Abstract
Diabetic retinopathy (DR) is a severe microvascular complication of diabetes and the primary cause of vision loss in diabetic patients. Previous research has revealed that long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) play pivotal roles in the pathogenesis of DR. However, the roles of lncRNA-miRNA-mRNA interactions in DR are poorly understood. In the present study, we aimed to compute a global triple network of competitive endogenous RNAs (ceRNAs) in order to pinpoint essential molecules. We found that there were 802 nodes (121 lncRNA nodes, 17 miRNA nodes, and 664 mRNA nodes) and 949 edges in the ceRNA network. Further functional analysis suggested that some molecules were specifically related to DR. Surprisingly, these molecules were involved in visual perception, eye development, and lens development in camera-type eye. In summary, our study highlighted specific lncRNAs and miRNAs related to the pathogenesis of DR, which might be used as potential diagnostic biomarkers and therapeutic targets for DR.
Keywords: diabetic retinopathy, ceRNA, lncRNA, miRNA
Introduction
Diabetic retinopathy (DR) is a severe microvascular complication of diabetes. It is the main cause of loss of vision in diabetic patients (1). Diabetic patients are known to suffer from hyperglycemia due to abnormalities in insulin signaling pathways; these abnormal levels of sugar can cause various pathogenetic processes, resulting in the following complications in diabetic patients: i) microvascular defects of the retina and ii) dysfunction and degeneration of the neuro-retina (2). The progression of DR depends upon two major types of diabetic mellitus. Research has shown that DR is the most common vision-threatening lesion in patients with type I diabetes. Diabetic macular edema seems to be more prevalent in patients with type II diabetes (3). Several studies have tried to determine the causes and progression of DR in diabetic patients (4–6); however, scientists have not yet been successful in completely elucidating the mechanisms associated with the pathogenesis of DR. Currently, diabetes mellitus cannot be prevented or treated effectively in clinical practice.
In the field of molecular biology, scientists have made major breakthroughs in understanding non-coding RNAs (ncRNAs). The prognosis of diabetic patients has improved tremendously because of various path-breaking studies conducted by researchers in the field of molecular biology (7). Recent studies have reported that long non-coding RNAs (lncRNAs) are involved in various biological processes (7), and it was found that lncRNAs are widely involved in signaling pathways, which regulate numerous aspects of life processes (7). In the context of oncology, pan-cancer molecular portraits of human tumors have been associated with essential lncRNAs (8). The activity of essential oncogenic pathways was altered with dysregulation in the quantities of lncRNAs (8). lncRNAs impact post-translation processes partly based on the competitive endogenous RNA (ceRNA) mechanism, where lncRNA interacts with microRNA (miRNA) through miRNA-binding sites (MREs) and hence regulate the expression of certain genes (9). As a result, ceRNA regulation is highly associated with several pathogenic processes.
lncRNAs and miRNAs were previously demonstrated to be associated with the pathogenesis of DR (10–12). Previous findings highlight that the development of DR is associated with various key molecules, such as miR-1273g-3p, miR-21 and miR-29a (13–15). Research has elucidated how miRNAs play a pivotal role in the diagnosis of DR (16). For example, Yan et al found that the expression of lncRNAs was aberrant in the early stages of DR. In other words, DR may develop as follows. The aberrantly expressed lncRNAs play pivotal roles in modulating target molecules, and they regulate multiple pathogenetic pathways (11). Presently, scientists have reported that disease-related lncRNAs are associated with function-related messenger RNAs (mRNAs) and miRNAs (17,18). In many other diseases except DR, ceRNA mechanism have been investigated to date; this has radically transformed the approach used to understand the pathogenesis of several diseases (19), including cardiac hypertrophy (20), rheumatoid arthritis (21) and papillary renal cell carcinoma (22). However, the role played by the lncRNA-miRNA-mRNA network in the pathogenesis of DR remains unclear. We have preliminary knowledge of the function and physiological significance of ceRNAs.
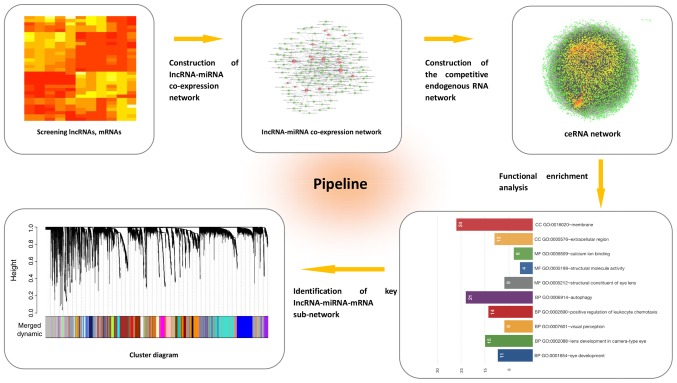
Here, based on ceRNA theory, we attempted to explore the crosstalk between lncRNA-miRNA-mRNA complexities. Key molecules were identified by performing Gene Ontology (GO) analysis and Weighted Gene Co-expression Network Analysis (WGCNA) (Fig. 1). In this study, ceRNA theory was used for the first time in DR to compute a systematic profile of the lncRNA-miRNA-mRNA network. Our results may be useful in comprehending the role played by ceRNAs in the pathogenesis of DR.
Figure 1.
The reconstruction pipeline of the lncRNA-miRNA-mRNA network. lncRNA, long non-coding RNA; miRNA, microRNA; mRNA, messenger RNA; ceRNA, competitive endogenous RNA.
Materials and methods
Data availability
Using eight week-old C57BL/6 mice, a mouse model of streptozotocin (STZ)-induced diabetes was constructed. Two months after administering STZ injection to diabetic mice, total RNAs were isolated from the retinas of the diabetic mice. The mice were considered to be diabetic when their blood glucose levels exceeded 250 mg/dl. Using TRIzol reagent, wild-type mice were matched according to age and sex. Agilent Mouse Gene Expression Microarrays (Product Number G4852A; Agilent Technologies, Santa Clara, CA, USA) were used to generate microarray data from mouse mRNA and lncRNA. The raw data were provided by Yan et al, which has been described in a previous study (11), and the ethics statement was previously provided. To generate microarray data of miRNAs from STZ-induced diabetic rats, we referred to a previous study and its supplementary material (12).
Screening of differentially expressed lncRNAs, miRNAs and mRNAs
As for the differential expression analysis, the Benjamini-Hochberg method was applied to calculate the adjusted P-value to minimize the false discovery rate (FDR) as was previously described (12). |Fold change|>2 and P<0.05 were set as cut-offs.
Prediction of target lncRNAs and mRNAs of miRNAs
The target lncRNAs of miRNAs was predicted by referring to Miranda (23). The minimum free energy (MFE) was calculated under the following condition: Max energy ≤-20 and score >160 were considered as cut-offs. The target mRNAs of miRNAs was predicted on the basis of miRTarBase (24), miRecords (25) and starBase version 2.0 (26).
Construction of the ceRNA network
The lncRNA-miRNA-mRNA network was reconstructed with ceRNA theory. The expression correlation of lncRNAs and mRNAs was determined with Pearson correlation coefficient (PCC). The mRNA-lncRNA pair was considered as the co-expressed one (P<0.05). The ceRNA network was constructed by assembling all the co-expressed competing triplets, and this network was visualized with Cytoscape software v3.5.1 (https://cytoscape.org/).
Function enrichment analysis
While performing GO analysis, these databases were used for annotation and visualization. Integration Discovery (DAVID) and Cytoscape plug-in BinGO were used to complete the GO analysis. To perform WGCNA analysis, we used R WGCNA package (https://cran.r-project.org/package=WGCNA). The cut height value was set to 0.992. We only considered modules in which the distance between two consecutive modules was <0.1. All the modules were then combined into a single module. The heat map was drawn by using pheatmap R package (https://cran.r-project.org/web/packages/pheatmap/).
Results
Screening of lncRNAs, mRNAs and miRNAs is associated specifically with DR
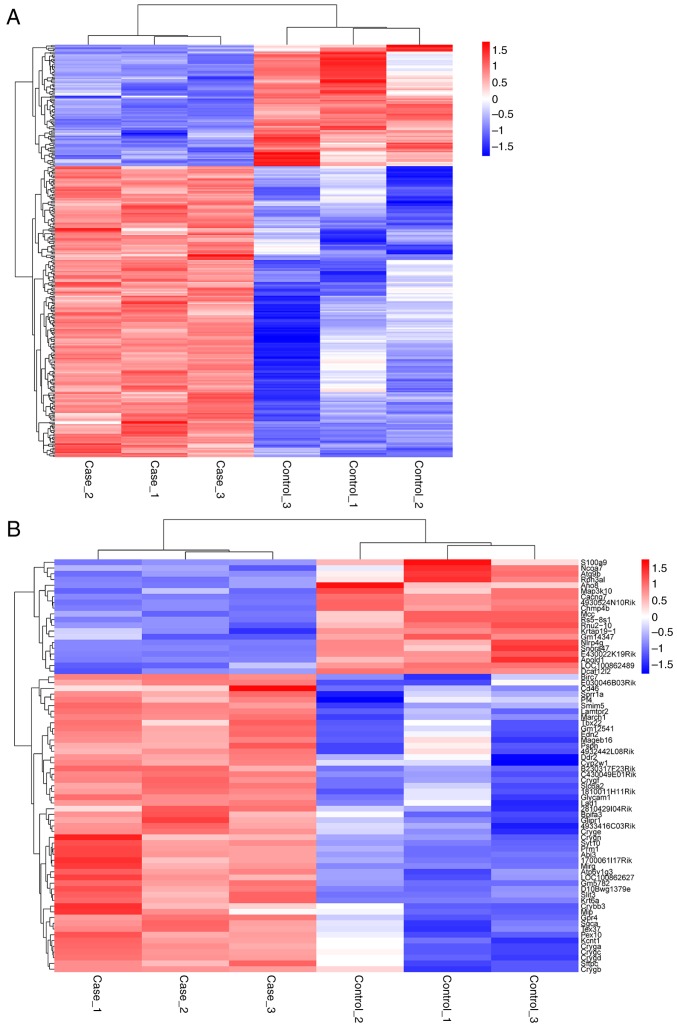
We compared expression profiles of lncRNAs, miRNAs and mRNAs, which were obtained from the retinas of different diabetic mice. To determine the key RNA molecules, differential expression analysis was performed (2). In this study, 305 differentially expressed lncRNAs and 17 differentially expressed miRNAs (diabetic vs. non-diabetic ones) were observed. Among them, 89 lncRNAs and 14 miRNAs showed upregulated expression, whereas 214 lncRNAs and 3 miRNAs showed downregulated expression in the diabetic retina samples compared with the controls (Fig. 2). The occurrence and development of DR was regulated by the differentially expressed genes.
Figure 2.
(A) Heat map of the differentially expressed lncRNAs for the DR group and control group. (B) Heat map of differentially expressed mRNAs for the DR group and control group. All the values were normalized to t values. In diabetic retina samples, 89 lncRNAs and 14 miRNAs were upregulated, whereas 214 lncRNAs and 3 miRNAs were downregulated. lncRNAs, long non-coding RNAs; DR, diabetic retinopathy; mRNAs, messenger RNAs; miRNAs, microRNAs.
Construction of lncRNA-miRNA and mRNA-miRNA co-expression network
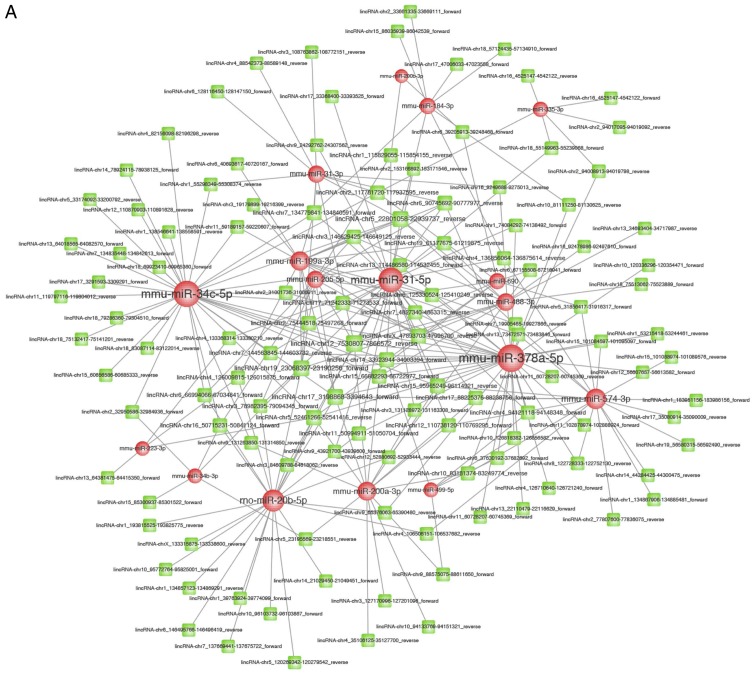
lncRNAs can competitively interact with miRNAs, and they may function as ceRNAs. With the help of miRanda, the interactions between lncRNAs and miRNAs were predicted. The interactions were predicted by referring to the minimal free energy of miRNA-lncRNA duplexes (max energy ≤-20 and score >160). In total, we predicted 246 pairs of lncRNA-miRNA; 17 miRNAs and 121 lncRNAs were involved in these 246 pairs (Fig. 3A). Using miRTarBase, miRecords and starBase version 2.0, we identified the target mRNAs of miRNAs. Thus, we identified 664 target mRNAs of 14 miRNAs (Fig. 3B).
Figure 3.
(A) The lncRNA-miRNA co-expression network. In total, 246 lncRNA-miRNA pairs were predicted; 17 miRNAs and 121 lncRNAs were involved in 246 pairs. lncRNA, long non-coding RNA; miRNA, microRNA. (B) The mRNA-miRNA co-expression network. The computations of miRTarBase, miRecords, and starBase pinpointed the target mRNAs of miRNAs; moreover, we identified 664 target mRNAs of 14 miRNAs. miRNA, microRNA; mRNA, messenger RNA.
Construction of the competitive endogenous RNA network
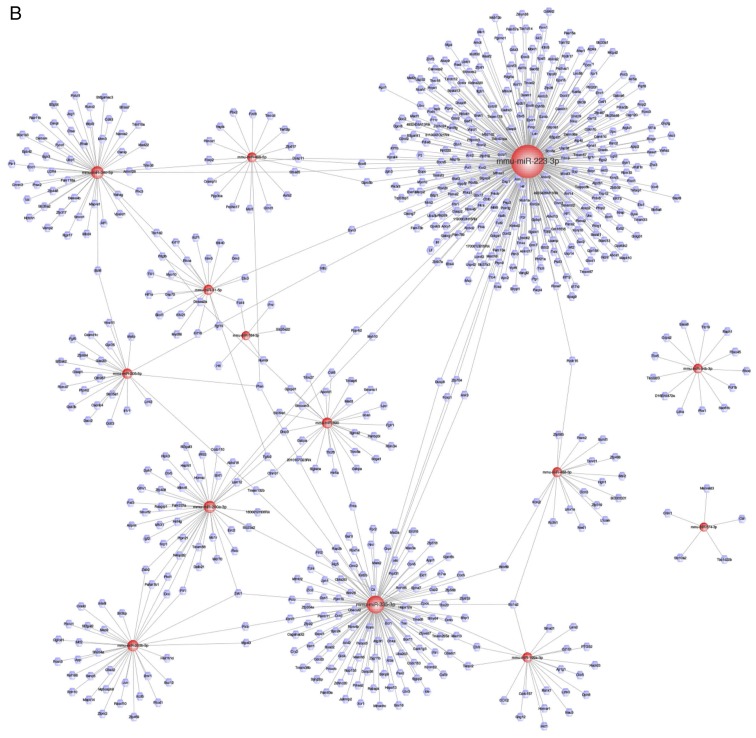
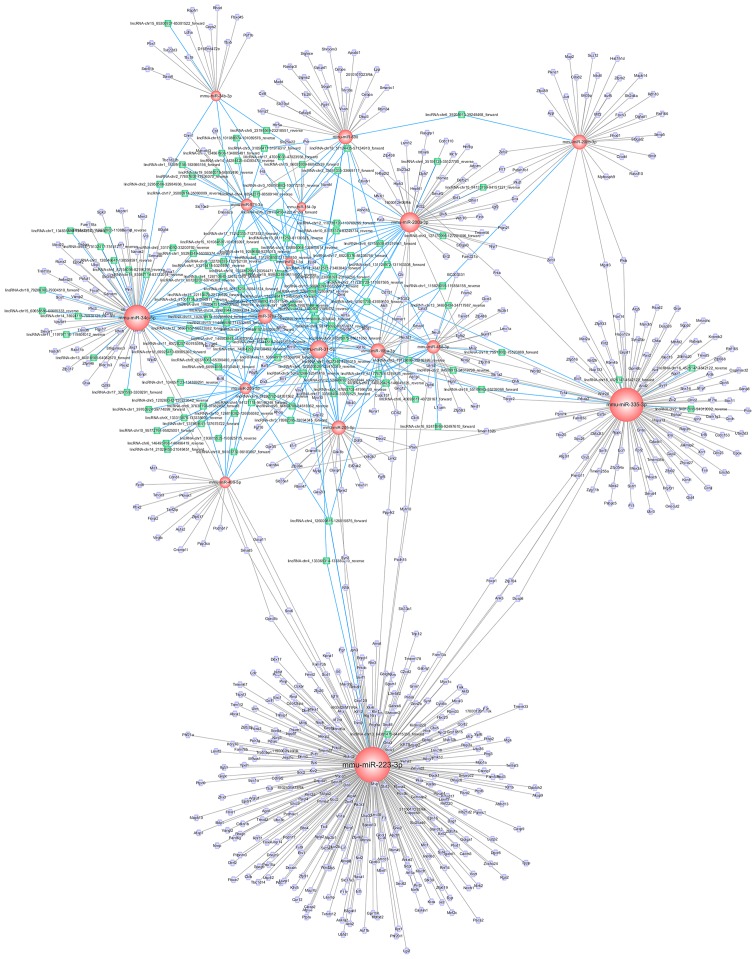
Since lncRNAs were found to be specifically associated with DR, we further investigated them with Cytoscape software. Then, the results of the analyses were used to construct the ceRNA network. Based on previous results of the mRNA-miRNA and lncRNA-miRNA co-expression network, the interactions between the lncRNA-miRNA-mRNA network were visualized (Fig. 4). In total, 802 nodes (121 lncRNA nodes, 17 miRNA nodes, and 664 mRNA nodes) and 949 edges were found in this network. The key miRNAs were as follows: miR-223-3p, miR-34c-5p, and miR-200b-3p; these miRNAs were essential hubs in the entire network. Moreover, these miRNAs potentially play an important role in the pathogenesis of DR.
Figure 4.
The ceRNA network based on the mRNA-miRNA and lncRNA-miRNA co-expression pair network by using Cytoscape. ceRNA, competitive endogenous RNA; mRNA, messenger RNA; miRNA, microRNA; lncRNA, long non-coding RNA.
Functional evaluation of the diabetic retinopathy-specific lncRNAs
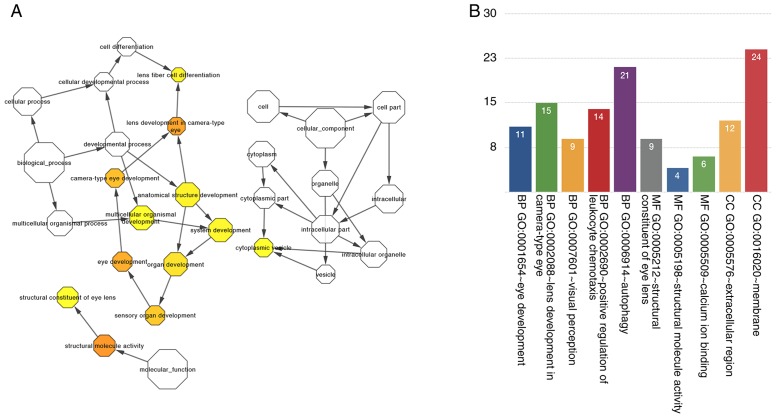
We observed that mRNAs were connected to lncRNAs in the network. To comprehend the functions of every lncRNA, the functions of connected mRNAs were assessed. Thus, we deciphered how gene products can be enriched in biological processes, cellular components, and molecular functions. Surprisingly, the results of GO analysis indicate that enriched GO terms were associated with the following events: the development of eyes and lens in camera-type eye, visual perception, structural constituents of eye lens, structural molecule activity and calcium-binding ions (Fig. 5A). We visualized gene function enrichment in the network by using BinGO (Fig. 5B).
Figure 5.
Gene Ontology (GO) function enrichment analysis of mRNAs: (A) GO terms displayed as an interaction network by using the plug-in BinGO of Cytoscape software. (B) The significant changes in the GO of lncRNA-related mRNAs. mRNAs, messenger RNAs; lncRNA, long non-coding RNA.
Key lncRNA-miRNA-mRNA sub-network based on the module analysis
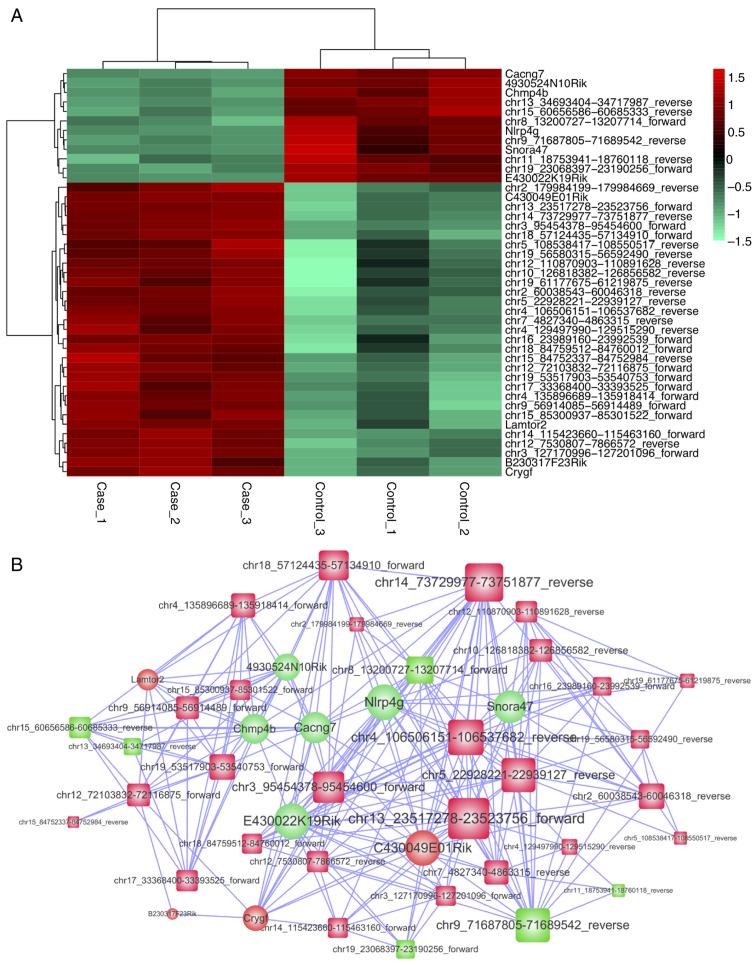
To investigate the cross-talks between mRNAs and lncRNAs, key lncRNAs were extracted; moreover, the correlation between mRNAs and miRNAs was determined by WGCNA analysis. Thus, we constructed a novel sub-network of lncRNA-miRNA-mRNA. We discovered a module that was significantly related to DR (P<0.001, R=0.980). In this module, 33 lncRNAs and 10 mRNAs were involved (Fig. 6).
Figure 6.
(A) Heat map of key genes, including 33 lncRNAs and 10 mRNAs. (B) The interaction network of key genes, including 33 lncRNAs and 10 mRNAs. lncRNAs, long non-coding RNAs; mRNAs, messenger RNAs.
Discussion
In the past few decades, many studies have been conducted to determine the molecular mechanisms underlying diabetic retinopathy (DR). Protein-coding genes or miRNAs have been previously identified. Yet, very few studies have determined the function of lncRNAs which are associated with the pathogenesis of DR. Scientists have not been successful in elucidating the functions of the lncRNA-miRNA-mRNA network in DR to date. Here, we report certain lncRNAs which can be used as miRNA sponges, where these sponges are involved in the pathogenesis of DR. To the best our knowledge, this is the first study to determine whether ceRNAs play a pivotal role in the development of DR.
In the present study, we performed GO analysis to determine the possible functions of the differentially expressed lncRNAs. We found that the functions of lncRNAs were completely related to the functions of the connected mRNAs; this notion was used to determine gene product enrichment in biological processes, cellular components and molecular functions. By performing GO analysis of mRNAs, we found that differentially expressed genes were involved in the following ophthalmic functions: Eye development, lens development in camera-type eye, visual perception and structural constituents of eye lens. This indicates that disease-related lncRNAs play important roles in the pathogenesis of DR. Furthermore, we also performed KEGG analysis; however, the results indicated that disease-specific genes were not significantly associated with certain pathways.
It has been established that WGCNA is a widely-used bioinformatic method that describes correlation patterns among genes of microarray samples. We performed WGCNA analysis for the following purposes: i) to identify clusters (modules) of highly correlated genes, ii) to summarize clusters (module) by using eigen gene or an intramodular hub gene, iii) to relate modules with one another and external sample traits using eigen gene network methodology, and iv) to calculate module membership measures. Although the analysis was performed with utmost diligence, we obtained a paradoxical result: We found that only one module was specifically related to DR. This module included 33 lncRNAs and 10 mRNAs. These 33 lncRNAs might serve as potential diagnostic biomarkers and therapeutic targets for DR.
Importantly, previous studies have reported that some key hub miRNAs, depicted in this study, play an important role in the pathogenesis of different diseases. For example, Bozec et al investigated the functions of miR-223-3p, reporting that this molecule is associated with the angiogenesis of head and neck squamous cell carcinoma (27). This indicates that miR-223-3p plays a pivotal role in the development of DR. In another study, miR-223-3p was found to be involved in the pathogenesis and progression of diabetic kidney disease (28). Another study reported that miR-34c-5p is an inflammation-related miRNA, which is associated with the vascular repair factor HGF and miR-574-3p. It is upregulated in type 2 diabetes monocytes, implying that anti-inflammatory cells play a pivotal role in adhesion, vascular repair and invasion (29). Moreover, miR-34c-5p is also a tumor suppressor (30), and it is associated with apoptosis and differentiation of cancer cells. Furthermore, miR-200b-3p is also a hub molecule in ceRNA network. It was found that miR-200b-3p is associated with several types of cancers, dysregulating monocyte/macrophage (31) and differentiating epithelial-to-mesenchymal transition of glioma cancer cells (32).
Our study has several limitations. A major concern is the insufficient data and a relatively small sample size. Owing to the limited sample size, the miRNA-lncRNA-mRNA network might be restricted. Moreover, the type of diabetes was not reported by study participants. Another major concern of this research study is that computational results might be noises and false-positive results; however, we set cut-offs to the most used values. Moreover, we used the widely accepted methods of computation. For example, the target mRNAs of miRNAs were predicted on the basis of three widely used databases (miRTarBase, miRecords and starBase). These results need further confirmation in terms of cell lines and animal models.
In summary, we constructed a competitive endogenous RNA network and identified several potential key molecules. We described the important role played by non-coding RNAs in the pathogenesis of DR. Our study highlighted specific lncRNAs and miRNAs related to the pathogenesis of DR, which might be used as novel diagnostic biomarkers and therapeutic targets for DR.
Acknowledgements
Not applicable.
Funding
This study was supported by a grant from the National Natural Science Foundation of China (81501559 and 61671255), the Jiangsu Overseas Research and Training Program for University Prominent Young and Middle-aged Teachers and Presidents 2016, and the Graduate Research and Innovation Plan Project of Nantong University (YKC15056, YKC16072), the Innovative Training Program of Undergraduate Students of Nantong University (2018144) and the National Key R&D Program of China (2018YFC1314902).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YW and HW designed the study, analyzed and interpreted the data, and wrote the manuscript. KJ, AS, LW, LS, KJ and JD conducted the data analysis and wrote the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Klein R, Klein BE, Moss SE, Davis MD, DeMets DL. The Wisconsin epidemiologic study of diabetic retinopathy. II. Prevalence and risk of diabetic retinopathy when age at diagnosis is less than 30 years. Arch Ophthalmol. 1984;102:520–526. doi: 10.1001/archopht.1984.01040030398010. [DOI] [PubMed] [Google Scholar]
- 2.Brownlee M. The pathobiology of diabetic complications: A unifying mechanism. Diabetes. 2005;54:1615–1625. doi: 10.2337/diabetes.54.6.1615. [DOI] [PubMed] [Google Scholar]
- 3.Ding J, Wong TY. Current epidemiology of diabetic retinopathy and diabetic macular edema. Curr Diab Rep. 2012;12:346–354. doi: 10.1007/s11892-012-0283-6. [DOI] [PubMed] [Google Scholar]
- 4.Wu H, Wu H, Shi L, Yuan X, Yin Y, Yuan M, Zhou Y, Hu Q, Jiang K, Dong J. The association of haptoglobin gene variants and retinopathy in type 2 diabetic patients: A meta-analysis. J Diabetes Res. 2017;2017:2195059. doi: 10.1155/2017/2195059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu H, Geng X, Zhang X, Qiu M, Jiang K, Tang L, Dong J. A self-adaptive distance regularized level set evolution method for optical disk segmentation. Biomed Mater Eng. 2014;24:3199–3206. doi: 10.3233/BME-141141. [DOI] [PubMed] [Google Scholar]
- 6.Wu HQ, Wu H, Shi LL, Yu LY, Wang LY, Chen YL, Geng JS, Shi J, Jiang K, Dong JC. The association between retinal vasculature changes and stroke: A literature review and Meta-analysis. Int J Ophthalmol. 2017;10:109–114. doi: 10.18240/ijo.2017.01.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: Past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiu HS, Somvanshi S, Patel E, Chen TW, Singh VP, Zorman B, Patil SL, Pan Y, Chatterjee SS, Sood AK, et al. Cancer Genome Atlas Research Network: Pan-cancer analysis of lncRNA regulation supports their targeting of cancer genes in each tumor context. Cell Rep. 2018;23:297–312.e12. doi: 10.1016/j.celrep.2018.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: Possible functions and clinical implications. J Med Genet. 2015;52:710–718. doi: 10.1136/jmedgenet-2015-103334. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, Sun X, Icli B, Feinberg MW. Emerging roles for MicroRNAs in diabetic microvascular disease: Novel targets for therapy. Endocr Rev. 2017;38:145–168. doi: 10.1210/er.2016-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan B, Tao ZF, Li XM, Zhang H, Yao J, Jiang Q. Aberrant expression of long noncoding RNAs in early diabetic retinopathy. Invest Ophthalmol Vis Sci. 2014;55:941–951. doi: 10.1167/iovs.13-13221. [DOI] [PubMed] [Google Scholar]
- 12.Kovacs B, Lumayag S, Cowan C, Xu S. MicroRNAs in early diabetic retinopathy in streptozotocin-induced diabetic rats. Invest Ophthalmol Vis Sci. 2011;52:4402–4409. doi: 10.1167/iovs.10-6879. [DOI] [PubMed] [Google Scholar]
- 13.Zhang LQ, Cui H, Wang L, Fang X, Su S. Role of microRNA-29a in the development of diabetic retinopathy by targeting AGT gene in a rat model. Exp Mol Pathol. 2017;102:296–302. doi: 10.1016/j.yexmp.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 14.Chen Q, Qiu F, Zhou K, Matlock HG, Takahashi Y, Rajala RV, Yang Y, Moran E, Ma JX. Pathogenic role of microRNA-21 in diabetic retinopathy through downregulation of PPARα. Diabetes. 2017;66:1671–1682. doi: 10.2337/db16-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ye Z, Li ZH, He SZ. miRNA-1273g-3p involvement in development of diabetic retinopathy by modulating the autophagy-lysosome pathway. Med Sci Monit. 2017;23:5744–5751. doi: 10.12659/MSM.905336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomaa AR, Elsayed ET, Moftah RF. MicroRNA-200b expression in the vitreous humor of patients with proliferative diabetic retinopathy. Ophthalmic Res. 2017;58:168–175. doi: 10.1159/000475671. [DOI] [PubMed] [Google Scholar]
- 17.Yang C, Wu D, Gao L, Liu X, Jin Y, Wang D, Wang T, Li X. Competing endogenous RNA networks in human cancer: Hypothesis, validation, and perspectives. Oncotarget. 2016;7:13479–13490. doi: 10.18632/oncotarget.7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Hou J, He D, Sun M, Zhang P, Yu Y, Chen Y. The emerging function and mechanism of ceRNAs in cancer. Trends Genet. 2016;32:211–224. doi: 10.1016/j.tig.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song C, Zhang J, Liu Y, Pan H, Qi HP, Cao YG, Zhao JM, Li S, Guo J, Sun HL, Li CQ. Construction and analysis of cardiac hypertrophy-associated lncRNA-mRNA network based on competitive endogenous RNA reveal functional lncRNAs in cardiac hypertrophy. Oncotarget. 2016;7:10827–10840. doi: 10.18632/oncotarget.7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang H, Ma R, Zou S, Wang Y, Li Z, Li W. Reconstruction and analysis of the lncRNA-miRNA-mRNA network based on competitive endogenous RNA reveal functional lncRNAs in rheumatoid arthritis. Mol BioSyst. 2017;13:1182–1192. doi: 10.1039/C7MB00094D. [DOI] [PubMed] [Google Scholar]
- 22.Huang C, Yuan N, Wu L, Wang X, Dai J, Song P, Li F, Xu C, Zhao X. An integrated analysis for long noncoding RNAs and microRNAs with the mediated competing endogenous RNA network in papillary renal cell carcinoma. OncoTargets Therapy. 2017;10:4037–4050. doi: 10.2147/OTT.S141951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chou CH, Chang NW, Shrestha S, Hsu SD, Lin YL, Lee WH, Yang CD, Hong HC, Wei TY, Tu SJ, et al. miRTarBase 2016: Updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016;44:D239–D247. doi: 10.1093/nar/gkv1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: An integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37:D105–D110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li JH, Liu S, Zhou H, Qu LH, Yang JH. StarBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bozec A, Zangari J, Butori-Pepino M, Ilie M, Lalvee S, Juhel T, Butori C, Brest P, Hofman P, Vouret-Craviari V. MiR-223-3p inhibits angiogenesis and promotes resistance to cetuximab in head and neck squamous cell carcinoma. Oncotarget. 2017;8:57174–57186. doi: 10.18632/oncotarget.19170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L, Li R, He J, Yang Q, Wu Y, Huang J, Wu B. Co-expression analysis among microRNAs, long non-coding RNAs, and messenger RNAs to understand the pathogenesis and progression of diabetic kidney disease at the genetic level. Methods. 2017;124:46–56. doi: 10.1016/j.ymeth.2017.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baldeón Rojas L, Weigelt K, de Wit H, Ozcan B, van Oudenaren A, Sempértegui F, Sijbrands E, Grosse L, van Zonneveld AJ, Drexhage HA, Leenen PJ. Study on inflammation-related genes and microRNAs, with special emphasis on the vascular repair factor HGF and miR-574-3p, in monocytes and serum of patients with T2D. Diabetol Metab Syndr. 2016;8:6. doi: 10.1186/s13098-015-0113-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li F, Chen H, Huang Y, Zhang Q, Xue J, Liu Z, Zheng F. miR-34c plays a role of tumor suppressor in HEC1-B cells by targeting E2F3 protein. Oncol Rep. 2015;33:3069–3074. doi: 10.3892/or.2015.3894. [DOI] [PubMed] [Google Scholar]
- 31.Yu X, Wang QL, Li YF, Wang XD, Xu A, Li Y. A novel miR-200b-3p/p38IP pair regulates monocyte/macrophage differentiation. Cell Discov. 2016;2:15043. doi: 10.1038/celldisc.2015.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu J, Cui H, Zhu Z, Wang L. MicroRNA-200b-3p suppresses epithelial-mesenchymal transition and inhibits tumor growth of glioma through down-regulation of ERK5. Biochem Biophys Res Commun. 2016;478:1158–1164. doi: 10.1016/j.bbrc.2016.08.085. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Using eight week-old C57BL/6 mice, a mouse model of streptozotocin (STZ)-induced diabetes was constructed. Two months after administering STZ injection to diabetic mice, total RNAs were isolated from the retinas of the diabetic mice. The mice were considered to be diabetic when their blood glucose levels exceeded 250 mg/dl. Using TRIzol reagent, wild-type mice were matched according to age and sex. Agilent Mouse Gene Expression Microarrays (Product Number G4852A; Agilent Technologies, Santa Clara, CA, USA) were used to generate microarray data from mouse mRNA and lncRNA. The raw data were provided by Yan et al, which has been described in a previous study (11), and the ethics statement was previously provided. To generate microarray data of miRNAs from STZ-induced diabetic rats, we referred to a previous study and its supplementary material (12).
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.