Abstract
We previously reported that among the 37 RING finger protein (RNF) family members, RNF183 mRNA is specifically expressed in the kidney under normal conditions. However, the mechanism supporting its kidney-specific expression pattern remains unclear. In this study, we elucidated the mechanism of the transcriptional activation of murine Rnf183 in inner-medullary collecting duct cells. Experiments with anti-RNF183 antibody revealed that RNF183 is predominantly expressed in the renal medulla. Among the 37 RNF family members, Rnf183 mRNA expression was specifically increased in hypertonic conditions, a hallmark of the renal medulla. RNF183 up-regulation was consistent with the activation of nuclear factor of activated T cells 5 (NFAT5), a transcription factor essential for adaptation to hypertonic conditions. Accordingly, siRNA-mediated knockdown of NFAT5 down-regulated RNF183 expression. Furthermore, the −3,466 to −3,136-bp region upstream of the mouse Rnf183 promoter containing the NFAT5-binding motif is conserved among mammals. A luciferase-based reporter vector containing the NFAT5-binding site was activated in response to hypertonic stress, but was inhibited by a mutation at the NFAT5-binding site. ChIP assays revealed that the binding of NFAT5 to this DNA site is enhanced by hypertonic stress. Of note, siRNA-mediated RNF183 knockdown increased hypertonicity-induced caspase-3 activation and decreased viability of mIMCD-3 cells. These results indicate that (i) RNF183 is predominantly expressed in the normal renal medulla, (ii) NFAT5 stimulates transcriptional activation of Rnf183 by binding to its cognate binding motif in the Rnf183 promoter, and (iii) RNF183 protects renal medullary cells from hypertonicity-induced apoptosis.
Keywords: kidney, NFAT transcription factor, promoter, transcription regulation, ubiquitin ligase, hypertonic stress, NFAT5, renal cell, RNF family, RNF183
Introduction
The renal medulla is the only tissue that is constantly under hypertonic conditions to concentrate urine through water reabsorption in mammals (1). The driving forces for water reabsorption are elevated concentrations of sodium chloride and urea, which create exceptionally hypertonic conditions (600 and 2,000 mosmol/kg H2O under conditions of diuresis and antidiuresis in rodents, respectively) (2). Renal cells, particularly renal medullary cells, partly adapt to the hypertonic conditions by accumulating intracellular organic osmolytes that reduce intracellular ionic strength. Nuclear factor of activated T cells 5 (NFAT5)3/tonicity-responsive enhancer binding protein plays a key role in the adaptation to hypertonic conditions by binding to the consensus DNA-binding element RNRNNTTTCCA (where N is any nucleotide, and R is any purine) (3, 4). NFAT5 stimulates the transcription of aldose reductase (AR/AKR1B1) (5, 6), sodium- and chloride-dependent betaine transporter (BGT1/SLC6A13) (4, 6), sodium myo-inositol co-transporter (SMIT/SLC5A3) (3, 6), and heat shock protein 70 (HSP70/HSPA1B) (7, 8), which mediate the intracellular accumulation of sorbitol, betaine, and myo-inositol and function as an intracellular molecular chaperone, respectively. In addition to these genes, many downstream genes of NFAT5 are known (9–15). However, a ubiquitin ligase that is directly activated by NFAT5 has not yet been reported.
Ubiquitin ligases are involved in a wide variety of cellular events through ubiquitination (16, 17). RING finger protein 183 (RNF183) is a member of the RNF family, which functions as a ubiquitin ligase (18). Previous studies have demonstrated that RNF183 mRNA expression in the colon of patients with inflammatory bowel disease (IBD) and colorectal cancers was ∼5- and 2-fold higher than that in control subjects; in these patients, RNF183 promotes intestinal inflammation (19) and proliferation and metastasis of cancers (20), respectively. On the contrary, we previously demonstrated that RNF183 was specifically expressed in human and mouse kidney, and that mouse Rnf183 mRNA expression in the kidney was ∼324-fold higher than in the colon (21). To date, however, the reason why Rnf183 mRNA is selectively expressed in the kidney remains unclear.
In this study, we demonstrated that RNF183 is dominantly expressed in the renal medulla and that NFAT5 regulates Rnf183 transcription in mouse inner-medullary collecting duct (mIMCD-3) cells.
Results
RNF183 is predominantly expressed in the renal medulla
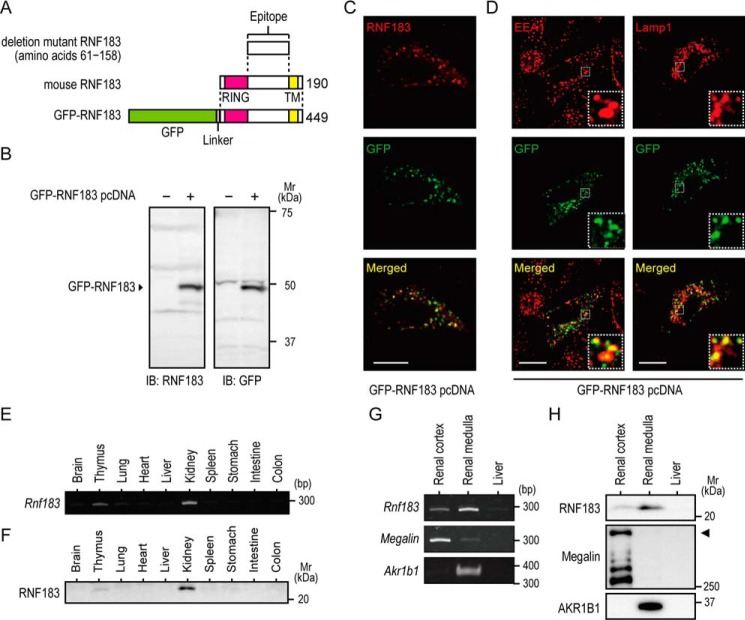
RNF183 has been described as a ubiquitin ligase, which is expressed in normal colonic epithelial cells and colorectal cancer cells (19, 20). In addition, we previously reported that Rnf183 mRNA expression in the kidney is ∼324-fold higher than in the colon (21); however, RNF183 protein expression in the kidney remains unclear. To detect RNF183 protein expression, we generated an affinity-purified antibody using recombinant deletion mutant RNF183 (amino acids 61–158) lacking a RING finger domain at its N terminus and a transmembrane domain at its C terminus (Fig. 1A). The antibody specifically recognized GFP-tagged RNF183 at ∼50 kDa, which was transiently transfected into human embryonic kidney 293 (HEK293) cells (Fig. 1B). Immunofluorescence staining using HeLa cells transiently transfected with GFP-RNF183 showed that punctate signals of GFP-RNF183 were recognized by the anti-RNF183 antibody (Fig. 1C). GFP signals were localized in the late endosome/lysosome (Lamp1) and early endosome (EEA1), with additional weak signal in the recycling endosome (transferrin receptor) and Golgi complex (GM130) but not in the endoplasmic reticulum (calnexin) or mitochondria (COX IV) (Fig. 1D and Fig. S1). To evaluate endogenous RNF183 protein expression, we performed RT-PCR and Western blot analysis using tissue extracts in 4-week-old mice. Western blotting revealed that endogenous RNF183 protein was expressed markedly in the kidney, particularly in the renal medulla, and in the thymus (Fig. 1, F and H), consistent with Rnf183 mRNA expression (Fig. 1, E and G). Megalin and AKR1B1 were used as positive controls for the renal cortex and medulla, respectively.
Figure 1.
RNF183 is predominantly expressed in the renal medulla. A, schematic representation of the predicted domain organization of mouse RNF183 (middle), deletion mutant RNF183 (amino acids 61–158) (top), and GFP-RNF183 (bottom) used in this study. Anti-RNF183 antibody was generated using deletion mutant RNF183. The number on the right indicates the peptide length. B, Western blotting of HEK293 cells transfected with GFP-RNF183. The anti-RNF183 and GFP antibody recognized a GFP-RNF183 at ∼50 kDa. C, confocal images of immunofluorescence with anti-RNF183 antibody (top, red) and GFP signals (middle, green) in HeLa cells transfected with GFP-RNF183. Bar, 10 μm. D, confocal images of immunofluorescence with GFP signals (middle panels, green) and various antibodies for organelle markers (EEA1 and Lamp1; top panels, red) in HeLa cells transfected with GFP-RNF183. Bars, 10 μm. E, RT-PCR analysis of Rnf183 mRNA in 10 tissues from mice. F, tissue immunoblot analysis of RNF183. Tissue lysates containing equal amounts of total protein were analyzed by Western blotting. G and H, expression patterns of RNF183 mRNA and protein in the renal cortex and medulla. Mouse tissue lysates were analyzed by RT-PCR (G) and Western blotting (H). Liver was used as a negative control. Megalin and AKR1B1 were used as positive controls for the renal cortex and medulla, respectively. Arrowhead, full-length megalin (H, middle). The results shown are representative of at least three replicates in independent observations.
RNF183 expression is up-regulated in response to hypertonic stress
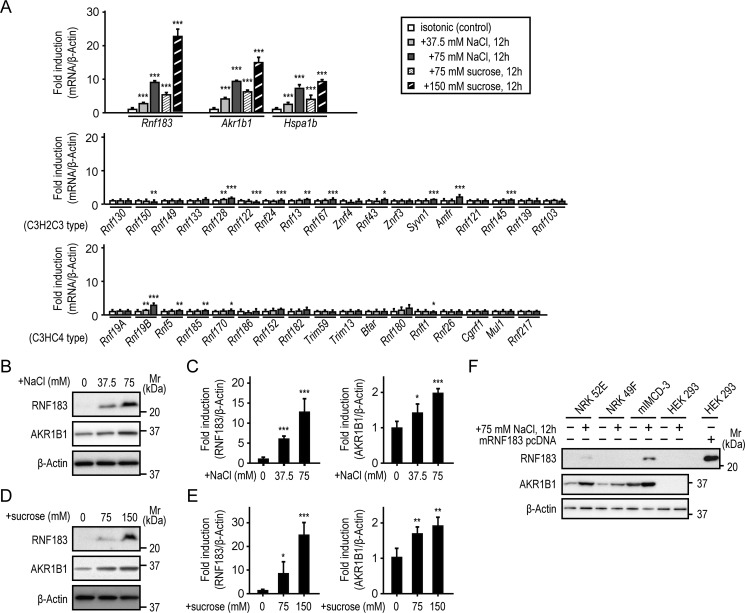
Renal medullary cells are constantly exposed to extreme hypertonic stress to concentrate the urine (1). Therefore, using quantitative real-time RT-PCR (qRT-PCR), we examined whether RNF183 expression and the other transmembrane RNF family members, those containing RING finger (C3H2C3 or C3HC4) and transmembrane domains, are up-regulated in response to hypertonic stress. Mouse IMCD-3 cells were incubated with isotonic medium or NaCl- or sucrose-supplemented medium. The extracellular tonicity was increased by ∼150 mosmol/kg H2O (final osmolality: 450 mosmol/kg H2O) by adding 75 mm NaCl or 150 mm sucrose. Akr1b1 (5, 6) and Hapa1b (7, 8) were used as positive controls for tonicity dependence. We found that Rnf183 mRNA expression was markedly up-regulated in a tonicity-dependent manner in both hypertonic NaCl- and sucrose-treated cells compared with that in isotonic control cells (Fig. 2A), which was consistent with the Akr1b1 and Hapa1b up-regulation patterns (Fig. 2A). Rnf128 and Rnf19B were modestly up-regulated in a tonicity-dependent manner. Further, Rnf24, Rnf13, Rnf167, Rnf43, Syvn1, Amfr, Rnf145, Rnf5, Rnf185, and Rnf170 were slightly up-regulated in cells treated with only 75 mm NaCl; the other RNF family members were not up-regulated (Fig. 2A). Western blot analysis showed that RNF183 and AKR1B1 protein expression was up-regulated in a tonicity-dependent manner (Fig. 2, B–E). However, Rnf183 mRNA was not up-regulated under hypoxic conditions (oxygen concentration, 1 and 0.3%) (Fig. S2), which is another characteristic of the renal medulla. These results suggest that hypertonic conditions play a more important role in RNF183 expression than hypoxic conditions. Next, we examined whether RNF183 up-regulation was different between mIMCD-3 cells and other renal cell lines. Normal rat kidney (NRK)-52E (a rat kidney tubular epithelial cell line), NRK-49F (a rat kidney interstitial fibroblast cell line), mIMCD-3, and HEK293 cells were used. HEK293 cells transiently transfected with mouse RNF183 were used as a positive control. We found that RNF183 expression increased markedly in mIMCD-3 cells treated with hypertonic NaCl and increased slightly in NRK-52E cells, whereas no expression was detected in NRK-49F and HEK293 cells (Fig. 2F). Interestingly, the difference in RNF183 expression among renal cell lines was similar to that in AKR1B1 expression. The similarity between RNF183 and AKR1B1 up-regulation in response to hypertonic stress suggests that NFAT5 regulates RNF183 expression as well as AKR1B1 expression.
Figure 2.
RNF183 is up-regulated in response to hypertonic stress. A, the hypertonic response of Rnf183 (top) and the other transmembrane RNF family member (middle, 18 of the C3H2C3 type; bottom, 17 of the C3HC4 type) mRNA levels in mIMCD-3 cells. Akr1b1 and Hspa1b were used as tonicity-dependent positive controls (top). Cells were treated with isotonic or the indicated NaCl- or sucrose-supplemented medium for 12 h and analyzed by qRT-PCR (n = 5). B and D, the tonicity-dependent induction of RNF183 and AKR1B1 protein in mIMCD-3 cells. Cells were cultured under hypertonic conditions by adding the indicated NaCl (B) or sucrose (D) supplementation for 12 h and analyzed by Western blotting. C and E, quantitative analysis of RNF183 (left) and AKR1B1 (right) protein expression in NaCl (B) and sucrose (D) (n = 5). F, the up-regulation patterns of RNF183 and AKR1B1 proteins in four renal cell lines in response to hypertonic stress. NRK-52E, NRK-49F, mIMCD-3, and HEK293 cells were treated with isotonic or 75 mm NaCl–supplemented medium for 12 h and analyzed by Western blotting. HEK293 cells transfected with mouse RNF183 were used as a positive control. Data were analyzed by one-way ANOVA, followed by post hoc tests using t tests with Bonferroni correction. Values represent mean ± S.D. (error bars). *, p < 0.05; **, p < 0.01; ***, p < 0.001 (versus isotonic control).
RNF183 expression is up-regulated concurrently with NFAT5 activation
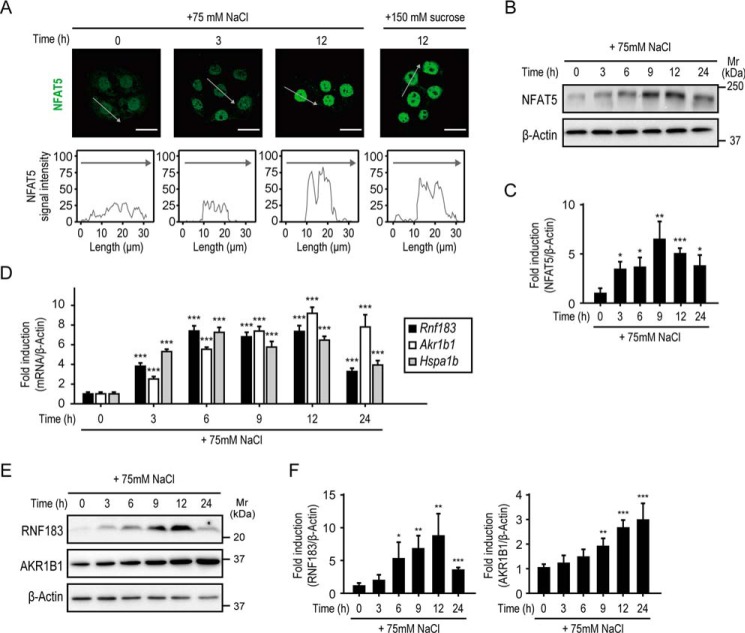
The NFAT5 transcription factor is the master regulator for hypertonic stress in mammals (22). In response to hypertonic stress, NFAT5 activation is achieved by a combination of NFAT5 induction and localization into the nucleus (4, 23, 24). Thus, we evaluated the effects of hypertonicity on NFAT5 activation in mIMCD-3 cells. We performed immunofluorescence staining of NFAT5 and plotted the fluorescence intensity along a line drawn through the nucleus. These analyses showed that NFAT5 was present in both the cytoplasm and nucleus under isotonic conditions (Fig. 3A). Hypertonic stimulation for 3 h reduced NFAT5 cytoplasmic expression and slightly enhanced nuclear expression, and stimulation for 12 h drastically enhanced nuclear expression (Fig. 3A). Western blot analysis demonstrated that the total abundance of NFAT5 protein significantly increased after 3 h of hypertonic stimulation and peaked at 9 h (Fig. 3, B and C), consistent with NFAT5 localization (Fig. 3A). Moreover, qRT-PCR analysis revealed that the expression of Rnf183 and NFAT5 downstream genes, Akr1b1 (5, 6) and Hspa1b (7, 8), concurrently increased after 3 h of hypertonic stimulation (Fig. 3D). The RNF183 and AKR1B1 protein levels significantly increased after 6 and 9 h of hypertonic stimulation (Fig. 3, E and F), respectively. In contrast to the downstream genes of NFAT5, the Tnfa mRNA level, which is that of NF-κB, peaked after 3 h of hypertonic stimulation and decreased after 6 h (Fig. S3). These results demonstrated that RNF183 expression was up-regulated under hypertonic conditions and was accompanied by NFAT5 activation and the up-regulation of NFAT5 downstream genes, suggesting that RNF183 expression is directly mediated by NFAT5.
Figure 3.
RNF183 is up-regulated concurrently with NFAT5 activation. A, nuclear translocation of NFAT5 in response to hypertonic stress. Mouse IMCD-3 cells were treated with hypertonic medium (75 mm NaCl– or 150 mm sucrose–supplemented medium) for the indicated time and subjected to immunofluorescence staining with anti-NFAT5 antibody (top panels, green). Bars, 20 μm. The NFAT5 fluorescence intensities of cells were plotted on line graphs (bottom panels). Gray lines correspond to the relative fluorescence of cells marked with gray arrows. B, increase in total NFAT5 protein abundance in response to hypertonic stress. Mouse IMCD-3 cells were treated with 75 mm NaCl–supplemented medium for the indicated time and analyzed by Western blotting. C, quantification of data from B (n = 4). D and E, time-course analysis of mRNA and protein in response to hypertonic stress. Mouse IMCD-3 cells were treated with NaCl-supplemented medium for the indicated time and analyzed by qRT-PCR (D; n = 6) or by Western blotting (E). F, quantitative analysis of RNF183 (left) and AKR1B1 (right) expression in E (n = 5). Data were analyzed by one-way ANOVA, followed by post hoc tests using t tests with Bonferroni correction. Values represent mean ± S.D. (error bars). *, p < 0.05; **, p < 0.01; ***, p < 0.001 (versus isotonic control).
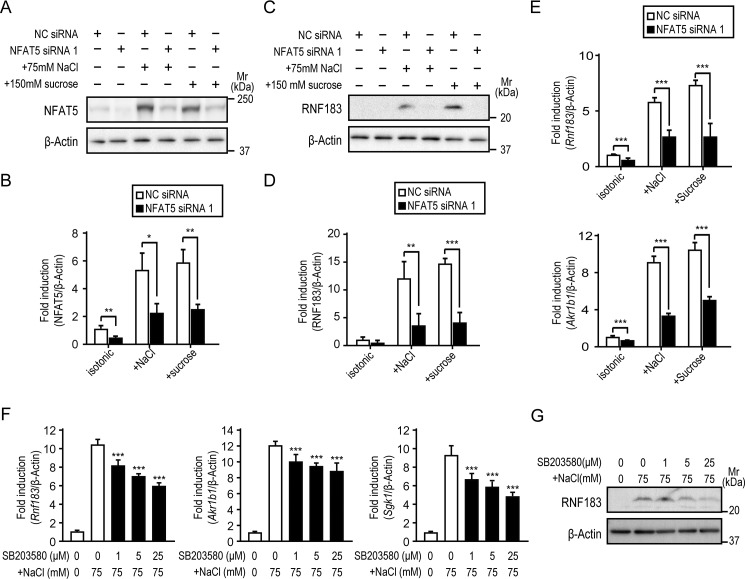
NFAT5 knockdown and p38/MAPK inhibitor SB203580 attenuate hypertonicity-induced RNF183 expression
To determine the effect of NFAT5 inhibition on RNF183 expression, mIMCD-3 cells were transfected with siRNA oligonucleotides targeting NFAT5 (NFAT5 siRNA 1) or negative control siRNA (NC siRNA). After 12 h of hypertonic stimulation, the NFAT5 protein levels were significantly reduced in mIMCD-3 cells transfected with NFAT5 siRNA 1 (Fig. 4, A and B). NFAT5 knockdown significantly inhibited the hypertonicity-induced RNF183 protein and mRNA expression of both Rnf183 and Akr1b1 (Fig. 4, C–E). Another NFAT5 siRNA (NFAT5 siRNA 2) produced similar results (Fig. S4, A–C). The hypertonic stress signal is mediated through p38/MAPK, which regulates NFAT5 activation in mammals (12, 13, 25, 26). Thus, to determine the effect of p38/MAPK inhibition on RNF183 expression, mIMCD-3 cells were treated with 75 mm NaCl–supplemented medium in the presence of the indicated concentration of SB203580. The hypertonicity-induced protein and mRNA expression of RNF183 and NFAT5 downstream genes Akr1b1 and Sgk1 were concurrently inhibited by SB203580 in a dose-dependent manner (Fig. 4, F and G). These results demonstrated that RNF183 expression depends on NFAT5.
Figure 4.
NFAT5 knockdown and p38/MAPK inhibitor SB203580 attenuate hypertonicity-induced RNF183 expression. A, inhibition of NFAT5 expression using siRNA. Mouse IMCD-3 cells transfected with NFAT5 siRNA 1 or NC siRNA were treated with isotonic or 75 mm NaCl– or 150 mm sucrose–supplemented medium for 12 h and analyzed by Western blotting. B, quantification of data from A (n = 4). C, effect of NFAT5 knockdown on RNF183 protein expression induced by hypertonicity. D, quantitative analysis of RNF183 in C (n = 4). E, effect of NFAT5 knockdown on Rnf183 (left) and Akr1b1 (right) mRNA (n = 5). F and G, effect of p38/MAPK inhibitor SB203580 on mRNA expression of Rnf183, Akr1b1, and Sgk1 (F) and protein expression of RNF183 (G). Cells were treated with 75 mm NaCl for 12 h in the presence of the indicated concentrations of SB203580 (1 h of preincubation) and analyzed by qRT-PCR (F) (n = 6) or Western blotting (G). Data were analyzed by one-way ANOVA, followed by post hoc tests using t tests with Bonferroni correction. Values represent mean ± S.D. (error bars). *, p < 0.05; **, p < 0.01; ***, p < 0.001 (versus NC).
Identification of the Rnf183 enhancer region that responds to hypertonic stress
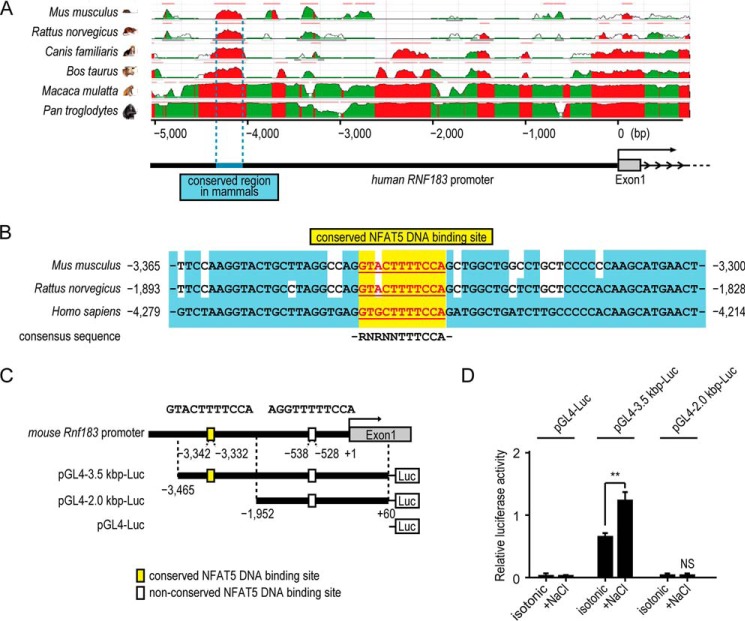
To identify the putative enhancer region in the Rnf183 promoter, we searched for an evolutionarily conserved region (ECR) within 5,000 bp upstream of the human RNF183 gene using the ECR browser (27). The 319-bp region (−4,367 to −4,049-bp region; numbers indicate nucleotide positions relative to the transcription start site) upstream of the human RNF183 gene was identified as conserved in mammals (blue; Fig. 5A). Although its actual position from the transcription start site varies among the species (Fig. 5B), it shows high sequence identity with the corresponding region in the following mammals: Mus musculus, 71%; Rattus norvegicus, 73%; Canis familiaris, 81%; Bos taurus, 80%; Macaca mulatta, 94%; and Pan troglodytes, 98%. We next searched for potential binding sites for various transcription factors within the mammalian conserved region of RNF183 genes (−3,466 to −3,136, mouse (Table S1); −1,978 to −1,664, rat (top right); −4,367 to −4,049, human (bottom right)) using the JASPAR database (28). This analysis identified one binding site for NFAT proteins conserved among mouse, rat, and human (Table 1 and Table S1). The sequences (GTACTTTTCCA (−3,342 to −3,332; numbers indicate the nucleotide position relative to the transcription start site of mouse Rnf183), GTACTTTTCCA (−1,870 to −1,860; numbers indicate the nucleotide position relative to the transcription start site of rat RNF183), and GTGCTTTTCCA (−4,256 to −4,246; numbers indicate the nucleotide position relative to the transcription start site of human RNF183)) were completely homologous with the NFAT5 consensus sequence (RNRNNTTTCCA) (3, 4) (Fig. 5B). Furthermore, the sequences around the binding site were highly conserved among mouse, rat, and human (blue background). Between the conserved region and the transcription start site of the mouse Rnf183 gene (−3,135 to −1), we found another site (AGGTTTTTCCA; −538 to −528) that was completely homologous with the NFAT5 consensus sequence, but not conserved among the species. To determine whether NFAT5 acts on the Rnf183 promoter containing these sites to activate Rnf183 transcription, we performed luciferase reporter assays using the promoter region in mIMCD-3 cells. A 3.5-kbp (containing both NFAT5 DNA–binding sites) or 2.0-kbp (containing the nonconserved NFAT5 DNA–binding site) promoter region of the mouse Rnf183 gene (pGL4–3.5 kbp-Luc and pGL4–2.0 kbp-Luc, respectively; Fig. 5C) was used. The reporter activity of pGL4–3.5 kbp-Luc, but not that of pGL4–2.0 kbp-Luc or pGL4-Luc, was considerably induced by hypertonic stress (Fig. 5D). These results demonstrated that the putative Rnf183 enhancer region has responsiveness to hypertonic conditions.
Figure 5.
Identification of the Rnf183 enhancer region that responds to hypertonic stress. A, screen capture from the ECR Browser website (http://ecrbrowser.dcode.org/) (57) (please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site) of the human RNF183 gene with ECR in the genomes of mouse, rat, canis, cattle, rhesus, and chimpanzee. The peaks in red show comparative pairwise ECR between human and the indicated mammals. The 319-bp region (−4,367 to −4,049 region; numbers indicate nucleotide positions relative to the transcription start site) is conserved in mammals (blue). B, location and sequence of the conserved NFAT5 DNA–binding site (yellow background with underline; mouse, −3,342 to −3,332 bp; rat, −1,870 to −1,860 bp; human, −4,256 to −4,246 bp). Conserved bases around the binding site are indicated as blue background. NFAT5 consensus sequence (RNRNNTTTCCA) is indicated below the site. C, scheme of the mouse Rnf183 promoter region and reporter constructs. Two NFAT5 DNA–binding sites are indicated in 3.5 kbp upstream of the mouse Rnf183 start site. 3.5 kbp-Luc and 2.0 kbp-Luc constructs consist of ∼3.5 kbp and 2.0 kbp upstream of the mouse Rnf183 transcription start site, respectively. D, effect of hypertonic stress on luciferase activities. Mouse IMCD-3 cells transfected with 3.5 kbp-Luc or 2.0 kbp-Luc constructs were treated with isotonic or 75 mm NaCl–supplemented medium for 24 h. The pGL4-Luc without the promoter was used as a control. Firefly luciferase activities were normalized to Renilla luciferase signals (n = 3). Data were analyzed by one-way ANOVA, followed by post hoc tests using t tests with Bonferroni correction. Values represent mean ± S.D. (error bars). **, p < 0.01; N.S., p > 0.05.
Table 1.
Conserved putative transcription factor binding sites in the RNF183 enhancer region
Transcription factor–binding sites predicted using the JASPAR software are shown in the mammalian conserved region of RNF183 genes (mouse, −3,466 to −3,136; rat, −1,978 to −1,664; human, −4,367 to −4,049). Conserved binding sites from mice, rats, and humans were selected from the potential binding sites with a >95% chance (relative score) of binding to any of the listed transcription factors (Table S1). The conserved NFAT5 DNA–binding site is indicated in boldface type. NFIC, nuclear factor 1 C-type; ZNF354C, zinc finger protein 354C; NFE2L1-MafG, nuclear factor erythroid 2–related factor 1 and V-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G complex; NFAT, nuclear factor of activated T cells; Atoh1, atonal homolog1; Prrx2, paired-related homeobox 2.
| Species | Transcription factor | JASPAR score | Relative score | Start | End | Strand | Predicted site sequence |
|---|---|---|---|---|---|---|---|
| % | |||||||
| M. musculus | NFIC | 8.520 | 96.1 | −3407 | −3402 | 1 | TTGGCT |
| ZNF354C | 8.723 | 99.2 | −3398 | −3393 | 1 | CTCCAC | |
| NFE2L1–MafG | 8.072 | 96.8 | −3385 | −3380 | 1 | TATGAC | |
| NFAT | 11.360 | 100.0 | −3338 | −3332 | 1 | TTTTCCA | |
| Atoh1 | 13.740 | 99.6 | −3333 | −3326 | 1 | CAGCTGGC | |
| Prrx2 | 9.124 | 100.0 | −3299 | −3295 | 1 | AATTA | |
| R. norvegicus | NFIC | 8.520 | 96.1 | −1935 | −1930 | 1 | TTGGCT |
| ZNF354C | 8.723 | 99.2 | −1926 | −1921 | 1 | CTCCAC | |
| NFE2L1–MafG | 8.812 | 100.0 | −1913 | −1908 | 1 | CATGAC | |
| NFAT | 11.360 | 100.0 | −1866 | −1860 | 1 | TTTTCCA | |
| Atoh1 | 13.740 | 99.6 | −1861 | −1854 | 1 | CAGCTGGC | |
| Prrx2 | 9.124 | 100.0 | −1826 | −1822 | 1 | AATTA | |
| Homo sapiens | NFIC | 8.520 | 96.1 | −4322 | −4317 | 1 | TTGGCT |
| ZNF354C | 8.723 | 99.2 | −4313 | −4308 | 1 | CTCCAC | |
| NFE2L1–MafG | 8.812 | 100.0 | −4300 | −4295 | 1 | CATGAC | |
| NFAT | 11.360 | 100.0 | −4252 | −4246 | 1 | TTTTCCA | |
| Atoh1 | 14.033 | 100.0 | −4247 | −4240 | 1 | CAGATGGC | |
| Prrx2 | 9.124 | 100.0 | −4208 | −4204 | 1 | AATTA |
Rnf183 enhancer region depends on NFAT5
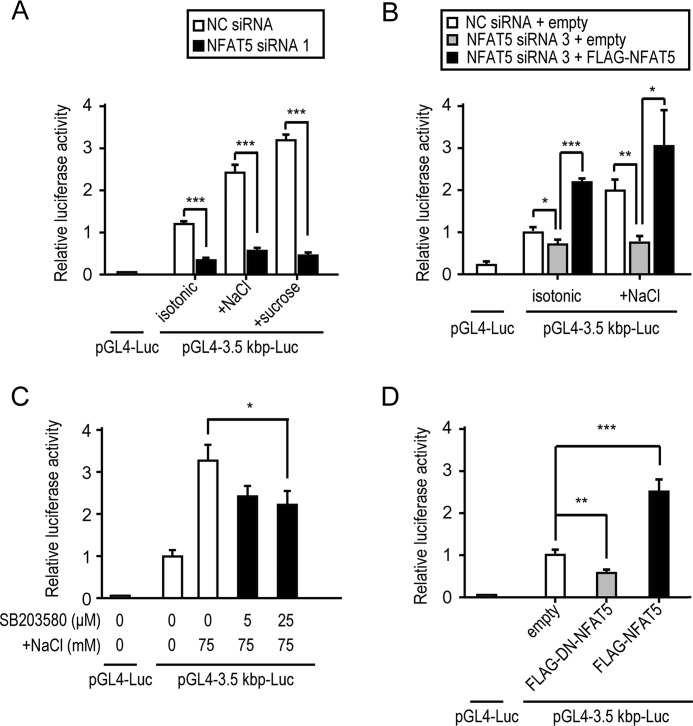
To determine whether the putative Rnf183 enhancer region depended on NFAT5, we examined the effects of NFAT5 knockdown, p38/MAPK inhibition, and the dominant negative (DN) form of NFAT5 on luciferase activities. NFAT5 knockdown markedly reduced the reporter activity of pGL4–3.5 kbp-Luc (Fig. 6A and Fig. S4D), whereas FLAG-NFAT5 overexpression fully rescued the down-regulated reporter activity (Fig. 6B and Fig. S5). p38/MAPK inhibitor SB203580 significantly inhibited the hypertonicity-induced Rnf183 luciferase activities (Fig. 6C), which is consistent with mRNA and protein expression of RNF183 (Fig. 4, F and G). Moreover, Rnf183 luciferase activities were inhibited by overexpression of DN-NFAT5, whereas they were up-regulated by that of the full-length NFAT5 (Fig. 6D). These results suggest that the putative Rnf183 enhancer region depends on NFAT5.
Figure 6.
Rnf183 enhancer region depends on NFAT5. A, effect of NFAT5 knockdown on hypertonicity-induced luciferase activities. Cells cotransfected with 3.5 kbp-Luc and siRNA (NFAT5 1 or NC) were treated with isotonic or 75 mm NaCl– or sucrose–supplemented medium for 24 h (n = 3). B, effect of rescue with FLAG-NFAT5 on luciferase activities. Cells pretransfected with siRNA (NFAT5 3 or NC) were cotransfected with 3.5 kbp-Luc and either empty or pFLAG-NFAT5 vectors after 12 h of knockdown. After 24 h of cotransfection, cells were treated with isotonic or 75 mm NaCl–supplemented medium for an additional 24 h (n = 4). C, effect of p38/MAPK inhibitor SB203580 on Rnf183 luciferase activities. Cells transfected with 3.5 kbp-Luc were treated with 75 mm NaCl for 24 h in the presence of the indicated concentrations of SB203580 (1 h of preincubation) (n = 3). D, effect of overexpression with FLAG-DN-NFAT5 and FLAG-NFAT5 on luciferase activities. Cells cotransfected with 3.5 kbp-Luc and either empty, pFLAG-DN-NFAT5, or pFLAG-NFAT5 vectors were analyzed after 48 h of transfection (n = 4). Data were analyzed by one-way ANOVA, followed by post hoc tests using t tests with Bonferroni correction. Values represent mean ± S.D. (error bars). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Rnf183 is a direct target of NFAT5
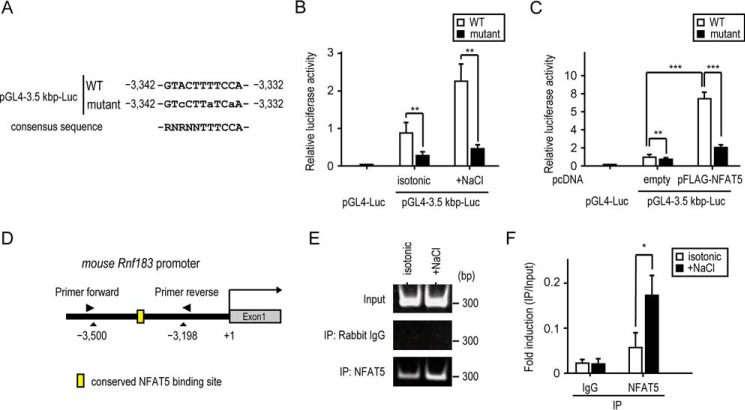
We next performed luciferase reporter assays using pGL4–3.5 kbp-Luc of WT and mutant having a mutation in the conserved NFAT5 DNA–binding site (Fig. 7A). In mIMCD-3 cells transfected with the mutant, reporter activities were markedly reduced even in hypertonic conditions (Fig. 7B). Consistent with this, NFAT5 overexpression markedly induced reporter activity of WT, which was inhibited by the mutation (Fig. 7C). To confirm whether NFAT5 directly binds to the conserved binding site in the Rnf183 promoter, we performed ChIP assays using mIMCD-3 cells under hypertonic conditions (Fig. 7D) and detected a high level of NFAT5 binding to the conserved binding site in the endogenous Rnf183 promoter (Fig. 7, E and F). These results indicated that NFAT5 directly acts on the conserved NFAT5 DNA–binding site within the Rnf183 promoter and facilitates its transcription in mIMCD-3 cells.
Figure 7.
Rnf183 is a direct target of NFAT5. A, location and site-directed mutagenesis of the conserved NFAT5 DNA–binding site. The sequences from the pGL4–3.5 kbp-Luc of the WT and mutant are shown. Mutagenized bases are indicated by lowercase letters. B, effect of conserved site mutation on luciferase activities. Cells transfected with WT or mutant were cultured in isotonic or 75 mm NaCl–supplemented medium for 24 h. The pGL4-Luc was used as a control. Firefly luciferase activities were normalized to Renilla luciferase signals (n = 4). C, effect of NFAT5 overexpression on luciferase activities. Cells transfected with WT, mutant, empty, or pFLAG-NFAT5 were cultured in an isotonic medium for 36 h (n = 5). D, schematic representation of the Rnf183 promoter and the annealing sites of the primer set used in the ChIP assays. E, ChIP assay analysis of the NFAT5 binding to the Rnf183 promoter in mIMCD-3 cells. ChIP assays were performed with the indicated antibodies. Immunoprecipitated (IP) DNA and input DNA were subjected to RT-PCR. IgG was used as a control. F, quantitative analysis of RT-PCR in E. Results were normalized to input DNA (n = 3). Data were analyzed by one-way ANOVA, followed by post hoc tests using t tests with Bonferroni correction. Values represent mean ± S.D. (error bars). *, p < 0.05; **, p < 0.01; ***, p < 0.001 (versus isotonic control).
RNF183 expression provides protection against hypertonicity-induced apoptosis
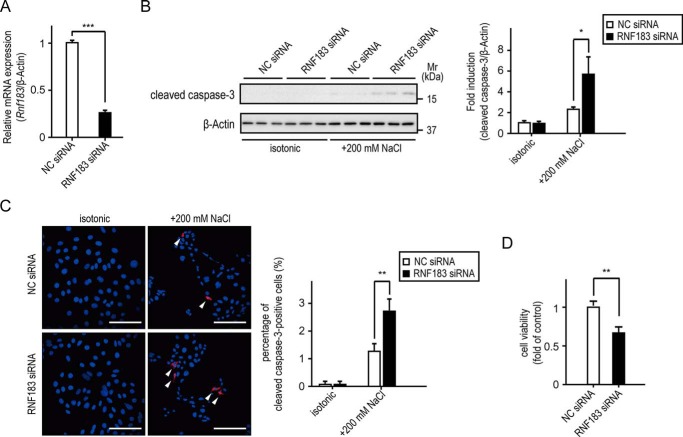
Stress induced by an elevated NaCl concentration mediates apoptosis in mIMCD3 cells when osmolality is acutely raised to >600 mosmol/kg (29–31). Moreover, several NFAT5 downstream genes show a protective effect against hypertonicity-induced apoptosis (9, 32, 33). Thus, to determine whether RNF183 expression affects the osmotic tolerance of mIMCD-3 cells to high NaCl levels, mIMCD-3 cells were transfected with siRNA oligonucleotides targeting RNF183 (RNF183 siRNA) or NC siRNA. RNF183 siRNA effectively reduced the Rnf183 mRNA levels compared with NC siRNA (Fig. 8A). After 4 h of exposure to 200 mm NaCl–supplemented medium, cleaved caspase-3 protein levels and cleaved caspase-3–positive cells (which are indexes of apoptosis) significantly increased in mIMCD-3 cells transfected with RNF183 siRNA (Fig. 8, B and C). Crystal violet assays revealed that RNF183 knockdown significantly reduces cell viability under hypertonic conditions (Fig. 8D). In contrast, RNF183 knockdown did not promote caspase activation upon treatment of mIMCD-3 cells with the apoptosis inducer staurosporine (Fig. S6). These results demonstrate that RNF183 protects inner-medullary cells from hypertonicity-induced apoptosis.
Figure 8.
RNF183 expression provides protection against hypertonicity-induced apoptosis. A, inhibition of Rnf183 expression using siRNA. Mouse IMCD-3 cells transfected with RNF183 siRNA or NC siRNA were analyzed using qRT-PCR (n = 3). B and C, effect of RNF183 knockdown on hypertonicity-induced apoptosis. Cells transfected with the indicated siRNAs were treated with isotonic or 200 mm NaCl–supplemented medium for 4 h and analyzed using Western blotting (B) or immunofluorescence staining (C) using anti-cleaved caspase-3–specific antibody. B, the accompanying bar graph summarizes the quantification of relative amounts of cleaved caspase-3 (n = 3). C, the cleaved caspase-3 fluorescence–positive cells (red) are marked with white arrows. Bars, 100 μm. The number of cleaved caspase-3–positive cells was counted in five different fields of view and divided by the number of DAPI-stained cells (blue) to yield the ratio of cleaved caspase-3 positive cells; the results are summarized in the accompanying bar graph (n = 3). D, effect of RNF183 knockdown on cell viability under hypertonic conditions. Cells transfected with the indicated siRNAs were treated with isotonic or 200 mm NaCl–supplemented medium for 12 h and analyzed using a crystal violet assay (n = 3). Data were analyzed using a t test (A and D) or one-way ANOVA, followed by post hoc tests using t tests with Bonferroni correction (B and C). Values represent mean ± S.D. (error bars). *, p < 0.05; **, p < 0.01; ***, p < 0.001 (versus NC).
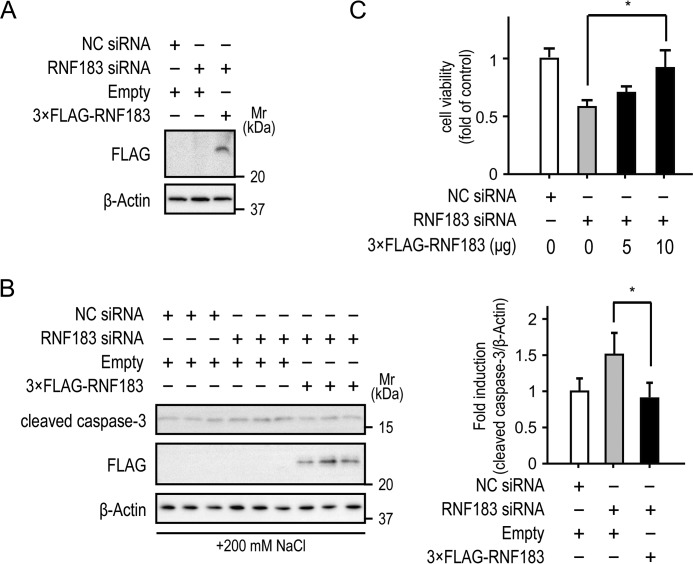
Rescue of RNF183 knockdown attenuates hypertonicity-induced apoptosis
To eliminate the possibility of siRNA off-target effects, mIMCD-3 cells transfected with RNF183 siRNA or NC siRNA were rescued with siRNA-resistant 3×FLAG-RNF183, which was expressed in mIMCD-3 cells under RNF183 knockdown (Fig. 9A). After 4 h of exposure to 200 mm NaCl–supplemented medium, cleaved caspase-3 protein levels significantly decreased in mIMCD-3 cells rescued with 3×FLAG-RNF183 (Fig. 9B). Crystal violet assays showed that the rescue of RNF183 knockdown significantly improved cell viability under hypertonic conditions (Fig. 9C). These results confirm that RNF183 protects inner-medullary cells from hypertonicity-induced apoptosis.
Figure 9.
Rescue of RNF183 knockdown attenuates hypertonicity-induced apoptosis. A, resistance of 3×FLAG-RNF183 to RNF183 siRNA. Mouse IMCD-3 cells were transfected with RNF183 siRNA or NC siRNA. After 12 h, the cells were electroporated with empty or 3×FLAG-RNF183-pcDNA vectors and were analyzed by Western blotting 48 h after electroporation. B and C, effect of rescue of RNF183 knockdown on hypertonicity-induced apoptosis (B) and cell viability (C). Cells transfected with RNF183 siRNA or NC siRNA were electroporated with empty or 3×FLAG-RNF183-pcDNA vectors. After 48 h of electroporation, cells were treated with 200 mm NaCl–supplemented medium for 4 (B) and 12 h (C) and then analyzed using Western blotting (B) and a crystal violet assay (C) (n = 3), respectively. The accompanying bar graph summarizes the quantification of the relative amounts of cleaved caspase-3 (n = 3). Data were analyzed by one-way ANOVA, followed by post hoc tests using t tests with Bonferroni correction. Values represent mean ± S.D. (error bars). *, p < 0.05.
Discussion
We identified the conserved NFAT5 DNA–binding site in the Rnf183 enhancer region and found that this site induced RFN183 reporter activity under hypertonic conditions, whereas mutation at the site resulted in almost complete inhibition of Rnf183 reporter activity. In addition, ChIP assays revealed a high level of NFAT5 binding to the site. Thus, these results demonstrated a mechanism of RNF183 up-regulation under hypertonic conditions by which NFAT5 directly binds to the conserved NFAT5 DNA–binding site in the Rnf183 enhancer region and leads to its transcriptional activation. This is the first study to demonstrate that ubiquitin ligase can be directly regulated by NFAT5. However, Yu et al. (19) demonstrated that microRNA-7 (miR-7) directly bound to the 3′-UTR of RNF183 mRNA, leading to RNF183 degradation and translational inhibition, and that down-regulation of miR-7 induced RNF183 elevation in inflamed colon tissues of IBD patients and colitic mice. Of the two regulators of RNF183 expression, we considered NFAT5 to be more important than miR-7 in a normal kidney because the tissue distribution pattern of RNF183 is different from that of miR-7 (34). The expression of miR-7 is restricted to specific tissues of the brain, colon, and thymus, whereas it is extremely low in the kidney, heart, liver, lung, spleen, and stomach (34), which is not consistent with our results on RNF183 expression (Fig. 1, E and F). Whereas NFAT5 is broadly expressed as an essential factor during exposure to the hypertonic environment (4), systemic or immature thymocyte-specific NFAT5 knockout mice display profound defects in the renal medulla and reduced thymocyte compartment in the thymus, respectively (6, 35, 36). This is because NFAT5 is activated by hypertonicity-dependent and -independent pathways in the renal medulla (6, 37, 38) and thymus (36), respectively. Consistent with these findings, RNF183 was highly expressed in the renal medulla and was also expressed in the thymus (Fig. 1, E–H). Therefore, we hypothesized a mechanism by which high RNF183 expression in the normal kidney and thymus is predominantly regulated by the NFAT5 activation, but not miR-7.
We observed an ∼2–3-fold increase in the induction of Rnf183 reporter activity following hypertonic stimulation. One possible explanation for this mild induction of the Rnf183 promoter is that only a short sequence of the Rnf183 promoter was investigated. Consistent with our results, Hasler et al. (39) and Chen et al. (12) have demonstrated that the activity of the AQP2 and SGK1 promoters (each containing one NFAT5-binding element) increased ∼1.5- and 5-fold, respectively, following hypertonic stimulation. However, at least five NFAT5-binding elements spread over 50-kb pairs, which participate in NFAT5-mediated transcriptional stimulation, were revealed in the SMIT promoter (3). Similarly, three NFAT5-binding sequences that were identified in the AKR1B1 promoter may collectively participate in the hyperosmotic response (5). Therefore, other NFAT5-binding elements may be present in regions of the Rnf183 promoter that are further upstream or downstream. Moreover, we demonstrated that Rnf183 reporter activity and Rnf183 and Akr1b1 mRNA expression were concurrently reduced by NFAT5 knockdown under isotonic conditions. Several previous studies have shown that, to some extent, NFAT5 knockdown or dominant negative NFAT5 expression inhibited mRNA expression or promotor activity of its downstream genes under isotonic conditions (7, 9, 39, 40). In contrast, two studies have shown that NFAT5 inhibition did not affect the basal expression of its downstream genes under isotonic conditions (12, 41). This difference in NFAT5 inhibition may result from variation in the basal NFAT5 protein levels because basal AKR1B1 protein expression levels were high in mIMCD-3 cells compared with those in other renal cell lines (Fig. 2F).
We further examined whether RNF183 was up-regulated by hypoxia in mIMCD-3 cells because the renal medulla is characteristically hypoxic (42). Although vascular endothelial growth factor A (Vegfa) mRNA, which is downstream of the hypoxic master regulator hypoxia-inducible factor 1 (HIF-1) (43), was significantly up-regulated under hypoxia (oxygen concentration, 1 and 0.3%), Rnf183 was not up-regulated (Fig. S2). Moreover, the HIF-1 DNA–binding site (ACGTG) was not observed in the Rnf183 enhancer region (Table S1). These results suggest that the high RNF183 expression in the renal medulla does not result from hypoxia.
Our results demonstrated that Rnf183 mRNA was markedly up-regulated compared with the other transmembrane RNF family members under hypertonic conditions and that the siRNA-induced knockdown of RNF183 exacerbates apoptosis and reduces cell viability in mIMCD-3 cells at ∼700 mosmol/kg. Although the effects of RNF183 on adaptation to hypertonicity appear to be mild, we consider RNF183 to be as important as other NFAT5-regulated genes. Lee et al. (44) demonstrated up to a 1.5-fold increase in the hypertonicity-induced LDH release in MEF cells transfected with siRNA targeting SMIT, BGT1, AKR1B1, HSPA1B, etc. Shim et al. (33) have also demonstrated an ∼20–30% decrease in cell viability following 100 mm NaCl supplementation in HSPA1B-knockout MEF cells. Their effects on adaptation to hypertonicity appear to be similar to that of RNF183 in the present study. Therefore, RNF183 expression is induced by hypertonic stress and plays an important role in the kidney's osmoprotective function under hypertonic conditions. However, we could not clarify the substrate of RNF183 under hypertonic conditions. To date, three previous studies have demonstrated the different functions of RNF183 in cancers and the colon as follows. (i) In tumors, RNF183 interacts with fetal and adult testis–expressed 1 (FATE1), which is a cancer/testis antigen, and negatively regulates the apoptotic effector Bcl-2–interacting killer (BIK), a component of FATE1 complex, leading to increased tumor cell viability (45). (ii) In the colon, RNF183 degrades IκBα, thereby inducing NF-κB activation, which might contribute the pathogenesis of IBD (19). (iii) In the colorectal cancers, RNF183 induces the activation of NF-κB and expression of its downstream gene, IL8, resulting in increased proliferation and metastasis of colorectal cancer cells (20). However, FATE1 expression is restricted to cancer or testis and is absent in the other tissues, such as the kidney (45). Moreover, NF-κB activation by hypertonicity is transient and starts decreasing after 3 h of hypertonic stimulation in cortical collecting duct cells due to the restoration of IκBα protein levels (46, 47). Consistent with this, the mRNA level of Tnfa, an NF-κB downstream gene, peaked after 3 h of hypertonic stimulation and decreased after 6 h at the time of RNF183 protein level elevation (Fig. S3). Hence, these results suggest that RNF183 ubiquitylates other substrates under hypertonic conditions as there is a contradiction on whether the NFAT5-RNF183 axis regulates BIK and IκBα expression.
RNF183 expression in response to hypertonicity is mediated by NFAT5, which is essential for long-term adaptation to hypertonic conditions (48). Short-term adaptation involves rapid influx of electrolytes and amino acids through various transporters, such as Na+-K+-Cl− cotransporters, Na+/H+ exchangers, Cl−/HCO3− exchangers (49), and sodium-coupled neutral amino acid transporter 2 (50). Long-term adaptation involves delayed replacement of ionic osmolytes with uncharged small organic osmolytes via expression of various enzyme and transporters, such as AKR1B1 (5, 6), BGT1 (4, 6), and SMIT (3, 6), mediated by NFAT5. However, in long-term adaptation, the mechanism by which the dispensable transporters of ionic osmolytes are down-regulated remains unclear. Here, the punctate signals of RNF183 were predominantly localized to endosomes and lysosomes (Fig. 1D). Early or late endosome-localized ubiquitin ligases can sort internalized cell-surface receptors from the recycling pathway to lysosomal degradation through ubiquitination (51–53). Therefore, we speculated that RNF183 plays a role in sorting internalized dispensable transporters to lysosomes for long-term adaptation to hypertonic conditions.
In summary, NFAT5 directly binds to the conserved DNA binding site in the Rnf183 enhancer region and leads to Rnf183 transcriptional activation, thereby inducing its high expression in the renal medulla. Among the transmembrane RNF family members, Rnf183 is specifically up-regulated by hypertonic stress and protects the cells from hypertonicity-induced apoptosis. However, our study did not include identification of substrate for RNF183 and validation by animal experiments under hypertonic conditions. Further study is warranted to elucidate the localization and function of RNF183 in the kidney.
Experimental procedures
Plasmids
A full-length mouse RNF183 construct with a stop codon was transferred from pENTR/D-TOPO (catalog no. 45-0218, Invitrogen) (54) into the Vivid Colors pcDNA6.2/N-EmGFP-DEST vector (catalog no. 35-1017, Invitrogen) containing a N-terminal emerald GFP and into pcDNA6.2-DEST (catalog no. 35-1264, Invitrogen) using Gateway LR Clonase II enzyme mix (catalog no. 12538-120, Invitrogen). The 3×FLAG-RNF183 construct synthesized using GeneArt Strings DNA fragments (Invitrogen) was cloned into the pENTR/D-TOPO (Invitrogen). The entry clone product was recombined into the pcDNA6.2-DEST vector using LR Clonase II enzyme mix. The deletion mutant mouse RNF183 (amino acids 61–158) with a stop codon was amplified by PCR using the following primer sets and was subcloned into pENTR/D-TOPO: 5′-CACCATGCAGCCCACCGT-3′ (forward) and 5′-TCAGAAGTGAGGGTTGCGGACACA-3′ (reverse). Subsequently, using Gateway LR Clonase II enzyme mix, the sequence was transferred into the pDest-565 vector (catalog no. 11520, Addgene), containing a GSH S-transferase (GST) at the N terminus of insert sequences and into the pDest-566 vector (catalog no. 11517, Addgene) containing a maltose-binding protein (MBP) at the N terminus of the insert sequences. pFLAG-NFAT5, the expression vector of NFAT5, was gifted to us by Dr. B. C. Ko (23) and Dr. Takashi Ito (9). The pFLAG-DN-NFAT5 was a generous gift from Dr. Takashi Ito (9).
Rnf183 promoter fragments were amplified from C57BL/6 murine genomic DNA by PCR using the following primer sets: 3,525 bp (−3,465 to +60 region; numbers indicate the nucleotide position relative to the transcription start site), 5′-GGTACCTTTCCCAGAAAGCCCATCCC-3′ (forward) and 5′-TTTCTCGAGTGTGAGTCACCCCTGCTTTC-3′ (reverse); 2,012 bp (−1,952 to +60 region), 5′-CTGGTACCTCAAGAGCCATTGAAAGAACACT-3′ (forward) and 5′-TTTCTCGAGTGTGAGTCACCCCTGCTTTC-3′ (reverse). Subsequently, the purified products were subcloned into the pGL4.10[luc2] vector (catalog no. 9PIE665, Promega) between KpnI and XhoI sites, respectively. The resulting vectors were named pGL4–3.5 kbp-Luc and pGL4–2.0 kbp-Luc, respectively. Mutagenesis of the conserved NFAT5 DNA–binding site (−3,358 GTACTGCTTAGGCCAGGTACTTTTCCAGCTGGCTGGCCTGC; NFAT5 DNA–binding site is underlined) was performed using the QuikChange Lightning site-directed mutagenesis kit (catalog no. 210518, Agilent Technologies) and the following series of oligonucleotides (sequence represents antisense strand, with mutagenized bases represented in lowercase type): mutant, −3,358 GTACTGCTTAGGCCAGGTcCTTaTCaAGCTGGCTGGCCTGC.
Antibodies
For obtaining the recombinant deletion mutant, plasmids containing RNF183 (amino acids 61–158) fused to GST (pDEST-565) or MBP (pDEST-566) were transformed into the Escherichia coli strain BL21 (DE3) (catalog no. 69450-3, Millipore, Tokyo, Japan); protein expression was induced by isopropyl β-d-1-thiogalactopyranoside (catalog no. R0392, Thermo Fisher Scientific). Rabbit antiserum was raised against the GST-tagged protein, and affinity purification was performed using a HiTrap NHS-activated HP column (catalog no. 17-0716-01, GE Healthcare) coupled with MBP-tagged protein.
Animals
Four-week-old C57BL/6 mice were used in this study. All animal experiments were performed in accordance with the National Institutes of Health guidelines for the care and use of laboratory animals and were approved by the Committee of Animal Experimentation, Hiroshima University. The mice were maintained in a room at 23 °C and a constant day–night cycle and were provided food and water ad libitum.
Cell culture, treatments, and transfection
HEK293, HeLa, mIMCD-3, NRK-49F, and NRK-52E cells were maintained in Dulbecco's modified Eagle's medium (DMEM; catalog no. 12800-017, Gibco) or DMEM/nutrient mixture F-12 (DMEM/F-12, catalog no. 12500-062, Gibco) supplemented with 10% (v/v) heat-inactivated fetal bovine serum at 37 °C in a 5% CO2 and 95% humidified air atmosphere. To induce hyperosmotic stress, cells received equal volumes of DMEM/F-12 (control) or 75 mm NaCl or 150 mm sucrose in DMEM/F-12 for the indicated time, according to the method reported by Bell et al. (55). For knockdown experiments, mIMCD-3 cells were transfected with three types of NFAT5-targeting Silencer siRNA (catalog no. 4390771 (ID: s21069 and s203948) and catalog no. 4399665 (ID: s550857), Invitrogen) or an RNF183-targeting Silencer siRNA (catalog no. 4390771 (ID: s94062), Invitrogen) or a negative control Silencer siRNA (catalog no. 4390843, Invitrogen) as a control using ScreenFect siRNA (catalog no. 295-75003, Wako Pure Chemical Industries, Osaka, Japan) in 6-well plates (12.5 pmol/well). HeLa or HEK293 cells were transfected with each expression plasmid using ScreenFect A (catalog no. 297-73204, Wako Pure Chemical Industries). For inhibition of p38/MAPK, mIMCD-3 cells were preincubated with SB203580 at 0–25 μm (catalog no. 199-16551, Wako Pure Chemical Industries) for 1 h and treated with 75 mm NaCl–supplemented medium for the indicated time, according to the method reported by Chen et al. (12).
Analysis of mRNA levels in murine tissues
Total murine tissue RNA was extracted from C57BL/6 mice using ISOGEN (catalog no. 311-02501, Nippon Gene, Toyama, Japan) according to the manufacturer's protocol. Reverse transcription was performed with ReverTra Ace (catalog no. TRT-101, Toyobo Co., Ltd., Osaka, Japan). PCR was performed using the following primer sets in a total volume of 20 μl containing 0.2 μm of each primer, 0.2 mm dNTPs, 1 unit of Paq5000 DNA polymerase, and 10× PCR buffer (catalog no. 600680, Agilent Technologies): mouse Rnf183, 5′-GACCAGCCCAAGAGCCGCTA-3′ (forward) and 5′-CCCCAAAAGAACTGCTTAGTCCA-3′ (reverse); mouse Megalin, 5′-CTCCTGCAAGTCGGTCCATT-3′ (forward) and 5′-GACCGCCCAGTAGAGCTTTT-3′ (reverse); mouse Akr1b1, 5′-AGAGCATGGTGAAAGGAGCC-3′ (forward) and 5′-TGGCACTCGATCTGGTTCAC-3′ (reverse). The PCR conditions were as follows: 94 °C for 2 min, 22–26 cycles at 98 °C for 10 s, 60 °C for 30 s, 72 °C for 30 s, and 72 °C for 3 min. The PCR products were resolved by electrophoresis on a 4.8% acrylamide gel. The density of each band was quantified using the Adobe Photoshop Elements version 2.0 program (Adobe Systems Inc.).
Analysis of mRNA levels in culture cells
Total RNA from mIMCD-3 cells was extracted using ISOGEN. Reverse transcription was performed with ReverTra Ace. The reverse-transcribed cDNA was measured by TaqMan-based real-time PCR assay using the ΔΔCt method. The following TaqMan primer and probe sets, purchased from Integrated DNA Technologies (Coralville, IA) were used: mouse Rnf183 (assay ID: Mm.PT.58.42013627), mouse Akr1b1 (assay ID: 214320988), mouse Hspa1b (assay ID: Mm.PT.58.31570020.g), mouse Vegfa (assay ID: Mm.PT.58.31754187), mouse Tnfa (assay ID: Mm.PT.58.29509614), mouse Rnf130 (assay ID: Mm.PT.58.9219868), mouse Rnf150 (assay ID: Mm.PT.58.10273312), mouse Rnf149 (assay ID: Mm.PT.58.11306683), mouse Rnf133 (assay ID: Mm.PT.58.43776563.g), mouse Rnf128 (assay ID: Mm.PT.56a.8462982), mouse Rnf122 (assay ID: Mm.PT.58.8505793), mouse Rnf24 (assay ID: Mm.PT.58.9635653), mouse Rnf13 (assay ID: Mm.PT.58.11306683), mouse Rnf167 (assay ID: Mm.PT.58.32670381), mouse Znf4 (assay ID: Mm.PT.58.32478833.g), mouse Rnf43 (assay ID: Mm.PT.56a.31556976), mouse Znf3 (assay ID: Mm.PT.56a.5645486), mouse Syvn1 (assay ID: Mm.PT.58.14132695), mouse Amfr (assay ID: Mm.PT.58.9856954), mouse Rnf121 (assay ID: Mm.PT.58.6378114), mouse Rnf145 (assay ID: Mm.PT.58.10110397), mouse Rnf139 (assay ID: Mm.PT.58.45812642), mouse Rnf103 (assay ID: Mm.PT.58.9763784), mouse Rnf19A (assay ID: Mm.PT.58.28600163), mouse Rnf19B (assay ID: Mm.PT.58.28736280), mouse Rnf5 (assay ID: Mm.PT.58.14209781), mouse Rnf185 (assay ID: Mm.PT.56a.11642500), mouse Rnf170 (assay ID: Mm.PT.56a.13272002), mouse Rnf186 (assay ID: Mm.PT.56a.29338718.g), mouse Rnf152 (assay ID: Mm.PT.58.29277956.g), mouse Rnf182 (assay ID: Mm.PT.58.30902864.g), mouse Trim59 (assay ID: Mm.PT.58.30417218), mouse Trim13 (assay ID: Mm.PT.56a.9751937), mouse Bfar (assay ID: Mm.PT.56a.28494992), mouse Rnf180 (assay ID: Mm.PT.58.45835696), mouse Rnft1 (assay ID: Mm.PT.58.6836835), mouse Rnf26 (assay ID: Mm.PT.58.5764306.g), mouse Cgrnf1 (assay ID: Mm.PT.58.5664724), mouse Mul1 (assay ID: Mm.PT.58.11670267), and mouse Rnf217 (assay ID: Mm.PT.56a.33323466). Mouse β-actin TaqMan primer and probe set was as follows: 5′-GCGGTTCCGATGCCCT-3′ (forward), 5′-CATGGATGCCACAGGATTCC-3′ (reverse), and 5′-AGGCTCTTTTCCAGCCTTCCTTCTTGG-3′ (probe).
Western blotting
Tissue or cell lysates were prepared in 1× Laemmli sample buffer. Equivalent protein samples were loaded onto SDS-polyacrylamide gels. Gels were transferred to Immobilon-P polyvinylidene difluoride membranes (catalog no. IPVH00010, Millipore) and blocked in 5% nonfat dry milk at room temperature for 1 h. The membrane was incubated with primary antibody at 4 °C overnight and with secondary antibody at room temperature for 1 h. The following primary antibodies were used: anti-β-actin (catalog no. sc47778, Santa Cruz Biotechnology, Inc.), anti-aldose reductase (catalog no. sc271007, Santa Cruz Biotechnology), anti-cleaved caspase-3 (catalog no. 9661, Cell Signaling Technology), anti-GFP (catalog no. 598, Medical Biological Laboratories, Nagoya, Japan), anti-megalin (catalog no. 101-M144, ReliaTech GmbH, Germany), anti-NFAT5 (catalog no. ab3446, Abcam, UK), and anti-RNF183. The secondary antibodies used were as follows: horseradish peroxidase–conjugated goat anti-mouse IgG antibody (catalog no. 1031-05 Southern Biotech) and horseradish peroxidase–conjugated goat anti-rabbit antibodies (catalog no. 4050-05 Southern Biotech). Images were obtained using a WSE-6100 LuminoGraph (ATTO Corp., Tokyo, Japan).
Immunofluorescence
Cells were fixed in 4% paraformaldehyde at room temperature for 10 min and then permeabilized in methanol at −20 °C for 10 min. Cells were blocked with 5% anti-goat serum at room temperature for 30 min and incubated with primary antibody at 4 °C overnight and then with Alexa Fluor–conjugated secondary antibodies at room temperature for 30 min. The following primary antibodies were used: anti-GFP (catalog no. 598, Medical Biological Laboratories), anti-NFAT5 (catalog no. ab3446, Abcam), anti-cleaved caspase-3 (catalog no. 9661, Cell Signaling Technology), anti-calnexin (catalog no. 2679, Cell Signaling Technology), anti-COX IV (catalog no. 4850, Cell Signaling Technology), anti-EEA1 (catalog no. 3288, Cell Signaling Technology), anti-GM130 (catalog no. 12480, Cell Signaling Technology), anti-transferrin receptor (catalog no. 13-6800, Invitrogen), and anti-Lamp1 (catalog no. 9091, Cell Signaling Technology). The secondary antibodies used were as follows: goat anti-mouse Alexa Fluor 555 (catalog no. A-21422, Invitrogen), goat anti-rabbit Alexa Fluor 568 (catalog no. A-11036, Invitrogen), goat anti-mouse Alexa Fluor 488 (catalog no. A-11001, Invitrogen), and goat anti-rabbit Alexa Fluor 488 (catalog no. A-11070, Invitrogen). ProLong Diamond Antifade Mountant with DAPI (catalog no. P36962, Invitrogen) was used to mount slips on glass slides. Fluorescence images were acquired using an Olympus FluoView FV1000 confocal microscope (Olympus, Tokyo, Japan). Apoptotic cells were counted in five different fields of view, and the ratio of the number of cleaved caspase 3–positive cells to that of DAPI-stained cells was calculated.
Luciferase assay
Mouse IMCD-3 cells were transiently transfected with 0.2 μg of pGL4.10[luc2] carrying both the firefly luciferase gene and the referenced constructs and 0.02 μg of pGL4.74[hRluc/TK] carrying the Renilla luciferase gene under the control of the herpes simplex virus thymidine kinase enhancer and promoter (catalog no. 9PIE692, Promega) in the presence of NFAT5 siRNA 1, NFAT5 siRNA 2 (catalog no. 4390771, Invitrogen), NFAT5 siRNA 3 (catalog no. 4399665, Invitrogen), NC siRNA (catalog no. 4390843, Invitrogen), or empty, pFLAG-DN-NFAT5, or pFLAG-NFAT5 vectors; the transfection was performed using ScreenFect A (catalog no. 297-73204, Wako Pure Chemical Industries) under conditions recommended by the manufacturer. For rescue of NFAT5 knockdown, cells pretransfected with siRNA (NFAT5 siRNA 3 or NC) using ScreenFect siRNA (catalog no. 295-75003, Wako Pure Chemical Industries) were cotransfected with 3.5 kbp-Luc and either empty or pFLAG-NFAT5 vectors after 12 h of knockdown. Twenty-four hours after transfection, cells were treated with 75 mm NaCl or 150 mm sucrose in DMEM/F-12 for 24 h. Cells were washed twice with PBS and subsequently lysed with Passive Lysis Buffer (catalog no. 194A, Promega). Luciferase activities were measured with the Dual-Luciferase Reporter Assay System (catalog no. 1960, Promega) and a GloMax Multi+ Detection System (Promega), according to the manufacturer's protocol. Relative activity was defined as the ratio between the activities of firefly luciferase and Renilla luciferase. The vector pGL4.10[luc2] without the promoter (pGL4-Luc) was used as a negative control.
ChIP assay
Two 10-cm plates of ∼80% confluent mIMCD-3 cells were cultured for 24 h and treated with 75 mm NaCl for 4 h prior to cross-linking. Cells were treated with 1% (v/v) formaldehyde for 15 min at 37 °C, followed by an additional 5 min in 150 mm glycine. Subsequently, cells were washed with cold PBS and lysed in nuclear lysis buffer (50 mm Tris-HCl (pH 8.0), 10 mm EDTA, 1% SDS, and protease inhibitor mixture) for 10 min on ice. Nuclear lysates were sonicated to a final average fragment length of 1,000 bp using a Bioruptor II (BM Equipment, Tokyo, Japan). Sonicated chromatin was cleared by centrifugation at 13,000 rpm for 10 min and then diluted 10-fold with dilution buffer (16.7 mm Tris-HCl (pH 8.0), 1.2 mm EDTA, 0.01% SDS, 1.1% Triton X-100, 167 mm NaCl, and protease inhibitor mixture). The soluble chromatin was incubated with 2 μg of normal IgG rabbit (catalog no. 2729, Cell Signaling Technology) or anti-NFAT5 antibody (catalog no. ab3446, Abcam) on a rotating platform at 4 °C overnight, followed by incubation with 20 μl of Magna ChIP Protein A + G magnetic beads (catalog no. 16-663, Millipore) for 1 h. Subsequently, the beads were washed with the following four buffers: low-salt buffer (20 mm Tris-HCl (pH 8.0), 2 mm EDTA, 0.1% SDS, 1% Triton X-100, 150 mm NaCl), high-salt buffer (same as low-salt buffer with 500 mm NaCl), LiCl buffer (10 mm Tris-HCl (pH 8.0), 1 mm EDTA, 0.25 m LiCl, 1% Nonidet P-40, 1% sodium deoxycholate), and TE buffer (10 mm Tris-HCl (pH 8.0), 1 mm EDTA). Precipitated complexes were eluted twice from the beads in fresh elution buffer (1% SDS, 0.1 m NaHCO3) for 30 min each at room temperature and subsequently heated at 65 °C overnight to reverse formaldehyde-induced cross-linking. The DNA was purified by phenol–chloroform extraction and ethanol precipitation. The purified DNA was amplified by PCR in a total volume of 20 μl containing a 0.45 μm concentration of each primer, 0.4 mm dNTPs, 1 unit of KOD FX DNA polymerase, and 2× PCR buffer (catalog no. KFX-101, Toyobo). The following primer set was used, yielding a 303-bp product: 5′-GTTTTACCCTTTTGCCTGTTTCCTT-3′ (forward) and 5′-CAGGCTAAGAAGTCCTTGGTAAACA-3′ (reverse). The PCR conditions were as follows: 94 °C for 5 min, 33 cycles at 94 °C for 15 s, 60 °C for 30 s, 68 °C for 20 s, and 68 °C for 3 min. The PCR products were resolved by electrophoresis on a 4.8% acrylamide gel. The density of each band was quantified using the Adobe Photoshop Elements version 2.0 program (Adobe Systems Inc.).
Cell viability assay
mIMCD-3 cells were transfected with RNF183 or NC siRNA in 6-well plates (12.5 pmol/well). After 24 h, the cells were reseeded in a 24-well plate and cultured in 5% CO2 at 37 °C for an additional 24 h. Subsequently, cells were treated with isotonic or 200 mm NaCl–supplemented medium in 5% CO2 at 37 °C for 12 h and washed with PBS and then stained with crystal violet. The wells were washed with water and air-dried, and the dye was eluted with water-containing 0.5% SDS. The absorbance was measured at 590 nm using a Bio-Rad SmartSpec 3000 spectrophotometer, according to the method reported by Omura et al. (56).
Electroporation
For RNF183 rescue experiments using electroporation, mIMCD-3 cells were transfected with RNF183 or NC siRNA in 10-cm dishes (62.5 pmol/dish). After 12 h, the cells were trypsinized and resuspended in DMEM/F-12. They were then pelleted at 1,000 rpm for 5 min and resuspended in Opti-MEM at a working concentration of ∼1.0 × 107 cells/ml. One hundred microliters of cell suspension was added to 10 μg of empty or 3×FLAG-RNF183-pcDNA vectors in electroporation cuvettes (catalog no. SE-202, BEX Co., Ltd., Tokyo, Japan) and then transfected by one direct-current 150-V electrical pulse and five direct-current 20-V electrical pulses at intervals of 50 ms using an electroporator (CUY 21 Vitro-EX, BEX Co.). After 48 h, cells were treated with 200 mm NaCl in DMEM/F-12 for the indicated time and analyzed using Western blotting or a crystal violet assay.
Statistical analysis
Results are expressed as the mean ± S.D. Statistical evaluation was performed using JMP statistical software (version Pro 12). Comparisons between two groups were analyzed using the two-tailed t test. For multiple-group comparisons, one-way analysis of variance (ANOVA), followed by two-tailed Student's t test with Bonferroni correction were applied. Significant p values are as follows: *, p < 0.05; **, p < 0.01; and ***, p < 0.001. Nonsignificant p values are shown as N.S. (p > 0.05).
Author contributions
M. K. and Y. M. conceptualized the study; Y. M. performed formal analysis; M. K. and K. I. acquired funding; Y. M. performed the investigation; S. K., T. O., A. S., R. A., and K. M. designed the methodology; K. I. oversaw project administration; T. M. supervised the study; Y. M., Y. W., and X. G. performed data validation; Y. M. performed visualization of cells and tissues; Y. M. and M. K. wrote the original draft; and K. I., T. O., and S. K. reviewed and edited the manuscript.
Supplementary Material
Acknowledgments
We appreciate the advice and expertise of Isao Naguro, Shigehiro Doi, Ayumu Nakashima, Toshiki Doi, Shuma Hirashio, and Kensuke Sasaki. We sincerely appreciate Keiji Tanimoto for handling equipment for the hypoxia experiment. Sincere appreciation is extended to Ryoji Kojima for supplying mIMCD-3 cells. Gratitude is expressed to Dr. Takashi Ito and Dr. B. C. Ko for the generous donation of the expression vector pFLAG-NFAT5 and pFLAG-DN-NFAT5. We are grateful to Takeshi Ike and Yui Tanita for assistance. Part of this work was performed at the Analysis Center of Life Science, Natural Science Center for Basic Research and Development, Hiroshima University. We thank Enago for providing an English language review.
This study was supported by Grants-in-Aid for Scientific Research (KAKENHI) 17H06416, 15K21706, and 26460099 from the Ministry of Education, Culture, Sports, Science, and Technology, Japan and also supported by the Takeda Science Foundation. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1 and Figs. S1–S6.
- NFAT5
- nuclear factor of activated T cells 5
- RNF
- RING finger protein
- AR
- aldose reductase
- BGT1
- sodium- and chloride-dependent betaine transporter
- SMIT
- sodium myo-inositol co-transporter
- HSP70
- heat shock protein 70
- IBD
- inflammatory bowel disease
- HEK293
- human embryonic kidney 293
- qRT-PCR
- quantitative real-time RT-PCR
- mIMCD-3
- mouse inner-medullary collecting duct
- NRK
- normal rat kidney
- NC
- negative control
- ECR
- evolutionarily conserved region
- miR-7
- microRNA-7
- VEGFA
- vascular endothelial growth factor A
- HIF-1
- hypoxia-inducible factor 1
- FATE1
- fetal and adult testis expressed 1
- BIK
- Bcl-2–interacting killer
- DMEM
- Dulbecco's modified Eagle's medium
- DN
- dominant negative
- GST
- glutathione S-transferase
- MBP
- maltose-binding protein
- DAPI
- 4′,6-diamidino-2-phenylindole
- ANOVA
- analysis of variance
- MAPK
- mitogen-activated protein kinase.
References
- 1. Bankir L., Bouby N., and Trinh-Trang-Tan M. M. (1989) The role of the kidney in the maintenance of water balance. Baillieres Clin. Endocrinol. Metab. 3, 249–311 10.1016/S0950-351X(89)80005-9 [DOI] [PubMed] [Google Scholar]
- 2. Jamison R. L., and Kriz W. (1982) Urinary Concentrating Mechanism: Structure and Function, Oxford University Press, Oxford, UK [Google Scholar]
- 3. Rim J. S., Atta M. G., Dahl S. C., Berry G. T., Handler J. S., and Kwon H. M. (1998) Transcription of the sodium/myo-inositol cotransporter gene is regulated by multiple tonicity-responsive enhancers spread over 50 kilobase pairs in the 5′-flanking region. J. Biol. Chem. 273, 20615–20621 10.1074/jbc.273.32.20615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Miyakawa H., Woo S. K., Dahl S. C., Handler J. S., and Kwon H. M. (1999) Tonicity-responsive enhancer binding protein, a rel-like protein that stimulates transcription in response to hypertonicity. Proc. Natl. Acad. Sci. U.S.A. 96, 2538–2542 10.1073/pnas.96.5.2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ko B. C., Ruepp B., Bohren K. M., Gabbay K. H., and Chung S. S. (1997) Identification and characterization of multiple osmotic response sequences in the human aldose reductase gene. J. Biol. Chem. 272, 16431–16437 10.1074/jbc.272.26.16431 [DOI] [PubMed] [Google Scholar]
- 6. López-Rodríguez C., Antos C. L., Shelton J. M., Richardson J. A., Lin F., Novobrantseva T. I., Bronson R. T., Igarashi P., Rao A., and Olson E. N. (2004) Loss of NFAT5 results in renal atrophy and lack of tonicity-responsive gene expression. Proc. Natl. Acad. Sci. U.S.A. 101, 2392–2397 10.1073/pnas.0308703100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Woo S. K., Lee S. D., Na K. Y., Park W. K., and Kwon H. M. (2002) TonEBP/NFAT5 stimulates transcription of HSP70 in response to hypertonicity. Mol. Cell. Biol. 22, 5753–5760 10.1128/MCB.22.16.5753-5760.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Heo J. I., Lee M. S., Kim J. H., Lee J. S., Kim J., Park J. B., Lee J. Y., Han J. A., and Kim J. I. (2006) The role of tonicity responsive enhancer sites in the transcriptional regulation of human hsp70–2 in response to hypertonic stress. Exp. Mol. Med. 38, 295–301 10.1038/emm.2006.35 [DOI] [PubMed] [Google Scholar]
- 9. Ito T., Fujio Y., Hirata M., Takatani T., Matsuda T., Muraoka S., Takahashi K., and Azuma J. (2004) Expression of taurine transporter is regulated through the TonE (tonicity-responsive element)/TonEBP (TonE-binding protein) pathway and contributes to cytoprotection in HepG2 cells. Biochem. J. 382, 177–182 10.1042/BJ20031838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. López-Rodríguez C., Aramburu J., Jin L., Rakeman A. S., Michino M., and Rao A. (2001) Bridging the NFAT and NF-κB families: NFAT5 dimerization regulates cytokine gene transcription in response to osmotic stress. Immunity 15, 47–58 10.1016/S1074-7613(01)00165-0 [DOI] [PubMed] [Google Scholar]
- 11. Buxadé M., Lunazzi G., Minguillón J., Iborra S., Berga-Bolaños R., Del Val M., Aramburu J., and López-Rodríguez C. (2012) Gene expression induced by Toll-like receptors in macrophages requires the transcription factor NFAT5. J. Exp. Med. 209, 379–393 10.1084/jem.20111569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen S., Grigsby C. L., Law C. S., Ni X., Nekrep N., Olsen K., Humphreys M. H., and Gardner D. G. (2009) Tonicity-dependent induction of Sgk1 expression has a potential role in dehydration-induced natriuresis in rodents. J. Clin. Invest. 119, 1647–1658 10.1172/JCI35314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kleinewietfeld M., Manzel A., Titze J., Kvakan H., Yosef N., Linker R. A., Muller D. N., and Hafler D. A. (2013) Sodium chloride drives autoimmune disease by the induction of pathogenic TH17 cells. Nature 496, 518–522 10.1038/nature11868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ito T., Asakura K., Tougou K., Fukuda T., Kubota R., Nonen S., Fujio Y., and Azuma J. (2007) Regulation of cytochrome P450 2E1 under hypertonic environment through TonEBP in human hepatocytes. Mol. Pharmacol. 72, 173–181 10.1124/mol.106.033480 [DOI] [PubMed] [Google Scholar]
- 15. Machnik A., Neuhofer W., Jantsch J., Dahlmann A., Tammela T., Machura K., Park J. K., Beck F. X., Müller D. N., Derer W., Goss J., Ziomber A., Dietsch P., Wagner H., van Rooijen N., Kurtz A., Hilgers K. F., Alitalo K., Eckardt K. U., Luft F. C., Kerjaschki D., and Titze J. (2009) Macrophages regulate salt-dependent volume and blood pressure by a vascular endothelial growth factor-C-dependent buffering mechanism. Nat. Med. 15, 545–552 10.1038/nm.1960 [DOI] [PubMed] [Google Scholar]
- 16. Hershko A., and Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 10.1146/annurev.biochem.67.1.425 [DOI] [PubMed] [Google Scholar]
- 17. Grabbe C., Husnjak K., and Dikic I. (2011) The spatial and temporal organization of ubiquitin networks. Nat. Rev. Mol. Cell Biol. 12, 295–307 10.1038/nrm3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nakamura N. (2011) The role of the transmembrane RING finger proteins in cellular and organelle function. Membranes (Basel) 1, 354–393 10.3390/membranes1040354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu Q., Zhang S., Chao K., Feng R., Wang H., Li M., Chen B., He Y., Zeng Z., and Chen M. (2016) E3 ubiquitin ligase RNF183 is a novel regulator in inflammatory bowel disease. J. Crohns Colitis 10, 713–725 10.1093/ecco-jcc/jjw023 [DOI] [PubMed] [Google Scholar]
- 20. Geng R., Tan X., Wu J., Pan Z., Yi M., Shi W., Liu R., Yao C., Wang G., Lin J., Qiu L., Huang W., and Chen S. (2017) RNF183 promotes proliferation and metastasis of colorectal cancer cells via activation of NF-κB-IL-8 axis. Cell Death Dis. 8, e2994 10.1038/cddis.2017.400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaneko M., Iwase I., Yamasaki Y., Takai T., Wu Y., Kanemoto S., Matsuhisa K., Asada R., Okuma Y., Watanabe T., Imaizumi K., and Nomura Y. (2016) Genome-wide identification and gene expression profiling of ubiquitin ligases for endoplasmic reticulum protein degradation. Sci. Rep. 6, 30955 10.1038/srep30955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Woo S. K., Lee S. D., and Kwon H. M. (2002) TonEBP transcriptional activator in the cellular response to increased osmolality. Pflugers Arch. 444, 579–585 10.1007/s00424-002-0849-2 [DOI] [PubMed] [Google Scholar]
- 23. Ko B. C., Turck C. W., Lee K. W., Yang Y., and Chung S. S. (2000) Purification, identification, and characterization of an osmotic response element binding protein. Biochem. Biophys. Res. Commun. 270, 52–61 10.1006/bbrc.2000.2376 [DOI] [PubMed] [Google Scholar]
- 24. Dahl S. C., Handler J. S., and Kwon H. M. (2001) Hypertonicity-induced phosphorylation and nuclear localization of the transcription factor TonEBP. Am. J. Physiol. Cell. Physiol. 280, C248–C253 10.1152/ajpcell.2001.280.2.C248 [DOI] [PubMed] [Google Scholar]
- 25. Shapiro L., and Dinarello C. A. (1995) Osmotic regulation of cytokine synthesis in vitro. Proc. Natl. Acad. Sci. U.S.A. 92, 12230–12234 10.1073/pnas.92.26.12230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jantsch J., Schatz V., Friedrich D., Schröder A., Kopp C., Siegert I., Maronna A., Wendelborn D., Linz P., Binger K. J., Gebhardt M., Heinig M., Neubert P., Fischer F., Teufel S., et al. (2015) Cutaneous Na+ storage strengthens the antimicrobial barrier function of the skin and boosts macrophage-driven host defense. Cell Metab. 21, 493–501 10.1016/j.cmet.2015.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ovcharenko I., Nobrega M. A., Loots G. G., and Stubbs L. (2004) ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 32, W280–W286 10.1093/nar/gkh355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bryne J. C., Valen E., Tang M. H., Marstrand T., Winther O., da Piedade I., Krogh A., Lenhard B., and Sandelin A. (2008) JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 36, D102–D106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Santos B. C., Chevaile A., Hébert M. J., Zagajeski J., and Gullans S. R. (1998) A combination of NaCl and urea enhances survival of IMCD cells to hyperosmolality. Am. J. Physiol. 274, F1167–F1173 [DOI] [PubMed] [Google Scholar]
- 30. Michea L., Ferguson D. R., Peters E. M., Andrews P. M., Kirby M. R., and Burg M. B. (2000) Cell cycle delay and apoptosis are induced by high salt and urea in renal medullary cells. Am. J. Physiol. Renal Physiol. 278, F209–F218 10.1152/ajprenal.2000.278.2.F209 [DOI] [PubMed] [Google Scholar]
- 31. Dmitrieva N., Kultz D., Michea L., Ferraris J., and Burg M. (2000) Protection of renal inner medullary epithelial cells from apoptosis by hypertonic stress-induced p53 activation. J. Biol. Chem. 275, 18243–18247 10.1074/jbc.M000522200 [DOI] [PubMed] [Google Scholar]
- 32. Alfieri R. R., Cavazzoni A., Petronini P. G., Bonelli M. A., Caccamo A. E., Borghetti A. F., and Wheeler K. P. (2002) Compatible osmolytes modulate the response of porcine endothelial cells to hypertonicity and protect them from apoptosis. J. Physiol. 540, 499–508 10.1113/jphysiol.2001.013395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shim E. H., Kim J. I., Bang E. S., Heo J. S., Lee J. S., Kim E. Y., Lee J. E., Park W. Y., Kim S. H., Kim H. S., Smithies O., Jang J. J., Jin D. I., and Seo J. S. (2002) Targeted disruption of hsp70.1 sensitizes to osmotic stress. EMBO Rep. 3, 857–861 10.1093/embo-reports/kvf175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Choudhury N. R., de Lima Alves F., de Andrés-Aguayo L., Graf T., Cáceres J. F., Rappsilber J., and Michlewski G. (2013) Tissue-specific control of brain-enriched miR-7 biogenesis. Genes Dev. 27, 24–38 10.1101/gad.199190.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Berga-Bolaños R., Drews-Elger K., Aramburu J., and López-Rodríguez C. (2010) NFAT5 regulates T lymphocyte homeostasis and CD24-dependent T cell expansion under pathologic hypernatremia. J. Immunol. 185, 6624–6635 10.4049/jimmunol.1001232 [DOI] [PubMed] [Google Scholar]
- 36. Berga-Bolaños R., Alberdi M., Buxadé M., Aramburu J., and López-Rodríguez C. (2013) NFAT5 induction by the pre-T-cell receptor serves as a selective survival signal in T-lymphocyte development. Proc. Natl. Acad. Sci. U.S.A. 110, 16091–16096 10.1073/pnas.1215934110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cha J. H., Woo S. K., Han K. H., Kim Y. H., Handler J. S., Kim J., and Kwon H. M. (2001) Hydration status affects nuclear distribution of transcription factor tonicity responsive enhancer binding protein in rat kidney. J. Am. Soc. Nephrol. 12, 2221–2230 [DOI] [PubMed] [Google Scholar]
- 38. Sheen M. R., Kim J. A., Lim S. W., Jung J. Y., Han K. H., Jeon U. S., Park S. H., Kim J., and Kwon H. M. (2009) Interstitial tonicity controls TonEBP expression in the renal medulla. Kidney Int. 75, 518–525 10.1038/ki.2008.601 [DOI] [PubMed] [Google Scholar]
- 39. Hasler U., Jeon U. S., Kim J. A., Mordasini D., Kwon H. M., Féraille E., and Martin P. Y. (2006) Tonicity-responsive enhancer binding protein is an essential regulator of aquaporin-2 expression in renal collecting duct principal cells. J. Am. Soc. Nephrol. 17, 1521–1531 10.1681/ASN.2005121317 [DOI] [PubMed] [Google Scholar]
- 40. Na K. Y., Woo S. K., Lee S. D., and Kwon H. M. (2003) Silencing of TonEBP/NFAT5 transcriptional activator by RNA interference. J. Am. Soc. Nephrol. 14, 283–288 10.1097/01.ASN.0000045050.19544.B2 [DOI] [PubMed] [Google Scholar]
- 41. Lanaspa M. A., Andres-Hernando A., Li N., Rivard C. J., Cicerchi C., Roncal-Jimenez C., Schrier R. W., and Berl T. (2010) The expression of aquaporin-1 in the medulla of the kidney is dependent on the transcription factor associated with hypertonicity, TonEBP. J. Biol. Chem. 285, 31694–31703 10.1074/jbc.M109.093690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brezis M., and Rosen S. (1995) Hypoxia of the renal medulla—its implications for disease. N. Engl. J. Med. 332, 647–655 10.1056/NEJM199503093321006 [DOI] [PubMed] [Google Scholar]
- 43. Forsythe J. A., Jiang B. H., Iyer N. V., Agani F., Leung S. W., Koos R. D., and Semenza G. L. (1996) Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol. Cell. Biol. 16, 4604–4613 10.1128/MCB.16.9.4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lee S. D., Choi S. Y., Lim S. W., Lamitina S. T., Ho S. N., Go W. Y., and Kwon H. M. (2011) TonEBP stimulates multiple cellular pathways for adaptation to hypertonic stress: organic osmolyte-dependent and -independent pathways. Am. J. Physiol. Renal Physiol. 300, F707–F715 10.1152/ajprenal.00227.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Maxfield K. E., Taus P. J., Corcoran K., Wooten J., Macion J., Zhou Y., Borromeo M., Kollipara R. K., Yan J., Xie Y., Xie X. J., and Whitehurst A. W. (2015) Comprehensive functional characterization of cancer-testis antigens defines obligate participation in multiple hallmarks of cancer. Nat. Commun. 6, 8840 10.1038/ncomms9840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hasler U., Leroy V., Jeon U. S., Bouley R., Dimitrov M., Kim J. A., Brown D., Kwon H. M., Martin P. Y., and Féraille E. (2008) NF-κB modulates aquaporin-2 transcription in renal collecting duct principal cells. J. Biol. Chem. 283, 28095–28105 10.1074/jbc.M708350200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Roth I., Leroy V., Kwon H. M., Martin P. Y., Féraille E., and Hasler U. (2010) Osmoprotective transcription factor NFAT5/TonEBP modulates nuclear factor-κB activity. Mol. Biol. Cell 21, 3459–3474 10.1091/mbc.e10-02-0133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cheung C. Y., and Ko B. C. (2013) NFAT5 in cellular adaptation to hypertonic stress—regulations and functional significance. J. Mol. Signal. 8, 5 10.1186/1750-2187-8-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lang F., Busch G. L., Ritter M., Völkl H., Waldegger S., Gulbins E., and Häussinger D. (1998) Functional significance of cell volume regulatory mechanisms. Physiol. Rev. 78, 247–306 10.1152/physrev.1998.78.1.247 [DOI] [PubMed] [Google Scholar]
- 50. Petronini P. G., Alfieri R. R., Losio M. N., Caccamo A. E., Cavazzoni A., Bonelli M. A., Borghetti A. F., and Wheeler K. P. (2000) Induction of BGT-1 and amino acid system A transport activities in endothelial cells exposed to hyperosmolarity. Am. J. Physiol. Regul. Integr. Comp. Physiol. 279, R1580–R1589 10.1152/ajpregu.2000.279.5.R1580 [DOI] [PubMed] [Google Scholar]
- 51. Hicke L. (2001) A new ticket for entry into budding vesicles-ubiquitin. Cell 106, 527–530 10.1016/S0092-8674(01)00485-8 [DOI] [PubMed] [Google Scholar]
- 52. Hong J. H., Kaustov L., Coyaud E., Srikumar T., Wan J., Arrowsmith C., and Raught B. (2015) KCMF1 (potassium channel modulatory factor 1) links RAD6 to UBR4 (ubiquitin N-recognin domain-containing E3 ligase 4) and lysosome-mediated degradation. Mol. Cell. Proteomics 14, 674–685 10.1074/mcp.M114.042168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Holleman J., and Marchese A. (2014) The ubiquitin ligase deltex-3l regulates endosomal sorting of the G protein-coupled receptor CXCR4. Mol. Biol. Cell 25, 1892–1904 10.1091/mbc.e13-10-0612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wu Y., Guo X. P., Kanemoto S., Maeoka Y., Saito A., Asada R., Matsuhisa K., Ohtake Y., Imaizumi K., and Kaneko M. (2018) Sec16A, a key protein in COPII vesicle formation, regulates the stability and localization of the novel ubiquitin ligase RNF183. PLoS One 13, e0190407 10.1371/journal.pone.0190407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bell L. M., Leong M. L., Kim B., Wang E., Park J., Hemmings B. A., and Firestone G. L. (2000) Hyperosmotic stress stimulates promoter activity and regulates cellular utilization of the serum- and glucocorticoid-inducible protein kinase (Sgk) by a p38 MAPK-dependent pathway. J. Biol. Chem. 275, 25262–25272 10.1074/jbc.M002076200 [DOI] [PubMed] [Google Scholar]
- 56. Omura T., Kaneko M., Okuma Y., Orba Y., Nagashima K., Takahashi R., Fujitani N., Matsumura S., Hata A., Kubota K., Murahashi K., Uehara T., and Nomura Y. (2006) A ubiquitin ligase HRD1 promotes the degradation of Pael receptor, a substrate of Parkin. J. Neurochem. 99, 1456–1469 10.1111/j.1471-4159.2006.04155.x [DOI] [PubMed] [Google Scholar]
- 57. Ovcharenko I., Nobrega M. A., Loots G. G., and Stubbs L. (2004) ECR Browser: A tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 32, W280–W286 10.1093/nar/gkh355 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.