge Beckman in SW40TI rotor at 28000g in 20°C for 16h with maximum acceleration and minimum deceleration. The quality and reproducibility of size separation of genomic DNA can be assessed using an alkaline 1.5% agarose gel. Only fractions containing DNA fragments no longer than 200 bp were collected and concentrated. The EdU-labeled DNA was then biotinylated in standard Click-it reaction. The purified DNA was then subjected to RNA hydrolysis with 250mM NaOH for 30 min at 37°C. DNA fragments were then phosphorylated with PNK kinase prior to ligation of adaptors using a two-step procedure (before and after Streptavidin pulldown). The biotinylated DNA was captured on MyOne T1 Streptavidin dynabeads (Thermo Fisher Scientific) in 10mM Tris (pH-7.5), 1mM EDTA, 2M NaCl, and 0.1% (vol/vol) Tween 20 buffer. PCR was used to survey Okazaki fragments library and primer removal prior to analysis by TapeStation (Agilent). Libraries were sequenced using the HiSeq-2500 platform using one lane per sequencing reaction, and reads were aligned to the hg19 build of the human genome using bowtie2, excluding reads that did not map uniquely. Data were binned into 1 kb bins prior to analysis.
Data analysis
TSS and TTS locations were obtained from the UCSC genome browser http://genome.ucsc.edu. Genes were separated by quartile based on RNA-seq read density, volume (FPKM × length) or RNAP2 Chip-seq in the appropriate cell type. Genes with missing RNA-seq FPKM values were dropped from the analysis. Genes were further separated by length or DRIP-seq read density in HeLa cells, as described in the main text. Enhancer regions were separated by RNAP2 Chip-seq read density and sorted into bins based on distance from nearest TSS using a custom python script. Random enhancer sites were generated by a python script that matches the distance distribution of enhancers from nearest TSS while choosing a random gene for placing each site. All plots and heat maps were produced using python matplotlib and seaborn packages.
Statistics and reproducibility
Two biological replicates were obtained for all datasets. Data from one replicate are presented in Figs 1–6: data from the second replicate showed excellent agreement and are presented in the supplement.
Efficiencies and Statistics were calculated for strand bias curves by taking the average difference between ranges downstream and upstream of the site being analyzed. These ranges were: 1–11kb downstream and 50–30kb upstream of annotated transcription start sites, ±1–10kb around annotated transcription termination sites, and ±20–40kb around midpoints of enhancer regions. Efficiencies were calculated from the averaged data as shown in the figures. P-values were calculated using the Kruskal-Wallis H-Test for the indicated gene lists. The Kruskal-Wallis H-test tests the null hypothesis that the population median of all of the groups are equal. It is a non-parametric version of ANOVA.
When genes were binned by FPKM, n values were:
Q1, Q2 & Q3 n= 4541, Q4 n=4542
When genes with FPKM > median were binned by length, n values were:
Q1 & Q4 n=2271, Q2&Q3 n=2270
When genes were binned by transcriptional volume or RNAP2 ChIP-seq signal, n values were:
Q1 & Q4 n=4563, Q2&Q3 n=4562
For genes transcribed above median levels in RPE-1 and below median levels in GM06990, n=324.
For genes transcribed below median levels in RPE-1 and below median levels in GM06990, n=302.
For enhancers > 50 kb from the nearest TSS, n=10959. 4628 of these are transcribed above the median level for all enhancers (high RNAP2); 6331 are transcribed below the median level for all enhancers (low RNAP2). These enhancers were compared to 10959 random sites located > 50 kb from a TSS.
When genes transcribed above median levels in RPE-1 were binned by R-loop levels in HeLa, 6078 show high DRIP signal (high DRIP TTS) and 2572 show low DRIP signal (low DRIP TTS).
P values between adjacent quartiles, calculated by Kruskal-Wallis from the first replicate in each case, are shown in figure panels and listed below. P values below 1e-04 are shown to one significant figure.
By FPKM: Q1–Q2 p = 7e-84; Q2–Q3 p = 8e-81; Q3–Q4 p = 5e-06.
By gene length: Q1–Q2 p = 1e-10; Q2–Q3 p = 7e-10; Q3–Q4 p = 1e-31.
By FPKM × gene length: Q1–Q2 p = 2e-63; Q2–Q3 p = 5e-53; Q3–Q4 p = 4e-92.
By RNAP2 ChIP-seq: Q1–Q2 p = 7e-53; Q2–Q3 p = 3e-17; Q3–Q4 p = 5e-18
RPE-1 Ok-seq, RPE-specific genes vs GM06990-specific genes: p = 2e-18
GM06990 Ok-seq, RPE-specific genes vs GM06990-specific genes: p = 7e-26
High RNAP2 vs low RNAP2; p = 5e-07
Low RNAP2 vs random sites; p = 2e-54
By FPKM × gene length:
Q1 no HU vs Q1 0.2mM HU p = 0.27;
Q2 no HU vs Q2 0.2mM HU p = 0.55;
Q3 no HU vs Q3 0.2mM HU p = 0.05;
Q4 no HU vs Q4 0.2mM HU p = 1e-06;
Q1 FANCI RNAi 0.2 mM HU vs Q1 0.2mM HU p = 0.78;
Q2 FANCI RNAi 0.2 mM vs Q2 0.2mM HU p = 0.02;
Q3 FANCI RNAi 0.2 mM vs Q3 0.2mM HU p = 0.001;
Q4 FANCI RNAi 0.2 mM vs Q4 0.2mM HU p = 2e-06;
Q1 FANCD2 RNAi 0.2 mM HU vs Q1 0.2mM HU p = 0.003;
Q2 FANCD2 RNAi 0.2 mM vs Q2 0.2mM HU p = 0.04;
Q3 FANCD2 RNAi 0.2 mM vs Q3 0.2mM HU p = 0.001;
Q4 FANCD2 RNAi 0.2 mM vs Q4 0.2mM HU p = 0.23;
By FPKM: Q1–Q2 p = 0.01; Q2–Q3 p = 1e-18; Q3–Q4 p = 7e-08.
High DRIP vs low DRIP; p = 0.85
By FPKM:
Q1 no HU vs Q1 0.2mM HU p = 2e-05;
Q2 no HU vs Q2 0.2mM HU p = 1e-12;
Q3 no HU vs Q3 0.2mM HU p = 1e-07;
Q4 no HU vs Q4 0.2mM HU p = 3e-05;
DATA AVAILABILITY STATEMENT
Data, including raw sequencing reads and tables used to generate source data for graphs in figures 1–6, are publicly available under accession number GSE114017. Custom scripts are available upon request from the corresponding authors.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the NYU Genome Technology Center for assistance with TapeStation and sequencing. We thank D. Remus, I. Whitehouse, E. Mazzoni, and H. Klein for helpful discussions, and G Sanchez for sharing RPE enhancer data. Y-H.C. was funded in part by the Molecular Oncology and Immunology NCI training program through NYU School of Medicine (5T32CA009161–40). Work in T.T.H. laboratory is supported by grants from the NIH (ES025166), V Foundation BRCA Research and Basser Innovation Award. Work in D.J.S. laboratory is supported by grants from the NIH (GM127336, GM114340) and the Searle Scholars Program.
Footnotes
ACCESSION CODES
All data have been submitted to the GEO under accession number GSE114017
COMPETING INTERESTS
No competing interests exist.
REFERENCES
- 1.Stinchcomb DT, Struhl K & Davis RW Isolation and characterisation of a yeast chromosomal replicator. Nature 282, 39–43 (1979). [DOI] [PubMed] [Google Scholar]
- 2.Hyrien O Peaks cloaked in the mist: the landscape of mammalian replication origins. J Cell Biol 208, 147–160 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prioleau MN & MacAlpine DM DNA replication origins-where do we begin. Genes Dev 30, 1683–1697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Besnard E et al. Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat Struct Mol Biol 19, 837–844 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Dellino GI et al. Genome-wide mapping of human DNA-replication origins: levels of transcription at ORC1 sites regulate origin selection and replication timing. Genome Res 23, 1–11 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langley AR, Gräf S, Smith JC & Krude T Genome-wide identification and characterisation of human DNA replication origins by initiation site sequencing (ini-seq). Nucleic Acids Res 44, 10230–10247 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petryk N et al. Replication landscape of the human genome. Nat Commun 7, 10208 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donovan S, Harwood J, Drury LS & Diffley JF Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci U S A 94, 5611–5616 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwards MC et al. MCM2–7 complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in Xenopus egg extracts. J Biol Chem 277, 33049–33057 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Ge XQ, Jackson DA & Blow JJ Dormant origins licensed by excess Mcm2–7 are required for human cells to survive replicative stress. Genes Dev 21, 3331–3341 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rocha EPC Gene essentiality determines chromosome organisation in bacteria. Nucleic Acids Research 31, 6570–6577 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osmundson JS, Kumar J, Yeung R & Smith DJ Pif1-family helicases cooperatively suppress widespread replication-fork arrest at tRNA genes. Nat Struct Mol Biol 24, 162–170 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pourkarimi E, Bellush JM & Whitehouse I Spatiotemporal coupling and decoupling of gene transcription with DNA replication origins during embryogenesis in C. elegans. Elife 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamperl S, Bocek MJ, Saldivar JC, Swigut T & Cimprich KA Transcription-Replication Conflict Orientation Modulates R-Loop Levels and Activates Distinct DNA Damage Responses. Cell 170, 774–786.e19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tran PLT et al. PIF1 family DNA helicases suppress R-loop mediated genome instability at tRNA genes. Nat Commun 8, 15025 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGuffee SR, Smith DJ & Whitehouse I Quantitative, Genome-Wide Analysis of Eukaryotic Replication Initiation and Termination. Mol Cell 50, 123–135 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tubbs A et al. Dual Roles of Poly(dA:dT) Tracts in Replication Initiation and Fork Collapse. Cell 174, 1127–1142.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith DJ & Whitehouse I Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature 483, 434–438 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harenza JL et al. Transcriptomic profiling of 39 commonly-used neuroblastoma cell lines. Sci Data 4, 170033 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macheret M & Halazonetis TD Intragenic origins due to short G1 phases underlie oncogene-induced DNA replication stress. Nature 555, 112–116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Almeida R et al. Chromatin conformation regulates the coordination between DNA replication and transcription. Nat Commun 9, 1590 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanchez GJ et al. Genome-wide dose-dependent inhibition of histone deacetylases studies reveal their roles in enhancer remodeling and suppression of oncogenic super-enhancers. Nucleic Acids Res 46, 1756–1776 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blow JJ, Ge XQ & Jackson DA How dormant origins promote complete genome replication. Trends Biochem Sci 36, 405–414 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karnani N & Dutta A The effect of the intra-S-phase checkpoint on origins of replication in human cells. Genes Dev 25, 621–633 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ceccaldi R, Sarangi P & D’Andrea AD The Fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Biol 17, 337–349 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Michl J, Zimmer J & Tarsounas M Interplay between Fanconi anemia and homologous recombination pathways in genome integrity. EMBO J 35, 909–923 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen YH et al. ATR-mediated phosphorylation of FANCI regulates dormant origin firing in response to replication stress. Mol Cell 58, 323–338 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Proudfoot NJ Transcriptional termination in mammals: Stopping the RNA polymerase II juggernaut. Science 352, aad9926 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanz LA et al. Prevalent, Dynamic, and Conserved R-Loop Structures Associate with Specific Epigenomic Signatures in Mammals. Mol Cell 63, 167–178 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merrikh H Spatial and Temporal Control of Evolution through Replication-Transcription Conflicts. Trends Microbiol 25, 515–521 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paul S, Million-Weaver S, Chattopadhyay S, Sokurenko E & Merrikh H Accelerated gene evolution through replication-transcription conflicts. Nature 495, 512–515 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Srivatsan A, Tehranchi A, MacAlpine DM & Wang JD Co-orientation of replication and transcription preserves genome integrity. PLoS Genet 6, e1000810 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hansen RS et al. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc Natl Acad Sci U S A 107, 139–144 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anderson JD & Widom J Poly(dA-dT) promoter elements increase the equilibrium accessibility of nucleosomal DNA target sites. Mol Cell Biol 21, 3830–3839 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vashee S et al. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev 17, 1894–1908 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glover-Cutter K, Kim S, Espinosa J & Bentley DL RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol 15, 71–78 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mirkin EV, Castro Roa D, Nudler E & Mirkin SM Transcription regulatory elements are punctuation marks for DNA replication. Proc Natl Acad Sci U S A 103, 7276–7281 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lang KS et al. Replication-Transcription Conflicts Generate R-Loops that Orchestrate Bacterial Stress Survival and Pathogenesis. Cell 170, 787–799.e18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paulsen RD et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell 35, 228–239 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stirling PC et al. R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev 26, 163–175 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gros J, Devbhandari S & Remus D Origin plasticity during budding yeast DNA replication in vitro. EMBO J 33, 621–636 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gros J et al. Post-licensing Specification of Eukaryotic Replication Origins by Facilitated Mcm2–7 Sliding along DNA. Mol Cell 60, 797–807 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Douglas ME, Ali FA, Costa A & Diffley JFX The mechanism of eukaryotic CMG helicase activation. Nature 555, 265–268 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Helmrich A, Ballarino M & Tora L Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol Cell 44, 966–977 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Klein K et al. Genome-Wide Identification of Early-Firing Human Replication Origins by Optical Replication Mapping. https://www.biorxiv.org/content/early/2017/11/06/214841
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data, including raw sequencing reads and tables used to generate source data for graphs in figures 1–6, are publicly available under accession number GSE114017. Custom scripts are available upon request from the corresponding authors.