Abstract
Tissue inhibitor of metalloprotease-2 (TIMP-2) is a secreted 21 kDa multifunctional protein first described as an endogenous inhibitor of matrix metalloproteinases (MMPs) that prevents breakdown of the extracellular matrix often observed in chronic diseases. TIMP-2 diminishes the level of growth factor-mediated cell proliferation in vitro, as well as neoangiogenesis and tumor growth in vivo independent of its MMP inhibitory activity. These physiological properties make TIMP-2 an excellent candidate for further preclinical development as a biologic therapy of cancer. Here we present a straightforward bioprocessing methodology that yields >35 mg/L recombinant human TIMP-2 6XHis-tagged protein (rhTIMP-2) from suspension cultures of HEK-293-F cells. Enhanced rhTIMP-2-6XHis yields were achieved by optimization of both TIMP-2 cDNA codon sequence and cell culture conditions. Using a two-step chromatographic process, we achieved >95% purity with minimal processing losses. Purified rhTIMP-2-6XHis was free of mouse antigen contamination. Circular dichroism spectroscopy indicated a well-folded rhTIMP-2-6XHis that is highly stable and refractory to pH changes. Two-dimensional heteronuclear single-quantum coherence nuclear magnetic resonance of full length rhTIMP-2-6XHis also indicated a monodisperse, well-folded protein preparation. Purified rhTIMP-2-6XHis inhibited MMP-2 enzymatic activity in a dose-dependent fashion with an IC50 of ~1.4 nM. Pretreatment of A549 lung cancer and JygMC(A) triple-negative breast cancer cells with rhTIMP-2-6XHis in low-nanomolar amounts inhibited EGF-induced proliferation to basal (unstimulated) levels. This study therefore not only offers a robust bioprocess methodology for rhTIMP-2 production but also characterizes critical physicochemical and biological attributes that are useful for monitoring quality control of the production process.
Graphical Abstract
Aberrant turnover of the extracellular matrix (ECM) is a key feature in the development and progression of several diseases such as metastatic cancer, macular degeneration, myocardial infarction, and rheumatoid arthritis, as well as the neovascularization associated with many of these conditions.1 One group of key players in ECM turnover consists of the zinc-dependent endoproteases that belong to the metzincin superfamily, which are further subclassified as matrix metalloproteinases (MMPs) or matrixins, the Astacins, a disintegrin, and metalloproteinases (ADAMs), and ADAMS with throm-bospondin motifs (ADAM-TS).2,3 In early studies of tumor cell invasion, it was thought that members of these protease families resulted in ECM degradation that directly contributed to the invasive behavior of malignant cells.4 This led to pharmaceutical companies’ development of several, synthetic small molecule MMP inhibitors, most based on a collagen-mimicking hydoxamate structure, that were subsequently tested in clinical trials. Unfortunately, these trials demonstrated a lack of efficacy, showed detrimental musculoskeletal side effects, and showed no significant therapeutic effect.5–8 However, it was never determined if these small molecule MMP inhibitors effectively targeted MMP protease activity in vivo. Despite these setbacks, enhanced MMP expression continues to be associated with a poor prognosis in the chronic diseases listed previously and thus remains an attractive therapeutic target.
Tissue inhibitors of metalloproteases (TIMPs) are multifunctional proteins first described as endogenous inhibitors of MMPs that form 1:1 stoichiometric complexes. The mammalian TIMP family has four known isoforms (1–4) that have similar but not identical protease inhibitory profiles. The properties of TIMP family members and their MMP inhibitory effects have been previously reviewed.9,10 In addition to their metalloproteinase inhibitory activity, TIMPs also demonstrate antitumor and anti-angiogenic activity. These effects appear to be mediated by activation of cell signaling pathways that are initiated by direct binding to cell surface receptors. One member of the TIMP family, TIMP-2, has several distinct properties and functions that are independent of protease inhibitory activity and have been reviewed in detail elsewhere.11,12 These properties include inhibition of tyrosine kinase receptor signaling by binding to α3β1 integrin on the cell surface and activation of Shp-1 protein tyrosine phosphatase activity. This mechanism of heterologous receptor inactivation results in inhibition of growth factor-mediated proliferation of both normal (endothelial cells and fibroblasts) and tumor cells.12 Forced expression of TIMP-2 in tumor cells results in inhibition of tumor cell growth in vivo, as well as suppression of the epithelial-to-mesenchymal transition (EMT) that is associated with tumor aggressiveness and metastasis.13 These findings suggest a potential therapeutic benefit in the administration of exogenous TIMP-2 to prevent tumor growth and metastasis without the significant side effects associated with cytotoxic chemotherapies.
A major hindrance in the preclinical development of TIMP-2 has been the development of an expression platform and bioprocess to produce human TIMP-2 in high yield and purity. The goal of this study is to outline a straightforward, efficient, and robust method that can now be used for the production and purification of recombinant human TIMP-2 (rhTIMP-2) for future in-depth biophysical and preclinical studies of antitumor and anti-metastatic activity. It also aims to characterize the biophysical, biochemical, and biological properties of the protein that is obtained in high purity and yield, as a means of quality control for this process. We describe a bioprocess system with enhanced rhTIMP-2-6XHis expression (>35 mg/L), with a >95% purity using a simple two-step chromatographic process. Furthermore, we demonstrate that these rhTIMP-2-6XHis preparations show biophysical properties characteristic of a well-folded protein, as well as biochemical and biologic activities that can be utilized to monitor quality control.
MATERIALS AND EXPERIMENTAL DETAILS
Preparation of Wild-Type TIMP-2-EK-6XHis cDNA.
Using the coding region of wild-type (wt) TIMP-2, also designated as the pT2MO1 coding region described previously,14 a TIMP-2-EK-6XHis expression construct was prepared by polymerase chain reaction (PCR) with an enterokinase cleavage site15,16 inserted between the C-terminus of TIMP-2 and a six-histidine tag (6XHis) for recombinant protein purification. The PCR was performed using the pT2MO1 cDNA as a template with the forward 5’-ATAGTCGACATGGGAGCTGCTGCTCGCAC-3’ and reverse 5’-GACGGTATCGATAAGCTTTCAATGGTGATGGTGATGATGCTTGTCGTCGTCATCTGGGTCCTCGATGTCGAGAAA-3’ primers using High Fidelity PCR EcoDryTM Premix (Clontech). The amplified cDNA fragment was inserted into the pcDNA 3.3 Topo TA vector using a Topo TA cloning kit (both from Themo Fisher Scientific). The sequence of the cDNA construct was confirmed by the NIH DNA sequencing core.
Codon Optimization of TIMP-2-6XHis cDNA.
Codon optimization was performed on the coding region of the wt TIMP-2 cDNA sequence using a proprietary algorithm by Neo Biolabs (Boston, MA). In addition to codon bias, optimization criteria included reductions or elimination of the following sequences: GC content, premature polyA sites, internal χ sites and ribosomal binding sites, RNA instability motif (ARE), mRNA secondary structure, CpG dinucleotide content, and repeat sequences (direct repeat, inverted repeat, and dyad repeats). The codon-optimized sequence was prepared with a 3’ extension encoding a 6XHis tag by direct oligonucleotide synthesis and inserted in frame into the pcDNA 3.1(+) (Thermo Fisher Scientific, catalog no. V790–20) expression vector. All sequences were verified by the NIH DNA sequencing core and Neo Biolabs.
Construction of the in Silico Model of the mRNA Lowest-Energy Conformer.
Sequences of wt TIMP-2 (pT2MO1 coding region) and the codon-optimized TIMP-2-6XHis epitope tagged were obtained from NCBI CCDS11758.1 and Neo Biolabs, respectively. cDNA sequences of wt TIMP-2 and codon-optimized TIMP-2-6XHis cDNA were converted to mRNA sequences using the online transcription and translation tool (http://www.attotron.com/cybertory/analysis/trans.htm). The resulting mRNA sequences were uploaded to the online oligonucleotide model builder, RNA structure (http://rna.urmc.rochester.edu/RNAstructureWeb/Servers/Predict1/Predict1.html), for the generation of the lowest-energy mRNA conformer.
Cell Culture and Reagents.
HEK-293-F (FreeStyleTM 293-F, catalog no. R790–07) and CHO-S (FreeStyleTM CHOS, catalog no. R800–07) cells were obtained from Thermo Fisher Scientific (formerly Life Technologies). HEK-293-F and CHO-S cells were cultured and expanded in FreeStyle 293-F SFM (catalog no. 12338018) and FreeStyle CHO (catalog no. 12561014) expression medium, respectively. FreeStyle Max transfection reagent (catalog no. 94764), OptiPRO serum free transfection media (catalog no. 12309–050), Geneticin (catalog no. 10131035), and anti-clumping agent (catalog no. 010057AE) were all obtained from Thermo Fisher Scientific.
Transient Transfections for Upstream Bioprocessing Development.
HEK-293-F and CHO-S cells (>98% viability) were inoculated at a density of 0.5 × 106 cells/mL and cultured for 24 h in serum free medium. Cells were cultured on a shaker at 120 rpm in vented Erlenmeyer flasks (Corning Inc., Corning, NY) at 37 °C and 5% CO2. TIMP-2-6XHis plasmid DNA and FreeStyle Max transfection reagent were each dissolved in Serum Free OptiPRO medium at a ratio of 1 μg/μL, before being mixed together in a 1:1 (v/v) ratio and incubated for 10 min at room temperature. Following this initial incubation, the mixture was added dropwise directly to the cell culture medium, at a final ratio of transfection mixture to cell culture media of 1:25 (v/v) with either shaking or stirring. For larger scale transfection conditions, the same procedures were utilized and the ratio of the transfection mix to cell culture medium was the same as described above.
Stable HEK-293-F Cell Line Development for Upstream Bioprocessing.
For stable cell line development, HEK-293-F or CHO-S cell lines were transfected with the codon-optimized TIMP-2-6XHis plasmid per the procedure described above. Cells were then diluted and plated at a density of one cell per well in 96-well plates and selected in FreeStyle 293-F SFM or FreeStyle CHO medium supplemented with 500 jMg/mL G418 (Neomycin). Healthy colony-forming cells were selected from the wells and transferred to 24-, 12-, and 6-well plates successively. Spent media per well from 6-well plates were collected and analyzed with an anti-TIMP-2 enzyme- linked immunosorbent assay (ELISA) kit (catalog no. QIA 40, Calbiochem). Clones expressing the highest levels of TIMP-2-6XHis were further expanded in shaker flask cultures. Stable clones were cultured and maintained in FreeStyle HEK-293-F SFM or FreeStyle CHO medium supplemented with 250 μg/mL G418. Recombinant protein concentrations in all steps were determined by either a BCA assay (Thermo Scientific BCA Protein Assay Kit, catalog no. 23225), 280 nm absorbance measurements (ε = 33180 M−1 cm−1), a TIMP-2 sandwich ELISA (Ray Biotech, catalog no. ELH-TIMP-2), or all three procedures.14
Cell Culture and Process Scale-up.
Cells expressing rhTIMP-2-6XHis were cultured in sequentially expanding culture volumes from 30 to 60 to 350 mL at 37 °C in shaker, vented cap Erlenmeyer flasks (shaker flasks) prior to being transferred to 1 L spinner flasks. Cells were cultured in FreeStyle HEK-293-F SFM medium supplemented with 250 μg/mL G418 and 1:100 diluted anti-clumping agent. Cultures were harvested, and the spent medium was filtered through a 0.22 μm filter before purification or stored frozen at −20 °C. Further scale-up was conducted using highly viable, mid-log phase cultures of stably transfected rhTIMP-2-6XHis-express- ing HEK-293-F cells grown in 2 or 5 L wave bags (GE Healthcare Cell Bag Disposable Bioreactor, catalog no. CB0002L/5L 10–1). Cells were seeded at a density of 0.5 × 106 cells/mL in 1 or 2 L of medium and cultured in a Wave Bioreactor (Wave Bioreactor, Biodirect System 20/50 EH). Cells were cultured in the wave bag for 7 days post-inoculation with a rocking speed of 15–20 rpm, at a maximum angle of 7°. The Bioreactor platform temperature was maintained at 37 °C, and the wave bag was linked to a 0.22 μm filter 5% CO2 from a CO2 Mix-20 system.
Downstream Bioprocess Development for rhTIMP-2-6XHis Production in HEK-293-F and CHO-S Cell Lines.
Following culture termination, spent culture medium was collected and centrifuged at 4 °C and 10800g for 15 min, and the clarified spent medium was collected by decantation. Spent medium was supplemented with 1:1000 (v/v) protease inhibitor (protease inhibitor cocktail, catalog no. P8340–5ML, Sigma-Aldrich) and then filtered through a 0.22 μm filter. The spent medium was then purified by chromatography, as described below, or stored at −20 °C until it was used.
Two-Step Purifications of rhTIMP-2-6XHis and Its Analogues.
FPLC purification of the clarified conditioned media from codon-optimized TIMP-2-6XHis-transfected cell cultures was performed using an AKTA Explorer 100 Air instrument (GE Healthcare) equipped with a direct loading P-960 sample pump at 4 °C. The sample was loaded into a preequilibrated 5 mL His Trap column (catalog no. 17-5247-01, GE Healthcare) pre-equilibrated and washed back to baseline UV with loading buffer [20 mM sodium phosphate and 500 mM NaCl (pH 7.4)]. Nonspecifically bound contaminants and the final His-tagged protein were eluted by a stepwise increase in the imidazole concentration to 25 and 250 mM, respectively. All fractions for the chromatographic process were saved in aliquots for further characterization. Samples were loaded and separated on a 4 to 20% gradient sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gel. Separated proteins were blotted and probed with the murine anti-TIMP-2 antibody (catalog no. MAB 9711, R&D Systems) and anti-His-6 (catalog no. MA1–21315, Pierce). Fractions containing rhTIMP-2-6XHis (30 mL total) were pooled together and dialyzed against 4 L of 20 mM Tris-HCl overnight at 4 °C. Pooled samples were frozen at −80 °C and lyophilized for further storage or downstream processing.
Purified rhTIMP-2-6XHis was further purified by preparative reverse phase high-performance liquid chromatography (HPLC) using a Waters Delta Prep HPLC instrument. The lyophilized sample was resuspended in a water/acetonitrile (9:1) mixture with 0.1% trifluoroacetic acid (all HPLC grade reagents). rhTIMP-2-6XHis was purified with a POROS R1/10 16 mm × 100 mm reverse phase (catalog no. 1-1012-56, Applied Biosystems) column using a 10 to 60% acetonitrile gradient from 2 to 6 min post-injection at a flow rate of 36 mL/min. Fractions (6 mL) were collected every 10 s, frozen and lyophilized, resuspended in Milli-Q water, and frozen and lyophilized again for long-term storage.
Antigenicity Testing of rhTIMP-2-6XHis and Other Analogues.
Batches of rhTIMP-2-6XHis purified protein were tested for the presence of mouse antigen (MAP/RAP) at the Animal Health Diagnostic Laboratory of Frederick National Laboratory. Purified proteins were tested for purity and identity by SDS–PAGE as well as anti-TIMP-2 and the anti-6XHis epitope tag Western blots.
Circular Dichroism of rhTIMP-2-6XHis.
Circular dichroism (CD) spectra of rhTIMP-2-6XHis were obtained under different pH conditions (pH 4, 7, and 10). rhTIMP-2-6XHis was formulated at pH 4.0 (acetate and NaCl), pH 7.0 (Bis-Tris propane and NaF), and pH 10.0 (borate and NaCl). Spectra were recorded using an AVIV spectropolarimeter (AVIV Biomedical) with 300 μL (100 μg/mL) of the purified rhTIMP-2-6XHis sample in a quartz cuvette with a 1 mm path length. Spectral scans were obtained between 190 and 260 nm. The amounts of salt were kept at a minimum (150 mM) to reduce the fluctuation of the dynode voltage interfering with the spectral scan. All scans were performed at 37 °C, and the temperature of the cell was maintained with a Peltier heating system.
Nuclear Magnetic Resonance (NMR) Spectroscopy.
rhTIMP-2-6XHis was buffer exchanged four times into 50 mM sodium chloride and 20 mM sodium deuteroacetate (pH 5.0) with a 5% D2O/95% ultrapure H2O mixture using a 10 kDa molecular weight cutoff Amicon centrifugal filter (EMD Millipore Corp.). The final protein concentration was 207 μM.
Two-dimensional (2D) 15NH, 1HN and 13C, 1H selective optimized flip angle short transient (SOFAST) heteronuclear multiple-quantum coherence (HMQC) experiments17 were performed on a 900 MHz NMR spectrometer (Bruker, Inc.) equipped with a triple-resonance cryogenically cooled TCI probe with a triple axis gradient system. Both spectra were recorded with 50% non-uniform sampling18 with the NUS schedule determined by the Poisson gap algorithm.19 For the implementation of the 15NH, 1HN SOFAST-HMQC experiment, a Pc9 90° excitation pulse20 and a Reburp refocusing pulse21 were centered at 8.25 ppm with an excitation region of 5.55 ppm. The spectral width was 34 ppm × 14 ppm, corresponding to acquisition times of 20.6 ms (120 total reconstructed points) and 50 ms (1260 total points) in the 15N and 1H dimensions, respectively. The 15N transmitter was placed at 117 ppm. The interscan delay was 0.5 s, and 3891 scans per transient were collected. For the 13C, 1H SOFAST-HMQC experiment, the Pc9 90° excitation pulse and the Reburp refocusing pulse were centered at 0.00 ppm with an excitation region of 4.00 ppm. The spectral width was 40 ppm × 14 ppm, corresponding to acquisition times of 7.1 ms (128 total reconstructed points) and 50 ms (1260 total points) in the 13C and 1H dimensions, respectively. The 13C transmitter was placed at 15 ppm. The interscan delay was 0.5 s, and 380 scans per transient were collected.
The spectra were reconstructed using the iterative soft thresholding algorithm22 within nmrPipe version 8.9.23 Data were apodized with a shifted sine-square bell function and zerofilled. The resulting spectra were visualized using NMRFAM-SPARKY version 1.4.24
MMP-2 Inhibition Assay.
The MMP-2 inhibition assay was performed using a colorimetric MMP-2 inhibitor screening assay kit (catalog no. ab139446, Abcam). This kit utilizes the purified catalytic domain of MMP-2 (40 kDa; Abcam, personal communication from R. Poerschke, Abcam Inc.). MMP-2 inhibitors prevent, in a dose-dependent fashion, formation of the 2-nitro-5-thiobenzoic acid that can be detected by its absorbance at 412 nm. The assay was conducted using rhTIMP-2-6XHis and Ala+TIMP-2-6XHis, a TIMP-2 analogue that lacks MMP-2 inhibitory activity,14 in dose ranges of 0.25–281.6 nM in triplicate (n = 3). N-Isobutyl-N-(4-methoxyphenylsulfonyl) glycyl hydroxamic acid (NNGH), a potent small molecule inhibitor of MMP-2, was used as a positive control. Reaction mixtures containing active MMP-2 enzyme (stock concentration of 25 nM, personal communication from M. Norton, Abcam Inc.) and inhibitors as well as appropriate controls were preincubated at 37 °C for 1 h prior to the addition of the substrate and measurement in a Tecan Infinite M1000 pro plate reader.
Reduction of Cancer Cell Line EGF-Stimulated Growth.
These experiments were conducted as previously reported using A549 lung cancer cells (ATCC CCL-185)25 grown in DMEM-F12 medium (Lonza, catalog no. 12–719F) supplemented with 10% heat-inactivated FBS and 0.01% PennStrep and with JygMC(A) triple negative breast cancer cells (a gift from David Salomon, National Cancer Institute) grown in RPMI 1640 medium (Lonza, catalog no. 12–702F) supplemented with 10% FBS and 0.01% PennStrep. Cells were grown in 96-well cell culture plates coated with 200 μL of a 0.1% porcine gelatin solution (catalog no. ES-006-B, EMD Millipore) per well. To coat the plates, the gelatin solution was left to stand in the plates for 30 min prior to aspiration from the individual wells. Plates were then sealed under aseptic conditions and stored at room temperature for future use. A549 and JygMC(A) cells were plated in gelatin-coated plates at a density of 20 × 103 cells/well in complete medium (with 10% FBS) for 24 h. Following serum starvation for the next 24 h, the cells were pretreated with rhTIMP-2-6XHis (6.25–100 nM) for 30 min followed by 100 ng/mL rhEGF treatment. At 24, 48, or 72 h post-treatment, 40 μL of 5 μg/μL Thiazolyl Blue Tetrazolium Bromide (catalog no. M5655, Sigma-Aldrich) reagent dissolved in PBS was added per well and incubated for 3 h at 37 °C and 5% CO2. The medium was then aspirated from the wells, and 100 μL of dimethyl sulfoxide was added to each well and the plate incubated on a shaker for 10 min. Color development was read at 560 nm in a Tecan Infinite M1000 pro plate reader after the plate had been shaken for 5 s at low amplitude. For data analysis, mean absorbance data at respective treatments from the MTT assay absorbance values were converted to % max EGF proliferation using the formula
where basal Abs560 and EGF Abs560 refer to the mean absorbance from cells with no treatment and those treated with EGF alone (with no inhibitor treatment), respectively. Analysis of the statistical significance of the inhibition, between the untreated and rhTIMP-2-6XHis pretreatment doses, was performed on triplicate % max EGF proliferation analysis using one-tailed, type 2 Student’s t tests.
RESULTS
rhTIMP-2-6XHis Codon Optimization and Protein Expression.
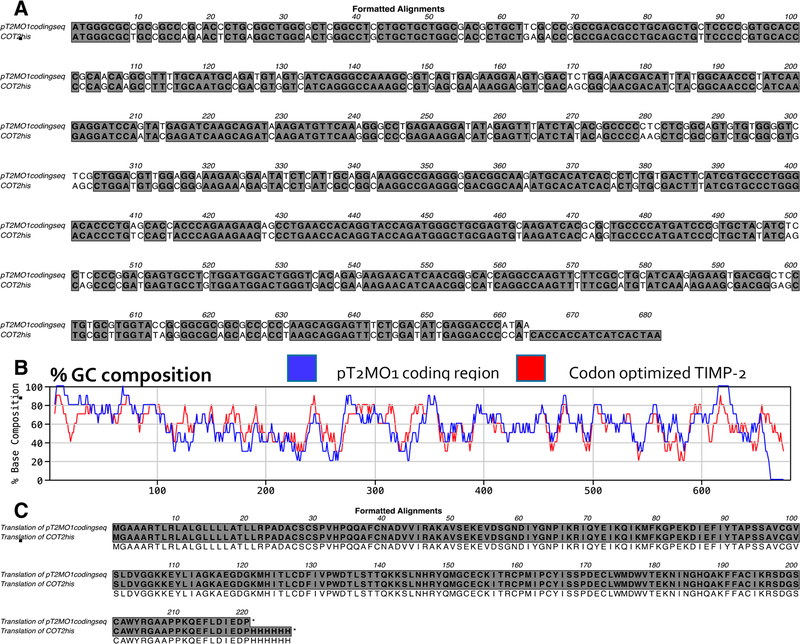
Comparison of the cDNA sequences for wt TIMP-2 and codon-optimized rhTIMP-2-6XHis shows no significant reduction (1%) in total GC content, 59.3 and 58.4%, respectively (Figure 1A). However, the plot of % GC content when examined using a 10-nucleotide window across the entire cDNA sequence shows a significant decrease in at least two critical areas that could explain the enhanced translation efficiency observed in our protein expression experiments. These areas are in the proximal 5’ region and the penultimate 3’ region of the codon-optimized rhTIMP-2 cDNA sequence (Figure 1B). Expression difficulties observed for rhTIMP-2-EK-6XHis and rhTIMP-2-6XHis in HEK-293 or CHO-S cells may arise from the high GC content within the first 90 bp of the wt TIMP-2 cDNA, resulting in the formation of significant stem–loop structures as observed in the converted mRNA structure (as shown in Supporting Information S1). This structured region in the mRNA may stall or prevent engagement of the 40S subunit of the ribosome and could explain the decreased expression levels observed with wt TIMP-2 cDNA constructs.26 The reduction in the level of mRNA secondary structure and the increase in mRNA half-life both correlate with the increased translational efficiency of the mature protein.27,28 Although codon optimization of the TIMP-2 cDNA afforded no significant reduction in overall GC content (Figure 1A), codon optimization did result in a significant decrease in the levels of these stem–loop structures and simplification of the mRNA structure (shown in Supporting Information S1A). These sequence changes clearly altered mRNA secondary structure and influenced other parameters such as CpG dinucleotide content (~1% decrease) and possibly repeat sequences. However, no change in the predicted protein sequence from that of wt TIMP-2 was observed when using the codon-optimized rhTIMP-2-6XHis cDNA expression constructs (Figure 1C), which was confirmed by mass spectroscopic analyses (vide infra). These changes clearly resulted in an increase in the translational initiation and efficiency resulting in enhanced rhTIMP-2-6XHis production (vide infra).
Figure 1.
Comparison of wild-type (wt) TIMP-2 (pT2MO1 coding sequence) and codon-optimized (CO) rhTIMP-2-6XHis (COT2his) cDNA and protein sequences. (A) ClustalW (MacVector version 15.5.0) pairwise nucleotide alignment and comparison of the wt TIMP-2 (pT2MO1) and codon-optimized rhTIMP-2-6XHis (COT2his) cDNA sequences using a 10-nucleotide window indicating changes made to and from G and C nucleotides. (B) Comparison of the % GC content in the pT2MO1 (blue) and COT2his (red) lines in 10-nucleotide sequence windows using the MacVector software. (C) Alignment of the translated protein sequences from wt (pT2MO1) and codon-optmized rhTIMP-2-6XHis (COT2his) cDNA sequences.
Suspension cultures of HEK-293-F cell lines transfected with codon-optimized rhTIMP-2-6XHis cDNA were selected by limiting dilution and maintained using 500 μg/mL Geneticin in the culture medium. We examined rhTIMP-2-6XHis expression levels under a variety of culture conditions, using both HEK-293-F and CHO-S cells. rhTIMP-2-6XHis protein levels were determined by an ELISA and compared as yields in milligrams per liter (Table 1). Using either transient transfection or stably transfected cell lines (HEK-293 monolayer, as well as a suspension HEK-293-F and CHO-S cell cultures), suspension cultures in shaker flasks demonstrated yields of rhTIMP-2– 6XHis protein as great as ~21 mg/L, a significant increase over the yield of 3 mg/L obtained from monolayer HEK-293 or HEK-293-F suspension cultures with transient or stable transfection of the rhTIMP-2-EK-6XHis cDNA construct. Still higher yields of 35–40 mg/L were obtained from cultures performed in spinner flasks or in single-use wave bags using stable, codon-optimized rhTIMP-2-6XHis stably transfected HEK-293-F cells. Kinetics of rhTIMP-2-6XHis production as well as the overall yield of rhTIMP-2-6XHis from HEK-293-F cells were monitored in pilot studies both by an ELISA and by a Western blot using antibodies against the 6XHis epitope tag. These preliminary experiments show increasing levels of rhTIMP-2-6XHis that plateaued by day 7 following expansion to the final volume in spinner or wave bag cultures. Cells were always grown and maintained in serum free medium to diminish the chance of carryover of animal proteins from fetal bovine serum. Also, for suspension cultures at all culture volumes, growth media required supplementation with the anti-clumping agent. HEK-293-F cells demonstrated an increased level of clumping in the absence of the reagent, which was associated with reduced cell viability and diminished recombinant protein yields. Recombinant rhTIMP-2-6XHis produced from CHO cells in initial cultures showed reduced MMP-2 inhibitory activity as compared to those produced from the HEK-293-F cells (data not shown). These data demonstrate that use of a codon-optimized rhTIMP-2-6XHis cDNA construct to stably transfect HEK-293-F cells and optimization of culture conditions (use of anti-clumping agent in spinner flask or wave bioreactor cultures) results in an ~10-fold increase in the yield of recombinant protein over that obtained in a transient suspension culture or stable monolayer transfection of non-codon-optimized cDNA.
Table 1.
Effects of cDNA Construct (codon optimization), Transfection, Cell Line, and Culture Conditions on rhTIMP-2-EK-6XHis Expressiona
| sample | construct | cell | transfection | culture conditions | yield (mg/L) | standard deviation |
|---|---|---|---|---|---|---|
| 1 | TIMP-2-EK-6XHis | HEK-293-F | transient | suspension | 3 | 0.3 |
| 2 | TIMP-2-EK-6XHis | HEK-293 | stable | monolayer | 3 | 0.3 |
| 3 | TIMP-2-EK-6XHis | CHO-S | transient | suspension | 5.5 | 0.6 |
| 4 | TIMP-2-EK-6XHis | CHO-S | stable | suspension | 8 | 0.8 |
| 5 | coTIMP-2-6XHis | HEK-293-F | stable | suspension/shaker | 15 | 1.1 |
| 6 | coTIMP-2-6XHis | HEK-293-F | stable | suspension/shaker | 20.8 | 1.2 |
| 7 | coTIMP-2-6XHis | HEK-293-F | stable | suspension/spinner | 40 | 1.6 |
| 8 | coTIMP-2-6XHis | HEK-293-F | stable | suspension/wave | 35 | 1.7 |
Production yields are significantly higher in stably transfected HEK-293-F cells vs transient transfections and using codon-optimized TIMP-2. Yields are reported for all combinations of cDNA, cell lines, and culture conditions (n = 3).
rhTIMP-2-6XHis Protein Purification.
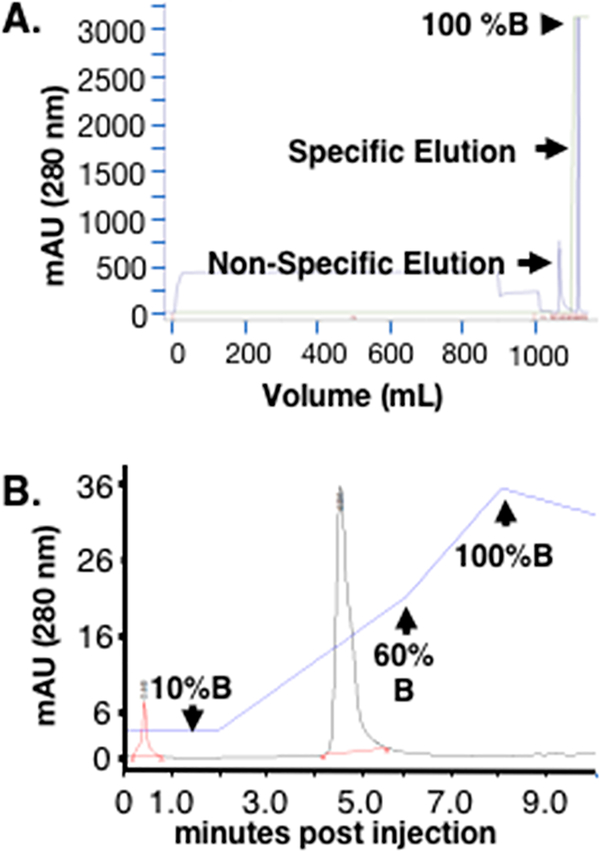
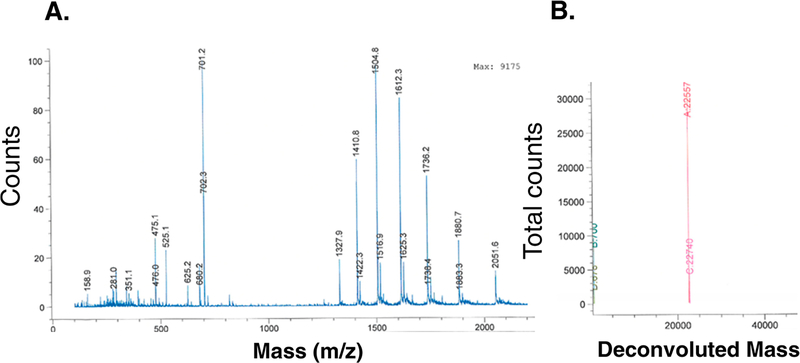
We have developed a straightforward two-step purification process for the initial capture and final polishing of rhTIMP-2-6XHis from the HEK-293-F harvested conditioned medium. Ni2+ immobilized metal ion affinity chromatography (IMAC) (HisTrap columns) yields a single peak upon specific elution with 250 mM imidazole (Figure 2A). SDS–PAGE analysis of this specificelution showed a predominant 22 kDa protein band with a few minor protein bands (Figure 3A,B). Reverse phase HPLC purification of the specific eluted protein from IMAC demonstrates a single main peak (Figure 2B). HPLC-purified rhTIMP-2-6XHis exhibits a single band in the SDS–PAGE gel and a single peak in ESI-TOF mass spectroscopy (Figure 4). Purified rhTIMP-2-6XHis was immunoreactive against anti-TIMP-2 and anti-6XHis antibodies (Figure 3C) and was >98% pure, as estimated by relative densitometric evaluation of the lane and bands by ImageLab software of the SDS–PAGE gel (Figure 3B). rhTIMP-2 was obtained at a purity of ~98% following all the purification steps and had an overall yield of ~82%. Larger losses were associated with the preparative RP-HPLC polishing step that can be attributed to the dilution of the sample and loss associated with the recollection step (Figure 3B and Table 1). Following HPLC purification, rhTIMP-2-6XHis samples were lyophilized and resuspended in autoclaved ultrapure water (resistivity of 18.2 MΩ cm−1 at 25 °C) or 25 mM Tris-HCl and stored at −80 °C as lyophilized powder. ESI TOF mass spectroscopic analysis revealed a single peak at 22.577 kDa that was identical with the calculated molecular weight of rhTIMP-2-6XHis (Figure 4A,B).
Figure 2.
Two-step downstream purification scheme for rhTIMP-2– 6XHis. (A) Immobilized metal ion affinity chromatography (HisTrap column) for batchwise elution yields a single sharp elution peak using a step gradient of 20 mM (nonspecific) and 250 mM (specific, buffer B) elution. (B) Preparative reverse phase HPLC also yields a single Gaussian peak. The blue line indicates the % solvent B (acetonitrile, 0.1% TFA) composition of the mobile phase, with initial isocratic elution using 10% solvent B until 2 min and then a 10 to 60% solvent B gradient between 2 and 6 min. The column was then washed with a gradient of 60 to 100% solvent B between 6 and 10 min, before reequilibration with 10% solvent B.
Figure 3.
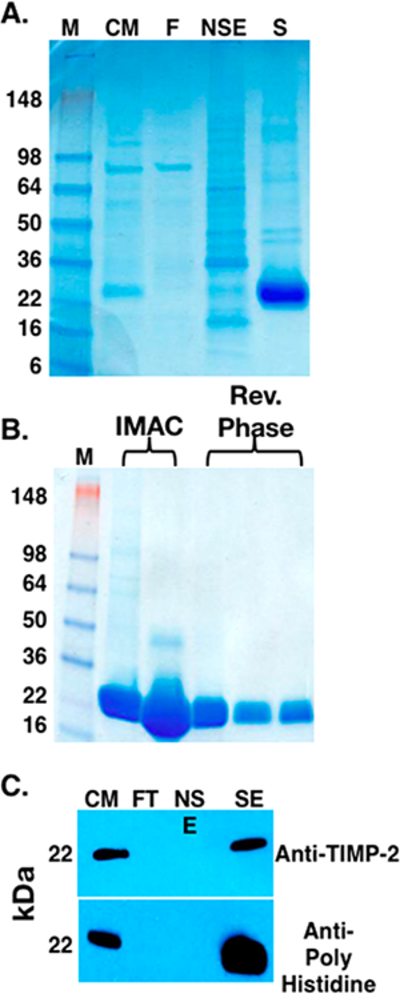
Analysis of two-step downstream purification of rhTIMP-2– 6XHis following bioprocess scale expression. Samples from the bioprocess purification were analyzed as shown in the (A) PageBlue Protein-stained SDS−PAGE gel of the fractions obtained from the IMAC (HisTrap column) purification step. Lane M contained the molecular weight standards (SeeBlue Plus 2 Prestained Standards, Invitrogen, catalog no. LC 5925). Lane CM contained the starting condition medium sample obtained from the HEK-293-F suspension culture. Lane FT contained the flow-through (unbound) fraction obtained during sample loading. Lane NSE contained the nonspecific elution obtained during step gradient elution with 20 mM imidazole. Lane SE contained the specific elution fraction obtained following 250 mM imidazole step elution. (B) PageBlue stained SDS−PAGE gel showing that the rhTIMP-2-6XHis-containing fractions from the IMAC (HisTrap) purification contain a predominant 22 kDa band with minor higher-molecular weight contaminants. The reverse phase HPLC purification effectively removed these higher-molecular weight contaminants, resulting in a single 22 kDa band with >95% purity as estimated by densitometry using a Bio-Rad ChemiDoc XRS+ instrument with Image Lab software. (C) Western Blot analysis of the IMAC fractions using anti-TIMP-2 (top) anti-6XHis tag (bottom) antibodies. The lanes are labeled the same as in panel A.
Figure 4.
(A) ESI-TOF MS of total component ions upon fragmentation. (B) Deconvoluted spectrum of the injection performed with rhTIMP-2-6XHis in a water/methanol mobile phase. Spectra show a molecular weight of 22577 Da that is identical to the theoretically calculated molecular weight of rhTIMP-2-6XHis (SwissProt ProtParam tool).
In preparation for use in future in vivo assays, we tested mouse antigen levels in the rhTIMP-2-6XHis preparations as described in Materials and Experimental Details. Purified rhTIMP-2-6XHis tested negative for the presence of mouse antigens (see Supporting Information S2 for a complete list of antigens tested).
Biophysical Characterization of Purified rhTIMP-2-6XHis by Circular Dichorism and 2D NMR Spectral Mapping.
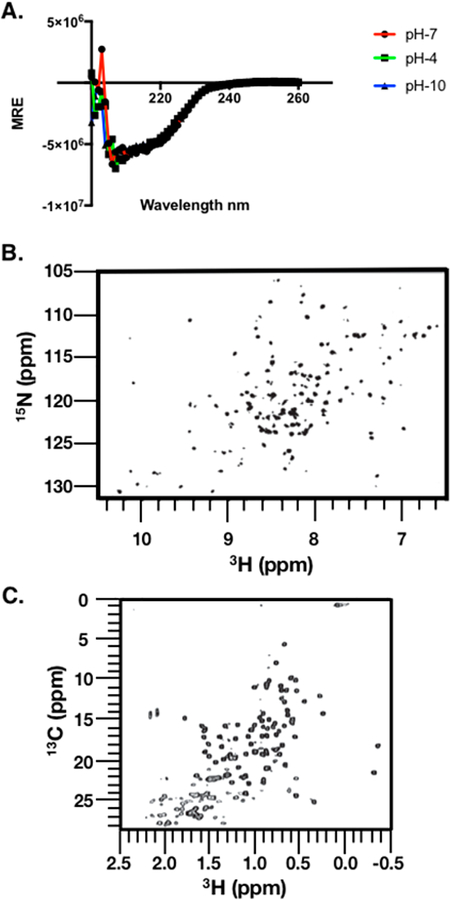
Circular dichroism (CD) spectroscopy indicated a well-folded protein with a very high degree of pH stability (Figure 5A). The spectral dip at ~200 nm indicates the presence of β-pleated sheet structures that can be organized into higher-order motifs to form individual functional domains,29 and these data are very similar to those from our previous reports on TIMP-2 and Ala+TIMP-2 obtained from prokaryotic expression systems.14 Our results are also highly consistent with the previous X-ray crystal structure of TIMP-2 at 2.1 Å resolution that reported N- and C-terminal domains composed of β-barrel and antiparallel stranded β-sheets.30 Calculation of the overall secondary structure of rhTIMP-2-6XHis from the CD data (delta ε) within the wavelength range of 190–240 nm using the K2D3 algorithm indicated an overall 32.39% a-helical and 15.24% β-stranded structure.31 The α-helical contribution in the spectra is likely derived from the helical coils that are present at the N-terminus, connecting interdomain sequence, and the C-terminal sequence detected in the TIMP-2 X-ray structure (Protein Data Bank entry 1BR9). Lyophilized rhTIMP-2-6XHis powder was resuspended to equal concentrations in buffers at pH 4, 7, and 10 in cuvettes of the same path length, and CD spectra were recorded for a wavelength range of 190–260 nm. Visually, no precipitation was observed at these pH values at a final concentration of 0.2 mg/mL. An overlay of the CD spectra of rhTIMP-2-6XHis at pH 4, 7, and 10 did not show any significant differences, indicating no pH-dependent perturbation of the secondary structure (Figure 5A). The pH stability of rhTIMP-2-6XHis would render to it greater stability as a biologic therapeutic within physiologic or charged formulation buffers or excipients.
Figure 5.
Spectral mapping of the rhTIMP-2-6XHis structure. (A) Circular dichorism spectroscopy (CD spectra) of purified rhTIMP-2-6XHis. Purified, lyophilized rhTIMP-2-6XHis was reconstituted in buffer at pH 4 (acetate and 150 mM NaCl), pH 7 (Bis-Tris Propane and 150 mM NaF), and pH 10 (borate and 150 mM NaCl). Spectral scans were recorded at 37 °C between 180 and 260 nm. The CD signal of the mean residue ellipticity (MRE) is plotted vs wavelength.Scans with rhTIMP-2-6XHis at pH 7 (red), pH 4 (green), and pH 10 (blue) overlap almost completely, indicating no significant effect of pH on the overall secondary structure. (B) Amide fingerprint region from the 15NH, 1HN SOFAST-HMQC spectrum recorded at 15N natural isotope abundance. (C) Methyl fingerprint region from the 13C, 1H region recorded at 13C natural isotope abundance. All spectra were recorded at 900 MHz and 25 °C in 50 mM sodium chloride, 20 mM sodium deuteroacetate (pH 5.0), and 5% D2O.
While CD spectroscopy has traditionally been used to access protein folding at low resolution, 2D heteronuclear NMR spectroscopy at natural isotope abundance is increasingly being applied to protein therapeutics.32 The atomic level resolution at each amino acid allows for facile evaluation of the folded state of the predominant conformational ensemble of the biologic. The 2D 15NH, 1HN and 13C, 1H spectral fingerprints, which were collected using rapid pulsing techniques,22 confirmed that rhTIMP-2-6XHis was indeed well-folded as determined from the spectral dispersion of the crosspeaks (Figure 5B,C). Of the 244 expected amide and side chain 15N−1H corrections, nearly 92% of peaks (224 peaks) were observed. While spectral assignments are known for the N-terminal domain (amino acids 1–127), these largely could not be plotted onto the spectral map of the full rhTIMP-2-6XHis because of vastly different experimental conditions.33 For the 13C methyl spectral map, approximately 89 of 93 peaks were counted. Taken together, the 15N and 13C spectral fingerprints confirm that the optimized expression system generates a well-folded rhTIMP-2– 6XHis protein preparation.
rhTIMP-2-6XHis Inhibits MMP-2 Enzymatic Activity.
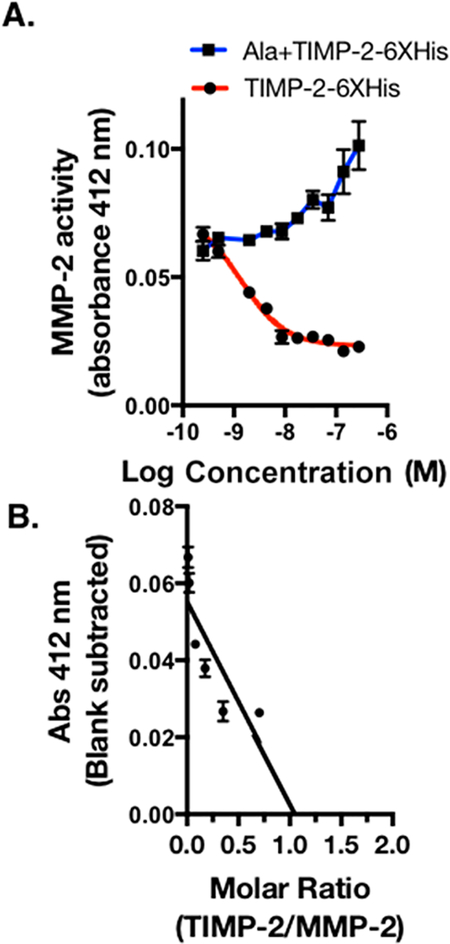
The inhibition of activated MMPs by TIMPs is dependent upon the correct tertiary structure. To determine if our recombinant protein retained this important biochemical function, we used linear regression analysis to assess the ability of rhTIMP-2-6XHis to inhibit recombinant MMP-2 activity. Assay quality was tightly controlled using both negative (no inhibitor) and positive (NNGH, a potent broad range inhibitor of various MMPs) controls.34,35 Recombinant TIMP-2-6XHis exhibited a dose-dependent inhibition of MMP-2 enzymatic activity from 0.25 to 281.6 nM, while no such inhibition was observed with Ala+TIMP-2-6XHis within the same dose range, consistent with previous reports.25 The reaction was monitored at 2 min intervals over a 20 min incubation period following the addition of MMP-2 substrate. The absorbance at 412 nm obtained following a 20 min reaction time was plotted against the log molar concentration of rhTIMP-2-6XHis, and the data were fitted to a standard one-site inhibitor dose–response nonlinear curve fit that exhibited an IC50 of 1.39 nM (R2 = 0.963) (red line), whereas no inhibitory effect of Ala+TIMP-2-6XHis treatment was seen (blue line) (Figure 6A). Comparable IC50 values of <2 nM were also observed for the reaction at 4, 10, and 16 min. It should also be noted that rhTIMP-2-6XHis completely blocks MMP-2 enzymatic activity at ~10 nM and therefore exhibited >1000-fold inhibition compared to NNGH.
Figure 6.
Inhibition of MMP-2 activity by rhTIMP-2-6XHis and Ala +TIMP-2 was monitored by colorimetric product formation at 412 nm. (A) MMP-2 enzymatic activity data plotted vs log molar concentration of rhTIMP-2-6XHis (red) and Ala+TIMP-2 (blue) were fitted to a nonlinear curve for the one-site enzyme inhibitor response. (B) Data from the linear range of the rhTIMP-2-6XHis inhibition curve were analyzed by linear regression analysis and extrapolation to the X-axis to estimate the molar ratio of rhTIMP-2-6XHis to MMP-2, resulting in complete inhibition of MMP-2 enzymatic activity (n = 3; mean ± standard error of the mean).
To estimate the stoichiometry of the fully inhibited rhTIMP-2-6XHis/MMP-2 complex, MMP-2 enzymatic activity was also plotted against rhTIMP-2-6XHis/MMP-2 molar ratios (Figure 6B). MMP-2 activity was sharply reduced to the baseline colorimetric absorbance value well within the rhTIMP-2-6XHis/MMP-2 molar ratio of ~1. The Ala+TIMP-2-6XHis mutant did not exhibit any MMP-2 inhibitory activity even at higher saturating concentrations. Linear regression analysis of the rhTIMP-2-6XHis/MMP-2 molar ratio indicated complete MMP-2 inhibition at a rhTIMP-2-6XHis/MMP-2 molar ratio of 1.064 as determined by the intersection of the best fit line at the X-axis. All tested rhTIMP-2-6XHis batches exhibited tight repeatable MMP-2 inhibition at similar IC50 values, indicating the consistent quality of the rhTIMP-2-6XHis produced. rAla +TIMP-2, which possesses no MMP-2 inhibitory activity, continues to serve as an excellent analogue for the analysis of TIMP-2’s MMP-2-independent activities.
rhTIMP-2-6XHis Suppresses EGF-Stimulated Growth of A549 Lung Cancer and JygMC(A) Breast Cancer Cell Lines.
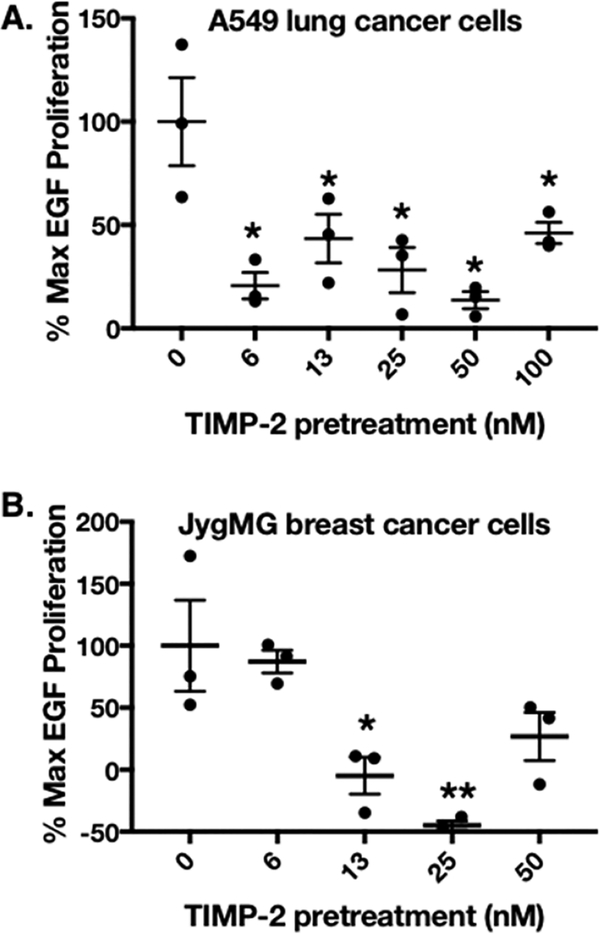
We have demonstrated an in vitro, dose-dependent effect of rhTIMP-2-6XHis on the inhibition of rhEGF-mediated proliferation of lung A549 and triple negative breast JygMC(A) cancer cell lines (panels A and B of Figure 7, respectively). Pilot experiments using A549 and JygMC(A) cells revealed an optimal stimulatory dose of 100 ng/mL EGF per 20000 cells per well (data not shown). Results of MTT assays performed at optimal time points [48 and 72 h post-EGF stimulation using A549 and JygMC(A) cells, respectively] are shown as percentages of maximal EGF stimulation (Figure 7A,B). Significant inhibition (>50%) of maximal EGF-stimulated cell growth was observed at all rhTIMP-2-6XHis concentrations tested for A549 cells. Similar results were obtained using JygMC(A) cells, except that no significant growth inhibition was observed at 6.25 nM rhTIMP-2-6XHis. Furthermore, the mean absorbance at 562 nm from several rhTIMP-2-6XHis concentrations demonstrates values below that obtained for the basal (control, no treatment) group. This may be due to intraassay variation, but no evidence of programmed cell death was observed in these experiments or similar investigations reported previously.25
Figure 7.
Inhibition of EGF-mediated proliferation by rhTIMP-2-6XHis preincubation. MTT assays determed the % max EGF proliferation as described in Materials and Experimental Details (n = 3; mean ± standard error of the mean). Proliferation data were plotted for (A) A549 NSCLC lung cancer cell lines and (B) JygMC(A) triple negative breast cancer cells in gelatin-coated cell culture plates. Proliferation was measured at optimal times following EGF stimulation, 48 and 72 h post-EGF stimulation for A549 and JygMC(A) cells, respectively.
Breakdown of the ECM within the normal adjacent tissue during cancer progression releases growth factors and proangiogenic factors, including EGF, VEGF, FGF, and PDGF.36,37 In several previous studies, A549 lung cancer cells have been used as a positive control for EGFR-expressing cells.38,39 Studies have also reported EGFR immunoreactivity as a poor prognostic indicator in the triple negative breast cancer phenotype.40 Previous investigations have shown that Shp-1 protein tyrosine phosphatase activity mediates TIMP-2 suppression of receptor tyrosine kinase signaling in normal (endothelial cells and fibroblasts) as well as A549 tumor cells. This study confirms the in vitro antiproliferative activity, in low- nanomolar amounts, of rhTIMP-2-6XHis produced via the bioprocess methodology described above.
SUMMARY
Earlier efforts aimed at the production and purification of rhTIMP-2 from Vaccinia virus-infected monkey kidney BSC-1 cells or refolded from Escherichia coli inclusion bodies could not be developed into a robust and reproducible method for large scale production of milligram quantities of rhTIMP-2.14,41 The new bioprocess and characterization methodology described here is a straightforward and robust bioprocessing method that is already yielding >35 mg/L rhTIMP-2-6XHis. The efficacy, cost effectiveness, robustness, repeatability, and purity of the final rhTIMP-2-6XHis protein are central to its preclinical development as all current and future studies depend on the continuous availability of significant quantities of “therapeutic grade” rhTIMP-2-6XHis. It is also essential for enabling future in-depth biophysical characterization experiments that will provide critical data and assist in engineering rhTIMP-2 to enhance specificity and efficacy via both MMP-dependent and -independent directed activities. The bioprocess methodology developed and reported here not only provides a strategy for rhTIMP-2 and its analogue libraries but also offers a method that can be used as a platform upon which to build for greater production and purification efficiencies. Investigation of the 2D structure and the “molecular fingerprint” of rhTIMP-2 by CD and 2D NMR establishes the monodispersed and well-folded characteristics of the rhTIMP-2 protein. Furthermore, the excellent Ph–CD profile, indicative of the overall stability of the rhTIMP-2 protein, is encouraging for further formulation of rhTIMP-2 as a biologic candidate for development efforts. Moreover, rhTIMP-2 exhibited an efficacy of MMP-2 inhibition at low nanomolar concentrations ~13-fold higher than that of NNGH, which is significant as upregulated MMPs are still considered therapeutic targets in a range of diseases. Similarly, low-nanomolar amounts (<50 nM) of rhTIMP-2 pretreatment effectively reduced the level of EGF-mediated proliferation of A549 lung and JygMC(A) triple negative breast cancer cells back to basal levels. This study enables us to pursue further indepth preclinical evaluation and biophysical characterization with rTIMP-2 that will broaden our understanding of TIMP-2 biology and allow us to move forward with the development of this very promising anticancer biologic.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Drs. Sandra Jensen and David Peeney for providing valuable suggestions and critical review of the manuscript.
Funding
The authors acknowledge National Institutes of Health Intramural Grants ZIA-SC-009179 and ZIA-BC-011204 (NCI/CCR Project 8386620) to W.G.S.-S.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.7b00700.
mRNA structure based on cDNA sequences for wt TIMP-2 (pT2MO1) and codon-optimized rhTIMP-2-6XHis (Supporting Information S1) and a table describing additional mouse pathogen testing of purified rhTIMP-2-6XHis (Supporting Information S2) (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Malemud CJ (2006) Matrix metalloproteinases (MMPs) in health and disease: an overview. Front. Biosci., Landmark Ed 11, 16961701. [DOI] [PubMed] [Google Scholar]
- (2).Murphy G (2017) Riding the metalloproteinase roller coaster. J. Biol. Chem 292, 7708–7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Apte SS, and Parks WC (2015) Metalloproteinases: A parade of functions in matrix biology and an outlook for the future. Matrix Biol. 44–46, 1–6. [DOI] [PubMed] [Google Scholar]
- (4).Stetler-Stevenson WG, Brown PD, Onisto M, Levy AT, and Liotta LA (1990) Tissue inhibitor of metalloproteinases-2 (TIMP-2) mRNA expression in tumor cell lines and human tumor tissues. J. Biol. Chem 265, 13933–13938. [PubMed] [Google Scholar]
- (5).Coussens LM, Fingleton B, and Matrisian LM (2002) Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science 295, 2387–2392. [DOI] [PubMed] [Google Scholar]
- (6).Levin M, Udi Y, Solomonov I, and Sagi I (2017) Next generation matrix metalloproteinase inhibitors - Novel strategies bring new prospects. Biochim. Biophys. Actay Mol. Cell Res 1864, 1927–1939. [DOI] [PubMed] [Google Scholar]
- (7).Liu J, and Khalil RA (2017) Matrix Metalloproteinase Inhibitors as Investigational and Therapeutic Tools in Unrestrained Tissue Remodeling and Pathological Disorders. Prog. Mol. Biol. Transl Sci 148, 355–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Overall CM, and Lopez-Otin C (2002) Strategies for MMP inhibition in cancer: innovations for the post-trial era. Nat. Rev. Cancer 2, 657–672. [DOI] [PubMed] [Google Scholar]
- (9).Cruz-Munoz W, and Khokha R (2008) The role of tissue inhibitors of metalloproteinases in tumorigenesis and metastasis. Crit. Rev. Clin. Lab. Sci 45, 291–338. [DOI] [PubMed] [Google Scholar]
- (10).Arpino V, Brock M, and Gill SE (2015) The role of TIMPs in regulation of extracellular matrix proteolysis. Matrix Biol. 44–46, 247–254. [DOI] [PubMed] [Google Scholar]
- (11).Stetler-Stevenson WG (2008) Tissue inhibitors of metalloproteinases in cell signaling: metalloproteinase-independent biological activities. Sci. Signaling 1, re6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Remillard TC, Bratslavsky G, Jensen-Taubman S, Stetler-Stevenson WG, and Bourboulia D (2014) Molecular mechanisms of tissue inhibitor of metalloproteinase 2 in the tumor microenvironment. Mol. Cell Ther 2, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Bourboulia D, Han H, Jensen-Taubman S, Gavil N, Isaac B, Wei B, Neckers L, and Stetler-Stevenson WG (2013) TIMP-2 modulates cancer cell transcriptional profile and enhances E-cadherin/beta-catenin complex expression in A549 lung cancer cells. Oncotarget 4, 163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Wingfield PT, Sax JK, Stahl SJ, Kaufman J, Palmer I, Chung V, Corcoran ML, Kleiner DE, and Stetler-Stevenson WG (1999) Biophysical and functional characterization of full-length, recombinant human tissue inhibitor of metalloproteinases-2 (TIMP-2) produced in Escherichia coli. Comparison of wild type and amino-terminal alanine appended variant with implications for the mechanism of TIMP functions. J. Biol. Chem 274, 21362–21368. [DOI] [PubMed] [Google Scholar]
- (15).Lin X, Xu S, Yang Y, Wu J, Wang H, Shen H, and Wang H (2009) Purification and characterization of anthranilate synthase component I (TrpE) from Mycobacterium tuberculosis H37Rv. Protein Expression Purif. 64, 8–15. [DOI] [PubMed] [Google Scholar]
- (16).Lin Y, Yu X, He Q, Li H, Li D, Song X, Wang Y, Wen H, Deng H, and Deng J (2009) Expression and functional characterization of chitribrisin, a thrombin-like enzyme, in the venom of the Chinese green pit viper (Trimeresurus albolabris). Protein Expression Purif. 67, 48–52. [DOI] [PubMed] [Google Scholar]
- (17).Schanda P, and Brutscher B (2005) Very fast two-dimensional NMR spectroscopy for real-time investigation of dynamic events in proteins on the time scale of seconds. J. Am. Chem. Soc 127, 80148015. [DOI] [PubMed] [Google Scholar]
- (18).Horiuchi K, Asaji T, and Ikeda R (1994) NQR and NMR studies on the electron-spin correlation time in paramagnetic insulators. Phys. Rev. B: Condens. Matter Mater. Phys 50, 6169–6173. [DOI] [PubMed] [Google Scholar]
- (19).Hyberts SG, Takeuchi K, and Wagner G (2010) Poisson- gap sampling and forward maximum entropy reconstruction for enhancing the resolution and sensitivity of protein NMR data. J. Am. Chem. Soc 132, 2145–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Kupce E, and Freeman R (1994) Wideband excitation with polychromatic pulses. J. Magn. Reson., Ser. A 108, 268–273. [Google Scholar]
- (21).Geen H, and Freeman R (1991) Band-Selective Radiofrequency Pulses. J. Magn. Reson 93, 93–141. [Google Scholar]
- (22).Hyberts SG, Milbradt AG, Wagner AB, Arthanari H, and Wagner G (2012) Application of iterative soft thresholding for fast reconstruction of NMR data non-uniformly sampled with multidimensional Poisson Gap scheduling. J. Biomol. NMR 52, 315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, and Bax A (1995) Nmrpipe: A Multidimensional Spectral Processing System Based on Unix Pipes. J. Biomol. NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- (24).Lee W, Tonelli M, and Markley JL (2015) NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Hoegy SE, Oh HR, Corcoran ML, and Stetler-Stevenson WG (2001) Tissue inhibitor of metalloproteinases-2 (TIMP-2) suppresses TKR-growth factor signaling independent of metal- loproteinase inhibition. J. Biol. Chem 276, 3203–3214. [DOI] [PubMed] [Google Scholar]
- (26).Reuter JS, and Mathews DH (2010) RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinf. 11, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Kozak M (1989) Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol. Cell. Biol 9, 5134–5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Kozak M (1986) Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc. Natl. Acad. Sci. U. S. A 83, 2850–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Greenfield NJ (2007) Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc 1, 2876–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Tuuttila A, Morgunova E, Bergmann U, Lindqvist Y, Maskos K, Fernandez-Catalan C, Bode W, Tryggvason K, and Schneider G (1998) Three-dimensional structure of human tissue inhibitor of metalloproteinases-2 at 2.1 A resolution. J. Mol. Biol 284, 1133–1140. [DOI] [PubMed] [Google Scholar]
- (31).Louis-Jeune C, Andrade-Navarro MA, and Perez-Iratxeta C (2012) Prediction of protein secondary structure from circular dichroism using theoretically derived spectra. Proteins: Struct., Funct., Genet 80, 374–381. [DOI] [PubMed] [Google Scholar]
- (32).Aubin Y, Gingras G, and Sauve S (2008) Assessment of the three-dimensional structure of recombinant protein therapeutics by NMR fingerprinting: demonstration on recombinant human granulocyte macrophage-colony stimulation factor. Anal. Chem 80, 26232627. [DOI] [PubMed] [Google Scholar]
- (33).Muskett FW, Frenkiel TA, Feeney J, Freedman RB, Carr MD, and Williamson RA (1998) High resolution structure of the N-terminal domain of tissue inhibitor of metalloproteinases-2 and characterization of its interaction site with matrix metalloproteinase-3. J. Biol. Chem 273, 21736–21743. [DOI] [PubMed] [Google Scholar]
- (34).Calderone V, Fragai M, Luchinat C, Nativi C, Richichi B, and Roelens S (2006) A high-affinity carbohydrate-containing inhibitor of matrix metalloproteinases. ChemMedChem 1, 598–601. [DOI] [PubMed] [Google Scholar]
- (35).Hu T, You Q, Chen D, Tong J, Shang L, Luo J, Qiu Y, Yu H, Zeng L, and Huang J (2016) Inhibiting Matrix Metalloproteinase 3 Ameliorates Neuronal Loss in the Ganglion Cell Layer of Rats in Retinal Ischemia/Reperfusion. Neurochem. Res 41, 1107–1118. [DOI] [PubMed] [Google Scholar]
- (36).Hanahan D, and Folkman J (1996) Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell 86, 353–364. [DOI] [PubMed] [Google Scholar]
- (37).Tosetti F, Benelli R, and Albini A (2002) The angiogenic switch in solid tumors: clinical implications. Suppl Tumori 1, S9–11. [DOI] [PubMed] [Google Scholar]
- (38).Li D, Ambrogio L, Shimamura T, Kubo S, Takahashi M, Chirieac LR, Padera RF, Shapiro GI, Baum A, Himmelsbach F, Rettig WJ, Meyerson M, Solca F, Greulich H, and Wong KK (2008) BIBW2992, an irreversible EGFR/HER2 inhibitor highly effective in preclinical lung cancer models. Oncogene 27, 4702–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Stabile LP, Lyker JS, Gubish CT, Zhang W, Grandis JR, and Siegfried JM (2005) Combined targeting of the estrogen receptor and the epidermal growth factor receptor in non-small cell lung cancer shows enhanced antiproliferative effects. Cancer Res. 65, 1459–1470. [DOI] [PubMed] [Google Scholar]
- (40).Viale G, Rotmensz N, Maisonneuve P, Bottiglieri L, Montagna E, Luini A, Veronesi P, Intra M, Torrisi R, Cardillo A,Campagnoli E, Goldhirsch A, and Colleoni M (2009) Invasive ductal carcinoma of the breast with the “triple-negative” phenotype: prognostic implications of EGFR immunoreactivity. Breast Cancer Res. Treat. 116, 317–328. [DOI] [PubMed] [Google Scholar]
- (41).Fridman R, Bird RE, Hoyhtya M, Oelkuct M, Komarek D, Liang CM, Berman ML, Liotta LA, Stetler-Stevenson WG, and Fuerst TR (1993) Expression of human recombinant 72 kDa gelatinase and tissue inhibitor of metalloproteinase-2 (TIMP-2): characterization of complex and free enzyme. Biochem. J 289 (Part 2), 411–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.