SUMMARY
Genomic imprinting is an allelic gene expression phenomenon primarily controlled by allele-specific DNA methylation at the imprinting control region (ICR), but the underlying mechanism remains largely unclear. N-α-acetyltransferase 10 protein (Naa10p) catalyzes N-α-acetylation of nascent proteins, and mutation of human Naa10p is linked to severe developmental delays. Here we report that Naa10-null mice display partial embryonic lethality, growth retardation, brain disorders, and maternal effect lethality, phenotypes commonly observed in defective genomic imprinting. Genome-wide analyses further revealed global DNA hypomethylation and enriched dysregulation of imprinted genes in Naa10p-knockout embryos and embryonic stem cells. Mechanistically, Naa10p facilitates binding of DNA methyltransferase 1 (Dnmt1) to DNA substrates, including the ICRs of the imprinted allele during S phase. Moreover, the lethal Ogden syndrome-associated mutation of human Naa10p disrupts its binding to the ICR of H19 and Dnmt1 recruitment. Our study thus links Naa10p mutation-associated Ogden syndrome to defective DNA methylation and genomic imprinting.
Graphical Abstract
In Brief
Lee et al. reveal an unexpected function for Naa10p, which is primarily known to acetylate nascent peptides from ribosomes, in maintaining global DNA methylation and marking the imprinted allele for genomic imprinting maintenance. Their results suggest that defects in DNA methylation and genomic imprinting may contribute to Naa10p-associated Ogden syndrome.
INTRODUCTION
Genomic imprinting, a parental allele-specific gene expression phenomenon, is caused by allelic DNA methylation of imprinting control regions (ICRs) (also known as differentially methylated regions [DMRs]) and plays an important role in development (Barlow and Bartolomei, 2014). Imprinting is established de novo during spermatogenesis and oogenesis by the members of the DNA methyltransferase family Dnmt3a, 3b, and 3l, maintained by Dnmt1 in pre-implantation embryos and later developmental stages, and demethylated by Tet1 (ten eleven translocation 1) dioxygenase in primordial germ cells so that gamete-specific DNA methylation can be reestablished (Barlow and Bartolomei, 2014; Yamaguchi et al., 2013). In addition to DNA methylation, specific histone modifications such as H3K9me2 (i.e., histone 3 dimethylated at lysine 9) (Zhang et al. 2016), H3K9me3, and H4K20me3 (Delaval et al., 2007; Regha et al., 2007) are known to decorate the ICRs/DMRs of the imprinted allele. A recent study revealed that H3K27me3 controls DNA methylation-independent genomic imprinting (Inoue et al., 2017) Defects in genomic imprinting often result in developmental defects (Peters, 2014).
To date, only a few proteins have been reported to be important for maintaining genomic imprinting during global DNA demethylation in the zygotes. One example is the maternal krüppel-associated box domain-zinc-finger binding protein 57 (Zfp57), which binds to the methylated TGCCGC motif in a subset of ICRs/DMRs and interacts with Dnmts via a corepressor, the tripartite motif-containing 28 (Trim28) (also called KRAB-ZFP-interacting protein [KAP1/TIF1β]) (Li et al., 2008; Quenneville et al., 2011; Zuoetal., 2012). Maternal-zygotic deletion of Zfp57 or deficiency of oocyte Trim28 results in partial loss of genomic imprints and embryonic lethality (Li et al., 2008; Messerschmidt et al., 2012). In addition, primordial germ cell protein 7 (PGC7, also called Stella/Dppa3) has been reported to bind to H3K9me2 to protect the imprinted loci from being demethylated by Tet3 in pre-implantation embryos (Nakamura et al., 2012). A full account of the factors involved in imprinting maintenance is still lacking.
Mammalian N-α-acetyltransferase 10 protein (Naa10p, also called Ard1), encoded by the X-linked gene Naa10, belongs to the highly conserved N-terminal acetyltransferase (NAT) family of proteins that co- or post-translationally acetylate the N-terminal α-amino group of more than 80% of human proteins (Dörfel and Lyon, 2015). Both human and mouse Naa10p contain 235 amino acid residues (96% identity), with the acetyltransferase domain located at residues 42–110 (Sánchez-Puig and Fersht, 2006). Although Naa10p was originally shown to associate with ribosomes in the NatA complex with Naa15p and Huntingtin-interacting protein K, it has also been reported to localize in the nucleus in a variety of cell types (Aksnes et al., 2015; Dörfel and Lyon, 2015; Park et al., 2014). Naa10p regulates cell proliferation and cancer formation, DNA damage response, hypoxia, bone formation, and neuronal development in enzymatic activity-dependent and -independent manners (Dörfel and Lyon, 2015). Missense mutations such as S37P (Rope et al., 2011), R116W (Popp et al., 2015), V107F (Popp et al., 2015), Y43S (Casey et al., 2015), R83C and F128L/I (Saunier et al., 2016), as well as a splice donor mutation (Esmailpour et al., 2014) in human Naa10p (hNaa10p), have been associated with severe developmental delays. Among these, S37P-associated Ogden syndrome has the most severe phenotypes, which include postnatal growth failure, minimal subcutaneous fat, intellectual disability, premature aging, brain disorders, and lethality before 1.5 years of age. Although loss of hNaa10p NAT activity has been suggested to account for pathogenic phenotypes (Myklebust et al., 2015; Popp et al., 2015; Rope et al., 2011), the R116W mutation only barely affects the NAT activity of hNaa10p (Popp et al., 2015), suggesting the existence of NAT-independent mechanisms. We previously showed that hNaa10p binds directly to DNMT1 to promote lung tumorigenesis in an hNaa10p NAT activity-independent manner (Lee et al., 2010). Whether dysregulation of DNMT1 contributes to Naa10p mutation-caused developmental delays remains an intriguing question.
To characterize the role of Naa10p in DNA methylation and development, we generated a Naa10p-knockout (KO) mouse model. Our results show that Naa10 KO leads to defects in DNA methylation and genomic imprinting and that the Ogden syndrome-causing Naa10p S37P mutation disrupts ICR binding of Naa10p and Dnmt1.
RESULTS
Naa10p KO Results in Embryonic and Postnatal Developmental Defects
To understand the developmental function of Naa10p, we generated Naa10-KO mice (Figure S1A). Examination of the pups revealed that Naa10 KO resulted in lower birthrates (Table S1). Consistently, 35% of Naa10-KO male and 27% of Naa10-KO female pups died with developmental defects during embryonic days 12.5–14.5. 18% embryonic lethality was observed for heterozygous Naa10−/x embryos (the Naa10 allele on the maternally inherited X chromosome is shown first), but no lethality was observed for heterozygous Naa10x/− embryos (Figure 1A; Figure S1B; Table S2). The lethality of the Naa10−/x embryos was likely caused by preferential inactivation of the paternal X chromosome carrying the WT Naa10 allele in the placental tissues (Takagi and Sasaki, 1975).
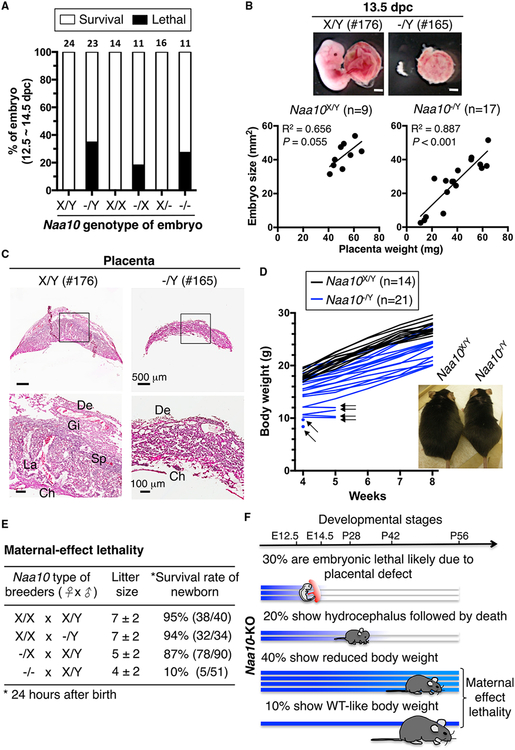
Figure 1. Phenotypes of Naa10-KO Mice.
(A) Naa10p deficiency causes partial embryonic lethality. Survival of Naa10-KO embryos at 12.5–14.5 days post-coitum (dpc) is shown. Naa10X/Y and Naa10−/Y are from Naa10−/X females crossed with WT or Naa10-KO males. Naa10X/X and Naa10−/X are from Naa10−/X females crossed with WT males. Naa10X/− and Naa10−/− are from Naa10−/X females crossed with Naa10-KO males. Embryos alive or dead were determined by heartbeat and embryo morphology. Embryo numbers are indicated above each bar.
(B) Positive correlation between embryo size and placenta weight in defective Naa10-KO embryos. Embryo size and placenta weight were measured for embryonic day 13.5 embryos of the WT (Naa10X/Y, n = 9) and mutant (Naa10−/Y, n = 17), derived from Naa10−/X females crossed with WT males. Representative images of embryos (176 and 165) and their placentae are shown. Embryo size is plotted versus placenta weight for each individual. The correlation (R2) and the p value for the Pearson analysis are shown.
(C) Naa10 KO results in placental defects. Representative images of H&E-stained WT (176) and Naa10-KO (165) placentae at 13.5 dpc are shown. Regions in black rectangles are enlarged at the bottom. De, decidual; Ch, chorionic plate; Gi, trophoblast giant cells; La; labyrinthine zone; Sp, spongioblast.
(D) Naa10-KO mice show growth retardation. The body weight of WT (Naa10X/Y, n = 14) and Naa10-KO (Naa10−/Y n = 21) mice was monitored for 4 weeks. Black arrows indicate mice that were found dead at the next time point. The image is representative of WT and Naa10-KO mice at the age of 25 weeks.
(E) Naa10-KO mice exhibit maternal effect lethality. Four types of breeding were carried out with parental strains of the indicated genotype. Pup survival was determined by heartbeat 24 hr after birth.
(F) Summary of phenotypes of Naa10-KO mice.
See also Figure S1 and Tables S1–S3.
Next we assessed whether the lethality of Naa10-KO embryos was caused by placental abnormality. We found that the sizes of Naa10-null embryos were highly correlated with placental weight (see Figure 1B for Naa10−/Y and Figure S1C for Naa10−/X and Naa10−/−), suggesting that the placental defect contributed to lethality. In agreement with the notion, histological analysis indicated that the trophoblast giant cells (Gis), spongioblasts (Sps), and labyrinthine zone (La) were lost in the placentae of abnormally smaller Naa10−/Y (Figure 1C) and Naa10−/− (Figure S1D) embryos. Given that these regions are involved in oxygen and nutrient exchanges between fetus and mother (Pijnenborg et al., 1981), the placental defect was likely a major cause of embryonic lethality and reduced litter size of Naa10-KO mice.
Of the mice that survived through embryonic development, ~34% of Naa10-KO male mice developed a dome-shaped head, which is a common phenotype of hydrocephalus (Rekate, 2009), 4–5 weeks after birth and died within 1 week after exhibiting the symptom (Figure S1E; Table S3). These mice exhibited enlarged lateral ventricles, a dilated third ventricle, and meningeal hemorrhage on the dome-shaped cranium, indicating increased brain pressure (Figure S1E). Notably, patients with Naa10p mutations also exhibit dilated brain ventricles (Rope et al., 2011).
We also noticed that 40% of the Naa10-KO mice survived past 6 weeks but only had 80% of the body weight of their wild-type (WT) littermates at the age of 8 weeks. This reduced body weight persisted throughout development. At the age of 25 weeks, their body weight was only 75% of controls, implying that Naa10-KO mice were postnatally growth-retarded (Figure 1D). Because Naa10p is highly expressed in the testes and ovaries, we analyzed the effect of Naa10p deficiency on fertility and found that Naa10-KO female mice (Naa10−/−/) generated 4 ± 2 pups per litter with only a 10% perinatal survival. Among the 51 newborns, only 5 (exclusively Naa10−/X) survived for 24 hr, which is much less than those generated from WT or Naa10-heterozygous females crossed with WT males (Figure 1E). These results indicated that Naa10 KO resulted in maternal effect lethality. Because WT female mice crossed with WT or Naa10-KO male mice generated a similar litter size and perinatal survival rate of pups, we suspected that Naa10p deficiency had little effect, if any, on spermatogenesis (Figure 1E). In summary, ~30% of Naa10-KO mice exhibited embryonic lethality, likely because of placental defects, 20% developed hydrocephalus followed by death, and 40% showed postnatal growth retardation with reduced body weight and maternal effect lethality (Figure 1F). Only 10% of Naa10-KO mice had a comparable body size as the WT; however, these mice also exhibited maternal effect lethality (Figure 1E).
Naa10p KO Alters the Expression of Imprinted Genes
The developmental phenotypes exhibited in Naa10-KO mice are frequently observed in mice with imprinting defects, including mice derived from Dnmt1-KO oocytes or Tet1-KO sperms (Howell et al., 2001; Yamaguchi et al., 2013). This observation, combined with the fact that Naa10p regulates DNA methylation in human cancer cells (Lee et al., 2010), prompted us to analyze possible imprinting defects. To this end, we crossed Naa10-heterozygous C57BL/6 female with WT JF1 male mice (Kobayashi et al., 2006) to generate C57BL/6-JF1 hybrid embryos so that the two parental alleles could be distinguished using the species-specific SNPs. The two Naa10-KO embryos (79 and 85) were smaller than their WT littermates (Figure 2A). qRT-PCR analysis revealed increased mRNA levels for both H19 and Kcnq1ot1 and reduced levels for Igf2 and Cdkn1c in the Naa10-KO embryos (Figure 2A). The results are consistent with the known reciprocal regulation of these pairs of genes, H19-Igf2 and Kcnq1ot1-Cdkn1c (Fitzpatrick et al., 2002; Thorvaldsen et al., 1998). To confirm that upregulation of H19 and Kcnq1ot1 was due to loss of silencing of the imprinted allele, H19 and Kcnq1ot1 RT-PCR products were digested with a JF1- or C57BL/6 SNP-sensitive endonuclease, respectively. As expected, only digestion of the 360-bp H19 PCR product from Naa10-KO embryos yielded fragments of 200 and 160 bp, indicating that Naa10p deficiency caused abnormal paternal expression of H19 (Figure 2B). Similar results were obtained for Kcnq1ot1 (Figure 2B). These data demonstrate that Naa10p controls allele-specific imprinted gene expression in mouse embryos.
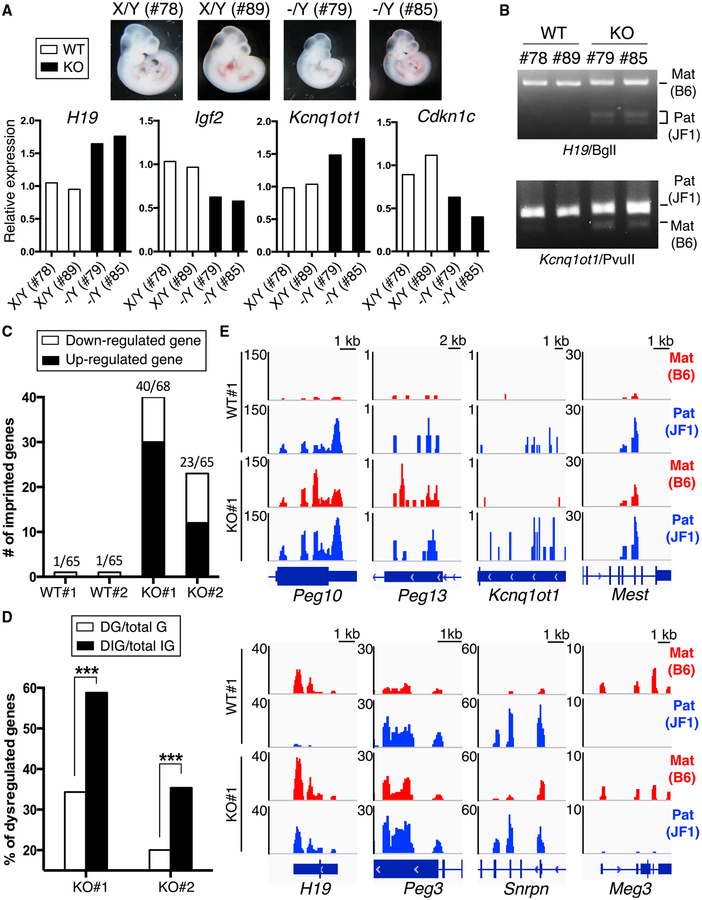
Figure 2. Dysregulation of Imprinted Genes in Naa10p-Null Mouse Embryos and ESCs.
(A) Naa10p KO dysregulatesthe expression of imprinted genes in embryos. Two WT(X/Y, 78 and 89) and two Naa10-KO (−/Y, 79 and 85) B6/JF1 hybrid embryos from Naa10−/X female mice (C57BL/6) crossed with WT male (JF1) mice were harvested at 10.5 dpc. The genotypes were determined by PCR, and the mRNA levels of H19, Igf2, Kcnq1ot1, and Cdkn1c were analyzed by qRT-PCR.
(B) Naa10p KO restores expression of the imprinted allele in embryos. RT-PCR products for H19 and Kcnq1ot1 from B6/JF1 hybrid embryos in (A) were digested with BglI and PvuII, respectively. The 360-bp and 200/160-bp bands of H19 fragments are from C57BL/6 and JF1, respectively. The 418- and 361-bp bands of Kcnq1ot1 are from JF1 and C57BL/6, respectively. Mat, maternal. Pat, paternal.
(C) RNA-seq identifies multiple dysregulated imprinted genes in Naa10p-KO ESCs. Two WT (X/Y) and two Naa10-KO (−/Y) B6/JF1 hybrid ESCs were used for RNA-seq analysis. Numbers of upregulated (black bar) and downregulated (white bar) imprinted genes with fold changes >1.5 and fragments per kilobase of exon per million fragments mapped (FPKM) >0.4 are shown. The numbers at the left above each bar indicate the numbers of dysregulated imprinted genes, and the numbers at the right indicate the numbers of total detectable imprinted genes (FPKM > 0.4). Fold change = FPKM of KO/average FPKM of WT1 and WT2.
(D) Imprinted genes are significantly enriched among the total number of genes for which expression changed in Naa10p-KO ESCs. The RNA-seq results in (C) were compared for dysregulated genes (DGs) among the total number of genes detected (total Gs) and dysregulated imprinted genes (DIGs) among the total number of imprinted genes detected (total IGs) assessed with the chi-square test. ***p < 0.001.
(E) Naa10p KO increases gene expression from the imprinted allele in ESCs. Allele-specific RNA-seq results of eight imprinted genes from WT#1 and KO#1 ESCs are shown. The reads were mapped to B6 and JF1 genomes using the allele-specific alignment pipeline (ASAP), and only the reads specific to either genome were retained as maternal B6 (red) or paternal JF1 (blue) reads. The RNA-seq density profiles were normalized to the density per million total reads with a resolution of 25 bp. For each gene, the genomic location is shown at the bottom.
To examine whether Naa10p has a general role in controlling imprinted gene expression, we carried out allele-specific RNA sequencing (RNA-seq) analysis of two clones of WT and Naa10-KO embryonic stem cells (ESCs) independently derived from different embryos of the same litter. Of note, Naa10p-deficient ESCs were similar to WT with regard to morphology (Figure S2A) and levels of pluripotent factors (Figure S2B). RNA-seq revealed that 1,542 genes involved in development and cell differentiation were differentially expressed in both KO ESCs (Figure S2C). Importantly, 40 and 23 of the 68 and 65 detectable imprinted genes, respectively, in the two KO clones were dysregulated (Figure 2C; Table S4). Statistical analysis showed that the imprinted genes were significantly enriched among the dysregulated genes in Naa10-KO ESCs (Figure 2D). Besides H19-Igf2 and Kcnq1ot1-Cdkn1c (Figure 2A), other dysregulated imprinted genes identified by RNA-seq included the well-characterized Peg10-Ppp1r9a pair (Ono et al., 2006; Table S4). As expected, expression of the maternally imprinted alleles of Peg10, Peg13, Kcnq1ot1, Mest, Peg3, and Snrpn and the paternally imprinted allele of H19 was increased in Naa10p-deficient ESCs (Figure 2E; Figure S2D). In KO#1, the paired imprinted genes Peg3-Zim3 (Kim et al., 2012) and Kcnq1ot1-Ascl2 (Fitzpatrick et al., 2002) were also dysregulated (Table S4). Notably, Naa10p deficiency affected both paternally and maternally imprinted genes (Table S4). These data indicate that loss of Naa10p function has a broad effect on imprinted gene expression, although Naa10p does not control all of them (e.g., Meg3).
Naa10p KO Causes Hypomethylation of ICRs
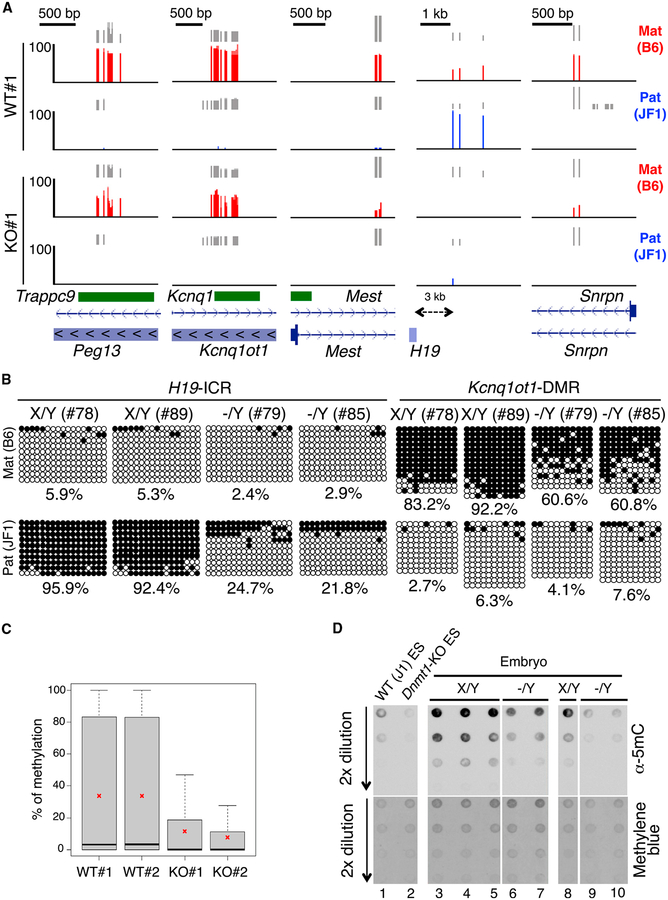
To understand the mechanism underlying the defect in imprinted gene expression, we analyzed the effect of Naa10p depletion on DNA methylation. Reduced representative bisulfite sequencing (RRBS) revealed that many ICRs/DMRs covered by RRBS were hypomethylated in Naa10p-null ESCs (Figure 3A). The hypomethylated regions included the maternally imprinted ICRs/DMRs of Trappc9-Peg13, Kcnq1-Kcnq1ot1, Mest, Snrpn, and Grb10 as well as the paternally imprinted H19-ICR (Figure 3A; Figure S3A). Because no SNPs on Peg10 and Peg3 DMRs were available, their methylation status could not be evaluated by RRBS (Figure 2E). Consistent with the RRBS results for ESCs, bisulfite sequencing indicated that the paternal H19-ICR and the maternal Kcnq1ot1-DMR were indeed hypomethylated in the two Naa10-KO embryos analyzed (Figure 3B). RRBS further revealed that hypomethylation was not limited to ICRs/DMRs but, rather, global (Figure 3C; Figure S3B). Consistently, dot blot analysis of genomic DNA from embryos showed that Naa10p KO greatly reduced the global 5-methylcytosine (5mC) level (Figure 3D, compare lanes 6 and 7 with lanes 3–5 and lanes 9 and 10 with lane 8). Collectively, these data indicate that Naa10p plays an important role in maintaining DNA methylation in mouse embryos and ESCs.
Figure 3. Decreased Allele-Specific Methylation in ICRs/DMRs of Naa10p-Null Mouse Embryos and ESCs.
(A) Naa10p KO dysregulates allele-specific methylation of ICRs/DMRs in ESCs. Allele-specific RRBS results of five ICRs/DMRs of WT#1 and KO#1 B6/JF1 hybrid ES clones are shown. RRBS reads were mapped to B6 and JF1 genomes using the ASAP, and only the reads specific to either genome were retained as maternal B6 (red) or paternal JF1 (blue) reads. Vertical gray lines represent the sequence read depth for each cytosine scored, and reads >10 are shown. The vertical red (B6) and blue (JF1) lines represent the percent methylation for each cytosine scored, which ranges from 0%–100%. For each gene, reference sequence (RefSeq) exon organization (blue) and location of CpG islands (green) are shown at the bottom.
(B) Naa10p KO causes hypomethylation of the imprinted allele in embryos. Allele-specific bisulfite sequencing was performed for H19-ICR and Kcnq1ot1-DMR in the same hybrid B6/JF1 embryos as in Figure 2. Open and closed circles indicate non-methylated and methylated, respectively, cytosines of CpGs examined. The numbers at the bottom of each panel represent the percentage of CpG methylation.
(C) RRBS analyses reveal global loss of DNA methylation in Naa10-null ESCs. Shown are the percentages of DNA methylation of two WT and two Naa10-KO ESCs used in (A). Asterisks indicate the mean of the DNA methylation.
(D) Naa10-KO embryos show DNA hypomethylation. Dot blot analyses are shown for the 5mC level using genomic DNAs from WT ESCs (lane 1), Dnmt1-KO ESCs (lane 2), embryos of Naa10X/Y (lanes 3–5 and lane 8), and Naa10−/Y mice (lanes 6 and 7 and lanes 9 and 10). Methylene blue staining shows equal loading in lanes 3–7 and lanes 8–10.
See also Figure S3.
Naa10p Deficiency Impairs Dnmt1 Activity and Recruitment to ICRs/DMRs
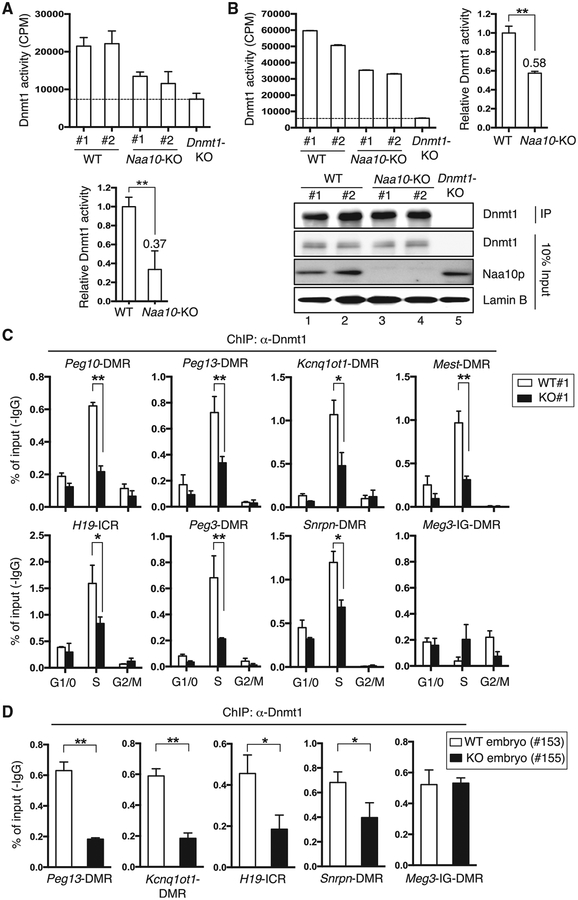
To understand how Naa10p KO downregulated DNA methylation, we first examined whether any of the proteins known to regulate DNA methylation were affected by Naa10p deficiency. Western blotting analysis indicated that the nuclear levels of Zfp57, Trim28, and components of the DNA methylation machinery (e.g., Dnmt1, Dnmt3a/b, and Uhrf1) and the 5mC oxidation machinery (e.g., Tet1 and Tet2) involved in DNA demethylation were not altered by Naa10p KO (Figures S3C, S3D, and S4A). Moreover, Naa10p KO did not disrupt the interaction between Dnmt1 and Uhrf1 (Figure S3D). Because hNaa10p can regulate DNMT1 activity in human lung cancer cells (Lee et al., 2010), we tested whether Naa10p KO affected Dnmt1 activity in mouse ESCs. Naa10p depletion caused an ~60% reduction in Dnmt1 activity toward a hemi-methylated DNA oligo substrate when nuclear extracts were used as the enzyme source (Figure 4A), whereas the activity was reduced by ~40% when endogenous Dnmt1 immunoprecipitated from the nuclear extracts of Naa10-KO ESCs was analyzed (Figure 4B). These results indicated that Naa10p positively modulates Dnmt1 activity in mouse ESCs. In contrast, the effect of Naa10p KO on the de novo DNA methyltransferase activities of Dnmt3a/3b seemed to be limited (Figure S4A). This result is consistent with our previous finding that hNaa10p does not associate with the full-length DNMT3a/3b (Lee et al., 2010). Thus, we conclude that Naa10p regulates DNA methylation mainly through Dnmt1.
Figure 4. Naa10p Loss Impairs Dnmt1 Activity and Binding to ICRs/DMRs.
(A and B) Naa10p KO reduces Dnmt1 activity in nuclear extracts of mouse ESCs. Nuclear extracts (A) or Dnmt1 immunoprecipitated (B) from two WT, two Naa10-KO, or Dnmt1-KO ESCs were incubated with hemi-methylated double-stranded DNA oligonucleotides and [3H]-S-adenosyl-L-methionine, followed by determination of radioactivity of the DNA oligonucleotides and western blotting. After subtracting the background signal of Dnmt1-KO cells, the relative Dnmt1 activity from each sample was normalized to the mean activity of WT replicates (A, bottom, and B, right). Data represent the mean ± SD from three independent experiments. Asterisks indicate statistical difference calculated by two-tailed t test: **p < 0.01. IP, immunoprecipitation.
(C) Naa10p KO impairs Dnmt1 binding to multiple ICRs/DMRs during S phase in mouse ESCs. ChIP-qPCR analyses of Dnmt1 occupancy at eight representative ICRs/DMRs in fluorescence-activated cell-sorted WT#1 and KO#1 ESCs are shown. Error bars represent SD of triplicate PCR reactions. Biological repeats using independent ESC clones WT#2 and KO#2 are shown in Figure S4C.
(D) Naa10p KO impairs Dnmt1 binding to multiple ICRs/DMRs in a mouse embryo. A similar experiment as in (C) was performed in mouse embryos. (C and D) Error bars represent SD determined from triplicate PCR reactions. Asterisks indicate a statistical difference calculated by two-tailed t test: *p < 0.05, **p < 0.01.
See also Figure S4.
We next explored the mechanism by which Naa10p maintains the global activity of Dnmt1. An electrophoretic mobility shift assay (EMSA) demonstrated that binding of Dnmt1 to the hemimethylated DNA oligo used in the activity assay was reduced in Naa10p-KO nuclear extracts (data not shown), suggesting that loss of Dnmt1 activity in the absence of Naa10p might be due to the decreased binding of Dnmt1 to its substrate. This observation was confirmed by chromatin immunoprecipitation (ChIP) analyses, which demonstrated that Naa10p KO greatly reduced the binding of Dnmt1 to ICRs/DMRs in S phase mouse embryonic stem cells (mESCs) (Figure 4C; Figures S4B and S4C). A similar effect was observed in Naa10p-KO embryos (Figure 4D). Notably, Naa10p depletion neither decreased Dnmt1 binding to Meg3-intergenic germline-derived (IG)-DMR (Figure 4C; Figures S4B and S4C) nor affected the binding of Zfp57 to these ICRs/DMRs (Figure S4D), supporting the specificity of Naa10p-mediated Dnmt1 recruitment. Furthermore, reduced binding of Dnmt1 to DNA was not limited to ICRs/DMRs but also occurred at non-imprinted loci (Figure S4E). Together, these results support that loss of Naa10p impairs global DNA methylation and genomic imprinting though decreased Dnmt1 recruitment to DNA.
Naa10p Co-localizes with Dnmt1 at ICRs/DMRs
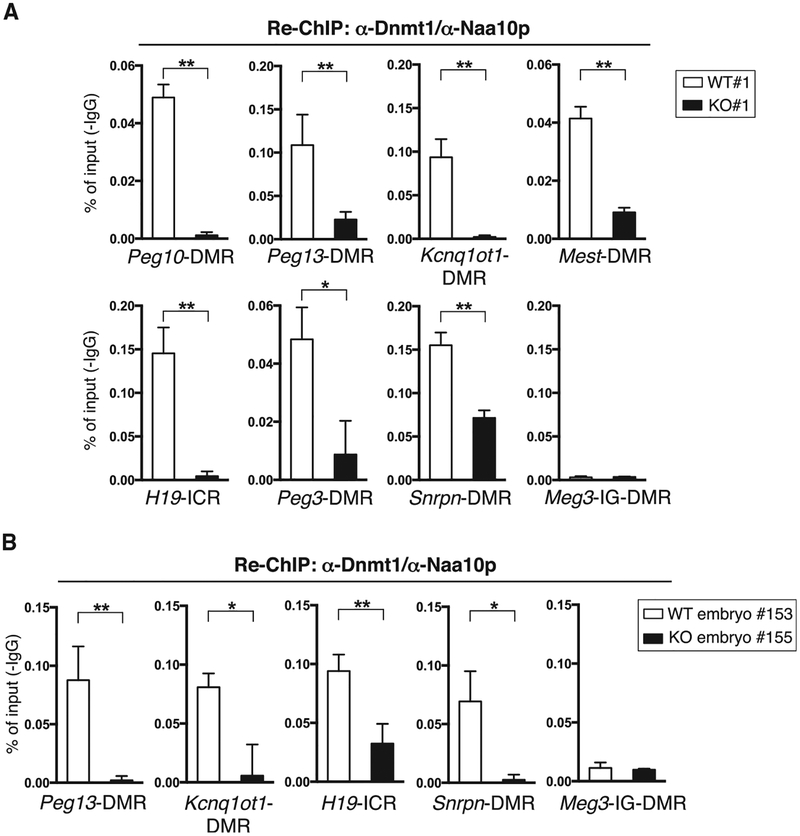
Given that hNaa10p associates with DNMT1 in vitro and in human lung cancer cells (Lee et al., 2010) and that ICR/DMR localization of Dnmt1 depends on Naa10p (Figures 4C and 4D; Figure S4C), we suspected that Naa10p and Dnmt1 might co-occupy ICRs/DMRs. To test this possibility, we performed a sequential ChIP analysis in which ChIP was initially carried out with a Dnmt1 antibody (Ab) (Figures 4C and 4D; Figure S4C), followed by ChIP with an Naa10p Ab. Indeed, Naa10p and Dnmt1 co-occupied ICRs/DMRs in both mouse ESC lines (Figure 5A; Figure S5) and an embryo (Figure 5B). The sequential ChIP signal was specific because very little binding was detected in the absence of Naa10p.
Figure 5. Naa10p and Dnmt1 Co-occupy ICRs/DMRs.
(A) Naa10p co-localizes with Dnmt1 at multiple ICRs/DMRs in WT mouse ESCs. ChIP with the Dnmt1 Ab from WT#1 and KO#1 ESC clones was carried out again with Naa10p Ab, followed by qPCR with primers against the ICRs/DMRs indicated. Biological repeats using independent ESC clones WT#2 and KO#2 are shown in Figure S5.
(B) Naa10p co-localizes with Dnmt1 at multiple ICRs/DMRs in a WT mouse embryo. ChIP with the Dnmt1 Ab in Figure 4D from embryos of WT (153) and Naa10-KO (155) was carried out again with Naa10p Ab, followed by qPCR with primers against the ICRs/DMRs indicated.
(A and B) Error bars represent SD determined from triplicate PCR reactions. Asterisks indicate a statistical difference calculated by two-tailed t test: *p < 0.05, **p < 0.01.
See also Figure S5.
Naa10p Selectively Binds to the Imprinted Allele at ICRs/DMRs
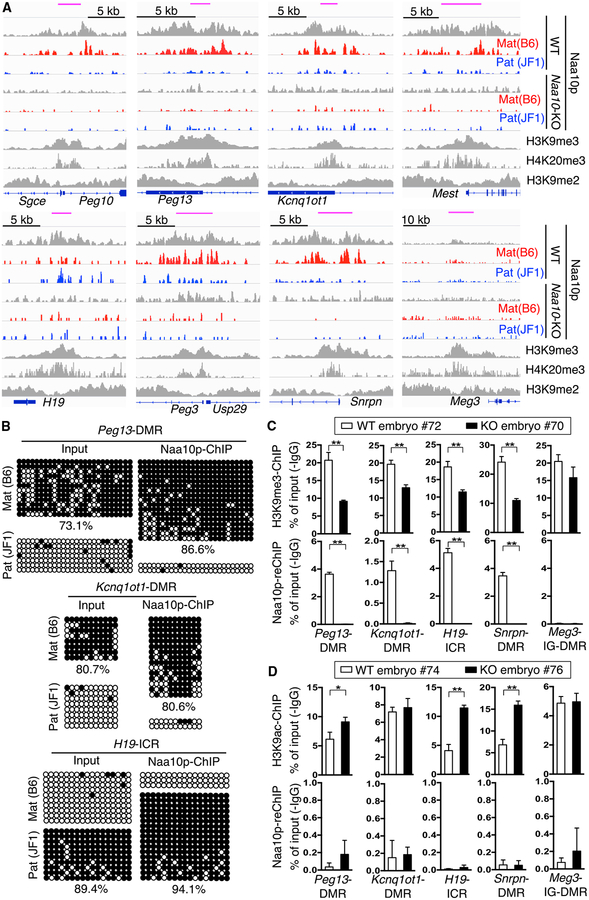
The observations that Naa10p KO reduces Dnmt1 binding to ICRs/DMRs and that Naa10p can be re-ChIPed with Dnmt1 on ICRs/DMRs raised the possibility that Naa10p might associate specifically with the imprinted allele to recruit Dnmt1 and/or stabilize its binding. To test this possibility, allele-specific ChIP-seq was performed in mESCs. The results demonstrated that Naa10p indeed selectively bound to the ICRs/DMRs of the imprinted allele of multiple imprinted genes except Meg3 (Figure 6A). Integrated analyses of the Naa10p ChIP-seq and the publicly available ChIP-seq data of H3K9me2 (Liu et al., 2015), H3K9me3 (Encode: ENCSR343RKY) and H4K20me3 (GEO: GSE26680) in mESCs indicated that Naa10p peaked together with H3K9me3 and H4K20me3 but not H3K9me2 at ICRs/DMRs (Figure 6A). ChIP-seq also revealed that Naa10p bound preferentially to distal intergenic regions (Figure S6A) and to both alleles at non-imprinted loci (Figure S6B). Analysis of the Naa10p ChIP-seq and RRBS data further demonstrated that Naa10p-bound regions were enriched for DNA methylation (Figure S6C). Consistently, ChIP-qPCR revealed that the ICRs/DMRs bound by Naa10p were enriched with SNPs that matched the imprinted allele (Figure S6D) with high levels of DNA methylation (Figure 6B). Furthermore, Naa10p co-existed with H3K9me3 (Figure 6C), but not H3 acetylated at lysine 9 (H3K9ac) (Figure 6D), at ICRs/DMRs in the WT but not the Naa10-KO embryo. These data indicate that Naa10p preferentially binds to the imprinted allele in mouse ESCs and embryos.
Figure 6. Naa10p Preferentially Binds to the Imprinted Allele at ICRs/DMRs.
(A) Naa10p binds to the imprinted allele and co-occupies with H3K9me3 and H4K20me3, but not H3K9me2, at ICRs/DMRs. A genome browser view of representative imprinted genes shows allele-specific Naa10p binding peaks in WT and Naa10-KO ESCs. The ChIP-seq density profiles shown were normalized to density per million total reads with 25-bp resolution. Blue boxes, genes; top pink bars, known ICRs/DMRs; red, maternal B6; blue, paternal JF1.
(B) ICRs/DMRs bound by Naa10p are enriched in DNA methylation. ICRs/DMRs ChIPed by Naa10p Ab from WT#1 ESCs were analyzed by bisulfite sequencing. Open and closed circles indicate non-methylated and methylated cytosines, respectively. The numbers at the bottom are the percentages of CpG methylation.
(C) Naa10p co-exists with H3K9me3 at multiple ICRs/DMRs. Sequential H3K9me3-Naa10p ChIP-qPCR assays in WT (72, Naa10X/Y) and Naa10-KO (70, Naa10−/Y) embryos were performed. Top, primary H3K9me3 ChIP. Bottom, secondary Naa10p ChIP.
(D) Naa10p does not co-localize with H3K9ac at ICRs/DMRs. Sequential H3K9ac-Naa10p ChIP-qPCR assays were performed with WT (74, Naa10X/Y) and Naa10-KO (76, Naa10−/Y) embryos. Top, primary H3K9ac ChIP. Bottom: secondary Naa10p ChIP.
(C and D) Data relative to input DNA are shown as the mean ± SD from triplicate PCR reactions. Asterisks indicate a statistical difference calculated by two-tailed t test: *p < 0.05, **p < 0.01.
See also Figure S6.
Ogden Syndrome-Causing Naa10p S37P Mutation Disrupts ICR-Binding of Naa10p and Dnmt1 In Vitro
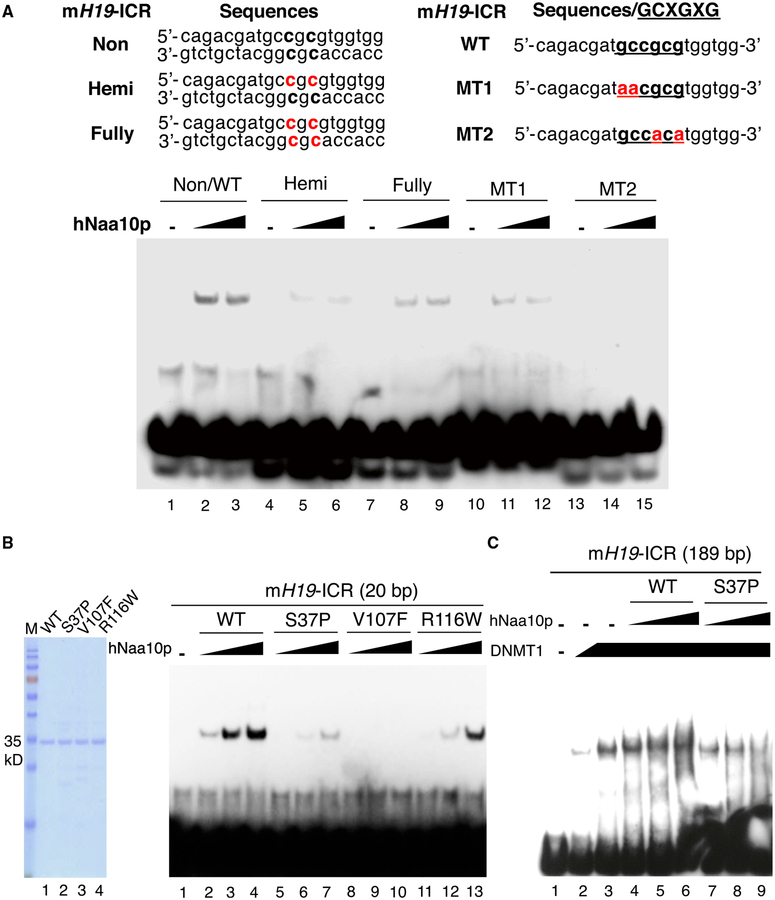
The fact that Naa10p harbors a putative homeobox DNA-binding domain at its C terminus (Whiteway and Szostak, 1985) prompted us to assess whether Naa10p could bind to ICRs/DMRs directly. EMSA analysis indicated that hNaa10p was able to bind specifically to a 189-bp mouse H19-ICR (Figure S7A). Moreover, Naa10p bound to a 20-bp region of the H19-ICR containing the GCXGXG motif, which shares consensus sequences with CTGT/GG/A identified by Naa10p ChIP-seq based on the STAMP (structural alignment of multiple proteins) analysis but failed to bind the fragment efficiently when the GCXGXG motif was mutated (Figure 7A, compare lanes 10–15 with lanes 1–3; Figure S7B). Because GCXGXG contains two potential CG sites, we further analyzed whether the methylation of these sites would affect Naa10p binding. Interestingly, hNaa10p had the greatest affinity for the non-methylated oligo (Figure 7A, compare lanes 1–3 with lanes 4–9).
Figure 7. WT Naa10p, but Not the Human Disease-Associated Mutants, Directly Binds to the Non-methylated GCXGXG Motif of H19-ICR and Facilitates DNMT1 Binding.
(A) Naa10p preferentially binds to the non-methylated GCXGXG motif at H19-ICR. The binding of recombinant hNaa10p to the biotin-labeled 20-mer oligo of mouse H19-ICR with or without methylated cytosines or mutations (indicated in red) was analyzed by EMSA. hNaa10p amount: 2 μg (lanes 2,5,8,11, and 14) and 4 μg (lanes 3, 6, 9, 12, and 15).
(B) Clinically relevant mutations disrupt the binding of Naa10p to the H19-ICR fragment. Left: Coomassie blue staining of recombinant WT and mutant hNaa10p used for the EMSA. Right: EMSA of WT and mutant hNaa10p binding to the biotin-labeled 20-mer oligo of mouse H19-ICR. hNaa10p amount: 1 μg (lanes 2,5, 8, and 11), 2 μg (lanes 3, 6, 9, and 12), and 4 μg (lanes 4, 7, 10, and 13).
(C) WT Naa10p, but not the Ogden syndrome-relevant S37P mutant, facilitates DNMT1 binding to H19-ICR.The biotin-labeled 189-bp ChIP region of mouse H19-ICR was incubated in the absence (lane 1) or presence of recombinant human DNMT1 (Hashimoto et al., 2012) (lane 2,0.5 μg, and lanes 3–9,1 μg) with WT human Naa10p (lanes 4–6) or its S37P mutant (lanes 7–9) for EMSA. hNaa10p amount: 0.5 μg (lanes 4 and 7), 1 μg (lanes 5 and 8), and 2 μg (lanes 6 and 9).
See also Figure S7.
Next we analyzed whether clinically relevant mutations could disrupt the DNA binding activity of Naa10p. The EMSA results indicated that the Ogden syndrome-associated mutation S37P, as well as another two clinically relevant mutations, V107F and R116W, markedly reduced the binding of Naa10p to the 20-bp fragment of mouse H19-ICR (Figure 7B) or nucleosomes reconstituted onto the 216-bp Widom-601 sequence (Juan et al., 1994), which contains four GCXGXG motifs (Figure S7C). Furthermore, WT Naa10p, but not the S37P mutant, could enhance the binding of purified DNMT1 to mouse H19-ICR (Figure 7C). Indeed, the S37P mutant disrupted DNMT1 binding (Figure 7C, compare lanes 7–9 with lane 3). Together, these data suggest that failure of the Naa10p mutant in binding to ICRs/DMRs affects DNMT1 recruitment.
DISCUSSION
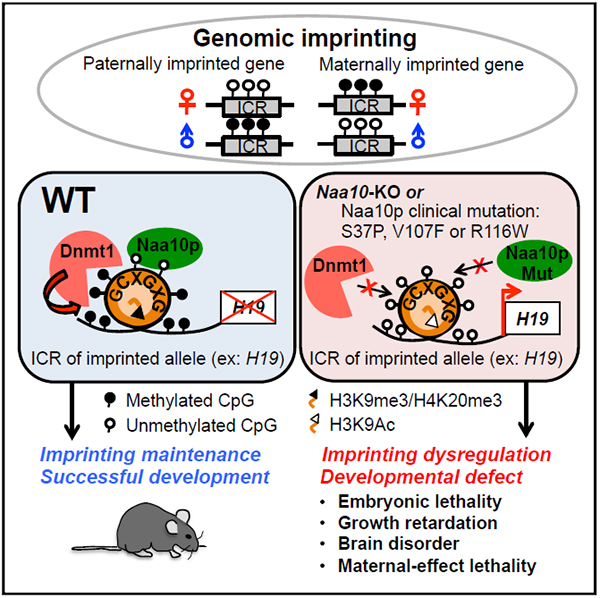
This study not only reveals an unexpected function for Naa10p, a NAT primarily known to acetylate nascent peptides from ribosomes, in maintaining global DNA methylation and marking the imprinted allele for genomic imprinting maintenance but also suggests that defects in DNA methylation and genomic imprinting may contribute to Naa10p-associated human diseases.
Naa10p Is a DNA Binding Protein that Marks the Imprinted Allele
The finding that Naa10p preferentially recognizes non-methylated GCXGXG in vitro (Figure 7A; Lee et al., 2010) and selectively binds to methylated ICRs/DMRs at the imprinted allele in vivo (Figure 6B) is particularly intriguing. Given that GCXGXG is highly enriched in ICRs/DMRs (Figure S7D), it is likely that Naa10p first binds to unmethylated GCXGXG in the ICRs/DMRs of the imprinted allele and then recruits Dnmt1 at S phase for methylation. Currently, it is unclear how Naa10p selectively binds to the imprinted allele. The fact that Naa10p peaked together with H3K9me3 and H4K20me3, but not H3K9me2, at ICRs/DMRs in mESCs (Figure 6A) and that the H3K9me3 associated with ICRs/DMRs was significantly decreased in the Naa10-KO embryo (Figure 6C) suggest a role for the imprinted allele-specific H4K20me3 in the recruitment of Naa10p, which remains to be shown. In addition, DNA methylation was reduced in both Naa10p-bound and -unbound regions when Naa10p was knocked out (data not shown), suggesting that, although Naa10p exhibits sequence-specific effects, it does have a broader role in DNA methylation.
Naa10p and Zfp57
Prior to our study, Zfp57 was the only known DNA binding protein confirmed by allele-specific SNPs to selectively associate with the imprinted allele for genomic imprinting maintenance (Strogantsev et al., 2015). By analyzing the Zfp57 ChIP-seq data of mESCs (Quenneville et al., 2011; Strogantsev et al., 2015), we found that Zfp57 could bind to all ICRs/DMRs occupied by Naa10p revealed in our study (Figure S4D). Nevertheless, Zfp57 binding to ICRs/DMRs of Peg13, Kcnq1ot1, H19, and Snrpn was not altered in Naa10-KO ESCs despite disruption of imprinting under our experimental conditions (Figure S4D). Thus, it is unlikely that disruption of genomic imprinting in Naa10p-KO ESCs was due to a loss of Zfp57 function. Instead, the results suggest that, in the absence of Naa10p, binding of Zfp57 to ICRs/DMRs is not sufficient to achieve genomic imprinting. We also analyzed the list of dysregulated imprinted genes in a Zfp57-KO mESC clone (Quenneville et al., 2011) and two Trim28-KO mESC lines (Cheng et al., 2014) and found that only eight targets are commonly regulated by Naa10p and Zfp57. Interestingly, Naa10p and Trim28 appear to share more imprinted gene targets (Figure S2E).
Naa10p Is Not Likely to Acetylate Dnmt1
As an acetyltransferase, Naa10p may regulate Dnmt1 activity and its binding to DNA by acetylating Dnmt1 at its N terminus or internal lysines. However, this is unlikely to be the case. We have demonstrated previously that the acetyltransferase activity is dispensable for Naa10p to stimulate DNMT1 activity (Lee et al., 2010). Moreover, the N-terminal amino acid sequence of Dnmt1 does not match the consensus acetylation sequence of Naa10p (Arnesen et al., 2009; Van Damme et al., 2011). Consistently, DNMT1 is not one of the human Naa10p substrates identified by proteomic approaches (Arnesen et al., 2009; Van Damme et al., 2011). Finally, purified (and active) Naa10p failed to acetylate the N-terminal amino acid nor internal lysine residues of a Dnmt1 peptide in vitro (data not shown), which is consistent with a previous study showing that Naa10p does not acetylate internal lysine (Magin et al., 2016). Together, these data suggest that Naa10p is not likely to regulate DNA methylation by acetylating Dnmt1.
The Role of Naa10p in Development
Although Naa10p-KO-mediated growth retardation is consistent with reports of other Naa10p-deficient organisms (Chen et al., 2014; Ree et al., 2015; Wang et al., 2010; Whiteway and Szostak, 1985; Yoon et al., 2014), our data indicate that Naa10p-KO phenotypes are not fully penetrant (Figure 1). This could be due to general effects of imprinting disorders similar to those reported in Zfp57 KO (Li et al., 2008), Trim28 KO (Messerschmidt et al., 2012), oocyte Dnmt1 KO (Howell et al., 2001), and Tet1 KO (Yamaguchi et al., 2013). In addition, some of the Naa10p-KO mice could still develop to term. This is likely because DNA methylation is not completely lost in Naa10-KO mice. The remaining 20%–50% of DNA methylation at different regions in different Naa10p-KO mice may explain the stochastic nature of the phenotypic effect.
Several Naa10p-regulated imprinted genes may partly explain the phenotypes such as growth retardation and neuronal defects shared by Naa10-KO mice and patients with Naa10p mutations. For example, Naa10p KO results in upregulation of H19 and Peg3. H19-KO embryos are abnormally large (Constância et al., 2002; Leighton et al., 1995), suggesting that elevated H19 expression inhibits growth. Peg3 overexpression can cause neuronal defects by inducing apoptosis of neurons through a p53-dependent mechanism (Johnson et al., 2002). Imprinted genes such as Igf2, Ascl2, and Grb10 that are downregulated by Naa10p deficiency may also contribute to the phenotypes. Consistent with our study, Igf2-null embryos are smaller (Constância et al., 2002), and loss of Igf2 reduces proliferation of neuron progenitor cells in the adult hippocampus (Bracko et al., 2012). Ascl2-KO embryos die from placental failure on embryonic day 10.5. In mutant placentae lacking Ascl2 expression, spongiotrophoblast cells and their precursors are absent, and the chorionic ectoderm is reduced (Guillemot et al., 1994), which is similar to what we observed in Naa10-KO placentae. Furthermore, a low level of Grb10 correlates with growth and neuronal defects in mice (Plasschaert and Bartolomei, 2015) and a small placenta and low birth weight in humans (Mukhopadhyay et al., 2015). Besides these imprinted genes, other non-imprinted genes or protein acetylation dysregulated by Naa10p KO may also contribute to some of the observed phenotypes.
In summary, our study not only reveals a regulator of genomic imprinting but also links Naa10p mutation-associated syndromes to defects in DNA methylation and genomic imprinting.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Li-Jung Juan (ljjuan@gate.sinica.edu.tw).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All animal studies were performed according to procedures approved by Institutional Animal Care and Use Committee of Academia Sinica. Naa10-KO mice were generated as follows. Three C57BL/6 blastocysts were each injected with one Naa10-engineered (see Figure S1A) R1 ESC (129 Sv/Ev) clone and surgically implanted into pseudopregnant female mice (Swiss Webster) at 2.5 dpc to generate chimeras according to standard procedures. The chimeric males were crossed with C57BL/6 females. Southern blot and PCR were performed to screen for litters with agouti coat color germline transmission carrying floxed Naa10 allele (Naa10X/flox). Naa10X/flox female mice were next crossed with the EIIa-Cre transgenic male mice (Lakso et al., 1996) to generate mice carrying deleted allele at one of two Naa10 alleles. Since Naa10 is located on X chromosome, the Naa10-KO male mice were maintained by crossing Naa10-heterozygous females (Naa10−/X) with WT C57BL/6 males (Naa10X/Y, Mus musculus domesticus) and used for experiments after at least six generations of backcross with C57BL/6 mice. JF1 strain (Mus musculus molossinus) was obtained from National Institute of Genetics (NIG), Japan. Single nucleotide polymorphisms between C57BL/6 and JF1 strains were identified using NIG mouse genome database (http://molossinus.lab.nig.ac.jp/msmdb/index.jsp).
Mouse ESC Culture
The C57BL/6-JF1 hybrid-ESCs were derived from embryos of Naa10−/X C57BL/6 female mice crossed with WT JF1 male mice according to a previous report (Bryja et al., 2006) with the following modifications. Naa10-KO and littermate WT ESCs were cultured in N2B27 supplemented with the Mek inhibitor PD325901 (1 μM, Stemgen), Gsk3 inhibitor Chir99021 (3 μM, Stemgen), and leukemia inhibitory factor (LIF) (1×103 U/ml, ESGRO, Millipore) in the presence of irradiated mouse embryonic fibroblast (MEF) feeder cells. Dnmt1 -KO and J1 WT ESCs, gifts from Dr. En Li (Novartis Institutes for BioMedical Research, China) (Lei et al., 1996), and Dnmt-TKO ESC from RIKEN (Tsumuraet al., 2006) were cultured in standard DMEM supplemented with 15% fetal bovine serum and LIF (1×103 U/ml) in the presence of irradiated MEF feeders. All experiments with ESCs were performed in cells with five passages free of MEFs.
METHOD DETAILS
Histology Analysis
Whole-mount embryos at 13.5 dpc and placentae were placed in phosphate-buffered saline, examined, and photographed with a dissecting microscope (Olympus). For placental histology, placental tissues were fixed in cold paraformaldehyde (4%), dehydrated in sucrose (30%), embedded in OCT-compound (TissueTek), and sectioned into 8 μm in thickness, followed by hematoxylin and eosin staining. For brain histology, whole mouse head was obtained, fixed in cold paraformaldehyde (4%), decalcified in EDTA (10%), embedded in OCT-compound (TissueTek), coronally sectioned into 8 μm in thickness, followed by hematoxylin and eosin staining.
RNA-Seq, Data Analysis, and RT-PCR
Total RNAs purified from mouse embryonic stem cells using RNeasy Plus Mini Kit (QIAGEN) were treated with DNase I to remove residual genomic DNA, enriched by oligo-dT magnetic beads, and sonicated (Bioruptor, Diagenode) into roughly 200 bp fragments. cDNAs were synthesized with random hexamer-primers, purified by silicon-coated magnetic beads, blunted in the ends, added with a single adenine at 3′ end, ligated with sequencing adaptors and PCR-amplified. The quality and quantity of the resulting library were determined by Agilent 2100 Bioanaylzer and ABI StepOnePlus Real-Time PCR System, respectively. The library products were sequenced by Illumina HiSeq 2000. RNA-seq data analysis was performed with Tophat and Cufflinks using UCSC mm9 annotation (Trapnell et al., 2012). Functional annotation of the dysregulated genes was performed with DAVID (https://david.ncifcrf.gov/) (Huang et al., 2009). Allele specific analysis of RNA-seq was performed with Allele-specific Alignment Pipeline (ASAP) (https://www.bioinformatics.babraham.ac.uk/projects/ASAP/). The reads were mapped to B6 and JF1 genomes and only reads specific to either genome were retained as maternal B6 or paternal JF1 reads. The RNA-seq density profiles were normalized to the density per million total reads with a resolution of 25 bp using IGVtools. For RT-PCR, total RNAs were collected using RNeasy Plus Mini Kit (QIAGEN) and converted into cDNA with SuperScript III reverse transcriptase (Invitrogen) and poly-dT(12–18) oligo. Real-time PCR was performed using the LightCycler 480 (Roche) according to the manufacturer’s instructions. All real-time PCR was carried out in triplicate.
Allelic Gene Expression Analysis
Total RNAs of embryos at 10.5 dpc from Naa10-heterozygous female mice crossed with JF1 male mice were obtained using RNeasy Plus Mini Kit (QIAGEN) according to manufacturer’s instruction, followed by cDNA synthesis using oligo-dT primer and SuperScript III reverse transcriptase (Invitrogen). RT-PCR products of H19 (360 bp) and Kcnq1ot1 (418 bp) (for primers see Table S5) were digested with BglI and PvuII which cut at JF1- or B6-specific SNP, respectively, and separated by agarose gel eletrophoresis.
Bisulfite Sequencing
Conversion of non-methylated cytosine to uracil by Na-bisulfite treatment was carried out with embryonic genomic DNA using EpiTect Fast Bisulfite kit (QIAGEN) according to manufacturer’s instruction. PCR products (for primers see Table S7) were cloned into pCR2.1-Topo vector (Invitrogen) and the resulting clones were randomly picked up for sequencing.
Reduce Representation Bisulfite Sequencing (RRBS)
The RRBS libraries were generated from MspI-digested genomic DNA and sequenced on an Illumina Hiseq 2500 as previously described (Gu et al., 2011), with some modifications. All obtained reads were trimmed and mapped against the mouse genome (mm9 build) with Bismark v0.7.12, and the methylation call for every single CpG was extracted by bismark methylation extractor script. The CpGs covered by at least 8 reads were analyzed.
5mC Dot Blot Assay
Genomic DNA was isolated using a genomic DNA extraction kit (QIAGEN), serially diluted and denatured in 0.4 M NaOH, 10 mM EDTA at 95°Cfor 10 min. Denatured DNA samples were spotted onto Zeta probe blotting membrane (Bio-Rad) with a Bio-Dot microfiltration apparatus (Bio-Rad) according to manufacturer’s instructions. Each membrane was then washed with 2× saline-sodium citrate buffer, air-dried, crosslinked with UV at 5 mJ/cm2, and blocked with 5% skim milk in Tris-buffered saline containing 0.1% (v/v) Tween 20 (TBS-T), followed by incubation with the 5mC Ab (2 μg/ml, Eurogentec) for 1 hr at room temperature. Each membrane was then washed with TBS-T and probed with horseradish peroxidase-conjugated secondary Ab (goat anti-mouse IgG, 0.2 μg/ml, Santa Cruz Biotechnology) and detected with ECL reagent (Thermo Fisher Scientific). To measure the relative amount of each sample, the same blot was stained with 0.04% methylene blue in 0.3 M sodium acetate (pH 5.2).
Preparation of Nuclear Extract
ESCs were scraped from 10 cm plate in cold PBS and centrifuged at 3000 rpm, 4°C for 3 min. The cell pellets were resuspended in 500 μL buffer A (10 mM HEPES pH 8.0,1.5 mM MgCl2,10 mM KCl and 0.5% NP40 with protease inhibitors) and incubated on ice for 15 min. After centrifugation at 3000 rpm, 4°C for 5 min, the pellets were washed by buffer A twice and resuspended in 200 μL buffer C (20 mM HEPES pH 8.0, 25% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM PMSF and 0.5 mM DTT with protease inhibitors) and further incubated on ice for 30 min. The samples were then centrifuged at 13000 rpm, 4°C for 10 min and the supernatant was collected as nuclear extract. The extracts were further dialyzed against buffer containing 20 mM HEPES pH 8.0, 20% glycerol, 100 mM KCl, 0.2 mM EDTA, 0.5 mM PMSF and 0.5 mM DTT at 4°C. The extracts were stored at 80°C for further experiments.
Western Blot Analysis
Western blotting was performed according to the standard procedures with modifications. Cells were lysed in NP-40 buffer (50 mM Tris-HCl, 120 mM NaCl, and 1% NP-40) containing protease inhibitor mixture (Sigma-Aldrich).
DNA Methyltransferase Activity Assay
Nuclear extract (10 μg) of ESCs or 5 μL of beads containing immunoprecipitated Dnmt1 was used for western blotting or the Dnmt1 activity assay by mixing with 1 μg non-methylated or hemi-methylated double stranded DNA oligo (5′-GATCCGACGAC GACGCGCGCGCGACGACGAGATC, 34 bp, underlined Cs are hemi-methylated, PURIGO Biotech) (Yokochi and Robertson, 2002) with 3 μCi [methyl-3H]-S-adenosyl-L-methionine (PerkinElmer) in 20 μL of buffer D (20 mM HEPES pH 8.0, 100 mM KCl, 0.2 mM EDTA, 0.5 mM PMSF and 20% glycerol) at 37°C for 2 hr. Then, 40 μL Stop solution (1% SDS, 2 mM EDTA, 5% butanol, 0.25 mg/ml salmon sperm DNA, 125 mM NaCl and 1 mg/ml proteinase K) was added to each sample and incubated for 30 min at 37°C. DNA was precipitated with ethanol on ice for 30 min, then resuspended in 30 μL of 0.3 N NaOH and spotted onto a GF/C filter (Whatman). Each membrane was washed with 5% trichloroacetic acid and then 70% ethanol. The radioactivity on dried membranes was measured with a Beckman liquid scintillation counter. All reactions were carried out in triplicate.
Dnmt1 Immunoprecipitation (IP) for Dnmt1 Activity Assay
The IP-Dnmt1 activity assay was modified from a previous study (Zhang et al., 2015). 400 μg of ESC nuclear extracts in 200 μL of IP buffer [150 mM NaCl, 25 mM Tris (pH 7.4), 1 mM EDTA, 1% NP-40, 5% glycerol and protease inhibitors] were incubated with 2 μg of Dnmt1 antibody (rabbit polyclonal, ab87654) with rotation at 4°C overnight, followed by addition of 20 μL of protein G magnetic beads (Thermo Fisher Scientific) for additional 4 hr with rotation at 4°C. The beads were then collected, washed for three times with IP buffer and resuspended in 20 μL of IP buffer. 5 μL of beads containing antibody/Dnmt1 complexes were used for western blotting analysis or Dnmt1 activity assay.
Cell Sorting by FACS for ChIP
ESCs grown to 80% confluent were harvested, fixed with 1% formaldehyde for 10 min and quenched with 0.125 M glycine for 5 min. Fixed ESCs were then treated with RNase A (1 mg/ml) for 30 min followed by staining of propidium iodide (PI, 10 μg/ml). Sorting G1/0, S, and G2/M-phased ESCs was performed with FACS Aria II (BD Bioscience) according to cell size and DNA contents determined by forward scatter (FSC) and PI fluorescence, respectively. Ten million G1/0, S or G2/M-ESCs were used for ChIP.
Chromatin Immunoprecipitation (ChIP) and Sequential ChIP
Chromatin immunoprecipitation and sequential ChIP were carried out using CHIP-IT high sensitivity and Re-ChIP-IT, respectively, from ActiveMotif according to the manufacturer’s instructions. The immunoprecipitated DNA was quantified by qPCR using LightCycler 480II (Roche). For PCR primers used please see Table S6.
ChIP-Seq Sample Preparation
For Naa10p ChIP, mESCs were fixed with 1% formaldehyde for 10 min, followed by 0.125 M glycine quenching for 5 min. Cells were lysed in buffer containing 1% SDS, 10 mM EDTA and 50 mM Tris-HCl (pH 8.0) and the DNA was fragmented by sonication to approximately 150–300 bp (Bioruptor, Diagenode). Immunoprecipitation was performed with 10 μg of rabbit polyclonal Naa10p for overnight at 4°C. Antibody-bound samples were isolated by protein G agarose beads (Active Motif), washed, eluted, and reverse cross-linked. ChIPed DNA was extracted by phenol/chloroform, ethanol precipitated and quantified by Qubit assay (Invitrogen).
ChIPed DNA Preparation for NextSeq 500 Sequencing
The ChIPed DNA library for NextSeq 500 sequencing was constructed using the NEBNext DNA library Prep Master Mix Set for Illumina (New England BioLabs) according to the manufacturer’s instruction. Briefly, the ChIPed DNA was blunt ended and a dA tail was added. The DNA with dA overhangs was ligated with multiplex oligos for illumina with individual index sequences (New England BioLabs). Adaptor-ligated DNA was amplified by PCR for 12 cycles, followed by size selection using magnetic beads (KAPA Pure Beads, KAPA Biosystem). The purified DNA was quantified by Qubit assay (Invitrogen) and qualified by Agilent Bioanalyzer. Sequencing on a NextSeq 500 instrument for 75 bp single end was carried out by the BGI (Shenzhen).
ChIP-Seq Data Analysis
All sequencing reads (75 bp in length) were mapped to NCBI build 37 (mm9) of the mouse genome using the software Bowtie. Mapped reads were subject to the MACS program and bound regions (peaks) were determined using sequencing reads from Naa10-KO ESC as negative controls. When multiple reads mapped to the same genomic position, a maximum of two reads were retained. The statistical cutoff used for peak calling was p < 10−5 and > 5-fold enrichment over the Naa10-KO control. Allele-specific analysis of ChIP-seq data were performed with Allele-specific Alignment Pipeline (ASAP) (https://www.bioinformatics.babraham.ac.uk/projects/ASAP/). The reads were mapped to B6 and JF1 genomes and only reads specific to either genome were retained as maternal B6 or paternal JF1 reads. The ChIP-seq density profiles were normalized to the density per million total reads with a resolution of 25 bp using IGV tools. ChIP-seq read counts for each ChIP-seq experiment were visualized in the IGV genome browser. ChIP-seq data for H3K9me2, H3K9me3 and H4K20me3 were obtained from a previous publication (Liu et al., 2015), Encyclopedia of DNA Elements (Encode: ENCSR343RKY) and Gene Expression Omnibus (GEO: GSE26680), respectively. ChIP-seq data for histone modifications were reanalyzed with Bowtie (http://bowtie-bio.sourceforge.net/index.shtml) and MACS (https://github.com/taoliu/MACS) using parameters identical to Naa10p ChIP-seq. Genomic occupancy of Naa10p binding sites were analyzed with criteria, distal promoter, 1–3 kb upstream of transcription start site (TSS), proximal promoter, < 1 kb upstream of TSS, immediate downstream, < 1 kb downstream of transcription end site (TES), proximal downstream, 1–3 kb downstream of TES, using PAVIS (https://manticore.niehs.nih.gov/pavis2/). The genomic distribution of GCXGXG motif was performed via extracting GCXGXG motif from ICRs/DMRs and 10 kb-flanked regions followed by intersection using Bedtools. Numbers of motif were divided by genomic length (per kb) for calculating motif frequency. Motif for Naa10p-binding sites was analyzed with PeakAnalyzer for splitting peaks with separation float, 0.5, and minimum height, 5, followed by running MEME-ChIP (http://meme-suite.org/tools/meme-chip).
Protein Purification and Electrophoretic Mobility Shift Assay (EMSA)
Escherichia coli-expressed His-tagged hNaa10p and its mutants were purified using Ni-Sepharose (Bioman) according to the supplier’s instructions, dialyzed against buffer containing 20 mM HEPES, pH 8.0, 10% glycerol, 100 mM KCl, 0.2 mM EDTA and 0.5 mM DTT, concentrated to 10 mg/ml by VIVASPIN (Sartorius) and stored at −80°C. DNMT1 was purified according to a previous study (Hashimoto et al., 2012). EMSA was performed by incubating bovine serum albumin (control) or the indicated protein with the biotin-labeled probes (PURIGO Biotech) in 20 μL reaction buffer containing 5× Q buffer (20 mM HEPES, pH 8.0, 8% glycerol, 0.5 mM EDTA, 0.1M KCl, 0.5 mM DTT and 0.5 mM PMSF) and 150 ng dIdC. Reactions were incubated at 30°C for 30 min and separated on a 6% native gel (for nucleosomal templates and Dnmt1 binding) or 10% native gel in 0.5× Tris-borate-EDTA buffer containing 5% glycerol at 4°C and further analyzed with streptavidin-coupled horseradish peroxidase (Jackson ImmunoResearch).
Nucleosomal Templates for EMSA
216 bp DNA fragment containing 601 nucleosome position sequences was amplified from pGEM-3Z-601 (a gift provided by Dr. Jerry Workman) by PCR. The sequences of the primers are: 5′-biotin-CGACTGGCACCGGCAAGGT (forward) and 5′-TCCCTTATGTGATG GACCCTA (reverse). The amplified DNA probes were purified by gel extraction kit (QIAGEN). The oligonucleosomes from HeLa cells and nucleosome reconstitution by octamer transfer were prepared as described (Juan et al., 1994) with modifications, analyzed by agarose gel eletrophoresis and stored at −80°C for further experiments. For nucleosome reconstitution, the biotin-labeled DNA probe was incubated with HeLa oligonucleosomes in buffer D (10 mM Tris-Cl pH 7.4, 1 mM EDTA, 5 mM DTT and 0.5 mM PMSF) containing 1 M NaCl. NaCl concentration was further diluted to 0.8 M, 0.6 M, 0.4 M and 0.2 M by buffer D. Finally, the mixture was diluted to 0.1 M by buffer containing 10 mM Tris-Cl pH 7.4, 1 mM EDTA, 5 mM DTT, 0.5 mM PMSF, 0.1% NP-40, 20% glycerol and 200 μg/mL of BSA. The samples were incubated at 37°C for 15 min between each dilution. Freshly reconstituted nucleosomes were subject to EMSA assays.
Primers
Primers for genotyping, quantitative PCR, chromatin immunoprecepitation, bisulfite sequencing, the DNA probes and site-directed mutagenesis for EMSA are listed in Tables S5–S7.
Antibodies
Naa10p Ab for ChIP and western in Figures 4, 5, 6, and S3–S7 is a rabbit polyclonal Ab antigen affinity-purified from serum of rabbits immunized with E. coli-purified mouse Naa10p (235 aa) by GenScript, USA. Abs against mouse Dnmt1 for ChIP and western in Figures 4, 5, and S3–S5 (Wang et al., 2009), Dnmt3a (Ge et al., 2004), Dnmt3b (Ge et al., 2004) and Tet1 (Ito et al., 2010) were described previously. Other Abs used are against 5mC (BI-MECY-0500, Eurogentec), Dnmt1 for IP in Figure 4B (ab87654, Abcam), Naa10p for western in figure S1A (sc-33820, Santa Cruz), H3K9me3 (ab8898, Abcam), H3K9ac (07–352, Millipore), β-tubulin (MAB3408, Millipore), Tet2 (mAb-179–050, Diagenoda), Zfp57 (ab45341, Abcam), Trim28 (ab10483, Abcam), Lamin B (sc-6217, Santa Cruz), Uhrf1 (#12387, Cell Signaling) and α-tubulin (T5168, Sigma). IgG is from Millipore (12–370).
QUANTIFICATION AND STATISTICAL ANALYSIS
Embryo size (mm2) in Figure 1B and Figure S1C was calculated by multiplying the length of long axis from head to tail by the length of short axis from abdomen to dorsum using embryo photos. All statistical analyses were performed with Prism Graphpad software and indicated in figure legends.
DATA AND SOFTWARE AVAILABILITY
The accession numbers for the RNA-seq, RRBS, and ChIP-seq reported in this paper are GEO: GSE89055, GSE83206, and GSE102224, respectively. Raw data of western, dot blots and EMSA are also deposited in Mendeley data under link http://dx.doi.org/10.17632/k7xnhjhhsr.1.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-Naa10p | This paper | N/A |
| Rabbit polyclonal anti-hNaa10p | Santa Cruz | sc-33820; RRID: AB_2060656 |
| Rabbit polyclonal anti-Dnmt1 | Abcam | ab87654; RRID: AB_2041077 |
| Rabbit polyclonal anti-Dnmt1 | Wang et al., 2009 | N/A |
| Rabbit polyclonal anti-Dnmt3a | Ge et al., 2004 | N/A |
| Rabbit polyclonal anti-Dnmt3b | Ge et al., 2004 | N/A |
| Rabbit polyclonal anti-Tet1 | Ito et al., 2010 | N/A |
| Mouse monoclonal anti-Tet2 | Diagenode | MAB-179–050 |
| Rabbit monoclonal anti-Uhrf1 | Cell Signaling | 12387 |
| Rabbit polyclonal anti-Zfp57 | Abcam | ab45341; RRID: AB_946192 |
| Rabbit polyclonal anti-KAP1/Trim28 | Abcam | ab10483; RRID: AB_297222 |
| Mouse monoclonal anti-5-methylcytosine | Eurogentec | BI-MECY-0500 |
| Rabbit polyclonal anti-Histone H3K9me3 | Abcam | ab8898; RRID: AB_306848 |
| Rabbit polyclonal anti-Histone H3K9ac | Millipore | 07–352; RRID: AB_310544 |
| Mouse monoclonal anti-α-Tubulin | Sigma-Aldrich | T5168; RRID: AB_477579 |
| Mouse monoclonal anti-β-Tubulin | Millipore | MAB3408; RRID: AB_94650 |
| Goat polyclonal anti-Lamin B | Santa Cruz | sc-6217; RRID: AB_648158 |
| Rabbit polyclonal IgG | Millipore | 12–370; RRID: AB_145841 |
| Streptavidin HRP-conjugated | Jackson ImmunoResearch | 016-030-084; RRID: AB_2337238 |
| Bacterial and Virus Strains | ||
| ECOS 21 Competent Cells [BL21(DE3)] | Yeastern Biotech | FYE207–40VL |
| Economics ECOS 101 [DH5a] | Yeastern Biotech | FYE607–80VL |
| Chemicals, Peptides, and Recombinant Proteins | ||
| MEK inhibitor (PD0325901) | Stemgent | 04–0006 |
| Gsk3 inhibitor (Chir99021) | Stemgent | 04–0004 |
| ESGRO Leukemia Inhibitory Factor (LIF) | Millipore | ESG1106 |
| SuperScript III reverse transcriptase | Invitrogen | 18080093 |
| LightCycler 480 SYBR Green I Master | Roche | 04707516001 |
| SIGMAF/ASr Protease Inhibitor Cocktail Tablets | Sigma-Aldrich | S8830 |
| Protease Inhibitor Cocktail | Sigma-Aldrich | P8340 |
| [3H]-Acetyl Coenzyme A | PerkinElmer | NET290050UC |
| Protein G magnetic beads | Thermo Fisher Scientific | 88847 |
| Protein G agarose beads | Active Motif | 53039 |
| Ni-Sepharose | Bioman Scientific | PBP001.100 |
| VIVASPIN | Satorius | VS2002 |
| His-hNaa10p and its mutants | This paper | N/A |
| Full-length DNMT1 | Hashimoto et al., 2012 | N/A |
| Critical Commercial Assays | ||
| Genomic DNA Extraction Kit | QIAGEN | 51304 |
| RNeasy Plus Mini Kit | QIAGEN | 74136 |
| EpiTect Fast Bisulfite Kit | QIAGEN | 59826 |
| CHIP-IT High Sensitivity | Active Motif | 53040 |
| Re-ChIP-IT | Active Motif | 53016 |
| Qubit assay | Invitrogen | Q32851 |
| NEBNext DNA library Prep Master Mix Set for Illumina | New England BioLabs | E6040L |
| NEBNext Multiplex Oligos for illumina | New England BioLabs | E7335L |
| KAPA Pure Beads | Kapa Biosystems | KK8000 |
| Gel Extraction Kit | QIAGEN | 28706 |
| Chemiluminescent Nucleic Acid Detection Module Kit | Thermo Fisher Scientific | 89880 |
| Deposited Data | ||
| Raw and processed data: WT and Naa10-KO RNA-seq | This paper | GEO: GSE89055 |
| Raw and processed data: WT and Naa10-KO RRBS | This paper | GEO: GSE83206 |
| Raw and processed data: WT and Naa10-KO Naa 10p ChIP-seq | This paper | GEO: GSE102224 |
| Western, dot blots, and EMSA | This paper; Mendeley Data | http://dx.doi.org/10.17632/k7xnhjhhsr.1 |
| C57BL/6-JF1 SNPs | National Institute of Genetics Mouse Genome Database | http://molossinus.lab.nig.ac.jp/msmdb/index.jsp |
| Mouse Reference Genome NCBI build 37 (mm9) | Genome Reference Consortium | https://www.ncbi.nlm.nih.gov/grc/mouse |
| ChIP-seq data for H3K9me2 | Liu et al., 2015 | GEO: GSE54412 |
| ChIP-seq data for H3K9me3 | Bing Ren Lab at UCSD | Encode: ENCSR343RKY |
| ChIP-seq data for H4K20me3 | Richard A. Young Lab at Whitehead Institute for Biomedical Research | GEO: GSE26680 |
| Experimental Models: Cell Lines | ||
| C57BL/6-JF1 hybrid-ESCs | This paper | N/A |
| Dnmt1-KO mouse ESCs | Lei et al., 1996 | N/A |
| J1 WT mouse ESCs | Lei et al., 1996 | N/A |
| Dnmt-TKO mouse ESCs | Tsumura et al., 2006 | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6JNarl | National Laboratory Animal Center, Taiwan | RMRC11005 |
| Mouse: JF1/Ms strain | National Institute of Genetics, Japan | N/A |
| Mouse: Naa10-KO | This paper | N/A |
| Oligonucleotides | ||
| Primers for genotyping, quantitative PCR, chromatin immunoprecepitation, bisulfite sequencing, EMSA DNA probes and site-directed mutagenesis, see Tables S5–S7 | This paper; Sharif et al., 2007; Di Giacomo et al., 2014 | N/A |
| Primer: Widom-601 (216 bp) Biotin-5’-CGACTGGCACCGGCAAGGT (forward) and 5’-TCCCTTATGTGATGGACCCTA-3’ (reverse) | Juan et al., 1994 | N/A |
| WT H19-ICR (20 bp) Biotin-5’-cagacgatgccgcgtggtgg | This paper | N/A |
| WT H19-ICR-MT1 Biotin-5’-cagacgataacgcgtggtgg | This paper | N/A |
| WT H19-ICR-MT2 Biotin-5’-cagacgatgccacatggtgg | This paper | N/A |
| WT H19-ICR-MT3 Biotin-5’-gacacgatgccgcgtggtgg | This paper | N/A |
| Recombinant DNA | ||
| pET-11d | Novagen | 69439–3CN |
| pGEM-3Z-601 | Dr. Jerry Workman, Stowers Institute for Medical Research | N/A |
| pCR2.1-Topo vector | Invitrogen | K4500 |
| Software and Algorithms | ||
| GraphPad | Prism | https://www.graphpad.com/scientific-software/prism/ |
| Tophat | Trapnell et al., 2012 | http://tophat.cbcb.umd.edu/ |
| Cufflinks | Trapnell et al., 2012 | http://cuffiinks.cbcb.umd.edu/ |
| DAVID for functional annotation | Huang et al., 2009 | https://david.ncifcrf.gov |
| Bowtie | Johns Hopkins University | http://bowtie-bio.sourceforge.net/index.shtml |
| Bismark | Babraham Bioinformatics | http://www.bioinformatics.babraham.ac.uk/projects/bismark/ |
| Samtools | GitHub | https://github.com/samtools/ |
| Model-based analysis of ChIP-seq (MACS) | Xiaole Shirley Liu’s Lab at Harvard University | https://github.com/taoliu/MACS/ |
| PAVIS tool | National Institute of Environmental Health Sciences | https://manticore.niehs.nih.gov/pavis2/ |
| Allele-specific Alignment Pipeline (ASAP) | Strogantsev et al., 2015 | https://www.bioinformatics.babraham.ac.uk/projects/ASAP/ |
| Bedtools | Quinlan laboratory at the University of Utah | http://bedtools.readthedocs.io/en/latest/index.html |
| IGV tools | Broad Institute | https://software.broadinstitute.org/software/igv/igvtools |
| PeakAnalyzer | EMBL-EBI | http://www.ebi.ac.uk/research/bertone/software |
| MEME-ChIP | The MEME Suite | http://meme-suite.org/tools/meme-chip |
| STAMP | Benos Lab at University of Pittsburg | http://www.benoslab.pitt.edu/stamp/ |
Highlights.
Naa10p KO causes defects in genomic imprinting and embryonic development
Naa10p maintains Dnmt1 activity by facilitating Dnmt1 binding to DNA substrate
Naa10p selectively binds to ICRs of the imprinted allele via non-methylated GCXGXG
Ogden syndrome-causing Naa10p mutation disrupts ICR binding of Naa10p and Dnmt1
ACKNOWLEDGMENTS
We thank S. Yamaguchi, E. Li, A.C. Ferguson-Smith, G. Kelsey, and S.-C. Lee for discussions, E. Li for materials, T.-J. Chuang for statistic consulting, the Transgenic Mouse Models Core Facility of NRPGM for generating Naa10-KO mice, Y.-C. Hsiao for taking care of the mice, and R. Devaraj and C.-H. Chen for the in vitro acetyltransferase assay. The research was supported by NHRI-EX101–10110BI, MOST103–2311-B-001–028-MY3, a career development grant and investigator award from Academia Sinica (to L.-J.J.), and The Howard Hughes Medical Institute (to Y.Z.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and seven tables and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2017.08.025.
SUPPORTING CITATIONS
The following reference appears in the Supplemental Information: Lee et al. (2001).
REFERENCES
- Aksnes H, Van Damme P, Goris M, Starheim KK, Marie M, Støve SI, Hoel C, Kalvik TV, Hole K, Glomnes N, et al. (2015). An organellar nα-acetyltransferase, naa60, acetylates cytosolic N termini of transmembrane proteins and maintains Golgi integrity. Cell Rep 10, 1362–1374. [DOI] [PubMed] [Google Scholar]
- Arnesen T, Van Damme P, Polevoda B, Helsens K, Evjenth R, Colaert N, Varhaug JE, Vandekerckhove J, Lillehaug JR, Sherman F, and Gevaert K (2009). Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc. Natl. Acad. Sci. USA 106, 8157–8162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow DP, and Bartolomei MS (2014). Genomic imprinting in mammals. Cold Spring Harb. Perspect. Biol 6, a018382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracko O, Singer T, Aigner S, Knobloch M, Winner B, Ray J, Clemenson GD Jr., Suh H, Couillard-Despres S, Aigner L, et al. (2012). Gene expression profiling of neural stem cells and their neuronal progeny reveals IGF2 as a regulator of adult hippocampal neurogenesis. J. Neurosci 32, 3376–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryja V, Bonilla S, and Arenas E (2006). Derivation of mouse embryonic stem cells. Nat. Protoc 1, 2082–2087. [DOI] [PubMed] [Google Scholar]
- Casey JP, Støve SI, McGorrian C, Galvin J, Blenski M, Dunne A, Ennis S, Brett F, King MD, Arnesen T, and Lynch SA (2015). NAA10 mutation causing a novel intellectual disability syndrome with Long QT due to N-terminal acetyltransferase impairment. Sci. Rep 5, 16022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Zhang J, Minnerly J, Kaul T, Riddle DL, and Jia K (2014). daf-31 encodes the catalytic subunit of N alpha-acetyltransferase that regulates Caenorhabditis elegans development, metabolism and adult lifespan. PLoS Genet 10, e1004699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B, Ren X, and Kerppola TK (2014). KAP1 represses differentiation-inducible genes in embryonic stem cells through cooperative binding with PRC1 and derepresses pluripotency-associated genes. Mol. Cell. Biol 34, 2075–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constância M, Hemberger M, Hughes J, Dean W, Ferguson-Smith A, Fundele R, Stewart F, Kelsey G, Fowden A, Sibley C, and Reik W (2002). Placental-specific IGF-II is a major modulator of placental and fetal growth. Nature 417, 945–948. [DOI] [PubMed] [Google Scholar]
- Delaval K,Govin J,Cerqueira F,Rousseaux S, Khochbin S, and Feil R (2007). Differential histone modifications mark mouse imprinting control regions during spermatogenesis. EMBO J 26, 720–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giacomo M, Comazzetto S, Sampath SC, Sampath SC, and O’Carroll D (2014). G9a co-suppresses LINE1 elements in spermatogonia. Epigenetics Chromatin 7, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörfel MJ, and Lyon GJ (2015). The biological functions of Naa10—from amino-terminal acetylation to human disease. Gene 567, 103–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmailpour T, Riazifar H, Liu L, Donkervoort S, Huang VH, Madaan S, Shoucri BM, Busch A, Wu J, Towbin A, et al. (2014). A splice donor mutation in NAA10 results in the dysregulation of the retinoic acid signalling pathway and causes Lenz microphthalmia syndrome. J. Med. Genet 51, 185–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick GV, Soloway PD, and Higgins MJ (2002). Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat. Genet 32, 426–431. [DOI] [PubMed] [Google Scholar]
- Ge YZ, Pu MT, Gowher H, Wu HP, Ding JP, Jeltsch A, and Xu GL (2004). Chromatin targeting of de novo DNA methyltransferases by the PWWP domain. J. Biol. Chem 279, 25447–25454. [DOI] [PubMed] [Google Scholar]
- Gu H, Smith ZD, Bock C, Boyle P, Gnirke A, and Meissner A (2011). Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat. Protoc 6, 468–481. [DOI] [PubMed] [Google Scholar]
- Guillemot F, Nagy A, Auerbach A, Rossant J, and Joyner AL (1994). Essential role of Mash-2 in extraembryonic development. Nature 371, 333–336. [DOI] [PubMed] [Google Scholar]
- Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, Zhang X, and Cheng X (2012). Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res 40, 4841–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell CY, Bestor TH, Ding F, Latham KE, Mertineit C, Trasler JM, and Chaillet JR (2001). Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell 104, 829–838. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, and Lempicki RA (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Inoue A, Jiang L, Lu F, Suzuki T, and Zhang Y (2017). Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 547, 419–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, and Zhang Y. (2010) Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466, 1129–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MD, Wu X, Aithmitti N, and Morrison RS (2002). Peg3/Pw1 is a mediator between p53 and Bax in DNA damage-induced neuronal death. J. Biol. Chem 277, 23000–23007. [DOI] [PubMed] [Google Scholar]
- Juan LJ, Utley RT, Adams CC, Vettese-Dadey M, and Workman JL (1994). Differential repression of transcription factor binding by histone H1 is regulated by the core histone amino termini. EMBO J 13, 6031–6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Ekram MB, Kim H, Faisal M, Frey WD, Huang JM, Tran K, Kim MM, and Yu S (2012). Imprinting control region (ICR) of the Peg3 domain. Hum. Mol. Genet 21, 2677–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Suda C, Abe T, Kohara Y, Ikemura T, and Sasaki H (2006). Bisulfite sequencing and dinucleotide content analysis of 15 imprinted mouse differentially methylated regions (DMRs): paternally methylated DMRs contain less CpGs than maternally methylated DMRs. Cytogenet. Genome Res 113, 130–137. [DOI] [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, and Westphal H (1996). Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. USA 93, 5860–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, and Copeland NG (2001). A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73, 56–65. [DOI] [PubMed] [Google Scholar]
- Lee CF, Ou DS, Lee SB, Chang LH, Lin RK, Li YS, Upadhyay AK, Cheng X, Wang YC, Hsu HS, et al. (2010). hNaa10p contributes to tumorigenesis by facilitating DNMT1-mediated tumor suppressor gene silencing. J. Clin. Invest 120, 2920–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei H, Oh SP, Okano M, Jüttermann R, Goss KA, Jaenisch R, and Li, (1996). De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 122, 3195–3205. [DOI] [PubMed] [Google Scholar]
- Leighton PA, Ingram RS, Eggenschwiler J, Efstratiadis A, and Tilghman SM (1995). Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature 375, 34–39. [DOI] [PubMed] [Google Scholar]
- Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, and Ferguson-Smith C (2008). A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev. Cell 15, 547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Zhang Z, Wu H, Jiang Y, Meng L, Xiong J, Zhao Z, Zhou X, Li J, Li H, et al. (2015). Recognition of H3K9 methylation by GLP is required for efficient establishment of H3K9 methylation, rapid target gene repression, and mouse viability. Genes Dev 29, 379–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magin RS, March ZM, and Marmorstein R (2016). The N-terminal acetyltransferase Naa10/ARD1 does not acetylate lysine residues. J. Biol. Chem 291, 5270–5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerschmidt DM, de Vries W, Ito M, Solter D, Ferguson-Smith A, and Knowles BB (2012). Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science 335, 1499–1502. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay A, Ravikumar G, Dwarkanath P, Meraaj H, Thomas A, Crasta J, Thomas T, Kurpad AV, and Sridhar TS (2015). Placental expression of the insulin receptor binding protein GRB10: Relation to human fetoplacental growth and fetal gender. Placenta 36, 1225–1230. [DOI] [PubMed] [Google Scholar]
- Myklebust LM, Van Damme P, Støve SI, Dörfel MJ, Abboud A, Kalvik TV, Grauffel C, Jonckheere V, Wu Y, Swensen J, et al. (2015). Biochemical and cellular analysis of Ogden syndrome reveals downstream Nt-acetylation defects. Hum. Mol. Genet 24, 1956–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Liu YJ, Nakashima H, Umehara H, Inoue K, Matoba S, Tachibana M, Ogura A, Shinkai Y, and Nakano T (2012). PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos. Nature 486, 415–419. [DOI] [PubMed] [Google Scholar]
- Ono R, Nakamura K, Inoue K, Naruse M, Usami T, Wakisaka-Saito N, Hino T, Suzuki-Migishima R, Ogonuki N, Miki H, et al. (2006). Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat. Genet 38, 101–106. [DOI] [PubMed] [Google Scholar]
- Park JH, Seo JH, Wee HJ, Vo TT, Lee EJ, Choi H, Cha JH, Ahn BJ, Shin MW, Bae SJ, and Kim KW (2014). Nuclear translocation of hARD1 contributes to proper cell cycle progression. PLoS ONE 9, e105185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J (2014). The role of genomic imprinting in biology and disease: an expanding view. Nat. Rev. Genet 15, 517–530. [DOI] [PubMed] [Google Scholar]
- Pijnenborg R, Robertson WB, Brosens I, and Dixon G (1981). Review article: trophoblast invasion and the establishment of haemochorial placentation in man and laboratory animals. Placenta 2, 71–91. [DOI] [PubMed] [Google Scholar]
- Plasschaert RN, and Bartolomei MS (2015). Tissue-specific regulation and function of Grb10 during growth and neuronal commitment. Proc. Natl. Acad. Sci. USA 112, 6841–6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp B, Støve SI, Endele S, Myklebust LM, Hoyer J, Sticht H, Azzarello-Burri S, Rauch A, Arnesen T, and Reis A (2015). De novo missense mutations in the NAA10 gene cause severe non-syndromic developmental delay in males and females. Eur. J. Hum. Genet 23, 602–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenneville S, Verde G, Corsinotti A, Kapopoulou A, Jakobsson J, Offner S, Baglivo I, Pedone PV, Grimaldi G, Riccio A, and Trono D (2011). In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanu-cleotideto affect chromatin and DNA methylation of imprinting control regions. Mol. Cell 44, 361–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ree R, Myklebust LM, Thiel P, Foyn H, Fladmark KE, and Arnesen T (2015). The N-terminal acetyltransferase Naa10 is essential for zebrafish development. Biosci. Rep 35, e00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regha K, Sloane MA, Huang R, Pauler FM, Warczok KE, Melikant B, Radolf M, Martens JH, Schotta G, Jenuwein T, and Barlow DP (2007). Active and repressive chromatin are interspersed without spreading in an imprinted gene cluster in the mammalian genome. Mol. Cell 27, 353–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekate HL (2009). A contemporary definition and classification of hydrocephalus. Semin. Pediatr. Neurol 16, 9–15. [DOI] [PubMed] [Google Scholar]
- Rope AF, Wang K, Evjenth R, Xing J, Johnston JJ, Swensen JJ, Johnson WE, Moore B, Huff CD, Bird LM, et al. (2011). Using VAAST to identify an X-linked disorder resulting in lethality in male infants due to N-terminal acetyltransferase deficiency. Am. J. Hum. Genet 89, 28–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Puig N, and Fersht AR (2006). Characterization of the native and fibrillar conformation of the human Nalpha-acetyltransferase ARD1. Protein Sci 15, 1968–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunier C, Støve SI, Popp B, Gérard B, Blenski M, AhMew N, de Bie C, Goldenberg P, Isidor B, Keren B, et al. (2016). Expanding the phenotype associated with NAA10-related N-terminal acetylation deficiency. Hum. Mutat 37, 755–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. (2007). The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450, 908–912. [DOI] [PubMed] [Google Scholar]
- Strogantsev R, Krueger F, Yamazawa K, Shi H, Gould P, Goldman-Roberts M, McEwen K, Sun B, Pedersen R, and Ferguson-Smith AC (2015). Allele-specific binding of ZFP57 in the epigenetic regulation of imprinted and non-imprinted monoallelic expression. Genome Biol 16, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi N, and Sasaki M (1975). Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature 256, 640–642. [DOI] [PubMed] [Google Scholar]
- Thorvaldsen JL, Duran KL, and Bartolomei MS (1998). Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev 12, 3693–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, and Pachter L (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumura A, Hayakawa T, Kumaki Y, Takebayashi S, Sakaue M, Matsuoka C, Shimotohno K, Ishikawa F, Li E, Ueda HR, et al. (2006). Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells 11, 805–814. [DOI] [PubMed] [Google Scholar]
- Van Damme P, Evjenth R, Foyn H, Demeyer K, De Bock PJ, Lillehaug JR, Vandekerckhove J, Arnesen T, and Gevaert K (2011). Proteome-derived peptide libraries allow detailed analysis of the substrate specificities of N(alpha)-acetyltransferases and point to hNaa10p as the post-translational actin N(alpha)-acetyltransferase. Mol. Cell Proteomics 10, M110.004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, Su H, Sun W, Chang H, Xu G, et al. (2009). The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat. Genet 41, 125–129. [DOI] [PubMed] [Google Scholar]
- Wang Y, Mijares M, Gall MD, Turan T, Javier A, Bornemann DJ, Manage K, and Warrior R (2010). Drosophila variable nurse cells encodes arrest defective 1 (ARD1), the catalytic subunit of the major N-terminal acetyltransferase complex. Dev. Dyn 239, 2813–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteway M, and Szostak JW (1985). The ARD1 gene of yeast functions in the switch between the mitotic cell cycle and alternative developmental pathways. Cell 43, 483–492. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Shen L, Liu Y, Sendler D, and Zhang Y (2013). Role of Tet1 in erasure of genomic imprinting. Nature 504, 460–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokochi T, and Robertson KD (2002). Preferential methylation of unmethylated DNA by Mammalian de novo DNA methyltransferase Dnmt3a. J. Biol. Chem 277, 11735–11745. [DOI] [PubMed] [Google Scholar]
- Yoon H, Kim HL, Chun YS, Shin DH, Lee KH, Shin CS, Lee DY, Kim HH, Lee ZH, Ryoo HM, et al. (2014). NAA10 controls osteoblast differentiation and bone formation as a feedback regulator of Runx2. Nat. Commun 5, 5176. [DOI] [PubMed] [Google Scholar]
- Zhang G, Estève PO, Chin HG, Terragni J, Dai N, Corrêa IR Jr., and Pradhan S (2015). Small RNA-mediated DNA (cytosine-5) methyltransferase 1 inhibition leads to aberrant DNA methylation. Nucleic Acids Res 43, 6112–6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Termanis A, Özkan B, Bao XX, Culley J, de Lima Alves F, Rappsilber J, Ramsahoye B, and Stancheva I (2016). G9a/GLP complex maintains imprinted DNA methylation in embryonic stem cells. Cell Rep 15, 77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X, Sheng J, Lau HT, McDonald CM, Andrade M, Cullen DE, Bell FT, Iacovino M, Kyba M, Xu G, and Li X (2012). Zinc finger protein ZFP57 requires its co-factor to recruit DNA methyltransferases and maintains DNA methylation imprint in embryonic stem cells via its transcriptional repression domain. J. Biol. Chem 287, 2107–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.