Abstract
Obesity increases the risk of hormone receptor–positive breast cancer in postmenopausal women. Levels of aromatase, the rate-limiting enzyme in estrogen biosynthesis, are increased in the breast tissue of obese women. Both prostaglandin E2 (PGE2) and hypoxia-inducible factor 1α (HIF-1α) contribute to the induction of aromatase in adipose stromal cells (ASCs). Sirtuin 1 (SIRT1) binds, deacetylates, and thereby inactivates HIF-1α. Here, we sought to determine whether SIRT1 also plays a role in regulating aromatase expression. We demonstrate that reduced SIRT1 levels are associated with elevated levels of acetyl–HIF-1α, HIF-1α, and aromatase in breast tissue of obese compared with lean women. To determine whether these changes were functionally linked, ASCs were utilized. In ASCs, treatment with PGE2, which is increased in obese individuals, down-regulated SIRT1 levels, leading to elevated acetyl–HIF-1α and HIF-1α levels and enhanced aromatase gene transcription. Chemical SIRT1 activators (SIRT1720 and resveratrol) suppressed the PGE2-mediated induction of acetyl–HIF-1α, HIF-1α, and aromatase. Silencing of p300/CBP-associated factor (PCAF), which acetylates HIF-1α, blocked PGE2-mediated increases in acetyl–HIF-1α, HIF-1α, and aromatase. SIRT1 overexpression or PCAF silencing inhibited the interaction between HIF-1α and p300, a coactivator of aromatase expression, and suppressed p300 binding to the aromatase promoter. PGE2 acted via prostaglandin E2 receptor 2 (EP2) and EP4 to induce activating transcription factor 3 (ATF3), a repressive transcription factor, which bound to a CREB site within the SIRT1 promoter and reduced SIRT1 levels. These findings suggest that reduced SIRT1-mediated deacetylation of HIF-1α contributes to the elevated levels of aromatase in breast tissues of obese women.
Keywords: estrogen, obesity, adipose tissue, prostaglandin, sirtuin, hypoxia-inducible factor (HIF), aromatase
Introduction
Obesity is a risk factor for the development of estrogen receptor-positive (ER+)3 breast cancer in postmenopausal women (1–3). In addition to its impact on breast cancer risk, obesity is recognized to be a poor prognostic factor for patients with ER+ breast cancer (4–8). Cytochrome P450 aromatase, a product of the CYP19A1 gene, is the rate-limiting enzyme for the synthesis of estrogens from androgens (9). Increased levels of aromatase are found in the breast tissue of obese women, which helps to explain the link between obesity and ER+ breast cancer (10, 11). It is important, therefore, to elucidate the mechanisms that lead to elevated aromatase expression in the breast tissue of obese women. The expression of aromatase is tightly regulated, with its transcription controlled by several tissue-specific promoters. In normal breast adipose, aromatase is under the control of promoter I.4 and expressed at low levels. By contrast, in obesity, the coordinated activation of promoters I.3 and II leads to a significant increase in aromatase expression (11). The proximal promoters I.3 and II are located near each other, stimulated by activation of the cAMP → PKA → cAMP-response element–binding protein (CREB) pathway, and aided by many other regulators, including hypoxia inducible factor-1α (HIF-1α). Prostaglandin E2 (PGE2), a bioactive lipid that is elevated in obesity, can induce HIF-1α, contributing, in turn, to enhanced CYP19A1 transcription in adipose stromal cells (ASCs) (11–13). These cells are believed to be a major source of estrogen that acts in a paracrine manner to stimulate tumor formation and growth (12, 14, 15). Although HIF-1α plays a critical role in regulating aromatase expression in breast ASCs, the mechanisms that control HIF-1α activity are incompletely understood.
HIF-1α and HIF-1β heterodimerize to form HIF-1, which binds to core hypoxia response elements and induces the expression of genes responsible for metabolic reprogramming, angiogenesis, and metastasis (16–18). The activity of HIF-1 is primarily regulated by oxygen-dependent protein degradation and the transactivation function of HIF-1α. Under normoxic conditions, HIF-1α is hydroxylated, which triggers ubiquitination leading to rapid proteolysis. By contrast, under hypoxic conditions, HIF-1α becomes stable and active because of inactivation of oxygen-dependent hydroxylases. The longevity-associated protein sirtuin 1 (SIRT1) has protein deacetylase activity and targets many transcription factors, including HIF-1α (19–21). SIRT1 binds to HIF-1α and deacetylates it, thereby inactivating HIF-1α (22). Levels of SIRT1 are reduced in the adipose tissue of obese humans (23–26). Collectively, these findings suggest the possibility that the increased expression of aromatase found in the breast tissue of obese women could be due, in part, to reduced expression of SIRT1, resulting in elevated levels of acetylated HIF-1α and activation of CYP19A1 gene expression.
In the present study, we had two main objectives. The first was to determine whether SIRT1 levels were reduced in breast tissue from obese versus lean women and correlated with levels of acetylated HIF-1α, HIF-1α, and aromatase. A second goal was to elucidate the role of SIRT1 in PGE2-mediated induction of aromatase. We present evidence that SIRT1 levels are reduced in the breast tissue of obese women and inversely correlated with levels of acetylated HIF-1α, HIF-1α, and aromatase. Moreover, new insights are provided into the signaling pathway by which PGE2 inhibits SIRT1 gene expression, leading, in turn, to increased levels of acetylated HIF-1α and aromatase in ASCs.
Results
Levels of SIRT1 are decreased in the breast tissue of obese women and correlate with amounts of acetyl–HIF-1α, HIF-1α, and aromatase
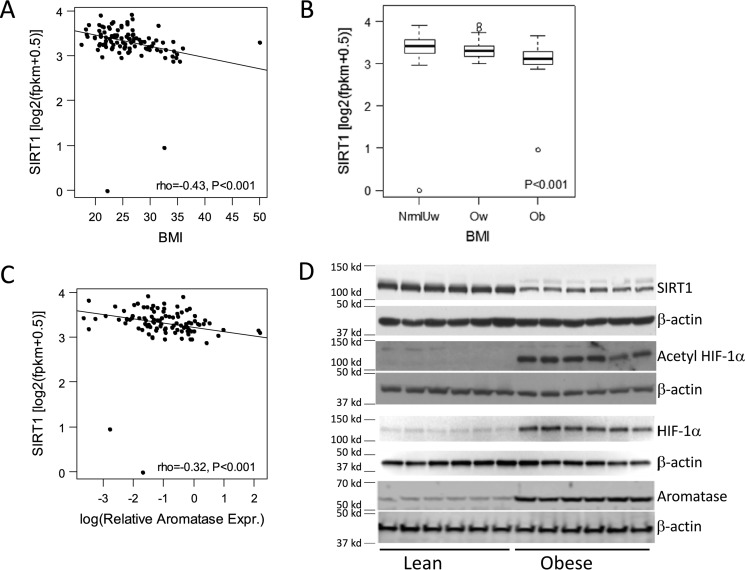
Initially, we correlated breast levels of SIRT1 mRNA with body mass index (BMI). SIRT1 mRNA levels negatively correlated with BMI (Fig. 1A) and were lowest in the breast tissue of obese women (Fig. 1B). To determine whether there was a potential relationship between SIRT1 and aromatase, levels of these two mRNAs were correlated. As shown in Fig. 1C, a negative correlation was found between levels of SIRT1 and aromatase mRNAs. Because SIRT1 can deacetylate and inactivate HIF-1α, a known regulator of CYP19A1 transcription, we also compared levels of SIRT1, acetyl–HIF-1α, HIF-1α, and aromatase proteins in the breast tissue of obese versus lean women. Following immunoprecipitation, Western blotting was performed and revealed reduced levels of SIRT1 in association with higher levels of acetylated HIF-1α, HIF-1α, and aromatase in breast tissue from obese versus lean women (Fig. 1D).
Figure 1.
Elevated BMI is associated with decreased expression of SIRT1 and increased acetyl–HIF-1α, HIF-1α, and aromatase in the breast. A, higher BMI correlates with decreased SIRT1 mRNA levels in breast tissue. B, obese women have decreased levels of SIRT1 mRNA in the breast compared with overweight or lean women (normal/underweight, n = 48; overweight, n = 33; obese, n = 19). Error bars, S.D. C, levels of SIRT1 and aromatase mRNAs are inversely correlated. RNA-Seq was used to quantify SIRT1 levels in A–C. D, immunoprecipitation followed by Western blotting demonstrated decreased levels of SIRT1 and increased levels of acetyl–HIF-1α, HIF-1α, and aromatase in breast tissue of obese (n = 6) versus lean (n = 6) women.
PGE2-mediated induction of aromatase depends on acetylation of HIF-1α
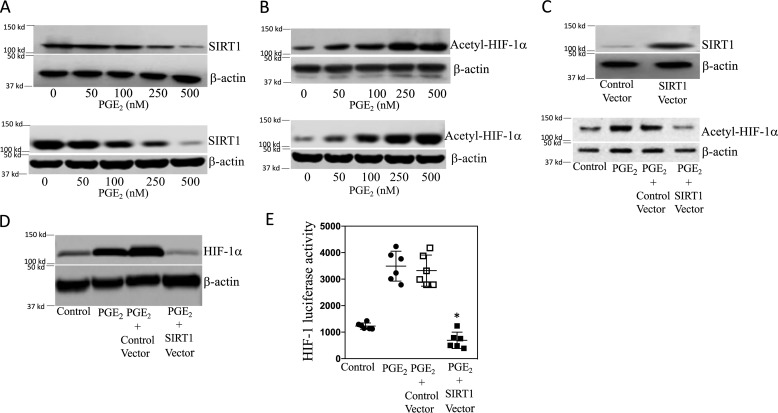
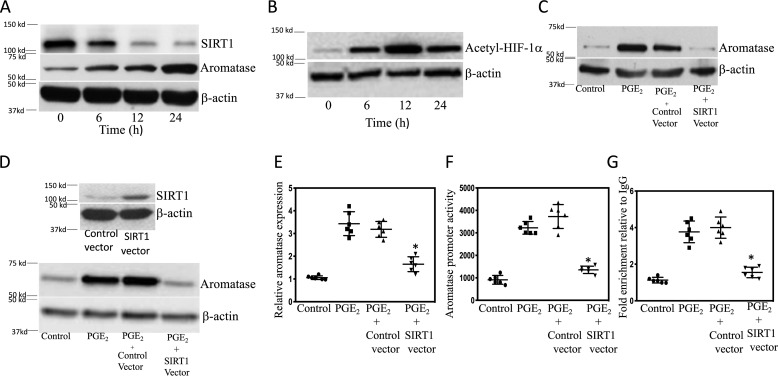
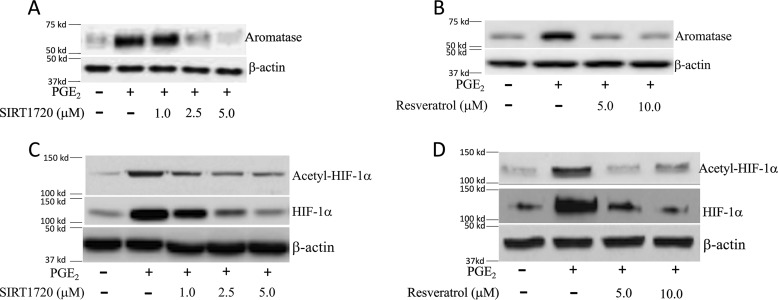
Although HIF-1α is important for PGE2-mediated induction of aromatase (12), the role of SIRT1 has not been investigated. We determined the effects of PGE2 on SIRT1 levels. Treatment of breast ASCs with PGE2 led to a dose-dependent decrease in SIRT1 protein levels (Fig. 2A). Because HIF-1α can be deacetylated by SIRT1, we measured levels of acetyl–HIF-1α following treatment with PGE2. Levels of acetyl–HIF-1α increased in response to PGE2 treatment and were inversely related to the levels of SIRT1 in both immortalized human breast ASCs (top panel in Fig. 2, A and B) and primary human breast ASCs (bottom panel in Fig. 2, A and B). To confirm the importance of SIRT1 in regulating levels of acetyl–HIF-1α, we next overexpressed SIRT1. As shown in Fig. 2C, the increase in acetyl–HIF-1α levels mediated by PGE2 was suppressed by overexpressing SIRT1. Similar changes in levels of HIF-1α were observed (Fig. 2D). To determine the functional consequences of PGE2-mediated induction of acetyl–HIF-1α, transient transfections were performed utilizing an HRE-luc reporter construct. Treatment with PGE2 stimulated HIF-1 activity, an effect that was abrogated by overexpressing SIRT1 (Fig. 2E). Because HIF-1α plays a role in PGE2-mediated induction of aromatase, we next evaluated levels of SIRT1 and aromatase over time following treatment with PGE2. Treatment with PGE2 led to a time-dependent decline in SIRT1 levels in association with a reciprocal increase in aromatase protein and acetyl–HIF-1α levels (Fig. 3, A and B). To determine whether the reduced levels of SIRT1 mediated by PGE2 were causally linked to the increase in aromatase, we overexpressed SIRT1 in ASCs. Overexpressing SIRT1 blocked PGE2-mediated induction of aromatase protein and mRNA (Fig. 3, C–E). To determine whether overexpressing SIRT1 altered aromatase transcription, transient transfections were performed. Overexpressing SIRT1 blocked PGE2-mediated stimulation of aromatase promoter activity (Fig. 3F). To investigate whether SIRT1 altered the binding of HIF-1α to the CYP19A1 promoter in response to PGE2, ChIP assays were performed. ChIP assays revealed increased binding of HIF-1α to the CYP19A1 promoter, an effect that was abrogated by overexpressing SIRT1 (Fig. 3G). To complement these studies, we also utilized small molecules that activate SIRT1. Consistent with the effects of overexpressing SIRT1, activators of SIRT1 (SIRT1720 and resveratrol) blocked PGE2-mediated induction of acetyl–HIF-1α, HIF-1α, and aromatase (Fig. 4).
Figure 2.
PGE2 increases HIF-1 activity by suppressing SIRT1 levels in human breast adipose stromal cells. A–E, human breast ASC cell line was used. A and B, the bottom panels represent primary human breast ASCs. A and B, cells were treated with the indicated concentrations of PGE2 for 24 h. Cells were then harvested, and lysates were subjected to Western blotting. C and D, cells were transfected with 2 μg of control vector or SIRT1 expression vector as indicated. C (top), cells were harvested, and Western blotting was performed for SIRT1 and β-actin to confirm overexpression. C (bottom) and D, cells were treated with vehicle or 500 nm PGE2 for 24 h. Cell lysates were prepared and subjected to Western blotting, and the blots were probed as indicated. E, cells were transfected as indicated with 0.9 μg of HRE-luciferase and 0.2 μg of psvβ-gal constructs. Cells labeled Control Vector also received 0.9 μg of expression vector; cells labeled SIRT1 Vector also received 0.9 μg of SIRT1 expression vector. 24 h after transfection, cells were treated with vehicle (control) or 500 nm PGE2 for 24 h. Cells were harvested, and luciferase activity was measured. Luciferase activity was normalized to β-gal activity. Means ± S.D. (error bars) are shown, n = 6. *, p < 0.001 versus PGE2-treated cells expressing control vector.
Figure 3.
Overexpression of SIRT1 inhibits PGE2-mediated induction of aromatase. Human breast ASC cell line was used except in D, where primary human breast ASCs were employed. A and B, cells were treated with 500 nm PGE2 for 0–24 h as indicated. C–E, cells were either untransfected (Control) or transfected as indicated. Cells were then treated with vehicle or 500 nm PGE2 for 24 h. A–D, cell lysates were subjected to Western blotting, and the blots were probed as indicated. E, aromatase mRNA levels were determined by quantitative PCR. F, cells were transfected with 0.9 μg of aromatase promoter PII-luciferase and 0.2 μg of psvβ-gal constructs. In addition, as indicated, cells received 0.9 μg of control expression vector or SIRT1 expression vector. Cells were treated with vehicle (control) or 500 nm PGE2 for 24 h. Cells were harvested, and luciferase activity was measured. Luciferase activity was normalized to β-gal activity. G, cells were either untransfected or transfected with 0.9 μg of control vector or SIRT1 expression vector. Subsequently, cells were treated with vehicle or 500 nm PGE2 for 3 h. ChIP assays were performed. Chromatin fragments were immunoprecipitated with antibodies against HIF-1α, and the aromatase promoter was amplified by real-time PCR. DNA sequencing was carried out, and the PCR products were confirmed to be the aromatase promoter. This promoter was not detected when normal IgG was used or when antibody was omitted from the immunoprecipitation step. In E–G, means ± S.D. (error bars) are shown (n = 6). *, p < 0.001.
Figure 4.
SIRT1 activators inhibit PGE2-mediated induction of acetyl–HIF-1α, HIF-1α and aromatase. The human breast ASC cell line was used. Cells were treated with vehicle, 500 nm PGE2, or PGE2 plus the indicated concentrations of SIRT1720 (A and C) or resveratrol (B and D) for 24 h. Cell lysates were subjected to Western blotting as indicated.
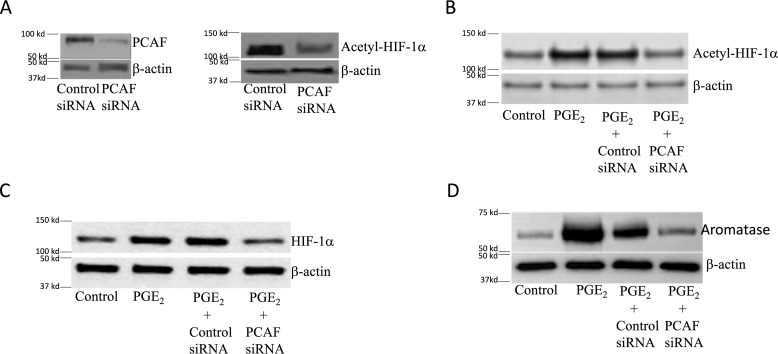
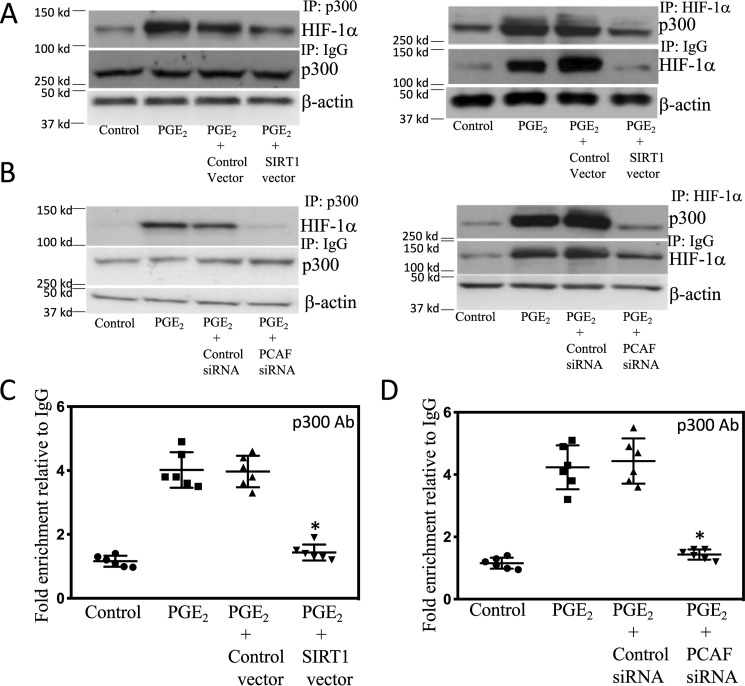
Because p300/CBP-associated factor (PCAF) can acetylate and activate HIF-1α (22), we next investigated whether silencing PCAF would attenuate PGE2-mediated induction of aromatase. First we showed that silencing PCAF led to a reduction in acetyl–HIF-1α (Fig. 5A). Moreover, the increase in levels of acetyl–HIF-1α and HIF-1α mediated by PGE2 was blocked by silencing PCAF (Fig. 5, B and C). Silencing PCAF also blocked the induction of aromatase by PGE2 (Fig. 5D). Acetylation of HIF-1α has been shown to enhance its interaction with p300 (22), a coactivator that plays a role in regulating aromatase expression (27). Next, we examined the role of acetylation of HIF-1α in its interaction with p300 in response to PGE2. As shown in Fig. 6 (A and B), treatment of cells with PGE2 increased the interaction of HIF-1α with p300, an effect that was abrogated when either SIRT1 was overexpressed or PCAF was silenced. Subsequently, ChIP assays were performed to determine the effects of overexpressing SIRT1 or silencing PCAF on PGE2-mediated induction of p300 binding to the CYP19A1 promoter. As shown in Fig. 6 (C and D), the increase in binding of p300 to the CYP19A1 promoter mediated by PGE2 was abrogated by overexpressing SIRT1 or silencing PCAF.
Figure 5.
Silencing PCAF inhibits PGE2-mediated induction of aromatase. A–D, human breast ASC cell line was used. Cells were either untransfected or transfected as indicated with 2 μg of siRNA to GFP (control siRNA) or PCAF siRNA. A, 48 h after transfection, cells were harvested, and Western blotting was performed for PCAF, acetyl–HIF-1α, and β-actin. B–D, cells were treated with vehicle (control) or 500 nm PGE2 for 24 h. Cell lysates were subjected to Western blotting, and the blots were probed as indicated.
Figure 6.
SIRT1 and PCAF regulate the binding of p300 to the CYP19A1 promoter. A–D, human breast ASC cell line was used. A, cells were either untransfected or transfected with 2 μg of control vector or SIRT1 expression vector for 48 h. Cells were then treated with vehicle (control) or 500 nm PGE2 for 3 h. B, cells were either untransfected or transfected with 2 μg of siRNA to GFP (control siRNA) or PCAF siRNA for 48 h. Cells were then treated with vehicle (control) or 500 nm PGE2 for 3 h. A and B, cell lysates were subjected to immunoprecipitation (IP) with p300 (left) or HIF-1α (right) antisera or IgG, and immunoprecipitates were subjected to Western blotting and probed as indicated. C and D, ChIP assays were performed. C, cells were either untransfected or transfected with 2 μg of control vector or SIRT1 expression vector for 48 h. D, cells were either untransfected or transfected with 2 μg of siRNA to GFP (control siRNA) or PCAF siRNA for 48 h. C and D, cells were then treated as indicated with vehicle (control) or 500 nm PGE2 for 3 h. Chromatin fragments were immunoprecipitated with antibodies against p300, and the aromatase promoter was amplified by real-time PCR. DNA sequencing was carried out, and the PCR products were confirmed to be the aromatase promoter. This promoter was not detected when normal IgG was used or when antibody was omitted from the immunoprecipitation step. In C and D, means ± S.D. (error bars) are shown (n = 6). *, p < 0.001.
PGE2 suppresses SIRT1 transcription via EP2 and EP4
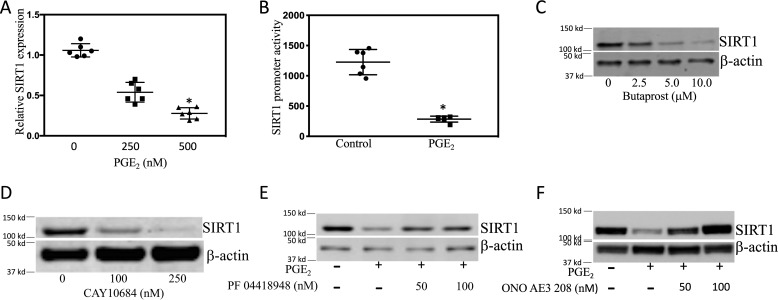
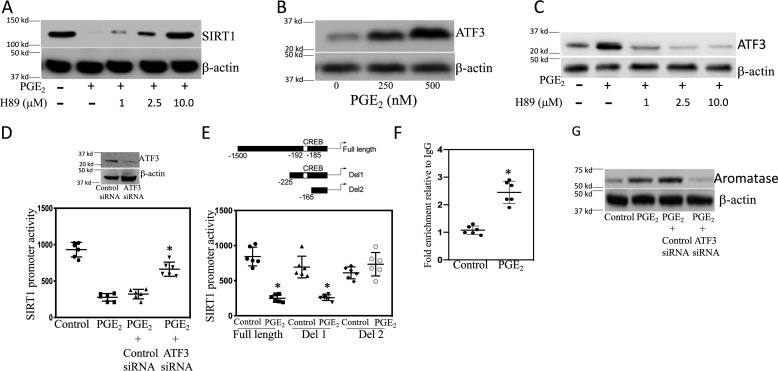
Having demonstrated that PGE2 down-regulated SIRT1 protein levels (Fig. 2A), we next explored the underlying mechanism. Treatment with PGE2 caused dose-dependent suppression of SIRT1 mRNA levels (Fig. 7A). Transient transfections were performed to determine the effect of PGE2 on SIRT1 promoter activity. As shown in Fig. 7B, treatment with PGE2 reduced SIRT1 promoter activity. PGE2 mediates its effects by binding to EP receptors. A combination of EP receptor agonists and antagonists was used to determine which EP receptors were responsible for PGE2-mediated suppression of SIRT1 transcription. Both butaprost, an EP2 receptor agonist, and CAY10684, an EP4 receptor agonist, down-regulated SIRT1 (Fig. 7, C and D). Moreover, PGE2-mediated down-regulation of SIRT1 was prevented by treatment with EP2 (PF04418948) and EP4 (ONO AE3 208) receptor antagonists (Fig. 7, E and F). Binding of PGE2 to EP2 or EP4 receptors can stimulate protein kinase A (PKA) activity. Hence, we next determined whether PKA was involved in reducing SIRT1 levels in response to PGE2. Treatment with H89, a PKA inhibitor, prevented the decrease in SIRT1 levels mediated by PGE2 (Fig. 8A). PGE2 also induced ATF3 in both immortalized human breast ASCs and primary human breast ASCs (Fig. 8B and Fig. S1A). Treatment with either an EP2 or EP4 receptor agonist also induced ATF3 (Fig. S1, B and C). Inhibition of PKA with H89 blocked PGE2-mediated induction of ATF3 (Fig. 8C). To determine whether this increase in ATF3 was important for explaining the reduction in SIRT1 levels, transient transfections were performed. Interestingly, PGE2-mediated suppression of SIRT1 promoter activity was relieved by silencing ATF3 (Fig. 8D). Next, we attempted to localize the site in the SIRT1 promoter that was responsible for mediating the suppressive effects of PGE2. Transient transfections were carried out utilizing a series of SIRT1 promoter deletion constructs (Fig. 8E). The suppressive effects of PGE2 were lost when promoter deletion construct 2 (Del2) that lacks the CRE site was used. ChIP assays were performed to determine whether ATF3 bound to the SIRT1 promoter in response to treatment with PGE2. In fact, PGE2 stimulated the binding of ATF3 to the SIRT1 promoter (Fig. 8F). Finally, we determined whether silencing ATF3 would block PGE2-mediated induction of aromatase. As shown in Fig. 8G, silencing ATF3 suppressed PGE2-mediated induction of aromatase. Taken together, these data suggest that PGE2 binds to EP2 and EP4 receptors, leading to down-regulation of SIRT1, enhanced acetylation of HIF-1α, and increased aromatase expression in ASCs.
Figure 7.
PGE2 down-regulates SIRT1 expression in human breast ASCs. A, cells were treated with the indicated concentrations of PGE2 for 24 h. Total RNA was isolated, and quantitative PCR for SIRT1 was performed. B, cells were transfected with 1.8 μg of full-length SIRT1 promoter and 0.2 μg of psv-β-gal constructs. 24 h after transfection, cells were treated with vehicle or 500 nm PGE2 for 24 h. Cells were harvested, and luciferase activity was measured. Luciferase activity was normalized to β-gal activity. C and D, cells were treated with vehicle (control) or the indicated concentrations of butaprost or CAY10684 for 24 h. E and F, cells were pretreated with vehicle, PF04418948, or ONO AE3 208 for 2 h. Subsequently, the cells were treated with vehicle, 500 nm PGE2, or 500 nm PGE2 plus the indicated concentrations of PF04418948 (E) or ONO AE3 208 (F) for 24 h. C–F, cell lysates were prepared and subjected to Western blotting. A and B, means ± S.D. (error bars) (n = 6). *, p < 0.001.
Figure 8.
ATF3 is important for PGE2-mediated down-regulation of SIRT1 in human breast ASCs. A–G, a human breast ASC cell line was used. A, cells were pretreated with vehicle or the indicated concentrations of H89, a PKA inhibitor, for 2 h and then treated with vehicle, 500 nm PGE2, or 500 nm PGE2 plus the indicated concentrations of H89 for 24 h. B, cells were treated with the indicated concentrations of PGE2 for 2 h. Cell lysates were subjected to Western blotting, and the blots were probed as indicated. C, cells were pretreated with vehicle or the indicated concentrations of H89 for 2 h and then treated with vehicle, 500 nm PGE2, or 500 nm PGE2 plus the indicated concentrations of H89 for 24 h. D, cells were transfected with 0.9 μg of full-length SIRT1 promoter and 0.2 μg of psv-β-gal constructs. In the column labeled Control siRNA, cells received 0.9 μg of control siRNA, and in the column labeled ATF3 siRNA, cells received 0.9 μg of ATF3 siRNA. 24 h later, cells were treated with vehicle (control) or PGE2 for 24 h. Cells were harvested, and luciferase activity was measured. Luciferase activity was normalized to β-gal activity. Inset, Western blotting was performed after transfecting ASCs with control or ATF3 siRNA. E, the top panel represents different deletions of SIRT1 promoter used. Full-length promoter and deletion 1 (Del1) contain a CRE site, whereas deletion 2 (Del2) lacks a CRE site. Cells were transfected with 1.8 μg of each of three SIRT1 promoter-luciferase constructs and 0.2 μg of psv-β-gal for 24 h. Subsequently, cells were treated with vehicle (control) or 500 nm PGE2 for 24 h. Cells were then harvested, and luciferase activity was measured. Luciferase activity was normalized to β-gal activity. F, ChIP assay was performed. Cells were treated with vehicle (control) or 500 nm PGE2 for 3 h. Chromatin fragments were immunoprecipitated with antibodies against ATF3, and the SIRT1 promoter was amplified by real-time PCR. DNA sequencing was carried out, and the PCR products were confirmed to be the SIRT1 promoter. This promoter was not detected when normal IgG was used or when antibody was omitted from the immunoprecipitation step. G, cells were either untransfected or transfected as indicated with 2 μg of siRNA to GFP (control siRNA) or ATF3 siRNA. 48 h after transfection, cells were treated with vehicle (control) or 500 nm PGE2 for 24 h. Cell lysates were subjected to Western blotting, and the blots were probed as indicated. D–F, means ± S.D. (error bars); n = 6. *, p < 0.001.
Discussion
The current study provides new insights into the mechanisms that underlie the elevation of aromatase expression in the breast tissue of obese women. Mechanistic studies were carried out in ASCs because obesity is associated with increased levels of both HIF-1α and aromatase in these cells (28). We focused on SIRT1 because of evidence that it can deacetylate and inactivate HIF-1α, raising the possibility that SIRT1 could modulate CYP19A1 gene expression and thereby aromatase levels (22).
Several lines of evidence suggest the importance of SIRT1 and the acetylation of HIF-1α as determinants of aromatase expression in the breast tissue of obese women. Reduced levels of SIRT1 correlated with increased levels of acetylated HIF-1α, HIF-1α, and aromatase in the breast tissue of obese versus lean women. Notably, the reduction in SIRT1 mRNA and protein in the breast tissue of obese women is consistent with prior evidence that SIRT1 levels are decreased in adipose tissue of obese humans (23–26). The reduction in SIRT1 levels has been attributed to hypoxia and an associated decrease in NAD+ levels (23). Saturated fatty acids and reactive oxygen species have also been suggested to play a role in down-regulating SIRT1 (23). We focused on PGE2 because levels of this bioactive lipid are increased in the breast tissue of obese women, and it is a known inducer of HIF-1α and aromatase in ASCs (11, 12, 28). More specifically, PGE2 is known to increase HIF-1α transcript and protein expression and binding to CYP19A1 promoter II, leading to increased aromatase levels. Here, we found that PGE2-mediated suppression of SIRT1 levels was associated with elevated levels of acetylated HIF-1α, HIF-1α, HIF-1 activity, and aromatase. Importantly, these inductive effects of PGE2, including the activation of CYP19A1 transcription, were abrogated by overexpressing SIRT1 or treatment with two activators of SIRT1 (SIRT1720 and resveratrol). Taken together, these results underscore the importance of acetylated HIF-1α as a determinant of aromatase expression.
The coactivator CBP/p300 possesses histone acetyltransferase activity and plays an important role in stabilizing HIF-1α protein and mediating the induction of aromatase in response to PGE2 treatment (27, 29). The acetylation status of HIF-1α is a determinant of its ability to bind to p300 and thereby regulate gene expression (22, 29). PCAF acetylates HIF-1α, whereas SIRT1 deacetylates it at Lys-674 (22). Thus, HIF-1α activity, including its ability to interact with p300, appears to be balanced by the opposing activities of PCAF and SIRT1. In the present study, we show for the first time that PCAF is important for PGE2-mediated induction of aromatase. Silencing PCAF suppressed PGE2-mediated induction of acetyl–HIF-1α, HIF-1α, and aromatase. PGE2 treatment led to enhanced interaction between p300 and HIF-1α, an effect that was reduced when either SIRT1 was overexpressed or PCAF was silenced. Moreover, overexpression of SIRT1 or silencing PCAF blocked PGE2-mediated recruitment of p300 to the CYP19A1 promoter. Because p300 can acetylate and stabilize HIF-1α protein, the increased interaction between p300 and HIF-1α in response to PGE2 can help to explain the observed enhanced levels of HIF-1α protein. The fact that changes in levels of acetylated HIF-1α were paralleled by levels of HIF-1α may be due, in part, to a p300-dependent mechanism.
Based on the finding that exogenous PGE2 down-regulated SIRT1, leading, in turn, to elevated levels of aromatase, we next focused on the regulation of SIRT1 gene expression. PGE2 exerts its effects by binding to four G protein–coupled receptors (EP1–EP4) (30). Both EP2 and EP4 activate cAMP signaling and induce aromatase (27). Here, we showed that PGE2 acted via EP2 and EP4 to down-regulate SIRT1 transcription. Binding of PGE2 to EP2 and EP4 can activate PKA. We showed that H89, a PKA inhibitor, blocked PGE2-mediated down-regulation of SIRT1. ATF3 is a member of the ATF/CREB family of basic leucine zipper transcription factors (31). It is induced by a variety of cellular stressors and can act as a transcriptional repressor. The ability of PGE2 to induce ATF3 was suppressed by inhibiting PKA. This finding is consistent with previous evidence that PKA can regulate the expression of ATF3 (32). The SIRT1 promoter contains a CRE element in its 5′-UTR (33), suggesting the possibility that the suppressive effects of PGE2 could be mediated via this site. Silencing ATF3 attenuated the ability of PGE2 to suppress SIRT1 promoter activity. When transient transfection studies were conducted using different SIRT1 promoter deletion constructs, PGE2 failed to suppress SIRT1 promoter activity when a construct that lacked the CRE site (bp −192 to −185) was utilized. ChIP assays indicated that treatment of cells with PGE2 caused an increase in ATF3 binding to the SIRT1 promoter. Taken together, these findings suggest that PGE2 via EP2 and EP4 activates the PKA → ATF3 pathway, resulting in reduced SIRT1 expression (Fig. 9). The decrease in SIRT1 leads, in turn, to increased acetyl–HIF-1α and enhanced transcription of CYP19A1, resulting in elevated levels of the estrogen synthase aromatase.
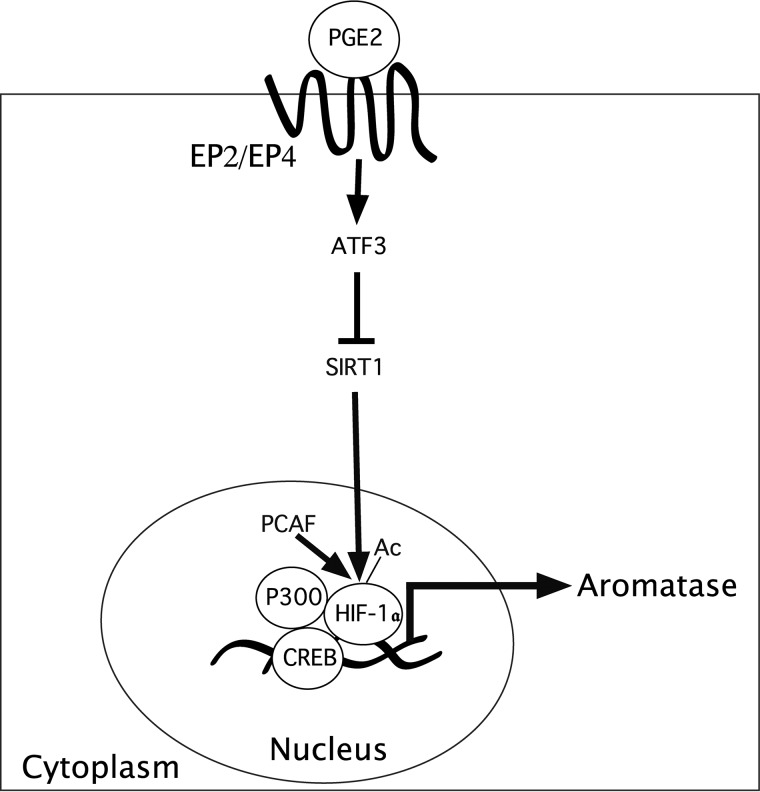
Figure 9.
SIRT1 is important for PGE2-mediated induction of aromatase expression. Obese women express lower levels of SIRT1 and higher levels of acetyl–HIF-1α, HIF-1α, and aromatase in breast tissue. PGE2 acted via the EP2 and EP4 receptors to induce ATF3, a repressive transcription factor, which bound to a CRE site within the SIRT1 promoter, resulting in reduced SIRT1 levels. The reduction in SIRT1, a deacetylase, leads to elevated levels of acetyl–HIF-1α and HIF-1α and enhanced aromatase transcription. PCAF acetylates HIF-1α and enhances its interaction with p300, a coactivator of CREB, leading to enhanced aromatase transcription. Collectively, these findings suggest the importance of SIRT1-mediated post-translational modification of HIF-1α as a determinant of the elevated levels of aromatase in the breast tissue of obese women.
In engineered mice, reduced levels of SIRT1 in adipose tissue have been associated with macrophage influx and the development of adipose inflammation (34, 35). A similar low-grade inflammatory process was recently linked to the pathogenesis of human breast cancer (36, 37). Taken together, the observed reduction in SIRT1 in the breast tissue of obese women may be causally linked to both subclinical inflammation and elevated aromatase expression, contributing to the increased risk of hormone receptor-positive breast cancer in obese postmenopausal women. We do note, however, that other studies have focused on SIRT1 in breast cancer cells and suggested its involvement in positively regulating aromatase expression and estrogen-induced tumor growth (37, 38). It is quite possible, therefore, that activators of SIRT1 will have different effects on the risk of estrogen-dependent breast cancer, depending on whether a woman is obese or lean. Although future studies will be needed to address this question, the current study is the first to demonstrate that post-translational modification of HIF-1α is a determinant of aromatase expression. Whether other post-translational modifications that affect HIF-1α levels also modulate aromatase expression should be considered.
Experimental procedures
Materials
Primers for aromatase, SIRT1720, and an antibody to β-actin were purchased from Sigma. Monoclonal aromatase antibody 677 was obtained from the Baylor College of Medicine (39). Antibodies to SIRT1 (1:1000; D1D7), p300 (1:1000; D8Z4E), ATF3 (1:1000; D2Y5W), and HIF-1α (1:1000; D2U3T) and microsomal isolation kits were bought from Cell Signaling Technology. Acetyllysine antibody was from Abcam (1:1000; ab80178). Control siRNA (GFP), and ATF3 and PCAF siRNAs were obtained from Thermo Fisher Scientific. Hypoxia-responsive element luciferase (HRE-luciferase), control vector, SIRT1 expression vector, and SIRT1 promoter constructs were from GeneCopoeia. PGE2, butaprost, PF04418948, CAY10684, resveratrol, ONO AE3208, and H89 were from Cayman. The aromatase promoter (CYP19PII)-luciferase construct was kindly provided by Dr. S. Chen (City of Hope, Duarte, CA).
Study population and samples
The study was approved by the institutional review boards of Weill Cornell Medical College and Memorial Sloan Kettering Cancer Center. Breast tissues used in the study were from women undergoing mastectomy at Memorial Sloan Kettering Cancer Center who were consented under a standard tissue acquisition protocol. This patient cohort has been described previously (40). Normal breast tissue from a quadrant uninvolved by tumor was collected and stored at −80 °C. BMI was calculated from height (in meters) and weight (kg) measurements obtained before surgery. Standard definitions were used to categorize BMI as underweight or normal weight (BMI < 25), overweight (BMI between 25.0 and 29.9), or obese (BMI ≥ 30).
Cell culture
An immortalized human mammary ASC line (HMS32-hTERT) was provided by Dr. Brittney-Shea Herbert (Indiana University School of Medicine) and grown as described previously (28, 41–43). Primary human breast ASCs and media were purchased from Zen-Bio.
Immunoprecipitation
Immunoprecipitation experiments were performed using a catch and release reversible immunoprecipitation system from Upstate Biotechnology. Cell or tissue lysate protein (500–1000 μg) was used for immunoprecipitation following the manufacturer's instructions.
Western blotting
Cells and tissues were sonicated in a lysis buffer (150 mm NaCl, 100 mm Tris, pH 8.0, 1% Tween 20, 50 mm diethyldithiocarbamate, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 10 μg/ml trypsin inhibitor, and 10 μg/ml leupeptin). Minute adipose tissue fractionation kits were purchased from Invent Biotechnologies Inc. and used to remove lipid from tissue lysates. The protein concentration was determined according to Lowry et al. (44). To measure the levels of aromatase, SIRT1, acetyl–HIF-1α, and HIF-1-α proteins, tissue lysates were prepared from frozen nontumorous breast tissue samples. For aromatase, microsomal protein was isolated from tissue lysates using differential centrifugation. The microsomal suspension was subjected to immunoprecipitation with antiserum to aromatase followed by Western blotting. SIRT1 and HIF-1α proteins were immunoprecipitated, and Western blotting was performed. For acetyl–HIF-1α, immunoprecipitation was performed first with an antibody to acetyllysine, and then the blots were probed with HIF-1α antibody. β-Actin levels were assessed in whole tissue lysates by Western blotting. For the in vitro studies, Western blots that are representative of a minimum of three independent experiments are shown.
RNA-Seq
Total RNA was isolated from nontumorous breast samples using Qiagen's RNeasy minikit. Sequencing libraries were constructed following the Illumina TrueSeq Stranded Total RNA Library preparation protocol with rRNA depletion. Next-generation sequencing was performed with paired-end 51 bp using the Illumina HiSeq4000 platform (Weill Cornell Medicine). Raw sequenced reads were aligned to the human reference genome (hg19) using STAR (version 2.4.2) aligner. Aligned reads were quantified against the reference annotation (hg19) to obtain fragments per kilobase per million (FPKM) and raw counts using CuffLinks (version 2.2.1) and HTSeq, respectively.
Quantitative real-time PCR
Total RNA was isolated from cells using the RNeasy minikit (Qiagen) and reverse-transcribed using murine leukemia virus reverse transcriptase and oligo(dT)16 primer. The resulting cDNA was then used for amplification. Primers for human aromatase have been described previously (11, 28, 43). The following SIRT1 primers were used: forward, 5′-CTGGACAATTCCAGCCATCT-3′; reverse, 5′-GGGTGGCAACTCTGACAAAT-3′. β-Actin (QT00095431) primers were obtained from Qiagen. β-Actin was used as an endogenous normalization control. Real-time PCR was done using 2× SYBR Green PCR master mix on a 7500 real-time PCR system (Applied Biosystems). Using the ΔΔCT (relative quantification) analysis protocol, relative -fold induction was determined.
Transient transfections
50–70% confluent cultures were grown in 6-well dishes. The cells were then transfected using Lipofectamine 2000 (Invitrogen) for 24 h. Following transfection, the medium was replaced with serum-free medium for another 24 h. Luciferase and β-gal enzyme activities were measured in cellular extracts. Luciferase activity in cell lysates was normalized to β-gal enzymatic activity. 2 μg of control vector or SIRT1 expression vector were transfected into cells using Lipofectamine 2000 (Invitrogen) grown to 60–70% confluence.
RNA interference
Cells were transfected with 2 μg of siRNA oligonucleotides using DharmaFECT 4 transfection reagent according to the manufacturer's instructions.
ChIP assays
The ChIP assay was performed using the EpiTect ChIPone-day kit from SA Bioscience. Approximately 4 × 106 cells were cross-linked in a 1% formaldehyde solution at 37 °C for 10 min. Cross-linked cells were then lysed and sonicated to generate 200–1000-bp DNA fragments. Cell lysates were subjected to centrifugation, and the cleared supernatant was incubated with 4 μg of the antibodies at 4 °C overnight. Immune complexes were precipitated, washed, and eluted as described in the manufacturer's protocol. DNA–protein cross-links were reversed by heating at 65 °C for 4 h, and the DNA fragments were purified and used as a template for PCR amplification. Quantitative real-time PCR was carried out. For ChIP analysis, the CYP19A1 oligonucleotide sequences for PCR primers were 5′-AACCTGATGAAGTCACAA-3′ (forward) and 5′-TCAGACATTTAGGCAAGACT-3′ (reverse). This primer set covers the CYP19A1 promoter I.3/II segment from nucleotide −302 to −38. For ChIP analysis, the SIRT1 oligonucleotide sequences for PCR primers were 5′-CTTCCAGCCCAGGCGGAGCG-3′ (forward) and 5′-GATTTAAACCCCATCACGTGACCCG-3′ (reverse). This primer set covers the human SIRT1 promoter region containing the CRE site from nucleotide −225 to −21. The primers were obtained from Sigma. PCR was performed at 94 °C for 30 s, 62 °C for 30 s, and 72 °C for 45 s for 35 cycles, and real-time PCR was performed at 95 °C for 15 s and 60 °C for 60 s for 40 cycles. The PCR product generated from the ChIP template was sequenced, and the identity of the CYP19A1 and SIRT1 promoters was confirmed.
Statistics
Comparisons between two independent groups were made by Student's t test. A difference between groups of p < 0.05 was considered significant. For the human study, correlation between SIRT1 expression in terms of log2-transformed FPKM values and each of the continuous variables, including BMI and the relative aromatase expression, was examined using Spearman's method. Differences in SIRT1 expression across BMI categories were examined using the nonparametric Kruskall–Wallis test. Pairwise comparisons were carried out using the Wilcoxon rank sum test, and p values were adjusted for multiple comparisons using the Bonferroni–Holm method.
Author contributions
K. S. and A. J. D. conceptualization; K. S., N. M. I., M. M., and A. J. D. resources; K. S., N. M. I., O. E., and X. K. Z. data curation; K. S. and X. K. Z. formal analysis; K. S. validation; K. S. investigation; K. S. visualization; K. S. methodology; K. S. and A. J. D. writing-original draft; K. S. and A. J. D. project administration; K. S., N. M. I., M. M., O. E., X. K. Z., and A. J. D. writing-review and editing; A. J. D. supervision; A. J. D. funding acquisition.
Supplementary Material
This work was supported, in whole or part, by National Institutes of Health Grants R01CA215797 and U54 CA210184, the Breast Cancer Research Foundation, and the Botwinick-Wolfensohn Foundation (in memory of Mr. and Mrs. Benjamin Botwinick). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Fig. S1.
- ER
- estrogen receptor
- CRE
- cyclic AMP-response element
- CREB
- CRE–binding protein
- HIF-1α
- hypoxia inducible factor-1α
- PGE2
- prostaglandin E2
- ASC
- adipose stromal cell
- SIRT1
- sirtuin 1
- BMI
- body mass index
- PCAF
- p300/CBP-associated factor
- PKA
- protein kinase A
- ATF3
- activating transcription factor 3
- EP
- prostaglandin E2 receptor
- HRE
- hypoxia-responsive element
- FPKM
- fragments per kilobase per million.
References
- 1. Munsell M. F., Sprague B. L., Berry D. A., Chisholm G., and Trentham-Dietz A. (2014) Body mass index and breast cancer risk according to postmenopausal estrogen-progestin use and hormone receptor status. Epidemiol. Rev. 36, 114–136 10.1093/epirev/mxt010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rohan T. E., Heo M., Choi L., Datta M., Freudenheim J. L., Kamensky V., Ochs-Balcom H. M., Qi L., Thomson C. A., Vitolins M. Z., Wassertheil-Smoller S., and Kabat G. C. (2013) Body fat and breast cancer risk in postmenopausal women: a longitudinal study. J. Cancer Epidemiol. 2013, 754815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Trentham-Dietz A., Newcomb P. A., Storer B. E., Longnecker M. P., Baron J., Greenberg E. R., and Willett W. C. (1997) Body size and risk of breast cancer. Am. J. Epidemiol. 145, 1011–1019 10.1093/oxfordjournals.aje.a009057 [DOI] [PubMed] [Google Scholar]
- 4. Iyengar N. M., Hudis C. A., and Dannenberg A. J. (2015) Obesity and cancer: local and systemic mechanisms. Annu. Rev. Med. 66, 297–309 10.1146/annurev-med-050913-022228 [DOI] [PubMed] [Google Scholar]
- 5. Calle E. E., Rodriguez C., Walker-Thurmond K., and Thun M. J. (2003) Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. N. Engl. J. Med. 348, 1625–1638 10.1056/NEJMoa021423 [DOI] [PubMed] [Google Scholar]
- 6. Ewertz M., Jensen M. B., Gunnarsdóttir K. Á., Højris I., Jakobsen E. H., Nielsen D., Stenbygaard L. E., Tange U. B., and Cold S. (2011) Effect of obesity on prognosis after early-stage breast cancer. J. Clin. Oncol. 29, 25–31 10.1200/JCO.2010.29.7614 [DOI] [PubMed] [Google Scholar]
- 7. Protani M., Coory M., and Martin J. H. (2010) Effect of obesity on survival of women with breast cancer: systematic review and meta-analysis. Breast Cancer Res. Treat. 123, 627–635 10.1007/s10549-010-0990-0 [DOI] [PubMed] [Google Scholar]
- 8. Sparano J. A., Zhao F., Martino S., Ligibel J. A., Perez E. A., Saphner T., Wolff A. C., Sledge G. W. Jr., Wood W. C., and Davidson N. E. (2015) Long-term follow-up of the E1199 phase III trial evaluating the role of taxane and schedule in operable breast cancer. J. Clin. Oncol. 33, 2353–2360 10.1200/JCO.2015.60.9271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Simpson E. R., Mahendroo M. S., Means G. D., Kilgore M. W., Hinshelwood M. M., Graham-Lorence S., Amarneh B., Ito Y., Fisher C. R., Michael M. D., Mendelson C. R., and, Bulun S. E. (1994) Aromatase cytochrome P450, the enzyme responsible for estrogen biosynthesis. Endocr. Rev. 15, 342–355 10.1210/edrv-15-3-342 [DOI] [PubMed] [Google Scholar]
- 10. Morris P. G., Hudis C. A., Giri D., Morrow M., Falcone D. J., Zhou X. K., Du B., Brogi E., Crawford C. B., Kopelovich L., Subbaramaiah K., and Dannenberg A. J. (2011) Inflammation and increased aromatase expression occur in the breast tissue of obese women with breast cancer. Cancer Prev. Res. (Phila.) 4, 1021–1029 10.1158/1940-6207.CAPR-11-0110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Subbaramaiah K., Morris P. G., Zhou X. K., Morrow M., Du B., Giri D., Kopelovich L., Hudis C. A., and Dannenberg A. J. (2012) Increased levels of COX-2 and prostaglandin E2 contribute to elevated aromatase expression in inflamed breast tissue of obese women. Cancer Discov. 2, 356–365 10.1158/2159-8290.CD-11-0241 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12. Samarajeewa N. U., Yang F., Docanto M. M., Sakurai M., McNamara K. M., Sasano H., Fox S. B., Simpson E. R., and Brown K. A. (2013) HIF-1α stimulates aromatase expression driven by prostaglandin E2 in breast adipose stroma. Breast Cancer Res. 15, R30 10.1186/bcr3410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Morris P. G., Zhou X. K., Milne G. L., Goldstein D., Hawks L. C., Dang C. T., Modi S., Fornier M. N., Hudis C. A., and Dannenberg A. J. (2013) Increased levels of urinary PGE-M, a biomarker of inflammation, occur in association with obesity, aging, and lung metastases in patients with breast cancer. Cancer Prev. Res. (Phila.) 6, 428–436 10.1158/1940-6207.CAPR-12-0431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bulun S. E., Price T. M., Aitken J., Mahendroo M. S., and Simpson E. R. (1993) A link between breast cancer and local estrogen biosynthesis suggested by quantification of breast adipose tissue aromatase cytochrome P450 transcripts using competitive polymerase chain reaction after reverse transcription. J. Clin. Endocrinol. Metab. 77, 1622–1628 10.1210/jcem.77.6.8117355 [DOI] [PubMed] [Google Scholar]
- 15. Bulun S. E., Sharda G., Rink J., Sharma S., and Simpson E. R. (1996) Distribution of aromatase P450 transcripts and adipose fibroblasts in the human breast. J. Clin. Endocrinol. Metab. 81, 1273–1277 10.1210/jcem.81.3.8772611 [DOI] [PubMed] [Google Scholar]
- 16. Lin N., and Simon M. C. (2016) Hypoxia-inducible factors: key regulators of myeloid cells during inflammation. J. Clin. Invest. 126, 3661–3671 10.1172/JCI84426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Majmundar A. J., Wong W. J., and Simon M. C. (2010) Hypoxia-inducible factors and the response to hypoxic stress. Mol. Cell 40, 294–309 10.1016/j.molcel.2010.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rey S., and Semenza G. L. (2010) Hypoxia-inducible factor-1-dependent mechanisms of vascularization and vascular remodelling. Cardiovasc. Res. 86, 236–242 10.1093/cvr/cvq045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rahman S., and Islam R. (2011) Mammalian Sirt1: insights on its biological functions. Cell Commun. Signal. 9, 11 10.1186/1478-811X-9-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chang H. C., and Guarente L. (2014) SIRT1 and other sirtuins in metabolism. Trends Endocrinol. Metab. 25, 138–145 10.1016/j.tem.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liang F., Kume S., and Koya D. (2009) SIRT1 and insulin resistance. Nat. Rev. Endocrinol. 5, 367–373 10.1038/nrendo.2009.101 [DOI] [PubMed] [Google Scholar]
- 22. Lim J. H., Lee Y. M., Chun Y. S., Chen J., Kim J. E., and Park J. W. (2010) Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1α. Mol. Cell 38, 864–878 10.1016/j.molcel.2010.05.023 [DOI] [PubMed] [Google Scholar]
- 23. de Kreutzenberg S. V., Ceolotto G., Papparella I., Bortoluzzi A., Semplicini A., Dalla Man C., Cobelli C., Fadini G. P., and Avogaro A. (2010) Downregulation of the longevity-associated protein sirtuin 1 in insulin resistance and metabolic syndrome: potential biochemical mechanisms. Diabetes 59, 1006–1015 10.2337/db09-1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kurylowicz A., Owczarz M., Polosak J., Jonas M. I., Lisik W., Jonas M., Chmura A., and Puzianowska-Kuznicka M. (2016) SIRT1 and SIRT7 expression in adipose tissues of obese and normal-weight individuals is regulated by microRNAs but not by methylation status. Int. J. Obes. (Lond.) 40, 1635–1642 10.1038/ijo.2016.131 [DOI] [PubMed] [Google Scholar]
- 25. Song Y. S., Lee S. K., Jang Y. J., Park H. S., Kim J. H., Lee Y. J., and Heo Y. S. (2013) Association between low SIRT1 expression in visceral and subcutaneous adipose tissues and metabolic abnormalities in women with obesity and type 2 diabetes. Diabetes Res. Clin. Pract. 101, 341–348 10.1016/j.diabres.2013.07.002 [DOI] [PubMed] [Google Scholar]
- 26. Pedersen S. B., Olholm J., Paulsen S. K., Bennetzen M. F., and Richelsen B. (2008) Low Sirt1 expression, which is upregulated by fasting, in human adipose tissue from obese women. Int. J. Obes. (Lond.) 32, 1250–1255 10.1038/ijo.2008.78 [DOI] [PubMed] [Google Scholar]
- 27. Subbaramaiah K., Hudis C., Chang S. H., Hla T., and Dannenberg A. J. (2008) EP2 and EP4 receptors regulate aromatase expression in human adipocytes and breast cancer cells: evidence of a BRCA1 and p300 exchange. J. Biol. Chem. 283, 3433–3444 10.1074/jbc.M705409200 [DOI] [PubMed] [Google Scholar]
- 28. Zahid H., Subbaramaiah K., Iyengar N. M., Zhou X. K., Chen I. C., Bhardwaj P., Gucalp A., Morrow M., Hudis C. A., Dannenberg A. J., and Brown K. A. (2018) Leptin regulation of the p53-HIF1α/PKM2-aromatase axis in breast adipose stromal cells: a novel mechanism for the obesity-breast cancer link. Int. J. Obes. (Lond.) 42, 711–720 10.1038/ijo.2017.273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Geng H., Liu Q., Xue C., David L. L., Beer T. M., Thomas G. V., Dai M. S., and Qian D. Z. (2012) HIF1α protein stability is increased by acetylation at lysine 709. J. Biol. Chem. 287, 35496–35505 10.1074/jbc.M112.400697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sugimoto Y., and Narumiya S. (2007) Prostaglandin E receptors. J. Biol. Chem. 282, 11613–11617 10.1074/jbc.R600038200 [DOI] [PubMed] [Google Scholar]
- 31. Thompson M. R., Xu D., and Williams B. R. (2009) ATF3 transcription factor and its emerging roles in immunity and cancer. J. Mol. Med. 87, 1053–1060 10.1007/s00109-009-0520-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Koivisto E., Jurado Acosta A., Moilanen A. M., Tokola H., Aro J., Pennanen H., Säkkinen H., Kaikkonen L., Ruskoaho H., and Rysä J. (2014) Characterization of the regulatory mechanisms of activating transcription factor 3 by hypertrophic stimuli in rat cardiomyocytes. PLoS One 9, e105168 10.1371/journal.pone.0105168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Noriega L. G., Feige J. N., Canto C., Yamamoto H., Yu J., Herman M. A., Mataki C., Kahn B. B., and Auwerx J. (2011) CREB and ChREBP oppositely regulate SIRT1 expression in response to energy availability. EMBO Rep. 12, 1069–1076 10.1038/embor.2011.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gillum M. P., Erion D. M., and Shulman G. I. (2011) Sirtuin-1 regulation of mammalian metabolism. Trends Mol. Med. 17, 8–13 10.1016/j.molmed.2010.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hui X., Zhang M., Gu P., Li K., Gao Y., Wu D., Wang Y., and Xu A. (2017) Adipocyte SIRT1 controls systemic insulin sensitivity by modulating macrophages in adipose tissue. EMBO Rep. 18, 645–657 10.15252/embr.201643184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Carter J. M., Hoskin T. L., Pena M. A., Brahmbhatt R., Winham S. J., Frost M. H., Stallings-Mann M., Radisky D. C., Knutson K. L., Visscher D. W., and Degnim A. C. (2018) Macrophagic “crown-like structures” are associated with an increased risk of breast cancer in benign breast disease. Cancer Prev. Res. (Phila.) 11, 113–119 10.1158/1940-6207.CAPR-17-0245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Holloway K. R., Barbieri A., Malyarchuk S., Saxena M., Nedeljkovic-Kurepa A., Cameron Mehl M., Wang A., Gu X., and Pruitt K. (2013) SIRT1 positively regulates breast cancer associated human aromatase (CYP19A1) expression. Mol. Endocrinol 27, 480–490 10.1210/me.2012-1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Elangovan S., Ramachandran S., Venkatesan N., Ananth S., Gnana-Prakasam J. P., Martin P. M., Browning D. D., Schoenlein P. V., Prasad P. D., Ganapathy V., and Thangaraju M. (2011) SIRT1 is essential for oncogenic signaling by estrogen/estrogen receptor α in breast cancer. Cancer Res. 71, 6654–6664 10.1158/0008-5472.CAN-11-1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sasano H., Edwards D. P., Anderson T. J., Silverberg S. G., Evans D. B., Santen R. J., Ramage P., Simpson E. R., Bhatnagar A. S., and Miller W. R. (2003) Validation of new aromatase monoclonal antibodies for immunohistochemistry: progress report. J. Steroid Biochem. Mol. Biol. 86, 239–244 10.1016/S0960-0760(03)00363-7 [DOI] [PubMed] [Google Scholar]
- 40. Iyengar N. M., Zhou X. K., Gucalp A., Morris P. G., Howe L. R., Giri D. D., Morrow M., Wang H., Pollak M., Jones L. W., Hudis C. A., and Dannenberg A. J. (2016) Systemic correlates of white adipose tissue inflammation in early-stage breast cancer. Clin. Cancer Res. 22, 2283–2289 10.1158/1078-0432.CCR-15-2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shay J. W., Tomlinson G., Piatyszek M. A., and Gollahon L. S. (1995) Spontaneous in vitro immortalization of breast epithelial cells from a patient with Li-Fraumeni syndrome. Mol. Cell. Biol. 15, 425–432 10.1128/MCB.15.1.425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Herbert B. S., Chanoux R. A., Liu Y., Baenziger P. H., Goswami C. P., McClintick J. N., Edenberg H. J., Pennington R. E., Lipkin S. M., and Kopelovich L. (2010) A molecular signature of normal breast epithelial and stromal cells from Li-Fraumeni syndrome mutation carriers. Oncotarget 1, 405–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Subbaramaiah K., Brown K. A., Zahid H., Balmus G., Weiss R. S., Herbert B. S., and Dannenberg A. J. (2016) Hsp90 and PKM2 drive the expression of aromatase in Li-Fraumeni syndrome breast adipose stromal cells. J. Biol. Chem. 291, 16011–16023 10.1074/jbc.M115.698902 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44. Lowry O. H., Rosebrough N. J., Farr A. L., and Randall R. J. (1951) Protein measurement with the folin phenol reagent. J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.