Abstract
Molecular tools to target RNA site-specifically allow recoding of RNA information and processing. SNAP-tagged deaminases, guided by a chemically stabilized guideRNA, enable the simultaneous editing of targeted adenosine to inosine in several endogenous transcripts, with high efficiency (up to 90%), high potency, sufficient duration, and high precision. We applied SNAP-ADARs for the efficient and concurrent editing of two disease-relevant signaling transcripts, KRAS and STAT1. We also show improved performance compared to the recently described Cas13b-ADAR.
Tools to manipulate RNA efficiently and precisely are highly desired.1 We recently introduced SNAP-ADARs to substitute adenosine by inosine in RNA in a rational and programmable way with a guideRNA (Supplementary Fig. 1).2,3 As inosine is interpreted as guanosine, RNA editing can alter splicing, START and STOP codons, miRNA action, and can reprogram the protein product.4 Manipulation at the RNA-level is tunable in yield and reversible in time. This might be particularly useful for substitutions that are either lethal or compensated when introduced at the DNA-level5, e.g. in signaling proteins.6 A further advantage is safety,7 as off-site RNA editing can be considered reversible. Current methods8–10 typically apply overexpression of (engineered) deaminases which results in massive global off-target editing. In contrast, deaminase and guideRNA are covalently linked in our SNAP-ADAR approach, allowing efficient RNA-targeting after single-copy, genomic integration of the editase. Here, we comprehensively define the SNAP-ADAR approach.
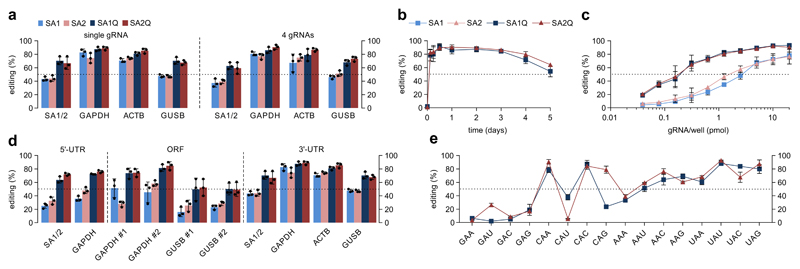
We validated four editases, SNAP-ADAR1 (SA1), SNAP-ADAR2 (SA2),2 and their hyper-active E→Q variants11 SA1Q and SA2Q. Editing was started by transfection of the short, chemically stabilized BG-guideRNA (Supplementary Fig. 1), and was analyzed for formal A-to-G conversion in cDNA at specific 5´-UAG triplets in the 3´-UTRs of the four targeted endogenous mRNAs: ACTB, GAPDH, GUSB, and SA1/2. For both wildtype enzymes (SA1/2), editing yields of 40-80% were achieved (Fig. 1a) depending on the target. Applying the hyperactive mutants (SA1Q/SA2Q) raised the yields to 65-90%, particularly the weaker edited transcripts GUSB & SA1/2 profited. The maximum editing yield (80-90%) was nearly obtained 3h post transfection (Fig. 1b), stayed constant for 3d, and then declined slowly, probably due to dilution of the guideRNA-enzyme conjugate by cell division. The activated enzymes (SA1Q&SA2Q) were up to 12fold more potent compared to the wildtype enzymes (SA1&SA2), achieving the half-maximum editing yield already with 0.15 pmol/well compared to 1-2 pmol/well (Fig. 1c). We tested the concurrent editing of all four transcripts by cotransfection of four guideRNAs. Notably, the yields stayed unchanged (Fig. 1a). We found similar results for the concurrent editing of three sites in the GAPDH mRNA (Supplementary Fig. 2). Editing yields were higher in the 3´-UTR compared to ORF and 5´-UTR (Fig. 1d), probably due to interference with translation. Accordingly, the faster enzymes (SA1Q & SA2Q) boosted the yields in the 5´-UTR from 25-50% to 60-75% and in the ORF from 15-60% to 50-85% (Fig. 1d). Furthermore, translation inhibition with puromycin increased ORF editing in SA1/2 cells to the level of 3´-UTR editing (Supplementary Fig. 3). To assess the codon scope, we targeted all 16 conceivable 5´-NAN triplets in the ORF of endogenous GAPDH for SA1Q and SA2Q. We obtained yields ranging from very little to almost quantitative reflecting the well-known preferences of ADARs (Fig. 1e).11,12 While editing was generally difficult for 5´-GAN triplets (<30%), significant yields (>50%) were achieved for 10/16 triplets. For 7/16 triplets, excellent editing yields (>70%) were obtained for at least one enzyme.
Figure 1. Editing performance of four SNAP-ADARs.
a) Engineered 293 cell lines expressing the respective SA enzyme were transfected with either a single gRNA or 4 gRNAs against 5´-UAG triplets in the indicated endogenous transcripts. b), c) Time- and dose-dependency of editing in the GAPDH transcript. d) Editing of 5´-UAG sites in various transcripts, 5´-UTR versus ORF and 3´-UTR. e) Comparative editing of all 16 triplets (5´-NAN) in the ORF of the endogenous GAPDH transcript. a) - e) Data are shown as the mean±SD, N=3 independent experiments, black dots represent individual data points.
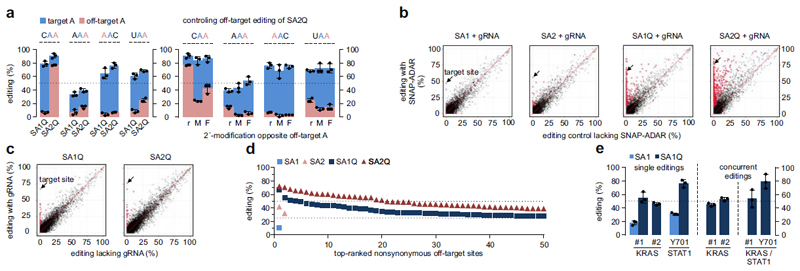
An important aspect is specificity. A major advantage of our strategy2 (compared to others8–10,13–15) is the suppression of off-site editing within the guideRNA/mRNA duplex by chemical modification of our guideRNA. Only for adenosine-rich triplets (AAC, AAA, UAA, CAA) some off-target editing was detected, mainly with SA2Q (5-75%) and mainly for the CAA triplet (Fig. 2a, left). Off-target editing was due to three natural nucleotides in the guideRNA opposite the targeted adenosine (Supplementary Fig. 4).2 Careful inclusion of further chemical modifications (2´-methoxy, 2´-fluoro, Fig. 2a, right) restricted off-target editing at the CAA triplet down to 20%, and limited off-target editing at all other sites to <10% without reducing on-target editing. Notably, for AAA, the additional modification even elevated the on-target yield from 40% to 50%. Global off-target editing is the main obstacle for RNA editing, in particularly under overexpression of editases.10,13,14,16 To test this for SNAP-ADARs under genomic expression, we conducted deep RNA sequencing when editing the ACTB transcript. We also assessed the role of guideRNA-dependent misguiding. The wildtype enzymes (SA1/2) were extremely precise. Among the 50.000 editing sites called (see Supplementary Data), only very few were significantly differently edited compared to the negative control (6 for SA1, 30 for SA2, Fig. 2b). Most of these sites are known17 sites in the 3´-UTRs (Table 1) and were edited below 25% (Supplementary Fig. 5a). For SA1, there was a single nonsynonymous editing (TMX3, 10%) that was guideRNA-dependent (Supplementary Table 1). For SA2, there were two nonsynonymous editings (AAGAB, 42% and CHFR, 32%) with the former being guideRNA-dependent. Off-targets were much more frequent with the hyper-active enzymes (835/1310 sites for SA1Q/SA2Q, Table 1, Fig. 2b), were caused by the free-floating enzyme, and comprised mainly of novel sites (74-85%). Only a small number of sites were edited guideRNA-dependent (for each editase ≈30 sites, Fig. 2c) A vast amount of sites was located in the ORF (347-496 sites) and gave rise to nonsynonymous editings (230-347 sites). However, none of the nonsynonymous editings exceeded that at the target site, and the majority was edited to low level. This was particularly true for SA1Q where only 4/227 sites were edited above 50% and 167/227 sites edited below 25% (Fig. 2d). For SA2Q, however, the average editing level was higher, with 20/344 sites above 50% and 240/344 below 25% editing yield. We found SA1Q and SA2Q to share only 414 of its off-target sites. SA1Q and SA2Q differ in their off-target codon preferences, with SA2Q accepting 5´-CAN triplets better (Supplementary Fig. 5b). All SNAP-ADAR cell lines behaved indistinguishable from normal 293 cells with respect to doubling times and morphology, and the FPKM analysis revealed no difference in gene expression due to the presence of (off-target) editing activity (Supplementary Fig. 6). As SA1(Q) shows a better balance of efficiency over specificity, we continued with SA1(Q).
Figure 2. Editing specificity and application.
a) Off-target editing of adjacent adenosines in A-rich triplets. r, M, F refers to the chemical modification opposite of the off-target A with r = natural ribonucleotide, M = 2´-methoxy, F = 2´-fluoro. b), c) Scatter plots of differential editing at ≈50.000 sites/experiment. The targeted site (ACTB) is indicated by an arrow. Significantly differently edited sites (p<0.01) are highlighted as red dots. In b), editing is compared to a control cell line lacking SA expression. c) shows editing in presence versus absence of the gRNA. d) Nonsynonymous off-target sites ranked by editing yields. e) Editing of signaling transcripts. Two 5´-UAG sites in the ORF of KRAS (#1, #2), and a 5´-UAU site in STAT1 (Tyr701) were targeted. For concurrent editing, two respective gRNAs were cotransfected into SA1Q cells. Data in a), e) are shown as the mean±SD, N=3 independent experiments, black dots represent individual data points. Significance in b), c) was tested by Fisher´s exact test (two-sided), N=2 independent experiments.
Table 1.
Global off-target editing. Given are the numbers of sites significantly differently edited compared to the related cell line lacking SA expression.
| location in mRNA |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| known | novel | coding regionb | ||||||||||
| enzymea | total | Alu | non-Alu | Alu | non-Alu | 5’-UTR | syn. | nonsyn. | 3’-UTR | othersc | ||
| SA1 | 6 | 2 | 1 | 0 | 3 | 0 | 0 | 1 | 3 | 2 | ||
| SA2 | 30 | 15 | 8 | 1 | 6 | 0 | 0 | 2 | 22 | 6 | ||
| SA1Q | 835 | 70 | 59 | 7 | 699 | 11 | 117 | 230 | 402 | 75 | ||
| SA2Q | 1310 | 267 | 71 | 24 | 948 | 13 | 149 | 347 | 637 | 164 | ||
Editings were carried out in cells expressing the respective SNAP-ADAR in presence of a BG-guideRNA targeting ACTB.
Syn. = synonymous, nonsyn. = nonsynonymous.
Others refers to editing in introns, intergenic regions, and ncRNA.
RNA editing would be particularly attractive for the manipulation of signaling networks. For illustration, we edited two 5´-UAG sites in KRAS (#1) and the Tyr701 site (5´-UAU) in STAT1, its most relevant phosphorylation site18 for signaling. With SA1Q, we achieved editing levels of 55±8% (KRAS site #1), 46±2% (KRAS site #2), and 76±6% (STAT1), see Fig. 2e. We found no detectable off-target editing in the guideRNA/mRNA duplex (Supplementary Fig. 7). Again, concurrent editing of either two sites on KRAS or on two transcripts (KRAS and STAT1) was possible without losing editing efficiency (Fig. 2e). The highly precise editase SA1 was less active, but still able to obtain yields of 18±3% (KRAS, site #1) and 31±2% (STAT1).
In summary, SNAP-ADARs are versatile tools due to their unique RNA-targeting mechanism. A single genomic copy is sufficient to achieve editing at endogenous transcripts after the transfection of one or several short guideRNAs. We demonstrated 11/16 codons to be editable in yields ranging from 50%- 90%. The editable codons (5´-UAG, UAC, UAU, UAA, and AAG) indicate amino acid substitutions (Thr-to-Ala, Tyr-to-Cys, Ser-to-Gly, and Lys-to-Arg (Supplementary Fig. 5c) highly attractive for the manipulation of signaling networks. This was highlighted by targeting the phosphorylation site Tyr701 that controls STAT1 signaling, and by editing two signaling transcripts concurrently. The chemical modification of our guideRNA restricts off-target editing in the mRNA/guideRNA duplex. The latter was problematic in two competing approaches (one based on Cas13b).10,13,14 Those approaches suffer from massive global off-target editing caused by the overexpressed editases (Supplementary Tables 2 and 3).10,16 For SNAP-ADARs, however, global off-target editing was restricted by genomic integration. It was almost eliminated for the precise editases SA1/2, while editing of endogenous targets was still sufficient for some codons (UAG, UAU). The performance of SA1 was also better than that of the “high-specificity variant” of Cas13b-ADAR (Supplementary Note 1).10 Notably, our integrated hyper-active SA1Q/2Q showed orders of magnitude less off-target editing compared to overexpressed Cas13b-ADAR version 110 or λN-deaminases16 (Supplementary Fig. 8). We showed that a further lowering of SA1Q/2Q expression (up to 25fold) is possible to further reduce off-target editing (Supplementary Fig. 9). One could further improve the guideRNA chemistry19 or the editase of our approach.10,11 Notably, we have tested the reported “high specificity variant” of Cas13b-ADAR (T375G) but in the context of SNAP-ADAR (Supplementary Fig. 10). Different from older claims,10 we found this mutant to be much less efficient than SA1Q/2Q, and even inferior to SA1/2. Compared to other approaches, our guideRNAs are extremely short (22 nt). Thus editing clearly depends on the targeting mechanism and will not interfere with endogenous ADARs.9 However, we found the long Cas13b-guideRNAs (85 nt) to recruit overexpressed human ADAR2 but also SA2Q to elicit editing of a co-transfected reporter to levels similar as Cas13b-ADAR does (Supplementary Fig. 11). This observation raises the question to which extent the claims done by Cox et al. are flawed by overexpression artefacts (online methods, Supplementary Note 2). Finally, the small size (20 kDa) and the human origin of the SNAP-tag provide further advantages over Cas13-ADAR. Together, we set a new benchmark for site-directed RNA editing and provide a tool ready for concurrent editing of endogenous transcripts.
Online Methods
BG-guideRNA synthesis
Synthesis of chemically modified BG-guideRNAs doesn´t require any chemistry equipment. All chemical modifications used all commercially available. The BG modification can be included by applying commercial amino or thiol reactive BG derivatives like BG-maleiimide (New England Biolabs). The sequences and chemical modification of all guideRNAs are given in Supplementary Table 4. For this study, all NH2-guideRNAs were purchased from Biospring (Germany) as HPLC-purified ssRNAs with a 5’-C6 amino linker. Alternative to commercial BG derivatives, our protocol can be used to introduce the BG moiety. Benzylguanine connected to a carboxylic acid linker2,3 (12 µl, 60 mM in DMSO) was in-situ activated as an OSu-ester by incubation with EDCI·HCl (12 µl, 17.4 mg/ml in DMSO) and NHS (12 µl, 17.8 mg/ml in DMSO) for 1 h at 30°C. Then, the NH2-guideRNA (25 µl, 6 µg/µl) and DIPEA (12 µl, 1:20 in DMSO) were added to the pre-activation mix and incubated (90 min, 30°C).20 The crude BG-guideRNA was purified from unreacted NH2-guideRNA by 20% urea PAGE and then extracted with H2O (700 µl, overnight at 4°C). RNA precipitation was done with sodium acetate (0.1 volumes, 3.0 M) and ethanol (3 volumes, 100%, overnight at -80°C). The BG-guideRNA was washed with ethanol (75%) and dissolved in water (60 µl).
SNAP-ADAR expressing cell lines
Each respective enzyme was integrated as a single copy under control of the dox-inducible CMV promotor at the FRT site into the genome of 293 Flip-In cells (R78007, Thermo Fisher scientific) as described before9. The exact cDNAs are given in the Supplementary Information (appendix). Enzyme expression of all four enzymes was inducible by doxycycline (10 ng/ml) to roughly comparable levels as validated by Western blot and fluorescence microscopy (Supplementary Fig. 12 and Supplementary Note 3). Also at the RNA level, the expression levels of SA1 (wt & Q) and SA2 (wt and Q) were roughly comparable with average FPKM values of 679 and 814 for SA1(Q) and SA2(Q), respectively. The E→Q mutation did not change the protein localization (Supplementary Note 3). SA1(Q) is localized to cytoplasm and nucleoplasm; SA2(Q) is mainly localized to cytoplasm. To determine the location of the different SNAP-ADAR proteins, 1 × 105 cells were seeded in 500 µl selection media with or without doxycline (10 ng/ml) on poly-d-lysine-coated cover slips in a 24-well format. After one day, BG-FITC labeling of the SNAP-tag and nuclear staining was done. To validate SNAP-ADAR protein amounts, Western blot analysis was used. For this, 3 × 105 cells were seeded in 500 µl selection media with or without doxycline (10 ng/ml) in a 24-well format for one day. Then, cells were lysed with urea buffer (8 M urea in 10 mM Tris, 100 mM NaH2PO4, pH 8.0). Protein lysate (5 µg) was separated by SDS-PAGE and transferred onto a PVDF membrane (Bio-Rad Laboratories, USA) for immunoblotting with primary antibodies against the SNAP-tag (1:1000, P9310S, New England Biolabs, USA) and ß-actin (1:40000, A5441, Sigma Aldrich, USA). Afterwards, the blot was incubated with HRP-conjugated secondary antibodies against rabbit (1:10000, 111-035-003, Jackson Immuno Research Laboratories, USA) and mouse (1:10000, 115-035-003, Jackson Immuno Research Laboratories, USA) and visualized by enhanced chemiluminescence.
RNA editing experiments
General. 293-Flip-In T-REx cells stably transfected with the respective SNAP-ADAR-expressing pcDNA5 vector were grown in DMEM + 10% FBS + 100 µg/ml hygromycin B + 15 µg/ml blasticidin S. For experiments, 3 × 105 cells/well were seeded in 24 well plates, and gene expression was induced by doxycycline (10 ng/ml) for one day. Then, 8 × 104 cells/well were resuspended in 100 µl DMEM + 10% FBS + 15 ng/ml doxycycline and reverse-transfected in 96 well format with the guideRNA transfection mixture (39 fmol – 40 pmol guideRNA + 0.75 µl Lipofectamine 2000 in 50 µl OptiMEM; the exact amounts guideRNA applied used in this study are given in Supplementary Table 4). After 24 h, cells were harvested for RNA isolation. When determining editing yields at later time points, cells were resuspended in DMEM + 10% FBS + 10 ng/ml doxycline and seeded into 24-well plates. 48 hours later, fresh medium containing 10% FBS + 10 ng/ml doxycline was again given to the cells. RNA was extracted using the RNeasy MinElute Kit (Qiagen, Germany) and treated with DNaseI. After DNA digestion, RNA was converted into cDNA for the subsequent amplification by Taq DNA PCR. The DNA was analyzed by Sanger sequencing (Eurofins Genomics, Germany). A-to-I editing yields were quantified by comparing the height of the resulting guanosine peak divided by the sum of the peak heights of the guanosine plus adenosine peaks at a respective site. In general, negative controls were run for all experiments and never showed detectable editing. Potential editing at the DNA versus RNA level. To check for potential A-to-I editing of the genomic DNA beside the RNA, the innuPREP DNA/RNA Mini Kit (Analytik Jena, Germany) was used to extract genomic DNA and RNA from the cells in parallel. We followed the manufacturer´s protocol. Cellular RNA was further reverse-transcribed as described above, the genomic DNA was immediately amplified by Taq DNA PCR and sequenced without reverse transcription. No A-to-G change at the DNA was detectable (Supplementary Fig. 13). Potency and time dependency. For the potency and the time-dependence experiments, RNA was isolated by using 500 µl of TRI Reagent (Sigma Aldrich, USA). Chloroform (100 µl) was added to extract the RNA for precipitation with isopropanol (350 µl) in presence of linear acryl amide (1.5 µl, 5 mg/ml). The RNA pellet was washed twice (500 µl 75% ethanol) and was finally dissolved in RNase-free water (30 µl). Furthermore, we tested if the editing efficiency and potency was dependent on the formation of the covalent bond between guideRNA and SNAP-ADAR. guideRNAs lacking the benzylguanine moiety could only elicit substantial editing with the hyperactive enzymes (up to 70% editing yield), but required ≈50fold higher guideRNA amounts (EC50 ca. 6-7 pmol/well, Supplementary Fig. 14). With the wild-type enzymes no substantial editing was obtained even at the highest guideRNA concentration (20 pmol/well). The target site in the potency screen was UAG site #2 in the ORF of endogenous GAPDH. The target in the time dependency screen was a 5´-UAG site in the 3´-UTR of endogenous GAPDH. Triplet scope. When studying the editing scope with all 16 5´-NAN triplets, we chose targets such that no amino acid change resulted. Only for four triplets, sites had to be chosen that elicit amino acid changes. Then, sites were selected that were expected not to interfere with GAPDH activity (see appendix in the Supplementary Information). Applicability. In terms of maximum yield (up to 90%), potency (≥1 pmol/well), and duration (several days), site-directed RNA editing behaves comparable to RNA interference with transfected siRNAs21 in cell culture and may allow numerous applications. However, it is difficult to reliably predict the outcome of an editing reaction from the triplet preference (Fig. 1e) alone. The accessibility of an arbitrary target might be limited by RNA secondary structure, RNA-binding proteins,22 low mRNA copy numbers, and short half-lives.
Off-target editing
Accurate analyses uncovered an example for off-target editing at the targeted transcript but outside the guideRNA/mRNA duplex. This was undetectable for SA1/2, but was found for SA1Q (50% editing of one AAG triplet in GAPDH) and for SA2Q (70% editing of a CAG site in GAPDH). These two strongly edited sites in GAPDH were predicted by mfold to be located in highly double-stranded regions of the transcript (Supplementary Fig. 15). In accordance, editing yields correlated with the proximity of the guideRNA binding site, a pattern reminiscent of the recently described TRIBE method to elucidate binding sites of RNA binding proteins.23
Next generation RNA sequencing experiments
The RNA editing was done by transfection of 5 pmol guideRNA against a 5´-UAG triplet in the 3´-UTR of ACTB into the respective Flip-In cell line as described above. Overall, seven settings were carried out, each with an independent duplicate. Those settings include: (1) empty lipofection into empty (not expressing SA) 293 Flip-In cells, (2) guideRNA lipofection into SA1 cells, (3) guideRNA transfection into SA2 cells, (4) empty transfection into SA1Q cells, (5) empty transfection into SA2Q cells, (6) guideRNA transfection into SA1Q cells, and (7) guideRNA transfection into SA2Q cells. RNA was isolated with the RNeasy MinElute Kit, treated with DNaseI and purified again with the RNeasy MinElute Kit. Purified RNA (1.2 µg) was delivered to CeGaT (Germany) for Poly(A)+ mRNA sequencing. The library was prepared from 100 ng RNA with the TruSeq Stranded mRNA Library Prep Kit (Illumina, USA), and sequenced with the HiSeq4000 (50 M reads, 2 × 100 bp paired end, Illumina, USA). Mapping of RNA-seq and reads: We adopted the previously published pipeline to accurately align RNA-seq reads onto the genome.24,25 We used BWA26 to align the reads to a combination of the reference genome sequences and exonic sequences surrounding known splicing junctions from known gene models. Each of the paired-end reads was mapped separately using the commands “bwa aln fastqfile” and “bwa samse -n4”. We then chose the length of the splicing junction to be slightly shorter than the RNA-seq reads to prevent redundant alignment (i.e. 95 bp for reads of 100 bp length). The reference genomes used were hg19 and the gene models were obtained through the UCSC Genome Browser for Gencode, RefSeq, Ensembl, and UCSC Genes. We only considered uniquely mapped reads with mapping quality q>10 and used samtools rmdup27 to remove clonal reads (PCR duplicates) mapped to the same location. Of these identical reads, only the read with the highest mapping quality was kept for downstream analysis. Unique and non-duplicate reads were subjected to local realignment and base score recalibration using the IndelRealigner and TableRecalibration from the Genome Analysis Toolkit (GATK)28. The above steps were applied separately to each of the RNA-seq samples. Identification of editing sites from RNA-seq data: We used the UnifiedGenotyper from GATK28 to call variants from the mapped RNA-seq reads. In contrast to the usual practice of variant calling, we identified the variants with relatively loose criteria by using the UnifiedGenotyper tool with options stand_call_conf 0, stand_emit_conf 0, and output mode EMIT_VARIANTS_ONLY. Variants from non-repetitive and repetitive non-Alu regions were required to be supported by at least three reads containing mismatches between the reference genome sequences and RNA-seq. Supporting of one mismatch read was required for variants in Alu regions. This set of variant candidates was subject to several filtering steps to increase the accuracy of editing site calling. We first removed all known human SNPs present in dbSNP (except SNPs of molecular type “cDNA”; database version 135; http://www.ncbi.nlm.nih.gov/SNP/), the 1000 Genomes Project, and the University of Washington Exome Sequencing Project (http://evs.gs.washington.edu/EVS/). To remove false positive RNA-seq variant calls due to technical artifacts, further filters were applied as previously described.24,25 In brief, we required a variant call quality Q>20,24,25 discarded variants if they occurred in the first six bases of a read,26 removed variants in simple repeats,27 removed intronic variants that were within 4bp of splice junctions, and28 discarded variants in homopolymers. Moreover, we removed sites highly similar regions of the genome by BLAT29. Finally, variants were annotated using ANNOVAR30 based on gene models from Gencode, RefSeq, Ensembl, and UCSC. The resulting sets of sites identified from RNA-seq data were compared with all sites available in the RADAR database17 and were subsequently referred to as ‘known’ sites if also found in RADAR, or ‘novel’ sites if not found. Identification of significantly differently edited sites: We quantified editing levels of edited sites with ≥ 50 reads coverage (combined coverage of both replicates) and performed Fisher’s exact tests (adjusted p-value < 0.01) to identify significantly differently edited sites across the samples (editing difference > 10%). Additional NGS quality data is given in the appendix of the Supplementary Information.
Benchmark with Cas13b-ADAR and λN-deaminases
The SNAP-ADAR approach was benchmarked to the recently published Cas13b-ADAR approach (Supplementary Notes 1 and 2, Supplementary Table 2, and Supplementary Fig. 10-11). First, we repeated the editing of KRAS site #1 and #2 with SA1 and SA1Q. SA1Q achieved better editing yields than Cas13b-ADAR version 1 (e.g. 50-65% compared to 15-25% for KRAS site #1), SA1 was better than Cas13b-ADAR version 2 (e.g. 18-20% versus ca. 12%), editing strictly depended on the targeting mechanism, there was no off-target editing in the mRNA/guideRNA duplex, see Supplementary Note 1. ADARs are known to edit dsRNA substrates of >30 bp readily. We wondering if the large Cas13-guideRNAs (85 nt, 50 bp duplex) are able to recruit human ADAR or any other ADAR fusion protein independent of a specific targeting mechanism. Indeed, we found such 50 bp guideRNAs to recruit overexpressed ADAR2 but also engineered SA2Q to elicit editing of a co-transfected reporter transcript to levels similar as Cas13-ADAR does (ca. 25-30%, Supplementary Fig. 11, Supplementary Note 2). This medium level editing was apparently due to self-targeting of the deaminase (domain) alone and independent of a specific targeting mechanism. Most of the experiments reported by Cox et al. are done under such co-overexpression conditions and it remains unclear to which extent those results rely on a true (Cas-dependent) targeting mechanism and which are overexpression artefacts (self targeting). The lacking codon preference reported for repairV1 (with 10-35% editing yields) could be impaired by this. Cox et al. argue that Cas-ADAR has a weak codon preference due to a tight binding of the Cas protein to the mRNA/gRNA complex but they don´t show sufficient data or controls to support this. In the worst case, a very stable long RNA duplex wrapped by Cas-ADAR could inhibit translation, in particular, when the Start codon is close or even included, as this is given for the KRAS transcript they mostly report on (Supplementary Note 1). As we have shown here in the context of SNAP-ADARs, translation inhibition with puromycin can indeed increase editing levels in the ORF (Supplementary Fig. 3). In this respect, it is notable that we have tested the mutation from their “high specificity” Cas-ADAR repair version2 (T375G), but in the context of SNAP-ADAR. For this, we genomically integrated SA2QG (E488Q + T375G) and tested it side-by-side with SA1 and SA2 for the editing of five codons in the ORF of GAPDH (UAG, CAA, CAG, AAG, GAU). SA2QG elicited only minor editing at the UAG codon (15%) and no significant yield with the other four codons (Supplementary Fig. 10). It was always less active than the two wildtype SA enzymes which gave editing at some of the codons (ca. 40% at UAG, 23-66% at CAA, 18% at CAG). In the ORF, SA2QG seems unable to edit even the preferred UAG codon sufficiently. However, editing was successful when targeting a UAG triplet in the 3´-UTR of GAPDH (80% SA2QG, 85-90% for wt SA). Unfortunately, Cox et al. missed to comprehensively characterize repairV2 and to show if and how it promotes the editing reaction. Notably, our data predicts that the wildtype deaminase would always be the better choice (than repairV2) to achieve decent editing at preferred codons with manageable off-target edits also in the context of Cas-ADAR. The true mechanism of Cas-ADAR directed RNA editing and how it is applied best remains partly unclear. We also provide a side-by-side comparison to the λN-deaminase approach (Supplementary Table 3) and reanalyzed their NGS data with our pipeline (Supplementary Fig. 8). In comparison, our wt SA1/SA2 enzymes are highly precise and provoke several hundred-fold less off-target editing. Our hyperactive enzymes SA1Q/2Q are less prone to off-target editing than the wildtype versions of the λN-deaminases and much less off-target prone than the hyperactive version of the λN-deaminases.
Supplementary Material
Acknowledgements
We gratefully acknowledge support from the Deutsche Forschungsgemeinschaft to T.S. (STA 1053/3-2; STA 1053/7-1). This work has received funding for T.S. from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement No 647328). This work is supported by National Institutes of Health (NIH) grants R01GM102484 and R01GM124215 to J.B.L.
Footnotes
Data availability. All original NGS data has been deposited to the NCBI GEO server under the data set identifier GSE112787. (please LINK to https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE112787) Our NGS data analysis is available online as Supplementary Data. All programs used are publically available. The gene sequences of all constructs are given in the Supporting Information, plasmids can be requested from T.S. Further information on experimental design is available in the Life Science Reporting Summary.
Author contributions. P.V., M.M., T.M., K.S. and T.S. conceived, performed and analyzed the wet lab experiments; Q.L. and J.B.L. analyzed and all authors interpreted NGS data, all authors contributed to writing the manuscript.
Competing financial interests. All authors declare no competing financial interest.
References
- 1.Frye M, Jaffrey SR, Pan T, Rechavi G, Suzuki T. RNA modifications: what have we learned and where are we headed? Nat Rev Gen. 2016;17:365–372. doi: 10.1038/nrg.2016.47. [DOI] [PubMed] [Google Scholar]
- 2.Vogel P, Schneider MF, Wettengel J, Stafforst T. Improving Site-Directed RNA Editing In Vitro and in Cell Culture by Chemical Modification of the GuideRNA. Angew Chem Int Ed. 2014;53:6267–6271. doi: 10.1002/anie.201402634. [DOI] [PubMed] [Google Scholar]
- 3.Stafforst T, Schneider MF. An Engineered Guide RNA-Dependent Deaminase Selectively Repairs Point Mutations. Angew Chem Int Ed. 2012;51:11166–9. doi: 10.1002/anie.201206489. [DOI] [PubMed] [Google Scholar]
- 4.Nishikura K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat Rev Mol Cell Biol. 2016;17:83–96. doi: 10.1038/nrm.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rossi A, et al. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524:230–233. doi: 10.1038/nature14580. [DOI] [PubMed] [Google Scholar]
- 6.Vogel P, Stafforst T. Site-Directed RNA Editing with Antagomir Deaminases — A Tool to Study Protein and RNA Function. ChemMedChem. 2014;9:2021–2025. doi: 10.1002/cmdc.201402139. [DOI] [PubMed] [Google Scholar]
- 7.Schaefer KA, Wu W-H, Colgan DF, Tsang SH, Bassuk AG, Mahajan VB. Unexpected mutations after CRISPR-Cas9 editing in vivo. Nat Meth. 2017;14:547–549. doi: 10.1038/nmeth.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Montiel-Gonzalez MF, Guillermo I, Yudowski A, Rosenthal JJC. Correction of mutations within the cystic fibrosis transmembrane conductance regulator by site-directed RNA editing. Proc Natl Acad Sci USA. 2013;110:18285–290. doi: 10.1073/pnas.1306243110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wettengel J, Reautschnig J, Geisler S, Kahle PJ, Stafforst T. Harnessing human ADAR2 for RNA repair – Recoding a PINK1 mutation rescues mitophagy. Nucl Acids Res. 2017;45:2797–2808. doi: 10.1093/nar/gkw911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, Zhang F. RNA editing with CRISPR-Cas13. Science. 2017 doi: 10.1126/science.aaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuttan A, Bass BL. Mechanistic insights into editing-site specificity of ADARs. Proc Natl Acad Sci USA. 2012;109:E3295–E3304. doi: 10.1073/pnas.1212548109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eggington JM, Greene T, Bass BL. Predicting sites of ADAR editing in double-stranded RNA. Nat Commun. 2011;2 doi: 10.1038/ncomms1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sinnamon JR, Kim SY, Corson GM, Song Z, Nakai H, Adelman JP, Mandel G. Site-directed RNA repair of endogenous Mecp2 RNA in neurons. Proc Natl Acad Sci USA. 2017 doi: 10.1073/pnas.1715320114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montiel-Gonzalez MF, Vallecillo-Viejo IC, Rosenthal JJC. An efficient system for selectively altering genetic information within mRNAs. Nucl Acids Res. 2016;44 doi: 10.1093/nar/gkw738. gkw738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukuda M, Umeno H, Nose K, Nishitarumizu A, Noguchi R, Nakagawa H. Construction of a guide-RNA for site-directed RNA mutagenesis utilising intracellular A-to-I RNA editing. Sci Rep. 2017;7 doi: 10.1038/srep41478. 41478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vallecillo-Viejo IC, et al. Abundant off-target edits from site-directed RNA editing can be reduced by nuclear localization of the editing enzyme. RNA Biol. 2017 doi: 10.1080/15476286.2017.1387711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramaswami G, Li JB. RADAR: a rigorously annotated database of A-to-I RNA editing. Nucl Acids Res. 2014;42:D109–D113. doi: 10.1093/nar/gkt996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bromberg J. Stat proteins and oncogenesis. J Clin Invest. 2002;109:1139–42. doi: 10.1172/JCI15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bennett CF, Baker BF, Pham N, Swayze E, Geary RS. Pharmacology of Antisense Drugs. Annu Rev Pharmacol Toxicol. 2017;57:81–105. doi: 10.1146/annurev-pharmtox-010716-104846. [DOI] [PubMed] [Google Scholar]
- 20.Hanswillemenke A, Kuzdere T, Vogel P, Jekely G, Stafforst T. Site-directed RNA editing in vivo can be triggered by the light-driven assembly of an artificial riboprotein. J Am Chem Soc. 2015;137:15875–81. doi: 10.1021/jacs.5b10216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim D-H, Behlke MA, Rose SD, Chang M-S, Choi S, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotech. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 22.Deffit SN, Hundley HA. To edit or not to edit: regulation of ADAR editing specificity and efficiency. WIREs RNA. 2016;7:113–127. doi: 10.1002/wrna.1319. [DOI] [PubMed] [Google Scholar]
- 23.McMahon AC, Rahman R, Jin H, Shen JL, Fieldsend A, Luo W, Rosbash M. TRIBE: Hijacking an RNA-Editing Enzyme to Identify Cell-Specific Targets of RNA-Binding Proteins. Cell. 2016;165:742–53. doi: 10.1016/j.cell.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramaswami G, Lin W, Piskol R, Tan MH, Davis C, Li JB. Accurate identification of human Alu and non-Alu RNA editing sites. Nat Meth. 2012;9:579–81. doi: 10.1038/nmeth.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramaswami G, et al. Identifying RNA editing sites using RNA sequencing data alone. Nat Meth. 2013;10:128–132. doi: 10.1038/nmeth.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinform. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinform. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucl. Acids Res. 2010;38:e164–e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.