Abstract
Model organism databases (MODs) have been collecting and integrating biomedical research data for 30 years and were designed to meet specific needs of each model organism research community. The contributions of model organism research to understanding biological systems would be hard to overstate. Modern molecular biology methods and cost reductions in nucleotide sequencing have opened avenues for direct application of model organism research to elucidating mechanisms of human diseases. Thus, the mandate for model organism research and databases has now grown to include facilitating use of these data in translational applications. Challenges in meeting this opportunity include the distribution of research data across many databases and websites, a lack of data format standards for some data types, and sustainability of scale and cost for genomic database resources like MODs. The issues of widely distributed data and application of data standards are some of the challenges addressed by FAIR data principles. The Alliance of Genome Resources is now moving to address these challenges by bringing together expertly curated research data from fly, mouse, rat, worm, yeast, zebrafish, and the Gene Ontology consortium. Centralized multi-species data access, integration, and format standardization will lower the data utilization barrier in comparative genomics and translational applications and will provide a framework in which sustainable scale and cost can be addressed. This article presents a brief historical perspective on how the Alliance model organisms are complimentary and how they have already contributed to understanding the etiology of human diseases. In addition, we discuss four challenges for using data from MODs in translational applications and how the Alliance is working to address them, in part by applying FAIR data principles. Ultimately, combined data from these animal models are more powerful than the sum of the parts.
Keywords: Model organism database, zebrafish, mouse, worm, fly, yeast, rat, alliance of genome resources, translational medicine
Introduction
The use of model organisms in research may mistakenly be considered a relatively modern phenomenon having origins in the 19th or 20th century. In fact, animal models were used as early as the 6th century BCE when Alcmaeon of Croton used dogs to establish that intelligence and sensory integration are rooted in the brain1. Over the subsequent centuries a diverse array of model organisms, including viruses, prokaryotes, protists, fungi, plants, vertebrates and invertebrates, has contributed immeasurably to our understanding of the functioning of living things ranging from basic cellular processes such as the cell cycle to the underpinnings of complex human diseases2. The reason for such diversity of models was well stated by August Krogh, the 1920 winner of the Nobel Prize in Physiology and Medicine, when he wrote “For a large number of problems there will be some animal of choice or a few such animals on which it can be [most] conveniently studied.”3 Many factors influence the choice of a research model, including the biological attributes of each species, previously published studies, status of the genome sequencing effort, feasibility of various research methods, and financial feasibility, among others. The past century has seen research focus increasingly on a subset of model organisms having attributes favorable for current basic and biomedical research questions.
As the volume, diversity, and complexity of new research data grew, better methods for storing, integrating, and accessing these data were needed. Advances in database technology in the last quarter of the 20th century resulted in the implementation of diverse data— and organism—centric scientific databases, including MODs. The 2018 Nucleic Acids Molecular Biology Database Collection contains 1737 databases4. The MODs have served their respective user communities as hubs for the integration of diverse data, access points to essential biological reagents, and shared infrastructure and standards to support data re-use and interoperability. Although the systems architecture and technologies for each of these databases has evolved independently, numerous collaborative initiatives over the years have resulted in the adoption of common software components and annotation standards. Examples include the widespread use of GBrowse/JBrowse among MODS for genome browsing and the implementation of the Gene Ontology (GO) for unified sharing of knowledge about the function of genes and gene products5,6.
In recent years there has been an increasing focus on translational research, applying the aggregate integrated knowledge from model organisms to understand and treat human disease. This need for data integration and translational application has driven increased collaboration between the MODS and model organism researchers and clinicians, leading to successful discovery of disease etiology of even rare diseases through efforts such as the Undiagnosed Disease Network7–9.
Fully realizing the translational application of model organism data and databases has been hampered by the distributed location, unique user interfaces, and in some cases lack of a data format standard for similar data types at each of the individual MODs. These challenges can prove to be especially difficult for users of model organism data who do not have a strong background in model organism research or data management. Further, the long-term sustainability of MODs has been called into question, leading to discussion and testing of new organizational and technological paradigms for these critical resources that could lead to operational efficiencies10,11.
To address these important issues, databases representing six of the major model organisms (fly, mouse, rat, worm, yeast, zebrafish) and the Gene Ontology Consortium joined together in 2016 to form the Alliance of Genome Resources (The Alliance; https://www.alliancegenome.org). This article reviews the characteristics of the model organisms that currently comprise the Alliance as well as the organism-specific knowledge bases that have been developed to support their use in basic and translational biomedical research. Four challenges are identified which hamper the application of model organism data to translational applications. We review how the Alliance is working to address these challenges, in part through application of FAIR data principles, and how integration of the different MODS as the Alliance brings the biomedical research community new capabilities in comparative genomics and translational medicine. These new capabilities are fundamental to advancing our understanding of the biological basis of human health and disease.
Model Organisms and Databases
Fly - FlyBase 2.0
FlyBase supports the community of researches that use Drosophila melanogaster (the fruit fly) as a model organism (FlyBase; http://flybase.org, MIR:00100050)12. Among the distinct advantages of the Drosophila genetic model system are its large brood sizes, fast generation time, and cost efficiency. Use of the fruit fly as a model has led to discovery of fundamental principles of inheritance and the genes and pathways that determine cellular identity. The subsequent discovery that these same pathways regulate development in all animals, including humans, led to a new appreciation for the unity of life on earth, and has been fundamental to understanding the molecular mechanisms of many diseases, including cancer13. A major milestone in Drosophila research was the use of mobile elements for DNA transformation into flies, the first in any multicellular animal14. The subsequent use of these mobile elements for insertional mutagenesis linked gene sequence to gene function, which motivated the designation of Drosophila as an official model of the human genome project15,16. Work in Drosophila has also led to breakthroughs in understanding of immunity, epigenetics, circadian rhythms, and stem cells, among other fundamental discoveries17. Thus, the Drosophila model system has contributed significantly to our understanding of inheritance, development, and disease.
Drosophila continues to be a major model for biological discovery and translational research18. Over 100 years of fly research has led to a large and growing collection of mutant gene alleles as the Drosophila community continues to define mutant phenotypes for genes of previously unknown function19. An important function of FlyBase is to keep up with this large and growing list of genetic variants and the corresponding fly strains that are available at the Bloomington Drosophila Stock Center (BDSC; https://bdsc.indiana.edu, MIR:00100426). FlyBase also must keep pace with the Drosophila research community’s rapid development of novel methods and fly strains that permit tagging, knockout, or over-expression of genes and the mosaic analysis of development20–22. These novel methods together with the low cost and ease of rearing large numbers of flies defines Drosophila as a powerful genetic model for translational research, including as an official model of the Undiagnosed Disease Network (UDN; https://undiagnosed.hms.harvard.edu), a national effort to model in flies and fish candidate disease-causing DNA polymorphisms from humans8. For example, missense polymorphisms in the human ortholog of the fly gene humpty dumpty are being modeled in flies to determine their contribution to microcephalic primordial dwarfism birth defects of children23–26. Drosophila genetics combined with high throughput drug screening (pharmacogenetics) is being used to develop new therapies that target specific disease pathways. This approach led to discovery of a drug that is highly effective against multiple endocrine neoplasia IIB (MEN2B), which has transformed clinical practice for this previously therapy-resistant cancer27,28. FlyBase is constantly evolving new ways to facilitate these translational research efforts. For example, by creating search functions that link fly genes to their human orthologs and associated diseases. Currently, the list contains >500 disease models and continues to grow (FlyDiseaseModel; http://flybase.org/lists/FBhh/)29.
In addition, FlyBase has collaborated with the groups of Norbert Perrimon and Hugo Bellen to develop new online tools that permit searching for orthologous gene function (Gene2Function; http://gene2function.org)30; gene interactions across organisms (MIST; http://fgrtools.hms.harvard.edu/mist)31; and the identification of model organism genes and disease models starting with a human gene symbol or sequence variant as the search entry point (MARRVEL; http://marrvel.org)32. These are just a few of the examples of how FlyBase is a rapidly evolving resource that is essential to support the Drosophila community’s foundational discoveries and translational research.
In summary, FlyBase has evolved over the last 25 years from a simple database into a powerful knowledge base12,33. In addition to its essential role to curate and disseminate fly data, FlyBase is continuing to develop new tools for discovery of gene expression patterns, interaction, and function across organisms, and their links to human disease. Many of the FlyBase tools and its back end architecture have been adopted by The Alliance of Genome Resources (The Alliance; https://alliancegenome.org) in its goal to increase the uniformity, accessibility, and power of model organism data for translational research. Going forward, FlyBase will continue to be essential to support the numerous data types specific to the fly research community (e.g. tools and strains) if we are to realize the full potential of Drosophila for translational research and the discovery of new biological pathways and principles, the identity of which we cannot now imagine.
Mouse - Mouse Genome Informatics (MGI)
The laboratory mouse (Mus musculus) is widely recognized as a premier vertebrate animal model for investigating genetic and cellular systems relevant to human biology and disease. A diverse array of experimental genetic resources is available for mouse, including unique inbred strains, complete and annotated genomes for more than 17 inbred lines34, and extensive genome variation data (e.g. SNPs). An international effort to generate targeted mutations in all protein-coding genes in mouse begun in 200735 is virtually complete36; the phenotyping phase to functionally characterize these knockout mouse strains is currently underway37. New resources including recombinant inbreds from the Collaborative Cross38,39 and heterogeneous populations such as Diversity Outbred mice40,41 are beginning to bear fruit in analysis of complex traits and multi-genic diseases42–44.
The laboratory mouse has been used in a variety of ways to understand the mechanisms, genetics, genomics, and environmental contributions to human disease. Thousands of mouse knockouts, induced and spontaneous mutations, conditional mutations and transgenic lines have been used extensively to study simple Mendelian diseases such as cystic fibrosis45, achondroplasia46, Charcot-Marie Tooth disease47 and more. Recently, genetic models that recapitulate symptoms of human disease have been developed, including the creation of or repair of mutations that mimic pathogenic human variants such as in retinitis pigmentosa48, mood disorders49 and alpha1-antitrypsin deficiency50. Genome editing technologies allow for unprecedented precision in the types of mutations that can be introduced into different genetic backgrounds and are key to assessing functional significance of human genome variation51–53.
Inbred mouse strains are used to model complex trait diseases such as autism54, schizophrenia55 and diabetes56. Each inbred mouse strain possesses unique characteristics, with some strains susceptible to environmentally-induced diseases whereas others are resistant. Inbred strains susceptible or resistant to infectious agents that cause human infectious disease have been identified57. Chemical or toxin treatment is used to induce autoimmune disease in susceptible strains, such as pristane induced lupus erythematosus58, streptozotocin induced diabetes59, pilocarpine or kainate-induced epilepsy60 and MPTP-induced Parkinson’s disease61. Western style high-fat or high-salt diets are used to compare inbred mice to study the genetics of susceptibility to obesity62 and hypertension63, and different mouse strains react differently upon exposure to addictive substances64–67. Identification of the molecular mechanisms underlying these strain differences has led to insights into effective treatments and therapeutics for these diseases.
Inbred strains are also used to discover genetic modifiers of disease. For example, mice carrying the multiple intestinal neoplasia mutation (ApcMin) mutation (MGI:1856318) develop numerous intestinal and colonic adenomas on the C57BL/6J inbred strain background, where the mutation was discovered, similar to humans carrying pathogenic mutations in the Apc gene (MGI:88039)68. However, the frequency of adenoma development is severely attenuated when the mutant mice are crossed once to the AKR inbred strain and is reduced further upon subsequent backcrosses69. The suppressor locus in AKR was identified as a variant in Pla2g2a, phospholipase A2 gene (MGI:104642) in AKR70,71. Many pharmacological avenues now exist to inhibit this and other secretory phospholipases and are actively studied as potential therapeutics for cancer and inflammatory disease72,73. Thus, identification of suppressive modifier alleles of disease in mice can provide insight into therapeutic approaches for protecting humans against disease.
In addition to genetic models, immunodeficient and humanized mouse strains are being used in preclinical settings to test novel cancer therapeutic strategies tailored to the genome properties of human tumors74. These Patient-Derived Xenograft (PDX) models are generated through the implantation of human tumor samples into profoundly immunodeficient strains such as NOD.Cg-Prkdc<scid> Il2rg<tm1Wjl>/SzJ (https://www.jax.org/strain/005557, MGI:3577020) (aka, NSG), or into humanized mouse hosts75. By passaging engrafted tumors, cohorts of tumor bearing mice from the same patient tumor can be established and used to test responses to single agent and combination therapies76–78. In some cases, the results from dosing studies in PDXs have been used successfully to guide patient therapy79–81.
The MGI resource is the community MOD for the laboratory mouse (http://www.informatics.jax.org, MIR:00100062)82. The earliest published mouse literature indexed in MGI dates back to 1909, and the full corpus of mouse research covers nearly 250,000 publications. MGI was launched in 1989 with the goal of integrating separate genetic mapping and phenotypic data resources. It was one of the first MODS to have a presence on the World Wide Web in the early 1990s. MGI hosts multiple databases and data resources including: Mouse Genome Database (MGD)82, Gene Expression Database (GXD)83, Mouse Tumor Biology database (MTB)84, and Gene Ontology (GO)85. MGI’s mission is to facilitate the use of the mouse as an experimental model for understanding the genetic and genomic basis of human health and disease. MGI is the authoritative source for key data types and information including: mouse gene, allele, and strain nomenclature; the comprehensive genome feature catalog for the C57BL/6J reference genome; phenotype annotations; functional annotations, developmental gene expression; and mouse models of human disease. The MGI resource serves as a catalog of all genetic mutations reported for the mouse and their phenotypic consequences. The database contains information on over 6285 mouse genetic models of 1498 human diseases and is updated as new models are reported.
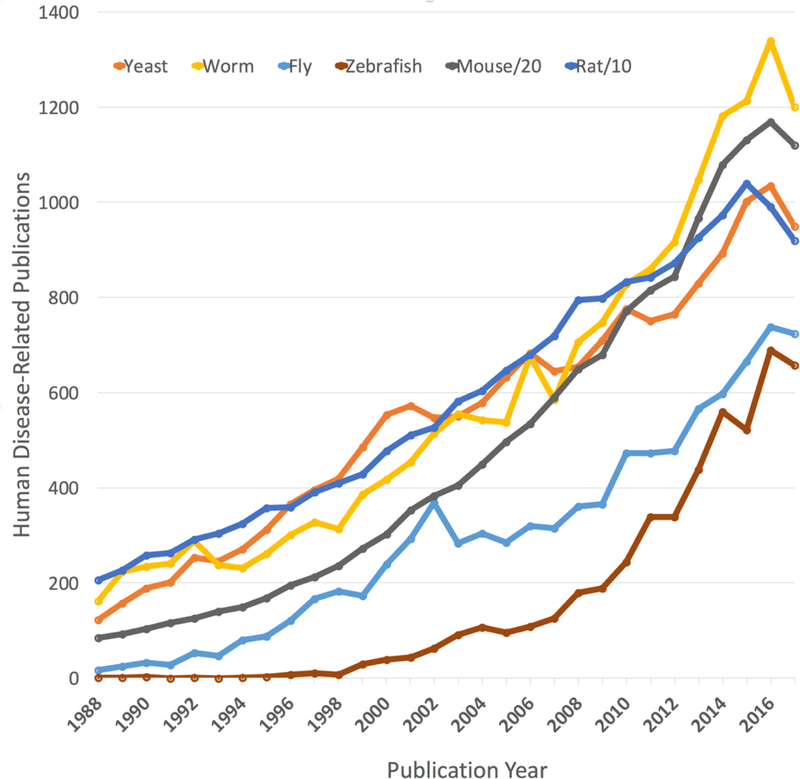
The experimental tractability of the mouse genome, well-established animal husbandry methods, and physiological similarities to human makes the laboratory mouse a versatile option for modeling human disease. All of these factors combined have resulted in a surge of translational applications of mouse models in recent years. In fact, an increase in human disease-related publication using model organisms is observed for all 6 of the Alliance model organisms (Figure 1). Given this exponential growth in human disease-related literature using model organisms, capturing this information and making it computationally accessible is critical to advancing the knowledge of human disease.
Figure 1. Disease-related journal publications using model organisms 1988–2017.
PubMed was searched for co-occurrence of “disease” or “syndrome” with each of the six current Alliance model organisms. The mouse and rat data have been divided by 20 and 10 respectively to keep the data on a similar scale with the other included organisms. The resulting publication counts per year were plotted for each model organism. Data were collected July 13, 2018.
Rat - Rat Genome Database (RGD)
The laboratory rat (Rattus norvegicus) has been used in scientific research for over 150 years. In the early 1800s rats were brought into laboratories for physiological studies, making it the first animal domesticated for the purpose of scientific research86. The primary contributions of the rat as a model organism are research in behavior, biochemistry, nutrition, pharmacology and physiology. More recently the rat has been used for the study of the genetics of hypertension, arthritis, diabetes, cancer and other diseases87–90. Specific strains have been selected or bred to serve the purpose of modeling human disease. The Rat Genome Database (RGD; https://rgd.mcw.edu, MIR:00000047) keeps records of more than 3,000 rat strains and sub-strains with the intent of providing researchers with information on any rat model known. Much of the rat genomic biomedical data until the early 2010s consisted of defining quantitative trait loci (QTLs) for many diseases in rat models. In the past ten years the increasing availability of rat strains with chemically generated gene mutations, targeted gene mutations, and genome edits has significantly expanded the importance of rat genomic and genetic studies in disease research86,91.
With the recent surge of genetic engineering techniques in rats, it is possible to take gene variants from clinical data and put them into rats to generate precision models of human disease. It will be important to use animal models to discover functional ramifications of new variants discovered in patients. Similar to the importance of the laboratory rat being used as a drug testing model in the pharmaceutical industry92, it is becoming a source of precision models for human disease from a genetic/genomics perspective.
Translational research flows in both directions between animal models and clinical medicine. Although it seems most logical to develop anti-disease strategies first in animal models before using those strategies on humans, sometimes the data comes first in humans, then on to animal models for further study. This was the case with modafinil, citalopram, and atomoxetine, three drugs for the treatment of ADHD93, where the rat data came after the human data and helped us to understand the mechanisms of action of these drugs. Other examples of translational success are the anti-estrogen drugs tamoxifen and raloxifene94. Having been used as anti-breast cancer drugs, there was concern of the drugs causing osteoporosis in postmenopausal women being treated for breast cancer. A study in rats showed that bone mineral density was maintained by both drugs, and raloxifene was later approved for the prevention of osteoporosis in postmenopausal women94.
Beyond the importance of the laboratory rat in testing drugs in preclinical research, rats are used as subjects of translational research in other biomedical areas such as orthodontics95. The orthodontic procedure of micro-osteoperforation was tested in the rat96, where it was shown to improve tooth movement as a supplement to controlled application of force. During the following decade it has become a popular and increasingly used technique in clinical practice.
Part of RGD’s goal has always been to facilitate research into the genetic and molecular basis of disease. Gene-, QTL-, and strain-based disease data for rat, mouse, and human has been a focus since the early years of RGD. That data will continue to be collected and analyzed at RGD as the rat continues to be a prominent model in translational medicine.
Yeast - Saccharomyces Genome Database (SGD)
While yeast has been the object of biochemical and cell biology studies since the 1800s, yeast genetics research began in full swing in the 1930s and 1940s, with a series of seminal works by Winge and Lindegren on the inheritance of mating type, nutritional requirements, metabolic pathways, and fermentation97,98. These studies led to the development of some of the first genetic and physical chromosome maps99,100. Decades later, the yeast community undertook the original genome project, producing the first complete eukaryotic genome sequence101. The availability of this sequence facilitated studies of chromosome structure, including that of centromeres, telomeres, and replication origins102–104. It also enabled, for the first time, new genomic surveys of different types of genes, including entire sets of transfer RNAs105 and small nucleolar RNAs106, complete list of cytoplasmic ribosomal protein genes107, and hundreds of retrotransposon insertions108. The field of genomics had been born and what soon followed helped establish yeast as the premier model organism for the fields of functional genomics and systems biology.
The Saccharomyces Genome Database (SGD; http://yeastgenome.org/, MIR:00000023) was established in 1989 to provide expert curation and management of data generated by the yeast research community. The yeast community developed the first genomic deletion libraries, sets of strains in which a single gene was replaced with a selectable marker109,110. These libraries, and those that came after, have proven indispensable for interrogating gene function on a genomic scale. Other collections, such as the GFP-fusion library, have been used to determine the cellular locations of entire proteomes111,112. While still others, for example, the synthetic gene arrays (SGA), have been used to determine phenotypes of all double mutants in the genome113,114. These technologies have provided at least some understanding of the functions of >85% of the genes and proteins of the budding yeast genome, the highest value for any eukaryote, making it the most thoroughly characterized model organism115. Yeast have now been used as a model system for mitochondrial diseases involving oxidative phosphorylation or metabolic disorders116. This knowledge is readily transferred to higher eukaryotes via the Gene Ontology (described below)117.
In the last several years, dozens of S. cerevisiae genomes have been sequenced, from natural isolates to industrial strains for beverages and bioethanol to opportunistic pathogens, with more to come118. Next-generation sequencing has become so common within the yeast community that entire genomes are being sequenced in bulk to answer specific questions regarding topics such as gene transfer and genome rearrangement119, nutrient utilization and fermentative capacity120, taxonomy and systematics121.
More recent uses of the yeast genome include both humanization122 and bacterialization123 of yeast proteins to understand other systems. While orthology is an imperfect tool, it remains valuable for predicting the functions of uncharacterized proteins124. Functional complementation studies, in which a gene from one species can successfully replace the function of a gene in another species, have proven invaluable for confirming conservation of function. In addition, identifying genes from other species that can functionally replace activities in yeast cells makes those genes amenable to study in the highly-tractable yeast genetics model system and thus utilizing all the power therein125.
Worm - WormBase
C. elegans is a cost effective pre-clinical model system with the following advantages: small size; transparent body; short generation time and lifespan (~3 days and 3 weeks respectively); large brood size; completely sequenced genome; ability to map out every cell lineage; and well developed genetic, molecular and imaging tools for study and ease of genetic manipulation.
C. elegans has been used as a model system to elucidate the genetic and cellular mechanisms underlying several disorders such as complex neurodegenerative diseases (Alzheimer’s disease (AD), Parkinson’s, Huntington’s Disease and tauopathies)126; neuromuscular diseases (spinal muscular atrophy, Duchenne muscular dystrophy, etc.)127,128; ciliary diseases (polycystic kidney disease, Bardet-Biedl syndrome, nephronophthisis)129; lysosomal storage diseases (Niemann-Pick disease, Batten disease and mucolipidosis IV); laminopathic diseases130; intestinal inflammatory diseases131; and obesity and aging132. C. elegans often bridges the gap between unicellular models such as yeast and complex models such as the mouse. WormBase (http://www.wormbase.org/, MIR:00000027) and its sister site, ParaSite (http://parasite.wormbase.org/) are the authoritative and comprehensive community resources for the genome, genetics, and biology of C. elegans and other nematode species, including several parasitic species133.
C. elegans has been particularly useful in elucidating mechanisms underlying the interplay between the aging process, cellular redox control and abnormal protein pathology seen in neurodegenerative diseases134. Several transgenic protein aggregation models have been generated in C. elegans135. Transgenic amyloid-beta-induced paralysis models such as strain CL2006, that expresses human amyloid-beta in body wall muscle, and strain CL2355, which exhibits neuronal expression of human amyloid-beta, provide a quantifiable behavioral output of amyloid-beta toxicity. Additionally, these lines bespeak the utility of examining direct modifiers of amyloid-beta toxicity, rather than modifiers of amyloid-beta production136,137. Experiments with the strain CL2006 have demonstrated the impact of aging and insulin-signaling on amyloid-beta neurotoxicity (mutational loss of daf-2 or RNAi in chronic A-beta paralyzed animals increased lifespan and attenuated paralysis)137.
C. elegans models of amyloid-beta toxicity also serve as platforms for bio-active compound and drug screening. Pharmacological modifiers such as caffeine, tannic acid and bacitracin, from a FDA-approved screen for drugs that protect against glucose-induced toxicity in primary neuronal cultures, attenuated amyloid-beta induced lifespan reduction in the worm137. Liuwei Dihuang and Dianxianning (from Chinese traditional medicine) reduce amyloid-beta toxicity through mechanisms involving antioxidant activity, heat shock proteins, reduced ROS and insulin signaling. Clioquinol and Dihydropyrimidine (DHPM-thione), two compounds identified in yeast amyloid-beta models for reducing amyloid-beta toxicity have been validated in the worm model to reduce neurodegeneration137.
Another area where the worm has contributed to a mechanistic understanding of pathogenesis is the study of kidney diseases. Worm models were used to discover the fundamental role of cilia in ciliopathies, including polycystic kidney disease, nephronophthisis, Meckel-Gruber syndrome, and Bardet-Biedl syndrome129. Evidence for a sterol-shortage and sterol-signaling defects in Niemann-Pick disease and evidence for involvement of ABC-transporters in mucolipidosis IV and Batten disease come from worm and fly models138,139.
Curation of disease relevant data is an ongoing project in WormBase. These data are displayed in the ‘Human Diseases’ section on gene pages. Disease data can also be accessed by searching for a disease name on the WormBase website. WormBase has recently started to curate data from screens for modifiers of disease such as drugs, herbals, etc. As the worm continues to fill an important niche in the model organism translational research landscape, WormBase has not only expanded to include translational research data but is also actively involved with other databases of the Alliance to define and formalize data standards.
Zebrafish - Zebrafish Information Network (ZFIN)
Zebrafish (Danio rerio) have long been used in research studies ranging from fisheries research140 to developmental and genomic research141. They are good laboratory research animals due to their optical clarity, ease of genetic manipulation, rapid external development, and high fecundity; traits which support their increasing use to investigate genes related to human disease142,143. The Zebrafish Information Network (ZFIN; https://zfin.org, MIR:00000079) serves as the central resource for genetic, genomic and phenotypic data that are the result of research studies using zebrafish144. With the increased use of zebrafish in translational research, ZFIN has also expanded to include data about zebrafish models of human disease145.
Scientists have used zebrafish in various ways to understand the mechanisms, genetics, genomics, and environmental contributions to human disease. Due to the high orthology between zebrafish and humans, genetic manipulation of zebrafish orthologs of human disease-associated genes has led to zebrafish models of many diseases including Duchenne muscular dystrophy146, Diamond Blackfan anemia147, epilepsy148, Rett syndrome149–151, and visceral heterotaxy152,153. Transgenic zebrafish are another form of genetic model where mutant human genes are expressed in zebrafish to understand disease etiology. For example, transgenic zebrafish that express mutant forms of the human γD-Crystallin gene have been created to understand the mechanisms involved in the development of cataracts154.
In addition to genetic models, zebrafish in which the experimental conditions, rather than the genetics, are manipulated have been created to recapitulate disease phenotypes. For example creating models through the application of chemicals to induce epilepsy155 and Parkinson’s disease156,157. Zebrafish are also becoming a tractable system for studying metabolic disorders158 with genetic159,160 as well chemical models161,162 created for obesity.
Translational science not only encompasses using model systems to understand the cellular and molecular function of genes and how their dysfunction contributes to disease states, it is also useful for elucidating potential therapeutics. Zebrafish are amenable to high throughput drug screening and discovery163–165 and have been used in drug screens for several diseases including leukemia166, melanoma167, and tuberculosis168. Zebrafish are also emerging as a valuable resource in personalized medicine169 where patient derived tumor cells are transplanted to create zebrafish xenograft models utilized to understand cancer biology and determine therapeutics for several cancers including gastric cancer170 and neuroendocrine tumors171. The combination of flexible genetic and chemical modeling of human disease promises a bright future for the zebrafish in translational medicine applications.
Gene Ontology - GO
The Gene Ontology (GO; http://geneontology.org, MIR:00000022) provides species-neutral definitions for different functional classes of gene products (i.e. proteins and RNAs, and complexes), and a comprehensive set of annotations across a wide range of organisms describing the role of their individual gene products using these classes. It is specifically designed to support the computational representation of biological systems. It currently covers hundreds of organisms, including (but not limited to) the Alliance model organisms and humans. Integral to GO are the principles of evolutionary biology. Because of our shared history, researchers can leverage the insights gained in one organism to shed light on the biology of other organisms, including human.
GO is most frequently used for gene set enrichment analyses. Given a set of genes, such as those co-expressed under a particular set of experimental conditions, the question is what GO functional grouping do these hold in common? There are thousands of published papers that include GO enrichment analysis as a key part of their experimental design. A number of recent independent resource valuation exercises determined that the GO is central to biological and medical research. GO is one of the top five resources (of 133) that together account for 47% of the total number of resource citations (the others are GenBank, UniProt, KEGG, and PDB) and its usage is growing172. Similarly, the report commissioned to evaluate the impact of the European Bioinformatics Institute (EBI; https://www.ebi.ac.uk) used GO as an exemplar of a successful scientific resource. GO was the fourth most utilized resource after Ensembl, UniProtKB, and Europe PubMed Central at the EBI173. More specifically, a recent example illustrates the growing use of GO in clinical research. In this case, GO was used for analysis and functional annotation of holoclones’ integration into epidermis regenerated using transgenic stem cells, which in turn led to regeneration of skin for the patient174. This reflects direct usage of GO for clinical research.
GO is also being used as a technical underpinning for phenotypic analyses. In the Human Phenotype Ontology (HPO) the underlying logical definitions used for reasoning rely (where appropriate) upon GO. HPO, plus the annotations associating specific HPO classes with specific genes/variants, has itself been used successfully for investigating rare diseases using the Exomiser software175. Exomiser, examining 11 previously diagnosed patients’ exomes and ranking the variant(s) for each, was successful in identifying the causative variant among the top 10. Additionally, Exomiser achieved a diagnosis for four of 23 cases undiagnosed by clinical evaluation175–177. These analyses include GO not only for logical definitions but also the annotations using GO, which are generated by expert curators at all six of the Alliance MODs. HPO is now the ontology for a number of major initiatives, including the Global Alliance for Genomics and Health (http://ga4gh.org/), Rare Disease Connect (https://rareconnect.org), DECIPHER (https://decipher.sanger.ac.uk), Monarch (https://monarchinitiative.org), ClinGen (https://clinicalgenome.org), Care for Rare (https://care4rare.ca), Centers for Mendelian Genomics (http://mendelian.org/)178, and others.
Integration - The Alliance of Genome Resources
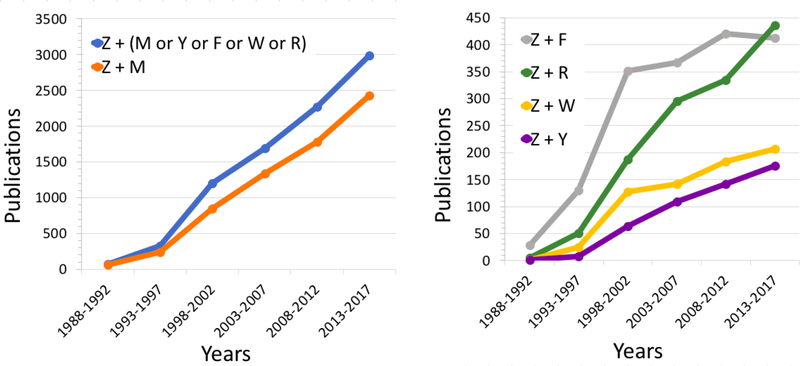
Research using individual model organisms has made great contributions to our understanding of basic biological mechanisms and disease states which result from their breakdown. Maximizing the translational application of this knowledge is now a key task which will be best achieved when these data are used together in an integrated fashion. For example, there are animal models of Amyotrophic Lateral Sclerosis (ALS) available in mice, rats, fruit flies, worms, zebrafish, dogs and pigs179, each providing an ideal model for unique aspects of this disease. Flies, mice, yeast, and zebrafish have all been used in chemical screens aimed at treating proteinopathies180, and as models of neuronal ceroid lipofuscinosis (Batten disease)181. Evidence of growing use of data simultaneously from multiple model organisms can be found in PubMed. For example, publications with co-occurrence of zebrafish and any of the other Alliance model organisms has consistently increased over the past 30 years with co-publication of zebrafish and mouse making up the majority (Figure 2). Although co-occurrence of species in publications does not necessarily indicate that both species were used experimentally, this trend is consistent with an increasing reliance of modern biomedical research on tools and data coming from multiple model organisms and highlights the need to optimize and streamline the combination of data from these data sources.
Figure 2. Increasing occurrence of publications involving both zebrafish and other model organisms.
PubMed searches were done for publications involving both zebrafish and each or any of the other Alliance model organisms. Z = zebrafish, M = mouse, F = fly, R = rat, W = worm, Y = yeast. Data were collected Feb. 22, 2018 at PubMed.
In 2016, best practices known as the FAIR principles for management and stewardship of scientific data were established182. FAIR stands for “Findable”, “Accessible”, “Interoperable”, and “Reproducible”. These principles are intended to support accessibility and reuse of scientific data by both machines and people. The following four significant challenges have been identified which hamper the combined utility of model organism data in a translational setting. Here we discuss how the Alliance of Genome Resources is working to address these challenges, in part through application of FAIR data principles182, with the aim of facilitating basic biomedical as well as translational research applications using model organism data.
Challenge 1: Distributed location of data
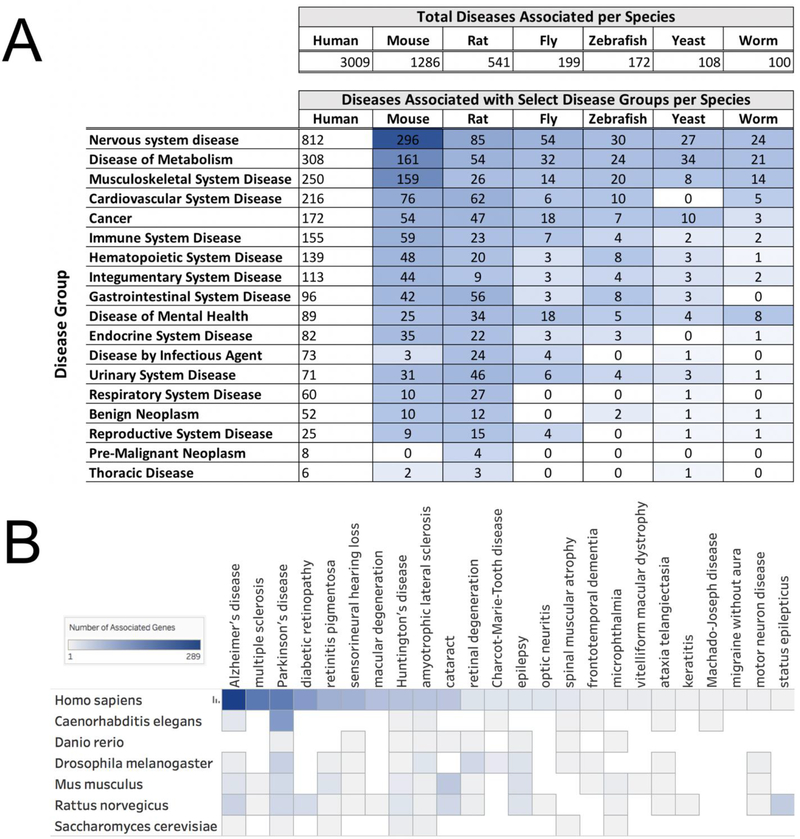
Each of the six Alliance model organisms has a dedicated organism-centric database and website to serve the needs of their specific research community. This is a strength in that the research communities each have unique needs which are best served by a dedicated resource. However, this model of discrete data storage has complicated research that is best done with data from multiple organisms. Where does one go to gather all the relevant data and how should they know when it has all been collected? For example, if a researcher wants to find out which model organisms have a model for a specific disease, they may visit each MOD and attempt to locate that information. Additional sites such as MARRVEL (http://marrvel.org/) and Gene2Function (http://gene2function.org/) bring together some of the necessary data and provide another good starting point for this search. Each model organism may or may not have a model to be found. This data aggregation step can be time consuming and error prone. To help address this challenge, the Alliance has now gathered information about human genetic diseases and related genetic models from fly, mouse, worm, yeast, rat, and zebrafish into a single location at https://alliancegenome.org (Figure 3). Having these data aggregated will facilitate searches for genetic models of human disease across these six model organisms. Human and model organism genes associated with specific human diseases and disease models can be found on the Alliance disease pages as well as individual species-specific gene pages at alliancegenome.org. These disease model data will be expanded in future releases to include experimental conditions such as treatment with alcohol in models of fetal alcohol syndrome, more complex genetic models, and model organism genotype and phenotype data.
Figure 3. The relationship between human diseases and associated genes in the Alliance model organism data.
A) The total count of human diseases with which each Alliance species has at least one gene associated in the Alliance human disease data set. Total associated diseases per species is shown at the top followed by a breakdown of the count of associated diseases in a selected subset of disease groups found in the Alliance disease search results. B) A drilldown into the ‘nervous system disease’ group showing a heat map of specific selected diseases and how many genes each species has associated via an “implicated_in” relationship. Data for figure A can be found in the “Disease Group” facet of the Disease category in the Alliance search results at https://www.alliancegenome.org/search?category=disease. Data for figure B can be downloaded from “nervous system disease” page at the Alliance: https://www.alliancegenome.org/disease/DOID:863. These data were collected from the Alliance website on March 5, 2018 and visualized using Tableau Professional Edition.
The Alliance also has a single consolidated set of orthologs including data from multiple computed and curated data sources. Several different levels of stringency are available to more or less strictly define the ortholog set. The shared orthology data view was developed as a new use of the DRSC Integrative Ortholog Prediction Tool (DIOPT)183. Upcoming releases of Alliance software and data are anticipated to include additional aggregated data types including wild type gene expression, phenotypes, genetic interactions, and genetic variants.
The Alliance MODs are also participating data providers in the NIH Data Commons Pilot Phase Consortium (DCPPC) (https://commonfund.nih.gov/bd2k/commons) which launched in December, 2017. One current aim of the DCPPC is centralized access to three major biomedical data sets: Genotype-Tissue Expression Project (GTEx; gtexportal.org, MIR:00100881)184, Trans-Omics for Precision Medicine Program (TOPMed; nhlbiwgs.org), and model organism data and tools. The GTEx project aims to facilitate the study of relationships between human genetic variation, gene expression, and additional molecular phenotypes. Summary statistics for the data included in GTEx are available at https://gtexportal.org/home/tissueSummaryPage. TOPMed aims to collect whole genome sequencing and additional -omics data to integrate with imaging, clinical, molecular, and environmental data. More extensive detail on the TOPMed program can be found here: https://www.nhlbi.nih.gov/science/trans-omics-precision-medicine-topmed-program. Discussions are currently underway in the DCPPC to establish data transfer standards and cloud storage locations for these data. Once this effort is successful, additional data sources will be added. Simplified and centralized access to biomedical data and tools is anticipated.
Metadata plays a critical role in making data sets findable by machines and people alike. The distributed location of MOD data sets and the inconsistent provision and format of metadata describing them is counter to the findability and reusability of these data as described by FAIR principles. Alliance data sets are tagged with metadata about the file contents including: date provided, data source, source release version, etc. In the future, the Alliance will participate in, and take advantage of, work done by the Data Commons on synchronizing metadata across data sets. For example, there are ongoing discussions around DATS (Data Tag Suite) meta data specifications and adapting our model as appropriate to use common frameworks185. Providing Alliance data sets in Big Data Bags (technology to aid in tagging data sets with verifiable file sizes, file manifests and defined meta-data) with Minids (lightweight identifiers that can be easily generated, dereferenced and validated globally) is also coming soon186.
Challenge 2: Unique user interfaces for similar data
The user interfaces of MOD websites evolved largely independently to best serve their specific research community. If researchers must go to six different websites to locate information, they must also learn to navigate six independently designed websites looking for similar kinds of data. To address this challenge, the Alliance aims to collect and present a variety of model organism data using standardized formats (see challenge 3) at a single location. The challenge presented by distinct user interfaces at each MOD will gradually be reduced as model organism data are brought together at the Alliance website. Researchers will increasingly be able to visit alliancegenome.org to obtain an overview of the data landscape for a particular data type for the alliance model organisms and then either obtain the desired data directly or be directed to the correct location at the MOD where the data, including organism-specific details, can be obtained. Another advantage of providing a single shared user interface is that all the model organism genes will have similar link outs to additional resources. For example, links to external resources such as MARRVEL and MIST are not implemented at every MOD. In the Alliance shared gene page interface, those links can be implemented once for all the MODs. In the future, new data displays developed for the Alliance may be implemented by other user interfaces, including the MOD websites, to display model organism data outside alliancegenome.org. Adoption of shared user interface components will help to reduce the number of unique views different websites use for similar MOD data types. Currently, the Alliance website includes pages for genes and human diseases, as well as searches for Gene Ontology terms and alleles which currently link back to the Gene Ontology Consortium website, AmiGO187, and the MOD allele pages respectively. Future releases of Alliance software are expected to include wild type gene expression data with support for comparative evaluation of these data across organisms. Additionally, shared views of genetic variant data, phenotype, and genetic/physical interaction data will be available.
Challenge 3: Lack of data format standards for certain data types
Although many data types collected at MODs are of the same type (e.g. gene expression, phenotypes, mutants, etc…), how those data have been gathered, stored, and shared has not always been as similar as one might hope. Originally, the roots (and funding) for each organism’s MOD was research-based, and consequently, each MOD independently adopted different curation methods and data structures through time for similar data types, complicating the integration of these data across species. One success story is how the Gene Ontology project started from the beginning as a consortium which included all the Alliance MODs. Consequently, these groups and many others who use the GO have always shared a single ontology for annotation and a single data exchange format. This has been an essential part of the broad success of the GO and illustrates the benefits of adopting data standardization at the outset of new projects.
Phenotype annotations may be the prototypical example of this issue. There are at least two major approaches to annotating phenotypic data188. One involves use of pre-composed terms to describe each phenotypic character. There are several pre-composed phenotype ontologies to cover various species. For example, mouse phenotypes may be recorded using terms from the pre-composed mammalian phenotype ontology, such as “abnormal otic placode morphology” (MP:0011173). Another phenotype annotation method, called “post-composition”, involves combining terms from several ontologies to describe a specific phenotype. Zebrafish morphological phenotypes are curated in this post-composition style utilizing the Zebrafish Anatomy Ontology (ZFA) and the Phenotype and Trait Ontology (PATO) ontology189. The pre-composed mouse “abnormal otic placode morphology” phenotype would be represented in a post-composed format for zebrafish by combining the ZFA term “otic placode” (ZFA:0000138) with the PATO term “morphology” (PATO:0000051) and the tag “abnormal”. This is currently displayed in ZFIN as “otic placode morphology, abnormal”.
The fundamental difference in how these data are curated and stored makes it exceedingly complex to combine and reason over the data sets correctly and without data loss. This challenge hampers correct and complete reuse of these important data. A solution to this issue has been the subject of research projects190–193, but maximizing utility of these data may be best achieved through adoption of a single shared and standardized format that is practical for general utilization. The Phenotype Exchange Format (PXF)194, a documented set of phenotype exchange standards, has been proposed to address this, but more work is needed to adapt this format to model organism phenotype data.
To address this issue in general, the Alliance has made data format standardization a high priority for all the data types being incorporated and integrated. Data types for which format standards are currently being generated include gene expression, phenotypes, disease models, alleles, genotypes, and genetic and physical interactions. Each data set submitted to the Alliance by the MODs conforms to an agreed upon standard data model. This means the data from each MOD is available in one format for each data type with the same attributes provided by all the MODs. Likewise, when retrieving data from the Alliance, all model organism data will follow the same agreed upon standards. For example, zebrafish researchers wishing to retrieve data about gene to disease relations in mouse and zebrafish, can be confident that their searches will return data from both organisms in the exact same way, with the same named attributes. Before this effort, it was possible that a search could retrieve gene to disease information from most of the MODs, but the data was provided from unique data models at each source. Each retrieval required data translation from one model to the next. Now, with one source of standardized attribute/value pairs, retrieval of data across model organisms will be much less error prone and data consumers will spend less time unifying the data. This standardization is a significant step towards supporting interoperability of these data as described by FAIR principles.
The data submission model for the Alliance is here: https://github.com/alliance-genome/agr_schemas. In addition, MOD data is being submitted to the Alliance tagged with common ontologies like the Disease Ontology, the Measurement Methods Ontology, the Sequence Ontology, and the Gene Ontology. Use of these common ontologies by all MODs facilitates interoperability among these and any other data which also use these ontologies. It is important to note that all IDs in the Alliance are CURIE (compact URI) IDs, resolvable at external resources using unique prefixes and existing identifiers, which are reused rather than re-minted. For example, ZFIN:ZDB-GENE-001103–1 is a CURIE ID for a zebrafish gene record at ZFIN.
Alliance data sets are accessible in a variety of formats. For those interested in accessing data programmatically, the Alliance provides several API endpoints, documented by swagger (https://alliancegenome.org/api/swagger-ui). Also provided are docker images of the Alliance data store and Elastic Search indexes from each Alliance release: https://hub.docker.com/r/agrdocker/agr_neo4j_qc_data_image/tags/ and https://hub.docker.com/r/agrdocker/agr_es_data_image/tags/. Raw data files, the product of data model unification across MODs, are provided in the Alliance Amazon AWS cloud storage at https://s3.amazonaws.com/mod-datadumps/. Data can also be downloaded directly from tabular data displays on the Alliance gene and disease pages. Lastly, genetic interaction data is downloadable in CSV format from the Alliance download page, accessible under the “Data” menu on the Alliance home page. It is anticipated that CSV file downloads for additional data types will be posted there in the future.
Once data format standardization is accomplished, multi-species analyses will be more tractable for both basic research and translational applications. In time, the number of MODs contributing data to the Alliance is anticipated to grow. Standardization of model organism research data formats will further reduce issues raised in Challenge 1 regarding use of distributed data.
Challenge 4: Scalability and Sustainability
Scalability and sustainability for model organism data are perhaps the most significant challenges for application of model organism data to translational medicine. The number of new publications involving Alliance model organisms continues to grow every year, with PubMed showing nearly 120,000 such publications in 2017 alone. This growth poses a scalability challenge for the model organism databases at a time when their resources are being reduced. This issue has been under active discussion for the past several years among the MOD community members and funding agencies. One part of the scalability and sustainability solution will be the Alliance itself, which aims to increase shared use of infrastructure and tools among its members whenever possible. For example, one of the early data sets to be combined in the Alliance project was genome sequence and gene model data. All the model organisms have these data and each has implemented a genome browser to display it at their MOD. Although the MODs have typically used versions of the same software (GBrowse or JBrowse) to accomplish this, effort could be conserved if all the MODs used a single centrally administered genome browser. The alliance has begun using such an instance of JBrowse in the Alliance website and standardization of genome data in GFF3 file format has been achieved. Work is ongoing to make it possible for individual MODs to move away from their individual genome browser installations if desired, in favor of using the centrally provided Alliance instance. This type of collaboration and consolidation will have the tripartite benefit of reducing the combined cost of MOD operations, facilitating development of shared data standards and curation tools, and supporting use of the same user interface at each of the MOD websites. As the work of the Alliance progresses, effort will be focused on emulating this type of shared data standard and UI when practical and possible. Improvements in scalability and sustainability will foster and increase the already large impact model organisms have had on translational medicine.
Another data type where efficiencies may be found is with the Gene Ontology data. Each of the Alliance model organisms is moving towards using the Gene Ontology curation tool, Noctua. Use of this shared curation interface will reduce the need for each model organism to support a Gene Ontology curation interface of its own and allow changes to GO curation policies and quality control of the annotations to be handled centrally by the GO consortium as part of the Alliance. This will further support standardization of these annotations across Alliance curators and make these same standards and quality controls available to any other MOD deciding to use Noctua for GO curation.
Efficiency of operations is only one aspect of the sustainability issue. The MODs are truly valuable global research resources. As such, there is an ongoing discussion on sustainable funding models for the MODS and other similar global research resources. There have been a range of potential options considered for sustainable funding including international funding mechanisms, inclusion of the MODs as part of the National Library of Medicine, funding by a broader range of NIH Institutes, and user-driven token or fee for service models. Regardless of how these genomic resources are funded, long-term stable funding for biomedical research data and infrastructure is critical for these resources to effectively plan for and meet the future needs of the research and translational medicine communities.
Discussion
Model organism research and MODs have played pivotal roles in furthering our understanding of normal functioning of biological systems as well as etiology of disease and modes of disease treatment. In addition to the six model organisms discussed here, there exist numerous others covering the complete taxonomic range. Some of these have dedicated MODs, such as Xenbase (http://xenbase.org, MIR:0100232) for Xenopus, dictyBase (http://dictybase.org, MIR:0100367) for Dictyostelium, the Arabidopsis Information Resource (TAIR; http://arabidopsis.org/, MIR:00000050) for Arabidopsis, Gramene (http://gramene.org, MIR:00000182) for over 50 crop and model plant species, and GEISHA (http://geisha.arizona.edu/geisha) for chicken, among many others4. As these MODs continue to collect, organize, and cross reference data, their value to both foundational and translational research grows. Although the focus here has been on maximizing utility of model organism data for translational research, it must not be forgotten that the MODs also provide an invaluable service to the foundational research community, from which so many novel and often unanticipated insights are derived195. Each model organism has strengths for specific types of studies and they each belong in the quiver of modern research tools. The contributions of model organism data to basic research and translational medicine are far from fully realized.
The challenges we have discussed for using model organism data in translational research include distributed data, dissimilar user interfaces for similar data, lack of data format standards, and sustainability and scalability. Complete and correct application of model organism data can be particularly challenging for users who lack the necessary expertise in model organism research and data manipulation. Addressing these challenges will have widely beneficial effects on the utility of model organism data in basic as well as translational research. The issues of distributed data, disparate user interfaces, and data format standards are all well within the scope of the model organism community to address. The Alliance has all of these as high priority items. Combining MOD data at the Alliance and committing to the application of FAIR data principles will together improve these challenges over time. The issue of scalability and sustainability is a global resource availability and allocation issue affecting all scientific data stores, not just model organism data. As such, establishment of stable and sustainable funding for critical biological databases, knowledgebases, and infrastructure is a high priority on a scale much larger than just for the Alliance. The NIH BD2K Data Commons Pilot Project is one example of new infrastructure being put in place to support these biomedical data for the future (https://commonfund.nih.gov/bd2k/commons). The challenges we have identified all point toward evolution past the time of relatively independent MODs and into a new era of synergizing “model organism data”. This new era will emphasize improved data integration, data access, data standards, shared infrastructure and tools, and translational application. The Alliance of Genome Resources is committed to this effort, driving forward towards the next step in the evolution and application of model organism data while continuing to serve the needs of each of the individual model organism research communities.
Acknowledgements
The authors would like to thank all the current and past members of FlyBase, WormBase, ZFIN, MGI, SGD, RGD, and the Gene Ontology Consortium for their dedication over the past 30 years. The resulting resources are truly amazing and have a real impact on human health.
Funding
ZFIN: National Human Genome Research Institute at the United States National Institutes of Health [U41 HG002659]
MGD: National Human Genome Research Institute at the United States National Institutes of Health [U41 HG000330, R25 HG007053]
WB: National Human Genome Research Institute at the US National Institutes of Health [U41 HG002223], the UK Medical Research Council and the UK Biotechnology and Biological Sciences Research Council.
FB: National Human Genome Research Institute at the United States National Institutes of Health [U41 HG000739]; British Medical Research Council.
SGD: National Human Genome Research Institute at the United States National Institutes of Health [U41 HG001315].
RGD: National Heart, Lung, and Blood Institute at the United States National Institutes of Health [RO1 HL64541]
GO: National Human Genome Research Institute at the United States National Institutes of Health [U41 HG002273]
Alliance of Genome Resources: National Human Genome Research Institute at the United States National Institutes of Health [U41 H002223]
Citations
- 1.Ericsson AC, Crim MJ & Franklin CL A brief history of animal modeling. Mo. Med 110, 201–5 [PMC free article] [PubMed] [Google Scholar]
- 2.Duronio RJ, O’Farrell PH, Sluder G & Su TT Sophisticated lessons from simple organisms: appreciating the value of curiosity-driven research. Dis. Model. Mech 10, 1381–1389 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krogh A THE PROGRESS OF PHYSIOLOGY. Science 70, 200–4 (1929). [DOI] [PubMed] [Google Scholar]
- 4.Rigden DJ & Fernández XM The 2018 Nucleic Acids Research database issue and the online molecular biology database collection. Nucleic Acids Res. 46, D1–D7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gene Ontology Consortium. Gene Ontology Consortium: going forward. Nucleic Acids Res. 43, D1049–56 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donlin MJ Using the Generic Genome Browser (GBrowse) Curr. Protoc. Bioinforma Chapter 9, Unit 9.9(2009). [DOI] [PubMed] [Google Scholar]
- 7.Chao H-T, Liu L & Bellen HJ Building dialogues between clinical and biomedical research through cross-species collaborations. Semin. Cell Dev. Biol 70, 49–57 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wangler MF et al. Model Organisms Facilitate Rare Disease Diagnosis and Therapeutic Research. Genetics 207, 9–27 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manolio TA et al. Bedside Back to Bench: Building Bridges between Basic and Clinical Genomic Research. Cell 169, 6–12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oliver SG, Lock A, Harris MA, Nurse P & Wood V Model organism databases: essential resources that need the support of both funders and users. BMC Biol. 14, 49(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poux S et al. On expert curation and scalability: UniProtKB/Swiss-Prot as a case study. Bioinformatics 33, 3454–3460 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gramates LS et al. FlyBase at 25: looking to the future. Nucleic Acids Res. 45, D663–D671 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wangler MF, Yamamoto S & Bellen HJ Fruit flies in biomedical research. Genetics 199, 639–53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rubin GM & Spradling AC Genetic transformation of Drosophila with transposable element vectors. Science 218, 348–53 (1982). [DOI] [PubMed] [Google Scholar]
- 15.Spradling AC et al. The Berkeley Drosophila Genome Project gene disruption project: Single P-element insertions mutating 25% of vital Drosophila genes. Genetics 153, 135–77 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bellen HJ et al. The Drosophila gene disruption project: progress using transposons with distinctive site specificities. Genetics 188, 731–43 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perrimon N, Bonini NM & Dhillon P Fruit flies on the front line: the translational impact of Drosophila. Dis. Model. Mech 9, 229–31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bilder D & Irvine KD Taking Stock of theDrosophilaResearch Ecosystem. Genetics 206, 1227–1236 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaufman TC A Short History and Description ofDrosophila melanogasterClassical Genetics: Chromosome Aberrations, Forward Genetic Screens, and the Nature of Mutations. Genetics 206, 665–689 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanca O, Bellen HJ & Schnorrer F Gene Tagging Strategies To Assess Protein Expression, Localization, and Function inDrosophila. Genetics 207, 389–412 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bier E, Harrison MM, O’Connor-Giles KM & Wildonger J Advances in Engineering the Fly Genome with the CRISPR-Cas System. Genetics 208, 1–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Germani F, Bergantinos C & Johnston LA Mosaic Analysis inDrosophila. Genetics 208, 473–490 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bandura JL et al. humpty dumpty is required for developmental DNA amplification and cell proliferation in Drosophila. Curr. Biol 15, 755–9 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Evrony GD et al. Integrated genome and transcriptome sequencing identifies a noncoding mutation in the genome replication factorDONSONas the cause of microcephaly-micromelia syndrome. Genome Res. 27, 1323–1335 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lesly S, Bandura JL & Calvi BR Rapid DNA Synthesis During EarlyDrosophilaEmbryogenesis Is Sensitive to Maternal Humpty Dumpty Protein Function. Genetics 207, 935–947 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reynolds JJ et al. Mutations in DONSON disrupt replication fork stability and cause microcephalic dwarfism. Nat. Genet 49, 537–549 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vidal M, Wells S, Ryan A & Cagan R ZD6474 suppresses oncogenic RET isoforms in a Drosophila model for type 2 multiple endocrine neoplasia syndromes and papillary thyroid carcinoma. Cancer Res. 65, 3538–41 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Dar AC, Das TK, Shokat KM & Cagan RL Chemical genetic discovery of targets and anti-targets for cancer polypharmacology. Nature 486, 80–4 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Millburn GH, Crosby MA, Gramates LS, Tweedie S & FlyBase Consortium. FlyBase portals to human disease research using Drosophila models. Dis. Model. Mech 9, 245–52 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu Y, Comjean A, Mohr SE, FlyBase Consortium N & Perrimon N Gene2Function: An Integrated Online Resource for Gene Function Discovery. G3 (Bethesda). 7, 2855–2858 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu Y et al. Molecular Interaction Search Tool (MIST): an integrated resource for mining gene and protein interaction data. Nucleic Acids Res. 46, D567–D574 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J et al. MARRVEL: Integration of Human and Model Organism Genetic Resources to Facilitate Functional Annotation of the Human Genome. Am. J. Hum. Genet 100, 843–853 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gelbart WM et al. FlyBase: a Drosophila database. The FlyBase consortium. Nucleic Acids Res. 25, 63–6 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keane TM et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477, 289–94 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.International Mouse Knockout Consortium, Collins FS, Rossant J & Wurst W. A mouse for all reasons. Cell 128, 9–13 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Bradley A et al. The mammalian gene function resource: the International Knockout Mouse Consortium. Mamm. Genome 23, 580–6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown SDM & Moore MW The International Mouse Phenotyping Consortium: past and future perspectives on mouse phenotyping. Mamm. Genome 23, 632–40 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collaborative Cross Consortium. The genome architecture of the Collaborative Cross mouse genetic reference population. Genetics 190, 389–401 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Threadgill DW & Churchill GA Ten years of the collaborative cross. G3 (Bethesda). 2, 153–6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Churchill GA, Gatti DM, Munger SC & Svenson KL The Diversity Outbred mouse population. Mamm. Genome 23, 713–8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Svenson KL et al. High-resolution genetic mapping using the Mouse Diversity outbred population. Genetics 190, 437–47 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Philip VM et al. Genetic analysis in the Collaborative Cross breeding population. Genome Res. 21, 1223–38 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Logan RW et al. High-precision genetic mapping of behavioral traits in the diversity outbred mouse population. Genes. Brain. Behav 12, 424–37 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chesler EJ Out of the bottleneck: the Diversity Outcross and Collaborative Cross mouse populations in behavioral genetics research. Mamm. Genome 25, 3–11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilke M et al. Mouse models of cystic fibrosis: phenotypic analysis and research applications. J. Cyst. Fibros 10 Suppl 2, S152–71 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Tsuji T & Kunieda T A loss-of-function mutation in natriuretic peptide receptor 2 (Npr2) gene is responsible for disproportionate dwarfism in cn/cn mouse. J. Biol. Chem 280, 14288–92 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Morelli KH et al. Severity of Demyelinating and Axonal Neuropathy Mouse Models Is Modified by Genes Affecting Structure and Function of Peripheral Nodes. Cell Rep. 18, 3178–3191 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu W-H et al. CRISPR Repair Reveals Causative Mutation in a Preclinical Model of Retinitis Pigmentosa. Mol. Ther 24, 1388–94 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Metzger MW et al. Heterozygosity for the Mood Disorder-Associated Variant Gln460Arg Alters P2X7 Receptor Function and Sleep Quality. J. Neurosci 37, 11688–11700 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bjursell M et al. Therapeutic Genome Editing With CRISPR/Cas9 in a Humanized Mouse Model Ameliorates α1-antitrypsin Deficiency Phenotype. EBioMedicine (2018). doi: 10.1016/j.ebiom.2018.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mali P et al. RNA-guided human genome engineering via Cas9. Science 339, 823–6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang H et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hara S & Takada S Genome editing for the reproduction and remedy of human diseases in mice. J. Hum. Genet 63, 107–113 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Molenhuis RT, Bruining H & Kas MJ Modelling Autistic Features in Mice Using Quantitative Genetic Approaches. Adv. Anat. Embryol. Cell Biol 224, 65–84 (2017). [DOI] [PubMed] [Google Scholar]
- 55.St Clair D & Johnstone M Using mouse transgenic and human stem cell technologies to model genetic mutations associated with schizophrenia and autism. Philos. Trans. R. Soc. Lond. B. Biol. Sci 373, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Attie AD, Churchill GA & Nadeau JH How mice are indispensable for understanding obesity and diabetes genetics. Curr. Opin. Endocrinol. Diabetes. Obes 24, 83–91 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skelton JK, Ortega-Prieto AM & Dorner M A Hitchhiker’s guide to humanized mice: new pathways to studying viral infections. Immunology (2018). doi: 10.1111/imm.12906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gunawan M et al. A Novel Human Systemic Lupus Erythematosus Model in Humanised Mice. Sci. Rep 7, 16642(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kitada M, Ogura Y & Koya D Rodent models of diabetic nephropathy: their utility and limitations. Int. J. Nephrol. Renovasc. Dis 9, 279–290 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leite JP, Garcia-Cairasco N & Cavalheiro EA New insights from the use of pilocarpine and kainate models. Epilepsy Res. 50, 93–103 (2002). [DOI] [PubMed] [Google Scholar]
- 61.Cenci MA & Crossman AR Animal models of l-dopa-induced dyskinesia in Parkinson’s disease. Mov. Disord (2018). doi: 10.1002/mds.27337 [DOI] [PubMed] [Google Scholar]
- 62.Kless C, Rink N, Rozman J & Klingenspor M Proximate causes for diet-induced obesity in laboratory mice: a case study. Eur. J. Clin. Nutr 71, 306–317 (2017). [DOI] [PubMed] [Google Scholar]
- 63.Combe R et al. How Does Circadian Rhythm Impact Salt Sensitivity of Blood Pressure in Mice? A Study in Two Close C57Bl/6 Substrains. PLoS One 11, e0153472(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dickson PE et al. Association of novelty-related behaviors and intravenous cocaine self-administration in Diversity Outbred mice. Psychopharmacology (Berl). 232, 1011–24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cervantes MC, Laughlin RE & Jentsch JD Cocaine self-administration behavior in inbred mouse lines segregating different capacities for inhibitory control. Psychopharmacology (Berl). 229, 515–25 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sittig LJ et al. Genetic Background Limits Generalizability of Genotype-Phenotype Relationships. Neuron 91, 1253–1259 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chesler EJ et al. Quantitative trait loci for sensitivity to ethanol intoxication in a C57BL/6J×129S1/SvImJ inbred mouse cross. Mamm. Genome 23, 305–21 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thompson MB The Min mouse: a genetic model for intestinal carcinogenesis. Toxicol. Pathol 25, 329–32 [DOI] [PubMed] [Google Scholar]
- 69.Dietrich WF et al. Genetic identification of Mom-1, a major modifier locus affecting Min-induced intestinal neoplasia in the mouse. Cell 75, 631–9 (1993). [DOI] [PubMed] [Google Scholar]
- 70.MacPhee M et al. The secretory phospholipase A2 gene is a candidate for the Mom1 locus, a major modifier of ApcMin-induced intestinal neoplasia. Cell 81, 957–66 (1995). [DOI] [PubMed] [Google Scholar]
- 71.Kennedy BP et al. A natural disruption of the secretory group II phospholipase A2 gene in inbred mouse strains. J. Biol. Chem 270, 22378–85 (1995). [DOI] [PubMed] [Google Scholar]
- 72.Quach ND, Arnold RD & Cummings BS Secretory phospholipase A2 enzymes as pharmacological targets for treatment of disease. Biochem. Pharmacol 90, 338–48 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yarla NS et al. Phospholipase A2 Isoforms as Novel Targets for Prevention and Treatment of Inflammatory and Oncologic Diseases. Curr. Drug Targets 17, 1940–1962 (2016). [DOI] [PubMed] [Google Scholar]
- 74.Shultz LD et al. Human Cancer Growth and Therapy in Immunodeficient Mouse Models. Cold Spring Harb. Protoc 2014, pdb.top073585–pdb.top073585 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang M et al. Humanized mice in studying efficacy and mechanisms of PD-1-targeted cancer immunotherapy. FASEB J. 32, 1537–1549 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pauli C et al. PersonalizedIn VitroandIn VivoCancer Models to Guide Precision Medicine. Cancer Discov. 7, 462–477 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dobrolecki LE et al. Patient-derived xenograft (PDX) models in basic and translational breast cancer research. Cancer Metastasis Rev. 35, 547–573 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Williams JA Using PDX for Preclinical Cancer Drug Discovery: The Evolving Field. J. Clin. Med 7, 41(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pan C et al. Development and Characterization of Bladder Cancer Patient-Derived Xenografts for Molecularly Guided Targeted Therapy. PLoS One 10, e0134346(2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Garralda E et al. Integrated next-generation sequencing and avatar mouse models for personalized cancer treatment. Clin. Cancer Res 20, 2476–84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hidalgo M et al. A Pilot Clinical Study of Treatment Guided by Personalized Tumorgrafts in Patients with Advanced Cancer. Mol. Cancer Ther 10, 1311–1316 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Smith CL et al. Mouse Genome Database (MGD)-2018: knowledgebase for the laboratory mouse. Nucleic Acids Res. 46, D836–D842 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Finger JH et al. The mouse Gene Expression Database (GXD): 2017 update. Nucleic Acids Res. 45, D730–D736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Krupke DM et al. The Mouse Tumor Biology Database: A Comprehensive Resource for Mouse Models of Human Cancer. Cancer Res. 77, e67–e70 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Drabkin HJ, Blake JA & Mouse Genome Informatics Database. Manual Gene Ontology annotation workflow at the Mouse Genome Informatics Database. Database 2012, bas045–bas045 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Aitman T, Dhillon P & Geurts AM A RATional choice for translational research? Dis. Model. Mech 9, 1069–1072 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jacob HJ et al. Genetic dissection of autoimmune type I diabetes in the BB rat. Nat. Genet 2, 56–60 (1992). [DOI] [PubMed] [Google Scholar]
- 88.Rapp JP Genetic analysis of inherited hypertension in the rat. Physiol. Rev 80, 135–72 (2000). [DOI] [PubMed] [Google Scholar]
- 89.Remmers EF et al. A genome scan localizes five non-MHC loci controlling collagen-induced arthritis in rats. Nat. Genet 14, 82–5 (1996). [DOI] [PubMed] [Google Scholar]
- 90.Shepel LA et al. Genetic identification of multiple loci that control breast cancer susceptibility in the rat. Genetics 149, 289–99 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jacob HJ, Lazar J, Dwinell MR, Moreno C & Geurts AM Gene targeting in the rat: advances and opportunities. Trends Genet. 26, 510–8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jacob H From rat pathophysiology to genomic medicine: an interview with Howard Jacob. Dis. Model. Mech 9, 1073–1077 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Robbins TW Cross-species studies of cognition relevant to drug discovery: a translational approach. Br. J. Pharmacol 174, 3191–3199 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jordan VC Proven value of translational research with appropriate animal models to advance breast cancer treatment and save lives: the tamoxifen tale. Br. J. Clin. Pharmacol 79, 254–67 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chou MY & Mani A A successful story of translational orthodontic research: Micro-osteoperforation-from experiments to clinical practice. APOS Trends Orthod 7, 6–11 (2017). [Google Scholar]
- 96.Teixeira CC et al. Cytokine expression and accelerated tooth movement. J. Dent. Res 89, 1135–41 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Winge Ø On haplophase and diplophase of some Saccharomycetes. C.R. Trav. Lab. Carlsberg, Ser. Physiol 21, 77–111 (1935). [Google Scholar]
- 98.Lindegren CC The Yeast Cell: Its Genetics and Cytology. (Education Publishers Inc., 1949). [Google Scholar]
- 99.LINDEGREN CC, LINDEGREN G, SHULT EE & DESBOROUGH S Chromosome maps of Saccharomyces. Nature 183, 800–802 (1959). [DOI] [PubMed] [Google Scholar]
- 100.Lindegren CC & Lindegren G Linkage relationships in Saccharomyces of genes controlling the fermentation of carbohydrates and the synthesis of vitamins, amino acids and nucleic acid components. Indian Phyt opahtol. 4, 11–20 (1951). [Google Scholar]
- 101.Goffeau A et al. Life with 6000 genes. Science 274, 546,563–567 (1996). [DOI] [PubMed] [Google Scholar]
- 102.Hieter P et al. Functional selection and analysis of yeast centromeric DNA. Cell 42, 913–921 (1985). [DOI] [PubMed] [Google Scholar]
- 103.Deshpande AM & Newlon CS The ARS consensus sequence is required for chromosomal origin function in Saccharomyces cerevisiae. Mol. Cell. Biol 12, 4305–4313 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Louis EJ, Naumova ES, Lee A, Naumov G & Haber JE The chromosome end in yeast: its mosaic nature and influence on recombinational dynamics. Genetics 136, 789–802 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lowe TM & Eddy SR tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lowe TM & Eddy SR A computational screen for methylation guide snoRNAs in yeast. Science 283, 1168–1171 (1999). [DOI] [PubMed] [Google Scholar]
- 107.Planta RJ & Mager WH The list of cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Yeast 14, 471–477 (1998). [DOI] [PubMed] [Google Scholar]
- 108.Kim JM, Vanguri S, Boeke JD, Gabriel A & Voytas DF Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 8, 464–478 (1998). [DOI] [PubMed] [Google Scholar]
- 109.Winzeler EA et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906 (1999). [DOI] [PubMed] [Google Scholar]
- 110.Giaever G et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391 (2002). [DOI] [PubMed] [Google Scholar]
- 111.Ghaemmaghami S et al. Global analysis of protein expression in yeast. Nature 425, 737–741 (2003). [DOI] [PubMed] [Google Scholar]
- 112.Huh W-K et al. Global analysis of protein localization in budding yeast. Nature 425, 686–691 (2003). [DOI] [PubMed] [Google Scholar]
- 113.Costanzo M et al. A global genetic interaction network maps a wiring diagram of cellular function. Science 353, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Costanzo M et al. The genetic landscape of a cell. Science 327, 425–431 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Botstein D & Fink GR Yeast: an experimental organism for 21st Century biology. Genetics 189, 695–704 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lasserre J-P et al. Yeast as a system for modeling mitochondrial disease mechanisms and discovering therapies. Dis. Model. Mech 8, 509–26 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dolinski K & Botstein D Orthology and functional conservation in eukaryotes. Annu. Rev. Genet 41, 465–507 (2007). [DOI] [PubMed] [Google Scholar]
- 118.Engel SR & Cherry JM The new modern era of yeast genomics: community sequencing and the resulting annotation of multiple Saccharomyces cerevisiae strains at the Saccharomyces Genome Database. Database (Oxford). 2013, bat012(2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Novo M et al. Eukaryote-to-eukaryote gene transfer events revealed by the genome sequence of the wine yeast Saccharomyces cerevisiae EC1118. Proc. Natl. Acad. Sci. U. S. A 106, 16333–16338 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wenger JW, Schwartz K & Sherlock G Bulk segregant analysis by high-throughput sequencing reveals a novel xylose utilization gene from Saccharomyces cerevisiae. PLoS Genet. 6, e1000942(2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Libkind D et al. Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc. Natl. Acad. Sci. U. S. A 108, 14539–14544 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kachroo AH et al. Evolution. Systematic humanization of yeast genes reveals conserved functions and genetic modularity. Science 348, 921–925 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kachroo AH et al. Systematic bacterialization of yeast genes identifies a near-universally swappable pathway. Elife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gabaldon T & Koonin EV Functional and evolutionary implications of gene orthology. Nature reviews. Genetics 14, 360–366 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Skrzypek MS et al. Saccharomyces genome database informs human biology. Nucleic Acids Res. 46, D736–D742 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Apfeld J & Alper S What Can We Learn About Human Disease from the Nematode C. elegans? Methods Mol. Biol 1706, 53–75 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Riessland M et al. Neurocalcin Delta Suppression Protects against Spinal Muscular Atrophy in Humans and across Species by Restoring Impaired Endocytosis. Am. J. Hum. Genet 100, 297–315 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Culetto E & Sattelle DB A role for Caenorhabditis elegans in understanding the function and interactions of human disease genes. Hum. Mol. Genet 9, 869–877 (2000). [DOI] [PubMed] [Google Scholar]
- 129.Ganner A & Neumann-Haefelin E Genetic kidney diseases: Caenorhabditis elegans as model system. Cell Tissue Res. 369, 105–118 (2017). [DOI] [PubMed] [Google Scholar]
- 130.Bank EM & Gruenbaum Y Caenorhabditis elegans as a model system for studying the nuclear lamina and laminopathic diseases. Nucleus 2, 350–357 (2011). [DOI] [PubMed] [Google Scholar]
- 131.Lin J & Hackam DJ Worms, flies and four-legged friends: the applicability of biological models to the understanding of intestinal inflammatory diseases. Dis. Model. Mech 4, 447–456 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Williams MJ, Almen MS, Fredriksson R & Schioth HB What model organisms and interactomics can reveal about the genetics of human obesity. Cell. Mol. Life Sci 69, 3819–3834 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Howe KL et al. WormBase 2016: expanding to enable helminth genomic research. Nucleic Acids Res. 44, D774–80 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kim D-K, Kim TH & Lee S-J Mechanisms of aging-related proteinopathies in Caenorhabditis elegans. Exp. Mol. Med 48, e263(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Alexander AG, Marfil V & Li C Use of Caenorhabditis elegans as a model to study Alzheimer’s disease and other neurodegenerative diseases. Front. Genet 5, 279(2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ma L et al. Caenorhabditis elegans as a model system for target identification and drug screening against neurodegenerative diseases. Eur. J. Pharmacol 819, 169–180 (2018). [DOI] [PubMed] [Google Scholar]
- 137.Griffin EF, Caldwell KA & Caldwell GA Genetic and Pharmacological Discovery for Alzheimer’s Disease Using Caenorhabditis elegans. ACS Chem. Neurosci 8, 2596–2606 (2017). [DOI] [PubMed] [Google Scholar]
- 138.Hindle S, Hebbar S & Sweeney ST Invertebrate models of lysosomal storage disease: what have we learned so far? Invert. Neurosci 11, 59–71 (2011). [DOI] [PubMed] [Google Scholar]
- 139.de Voer G, Peters D & Taschner PEM Caenorhabditis elegans as a model for lysosomal storage disorders. Biochim. Biophys. Acta 1782, 433–46 (2008). [DOI] [PubMed] [Google Scholar]
- 140.Laale H The biology and use of zebrafish Brachydanio rerio in fisheries research: a literature review. J. Fish Biol 10, 121–173 (1977). [Google Scholar]
- 141.Fishman MC Genomics. Zebrafish--the canonical vertebrate. Science 294, 1290–1291 (2001). [DOI] [PubMed] [Google Scholar]
- 142.Phillips JB & Westerfield M Zebrafish models in translational research: tipping the scales toward advancements in human health. Dis. Model. Mech 7, 739–43 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lieschke GJ & Currie PD Animal models of human disease: zebrafish swim into view. Nat. Rev. Genet 8, 353–367 (2007). [DOI] [PubMed] [Google Scholar]
- 144.Howe DGDG et al. ZFIN, the Zebrafish Model Organism Database: increased support for mutants and transgenics. Nucleic Acids Res. 41, D854–60 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bradford YM et al. Zebrafish Models of Human Disease: Gaining Insight into Human Disease at ZFIN. ILAR J. 58, 4–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Berger J & Currie PD Zebrafish models flex their muscles to shed light on muscular dystrophies. Dis. Model. Mech 5, 726–32 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Taylor AM & Zon LI Modeling Diamond Blackfan anemia in the zebrafish. Semin. Hematol 48, 81–88 (2011). [DOI] [PubMed] [Google Scholar]
- 148.Pena IA et al. Pyridoxine-Dependent Epilepsy in Zebrafish Caused by Aldh7a1 Deficiency. Genetics 207, 1501–1518 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cortelazzo A et al. Proteomic analysis of the Rett syndrome experimental model mecp2Q63Xmutant zebrafish. J. Proteomics 154, 128–133 (2017). [DOI] [PubMed] [Google Scholar]
- 150.Pietri T et al. The first mecp2-null zebrafish model shows altered motor behaviors. Front. Neural Circuits 7, 118(2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gao H et al. Mecp2 regulates neural cell differentiation by suppressing the Id1 to Her2 axis in zebrafish. J. Cell Sci 128, 2340–50 (2015). [DOI] [PubMed] [Google Scholar]
- 152.Noël ES et al. A Zebrafish Loss-of-Function Model for Human CFAP53 Mutations Reveals Its Specific Role in Laterality Organ Function. Hum. Mutat 37, 194–200 (2016). [DOI] [PubMed] [Google Scholar]
- 153.Cast AE, Gao C, Amack JD & Ware SM An essential and highly conserved role for Zic3 in left-right patterning, gastrulation and convergent extension morphogenesis. Dev. Biol 364, 22–31 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wu S-Y et al. Expression of Cataract-linked gamma-Crystallin Variants in Zebrafish Reveals a Proteostasis Network That Senses Protein Stability. J. Biol. Chem 291, 25387–25397 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hunyadi B, Siekierska A, Sourbron J, Copmans D & de Witte PAM Automated analysis of brain activity for seizure detection in zebrafish models of epilepsy. J. Neurosci. Methods 287, 13–24 (2017). [DOI] [PubMed] [Google Scholar]
- 156.Feng C-W et al. Effects of 6-hydroxydopamine exposure on motor activity and biochemical expression in zebrafish (Danio rerio) larvae. Zebrafish 11, 227–39 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Díaz-Casado ME et al. Melatonin rescues zebrafish embryos from the parkinsonian phenotype restoring the parkin/PINK1/DJ-1/MUL1 network. J. Pineal Res 61, 96–107 (2016). [DOI] [PubMed] [Google Scholar]
- 158.Seth A, Stemple DL & Barroso I The emerging use of zebrafish to model metabolic disease. Dis. Model. Mech 6, 1080–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chu C-Y et al. Overexpression of Akt1 enhances adipogenesis and leads to lipoma formation in zebrafish. PLoS One 7, e36474(2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Song Y & Cone RD Creation of a genetic model of obesity in a teleost. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol 21, 2042–2049 (2007). [DOI] [PubMed] [Google Scholar]
- 161.Oka T et al. Diet-induced obesity in zebrafish shares common pathophysiological pathways with mammalian obesity. BMC Physiol. 10, 21(2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Montalbano G et al. Morphological differences in adipose tissue and changes in BDNF/Trkb expression in brain and gut of a diet induced obese zebrafish model. Ann. Anat 204, 36–44 (2016). [DOI] [PubMed] [Google Scholar]
- 163.Chakraborty C, Hsu CH, Wen ZH, Lin CS & Agoramoorthy G Zebrafish: a complete animal model for in vivo drug discovery and development. Curr. Drug Metab 10, 116–124 (2009). [DOI] [PubMed] [Google Scholar]
- 164.Parng C, Seng WL, Semino C & McGrath P Zebrafish: a preclinical model for drug screening. Assay Drug Dev. Technol 1, 41–48 (2002). [DOI] [PubMed] [Google Scholar]
- 165.Williams CH & Hong CC Zebrafish small molecule screens: Taking the phenotypic plunge. Comput. Struct. Biotechnol. J 14, 350–356 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Deveau AP, Bentley VL & Berman JN Using zebrafish models of leukemia to streamline drug screening and discovery. Exp. Hematol 45, 1–9 (2017). [DOI] [PubMed] [Google Scholar]
- 167.White RM et al. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature 471, 518–22 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ordas A et al. Testing tuberculosis drug efficacy in a zebrafish high-throughput translational medicine screen. Antimicrob. Agents Chemother 59, 753–62 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Baxendale S, van Eeden F & Wilkinson R The Power of Zebrafish in Personalised Medicine. Adv. Exp. Med. Biol 1007, 179–197 (2017). [DOI] [PubMed] [Google Scholar]
- 170.Wu J-Q et al. Patient-derived xenograft in zebrafish embryos: a new platform for translational research in gastric cancer. J. Exp. Clin. Cancer Res 36, 160(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Gaudenzi G et al. Patient-derived xenograft in zebrafish embryos: a new platform for translational research in neuroendocrine tumors. Endocrine 57, 214–219 (2017). [DOI] [PubMed] [Google Scholar]
- 172.Duck G, Nenadic G, Brass A, Robertson DL & Stevens R bioNerDS: exploring bioinformatics’ database and software use through literature mining. BMC Bioinformatics 14, 194(2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Beagrie N & Houghton J The Value and Impact of the European Bioinformatics Institute. (2016). [Google Scholar]
- 174.Hirsch T et al. Regeneration of the entire human epidermis using transgenic stem cells. Nature 551, 327–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Smedley D et al. Next-generation diagnostics and disease-gene discovery with the Exomiser. Nat. Protoc 10, 2004–2015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bone WP et al. Computational evaluation of exome sequence data using human and model organism phenotypes improves diagnostic efficiency. Genet. Med 18, 608–617 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kohler S et al. The Human Phenotype Ontology in 2017. Nucleic Acids Res. 45, D865–D876 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Chong JX et al. The Genetic Basis of Mendelian Phenotypes: Discoveries, Challenges, and Opportunities. Am. J. Hum. Genet 97, 199–215 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Picher-Martel V et al. From animal models to human disease: a genetic approach for personalized medicine in ALS. Acta Neuropathol. Commun 4, 70(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Renna M, Jimenez-Sanchez M, Sarkar S & Rubinsztein DC Chemical inducers of autophagy that enhance the clearance of mutant proteins in neurodegenerative diseases. J. Biol. Chem 285, 11061–7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Bond M, Holthaus S-MK, Tammen I, Tear G & Russell C Use of model organisms for the study of neuronal ceroid lipofuscinosis. Biochim. Biophys. Acta 1832, 1842–65 (2013). [DOI] [PubMed] [Google Scholar]
- 182.Wilkinson MD et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. data 3, 160018(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hu Y et al. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics 12, 357(2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.GTEx consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet 45, 580–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Sansone S-A et al. DATS, the data tag suite to enable discoverability of datasets. Sci. data 4, 170059(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.K C. et al. I’ll take that to go: Big data bags and minimal identifiers for exchange of large, complex datasets. in IEEE International Conference on Big Data (Big Data) 319–328 (2016). [Google Scholar]
- 187.Carbon S et al. AmiGO: online access to ontology and annotation data. Bioinformatics 25, 288–289 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Gkoutos GV, Schofield PN & Hoehndorf R The anatomy of phenotype ontologies: principles, properties and applications. Brief. Bioinform (2017). doi: 10.1093/bib/bbx035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Sprague J et al. The Zebrafish Information Network: the zebrafish model organism database provides expanded support for genotypes and phenotypes. Nucleic Acids Res. 36, D768–72 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Köhler S et al. Clinical interpretation of CNVs with cross-species phenotype data. J. Med. Genet 51, 766–772 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Köhler S et al. Construction and accessibility of a cross-species phenotype ontology along with gene annotations for biomedical research. F1000Research 2, 30(2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Rodríguez-García MÁ, Gkoutos GV, Schofield PN & Hoehndorf R Integrating phenotype ontologies with PhenomeNET. J. Biomed. Semantics 8, 58(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Oliveira D & Pesquita C Improving the interoperability of biomedical ontologies with compound alignments. J. Biomed. Semantics 9, 1(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Haendel M Phenopackets: Making phenotype profiles FAIR++ for disease diagnosis and discovery. FigShare (2016). doi: 10.6084/m9.figshare.3180898.v1 [DOI] [Google Scholar]
- 195.Rine J A future of the model organism model. Mol. Biol. Cell 25, 549–553 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]