The gene responsible for the plenty hypernodulation phenotype in Lotus japonicus was identified. The enzymatic activity and Golgi localization of PLENTY demonstrated its function as a post-translational modification enzyme.
Keywords: Arabinosyltransferase, CLE peptide, glycosylation, legume–rhizobial symbiosis, Lotus japonicus, nodulation control, post-translational modification
Abstract
Legumes can survive in nitrogen-deficient environments by forming root-nodule symbioses with rhizobial bacteria; however, forming nodules consumes energy, and nodule numbers must thus be strictly controlled. Previous studies identified major negative regulators of nodulation in Lotus japonicus, including the small peptides CLAVATA3/ESR (CLE)-RELATED-ROOT SIGNAL1 (CLE-RS1), CLE-RS2, and CLE-RS3, and their putative major receptor HYPERNODULATION AND ABERRANT ROOT FORMATION1 (HAR1). CLE-RS2 is known to be expressed in rhizobia-inoculated roots, and is predicted to be post-translationally arabinosylated, a modification essential for its activity. Moreover, all three CLE-RSs suppress nodulation in a HAR1-dependent manner. Here, we identified PLENTY as a gene responsible for the previously isolated hypernodulation mutant plenty. PLENTY encoded a hydroxyproline O-arabinosyltransferase orthologous to ROOT DETERMINED NODULATION1 in Medicago truncatula. PLENTY was localized to the Golgi, and an in vitro analysis of the recombinant protein demonstrated its arabinosylation activity, indicating that CLE-RS1/2/3 may be substrates for PLENTY. The constitutive expression experiments showed that CLE-RS3 was the major candidate substrate for PLENTY, suggesting the substrate preference of PLENTY for individual CLE-RS peptides. Furthermore, a genetic analysis of the plenty har1 double mutant indicated the existence of another PLENTY-dependent and HAR1-independent pathway negatively regulating nodulation.
Introduction
Legumes have evolved the ability to make specialized root organs called nodules, in which nitrogen-fixing symbionts, rhizobial bacteria, reside. This symbiosis enables legumes to grow on nitrogen-limited soils; however, the beneficial nitrogen acquisition is balanced against the high energy inputs required to fuel cell division in the nodules, as well as the allocation of photoassimilates to the rhizobia (Udvardi and Poole, 2013). Legumes have therefore developed mechanisms for maintaining the symbiotic balance between forming nodules to satisfy their nitrogen requirements and sustaining the energy levels required for other biological processes; these mechanisms are termed the autoregulation of nodulation (AON) (Kosslak and Bohlool, 1984; Caetano-Anollés and Gresshoff, 1990, 1991). The potential AON mechanisms involving systemic signaling between the root and shoot were originally determined using split-root and grafting techniques (Kosslak and Bohlool, 1984; Delves et al., 1986), and the molecules participating in the negative control of nodulation have been elucidated (reviewed by (Magori and Kawaguchi, 2009; Reid et al., 2011b; Suzaki et al., 2015).
The current plausible model of AON via a systemic root-derived signal involves the expression of the systemic negative regulators, such as CLAVATA3/ESR (CLE)-RELATED-ROOT SIGNAL1 (CLE-RS1), CLE-RS2, and CLE-RS3 in Lotus japonicus, directly induced by transcription factors, NODULE INCEPTION (NIN) or NIN-like protein NITRATE UNRESPONSIVE SYMBIOSIS1 (NRSYM1) (Soyano et al., 2014; Nishida et al., 2016; Nishida and Suzaki, 2018; Nishida et al., 2018). Other related CLE peptides negatively regulating nodulation have also been found in L. japonicus and other legumes, including Medicago truncatula, pea (Pisum sativum), and soybean (Glycine max) (Mortier et al., 2010,
Other AON factors closely related to PLENTY, namely ROOT DETERMINED NODULATION1 (RDN1) and NODULATION3 (NOD3), were found in M. truncatula and in pea, respectively. MtRDN1 and PsNOD3 are orthologous and have been suggested as factors that function in the root but not in the shoot (Li et al., 2009; Novák, 2010; Schnabel et al., 2011; Osipova et al., 2012; Kassaw et al., 2017), but their molecular functions had been unclear. Prior to the functional identification of MtRDN1/PsNOD3, their Arabidopsis homologs were reported as hydroxyproline O-arabinosyltransferases (HPATs) and termed HPAT1 (At2g25260), HPAT2 (At5g13500), and HPAT3 (At5g25265), which have recently been classified into the GT95 glycosyltransferase family (Showalter and Basu, 2016). AtHPATs redundantly contribute to transferring an l-arabinosyl residue to the hydroxyl group of the hydroxyproline residues of several substrates, including extensin, AtCLE2, and Arabidopsis plant peptide containing sulfated tyrosine 1 (AtPSY1) (Ogawa-Ohnishi et al., 2013; MacAlister et al., 2016). This discovery raised the possibility that LjPLENTY, MtRDN1, or PsNOD3 mediates the arabinosylation of their CLE peptides functioning in AON in the respective species. Additionally, a recent study of the MtRDNs reported that, of the two CLE peptides, only MtCLE12 is required for a functional MtRDN1 to repress nodulation, suggesting that MtRDN1 post-translationally modifies MtCLE12 but not MtCLE13 (Kassaw et al., 2017).
The plenty mutant was previously isolated as a hypernodulator, but the gene responsible for this phenotype has not yet been identified (Yoshida et al., 2010). Here, we identify the plenty locus and characterize PLENTY as an ortholog of MtRDN1 and PsNOD3 (Postma et al., 1988; Sagan and Duc, 1996; Schnabel et al., 2011). We determine the localization of PLENTY to the Golgi and purified recombinant PLENTY protein to detect its HPAT activity. This biochemical assay using an artificially synthesized peptide as a substrate for arabinosylation provided direct molecular evidence that PLENTY acts as a post-translational modification HPAT enzyme. Furthermor e, the constitutive expression of CLE-RS1/2/3 in plenty suggested that PLENTY preferentially associates with CLE-RS3 peptides as substrate. Finally, plenty har1 double mutant analysis indicates the existence of an unknown substrate for PLENTY other than CLE-RS1/2/3, which is not accepted by HAR1. These findings provide a molecular clue for understanding how PLENTY regulates nodulation.
Materials and methods
Growth conditions
Lotus japonicus (Miyakojima MG-20) seeds were sown in sterilized vermiculite, to which Broughton and Dilworth (B&D) solution (Broughton and Dilworth, 1971) containing 0.5 mM KNO3 . Seeds were inoculated with Mesorhizobium loti MAFF303099, while the control seedlings were not inoculated. Seedlings were grown under 16 h light/8 h dark cycles at 24 °C.
Map-based cloning, genomic PCR, and 5'-/3'-RACE
The plenty mutants were backcrossed three times to the parental plant ‘Miyakojima MG-20’ and crossed with another accession, ‘Gifu B-129’. DNA polymorphisms in the genomes of the 1087 F2 progeny displaying the plenty mutant phenotype were identified using a series of genetic markers (http://www.kazusa.or.jp/lotus/markerdb_index.html, last accessed 25 October 2018) and two genetic markers newly developed in this study, EY004 and EY005 (see Supplementary Table S1 at JXB online). Genomic DNA was extracted with a DNeasy Plant Mini Kit (Qiagen). The ~16 kb deletion in the plenty mutant was identified using genomic PCR (Supplementary Fig. S1), with the sets of primers shown in Supplementary Table S2. The 5' and 3' ends of PLENTY, PLENTY2, and PLENTY3 were determined using a SMARTer® RACE cDNA Amplification Kit (Clontech), utilizing RNA extracted from whole roots inoculated with M. loti MAFF303099. The DDBJ accession numbers for the PLENTY, PLENTY2, and PLENTY3 mRNAs were LC010646–LC010648 (http://getentry.ddbj.nig.ac.jp/top-e.html, last accessed 25 October 2018).
Plasmid construction
A deletion series of PLENTY coding sequence (CDS), as well as a full-length CDS, were generated using reverse transcription and the appropriate primer sets (Supplementary Table S2), cloned into the pGEM®-T-Easy vector (Promega) using the TA strategy, and named pGEM-Full-PLENTY, pGEM-ΔN1-PLENTY, and pGEM-N-PLENTY. The full-length CDS clone starts at the seventh predicted codon of PLENTY compared with the previous report of MtRDN1 (Schnabel et al., 2011; Supplementary Appendix S2); at the time it was generated, that was the predicted start of the CDS. Next, each construct was digested with EcoRI and SpeI, inserted into the Gateway-based entry plasmid pJL-Blue (Suzaki et al., 2012), and named pJL-Blue-Full-PLENTY, pJL-Blue-ΔN1-PLENTY, and pJL-Blue-N-PLENTY, respectively. For the complementation test, full-length PLENTY CDS fragments were inserted into the Gateway site of pUB-GW-HYG using an LR recombination reaction (Invitrogen) to generate pUB-GW-Full-PLENTY (Maekawa et al., 2008). To produce PLENTY–green fluorescent protein (GFP) fusion proteins, the stop codon of each pJL-Blue-based entry vector was mutagenized using circular PCR with a phosphorylated set of primers (Supplementary Table S2), and the vector was then digested with DpnI and self-ligated. The subcellular localization of PLENTY was analyzed using a series of PLENTY fragments from the mutagenized pJL-Blue-based entry vectors, which were inserted into pUGW5 for particle bombardment into onion (Allium cepa) epidermal cells and L. japonicus roots. The PLENTY fragments were also inserted into pGWB5 for the Agrobacterium tumefaciens-mediated infiltration of Nicotiana benthamiana using an LR recombination reaction (Invitrogen). The two constructs, pUGW5 and pGWB5 (Nakagawa et al., 2007), were kindly provided by Dr Mano (National Institute for Basic Biology) and Dr Nakagawa (Shimane University), respectively. To detect the HPAT activities of the three PLENTY CDS fragments (full-length, ΔN1, and ΔN2), they were C-terminally fused to a FLAG tag using pGEM-Full-PLENTY as a template for the PCR (see Supplementary Table S2 for the list of primers). The fragments were inserted into pYES2 (Invitrogen) and linearized with BamHI in an In-Fusion reaction (Takara). The plasmids used for the constitutive expression of GUS (β-glucuronidase), CLE3, CLE-RS1/2 (Okamoto et al., 2009) and CLE-RS3 (Nishida et al., 2016) were previously described.
Hairy root and stable transformations of L. japonicus
Hairy root and stable transformations of L. japonicus were performed using the Agrobacterium rhizogenes strain AR1193 alone or in combination with the Agrobacterium tumefaciens strain AGL1 harboring the respective plasmids, as described previously (http://www.bio-protocol.org/wenzhang.aspx?id=795, http://www.bio-protocol.org/wenzhang.aspx?id=796, last accessed 25 October 2018). In the hairy root transformations constitutively expressing GUS, CLE3, and CLE-RS1/2/3, GFP fluorescence was used as a marker of transformation, and the successfully transformed roots were inoculated with M. loti MAFF303099. The nodule numbers and other phenotypes of the hairy roots and stably transformed roots were measured at 14 days after inoculation (DAI), while the growth phenotypes of the stable transformants were measured at 8 weeks after inoculation.
Phylogenetic analysis
Forty-one amino acid sequences were obtained from the study of MtRDN1 (Schnabel et al., 2011) and an additional 16 amino acid sequences of Brassica rapa Chiifu-401 v1.2, Carica papaya ASGPB v0.4, Gossypium raimondii v2.1, and Eucalyptus grandis v2.0 genomes were obtained from a BLAST search for the Phytozome website (http://www.phytozome.net/, last accessed 25 October 2018). The total 57 amino acid sequences were aligned using MUSCLE (Edgar, 2004). All positions containing gaps and missing data were eliminated, and the final data set comprised a total of 266 amino acid positions. Evolutionary analyses were conducted in MEGA6 (Tamura et al., 2013). The evolutionary history was inferred using the Maximum Likelihood method, based on the Le_Gascuel_2008 model (Le and Gascuel, 2008). The tree with the highest log likelihood (–7013.2051) is shown in Supplementary Fig. S2. The initial tree for the heuristic search was obtained by applying the Neighbor–Joining method to a matrix of pairwise distances estimated using the JTT model. A discrete Gamma distribution was used to model the evolutionary rate differences between the sites [five categories (+G, parameter=0.4454)].
Subcellular localization analysis
Onion epidermal cells and L. japonicus roots of 3-day-old seedlings grown on moistened filter paper were transformed with each construct (pUGW5-based series) via particle bombardment with a Helios Gene Gun (BIO-RAD), as described previously (Mano et al., 2006). To verify the co-localization of proteins with the Golgi network, A. tumefaciens AGL1 strains carrying each construct (pGWB5-based series) and an mCherry-fused Golgi marker construct (G-rb) (Nelson et al., 2007) were mixed and co-infiltrated into Nicotiana benthamiana leaves using a p19-harboring strain, as previously described (Voinnet et al., 2003; Kinoshita et al., 2010).
Microscopy observations
Bright-field and fluorescence images were generated using an SZX12/16 stereomicroscope or a BX50 microscope (Olympus) and a DP Controller (Olympus). Confocal images were generated using an A1 confocal laser-scanning microscope (Nikon) with a ×10 or ×20/0.75 NA objective lens.
Detection of HPAT activity in PLENTY
PLENTY proteins were expressed as a C-terminal FLAG-tag fusion in the yeast (Saccharomyces cerevisiae) strain INVSc1, which was transformed with pYES2 vectors (Invitrogen) harboring full-length PLENTY or one of the two N-terminally deleted PLENTY constructs. Their expression was detected using immunoblotting, and the proteins were collected within a pellet of microsomal membrane, as described previously (Ogawa-Ohnishi et al., 2013). HPAT activity assays were performed in 20 μl reaction mixtures containing 100 mM MOPS-KOH (pH 7.0) buffer, 1 mM MnCl2, 1.0% Triton X-100, 250 μM UDP-β-l-arabinofuranose (Peptide Institute, Inc.), 100 μM (PGVOOS)3 peptide, and 150 μg of total yeast microsomal membrane. The reaction mixture was incubated at 30 °C for 2 h, and then terminated by the addition of 100 μl of 0.1% formic acid. After centrifugation at 15000 rpm for 5 min, 40 µl aliquots of this solution were analyzed using LC/MS, as previously reported (Ogawa-Ohnishi et al., 2013). The mass spectra were obtained by scanning the selected ion [(PGVOOS)3+Ala1: m/z 1849.6] in zoom scan mode.
Double mutant analysis
To select the plenty har1-7 homozygous double mutant, each plant was checked for the presence of an amplified polymorphic sequence marker in har1-7 (a G1044A change in the HAR1 CDS causing a W348 stop codon) (Magori et al., 2009). Deletion of the plenty locus was detected using the primers listed in Supplementary Table S2. Nodules and other root phenotypes were counted and measured at 28 DAI.
Gene expression analysis
Total RNA was isolated from each plant tissue at selected time points using an RNeasy Plant Mini Kit (Qiagen), and the first-strand cDNA was prepared using a QuantiTect Reverse Transcription Kit (Qiagen). Reverse transcription–quantitative PCR analysis was performed using an ABI Prism 7000 (Applied Biosystems) with THUNDERBIRD SYBR qPCR Mix (Toyobo) or with a QuanTitect SYBR Green RT-PCR Kit (Qiagen), according to the manufacturers’ protocols. EF-1a (GNf095a12) expression was used as the reference (Groth et al., 2010). The relative expression levels were calculated using the ΔΔCt method (Livak and Schmittgen, 2001). The primers used in the expression analysis are shown in Supplementary Table S2. The data are presented as the mean ±SD of three biological replicates or three technical replicates.
Boxplot analysis
Boxplot analyses were performed in R using ggplot2 or basic R plotting commands. In the boxplots, the upper and lower ‘hinges’ correspond to the first and third quartiles. The upper whisker extends from the hinge to the highest value within 1.5× the interquartile range (IQR) between the first and third quartiles, while the lower whisker extends from the hinge to the lowest value within 1.5× IQR. Outliers (data beyond the end of the whiskers) are plotted as points.
Results
Identification of the PLENTY gene and phylogenetic analysis
The plenty mutant has two characteristic phenotypes, an increased number of nodules and short roots (Yoshida et al., 2010). We previously reported that the plenty locus is located between markers TM0002 and TM0324 on the long arm of chromosome II in L. japonicus. We narrowed down the region of interest using map-based cloning utilizing a larger mapping population of 1087 F2 plants (Supplementary Fig. S1). A genomic PCR analysis of the region between marker TM0308 and the newly developed marker EY005 (Supplementary Table S1) revealed an ~16 kb deletion spanning two protein-coding genes, CM0308.590.r2.d/Lj2g3v3022950 encoding a putative telomerase-binding protein and CM0308.600.r2.d/Lj2g3v3022970 (http://www.kazusa.or.jp/lotus/). The latter gene, which we named PLENTY, was orthologous to MtRDN1 (Supplementary Fig. S2; Schnabel et al., 2011). Similar to M. truncatula, two other PLENTY paralogs termed PLENTY2 and PLENTY3, were also identified in L. japonicus, and the three genes were phylogenetically divided into three related groups (Supplementary Fig. S2). There are no Arabidopsis homologs of LjPLENTY and MtRDN1; instead, there is a Brassicaceae-specific clade of PLENTY-like genes that are closely related to the group containing LjPLENTY3 and MtRDN3 (Schnabel et al., 2011; Ogawa-Ohnishi et al., 2013; Xu et al., 2015). To explore further the Brassicaceae-specific evolution of the PLENTY genes, we performed a phylogenetic analysis using genes from Brassica rapa, Carica papaya (papaya), Gossypium raimondii (cotton), and Eucalyptus grandis (eucalyptus), acquired from the Phytozome database (http://www.phytozome.net/), as well as the same data set used in the study of MtRDN1 (Schnabel et al., 2011). Among these plants, all PLENTY homologs from the eudicots, except for those from B. rapa, were classified into groups 1–3.
Nodule number and the non-symbiotic short-root phenotype of plenty were complemented by PLENTY expression
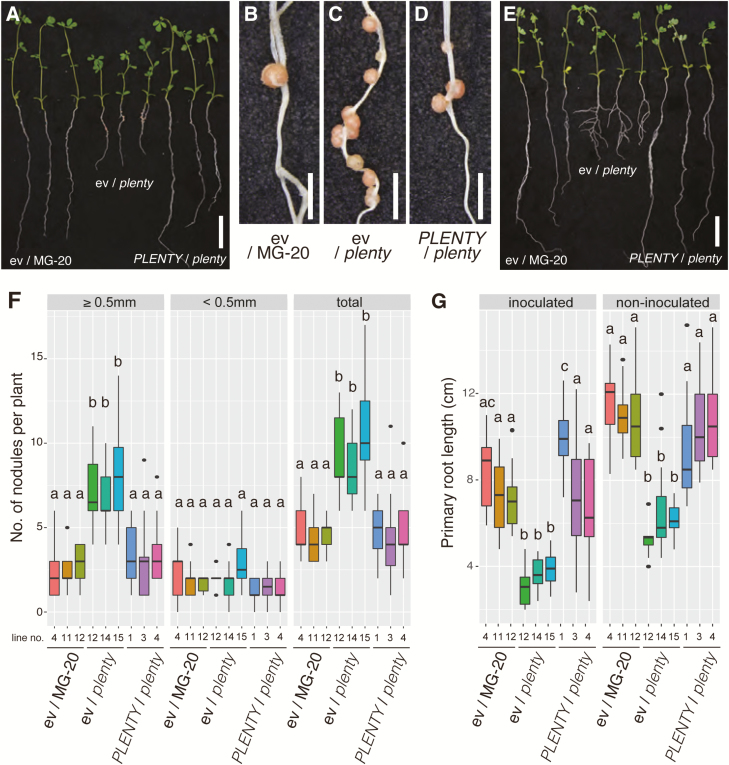
The plenty mutant has an increased number of nodules and shorter roots in both symbiotic and non-symbiotic conditions (Yoshida et al., 2010). We generated stably transformed plenty mutants harboring the PLENTY CDS under the control of the LjUBIQUITIN promoter or an empty vector (as the control) and observed their phenotypes in both inoculated and non-inoculated conditions. The significantly increased numbers of nodules and short primary roots of plenty were rescued by the expression of PLENTY CDS in all three independent T3 transgenic lines, but not by the empty vector control (Fig. 1). The rescued primary root length was also observed in the non-inoculated condition (Fig. 1E, G). There were no significant differences in the number of first-order lateral roots (i.e. lateral roots emerging from the primary root) in the complemented plants (Supplementary Fig. S3). These results indicate that PLENTY functions not only in inhibiting nodulation and but also in primary root elongation, in both the presence and absence of rhizobia. As previously reported, plenty tends to form increased numbers of larger nodules (Yoshida et al., 2010); therefore, we separately counted small nodules (<0.5 mm in diameter) and large nodules (>0.5 mm in diameter) to evaluate the complementation. The number of large nodules rather than small nodules was significantly reduced in the complemented lines (Fig. 1F).
Fig. 1.
Complementation of plenty. (A) Rhizobium-inoculated MG-20 plants stably transformed with the empty vector pUB-GW-GFP (ev/MG-20), a plenty mutant transformed with the empty vector (ev/plenty), and a plenty mutant transformed with pUB-GW-Full-PLENTY (PLENTY/plenty) at 14 days after inoculation (DAI) with M. loti MAFF303099. Magnified images of the nodulated regions of the ev/MG-20 (B), ev/plenty (C), and PLENTY/plenty (D) plants are shown. (E) Non-inoculated plants of ev/MG-20, ev/plenty, and PLENTY/plenty at 21 days after germination (DAG). (F) Boxplots of the nodule numbers [≥0.5 mm diameter (left), <0.5 mm diameter (middle), and total (right)] of the individual inoculated T3 transgenic lines (n≥10). (G) Boxplots of the primary root length of the inoculated T3 transgenic plants at 14 DAI (left) and the non-inoculated T3 transgenic plant at 21 DAG (right). Scale bars=2 cm in (A, E) and 2 mm in (B–D). Different lower case letters represent statistically significant differences (P<0.05; Tukey’s HSD). Experiments were performed in triplicate (n≥10 in each trial).
PLENTY is localized to the Golgi network
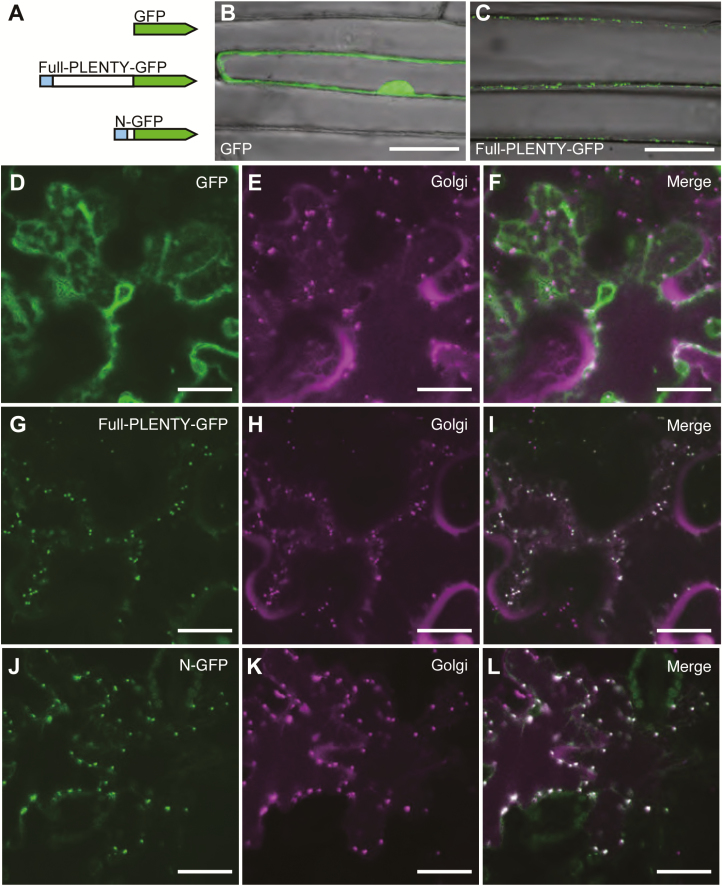
To assess the function of PLENTY, we next investigated its subcellular localization using a series of constructs expressing PLENTY–GFP fusion proteins. Since the N-terminal amino acid sequence of MtRDN1 was previously predicted to be a signal peptide (SP) for the secretory pathway (Schnabel et al., 2011), we generated three kinds of constructs fused to GFP: full-length PLENTY (Full-PLENTY–GFP), PLENTY lacking the first 46 amino acids of the N-terminal region including secretory peptides and the transmembrane domain (ΔN1-PLENTY–GFP), and only the first 58 amino acids of the N-terminal region of PLENTY (N–GFP) (Supplementary Appendix S2). We first examined the localization of GFP and Full-PLENTY–GFP constructs following their introduction into onion epidermal cells (Fig. 2B, C) and L. japonicus root cells (Supplementary Fig. S4) using particle bombardment. Full-PLENTY–GFP was visible in the intracellular punctate structures. We next examined the Agrobacterium infiltration of N. benthamiana leaves and observed the same punctate localization of Full-PLENTY–GFP and N–GFP. To analyze these localization patterns in detail, we compared them with the localization of a mCherry-fused cis-Golgi marker, soybean α-1,2-mannosidase I (Nebenführ et al., 1999; Saint-Jore-Dupas et al., 2006; Nelson et al., 2007). The localization of Full-PLENTY–GFP and N–GFP overlapped with that of the cis-Golgi marker (Fig. 2D–L), as well as the previous localizations reported for its orthologs AtHPAT1 and MtRDN1 (Ogawa-Ohnishi et al., 2013; Kassaw et al., 2017). In addition, the Golgi localization of N–GFP, which contrasts with the cytoplasmic or nuclear localization of ΔN1-PLENTY–GFP (Supplementary Fig. S4), suggests that the N-terminal (SP) region is necessary and sufficient for targeting PLENTY to the Golgi.
Fig. 2.
Subcellular localization of PLENTY–GFP fusion proteins. (A) Overview of the three GFP fusion protein constructs; GFP, Full-PLENTY–GFP, and N–GFP containing the first 58 amino acids of the N-terminal region of PLENTY. PLENTY has a putative secretory signal peptide at the N-terminus (shown in blue). (B–L) Confocal microscopic images of the localization of a series of PLENTY–GFP fusion proteins driven by the CaMV 35S promoter. (B, C) The transient expression in onion epidermal cells transformed using particle bombardment. (D–L) Transient expression in N. benthamiana pavement cells co-expressing the mCherry-fused cis-Golgi marker, transformed using A. tumefaciens infiltration. The constructs used for each analysis are shown in each panel. Merged images show the cytoplasmic localization of GFP (F) and the Golgi localization of Full-PLENTY–GFP and N–GFP (I, L). Scale bars=50 μm in (B, C) and 25 μm in (D–L). Similar GFP localization was observed in >10 transformed cells.
In vitro detection of PLENTY enzymatic activities
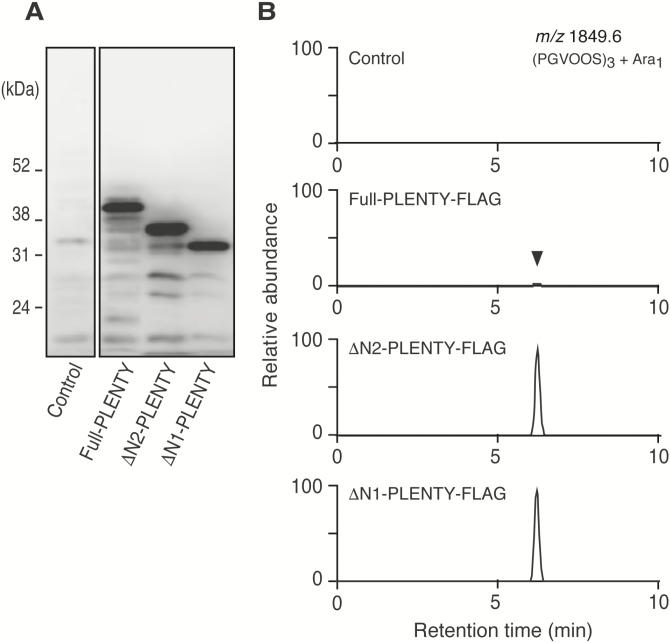
To determine whether PLENTY possess HPAT activity, we examined the enzymatic activity in vitro using yeast-expressed recombinant protein. A secretory signal and a transmembrane domain are predicted in the N-terminal region of PLENTY (Supplementary Appendix S2). Considering the difficulty in membrane protein purification, we designed not only Full-PLENTY but also two N-terminally deleted forms of PLENTY, ΔN1-PLENTY lacking the first 46 amino acids and ΔN2-PLENTY lacking the first 25 amino acids (Supplementary Appendix S2). Western blotting using an antibody to FLAG (anti-FLAG) demonstrated that all three recombinant proteins, Full-PLENTY (41.6 kDa), ΔN2-PLENTY (38.8 kDa), and ΔN1-PLENTY (36.4 kDa), were successfully expressed and collected in the microsomal membrane fractions (Fig. 3A). These fractions were then incubated with a synthetic tandem repeat peptide (PGVOOS)3, a previously developed substrate for detecting AtHPAT activity based on the native PSY1 peptide in Arabidopsis (Amano et al., 2007; Ogawa-Ohnishi et al., 2013), in the presence of UDP-β-l-arabinofuranose (Araf). A subsequent LC/MS analysis revealed that these PLENTY proteins catalyzed the arabinosylation of the peptide substrate (Fig. 3B).
Fig. 3.
Identification of the HPAT activity of PLENTY in vitro. (A) Western blots of microsomal proteins in yeast expressing C-terminally FLAG-fused PLENTY proteins [Full-PLENTY (41.6 kDa), ΔN2-PLENTY with the first 25 amino acids deleted (38.8 kDa), or ΔN1-PLENTY with the first 46 amino acids deleted (36.4 kDa)], probed using an anti-FLAG antibody. (B) Identification of HPAT activity of the three recombinant PLENTY proteins. The synthetic substrate peptide (PGVOOS)3 was incubated with the FLAG-tag-fused recombinant proteins in the presence of UDP-β-l-Araf and analyzed using LC/MS. The 1849.6 increase in m/z corresponds to the arabinosylation.
Hypernodulation of the plenty mutant was suppressed by the constitutive expression of CLE-RS1 and CLE-RS2 but not CLE-RS3
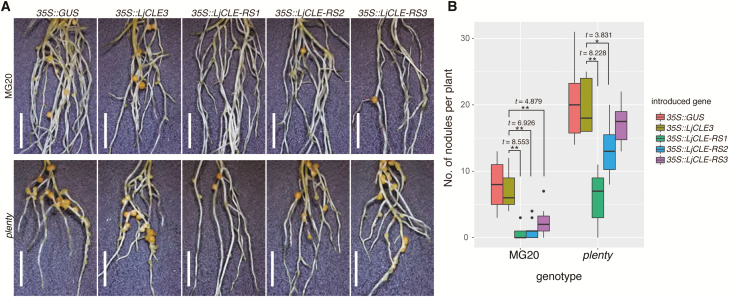
Although its native substrates have still not been identified, the HPAT activity of PLENTY led us to speculate that PLENTY modifies the CLE-RS1/2/3 peptides, because of their function in AON (Okamoto et al., 2009; Nishida et al., 2016). Hence, we investigated the nodule suppression effects introduced by the constitutive expression of CLE-RS1/2/3 in the plenty mutant background using hairy root transformation. We hypothesized that if PLENTY mediates the arabinosylation of CLE-RS1/2/3, and if this modification is critical for their activity, the nodule suppression effect arising from the constitutive expression of CLE-RS1/2/3 would be abolished in the plenty mutant, as was observed in har1 (Okamoto et al., 2009) and similarly for CLE12 expression in Mtrdn1 (Kassaw et al., 2017). All three 35S::CLE-RS (35S::CLE-RS1/2/3) constructs suppressed nodulation in the wild type in comparison with the negative controls 35S::GUS and 35S::CLE3. Unexpectedly, however, in plenty, the increased nodulation was significantly suppressed by two of the CLE-RS constructs, 35S::CLE-RS1 and 35S::CLE-RS2 (Fig. 4), although the degree of suppression was stronger in 35S::CLE-RS1 than in 35S::CLE-RS2. Thus, the constitutive expression of CLE-RS1/2 maintains sufficient biological activity to repress nodulation, even in plenty. On the other hand, the suppression effect of 35S::CLE-RS3 is completely abolished in plenty, indicating the involvement of PLENTY in the arabinosylation of CLE-RS3 rather than that of CLE-RS1/2.
Fig. 4.
Hypernodulation of plenty was strongly suppressed by CLE-RS1 and mildly suppressed by CLE-RS2 but not by CLE-RS3. (A) Stereoscopic images of transgenic hairy roots constitutively expressing GUS and LjCLE3 (as a control), LjCLE-RS1, LjCLE-RS2, or LjCLE-RS3. The constructs used for each analysis are shown in each panel. Scale bars=5 mm. (B) Boxplots of the number of nodules per individual transformed plant at 14 DAI with M. loti MAFF303099. The genotypes and introduced constructs are indicated on the graph. Statistical analyses were conducted using a two-tailed Welch’s t-test (**P<0.01, *P<0.05, n≥10). The black dots represent outliers. Experiments were performed in triplicate (n≥10 in each trial).
Increased nodulation is an additive phenotype in the plenty har1 double mutant, compared with the single mutants
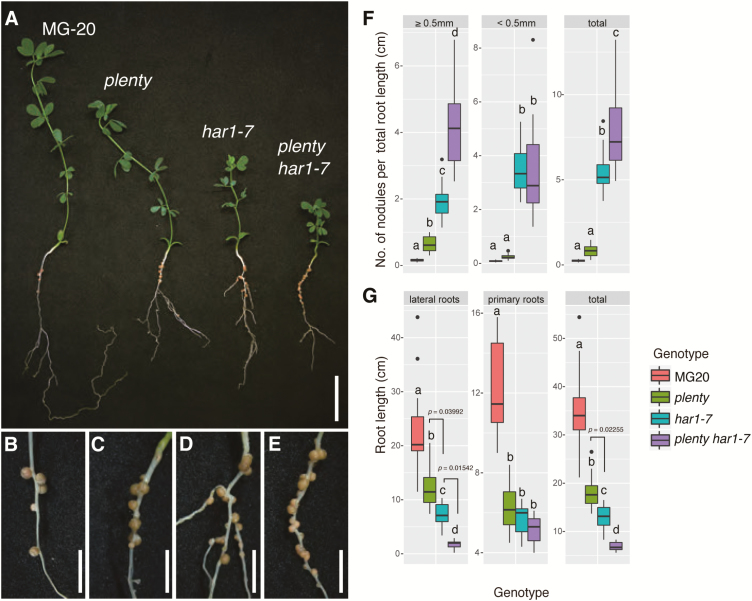
Although is it unclear whether the CLE-RS1/2 peptides are substrates of PLENTY, the ligand–receptor interaction between CLE-RS1/2 and HAR1 has been clearly defined previously (Okamoto et al., 2009, 2013; Sasaki et al., 2014), as the inhibition of nodulation introduced by the constitutive expression of CLE-RS1/2 occurs in a HAR1-dependent manner. To investigate the genetic interaction between PLENTY and HAR1 further, we generated a plenty har1-7 double mutant and determined the nodule numbers normalized to the total root length of each plant (Fig. 5). The har1-7 mutation resulted in the loss of both the transmembrane and kinase domains of HAR1; thus, har1-7 is a possible null mutant (Magori et al., 2009). The number of large nodules >0.5 mm in diameter and the total number of nodules in plenty har1-7 were significantly increased relative to those of the plenty or har1-7 single mutants (Fig. 5F). This indicates that PLENTY and HAR1 function in at least partially separate AON pathways. Notably, while the plenty, har1-7, and plenty har1-7 mutants all had similar primary root lengths, an additive effect was observed for the shortening of the lateral roots in plenty har1-7 (Fig. 5G), indicating their involvement in genetically non-overlapping pathways for lateral root elongation. Additionally, the number of first-order lateral roots in the inoculated condition was increased in har1-7, as previously reported (Szczyglowski et al., 1998; Wopereis et al., 2000), but decreased in plenty har1-7, suggesting that the plenty mutation suppressed not only lateral root elongation but also lateral root emergence in har1-7 (Supplementary Fig. S3).
Fig. 5.
Additive nodulation of the plenty har1-7 double mutant. (A) Nodulation in the wild-type (MG-20), plenty, har1-7, and plenty har1-7 double mutant plants. (B–E) Magnified images of nodulated roots of the wild type (MG-20) (B), plenty (C), har1-7 (D), and the plenty har1-7 double mutant (E). (F) Boxplot of the nodule number [≥0.5 mm diameter (left), <0.5 mm diameter (middle), total (right)], normalized by the total root length of each plant, counted at 21 DAI with M. loti MAFF303099. (G) Boxplot of the lengths of lateral loots (left), primary roots (middle), and total roots (right) of each plant, measured for normalization in (F). Scale bars=2 cm in (A) and 5 mm in (B–E). The values of the total nodule numbers were used for the statistical analysis in (F). Different lower case letters represent statistically significant differences (P<0.05; Tukey’s HSD; n=14). 0.01<P<0.05 are denoted on the graph. The black dots represent outliers. Experiments were performed in triplicate (n≥10 in each trial).
Discussion
In this study, we cloned LjPLENTY, an ortholog of MtRDN1 and PsNOD3, and a homolog of the three AtHPAT genes, and found that, like its orthologs, LjPLENTY localizes to the Golgi complex (Ogawa-Ohnishi et al., 2013; Kassaw et al., 2017). Hydroxyproline O-arabinosylation is widely observed in secreted Arabidopsis peptides (Shinohara and Matsubayashi, 2010; Matsubayashi, 2014; Kucukoglu and Nilsson, 2015), and studies of HPAT homologs in Arabidopsis, tomato (Solanum lycopersicum), and the moss Physcomitrella patens have shown that the substrates of these enzymes are involved in diverse aspects of plant development, such as cellular tip growth and meristem maintenance (Ogawa-Ohnishi et al., 2013; Xu et al., 2015; MacAlister et al., 2016). Additionally, the specific gene loss of the group 1 HPATs in the Brassicaceae (Supplementary Fig. S2) (Schnabel et al., 2011) may be associated with the loss of arbuscular mycorrhizal symbiosis in these species (Delaux et al., 2014).
Prior to the discovery of its molecular entity, PsNOD3 was hypothesized to be involved in generating an unknown root-derived systemic signal that inhibited nodulation based on the findings of a series of grafting experiments (Postma et al., 1988; Caetano-Anollés and Gresshoff, 1990; Li et al., 2009; Novák, 2010). In particular, the hypernodulation on the adventitious roots originating from the wild-type scions on nod3 rootstocks indicated that the decreased production of the systemic signal affected both wild-type and nod3 roots. These previous reports also support the subsequent identification of CLE peptides as the root-derived signal. The nodule suppression by the constitutive expression of MtCLE13 was found to be dependent on PsNOD3 (Osipova et al., 2012), and a separate study showed that the nodule suppression by 35S::MtCLE12, but not 35S::MtCLE13, was dependent on MtRDN1 (Kassaw et al., 2017). These findings suggest the hypothesis that LjPLENTY participates in the maturation of at least one of the LjCLE-RSs. In this study, stronger, milder, and no repression of nodulation was found by 35S::CLE-RS1, 35S::CLE-RS2, and 35S::CLE-RS3, respectively, in the plenty mutant background. Based on the differential nodule suppression levels, we can only state the order of likelihood that each CLE-RS is the substrate of PLENTY: CLE-RS3 >CLE-RS2 >CLE-RS1 (Supplementary Fig. S5).
The differential requirements for PLENTY between the CLE-RS genes are similar to the differential requirements for RDN1 in the functions of 35S::MtCLE12/13 in Medicago (Kassaw et al., 2017). The orthologous relationships of the CLEs are not clear because of their short amino acid sequences (Hastwell et al., 2015, 2017), and because the core sequences in the CLE domains of LjCLE-RS1/2 are shared more with MtCLE13 than with MtCLE12 (Imin et al., 2018). This means it is difficult to determine whether the differences in their enzyme–substrate specificities are caused by differences in their amino acid sequences. MtCLE13 did not suppress nodulation in nod3 plants, suggesting the requirement for PsNOD3 (Osipova et al., 2012); but further studies using PsCLEs rather than MtCLE13 are needed for an understanding of the substrate–enzyme specificity in pea. Despite the successful detection of the enzymatic activities of PLENTY using artificially synthesized peptides, it is still unknown whether the arabinosylation of CLE-RS1/2/3 was performed by PLENTY, because the relevant results were based solely on the constitutive expression analysis. So far, the arabinosylation has been detected successfully only in CLE-RS2 (Okamoto et al., 2013); therefore, whether CLE-RS1/3 are arabinosylated has also remained obscure (Supplementary Fig. S5). In conclusion, we cannot completely exclude the possibility that LjCLE-RS1/2 are the substrates of PLENTY, but the CLE-RS1/2/3-HAR1 signaling pathway can be divided into PLENTY-dependent and PLENTY-independent pathways, namely PLENTY-independent for CLE-RS1, partially dependent for CLE-RS2, and strongly dependent for CLE-RS3 peptides (Supplementary Fig. S5). To evaluate the contribution of various enzymes to the modification of the CLE peptides accurately, assays to detect the modification levels of in vivo native peptides in respective single, double, and triple mutants of the three PLENTY paralogs should be performed in future studies. Alternatively, a loss-of-function analysis of the CLE-RS genes together with the PLENTY paralogs will provide important information.
Based on the additive nodules of the plenty har1 double mutant, we propose that PLENTY and HAR1 at least partially function in separate AON pathways. First, this additive phenotype would be caused by the milder hypernodulation of plenty rather than that of har1. This milder phenotype may be affected by the functional redundancy among the three paralogs, as previously suggested for other species (Ogawa-Ohnishi et al., 2013; MacAlister et al., 2016; Kassaw et al., 2017). Thus, the PLENTY-independent nodule suppression by the constitutively expressed CLE-RS1/2 may be dependent on PLENTY2/3. As we expected, the expression patterns of PLENTY2 and PLENTY3 during nodulation were similar to that of PLENTY (Supplementary Figs S2, S6), as was previously shown for MtRDN2/3 (Schnabel et al., 2011). Nevertheless, the increased number of nodules in the plenty har1-7 double mutant raises the possibility that PLENTY provokes AON independently of HAR1. In fact, the MtSUNN-independent AON pathway has been discussed before (Kassaw et al., 2015), based on the persistent suppression of excessive nodule formation in Mtsunn; however, alternative receptors functioning in a completely HAR1-independent manner have not yet been identified. All known candidate receptors for CLE peptides, LjKLV, LjCLV2/MtCLV2/PsSYM28, and LjCRN/MtCRN, have consistently been thought to interact with LjHAR1/MtSUNN/PsSYM29 in the same genetic pathway (Miyazawa et al., 2010; Krusell et al., 2011; Crook et al., 2016). We therefore postulate that other unknown LRR-RLKs function in the PLENTY-dependent and HAR1-independent pathway (Supplementary Fig. S5).
Finally, we considered the potential substrates of PLENTY functioning in a HAR1-independent manner. Aside from CLE-RS1/2/3, the most plausible candidates are the other CLE peptides, including LjCLE40 (Nishida et al., 2016; Hastwell et al., 2017), the C-TERMINALLY ENCODED PEPTIDES (CEPs), and other related peptides found to be involved in nodulation in Medicago (de Bang et al., 2017; Patel et al., 2017); in particular, MtCEP1 functioning in promoting nodulation under nitrogen-limited conditions (Imin et al., 2013; Mohd-Radzman et al., 2016). Moreover, the tri-arabinosylation of the MtCEP1 proline reduced or eliminated the nodule-promoting effect of this peptide (Patel et al., 2017), which contrasts with the necessity of arabinosylation for LjCLE-RS2 or MtCLE12/13 activities (Okamoto et al., 2013; Imin et al., 2018). MtRDN1 may therefore keep MtCEP1 inactive to inhibit increased nodulation; thus, the MtCEP1 ortholog may actually be a substrate of PLENTY. Additionally, the inhibition of lateral root emergence by MtCEP was abolished by non-arabinosylated MtCEP (Patel et al., 2017). The reduced number of emerged lateral roots of the plenty har1-7 may be caused by accumulation of the non-arabinosylated CEP1-like peptide (Supplementary Fig. S3). Also, other unidentified substrates of PLENTY may have effects on the shorter primary root of plenty (Fig. 1) or the shorter and reduced lateral root of plenty har1-7 (Fig. 5; Supplementary Fig. S3). The arabinosylated peptides involved in root architecture are strong candidates to be these substrates (Corcilius et al., 2017; Patel et al., 2017; Oh et al., 2018). Interestingly, the MtCEP1 receptor, compact root architecture 2, acts on the shoot for nodulation but on the root for lateral root development through different pathways (Huault et al., 2014; Mohd-Radzman et al., 2016). LjHAR1 is also a common factor involved in both nodulation control and non-symbiotic root development (Wopereis et al., 2000), but its ligand and shoot/root dependency responding to symbiotic and non-symbiotic phenotype were unknown. Identifying substrates of PLENTY will lead to a further understanding of how nodulation and root architecture are controlled at the same time via HAR1 or other receptors.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Identification of the plenty locus.
Fig. S2. Phylogenetic tree of the PLENTY family in land plants.
Fig. S3. First-order lateral roots in the complementation test and the plenty har1-7 double mutant analysis.
Fig. S4. The N-terminal region of PLENTY is necessary and sufficient for localization to the Golgi.
Fig. S5. A working model of PLENTY in the negative control of nodulation.
Fig. S6. The gene expression patterns of the PLENTY paralogs.
Table S1. Newly developed genetic markers for the map-based cloning of PLENTY.
Table S2. Primers used in this study.
Appendix S1. FASTA file of amino acid sequences used for the phylogenetic analysis.
Appendix S2. The deletion series of PLENTY proteins used in this study.
Supplementary Material
Acknowledgements
We thank Shoji Mano, Michitaro Shibata, and the Spectrography and Bioimaging Facility of NIBB Core Research Facilities for their technical support; Norio Suganuma, Saori Tomisawa, Takema Sasaki, Shoji Mano, Tsuyoshi Nakagawa, Kenji Yamada, Shino Goto-Yamada, Kentaro Tamura, Mikio Nishimura, and Ikuko Hara-Nishimura for providing plant seeds or vectors; and Shoji Mano and Masanao Sato for their valuable comments. This work was supported by the Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for JSPS Fellow [grant nos 25-3940 to EY and 17J02948 to HN], and the Ministry of Education, Culture, Sports, Science and Technology Grants-in-Aid for Scientific Research [grant nos 25114519, 16H01457, and 18H04773 to TS, 25221105, 18H05274, and 15H05957 to YM and 22128006 to MK]. The authors have no conflicts of interest to declare.
References
- Amano Y, Tsubouchi H, Shinohara H, Ogawa M, Matsubayashi Y. 2007. Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proceedings of the National Academy of Sciences, USA 104, 18333–18338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton WJ, Dilworth MJ. 1971. Control of leghaemoglobin synthesis in snake beans. Biochemical Journal 125, 1075–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caetano-Anollés G, Gresshoff PM. 1990. Early induction of feedback regulatory responses governing nodulation in soybean. Plant Science 71, 69–81. [Google Scholar]
- Caetano-Anollés G, Gresshoff PM. 1991. Efficiency of nodule initiation and autoregulatory responses in a supernodulating soybean mutant. Applied and Environmental Microbiology 57, 2205–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcilius L, Hastwell AH, Zhang M, Williams J, Mackay JP, Gresshoff PM, Ferguson BJ, Payne RJ. 2017. Arabinosylation modulates the growth-regulating activity of the peptide hormone CLE40a from soybean. Cell Chemical Biology 24, 1347–1355. [DOI] [PubMed] [Google Scholar]
- Crook AD, Schnabel EL, Frugoli JA. 2016. The systemic nodule number regulation kinase SUNN in Medicago truncatula interacts with MtCLV2 and MtCRN. The Plant Journal 88, 108–119. [DOI] [PubMed] [Google Scholar]
- de Bang TC, Lundquist PK, Dai X, et al. . 2017. Genome-wide identification of medicago peptides involved in macronutrient responses and nodulation. Plant Physiology 175, 1669–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaux PM, Varala K, Edger PP, Coruzzi GM, Pires JC, Ané JM. 2014. Comparative phylogenomics uncovers the impact of symbiotic associations on host genome evolution. PLoS Genetics 10, e1004487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delves AC, Mathews A, Day DA, Carter AS, Carroll BJ, Gresshoff PM. 1986. Regulation of the soybean–Rhizobium nodule symbiosis by shoot and root factors. Plant Physiology 82, 588–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djordjevic MA, Oakes M, Wong CE, Singh M, Bhalla P, Kusumawati L, Imin N. 2011. Border sequences of Medicago truncatula CLE36 are specifically cleaved by endoproteases common to the extracellular fluids of Medicago and soybean. Journal of Experimental Botany 62, 4649–4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth M, Takeda N, Perry J, et al. . 2010. NENA, a Lotus japonicus homolog of Sec13, is required for rhizodermal infection by arbuscular mycorrhiza fungi and rhizobia but dispensable for cortical endosymbiotic development. The Plant Cell 22, 2509–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastwell AH, de Bang TC, Gresshoff PM, Ferguson BJ. 2017. Author Correction: CLE peptide-encoding gene families in Medicago truncatula and Lotus japonicus, compared with those of soybean, common bean and Arabidopsis. Scientific Reports 7, 15474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastwell AH, Gresshoff PM, Ferguson BJ. 2015. The structure and activity of nodulation-suppressing CLE peptide hormones of legumes. Functional Plant Biology 42, 229–238. [DOI] [PubMed] [Google Scholar]
- Huault E, Laffont C, Wen J, Mysore KS, Ratet P, Duc G, Frugier F. 2014. Local and systemic regulation of plant root system architecture and symbiotic nodulation by a receptor-like kinase. PLoS Genetics 10, e1004891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imin N, Mohd-Radzman NA, Ogilvie HA, Djordjevic MA. 2013. The peptide-encoding CEP1 gene modulates lateral root and nodule numbers in Medicago truncatula. Journal of Experimental Botany 64, 5395–5409. [DOI] [PubMed] [Google Scholar]
- Imin N, Patel N, Corcilius L, Payne RJ, Djordjevic MA. 2018. CLE peptide tri-arabinosylation and peptide domain sequence composition are essential for SUNN-dependent autoregulation of nodulation in Medicago truncatula. New Phytologist 218, 73–80. [DOI] [PubMed] [Google Scholar]
- Kassaw T, Bridges W Jr, Frugoli J. 2015. Multiple autoregulation of nodulation (AON) signals identified through split root analysis of Medicago truncatula sunn and rdn1 mutants. Plants 4, 209–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassaw T, Nowak S, Schnabel E, Frugoli J. 2017. ROOT DETERMINED NODULATION1 is required for M. truncatula CLE12, but not CLE13, peptide signaling through the SUNN receptor kinase. Plant Physiology 174, 2445–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkema M, Gresshoff PM. 2008. Investigation of downstream signals of the soybean autoregulation of nodulation receptor kinase GmNARK. Molecular Plant-Microbe Interactions 21, 1337–1348. [DOI] [PubMed] [Google Scholar]
- Kinoshita A, Betsuyaku S, Osakabe Y, Mizuno S, Nagawa S, Stahl Y, Simon R, Yamaguchi-Shinozaki K, Fukuda H, Sawa S. 2010. RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 137, 3911–3920. [DOI] [PubMed] [Google Scholar]
- Kosslak RM, Bohlool BB. 1984. Suppression of nodule development of one side of a split-root system of soybeans caused by prior inoculation of the other side. Plant Physiology 75, 125–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krusell L, Madsen LH, Sato S, et al. . 2002. Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature 420, 422–426. [DOI] [PubMed] [Google Scholar]
- Krusell L, Sato N, Fukuhara I, et al. . 2011. The Clavata2 genes of pea and Lotus japonicus affect autoregulation of nodulation. The Plant Journal 65, 861–871. [DOI] [PubMed] [Google Scholar]
- Kucukoglu M, Nilsson O. 2015. CLE peptide signaling in plants—the power of moving around. Physiologia Plantarum 155, 74–87. [DOI] [PubMed] [Google Scholar]
- Le SQ, Gascuel O. 2008. An improved general amino acid replacement matrix. Molecular Biology and Evolution 25, 1307–1320. [DOI] [PubMed] [Google Scholar]
- Li D, Kinkema M, Gresshoff PM. 2009. Autoregulation of nodulation (AON) in Pisum sativum (pea) involves signalling events associated with both nodule primordia development and nitrogen fixation. Journal of Plant Physiology 166, 955–967. [DOI] [PubMed] [Google Scholar]
- Lin YH, Ferguson BJ, Kereszt A, Gresshoff PM. 2010. Suppression of hypernodulation in soybean by a leaf-extracted, NARK- and Nod factor-dependent, low molecular mass fraction. New Phytologist 185, 1074–1086. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- MacAlister CA, Ortiz-Ramírez C, Becker JD, Feijó JA, Lippman ZB. 2016. Hydroxyproline O-arabinosyltransferase mutants oppositely alter tip growth in Arabidopsis thaliana and Physcomitrella patens. The Plant Journal 85, 193–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T, Kusakabe M, Shimoda Y, Sato S, Tabata S, Murooka Y, Hayashi M. 2008. Polyubiquitin promoter-based binary vectors for overexpression and gene silencing in Lotus japonicus. Molecular Plant-Microbe Interactions 21, 375–382. [DOI] [PubMed] [Google Scholar]
- Magori S, Kawaguchi M. 2009. Long-distance control of nodulation: molecules and models. Molecules and Cells 27, 129–134. [DOI] [PubMed] [Google Scholar]
- Magori S, Oka-Kira E, Shibata S, Umehara Y, Kouchi H, Hase Y, Tanaka A, Sato S, Tabata S, Kawaguchi M. 2009. Too much love, a root regulator associated with the long-distance control of nodulation in Lotus japonicus. Molecular Plant-Microbe Interactions 22, 259–268. [DOI] [PubMed] [Google Scholar]
- Mano S, Nakamori C, Nito K, Kondo M, Nishimura M. 2006. The Arabidopsis pex12 and pex13 mutants are defective in both PTS1- and PTS2-dependent protein transport to peroxisomes. The Plant Journal 47, 604–618. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y. 2014. Posttranslationally modified small-peptide signals in plants. Annual Review of Plant Biology 65, 385–413. [DOI] [PubMed] [Google Scholar]
- Miyazawa H, Oka-Kira E, Sato N, et al. . 2010. The receptor-like kinase KLAVIER mediates systemic regulation of nodulation and non-symbiotic shoot development in Lotus japonicus. Development 137, 4317–4325. [DOI] [PubMed] [Google Scholar]
- Mohd-Radzman NA, Laffont C, Ivanovici A, Patel N, Reid D, Stougaard J, Frugier F, Imin N, Djordjevic MA. 2016. Different pathways act downstream of the CEP peptide receptor CRA2 to regulate lateral root and nodule development. Plant Physiology 171, 2536–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortier V, Den Herder G, Whitford R, Van de Velde W, Rombauts S, D’Haeseleer K, Holsters M, Goormachtig S. 2010. CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiology 153, 222–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortier V, Fenta BA, Martens C, Rombauts S, Holsters M, Kunert K, Goormachtig S. 2011. Search for nodulation-related CLE genes in the genome of Glycine max. Journal of Experimental Botany 62, 2571–2583. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. 2007. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. Journal of Bioscience and Bioengineering 104, 34–41. [DOI] [PubMed] [Google Scholar]
- Nebenführ A, Gallagher LA, Dunahay TG, Frohlick JA, Mazurkiewicz AM, Meehl JB, Staehelin LA. 1999. Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiology 121, 1127–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. 2007. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. The Plant Journal 51, 1126–1136. [DOI] [PubMed] [Google Scholar]
- Nishida H, Handa Y, Tanaka S, Suzaki T, Kawaguchi M. 2016. Expression of the CLE-RS3 gene suppresses root nodulation in Lotus japonicus. Journal of Plant Research 129, 909–919. [DOI] [PubMed] [Google Scholar]
- Nishida H, Suzaki T. 2018. Nitrate-mediated control of root nodule symbiosis. Current Opinion in Plant Biology 44, 129–136. [DOI] [PubMed] [Google Scholar]
- Nishida H, Tanaka S, Handa Y, et al. . 2018. A NIN-LIKE PROTEIN mediates nitrate-induced control of root nodule symbiosis in Lotus japonicus. Nature Communications 9, 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura R, Hayashi M, Wu GJ, et al. . 2002. HAR1 mediates systemic regulation of symbiotic organ development. Nature 420, 426–429. [DOI] [PubMed] [Google Scholar]
- Novák K. 2010. Early action of pea symbiotic gene NOD3 is confirmed by adventitious root phenotype. Plant Science 179, 472–478. [DOI] [PubMed] [Google Scholar]
- Ogawa-Ohnishi M, Matsushita W, Matsubayashi Y. 2013. Identification of three hydroxyproline O-arabinosyltransferases in Arabidopsis thaliana. Nature Chemical Biology 9, 726–730. [DOI] [PubMed] [Google Scholar]
- Oh E, Seo PJ, Kim J. 2018. Signaling peptides and receptors coordinating plant root development. Trends in Plant Science 23, 337–351. [DOI] [PubMed] [Google Scholar]
- Okamoto S, Ohnishi E, Sato S, Takahashi H, Nakazono M, Tabata S, Kawaguchi M. 2009. Nod factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant & Cell Physiology 50, 67–77. [DOI] [PubMed] [Google Scholar]
- Okamoto S, Shinohara H, Mori T, Matsubayashi Y, Kawaguchi M. 2013. Root-derived CLE glycopeptides control nodulation by direct binding to HAR1 receptor kinase. Nature Communications 4, 2191. [DOI] [PubMed] [Google Scholar]
- Okamoto S, Suzuki T, Kawaguchi M, Higashiyama T, Matsubayashi Y. 2015. A comprehensive strategy for identifying long-distance mobile peptides in xylem sap. The Plant Journal 84, 611–620. [DOI] [PubMed] [Google Scholar]
- Osipova MA, Mortier V, Demchenko KN, Tsyganov VE, Tikhonovich IA, Lutova LA, Dolgikh EA, Goormachtig S. 2012. Wuschel-related homeobox5 gene expression and interaction of CLE peptides with components of the systemic control add two pieces to the puzzle of autoregulation of nodulation. Plant Physiology 158, 1329–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel N, Mohd-radzman NA, Corcilius L, Crossett B, Connolly A, Payne RJ, Djordjevic MA. 2017. Diverse peptide hormones affecting root growth identified in the Medicago truncatula secreted peptidome. Molecular & Cellular Proteomics 61, 1–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma JG, Jacobsen E, Feenstra WJ. 1988. 3 pea mutants with an altered nodulation studied by genetic-analysis and grafting. Journal of Plant Physiology 132, 424–430. [Google Scholar]
- Reid DE, Ferguson BJ, Gresshoff PM. 2011a. Inoculation- and nitrate-induced CLE peptides of soybean control NARK-dependent nodule formation. Molecular Plant-Microbe Interactions 24, 606–618. [DOI] [PubMed] [Google Scholar]
- Reid DE, Ferguson BJ, Hayashi S, Lin YH, Gresshoff PM. 2011b. Molecular mechanisms controlling legume autoregulation of nodulation. Annals of Botany 108, 789–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DE, Hayashi S, Lorenc M, Stiller J, Edwards D, Gresshoff PM, Ferguson BJ. 2012. Identification of systemic responses in soybean nodulation by xylem sap feeding and complete transcriptome sequencing reveal a novel component of the autoregulation pathway. Plant Biotechnology Journal 10, 680–689. [DOI] [PubMed] [Google Scholar]
- Sagan M, Duc G. 1996. Sym28 and Sym29, two new genes involved in regulation of nodulation in pea (Pisum sativum L.). Symbiosis 20, 229–245. [Google Scholar]
- Saint-Jore-Dupas C, Nebenführ A, Boulaflous A, Follet-Gueye ML, Plasson C, Hawes C, Driouich A, Faye L, Gomord V. 2006. Plant N-glycan processing enzymes employ different targeting mechanisms for their spatial arrangement along the secretory pathway. The Plant Cell 18, 3182–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Suzaki T, Soyano T, Kojima M, Sakakibara H, Kawaguchi M. 2014. Shoot-derived cytokinins systemically regulate root nodulation. Nature Communications 5, 4983. [DOI] [PubMed] [Google Scholar]
- Schnabel E, Journet EP, de Carvalho-Niebel F, Duc G, Frugoli J. 2005. The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Molecular Biology 58, 809–822. [DOI] [PubMed] [Google Scholar]
- Schnabel EL, Kassaw TK, Smith LS, Marsh JF, Oldroyd GE, Long SR, Frugoli JA. 2011. The ROOT DETERMINED NODULATION1 gene regulates nodule number in roots of Medicago truncatula and defines a highly conserved, uncharacterized plant gene family. Plant Physiology 157, 328–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle IR, Men AE, Laniya TS, Buzas DM, Iturbe-Ormaetxe I, Carroll BJ, Gresshoff PM. 2003. Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 299, 109–112. [DOI] [PubMed] [Google Scholar]
- Shinohara H, Matsubayashi Y. 2010. Arabinosylated glycopeptide hormones: new insights into CLAVATA3 structure. Current Opinion in Plant Biology 13, 515–519. [DOI] [PubMed] [Google Scholar]
- Showalter AM, Basu D. 2016. Extensin and arabinogalactan-protein biosynthesis: glycosyltransferases, research challenges, and biosensors. Frontiers in Plant Science 7, 814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano T, Hirakawa H, Sato S, Hayashi M, Kawaguchi M. 2014. NODULE INCEPTION creates a long-distance negative feedback loop involved in homeostatic regulation of nodule organ production. Proceedings of the National Academy of Sciences, USA 111, 14607–14612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki T, Yano K, Ito M, Umehara Y, Suganuma N, Kawaguchi M. 2012. Positive and negative regulation of cortical cell division during root nodule development in Lotus japonicus is accompanied by auxin response. Development 139, 3997–4006. [DOI] [PubMed] [Google Scholar]
- Suzaki T, Yoro E, Kawaguchi M. 2015. Leguminous plants: inventors of root nodules to accommodate symbiotic bacteria. International Review of Cell and Molecular Biology 316, 111–158. [DOI] [PubMed] [Google Scholar]
- Szczyglowski K, Shaw RS, Wopereis J, Copeland S, Hamburger D, Kasiborski B, Dazzo FB, de Bruijn FJ. 1998. Nodule organogenesis and symbiotic mutants of the model legume Lotus japonicus. Molecular Plant-Microbe Interactions 11, 684–697. [Google Scholar]
- Takahara M, Magori S, Soyano T, et al. . 2013. Too much love, a novel Kelch repeat-containing F-box protein, functions in the long-distance regulation of the legume–Rhizobium symbiosis. Plant & Cell Physiology 54, 433–447. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi M, Poole PS. 2013. Transport and metabolism in legume–rhizobia symbioses. Annual Review of Plant Biology 64, 781–805. [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. 2003. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. The Plant Journal 33, 949–956. [DOI] [PubMed] [Google Scholar]
- Wopereis J, Pajuelo E, Dazzo FB, Jiang Q, Gresshoff PM, De Bruijn FJ, Stougaard J, Szczyglowski K. 2000. Short root mutant of Lotus japonicus with a dramatically altered symbiotic phenotype. The Plant Journal 23, 97–114. [DOI] [PubMed] [Google Scholar]
- Xu C, Liberatore KL, MacAlister CA, et al. . 2015. A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nature Genetics 47, 784–792. [DOI] [PubMed] [Google Scholar]
- Yoshida C, Funayama-Noguchi S, Kawaguchi M. 2010. plenty, a novel hypernodulation mutant in Lotus japonicus. Plant & Cell Physiology 51, 1425–1435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.