SUMMARY
The mitochondrion is an organelle originating from an endosymbiotic event and playing a role in several fundamental processes such as energy production, metabolite syntheses, and programmed cell death. This organelle is delineated by two membranes whose synthesis requires an extensive exchange of phospholipids with other cellular organelles such as endoplasmic reticulum (ER) and vacuolar membranes in yeast. These transfers of phospholipids are thought to occur by a non-vesicular pathway at contact sites between two closely apposed membranes. In plants, little is known about the biogenesis of mitochondrial membranes. Contact sites between ER and mitochondria are suspected to play a similar role in phospholipid trafficking as in yeast, but this has never been demonstrated. In contrast, it has been shown that plastids are able to transfer lipids to mitochondria during phosphate starvation. However, the proteins involved in such transfer are still unknown. Here, we identified in Arabidopsis thaliana a large lipid-enriched complex called the mitochondrial transmembrane lipoprotein (MTL) complex. The MTL complex contains proteins located in the two mitochondrial membranes and conserved in all eukaryotic cells, such as the TOM complex and AtMic60, a component of the MICOS complex. We demonstrate that AtMic60 contributes to the export of phosphatidylethanolamine from mitochondria and the import of galactoglycerolipids from plastids during phosphate starvation. Furthermore, AtMic60 promotes lipid desorption from membranes, likely as an initial step for lipid transfer, and binds to Tom40, suggesting that AtMic60 could regulate the tethering between the inner and outer membranes of mitochondria.
Graphical Abstract
In Brief
Mitochondria membrane biogenesis requires exchanges of lipids within mitochondria and with other organelles. Michaud et al. identify a mitochondrial transmembrane lipoprotein complex containing Mic60 in plants. They show that Mic60 plays a key role in mitochondria lipid trafficking, probably by acting on membrane destabilization and contact site.
INTRODUCTION
In higher plants, mitochondria and plastids derive from two independent successive endosymbiotic events. These semi-autonomous organelles share many similarities in the way they were integrated within the host cell. Both organelles are delineated by two membranes, the outer (OM) and the inner (IM) membranes, but they have a completely different lipid profile: plastid membranes are mainly constituted of non-phosphorus galactoglycerolipids whereas mitochondria membranes mostly contain phospholipids. Whereas the biogenesis of plastid membranes has been extensively studied [1], little is known about the biogenesis of mitochondrial membranes in plants. Plastids play a fundamental role in lipid biogenesis in plants because they are responsible for the de novo fatty acids synthesis [2]. These fatty acids are incorporated into plastid glycerolipids to build photosynthetic membranes and also exported outside the plastids to fulfill organelle membrane biogenesis [2]. In all eukaryotes, the synthesis of most mitochondrial phospholipids occurs in the endoplasmic reticulum (ER) and these highly hydrophobic molecules are then imported into mitochondria [3]. Some phospholipids, such as cardiolipin (CL) and phosphatidylethanolamine (PE), can be synthesized directly in the IM of mitochondria, and significant amount of PE is exported outside of this organelle to be part of extra-mitochondrial membranes or metabolized [3]. Thus, a massive exchange of phospholipids continuously occurs between mitochondria and other organelles. In yeast and mammalian cells, this phospholipid exchange is thought to occur at contact sites between mitochondria and the ER or vacuolar membranes by non-vesicular pathways [3–7]. In plants, ER-mitochondrial junctions are suspected to play a similar role in mitochondrial membrane biogenesis, but this has never been demonstrated. In contrast, it has been shown that plastids are able to transfer lipids to mitochondria independently of the endomembrane network during phosphate (Pi) starvation [8]. At the whole-cell level, an extensive remodeling of membrane lipids is observed in this situation [9, 10]. This remodeling is characterized by a degradation of phospholipids, mainly phosphatidylcholine (PC) and PE, releasing Pi, which is necessary for other fundamental cellular processes. Phospholipids can be then recycled into digalactosyldiacylglycerol (DGDG), a galactoglycerolipid synthesized in the plastid envelope and present mainly in these membranes under standard growth conditions. DGDG from plastids is then exported toward the plasma membrane, the tonoplast, and mitochondrial membranes [8, 11], suggesting that, at least under Pi starvation, plastids can be involved in the biogenesis of mitochondrial membranes. However, the mechanism of DGDG transfer to extraplastidial membranes is still unknown. It was shown that the number of contact sites between plastids and mitochondria increases during Pi starvation and that isolated mitochondria can uptake DGDG from associated plastid envelope, suggesting that the lipid transfer operates at contact sites between organelles [8]. Furthermore, after entering the mitochondria, DGDG molecules have to be distributed between the two mitochondrial membranes whereas phospholipids have to be exported from this organelle for degradation. This huge lipid remodeling, required for plant adaptation to Pi starvation, involves extensive lipid exchanges between mitochondrial membranes mediated by protein factors that are still unknown.
Here, we took advantages of this specific lipid remodeling activated by Pi deprivation in Arabidopsis thaliana to point proteins involved in lipid trafficking in plant mitochondria. We identified a large lipoprotein complex containing DGDG and crossing the two mitochondrial membranes that we called the mitochondrial transmembrane lipoprotein (MTL) complex. This complex contains the TOM complex and Mic60, a conserved component of the MICOS complex. We demonstrated in Arabidopsis that AtMic60 is involved in mitochondrial lipid trafficking, attributing a new function to this protein. We propose a model in which AtMic60 could be involved in the regulation of lipid trafficking in mitochondria by promoting the tethering of the OM with the IM of mitochondria via Tom40 and by promoting the destabilization of membranes, likely as an early step for lipid transfer.
RESULTS
Identification and Characterization of a Mitochondrial Complex Enriched in DGDG
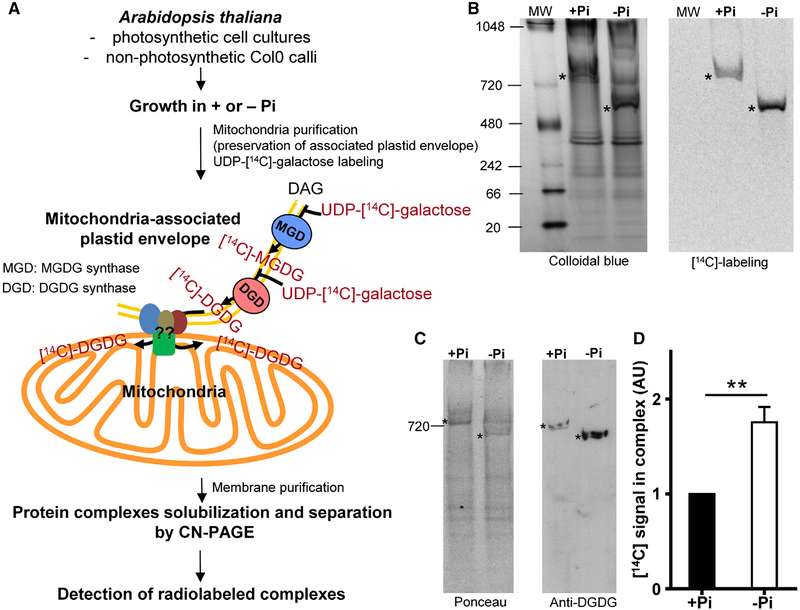
We used clear native PAGE (CN-PAGE) [12, 13] to identify protein complex(es) retaining [14C]-DGDG after in vitro lipid uptake performed on mitochondria isolated from A. thaliana cells grown in a medium with (+Pi) or without (−Pi) Pi (Figure 1A) [8]. In this strategy, mitochondria were purified on Percoll cushion, a condition which preserves the mitochondria-associated plastid envelope, and were incubated with UDP-[14C]-galactose. Galactose is incorporated into galactoglycerolipids by enzymes present in the plastid envelope, and [14C]-DGDG is then transferred to mitochondria. Mitochondrial complexes were solubilized with β-dodecylmaltoside (DDM) and separated by CN-PAGE. In calli grown 4 days in presence or absence of Pi, a single radioactive band was visible after electrophoresis (Figure 1B). To confirm this result, we performed the same experiment starting from mitochondria extracted from photosynthetic Arabidopsis cell cultures after 2 days of starvation and using two different concentrations of detergent (1.5 and 10 μg of DDM per μg of proteins) to solubilized complexes (Figure S1A). In this model, we also observed a single radioactive band that shifts to a lower molecular weight when DDM concentration is increased (Figure S1A). We analyzed total mitochondrial proteins and lipids present in the complex to demonstrate that the detected radiolabeling corresponded to galactoglycerolipids and that UDP-[14C]-galactose was not incorporated into proteins (Figures S1B and S1C). The presence of DGDG in the complex was also confirmed by western blots performed on CN-PAGE with anti-DGDG antibodies [14] (Figures 1C and S1D). Our results showed that this complex did not contain only proteins but was also enriched in lipids, at least DGDG. We named this lipid-enriched complex the MTL complex. Interestingly, we observed an increase in the amount of DGDG in this complex during Pi starvation (Figures 1B–1D, S1A, S1D, and S1E). In addition, a shift of the MTL complex size occurred in calli after 4 days of starvation whereas this shift was not observed in cell cultures grown only 2 days without phosphate (Figures 1B and S1A). These results showed that a remodeling of the lipids present in the MTL complex occurred during Pi starvation and that the migration pattern of this complex was dependent of the length of starvation and of the concentration of detergent used, suggesting that the migration of the MTL is probably dependent on its lipid environment.
Figure 1. Identification of a Mitochondrial Complex Enriched in DGDG during Pi Deprivation.
(A) Strategy used to identify a DGDG-enriched mitochondrial complex. We used the in vitro radiolabeled DGDG transfer assay designed in [8]. To identify a relevant complex, analyses were done with two different plant cell cultures. After a first purification, crude mitochondria with associated plastid envelope membranes are incubated with UDP-[14C]-galactose for 30 min at 22°C. UDP-[14C]-galactose is used as a substrate by the MGD and DGD enzymes present in plastid envelope co-purifying with mitochondria to synthesize radiolabeled galactoglycerolipids, which are then transferred into mitochondria. Then, mitochondrial membranes are retrieved. Protein complexes are solubilized with the non-ionic detergent β-dodecylmaltoside (DDM) and separated on clear native PAGE (CN-PAGE), which preserves the lipid environment of complexes. Gels are then colored, dried, and exposed to phosphorimager plate to detect radiolabeled complexes. DAG, diacylglycerol; MGDG, monogalactosyldiacylglycerol.
(B) Identification of one complex radiolabeled (*) with DGDG in mitochondria isolated from calli grown 4 days in presence (+Pi) or absence (−Pi) of Pi. Complexes were solubilized with 1.5 μg of DDM per mg of proteins and separated on CN-PAGE. MW, molecular weight (KDa).
(C) Western blot with antibodies raised against DGDG on CN-PAGE showing the presence of DGDG in the complex.
(D) Quantification of the radioactive signals obtained in (B). Signals were normalized to +Pi signal.
Data represent the mean value ± SEM; n = 3. See also Figure S1.
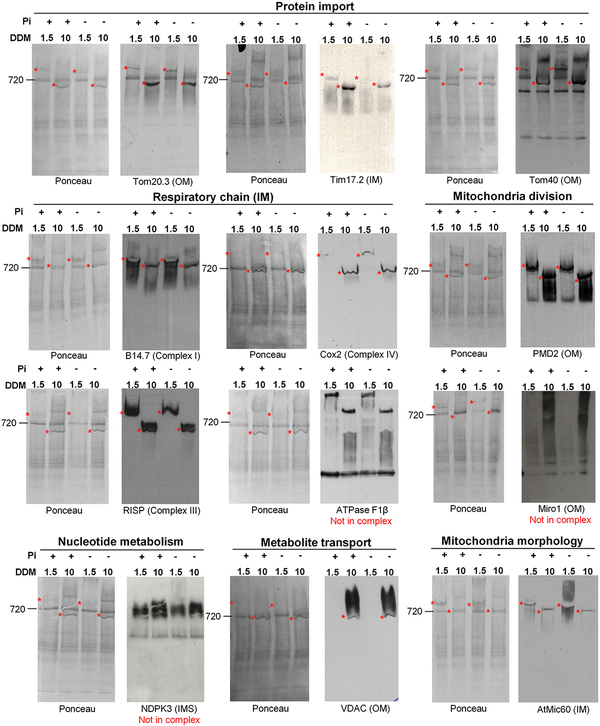
We next performed a proteomic analysis to determine the protein content of the MTL complex. To segregate proteins of the MTL complex from contaminants, proteomic analysis was carried out on three biological repeats of two different cell culture models (photosynthetic cells and calli) grown in +Pi and in −Pi and using two different concentrations of detergent for photosynthetic cells. A protein was considered to be part of the MTL complex if it was present in all the conditions analyzed (i.e., calli, cell cultures DDM 1.5, cell cultures DDM 10, and + and −Pi; see Supplemental Experimental Procedures and Table S1). With this strategy, we identified about 200 proteins, most of them located in mitochondria in either the OM or IM consistent with a location of the MTL complex at contact sites between both membranes (Table S1). The number of identified proteins was not consistent with the size of the complex determined on CN-PAGE (around 800 KDa), further supporting the idea that this complex was enriched in lipids and that its migration not only depends on its protein content but also on its lipid environment. Different mitochondrial functions were represented in this complex such as respiration, protein import, mitochondria division, and morphology or lipid metabolism. Using specific antibodies raised against putative components of the MTL complex, we confirmed the presence of several proteins identified by mass spectrometry in cell cultures using two different concentrations of DDM (Figure 2). However, some proteins, like Miro1, NDPK3, or the ATPase F1β, were found in the proteomic analysis, but we considered that they were not components of the MTL complex because they showed a smeary migration pattern that overlapped the MTL complex.
Figure 2. Confirmation by Western Blot of the Presence in the MTL Complex of Proteins Identified by Mass Spectrometry in Photosynthetic Cell Cultures Grown Two Days in +Pi or −Pi Condition.
20 μg of mitochondrial proteins were solubilized with 1.5 or 10 μg of β-DDM per mg of proteins and separated on CN-PAGE. The presence of a positive signal in the complex is indicated by an asterisk. A protein was considered as a component of the MTL complex if it was present in the MTL complex solubilized with 1.5 and 10 μg of DDM per μg of proteins. Some proteins (ATPase F1β, Miro1, and NDPK3) were detected by mass spectrometry because they migrate as a smear at the same level as the MTL complex on CN-PAGE, but they do not belong to it. The general function of the proteins analyzed and their mitochondrial localization are indicated. IM, inner membrane; IMS, intermembrane space; OM, outer membrane.
The TOM Complex and the Core Components of the MICOS Complex Are Part of the MTL Complex
Of particular interest, we detected in the MTL complex nearly all the subunits of the translocase of the outer membrane (TOM) complex and a putative homolog of the Mic60 protein (At4g39690; Table S1; Figure 2). The TOM complex plays a fundamental role in protein import into mitochondria [15]. It is composed of the conserved import channel Tom40, the receptors for precursor proteins, like Tom20, and three small proteins required for the assembly of the TOM complex [15]. Recently, it was reported that this complex interacts with the ER membrane complex (EMC) to facilitate lipid transfer from ER to mitochondria in yeast [7]. Therefore, the TOM complex could act as a possible gate for lipid trafficking at the OM. In yeast, Mic60 belongs to the well-characterized MICOS (mitochondria contact site cristae organizing system) complex [16]. The MICOS complex is involved in different functions such as mitochondria morphology, cristae organization, and protein import [17, 18]. MICOS is localized in the IM at contact sites between both mitochondrial membranes, and Mic60 has been shown to interact directly with several proteins of the OM like Tom40 [19, 20]. A role of MICOS in lipid trafficking has never been demonstrated until now, but its localization at the contact sites between the OM and IM makes Mic60 a candidate for lipid trafficking in mitochondria.
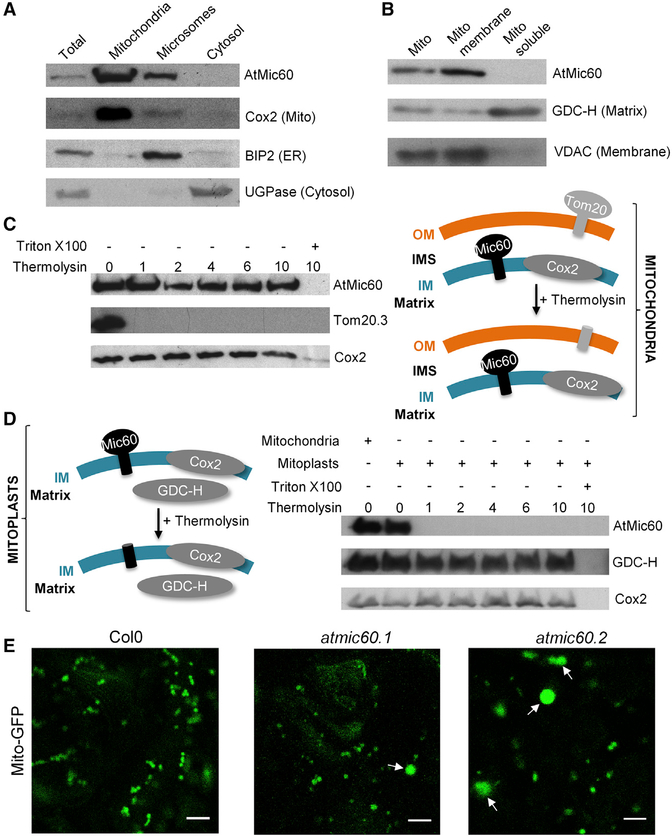
In plants, the MICOS complex has never been characterized. Recently, a phylogenetic analysis of the MICOS complex in eukaryotes suggested that only the core component Mic60 and Mic10 proteins were conserved in plants [21]. By sequence analyses, we also identified in A. thaliana genome only two putative homologs out of the six yeast MICOS subunits, corresponding to Mic60 and Mic10 (Figure S2A), which are both present in the MTL complex (Table S1). Despite a weak sequence similarity, At-Mic60 presents a conserved domain organization compared to yeast or human Mic60 with a predicted N-terminal mitochondrial targeting sequence (MTS) followed by a small transmembrane domain and a Mitofilin signature (Interpro IPR019133; Figures S2B and S2C). In yeast and mammals, Mic60 is anchored in the IM with its soluble C-terminal domain located in the intermembrane space (IMS) [22, 23]. To analyze the localization of AtMic60 in Arabidopsis, we performed western blot analyses on A. thaliana subcellular fractions and showed that AtMic60 is localized in mitochondria (Figure 3A) with an enrichment in the membrane fraction of this organelle (Figure 3B). Thermolysin treatments were performed on isolated mitochondria or mitoplasts (mitochondria with ruptured OM) to monitor the intra-mitochondrial localization of the soluble domain of AtMic60 (Figures 3C and3D). AtMic60 was insensitive to protease treatments achieved on isolated mitochondria, showing that the protein was not exposed at the surface of this organelle. However, AtMic60 was degraded when protease treatment was performed on mitoplasts (Figure 3D). These results showed that AtMic60 was also localized in the IM of mitochondria with its soluble domain protruding in the IMS.
Figure 3. AtMic60 Is Localized in the Inner Membrane of Mitochondria with Its Soluble Domain Protruding in the IMS and Plays a Role in Mitochondria Morphology.
(A) Col0 callus grown in +Pi were subfractioned into mitochondria (see Experimental Procedures), microsomes (pellet fraction after centrifugation at 100,000 g of the supernatant obtained after the pellet of mitochondria at 16,000 g), and cytosol (supernatant fraction after centrifugation at 100,000 g of the supernatant obtained after the pellet of mitochondria at 16,000 g). Twenty micrograms of proteins from each fraction were used for western blots against marker proteins of each compartment.
(B) Mitochondria were ruptured and subfractioned into the membrane and soluble fraction. Twenty micrograms of proteins from each fraction were used for western blots against marker proteins of each fraction.
(C and D) Thermolysin treatments of purified mitochondria (C) or mitoplasts (mitochondria with ruptured OM; D) from Col0 callus grown in +Pi. One hundred micrograms of mitochondria or mitoplasts were incubated with 0–10 μg of thermolysin. Pellets were resuspended in 50 μl of SDS-PAGE buffer, and 10 μl was used for western blot analyses. Triton was used to completely disrupt membranes and to show that proteins are sensitive to protease when accessible.
(E) Col0 and the two atmic60 KO lines (Figures S3A and S3B) were stably transformed with a GFP-targeted mitochondrial construction [24]. Observations were done in leaves of 10-day-old seedlings grown in vitro. The scale bar represents 5 μm. Arrows indicated bigger mitochondria observed in atmic60 KO lines.
See also Figure S2.
To investigate the functional homology between yeast and plant Mic60 proteins, we isolated two Arabidopsis knockout (KO) lines for this gene, atmic60.1 and atmic60.2 (Figures S3A and S3B) and analyzed their mitochondria morphology by confocal microscopy (Figure 3E). We observed an alteration of the mitochondria morphology in these two lines characterized by the presence of several bigger mitochondria with a round shape coexisting with mitochondria of normal shape and size. The phenotype of these big mitochondria is similar to the one obtained in yeast Δmic60 strains [23], demonstrating a role of At-Mic60 in the maintenance of mitochondria morphology. Taken together, our results showed that AtMic60 (At4g39690) is the homolog of the yeast and mammals Mic60 proteins.
AtMic60 and Tom40 Interact Together and Increase in the MTL Complex during Pi Starvation
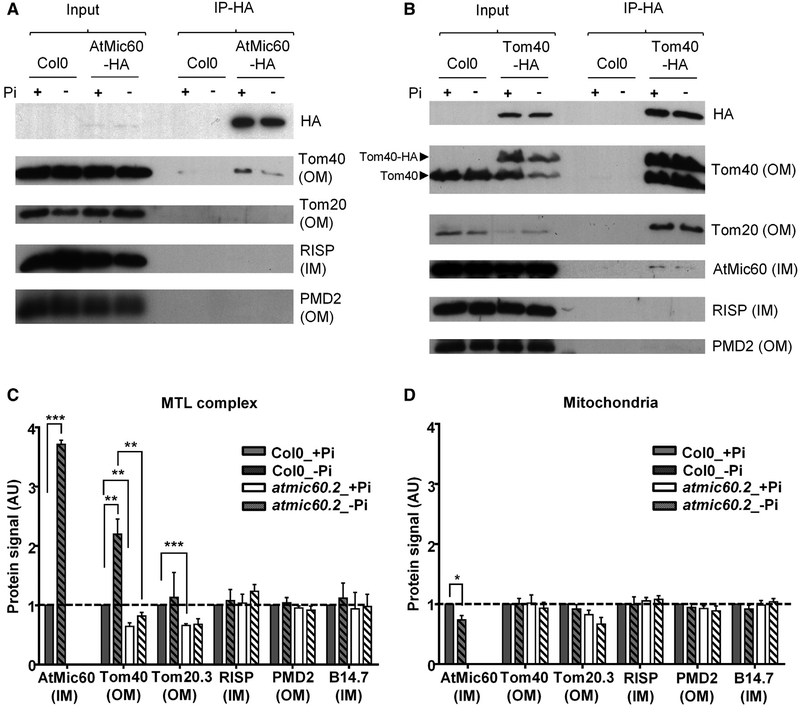
In yeast, Mic60 establishes contacts between the two mitochondrial membranes by interacting with several proteins of the OM such as Tom40 [19]. To check whether AtMic60 was also able to interact with Tom40 in Arabidopsis, we performed co-immunoprecipitation (coIP) experiments on a complemented atmic60.2 KO line stably transformed with a C-terminal HA-tagged version of AtMic60 (Figure S3C). The results obtained showed that Tom40 was co-immunoprecipitated with AtMic60-HA (Figure 4A). To confirm this interaction, coIPs were done on a Col0 line stably transformed with a C-terminal HA-tagged version of Tom40 (Figure S3D). AtMic60 co-eluted with Tom40-HA, confirming the interaction between these two proteins (Figure 4B). Thus, plant AtMic60 was also able to interact with Tom40, supporting its localization at contact sites between mitochondrial membranes. We also noticed that RISP and PMD2, two other proteins identified in the MTL complex, did not interact with AtMic60 or Tom40 (Figures 4A and 4B). These results suggested a modular organization of the MTL complex where not all the subunits are able to interact together.
Figure 4. AtMic60 and Tom40 Interact Together and Are More Abundant in the MTL Complex during Pi Starvation.
(A and B) Co-immunoprecipitation (coIP) experiments with anti-HA antibodies on mitochondria purified from calli grown 4 days in +Pi or in −Pi. CoIPs were done with 500 μg (A) or 300 μg (B) of proteins, and 3 μg of proteins was used for input.
(A) CoIPs with Col0 (control) and atmic60.2+AtMic60-HA mitochondria (Figure S3C) are shown.
(B) CoIPs with Col0 (control) and Col0+Tom40-HA mitochondria (Figure S3D) are shown.
(C and D) Quantification of the protein signals obtained in western blots in Figure S4 in the MTL complex (C) and in the whole mitochondria (D) of Col0 and atmic60.2 calli grown 4 days in + or in −Pi. The quantified signals were normalized with Col0 +Pi signal. Data represent the mean value ± SEM; n = 3.
See also Figures S3 and S4.
Remarkably, in −Pi condition, AtMic60 and Tom40 contents increased in the MTL complex whereas they remained constant or slightly decreased in whole mitochondria isolated from calli (Figures 4C, 4D, and S4; compare Col0_+Pi and Col0_−Pi). Because no variations were observed for other MTL proteins in the complex and in the mitochondria (Figures 4C, 4D, and S4; compare Col0_+Pi and Col0_−Pi), these data further suggest a role of AtMic60 and Tom40 in lipid remodeling during Pi starvation. To investigate the specific link between AtMic60 and Tom40, we analyzed the content of several proteins of the MTL complex in the atmic60.2 KO line. We observed a decrease of Tom40 in the complex of the mutant in both + and −Pi whereas its total level in the whole mitochondria did not present significant changes (Figures 4C, 4D, and S4; compare Col0 and atmic60.2). Some other MTL complex components were not affected by the absence of AtMic60 (Figures 4C, 4D, and S4; compare Col0 and atmic60.2). These results highlighted a correlation between the content of AtMic60 and Tom40 in the MTL complex. They also suggested that AtMic60 could regulate the localization of Tom40 into the MTL complex and in this way could impact the contact between the two mitochondrial membranes inside this complex.
AtMic60 Is Involved in Mitochondria Lipid Trafficking
To monitor the level of DGDG in the MTL complex of atmic60.2 during Pi starvation, we performed western blot analyses with anti-DGDG antibodies [14] on CN-PAGE (Figure 5A). In −Pi condition, the amount of DGDG was reduced in the MTL complex of atmic60.2 calli compared to Col0 and to the complemented atmic60.2+AtMic60-HA line, suggesting a perturbation of the lipid remodeling occurring in the complex during Pi starvation (Figure 5A). To confirm this hypothesis, we analyzed the lipid composition of the MTL complex of Col0 and atmic60.2 by mass spectrometry. We extracted lipids from three different bands cut on CN-PAGE: one band corresponding to the MTL complex and two other bands corresponding to complexes migrating at different levels on CN-PAGE (Figure S5A). We detected three lipid classes—i.e., PC and PE, the most abundant mitochondria lipids, and DGDG—solely in the MTL complex, supporting the high lipid content of this complex (Figure S5B). We next analyzed the proportion of these three glycerolipids in the MTL complex of Col0 and atmic60.2 calli grown in + or −Pi condition. The results presented in Figure 5B confirmed that the accumulation of DGDG during Pi starvation in the MTL complex was compromised in atmic60.2. Furthermore, in comparison with the +Pi condition, PE content of the MTL complex increased in the mutant in −Pi whereas it decreased in Col0 (Figure 5B). In contrast, no significant difference of PC content was detected in the MTL complex between Col0 and atmic60.2 (Figure 5B). During Pi starvation, PE and PC have to be exported from mitochondria for exchange with DGDG [8]. Thus, PE accumulation in the atmic60.2 MTL complex during Pi starvation suggested that, in absence of AtMic60, PE was blocked in this complex, leading to a defect in PE export and DGDG import. The presence of PC in the MTL complex indicated that PC trafficking might go through this complex as well but independently of AtMic60.
Figure 5. Lipid Composition of the MTL Complex and of Mitochondria Is Altered in atmic60.2.
(A) Western blots with anti-DGDG antibodies performed on CN-PAGE with 20 μg of mitochondrial membrane proteins of Col0 (c), atmic60.2 (m), or atmic60.2+AtMic60-HA (mC) calli grown 4 days in + or −Pi.
(B) Proportion of DGDG, PE, and PC present in the MTL complex of Col0 and atmic60.2 calli grown 4 days in + or −Pi determined by mass spectrometry. Data represent the mean value ± SEM; n = 3.
(C) Lipid analysis by 2D-TLC of mitochondria extracted from Col0, atmic60.2, and atmic60.2+AtMic60-HA calli grown 8 days in + or −Pi. Data represent the mean value ± SEM. MGDG, monogalactosyldiacylglycerol; PA, phosphatidic acid; PG, phosphatidylglycerol; PI, phosphatidylinositol; other abbreviations are used as in the text.
(D) Kinetic of incorporation of UDP-[14C]-galactose into galactoglycerolipids performed with total cellular extract of Col0 and atmic60.2 calli grown 4 days in + or −Pi. Data represent the mean value ± SEM; n = 3.
See also Figures S3 and S5.
To verify this hypothesis and determine whether the lipid differences observed in the atmic60.2 MTL complex impacted the entire mitochondria lipid composition, we performed lipid analysis of isolated mitochondria. In +Pi, we confirmed a reduction of the DGDG content and an accumulation of PE in atmic60.2 mitochondria compared to Col0 and atmic60.2+ AtMic60-HA, and these differences were amplified in −Pi (Figure 5C). In contrast, no significant difference was detected concerning PC content, supporting the idea that PC trafficking is not affected in atmic60.2 mitochondria.
We measured the efficiency of galactoglycerolipids synthesis in Col0 and atmic60.2 total cell extracts to verify that the decrease of DGDG observed in atmic60.2 mitochondria was not linked to a defect in galactoglycerolipid synthesis. As shown in Figure 5D, we observed an increase of the incorporation of UDP-[14C]-galactose in both Col0 and atmic60.2 calli in −Pi, but no significant differences were observed between these two lines, indicating a similar rate of synthesis of galactoglycerolipids in Col0 and atmic60.2. Thus, the accumulation of PE in mitochondria in +Pi and −Pi conditions suggested that the MTL complex via AtMic60 controls PE trafficking in mitochondria and particularly the export of PE stimulated by Pi starvation, directly impacting the level of DGDG imported from plastids.
AtMic60 Binds Lipids and Destabilizes Liposomes
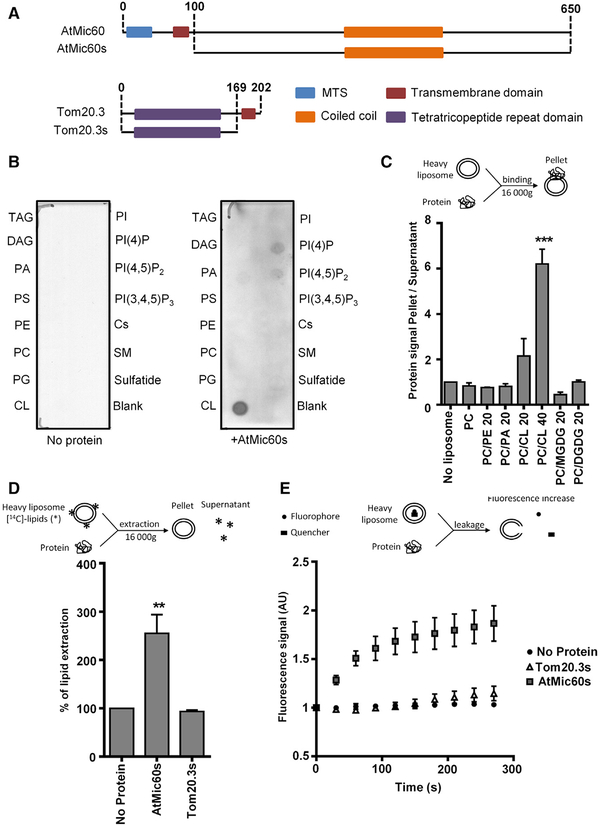
To analyze the role of AtMic60 in lipid trafficking, we investigated the ability of AtMic60 to bind lipids in vitro. To facilitate the overexpression and purification of AtMic60, we used a version of AtMic60 containing only its soluble IMS domain and deleted from its MTS and transmembrane domain and we called it AtMic60s (Figure 6A). Commercially available strips spotted with lipids were incubated with or without AtMic60s, and the binding of the protein to lipids was revealed by immunolabeling using anti-AtMic60 antibodies. As shown in Figure 6B, the addition of AtMic60s to the lipid strips led to the detection of a positive signal with CL. A weak signal was also observed for phosphatidylinositol-4-phosphate (PI(4)P) and phosphatidylinositol-4,5-diphosphate (PI(4,5)P2) (Figure 6B). These results showed that AtMic60 was able to bind glycerolipids with a preference for CL. To confirm these results and to study the ability of AtMic60 to bind galactoglycerolipids, we performed in vitro liposome-binding assays (Figure 6C) [25]. Liposomes are small vesicles composed of a lipid bilayer mimicking membranes. After incubation with the protein, liposomes were pelleted and the binding of the protein was monitored by the intensity of the protein signal in the pellet analyzed on SDS-PAGE. In our assay, no binding of AtMic60s was measured with liposomes containing only PC (Figure 6C). Addition of an increasing proportion of CL led to an increased binding of AtMic60s to liposomes (Figure 6C). No significant binding of AtMic60s could be detected with liposomes containing the MGDG or DGDG galactoglycerolipids (Figure 6C). Taken together, our results clearly demonstrated that AtMic60 was able to bind membranes enriched in CL.
Figure 6. AtMic60 Is Able to Bind Cardiolipin and to Destabilize Liposomes.
(A) Schematic representation of the soluble part of AtMic60 (AtMic60s) or Tom20.3 (Tom20.3 s) overexpressed and purified in E. coli to performed in vitro assays.
(B) Membrane Lipid Strips (Echellon Biosciences) were incubated without or with purified AtMic60s protein at 1 μg/ml. The binding of the protein to lipid spots was revealed by western blot using anti-AtMic60 antibodies. Cs, cholesterol; DAG, diacylglycerol; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PI(3,4,5)P3, phosphatidylinositol (3,4,5) triphosphate; PI(4)P, phosphatidylinositol (4) monophosphate; PI(4,5)P2, phosphatidylinositol (4,5) diphosphate; PS, phosphatidylserine; SM, sphingomyelin; TAG, triacylglycerol.
(C) In vitro liposome-binding assays of AtMic60s performed with liposomes composed of PC and the indicated percentage of other lipids.
(D) In vitro liposome extraction assays with AtMic60s or Tom20.3 s purified proteins. Liposomes are composed of 70% PC and 30% of [14C]-acetate radiolabeled glycerolipids from Arabidopsis cells. The percentage of lipid extraction corresponds to the percentage of the radioactivity present in the supernatant after incubation and centrifugation of liposomes.
(E) Liposome leakage assays with AtMic60s or Tom20.3 s proteins. Liposomes are composed of 70% PC and 30% of total glycerolipids from Arabidopsis cells, 12.5 mM of the fluorophore 8-aminonaphthalene-1,3,6-trisulfonic acid (ANTS), and 45 mM of the quencher p-xylene-bis-pridinium bromide (DPX). Fluorescence signals are normalized by the background signal at t = 0 s.
Data represent the mean value ± SEM; (C and E) n = 3; (D) n = 5 (no protein; AtMic60s), n = 3 (Tom20.3 s). See also Figure S6.
To investigate whether AtMic60s could extract lipids from membranes, we performed in vitro liposomes extraction assays [26]. In this assay, total lipids extracted from A. thaliana cell cultures labeled with [14C]-acetate were incorporated in liposomes and the lipid extraction activity of proteins was monitored by the radioactive signal present in the supernatant after removal of liposomes by high-speed centrifugation (Figure 6D). The results obtained showed a significant increase of the percentage of lipids extracted from liposomes when incubation was performed with AtMic60s (Figure 6D). As a control, the same assay was performed with the purified soluble domain of Tom20.3 called Tom20.3 s (Figure 6A) and no significant lipid extraction was detected with this protein (Figure 6D). To determine whether a specific class of lipids is extracted from liposomes by AtMic60s, we purified lipids from the supernatant phase obtained after in vitro extraction assay and we analyzed the radiolabeled lipids present by 2D high-performance thin-layer chromatography (2D-HPTLC). No specific lipid classes were enriched in this fraction compared to the initial composition of liposomes (Figure S6), suggesting that, rather than extracting specific lipids, AtMic60 probably destabilized liposomes.
To confirm this hypothesis, we performed liposome leakage assays in which a fluorescent probe and a quencher were encapsulated inside the liposomes [27]. If a protein is able to disrupt membranes, the fluorescent probe and the quencher are diluted in the medium leading to an increase of the fluorescence signal (Figure 6E). As shown in Figure 6E, the addition of AtMic60s, but not of Tom20.3 s, led to a fluorescence increase in the medium, demonstrating that AtMic60 is able to disrupt liposomes. Altogether, these results indicated that AtMic60 was able to bind CL and that AtMic60 destabilized membrane bilayers.
DISCUSSION
In this study, we identify a mitochondrial complex, the MTL complex, which contains DGDG. By a proteomic approach, we identified more than 200 proteins that are involved in protein import, respiratory chain, lipid metabolism, or in mitochondria division and morphology and that are localized in the OM and IM of mitochondria. Furthermore, we detected in the MTL complex three lipids classes: PC, PE, the main lipids of mitochondria, and also DGDG in Pi-deprivation condition. The high lipid content of this complex leads to a migration pattern on CN-PAGE that is not consistent with the molecular weight of the constitutive proteins. Altogether, our results show that the MTL complex is a lipoprotein multifunctional complex that is localized at contact sites between mitochondrial membranes.
In the MTL complex, we could detect important constituents of the TOM and MICOS complexes. In plants, the TOM complex is composed of six subunits [15, 28]. Five of these subunits were found in the MTL complex by mass spectrometry, and we confirmed the presence of two of them (Tom40 and Tom20.3) by western blots, supporting the presence of the entire TOM complex in the MTL complex. The plant MICOS complex has never been characterized until now, but a recent phylogenetic analysis suggested that the core component of the MICOS complex, constituted of two subunits, was conserved in almost all eukaryotes [21]. By sequence homology, we could identify in A. thaliana homologs for these two subunits, AtMic60 and AtMic10 (At4g39690 and At1g22520), which were detected by mass spectrometry in the MTL complex and by western blot for AtMic60. AtMic60, like its yeast homolog, is localized in the IM, is required to maintain mitochondrial morphology, and interacts with Tom40, likely at mitochondrial membranes contact sites. Interestingly, the two conserved MICOS subunits in plants have been shown to be core components of this complex in yeast and to play an essential role in the assembly of the MICOS complex [21, 29, 30] whereas the other subunits were proposed to play a regulatory role in the assembly and maintenance of the MICOS complex [30]. Therefore, in plants, the MICOS complex seems to be different than the one identified in yeast and contains Mic60 and Mic10, with the other subunits being absent or replaced by uncharacterized proteins [21].
Tom40 and AtMic60 constitute good candidates for lipid trafficking in mitochondria. Tom40 belongs to the well-known TOM complex involved in protein import across the OM. In yeast, the TOM complex was also shown to facilitate phosphatidylserine (PS) trafficking from ER to mitochondria [7]. By interacting with proteins of the EMC complex, the TOM complex is involved in the tethering of the ER and the OM of mitochondria, a pre-requisite for the traffic of lipids by non-vesicular pathway [7]. Because the TOM complex directly interacts with the translocase of inner membrane (TIM) complex in the IM [31], it was postulated that PS could traffic from the ER to the IM of mitochondria by a pathway involving the EMC, TOM, and TIM complexes [7]. As Tom40 is also able to interact with the MICOS complex [19], another pathway involving TOM and MICOS complexes can also be proposed for the transfer of lipids between the OM and IM. In addition, the MICOS complex also interacts with other complexes of the OM [18, 20, 32]. Notably, it was proposed that MICOS and ERMES, an ER-mitochondria-tethering complex involved in lipid exchanges [6], could interact together to form the ERMIONE (ER-mitochondria organizing network) complex that may be involved in lipid, metabolite, and ion trafficking; protein import; and mtDNA maintenance [18]. Whereas the role of ERMES in lipid trafficking between ER and mitochondria has been demonstrated [6], the role of the MICOS complex in intra-mitochondrial lipid transfer is still poorly documented.
The ERMES complex is not conserved in plants and mammals. However, the biogenesis of their mitochondrial membranes also requires exchange of lipids with other organelles, such as ER. It is therefore tempting to speculate that the MICOS complex could be involved in mitochondria lipid trafficking by interacting with other conserved OM partners such as Tom40. In line with this, in plants, the interaction between Tom40 and AtMic60 is conserved. In addition, during Pi starvation, the amount of Tom40 and AtMic60 is increased in the MTL complex whereas its level in the whole mitochondria decreases. Because no direct evidence was found on the role of MICOS in lipid trafficking, our work focused on the mechanism by which AtMic60 could act on lipid trafficking. We investigated the possibility that AtMic60 could directly transfer lipids from one membrane to another. We showed that AtMic60 is able to destabilize membranes in vitro. AtMic60 promotes a non-specific extraction of lipids from liposomes and is able to cause the releasing of soluble molecules present inside liposomes. Even if the exact mechanism by which AtMic60 acts on membranes is still unresolved, this membrane destabilization could facilitate the extrusion of lipids from membranes to facilitate their transfer to another one. We then addressed the specificity of AtMic60 for lipids and demonstrated that the soluble domain of AtMic60 is able to bind membranes enriched in CL. CL is mainly present in the mitochondria IM and is particularly enriched in cristae junction and contact sites between both mitochondrial membranes [3, 33]. The ability of yeast or mammals Mic60 to bind membranes and CL has never been demonstrated. Our results in plants feature novel properties of this protein. The binding of AtMic60 to CL could be a way to localize the MICOS complex in a specific region of the IM where this lipid is enriched. Another MICOS complex subunit not conserved in plants, Mic27, has been also shown to bind CL [34]. The presence of CL in mitochondrial membranes has been shown to be necessary for the assembly of Mic27 into a functional MICOS complex [35]. Thus, in absence of Mic27, plant Mic60 may have acquired during evolution the ability to bind CL, thus orchestrating the assembly and correct IM localization of the MICOS complex. In yeast, Mic60 protein is also involved in the formation of cristae junctions that are parts of the IM, presenting a high degree of curvature, and in plants, atmic60 KO mutants have bigger and rounder mitochondria. Thus, in cooperation with CL, a lipid-inducing negative curvature, AtMic60 might promote the curvature of the IM necessary for the formation of cristae junction. This property could also explain the destabilization of liposomes induced by AtMic60.
According to the localization of AtMic60 at contact sites between both mitochondrial membranes and the interaction between Tom40 and Mic60, we hypothesized that, in absence of AtMic60, the OM-IM contact capacity could be reduced, leading to a perturbation of lipid trafficking inside mitochondria. In line with this, we observed that the content of Tom40 is lowered in atmic60.2 MTL complex in both + and −Pi, suggesting that the contact sites between both mitochondria membranes might be altered in the MTL complex of this mutant. Furthermore, Tom40 content increases in the MTL complex during Pi starvation in correlation with the amount of AtMic60 and DGDG. These results suggest that, inside the MTL complex, Tom40, and, at a larger level, the TOM complex could act with AtMic60 in PE and DGDG trafficking in mitochondria. As KO plants for tom40 are embryolethal [36], we failed to verify this hypothesis by a classical reverse genetic approach.
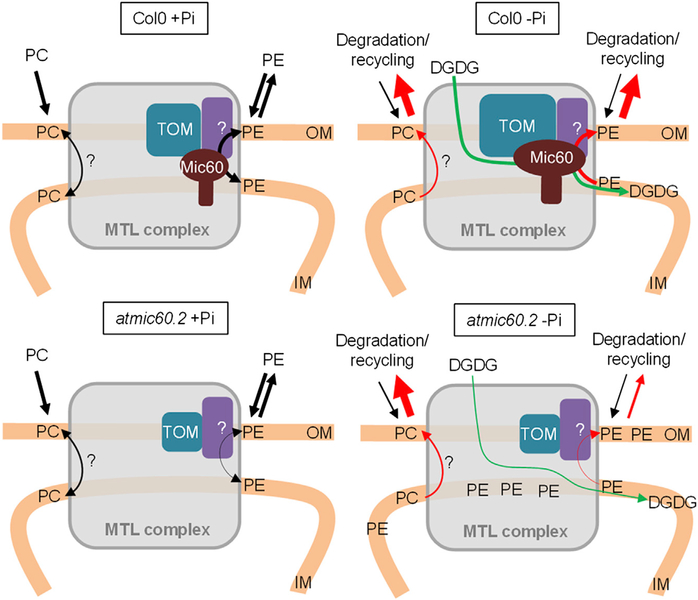
Finally, we show that, in absence of AtMic60, the lipid composition of mitochondria is altered in both + and −Pi conditions with a significant increase of PE. During Pi starvation, PE accumulation is correlated with a slowdown of DGDG import. Thus, the increased content of PE observed in the atmic60.2 MTL complex during Pi starvation is consistent with a model in which PE cannot be efficiently exported from the IM for degradation and therefore accumulates into the MTL complex (Figure 7). Thus, at the whole mitochondrial level, in the atmic60 KO line, PE is not efficiently exported outside for degradation and/or recycling into DGDG during Pi starvation and accumulates into mitochondria. Consequently, the import of DGDG is slowing down during Pi starvation (Figure 7). However, it is unclear between which membranes (i.e., plastid to OM and/or OM to IM) the DGDG traffic is reduced. In addition, our observations did not exclude the possibility that other proteins in the MTL complex could be involved in the trafficking of PC or other lipids, but the transfer of these lipids, if it occurs through the MTL complex, is independent of AtMic60.
Figure 7. Hypothetic Model for the Role of the MTL Complex and AtMic60 in PE and DGDG Trafficking in Mitochondria.
Our model proposes a role of AtMic60 in PE export and DGDG import into mitochondria with a putative role of Tom40 and the MTL complex in such trafficking. We do not rule out the possibility that the MTL complex could be involved in the traffic of other lipids (like PC) via other partners. In +Pi, in atmic60.2 mitochondria, the level of TOM complex decreases and the export of PE is reduced, leading to an accumulation of PE into mitochondria. During Pi starvation in Col0, the level of Mic60 and TOM complex increases in the MTL complex whereas the export of PE and PC for degradation and the import of DGDG from plastids are stimulated. In atmic60.2, PE accumulates into the MTL complex because its export is affected and the import of DGDG is slowed down. However, PC level is not affected in atmic60.2, showing that AtMic60 does not play a major role in PC trafficking.
Overall, our study has led to the identification of AtMic60 as a key player in the processes of PE export from mitochondria and of DGDG import from plastids, attributing a new function to this protein. As both Mic60 and PE export from mitochondria are conserved in non-plant organisms, our data open new perspectives on the possible involvement of Mic60 and the MICOS complex in mitochondrial lipid trafficking in other organisms. Our work suggests that the TOM complex could be a partner of Mic60 at the OM to mediate such trafficking. However, the ability of the MICOS complex to interact with a wide variety of OM complexes suggests that the versatile functions of this complex could be regulated according to its partners. Another remaining question is the identity of the proteins mediating the tethering and transfer of DGDG from plastids to mitochondria OM. Some proteins identified in the MTL complex are localized to plastid OM (Table S1) and could constitute good candidates to mediate such trafficking and need further investigation.
EXPERIMENTAL PROCEDURES
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Col0 plants were used as wild-type. Two T-DNA insertion lines in Col0 background (atmic60.1: SALK_007876c and atmic60.2: SALK_087650c) in ATMIC60 gene (At4g39690) were obtained from the SALK collection [37]. Growth conditions and callus obtaining are described further in Supplemental Experimental Procedures.
Mitochondria Purification, Mitoplast Preparation, and Protease Treatment
Mitochondria were purified from photosynthetic cell cultures or from calli according to the protocol described in [8] with a slight modification for the grinding of calli (30 ml of grinding buffer and 15 ml of sand for 64 g of material). Mitoplasts preparation and protease treatments were performed as described in Supplemental Experimental Procedures.
In Vitro Mitochondrial DGDG Transfer Assays
After a first purification on Percoll cushion, mitochondria were incubated 30 min at 22°C in washing buffer (0.3 M mannitol, MOPS 10 mM, 5 mM α-aminocaproic acid, and 1 mM benzamidine [pH 7.4]) containing 3 μg/ml of mitochondrial proteins, 1 mM DTT, 1 mM MgCl2, and 1 mM UDP-[14C]-galactose. For photosynthetic cells, mitochondria were centrifuged, resuspended in washing buffer, and further purified on a continuous Percoll gradient as described in [8]. For calli, after centrifugation, mitochondria were directly used for membrane isolation.
Mitochondrial Membrane Isolation, Complex Solubilization, and Separation on CN-PAGE
After purification, mitochondria were resuspended in washing buffer without mannitol (MOPS 10 mM, 5 mM α-aminocaproic acid, and 1 mM benzamidine [pH 7.4]) and vortexed. Membranes were retrieved by centrifugation and solubilized by addition of β-DDM. Samples are separated on a CN-PAGE. After migration, gels were stained by colloidal blue or transferred on nitrocellulose membrane for western blot. For a more detailed protocol, see Supplemental Experimental Procedures.
Western Blot Analyses
After migration on CN-PAGE or on denaturating SDS-PAGE, proteins were transferred on nitrocellulose membranes 1 hr at 90 V in transfer buffer (ethanol 20% [v/v], 25 mM Tris-HCl, and 0.19 M glycine [pH 8.3]). Membranes were colored with ponceau red (ponceau red 0.2% [w/v] and trichloroacetic acid 3% [v/v]) and used for western blots after decoloration. Immunodetection was performed with peroxidase-coupled antibodies. Quantification of signals was done using ImageJ software. Antibodies used in this study are described in Supplemental Experimental Procedures.
Proteomic Analysis by Mass Spectrometry
Band of the MTL complex in the CN-PAGE was cut and processed as described in Supplemental Experimental Procedures.
CoIP
CoIP experiments were done from purified mitochondrial membrane purified from Col0,atmic60.2+AtMic60-HA or Col0+Tom40-HA calli (4 days of growth in + or −Pi). Membrane was processed as described in Supplemental Experimental Procedures.
Protein Surexpression and Purification in Escherichia coli
The soluble domain of AtMic60 and Tom20.3 were cloned in the pET23d(+) and pET28a(+) (Novagen) expression vector, respectively, in frame with 6xHis in C-terminal or in N-terminal, respectively. Cloning and purification were performed as described in Supplemental Experimental Procedures.
Lipid Extractions
Lipid extractions were done differently according to the material they came from and are described in Supplemental Experimental Procedures.
Lipid Analysis by 2D-TLC
Lipid analysis by two-dimensional thin-layer chromatography (2D-TLC) was done according to [10]. For calli lipids, 400 μg were loaded on silica plates 20 × 20 cm (Merck Millipore). For mitochondrial lipids, 150 μg of lipids were loaded on high-performance TLC (HPTLC) silica plates 10 × 10 cm (Merck Millipore). After separation, each lipid spot was analyzed by quantification of the fatty acid methyl esters (FAMEs) using gas chromatography (GC) according to [10].
Lipid Analysis by Mass Spectrometry
Lipid quantifications were achieved by LC-MS/MS and are described in more detail in Supplemental Experimental Procedures.
Synthesis of Total [14C]-Labeled Lipids in A. thaliana Cell Cultures
[14C]-labeled lipids were obtained by feeding A. thaliana cell cultures with [14C]-acetate. Procedure is more detailed in Supplemental Experimental Procedures.
Galactoglycerolipid Synthase Activity
Synthesis of galactoglycerolipids was monitored by incubating total cellular extract from calli with UDP-[14C]-galactose. Procedure is more detailed in Supplemental Experimental Procedures.
Liposome Binding, Extraction, and Leakage Assays
Liposome binding, extraction, and leakage assays were adapted from [25–27]. For more details, see Supplemental Experimental Procedures.
Lipid Dot Blot Assay
Lipid dot blot assays were performed with the Membrane Lipid Strips according to the manufacturer instructions (Echelon Biosciences). Blocking and incubation with protein or antibodies were performed in TBS buffer (Tris-HCl 50 mM and NaCl 150 mM [pH 7.4]) supplemented with Tween-20 0.1% (v/v) and free fatty acid BSA (Sigma Aldrich) 3% (w/v). Proteins were incubated at a final concentration of 1 μg/ml.
Confocal Microscopy
For confocal imaging of mitochondria, Col0, atmic60.1, and atmic60.2 plants were stably transformed with a mitochondrial targeted GFP probe [24]. Observations were done with the F3 transgenic generation on leaves from 10-day-old seedlings. Seedlings were grown on MS medium supplemented with sucrose 3% (w/v) and cultivated on a 6 hr/18 hr light/dark cycle with a light intensity of 100 μmmol.m2.s−1. Images were acquired on a Leica TCS SP5 confocal laser scanning microscope with GFP excitation at 488 nm with an argon laser.
Statistical Treatment of Data
Statistical significance of data was evaluated with two-tailed unpaired t test using GraphPad software. In figures, statistical significance was represented by stars: *p < 0.05, **p < 0.01, ***p < 0.001.
Supplementary Material
Highlights.
AtMic60 is part of a mitochondrial transmembrane lipoprotein complex containing DGDG
AtMic60 is a key factor for mitochondria PE export and import of DGDG
AtMic60 could mediate mitochondria membrane contact site formation in the MTL complex
AtMic60 binds cardiolipin and destabilizes membranes
ACKNOWLEDGMENTS
This work was supported by the French National Research Agency (ANR-12-JSV2–001 Chloromitolipid; ANR-10-INBS-08 ProFI, Proteomics French Infrastructure; and ANR-10-LABEX-04 GRAL Labex, Grenoble Alliance for Integrated Structural Cell Biology) and EMBO (ASTF-638–2014). We thank Olivier Clerc and Grégoire Denay for assistance in MS and GC data processing, Anne-Marie Hesse and Thomas Burger for assistance in proteomic MS data processing, and Claude Alban for advice on protein production and purification.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, six figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.cub.2016.01.011.
REFERENCES
- 1.Boudière L, Michaud M, Petroutsos D, Rébeillé F, Falconet D, Bastien O, Roy S, Finazzi G, Rolland N, Jouhet J, et al. (2014). Glycerolipids in photosynthesis: composition, synthesis and trafficking. Biochim. Biophys. Acta 1837, 470–480. [DOI] [PubMed] [Google Scholar]
- 2.Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, DeBono A, Durrett TP, et al. (2013). Acyl-lipid metabolism. Arabidopsis Book 11, e0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horvath SE, and Daum G (2013). Lipids of mitochondria. Prog. Lipid Res. 52, 590–614. [DOI] [PubMed] [Google Scholar]
- 4.Elbaz-Alon Y, Rosenfeld-Gur E, Shinder V, Futerman AH, Geiger T, and Schuldiner M (2014). A dynamic interface between vacuoles and mitochondria in yeast. Dev. Cell 30, 95–102. [DOI] [PubMed] [Google Scholar]
- 5.Hönscher C, Mari M, Auffarth K, Bohnert M, Griffith J, Geerts W, van der Laan M, Cabrera M, Reggiori F, and Ungermann C (2014). Cellular metabolism regulates contact sites between vacuoles and mitochondria. Dev. Cell 30, 86–94. [DOI] [PubMed] [Google Scholar]
- 6.Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, and Walter P (2009). An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325, 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lahiri S, Chao JT, Tavassoli S, Wong AKO, Choudhary V, Young BP, Loewen CJR, and Prinz WA (2014). A conserved endoplasmic reticulum membrane protein complex (EMC) facilitates phospholipid transfer from the ER to mitochondria. PLoS Biol. 12, e1001969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jouhet J, Maréchal E, Baldan B, Bligny R, Joyard J, and Block MA (2004). Phosphate deprivation induces transfer of DGDG galactolipid from chloroplast to mitochondria. J. Cell Biol. 167, 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Härtel H, and Benning C (2000). Can digalactosyldiacylglycerol substitute for phosphatidylcholine upon phosphate deprivation in leaves and roots of Arabidopsis? Biochem. Soc. Trans 28, 729–732. [PubMed] [Google Scholar]
- 10.Jouhet J, Maréchal E, Bligny R, Joyard J, and Block MA (2003). Transient increase of phosphatidylcholine in plant cells in response to phosphate deprivation. FEBS Lett. 544, 63–68. [DOI] [PubMed] [Google Scholar]
- 11.Andersson MX, Larsson KE, Tjellström H, Liljenberg C, and Sandelius AS (2005). Phosphate-limited oat. The plasma membrane and the tonoplast as major targets for phospholipid-to-glycolipid replacement and stimulation of phospholipases in the plasma membrane. J. Biol. Chem 280, 27578–27586. [DOI] [PubMed] [Google Scholar]
- 12.Wittig I, and Schägger H (2008). Features and applications of blue-native and clear-native electrophoresis. Proteomics 8, 3974–3990. [DOI] [PubMed] [Google Scholar]
- 13.Wittig I, and Schägger H (2005). Advantages and limitations of clear-native PAGE. Proteomics 5, 4338–4346. [DOI] [PubMed] [Google Scholar]
- 14.Maréchal E, Azzouz N, de Macedo CS, Block MA, Feagin JE, Schwarz RT, and Joyard J (2002). Synthesis of chloroplast galactolipids in apicomplexan parasites. Eukaryot. Cell 1, 653–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michaud M, and Duchêne A-M (2012). Macromolecules trafficking to plant mitochondria In Advances in Botanical Research: Mitochondrial Genome Evolution, Maréchal-Drouard L, ed. (Elsevier; ), pp. 347–421. [Google Scholar]
- 16.Pfanner N, van der Laan M, Amati P, Capaldi RA, Caudy AA, Chacinska A, Darshi M, Deckers M, Hoppins S, Icho T, et al. (2014). Uniform nomenclature for the mitochondrial contact site and cristae organizing system. J. Cell Biol. 204, 1083–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zerbes RM, van der Klei IJ, Veenhuis M, Pfanner N, van der Laan M, and Bohnert M (2012). Mitofilin complexes: conserved organizers of mitochondrial membrane architecture. Biol. Chem 393, 1247–1261. [DOI] [PubMed] [Google Scholar]
- 18.van der Laan M, Bohnert M, Wiedemann N, and Pfanner N (2012). Role of MINOS in mitochondrial membrane architecture and biogenesis. Trends Cell Biol. 22, 185–192. [DOI] [PubMed] [Google Scholar]
- 19.von der Malsburg K, Müller JM, Bohnert M, Oeljeklaus S, Kwiatkowska P, Becker T, Loniewska-Lwowska A, Wiese S, Rao S, Milenkovic D, et al. (2011). Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Dev. Cell 21, 694–707. [DOI] [PubMed] [Google Scholar]
- 20.Xie J, Marusich MF, Souda P, Whitelegge J, and Capaldi RA (2007). The mitochondrial inner membrane protein mitofilin exists as a complex with SAM50, metaxins 1 and 2, coiled-coil-helix coiled-coil-helix domain-containing protein 3 and 6 and DnaJC11. FEBS Lett. 581, 3545–3549. [DOI] [PubMed] [Google Scholar]
- 21.Muñoz-Gómez SA, Slamovits CH, Dacks JB, Baier KA, Spencer KD, and Wideman JG (2015). Ancient homology of the mitochondrial contact site and cristae organizing system points to an endosymbiotic origin of mitochondrial cristae. Curr. Biol 25, 1489–1495. [DOI] [PubMed] [Google Scholar]
- 22.Gieffers C, Korioth F, Heimann P, Ungermann C, and Frey J (1997). Mitofilin is a transmembrane protein of the inner mitochondrial membrane expressed as two isoforms. Exp. Cell Res. 232, 395–399. [DOI] [PubMed] [Google Scholar]
- 23.Rabl R, Soubannier V, Scholz R, Vogel F, Mendl N, Vasiljev-Neumeyer A, Körner C, Jagasia R, Keil T, Baumeister W, et al. (2009). Formation of cristae and crista junctions in mitochondria depends on antagonism between Fcj1 and Su e/g. J. Cell Biol. 185, 1047–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Logan DC, and Leaver CJ (2000). Mitochondria-targeted GFP highlights the heterogeneity of mitochondrial shape, size and movement within living plant cells. J. Exp. Bot 51, 865–871. [PubMed] [Google Scholar]
- 25.Raychaudhuri S, Im YJ, Hurley JH, and Prinz WA (2006). Nonvesicular sterol movement from plasma membrane to ER requires oxysterol-binding protein-related proteins and phosphoinositides. J. Cell Biol. 173, 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schulz TA, Choi MG, Raychaudhuri S, Mears JA, Ghirlando R, Hinshaw JE, and Prinz WA (2009). Lipid-regulated sterol transfer between closely apposed membranes by oxysterol-binding protein homologues. J. Cell Biol. 187, 889–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roston R, Gao J, Xu C, and Benning C (2011). Arabidopsis chloroplast lipid transport protein TGD2 disrupts membranes and is part of a large complex. Plant J. 66, 759–769. [DOI] [PubMed] [Google Scholar]
- 28.Murcha MW, Kmiec B, Kubiszewski-Jakubiak S, Teixeira PF, Glaser E, and Whelan J (2014). Protein import into plant mitochondria: signals, machinery, processing, and regulation. J. Exp. Bot 65, 6301–6335. [DOI] [PubMed] [Google Scholar]
- 29.Harner M, Körner C, Walther D, Mokranjac D, Kaesmacher J, Welsch U, Griffith J, Mann M, Reggiori F, and Neupert W (2011). The mitochondrial contact site complex, a determinant of mitochondrial architecture. EMBO J. 30, 4356–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bohnert M, Zerbes RM, Davies KM, Mühleip AW, Rampelt H, Horvath SE, Boenke T, Kram A, Perschil I, Veenhuis M, et al. (2015). Central role of Mic10 in the mitochondrial contact site and cristae organizing system. Cell Metab. 21, 747–755. [DOI] [PubMed] [Google Scholar]
- 31.Dekker PJ, Martin F, Maarse AC, Bömer U, Müller H, Guiard B, Meijer M, Rassow J, and Pfanner N (1997). The Tim core complex defines the number of mitochondrial translocation contact sites and can hold arrested preproteins in the absence of matrix Hsp70-Tim44. EMBO J. 16, 5408–5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bohnert M, Wenz LS, Zerbes RM, Horvath SE, Stroud DA, von der Malsburg K, Müller JM, Oeljeklaus S, Perschil I, Warscheid B, et al. (2012). Role of mitochondrial inner membrane organizing system in protein biogenesis of the mitochondrial outer membrane. Mol. Biol. Cell 23, 3948–3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ardail D, Privat JP, Egret-Charlier M, Levrat C, Lerme F, and Louisot P (1990). Mitochondrial contact sites. Lipid composition and dynamics. J. Biol. Chem 265, 18797–18802. [PubMed] [Google Scholar]
- 34.Weber TA, Koob S, Heide H, Wittig I, Head B, van der Bliek A, Brandt U, Mittelbronn M, and Reichert AS (2013). APOOL is a cardiolipin-binding constituent of the Mitofilin/MINOS protein complex determining cristae morphology in mammalian mitochondria. PLoS ONE 8, e63683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friedman JR, Mourier A, Yamada J, McCaffery JM, and Nunnari J (2015). MICOS coordinates with respiratory complexes and lipids to establish mitochondrial inner membrane architecture. eLife 4, e07739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murcha MW, Wang Y, Narsai R, and Whelan J (2014). The plant mitochondrial protein import apparatus — the differences make it interesting. Biochim. Biophys. Acta 1840, 1233–1245. [DOI] [PubMed] [Google Scholar]
- 37.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.