Abstract
Rationale: The clinical utility of culture-independent testing of pediatric BAL specimens is unknown. In addition, the variability of the pediatric pulmonary microbiome with patient characteristics is not well understood.
Objectives: To compare testing with 16S rRNA gene–based sequencing to conventional cultures of BAL specimens in children
Methods: Study subjects were not more than 22 years old and underwent BAL from May 2013 to August 2015 as part of clinical care. DNA extracted from BAL specimens was used for 16S rRNA gene–based analysis, and results were compared with routine cultures from the same samples. Indices of microbial diversity and relative taxon abundances were compared on the basis of subject characteristics (diagnosis and antibiotic use).
Results: From 81 participants (male, 51%; median age, 9 yr), 89 samples were collected. The 16S rRNA genes of 77 samples (86.5%) from 70 subjects were successfully analyzed. These 70 subjects included 23 with cystic fibrosis, 19 who were immunocompromised, and 28 who were nonimmunocompromised. Of 68 organisms identified in culture, 16S rRNA gene–based analyses detected corresponding taxa in 66 (97.1%) and also identified potentially clinically significant organisms missed by cultures (e.g., Staphylococcus, Legionella, and Pseudomonas). Taxa that varied significantly with diagnosis and antibiotic use included Veillonella, Corynebacterium, Haemophilus, and Streptococcus. The microbiota of cystic fibrosis samples was less diverse. A “core” group of 15 taxa present in all three diagnosis groups was identified.
Conclusions: Culture-independent analysis was concordant with routine cultures and showed the potential to detect noncultured pathogens. Although culture-independent testing identified relative changes in organism abundance associated with clinical characteristics, distinct microbiome profiles associated with disease states were not identified.
Keywords: bronchoalveolar lavage, children, microbiome
Pediatric bronchoscopy has a yield for recovering bacterial pathogens in BAL specimens of 30–40% overall, and 28–68% in immunocompromised children (1, 2). At our institution, retrospective analyses of BAL results from immunocompromised children over a 12-year period showed a pathogen yield of 31%, consistent with historical studies (3). Direct amplification and sequencing of the bacterial 16S rRNA genes from BAL fluid offers the potential for more sensitive detection of potential pathogens as compared with conventional bacterial cultures, as well as an opportunity to examine the pulmonary microbiome more precisely (4–10). 16S rRNA gene–based, culture-independent (henceforward 16SRG-based) analysis can also estimate the relative abundance of various bacteria in the BAL sample (11). Whether 16SRG-based methods could detect potentially clinically relevant organisms in pediatric BAL specimens, as compared with routine bacterial cultures, has not been fully addressed. Previous studies using culture-independent approaches to assess the lung microbiota have also been done mostly in adults (9), or exclusively in pediatric patients with cystic fibrosis (12, 13).
In this study, we compare testing by 16SRG-based sequencing with conventional cultures of BAL specimens in children who underwent BAL at New York-Presbyterian Morgan Stanley Children’s Hospital (NYP-MSCH). We hypothesized that 1) culture-independent techniques would enhance the yield of pediatric BAL; 2) there would be concordance with culture-based methods; and 3) samples from patients grouped by diagnostic categories and by antibiotic exposure would demonstrate similar microbiota profiles.
Methods
Study Design
Study participants were a convenience sample of patients not more than 22 years old undergoing BAL at NYP-MSCH, a tertiary pediatric referral hospital in New York City, from May 2013 to July 2015. The Columbia University Medical Center Institutional Review Board approved this study with a waiver of assent, and informed consent was obtained from parents of children less than 18 or from patients 18 years of age and older. All BALs were done using sterile precautions and a standardized protocol by a team of pediatric pulmonologists as part of routine care.
Data Abstraction from Chart Reviews
We abstracted demographic and clinical data from the electronic medical record for each patient who underwent BAL. Clinical information included underlying diagnosis, indication for the BAL, antibiotic therapy (prophylactic and treatment) in the seven calendar days preceding the BAL, and the results of routine bacterial cultures. Subjects were categorized, by study investigators based on diagnosis at the time of BAL, into three groups: cystic fibrosis (CF); immunocompromised (IC), for example, use of immunosuppressive medication (e.g., tacrolimus) or diagnosis (e.g., leukemia); and nonimmunocompromised (nIC). Bronchoscopy was done most often for suspicion of a pulmonary infection, but occasionally for evaluation of anatomic anomalies (e.g., laryngotracheomalacia) in the nIC group.
Sample Collection and Laboratory Processing
Bacterial cultures were processed by the Columbia University Medical Center clinical microbiology laboratory according to routine protocols as per standard of care. BAL fluid was plated on chocolate, sheep blood, and MacConkey agar plates and incubated for 48 hours. For CF samples, mannitol salt and Burkholderia cepacia agar were used in addition to the three other media, and samples were incubated for 96 hours. Clinically significant bacteria were identified and reported if they grew at least 104 colony-forming units (cfu)/ml of a single organism. For CF samples, all gram-negative bacilli were identified and reported, but for other samples, clinically significant bacteria were identified and reported only when predominant growth was present.
The following organisms were considered normal microbiota and were not reported by the clinical microbiology laboratory: coagulase-negative Staphylococcus spp., α-hemolytic Streptococcus spp. (other than S. pneumoniae), γ-hemolytic Streptococcus spp., Micrococcus spp., Corynebacterium spp., Lactobacillus spp., Bacillus spp. (other than B. anthracis), Neisseria spp., Haemophilus spp. (other than H. influenzae), Eikenella spp., and Capnocytophaga spp.
BAL samples for genomic extraction were initially stored at 4°C and then frozen at −20°C within 24 hours. Genomic DNA was isolated with a DNeasy PowerSoil kit (Qiagen) with the following modifications to the manufacturer’s instructions: approximately 300 μl of BAL sample was used as starting material. Solution C1 was heated to 65°C before use and, after its addition, samples in the supplied PowerBead tubes were vortexed for 2 minutes. Eluted DNA was stored at −80°C before sequencing.
16S rRNA Gene Sequencing and Analysis
The V4 region of the 16S rRNA gene was amplified by PCR from BAL DNA in triplicate as described previously (14), using the primer pair 515f/806r. PCR amplicons were sequenced on an Illumina MiSeq at the University of Colorado Next Generation Sequencing Facility. The raw, paired-end reads were merged, quality-filtered, and clustered into operational taxonomic units (OTUs) at the ≥97% identity level, using the UPARSE pipeline (15). Taxonomy was assigned to each OTU, using the QIIME (16) implementation of the Ribosomal Database Project classifier (17) and the August 2013 release of the GreenGenes database (18). To account for variability in sequence depth across samples a standardization procedure called rarefaction was done. Communities were rarefied by randomly selecting 4,025 sequencing reads from each sample for further analysis. Where appropriate, OTUs identified as the same genus were grouped into a single taxon, for example, there were four different OTUs identified as Stenotrophomonas sp. that were grouped into a single Stenotrophomonas spp. taxon for some analyses. The relative abundance of each OTU is described as the number of sequencing reads corresponding to that OTU out of 4,025 sequences. Samples with fewer than 4,025 reads were thus excluded from analysis. α diversity (i.e., the distribution and variety of OTUs within a sample) was calculated using the Shannon-Weiner index. β diversity (i.e., how samples differ from each other) was calculated using the Bray-Curtis dissimilarity index. DNA sequences are available on the SRA public database (SRA accession: SRP144030).
Statistical Analysis
We first descriptively tabulated OTUs against organisms obtained by culture to test our hypothesis that 16SRG-based techniques would enhance the yield of pediatric BAL. We then tested our hypothesis that 16SRG-based results would be concordant with culture-based methods by comparing OTU identification with the bacteria identified by the clinical microbiology laboratory. OTUs classified only to the genus level of taxonomic resolution were assumed to correspond to the species belonging to the same genus identified in culture. For instance, an OTU determined to be Staphylococcus sp. was considered to correspond to S. aureus cultured from the same BAL sample.
We then determined whether organisms identified by routine cultures would rank among the more predominant microbiota identified by 16SRG-based analysis. OTUs were first ranked on the basis of relative abundance. We examined the likelihood that the organisms isolated in routine culture corresponded to either the most abundant OTU, or ranked within the top five OTUs by abundance (a number chosen to include a majority of the OTU reads within a sample).
To test our hypothesis that samples from patients grouped by diagnostic categories and by antibiotic exposure would demonstrate similar microbiota profiles we compared α diversity (Shannon-Weiner) between groups, using ANOVA corrected for multiple comparisons. For β diversity we used the PERMANOVA (permutational multivariate analysis of variance) statistical tests, using the vegan and ggplot2 packages in R version 3.2.3 (R Foundation for Statistical Computing).
The association of the relative abundance of select taxa with patients’ diagnosis group and antibiotic use during the 7 days before BAL was also determined. The mean relative abundance for each OTU was first compared between groups, using ANOVA. Significant differences were confirmed by multivariable regression. Log-transformed OTU counts for specific OTUs were modeled, using antibiotic treatment and diagnostic group as the predictor variables. The Benjamini-Hochberg procedure was used to correct for multiple comparisons and α error was set at 5%. Pearson’s correlation statistics were used to examine the relationship between α diversity and the abundance of specific taxa. ANOVA and Pearson’s correlation statistics were done in GraphPad Prism version 7.
To ascertain the impact of more than one BAL sample from a single patient, a sensitivity analysis was performed that included only the first collected sample from a specific patient.
An exploratory analysis was conducted to identify core species that were associated with patient groups (CF, IC, and nIC), as well as those OTUs that were present more universally. As described previously (19), we first confirmed the correlation of OTU abundance with prevalence. Indices of dispersion of OTUs were then plotted against prevalence to determine the species that were not randomly distributed in each diagnosis group. OTUs present in at least 30% of samples and present in all three groups were defined as the BAL core group of organisms.
Results
Recruited Study Population
A total of 81 patients were recruited (51% male) from whom 89 BAL samples were collected. The ages of recruited patients ranged from 5 months to 22 years (median, 9 yr; mean, 9.9 yr; SD, 6.3 yr) Demographic characteristics and diagnoses of recruited patients are shown (Table 1).
Table 1.
Characteristics of recruited subjects who underwent BAL
| Diagnosis (No. of Samples) |
|||
|---|---|---|---|
| CF (n = 27*) | IC (n = 26*) | nIC (n = 36) | |
| Age, yr† | |||
| Mean ± SD | 12.2 ± 5.8 | 11.8 ± 5.8 | 6.7 ± 5.9 |
| Range | 1–22 | 2–22 | 0–19 |
| Male sex, No. (%) | 9 (33.3%) | 12 (46.2%) | 21 (58.3%) |
| Diagnosis | CF (27) | Bone marrow transplant (9) | Asthma/wheezing/RAD (7) |
| Solid organ transplant (8) | Chronic lung disease (6) | ||
| Acute lymphocytic leukemia (2) | Chronic cough (5) | ||
| Astrocytoma (1) | Laryngotracheomalacia (4) | ||
| Hemophagocytic | Congenital heart disease (3) | ||
| lymphohistiocytosis (1) | Interstitial lung disease (3) | ||
| HIV (1) | Diabetes mellitus (2) | ||
| Hypogammaglobulinemia (1) | Kawasaki disease (1) | ||
| IgA deficiency (1) | Normal (1) | ||
| Neutropenia (1) | Obstructive sleep apnea (1) | ||
| Scleroderma (1) | Plastic bronchitis (1) | ||
| Recurrent pneumonia (1) | |||
| Vocal cord paralysis (1) | |||
| BAL done primarily to rule out infection, No. (%) | 27 (100%) | 25 (96.2%) | 21 (58.3%) |
| Treatment with antibiotics within 7 d of BAL, No. (%) | 15 (55.6%) | 14 (53.8%) | 6 (16.7%) |
Definition of abbreviations: BAL = bronchoalveolar lavage; CF = cystic fibrosis; IC = immunocompromised; nIC = nonimmunocompromised; RAD = reactive airway disease; SD = standard deviation.
One patient with CF had recently undergone organ transplantation, putting the patient in both the CF and IC categories. The patient is listed in the CF group in this table, but was not included in further analyses.
Patients with CF and IC patients were significantly older than nIC patients (P ≤ 0.001).
Twelve of the 89 samples (13.5%) were excluded from 16SRG-based analyses. Nine samples from nine patients (five nIC and four IC) were excluded because they did not have sufficient sequencing reads to attain rarefaction. Two of these samples were culture positive (one for methicillin-sensitive S. aureus and one for H. influenzae). Two samples from the nIC group were lost, and the sample from the patient with CF who had recently received an organ transplant was excluded. The final analysis included 77 samples from 70 patients: 26 CF samples (23 patients), 22 IC samples (19 patients), and 29 nIC samples (28 patients). In the CF and IC groups, there were two subjects who contributed three and two samples each, respectively. In the nIC group one patient provided two samples.
16SRG-PCR–based Analysis and Concordance with Culture
After rarefaction, 596 distinct OTUs were detected, with 398 (67%) identified to the genus level and 92 (15%) to the species level. The average number of OTUs per sample was 27.0 in the CF group (range, 8–60), which was significantly lower (P < 0.001) compared with 39.5 OTUs in the IC group (range, 6–90) and 56.4 OTUs in the nIC group (range, 8–112).
In all, 51 of 77 samples (66%) grew 68 clinically significant bacteria. The organism reported in culture corresponded to the most abundant OTU in 61% of samples (31 of 51). 16SRG-based analyses were concordant with culture results in 97.1% of cultured organisms (66 of 68), with 78% (53 of 68) present in the top five most abundant OTUs of those samples (Table 2). S. aureus was the most commonly detected organism in bacterial cultures overall (n = 28 samples) and in the CF and IC groups. Two samples had organisms detected by culture, which were not detected at the genus level with 16SRG-based results: Klebsiella pneumoniae and one instance of Stenotrophomonas maltophilia. However, OTUs from higher order taxonomic groups (Enterobacteriaceae and Proteobacteria, respectively) were identified in these samples with high relative abundance.
Table 2.
Bacterial species isolated from the 51 culture-positive BAL samples, and their corresponding operational taxonomic unit abundance rank by 16S rRNA PCR read counts
| Organism | Positive Culture | OTU Abundance Rank by 16S rRNA Read Counts |
|||
|---|---|---|---|---|---|
| Most Abundant | Top Five | Any Abundance | Absent | ||
| Staphylococcus aureus* | 28 | 14 | 21 | 28 | 0 |
| Stenotrophomonas maltophilia* | 14 | 9 | 12 | 13 | 1 |
| Haemophilus influenzae* | 7 | 4 | 6 | 7 | 0 |
| Pseudomonas aeruginosa* | 4 | 1 | 4 | 4 | 0 |
| Streptococcus pneumoniae* | 4 | 1 | 4 | 4 | 0 |
| Escherichia coli | 3 | 0 | 1 | 3 | 0 |
| Group G Streptococcus* | 1 | 0 | 0 | 1 | 0 |
| Proteus mirabilis* | 1 | 1 | 1 | 1 | 0 |
| Klebsiella pneumoniae† | 1 | 0 | 0 | 0 | 1 |
| Delftia sp. | 1 | 0 | 1 | 1 | 0 |
| Achromobacter sp. | 2 | 1 | 2 | 2 | 0 |
| Nocardia sp. | 1 | 0 | 1 | 1 | 0 |
| Serratia marcescens | 1 | 0 | 0 | 1 | 0 |
Definition of abbreviations: BAL = bronchoalveolar lavage; CF = cystic fibrosis; IC = immunocompromised; nIC = nonimmunocompromised; OTU = operational taxonomic unit; PCR = polymerase chain reaction.
A single organism was isolated from 34 samples (12 CF, 9 IC, and 13 nIC), two organisms were isolated from 17 samples (14 CF, 1 IC, 2 nIC), and 26 samples were culture-negative (12 IC and 14 nIC).
Not identified to species level by 16S rRNA analysis, but identified to corresponding genus and therefore assumed to be the cultured species.
Not identified to genus level by 16S rRNA analysis.
Eighteen bacteria were identified as the predominant taxa across the majority of samples. Ten of these were bacteria not routinely reported by the clinical laboratory. The predominant OTUs of two IC and two nIC samples were not identified to genus level (Table 3).
Table 3.
Number of samples in which each taxon was the most abundant by 16S rRNA read counts, separated by diagnostic group, with number of corresponding positive cultures in parentheses
| Predominant Taxon | No. Most Abundant by 16S rRNA Read Counts (No. identified by culture) |
||
|---|---|---|---|
| Cystic Fibrosis [n = 26] | Immunocompromised [n = 22] | Nonimmunocompromised [n = 29] | |
|
Routinely reported by microbiology laboratory | |||
| Staphylococcus sp. | 12 (12) | 9 (2) | 5 (0) |
| Delftia sp. | — | 1 (0) | — |
| Achromobacter sp. | 1 (1) | — | — |
| Stenotrophomonas sp. | 8 (8) | — | 1 (1) |
| Pseudomonas sp. | 1 (1) | 1 (0) | — |
| Proteus sp. | 1 (1) | — | — |
| Haemophilus sp. | 2 (1) | 3 (1) | 4 (2) |
| Streptococcus sp. | 0 | 2 (0) | 6 (1) |
|
Not routinely reported by microbiology laboratory | |||
| Neisseria subflava | — | 1 (0) | 3 (0) |
| Actinobacillus parahaemolyticus | — | — | 2 (0) |
| Corynebacterium sp. | — | 2 (0) | — |
| Prevotella spp. (2 OTUs) | 1 (0) | — | 1 (0) |
| Bacteroides sp. | — | — | 1 (0) |
| Pantoea agglomerans | — | — | 1 (0) |
| Granulicatella sp. | — | — | 1 (0) |
| Chryseobacterium sp. | — | — | 1 (0) |
| Acinetobacter rhizosphaerae | — | — | 1 (0) |
| Fusobacterium sp. | — | 1 (0) | — |
| Not identified to genus level | — | 2 (0) | 2 (0) |
Definition of abbreviation: OTU = operational taxonomic unit.
The total numbers of samples analyzed in each diagnostic group are listed in square brackets. Organisms “not routinely reported by microbiology laboratory” are those that are not reported in respiratory culture by our clinical laboratory.
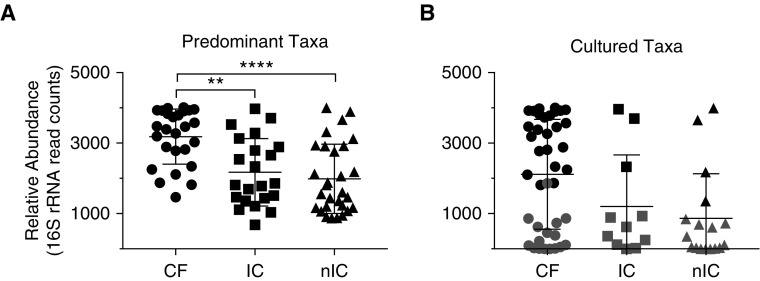
The predominant OTUs of CF samples tended to have higher relative abundances than those of IC or nIC samples (P < 0.01) (Figure 1A). These CF OTUs were also more likely to be isolated in culture, with the most abundant OTU corresponding to the organism cultured in 24 of 26 samples (92%), compared with only 14% of both IC (three of 22) and nIC (four of 29) samples. Overall culture positivity was also significantly higher (P < 0.01) in the CF group (26 of 26 samples, 100%) compared with the nIC group (15 of 29 samples, 52%) and IC group (10 of 22 samples, 45%) (Figure 1B).
Figure 1.
(A) Relative abundance, in operational taxonomic unit (OTU) read counts, of the most abundant taxon in each sample. The relative abundance of predominant cystic fibrosis (CF) OTUs was significantly higher than those of immunocompromised (IC) and nonimmunocompromised (nIC) samples following Kruskal-Wallis testing with Dunn’s multiple comparisons test. (B) Relative abundance of taxa that were isolated in the clinical laboratory. Solid points correspond to the most abundant taxa. Shaded points are nonpredominant taxa that were cultured. All samples were rarefied to 4,025 counts. **P = 0.0023, ****P < 0.0001.
A number of potential pathogens, many of which were among the top five most abundant taxa in all groups, were not identified by culture (Table 4 and Table E1 in the online supplement). These included, but were not limited to, Escherichia coli, Pseudomonas, Moraxella, Staphylococcus, Achromobacter, Legionella, Nocardia, and Mycoplasma (Tables 4 and E1).
Table 4.
All taxa included in top five operational taxonomic units by relative abundance of 16S rRNA read counts across all samples analyzed
| Taxon (No. of OTUs) | Identified by Routine Culture |
Not Identified by Routine Culture |
||||||
|---|---|---|---|---|---|---|---|---|
| No. of Samples |
Mean | No. of Samples |
Mean | |||||
| CF | IC | nIC | CF | IC | nIC | |||
| Staphylococcus spp. (2) | 18 | 2 | 1 | 1,788 | 7 | 15 | 18 | 954 |
| Streptococcus spp. (6) | — | 3 | 1 | 440 | 13 | 4 | 15 | 379 |
| Haemophilus sp. (1) | 1 | 1 | 4 | 2,531 | 2 | 4 | 9 | 997 |
| Stenotrophomonas spp. (4) | 11 | — | 1 | 1,566 | 4 | 1 | 3 | 164 |
| Escherichia coli (1) | — | 1 | — | 892 | 5 | 6 | 7 | 213 |
| Prevotella spp. (7)* | 9 | 5 | 6 | 330 | ||||
| Neisseria spp. (2) | 2 | 2 | 8 | 574 | ||||
| Actinobacillus parahaemolyticus (1) | 2 | 3 | 7 | 490 | ||||
| Veillonella dispar (2) | 4 | 3 | 5 | 137 | ||||
| Pseudomonas sp. (1) | 4 | — | — | 1,695 | 2 | 1 | 2 | 772 |
| Enterococcus sp. (1) | 2 | 1 | 5 | 261 | ||||
| Methylobacterium sp. (1) | 2 | 5 | 1 | 240 | ||||
| Fusobacterium sp. (1) | 1 | 3 | 2 | 505 | ||||
| Moraxella sp. (1) | 1 | 2 | 3 | 334 | ||||
| Brevibacillus sp. (1) | 1 | 4 | 1 | 138 | ||||
| Sphingomonas sp. (1) | 2 | 4 | — | 111 | ||||
| Acinetobacter spp. (2) | 1 | 2 | 2 | 800 | ||||
| Delftia sp. (1) | — | — | 1 | 851 | 1 | 1 | 2 | 775 |
| Corynebacterium spp. (3) | 1 | 3 | 1 | 585 | ||||
| Rothia mucilaginosa (1) | 4 | — | 1 | 89 | ||||
| Granulicatella sp. (1) | 2 | — | 2 | 731 | ||||
| Capnocytophaga spp. (2) | — | 1 | 3 | 244 | ||||
| Achromobacter sp. (1) | 1 | — | 1 | 2,049 | — | — | 1 | 1,275 |
| Parvimonas spp. (2) | 2 | 2 | — | 553 | ||||
| Pantoea agglomerans (1) | — | — | 2 | 1,962 | ||||
| Legionella sp. (1) | 1 | 1 | — | 403 | ||||
| Porphyromonas spp. (2) | 1 | — | 1 | 312 | ||||
| Campylobacter sp. (1) | — | 1 | 1 | 293 | ||||
| Aggregatibacter sp. (1) | — | 1 | 1 | 261 | ||||
| WAL_1855D (1) | 2 | — | — | 219 | ||||
| Meiothermus sp. (1) | — | 2 | — | 129 | ||||
| Leptotrichia spp. (2) | 1 | — | 1 | 64 | ||||
| Proteus sp. (1) | 1 | — | — | 2,338 | — | — | 1 | 38 |
| Nocardia spp. (2) | — | 1 | — | 936 | 1 | — | — | 22 |
| Other† | 5 | 5 | 9 | 258 | ||||
| Unidentified‡ | 20 | 21 | 20 | 331 | ||||
Definition of abbreviations: CF = cystic fibrosis; IC = immunocompromised; nIC = nonimmunocompromised; OTU = operational taxonomic unit.
In many cases, multiple OTUs of a single genus were identified, shown here in parentheses. Those that were present in the top five and also isolated in the clinical laboratory are listed under “identified by routine culture,” along with the mean 16S rRNA gene rarefied abundance for each taxon. Numerous taxa that were present in the top five by 16S rRNA abundance were not isolated in the clinical laboratory, listed here with mean abundances, under “not identified by routine culture.”
Table E1 (in the online supplement) is an extended form of this table, with each OTU listed.
The taxon Prevotella spp. includes two OTUs of genus Prevotella that belong to the family Paraprevotellaceae.
There were 19 taxa identified to genus level that were each present in the top five of only one sample.
Twenty-one different OTUs, detected in 61 samples, were not identified to genus level.
Comparison of Diagnostic Groups and Antibiotic Exposure by 16S rRNA Gene–based Amplification
The genus Staphylococcus had the highest relative abundance in all three broad diagnostic groups, but did not differ significantly between them. In multivariable regression analysis, Corynebacterium (P = 0.03) and Escherichia (P = 0.001) relative abundances were significantly decreased in the CF group compared with the nIC group (Figure E1A).
Samples from subjects exposed to antibiotics within 7 days of BAL had significantly lower Streptococcus (P = 0.04), Veillonella (P = 0.02), and Haemophilus (P = 0.01) abundance as compared with those samples from subjects without antibiotic exposure in multivariable analysis (Figure E1B).
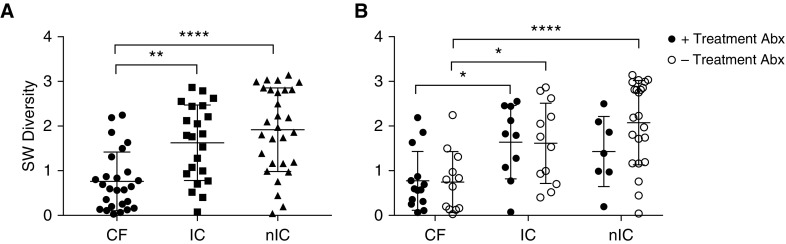
CF samples had significantly lower α diversity compared with IC and nIC samples (Figure 2A). This result is further demonstrated by the dominance of relatively fewer organisms. In the CF, IC, and nIC groups, 95%, 85%, and 79%, respectively, of the amplified 16S rRNA gene sequence reads belonged to taxa ranked in the top five by relative abundance. Differences in α diversity between groups were retained in samples with and without exposure to antibiotics, although these differences decreased with antibiotic exposure (Figure 2B).
Figure 2.
(A) α diversity (Shannon-Weiner [SW]) within groups was compared using the Kruskal-Wallis test with Dunn’s multiple comparisons test. Cystic fibrosis (CF) samples were significantly less diverse than immunocompromised (IC) and nonimmunocompromised (nIC) samples. (B) Influence of concurrent treatment antibiotics on diversity (Shannon-Weiner) within diagnosis groups by two-way ANOVA with Tukey’s multiple comparisons test. CF samples, with and without treatment antibiotics, were less diverse than IC samples. CF samples without treatment antibiotics were also less diverse than nIC samples without antibiotics. There was no significant reduction in diversity within diagnosis groups due to concurrent treatment antibiotic use. Abx = antibiotics.*0.05 > P > 0.01, **0.01 > P > 0.001, ****P < 0.0001.
When we controlled for diagnostic group, there were no significant differences in microbial α diversity associated with antibiotic exposure. In addition, a subset analysis of nIC samples for which there was a suspicion of infection versus no suspicion was similar in α diversity (P = 0.34).
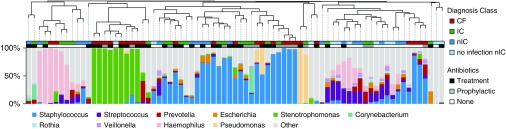
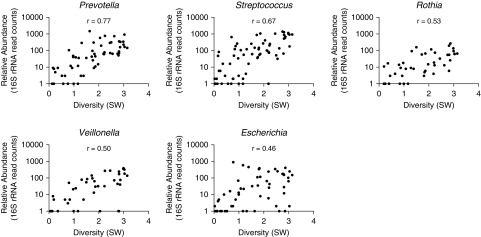
β diversity did not cluster by diagnosis or antibiotic exposure, but by the most abundant organism detected (Figure 3). In silico removal of the predominant OTU, followed by rescaling, did not enhance clustering by diagnostic group (data not shown). Abundances of certain OTUs were associated with increased diversity. For instance, the relative abundances of Prevotella (r = 0.77) and Streptococcus (r = 0.67) were associated with higher diversity (Figure 4).
Figure 3.
β diversity. Top: Dendrogram clustered by Bray-Curtis dissimilarity. Middle: Diagnosis class and concurrent antibiotic use of each sample. Bottom: Percent abundance of the top 10 most prevalent genera is shown in the corresponding stacked bar chart, with any remaining genera included in “other.” CF = cystic fibrosis; IC = immunocompromised; nIC = nonimmunocompromised.
Figure 4.
Correlation of α diversity (Shannon-Weiner [SW]) with relative abundance of the most prevalent genera. Genera shown are significantly correlated with P values less than 0.0001. Corynebacterium (r = 0.37) and Haemophilus (r = 0.35) were also significantly correlated with P = 0.0011 and 0.0016, respectively. r = Pearson correlation statistic.
Sensitivity Analysis
Repeating analysis including only the first sample obtained from patients who contributed multiple samples did not alter the findings.
BAL Core Microbiota and Nonrandomly Dispersed Organisms
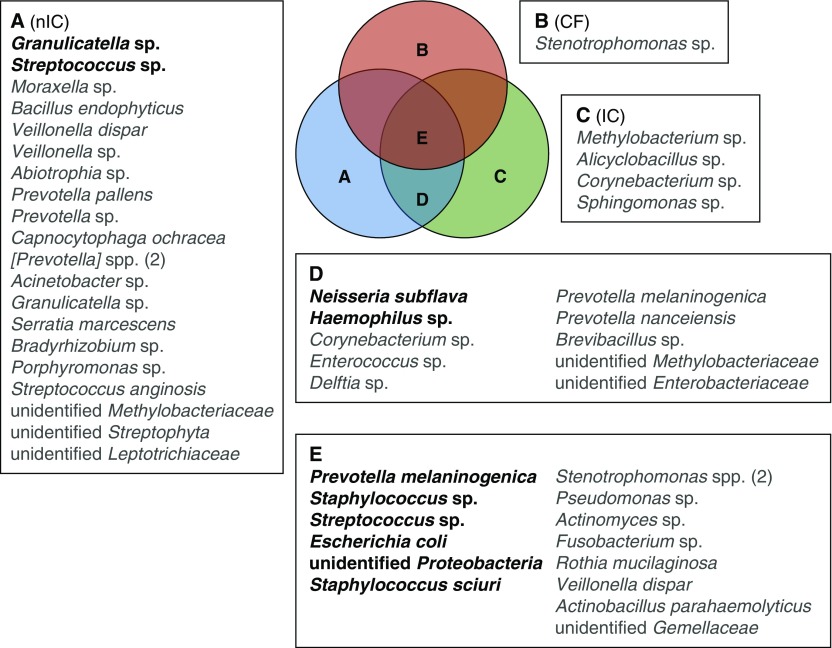
We explored identifying a core, shared microbiota across BAL samples. Across all three diagnostic groups, there were 51 prevalent (≥30%), nonrandomly dispersed taxa (Figures 5 and E2). One of these, a Stenotrophomonas OTU, was unique to the CF group, four OTUs were unique to the IC group, and 21 were unique to the nIC group. Ten OTUs were shared between IC and nIC samples, and 15 were common to all three diagnoses.
Figure 5.
BAL core taxa identified as nonrandomly distributed. Taxa listed were present in at least 30% (gray) or at least 50% (bold) of samples. There are two [Prevotella] operational taxonomic units (OTUs) (belonging to the family Paraprevotellaceae) included in group A and two Stenotrophomonas OTUs in group E. CF = cystic fibrosis; IC = immunocompromised; nIC = nonimmunocompromised.
Discussion
In this pilot study, 16SRG-based, culture-independent sequencing was a highly sensitive method for detecting bacteria present in BAL fluid in pediatric patients, similar to reports in adult patients (9). 16SRG-based sequencing identified clinically significant organisms, and rarely failed to identify organisms that grew in routine culture. In some instances, potentially pathogenic taxa including Staphylococcus, Stenotrophomonas, Escherichia, Moraxella, Mycoplasma, and Legionella, were identified using culture-independent sequencing only.
The increased sensitivity of 16SRG-based amplification has two implications when compared with routine cultures. First, it leads us to reconsider the clinical significance of organisms underreported by routine cultures either because of unique growth characteristics (e.g., obligate anaerobes), or labeling results “normal microbiota” or “mixed commensal flora.” Future work should revisit whether some of these organisms are true pathogens. Second, the ability of 16SRG-based sequencing to quantify the relative abundance of multiple taxa could be correlated with disease. The healthy lung has a relatively low bacterial burden compared with the oral cavity or gut (20). Thus, one could assume that relatively minor shifts in abundance could have more impact on pulmonary disease states. We did find that the large majority of cultured bacteria were also detected in the top five most abundant OTUs. However, the concordance of relative abundance to culture was not consistent, with high concordance in CF samples (>90%), but lower in the IC and nIC groups (<50%). Our findings confirm that routine culture techniques and reporting strategies may not provide a complete picture of the microbiota, which could result in missing true pathogens or not fully understanding the dysbiosis associated with disease.
The identification of additional organisms and tallying relative abundance by 16SRG-based amplification should be explored further for clinical utility as we found that taxa often regarded as commensal microbiota (e.g., Corynebacterium, Haemophilus, Streptococcus, Veillonella, and Prevotella) varied most with diagnosis and antibiotic use. Differences in similar taxa have been reported from the upper respiratory tract of infants with CF and control infants (21), and measuring these changes might represent an area of future studies describing the pathogenesis of pulmonary disease and therapeutic outcomes in children (22).
An unexpected finding was the lack of signature microbiota profiles associated with broad diagnosis groups or antibiotic use. Instead, predominant organisms drove microbial community similarity, with diverse clinical phenotypes having relatively similar microbial profiles. These findings highlight the limitations of cross-sectional studies of the pulmonary microbiome and underscore the importance of longitudinal data to elicit meaningful clinical information (23). Similarly, there were no striking differences in the metrics of diversity associated with antibiotic use within each diagnosis group. This could be due to the small number of patients in the non–antibiotic use group, but there is also emerging evidence that perturbations associated with antibiotic use may be lower than expected (24).
Contrary to other reports (21, 25, 26), the microbiome of CF samples in our study did not demonstrate a signature microbial profile. However, CF samples did have a lower average microbial diversity both in the number of unique taxa and the distribution of their relative abundance, consistent with other studies comparing CF and non-CF microbiota (25–28). The CF lung appears to have a microbiome that is strongly dominated by a small number of taxa likely because of enhanced fitness of specific microbes in the CF airway niche, the repeated use of antibiotics that kill other microbiota, or both.
Because definable microbial profiles for different disease states were lacking, we identified a “core” group of organisms detected across samples. These were characterized by genera such as Prevotella, Streptococcus, and Veillonella, similar to those found in healthy adult control subjects (22). In previous reports of healthy children and infants with CF, Prevotella, Streptococcus, Veillonella, and Gemella were the predominant contributors to the core group (12, 13, 27, 29), and this study extends this work to include non-CF subjects.
Our study has limitations. This was a single-center study with limited numbers of subjects, and not all samples could be included in the final analysis. Subjects within each broad clinical group had diverse characteristics, which limits the conclusions drawn from the group-level analysis. The 16SRG-based analysis does not detect viral or fungal pathogens, and the variable region (V4) used cannot provide species-level identification for many OTUs. Failure to assign a sequence to the genus or species level can occur for multiple reasons including 16S gene sequencing matches to multiple species or genera, or the existence of a novel lineage. This lack of resolution significantly limits the utility of this approach in a clinical setting. Because most comparisons were based on genus, correspondence between culture and culture-independent analyses could have been overestimated. We did not measure absolute abundance, which may be a better indicator of disease than relative abundance. Last, specimens from the lower respiratory tract could be contaminated with upper respiratory tract microbiota; simultaneous acquisition of upper respiratory swabs has been shown to improve the interpretation of lower respiratory samples (30–33).
In conclusion, we sought to determine whether 16S rRNA gene–based sequencing could be a useful diagnostic modality for testing BAL specimens in the pediatric population. Although we found that 16S rRNA gene–based sequencing techniques were extremely sensitive and rarely missed cultured microorganisms, this increased sensitivity comes at the cost of new diagnostic and therapeutic uncertainty. However, there is potential utility of culture-independent testing of BAL for identifying organisms not reported in culture, quantifying microbial relative abundance, and measuring deviations from core taxa. Longitudinal studies are better suited to correlate culture-independent results with diagnosis, outcomes, and prognosis.
Supplementary Material
Acknowledgments
Acknowledgment
The authors acknowledge Susan Whittier (Department of Pathology, Columbia University), and Tobin Hammer and Noah Fierer (Department of Ecology and Evolutionary Biology, University of Colorado, Boulder), for assistance with laboratory analysis of specimens.
Footnotes
Supported by a Thrasher Research Foundation Early Career Award (P.Z. and S.N.), National Institutes of Health T-32 Award (P.Z. and S.N.), and the following awards to P.J.P.: a Pediatric Infectious Disease Society-St. Jude Award, a Doris Duke Clinical Scientist Development Award, and a Cystic Fibrosis Foundation Pilot Grant. The funding agencies had no role in the study design; in the collection, analysis, and interpretation of the data; in the writing of the report; and in the decision to submit the article for publication.
Author Contributions: All authors contributed to the planning, conduct, and reporting of the work described in the article. This included study planning (P.Z., C.R., S.N., M.F., L.S., and P.J.P.), obtaining funding (S.N., L.S., M.F., and P.J.P.), patient recruitment/collection of clinical data/laboratory testing/analysis (P.Z., C.R., S.N., G.C., M.K., H.S., and P.J.P.); and writing the manuscript (P.Z., C.R., S.N., M.K., H.S., M.F., L.S., and P.J.P.). All authors have seen and approved this manuscript.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Park JR, Fogarty S, Brogan TV. Clinical utility of bronchoalveolar lavage in pediatric cancer patients. Med Pediatr Oncol. 2002;39:175–180. doi: 10.1002/mpo.10130. [DOI] [PubMed] [Google Scholar]
- 2.Efrati O, Gonik U, Bielorai B, Modan-Moses D, Neumann Y, Szeinberg A, et al. Fiberoptic bronchoscopy and bronchoalveolar lavage for the evaluation of pulmonary disease in children with primary immunodeficiency and cancer. Pediatr Blood Cancer. 2007;48:324–329. doi: 10.1002/pbc.20784. [DOI] [PubMed] [Google Scholar]
- 3.Nadimpalli S, Foca M, Satwani P, Sulis ML, Constantinescu A, Saiman L. Diagnostic yield of bronchoalveolar lavage in immunocompromised children with malignant and non-malignant disorders. Pediatr Pulmonol. 2017;52:820–826. doi: 10.1002/ppul.23644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Srinivasan L, Pisapia JM, Shah SS, Halpern CH, Harris MC. Can broad-range 16S ribosomal ribonucleic acid gene polymerase chain reactions improve the diagnosis of bacterial meningitis? A systematic review and meta-analysis. Ann Emerg Med. 2012;60:609–620.e2. doi: 10.1016/j.annemergmed.2012.05.040. [DOI] [PubMed] [Google Scholar]
- 5.Vondracek M, Sartipy U, Aufwerber E, Julander I, Lindblom D, Westling K. 16S rDNA sequencing of valve tissue improves microbiological diagnosis in surgically treated patients with infective endocarditis. J Infect. 2011;62:472–478. doi: 10.1016/j.jinf.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 6.Su G, Fu Z, Hu L, Wang Y, Zhao Z, Yang W. 16S ribosomal ribonucleic acid gene polymerase chain reaction in the diagnosis of bloodstream infections: a systematic review and meta-analysis. PLoS One. 2015;10:e0127195. doi: 10.1371/journal.pone.0127195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui L, Morris A, Huang L, Beck JM, Twigg HL, III, von Mutius E, et al. The microbiome and the lung. Ann Am Thorac Soc. 2014;11:S227–S232. doi: 10.1513/AnnalsATS.201402-052PL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pendleton KM, Erb-Downward JR, Bao Y, Branton WR, Falkowski NR, Newton DW, et al. Rapid pathogen identification in bacterial pneumonia using real-time metagenomics. Am J Respir Crit Care Med. 2017;196:1610–1612. doi: 10.1164/rccm.201703-0537LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dickson RP, Erb-Downward JR, Prescott HC, Martinez FJ, Curtis JL, Lama VN, et al. Analysis of culture-dependent versus culture-independent techniques for identification of bacteria in clinically obtained bronchoalveolar lavage fluid. J Clin Microbiol. 2014;52:3605–3613. doi: 10.1128/JCM.01028-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marsh RL, Kaestli M, Chang AB, Binks MJ, Pope CE, Hoffman LR, et al. The microbiota in bronchoalveolar lavage from young children with chronic lung disease includes taxa present in both the oropharynx and nasopharynx. Microbiome. 2016;4:37. doi: 10.1186/s40168-016-0182-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Props R, Kerckhof FM, Rubbens P, De Vrieze J, Hernandez Sanabria E, Waegeman W, et al. Absolute quantification of microbial taxon abundances. ISME J. 2017;11:584–587. doi: 10.1038/ismej.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boutin S, Graeber SY, Weitnauer M, Panitz J, Stahl M, Clausznitzer D, et al. Comparison of microbiomes from different niches of upper and lower airways in children and adolescents with cystic fibrosis. PLoS One. 2015;10:e0116029. doi: 10.1371/journal.pone.0116029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laguna TA, Wagner BD, Williams CB, Stevens MJ, Robertson CE, Welchlin CW, et al. Airway microbiota in bronchoalveolar lavage fluid from clinically well infants with cystic fibrosis. PLoS One. 2016;11:e0167649. doi: 10.1371/journal.pone.0167649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 16.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Gast CJ, Walker AW, Stressmann FA, Rogers GB, Scott P, Daniels TW, et al. Partitioning core and satellite taxa from within cystic fibrosis lung bacterial communities. ISME J. 2011;5:780–791. doi: 10.1038/ismej.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dickson RP, Erb-Downward JR, Huffnagle GB. The role of the bacterial microbiome in lung disease. Expert Rev Respir Med. 2013;7:245–257. doi: 10.1586/ers.13.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prevaes SM, de Winter-de Groot KM, Janssens HM, de Steenhuijsen Piters WA, Tramper-Stranders GA, Wyllie AL, et al. Development of the nasopharyngeal microbiota in infants with cystic fibrosis. Am J Respir Crit Care Med. 2016;193:504–515. doi: 10.1164/rccm.201509-1759OC. [DOI] [PubMed] [Google Scholar]
- 22.Man WH, de Steenhuijsen Piters WA, Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol. 2017;15:259–270. doi: 10.1038/nrmicro.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frayman KB, Armstrong DS, Carzino R, Ferkol TW, Grimwood K, Storch GA, et al. The lower airway microbiota in early cystic fibrosis lung disease: a longitudinal analysis. Thorax. 2017;72:1104–1112. doi: 10.1136/thoraxjnl-2016-209279. [DOI] [PubMed] [Google Scholar]
- 24.Fodor AA, Klem ER, Gilpin DF, Elborn JS, Boucher RC, Tunney MM, et al. The adult cystic fibrosis airway microbiota is stable over time and infection type, and highly resilient to antibiotic treatment of exacerbations. PLoS One. 2012;7:e45001. doi: 10.1371/journal.pone.0045001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Renwick J, McNally P, John B, DeSantis T, Linnane B, Murphy P Study of Host Immunity and Early Lung Disease in CF (SHIELD CF) The microbial community of the cystic fibrosis airway is disrupted in early life. PLoS One. 2014;9:e109798. doi: 10.1371/journal.pone.0109798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blainey PC, Milla CE, Cornfield DN, Quake SR. Quantitative analysis of the human airway microbial ecology reveals a pervasive signature for cystic fibrosis. Sci Transl Med. 2012;4:153ra130. doi: 10.1126/scitranslmed.3004458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Gast CJ, Cuthbertson L, Rogers GB, Pope C, Marsh RL, Redding GJ, et al. Three clinically distinct chronic pediatric airway infections share a common core microbiota. Ann Am Thorac Soc. 2014;11:1039–1048. doi: 10.1513/AnnalsATS.201312-456OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feigelman R, Kahlert CR, Baty F, Rassouli F, Kleiner RL, Kohler P, et al. Sputum DNA sequencing in cystic fibrosis: non-invasive access to the lung microbiome and to pathogen details. Microbiome. 2017;5:20. doi: 10.1186/s40168-017-0234-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zemanick ET, Wagner BD, Robertson CE, Ahrens RC, Chmiel JF, Clancy JP, et al. Airway microbiota across age and disease spectrum in cystic fibrosis. Eur Respir J. 2017;50:1700832. doi: 10.1183/13993003.00832-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dickson RP, Erb-Downward JR, Freeman CM, McCloskey L, Falkowski NR, Huffnagle GB, et al. Bacterial topography of the healthy human lower respiratory tract. MBio. 2017;8:e02287-16. doi: 10.1128/mBio.02287-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bassis CM, Erb-Downward JR, Dickson RP, Freeman CM, Schmidt TM, Young VB, et al. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio. 2015;6:e00037. doi: 10.1128/mBio.00037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. 2011;184:957–963. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charlson ES, Bittinger K, Chen J, Diamond JM, Li H, Collman RG, et al. Assessing bacterial populations in the lung by replicate analysis of samples from the upper and lower respiratory tracts. PLoS One. 2012;7:e42786. doi: 10.1371/journal.pone.0042786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.