Abstract
Mitochondria (MT), the major site of cellular energy production, are under dual genetic control by 37 mitochondrial DNA (mtDNA) genes and numerous nuclear genes (MT-nDNA). In the CHARGEmtDNA+ Consortium, we studied genetic associations of mtDNA and MT-nDNA associations with body mass index (BMI), waist-hip-ratio (WHR), glucose, insulin, HOMA-B, HOMA-IR, and HbA1c. This 45-cohort collaboration comprised 70,775 (insulin) to 170,202 (BMI) pan-ancestry individuals. Validation and imputation of mtDNA variants was followed by single-variant and gene-based association testing. We report two significant common variants, one in MT-ATP6 associated (p ≤ 5E−04) with WHR and one in the D-loop with glucose. Five rare variants in MT-ATP6, MT-ND5, and MT-ND6 associated with BMI, WHR, or insulin. Gene-based meta-analysis identified MT-ND3 associated with BMI (p ≤ 1E−03). We considered 2,282 MT-nDNA candidate gene associations compiled from online summary results for our traits (20 unique studies with 31 dataset consortia’s genome-wide associations [GWASs]). Of these, 109 genes associated (p ≤ 1E−06) with at least 1 of our 7 traits. We assessed regulatory features of variants in the 109 genes, cis- and trans-gene expression regulation, and performed enrichment and protein-protein interactions analyses. Of the identified mtDNA and MT-nDNA genes, 79 associated with adipose measures, 49 with glucose/insulin, 13 with risk for type 2 diabetes, and 18 with cardiovascular disease, indicating for pleiotropic effects with health implications. Additionally, 21 genes related to cholesterol, suggesting additional important roles for the genes identified. Our results suggest that mtDNA and MT-nDNA genes and variants reported make important contributions to glucose and insulin metabolism, adipocyte regulation, diabetes, and cardiovascular disease.
Keywords: mitochondria, mtDNA, MT-nDNA candidate genes, BMI, waist-to-hip ratio, glucose, insulin, HOMA-B, HOMA-IR, HbA1c
Introduction
Mitochondria (MT) are double membraned organelles that generate the majority of cellular adenosine triphosphate (ATP) and metabolites and play a central role in human health.1 MT reside within cells and contain a separate maternally inherited2, 3, 4, 5 ∼16.6 kb circular mtDNA genome, consisting of 37 genes (Figure 1). The mtDNA encodes 13 core catalytic peptides that form the oxidative phosphorylation complexes (I, III-V) as well as the machinery needed to translate these peptides consisting of 22 transport RNAs (tRNAs) and 2 ribosomal RNAs (rRNAs). In addition, a noncoding sequence, known as the displacement loop (D-loop), encompasses the replication origin(s) and promoters for mtDNA.
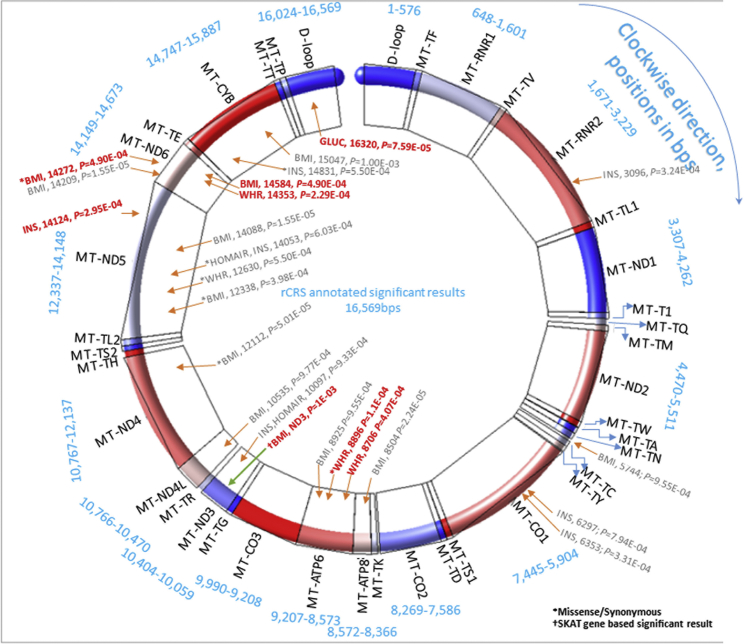
Figure 1.
Graphical Presentation of Single mtDNA- and Gene-Based Association Meta-Analysis Results
Significant associations (trait, MT position, and p value) at liberal threshold (p ≤ 1E−03) are annotated with red arrows and gray text for single SNV associations and with green arrow for gene-based association, and text in red color represents SNVs that passed the threshold p ≤ 5E−04. The asterisk (∗) marks missense or synonymous variants; a dagger (†) marks a SKAT gene-based significant association. The mtDNA is shown as one single molecule, by merging all genes from heavy and light MT strands. For visibility, gene colors rotate clockwise from first in blue in the D-loop to up to six coloring groups finishing in red at MT-TL1, and then recycling again from the first blue color at MT-ND1. The full information is summarized in Tables 1, 2, 3, and S2.a-S2.b. In blue text are shown bps-positions of 13 MT-coding genes (MT-ND1, MT-ND2, MT-CO1, MT-CO2, MT-ATP8, MT-ATP6, MT-CO3, MT-ND3, MT-ND4L, MT-ND4, MT-ND5, MT-ND6, MT-CYB), 2 MT-ribosomal RNAs (MT-RNR1 [12S RNA] and MT-RNR2 [16S RNA]), and 22 transport RNAs (MT-TF, TV, TL1, T1, TQ, TM, TW, TA, TN, TC, TY, TS1, TD, TK, TG, TR, TH, TS2, TL2, TE, TT, TP).
MT function is also dependent upon many nuclear genes that encode proteins involved in mtDNA transcription, replication, cell apoptosis and mitophagy, nucleotide biosynthesis, metabolism, and iron and calcium homeostasis. Here we evaluate 2,282 candidate nuclear genes that also may contribute to MT function. We refer to these genes as “MT-nDNA candidates,” curated for this study based upon protein co-localization within the MT and text mining.6, 7, 8 Unlike mtDNA-encoded proteins that are transcribed and translated within MT, MT-nDNA genes are translated in the cytoplasm and their proteins are transported into MT through porins or complexes such as translocases.
The dual genomic origin (mtDNA and MT-nDNA) of MT components and the fact that mtDNA has a much higher mutation rate compared to the nuclear genome9, 10 fuels our hypothesis that MT variants influence individual predisposition to complex diseases.11, 12 Because of heteroplasmy, i.e., the coexistence of mutated and wild-type mtDNA in the same cell, mutations in mtDNA may not result in disease until the copy number of mtDNA carrying the mutation relative to the wild-type mtDNA in the relevant tissue(s) exceeds a threshold.12 The mtDNA copy number may serve as an indicator of MT function13 and was recently shown to be associated with overall mortality14, 15 and various metabolic-related diseases, including cardiovascular disease (CVD).16, 17, 18 Studies have also shown that mitochondria have an important influence on multiple cardiovascular disease risk factors. For example, obesity impairs MT biogenesis.19, 20, 21 MT dysfunction has been also implicated in the development of insulin resistance.22, 23, 24 Thus, our study prioritized obesity and insulin resistance pathways to CVD.
In our study we analyzed the dual genomic influence of MT function on seven important metabolic traits (body mass index [BMI], waist-hip ratio [WHR], fasting glucose, fasting insulin, HOMA-B, HOMA-IR, and HbA1c) as major risk factors of cardiovascular disease (CVD) in participants of the CHARGEmtDNA+ 45 participating cohorts. The mtDNA associations were evaluated at both the single nucleotide variant (SNV) and at the gene-based levels for rare alleles (minor allele frequency [MAF] < 1%). The MT-nDNA candidates were studied for function, gene expression regulation, enrichment of pathways, and protein interactions. The overall goal was to illuminate MT-functioning and MT-disease causative variants.
Subjects and METHODS
CHARGE Cohorts
There were 45 participating cohorts with a total of 196,967 individuals (166,423 individuals were of the European [EA], 15,745 of African [AA], 1,567 of Asian [ASA], 11,307 of Hispanic/Latinos [HA], and 1,925 of Brazilian ancestry) who had at least one of the seven traits. CHARGE cohort summary descriptions are available online (see Supplemental Study Descriptions). The number of samples available from all cohorts differed by phenotype (Table S1). The resulting samples were smaller within each cohort when association tests were performed (by sub-setting individuals with genotyping available) to a total of 170,202 for BMI, 155,396 for WHR, 79,906K for fasting glucose, 70,778 for fasting insulin, 69,845 for HOMA-B, 69,926 HOMA-IR, and 101,218 for HbA1c. We use the acronym PA to refer to pan-ancestry. The procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and that proper informed consent was obtained.
Trait Harmonization
A total of seven traits were studied: BMI (kg/m2), WHR (waist-hip ratio), fasting plasma glucose (mg/dL), fasting insulin (μIUmL), HOMA-B, HOMA-IR, and glycated hemoglobin (HbA1c, %) indicative of chronically elevated glucose levels. The homeostasis model assessment was based on plasma levels of fasting glucose and fasting insulin. HOMA-B is an indicator of beta-cell function calculated as HOMA-B = (360 × fasting insulin) / (fasting glucose − 60). HOMA-IR is an indicator of insulin resistance calculated as HOMA-IR = (fasting glucose × fasting insulin) / 405. Glucose and insulin measures were required to have been measured with a minimum fasting time of 8 hr; otherwise, the measure was set to missing. Each study was responsible for ensuring that each trait was normally distributed. Natural-log transformation was implemented for insulin, power transformation was performed when no other solutions were available, and inverse normal transformation was used as a last resort for BMI25 (details in the Supplemental Material and Methods, section 1). The association test analysis was restricted to non-diabetic participants when studying glycemic traits, by setting values of glucose, insulin, and HbA1c to missing for T2D individuals with HbA1c > 6.5 or with fasting glucose > 126 mg/dL or using T2D medications. The participating cohorts were required to have at least one of the seven studied metabolic traits.
Preparation of Traits for Analyses
All cohorts used the same units for all traits for the raw measures. We observed differences in the raw measures of traits (Table S1). Thus, to harmonize results, all participating cohorts normalized trait distributions and produced standardized residuals, which are unit-less for each trait. Statistical regression via R / SAS programming languages was used to produce residuals fitting the following model:
Age2 was used to remove any age-quadratic trend from the response variable, and principal components (PCs) represent one or more genotype principal components, to minimize population substructure. The standardized residuals from this regression were the final response variables for each trait, used in the association tests with imputed dosages of mtDNA.
mtDNA Variant Harmonization
All mtDNA variants from each array (Table S14) were annotated to the nucleotide position according to the rCRS of the human mitochondrial DNA prior to analyses (see Web Resources). The probes used for each microarray were obtained from the manufacturer or dbSNP and aligned to the rCRS using Geneious 8.1.26 All probes were also submitted through the standard nucleotide basic local alignment search tool (BLAST) to ensure the probes bound with high specificity (≥90% identity) to the mitochondrial genome. In order to limit any potential binding to nuclear DNA segments, probes that bound to nuclear chromosomes with ≥80% matching were excluded from all analyses. The full lists of the validated mtDNA variants are available in the Supplemental Material and Methods, section 2 and Tables S15–S28.
mtDNA Imputation
The preparation of mtDNA data for imputation was done with PLINK.27, 28 When preparing data for imputation, a few heterozygotes that may have existed in the mtDNA genotypes were set to missing via PLINK. The prephasing of mtDNA scaffold haplotypes at the cohort level was done with SHAPEIT2 for the full mtDNA (see Web Resources).29 SHAPEIT2 helped to have the mtDNA genotypes of a cohort in the right format to be used in the imputation pipeline with IMPUTE2. SHAPEIT2 was also used for QC checks, such as evaluating missingness in markers. In order to combine analyses across different genotyping platforms, we performed imputation based on MT-1000G (see Supplemental Material and Methods, section 3). Imputations of mtDNA variants were implemented in IMPUTE2 at the cohort level in a window that included the full MT-chromosome (see Web Resources).30 Recoding of genotype probabilities into dosages was implemented via FCGENE (see Web Resources). Our reference MT-chromosome panel consisted of the 1000G data (PHASE 3, v.5) (see Web Resources). These data were based on sequence freeze and alignments, using only Illumina platform sequences and only sequences with read lengths of 70 bps or greater. There were 3,617 mtDNA markers in 1000G. The 1000G included 2,504 individuals representing 26 sampled populations (the “Cosmopolitan” reference panel).31 After imputation, cohorts had differing number of SNVs, up to 3,832 SNVs for BMI, 3,829 for WHR, 3,809 for glucose, 3,801 for insulin, 3,801 for HOMA-B, 3,801 for HOMA-IR, and up to 3,744 for HbA1c. Although the imputation quality was good for most of cohorts (see imputation accuracy in Table S14), a great number of the SNVs were filtered for INFO (imputation quality) < 0.30, monomorphic or very rare (MAF < 0.0001) at the cohort level before association tests. Other QC were genotype call per cell had to be p ≥ 0.90, a marker was dropped if its missing rate was >0.05 and MAC had to be ≥5, (which if it was autosomal could have corresponded to MAC ≥10). The MAC was calculated for haplotypes as MAF∗N. (See Supplemental Material and Methods, section 3 for a more detailed methodology.) After mtDNA imputation, before performing statistical associations, we set any heteroplasmic mutations to missing at the cohort level data. Details of QC via R Package: EasyQC (V10.0)32 (see Web Resources) and our internal programs and of the imputation steps can be found in the Supplemental Material and Methods, section 3. All filters implemented at the cohort level created non-uniform contributions of all cohorts in the meta-analysis. Many of the mtDNA variants were rare and per trait meta-analyses QQ-plots often showed an underestimation compared to expected association results (Figures S1–S7). The rarity of alleles was accompanied with lower quality of imputation and accordingly many of them were removed before any meta-analysis. Most studies had good imputation quality, given the rarity of mtDNA variants.
Variants MT-14272 and MT-14584 are opposite C/T versus T/C, but in full LD, while they are rare. The typical assumption is that rare variants are not in LD, but that does not have to hold for mtDNA. This is one more observation that the rare association findings in this study represent potential associations, until replicated from other future studies.
mtDNA Association Statistical Analyses
An additive genetic model was applied in association analyses using both self-developed regression models (the linear or linear mixed models written in R programming) and the SKAT (the prepScores() function in seqMeta R package: seqMeta package: Meta-Analysis of Region-Based Tests of Rare DNA Variants approaches33 (see Web Resources). Familial and maternal correlation structures34 were accounted for in the analysis of family data. Details of statistical association models 1–4 are described in the Supplemental Material and Methods, section 4.
mtDNA Meta-Analyses
Single-variant fixed-effects meta-analyses were conducted with METAL,35 and gene-based associations were performed in seqMeta (details in the Supplemental Material and Methods, sections 5–7) (Tables 1, 2, 3, S2.a, S2.b, and S13). The average allele frequency from Metal is an average of allele frequencies for a particular marker as contributed from each study and weighted by each study’s sample size as follows: , where k represents the indexing of contributing studies, wk is the sample size (number of individuals) per study, and fk is the coded allele frequency for a particular marker from a particular study. Our working group conducted an internal permutation test36 using ARIC study mtDNA data and determined that 49 independent mitochondrial variants represented an estimate of the number of independent genetic effects for mtDNA. Thus, 49 was used as a denominator for producing the Bonferroni corrected p significance threshold ≤ 0.05/49 ≤ 1E−03, a threshold for common variants (MAF ≥ 1%), but possibly liberal for rare variants (MAF < 1%) (Table 1). Furthermore, we considered p ≤ 0.05/(49 × 7 traits) ≤ 1.5E−04 as a conservative threshold when accounting for seven traits tested. We settled for a Bonferroni-threshold p ≤ 0.05/(49 × 2 domains) ≤ 5E−04, because the traits within a domain adipose/obesity (BMI and WHR) and glucose metabolism (glucose, insulin, HOMA-B, HOMA-IR, and HbA1c) are correlated and represent two domains. Because mtDNA is a small genome, the distributions of mtDNA QQ-plots with the existing samples do not always behave similarly to those observed for larger nuclear GWASs. While mtDNA variants MAF ≥ 1% formed relatively good QQ-plot distributions, the rare variants (MAF < 1%) were sometimes distributed with some deviation in the start (bottom-left) of the QQ-plot, as a result of meta-results in very rare alleles. Thus, the quality control of mtDNA QQ-plots were implemented by filtering at different rare MAF levels. We concluded that with the existing mtDNA meta-analysis results, QQ-plots with MAF > 0.8% were acceptable. Thus, out of 23 SNVs nominally (p ≤ 1E−03) associated with 6 metabolic traits (Figure 1), by adding the filter of MAC ≥ 5 for each cohort, then 7 variants passed p ≤ 5E−04 Bonferroni threshold.
Table 1.
mtDNA Variants Associated with BMI, WHR, Glucose, Insulin, HOMA-B, HOMA-IR, and HbA1c METAL Meta-Analysis Single-Variant Results
| No | Pos | rsID | Gene | Annotation | Trait | A1/2 | Freq1 WeightAve | FreqSE | MinFreq | MaxFreq | MAF | MAC | β(SE) | p | Dir | Het-p | N |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Meta-analysis Level Results Selected with MAF of Weighted Allele Average Frequency > 1% and Passing Meta-p-Threshold ≤ 5E−04 | |||||||||||||||||
| 1 | 8706 | – | MT-ATP6 | – | WHR | A/G | 0.9676 | 0.0295 | 0.9135 | 0.9974 | 0.0324 | 834 | −0.13 (0.04) | 4.1E−04 | ----+ | 9.44E−02 | 25,748 |
| 2 | 16320 | rs62581338 | D-loop | – | GLUC | T/C | 0.0158 | 0.0954 | 0.0028 | 0.2439 | 0.0158 | 301 | −0.21 (0.05) | 7.6E−05 | --+-- | 1.77E−01 | 19,046 |
| Meta-analysis Level Results Selected with MAF of Weighted Allele Average Frequency < 1% and Passing Meta-p-Threshold ≤ 5E−04 | |||||||||||||||||
| 1 | 8896 | rs202120082 | MT-ATP6 | missense | WHR | A/G | 0.0038 | 0.0045 | 0.0003 | 0.0121 | 0.0038 | 134 | 0.30 (0.08) | 1.12E−04 | ++++ | 8.80E−01 | 34,959 |
| 2 | 14124 | – | MT-ND5 | – | INS | T/C | 0.0035 | 0.0029 | 0.0022 | 0.0087 | 0.0035 | 21 | 0.57 (0.16) | 2.95E−04 | +++ | 1.62E−01 | 6,035 |
| 3 | 14272 | rs2853814 | MT-ND6 | missense | BMI | T/C | 0.0012 | 0.0007 | 0.0009 | 0.0022 | 0.0012 | 17 | 0.84 (0.24) | 4.90E−04 | ++ | 6.49E−01 | 13,636 |
| 4 | 14353 | – | MT-ND6 | – | WHR | T/C | 0.9916 | 0.0032 | 0.9880 | 0.9978 | 0.0084 | 120 | −0.33 (0.09) | 2.29E−04 | -- | 8.00E−01 | 14,315 |
| 5 | 14584 | – | MT-ND6 | – | BMI | T/C | 0.9988 | 0.0007 | 0.9978 | 0.9991 | 0.0012 | 17 | −0.84 (0.24) | 4.90E−04 | -- | 6.49E−01 | 13,636 |
Abbreviations and definitions: No, order number; Pos, MT position in bps; rsID, rsID name from NCBI dbSNP database when available; Gene, gene name or region; Annotation, role of the variants when available; Trait, one or more of seven traits studied; A1/2, the coded and non-coded alleles; Freq1 WeightAve, a weighted sample size average of allele frequency; FreqSE, standard error of allele frequency; MINFreq, a minimum allele frequency for contributing cohorts; MAXFreq, a maximum allele frequency for contributing cohorts; MAF, minor allele frequency; MAC, minor allele count, calculated as MAF × N; β(SE), beta coefficient and the corresponding standard error; p value, from single variant regression analysis; Dir, direction sign of contributing cohort’s beta; Het-p, heterogeneity p value test from METAL; N, individuals’ sample contributing in a particular marker meta-analysis (all results are of Pan-Ancestry).
Table 2.
Results of mtDNA Gene-Based Meta-analysis SKAT T1 and T5 Test (p ≤ 0.01)
| Trait | Ancestry | T | Gene | p | Qmeta | cMAF | nSNVs |
|---|---|---|---|---|---|---|---|
| HOMA-B | PA | 0.01 | MT-TF | 5.0E−03 | 4604374 | 0.022 | 5 |
| HOMA-B | PA | 0.05 | MT-TV | 8.0E−03 | 3341460 | 0.078 | 6 |
| HbA1c | EA | 0.05 | MT-TG | 9.0E−03 | 13674145 | 0.064 | 13 |
| BMI | PA | 0.05 | MT-TQ | 1.0E−02 | 21201188 | 0.075 | 12 |
| HOMA-B | PA | 0.05 | MT-RNR1 | 1.0E−02 | 69787232 | 1.323 | 119 |
Abbreviations and definitions: Trait, the trait used for a specific test; PA, Pan ancestry; EA, European ancestry; T, MAF-value threshold for selecting SNVs to be included in the gene-based association test; Gene, gene name from mtDNA; p, p value from the SKAT test; Qmeta, the SKAT Q statistics; , where w is a weight for SNVj and Uj is associated score statistics; cMAF, cumulative minor allele frequency; nSNVs, the number of SNVs used in the gene-based meta-analysis.
Table 3.
Results of mtDNA Gene-Based Burden T1 and T5 Meta-Analysis Test (p ≤ 0.01)
| Trait | Ancestry | T | Gene | p | Beta | SE | cMAFUsed | nSNVs |
|---|---|---|---|---|---|---|---|---|
| BMI | PA | 0.05 | MT-ND3 | 1.0E−03 | 0.007 | 0.002 | 0.290 | 82 |
| BMI | PA | 0.05 | MT-TQ | 2.0E−03 | 0.020 | 0.006 | 0.075 | 12 |
| BMI | PA | 0.05 | MT-CO2 | 5.0E−03 | 0.003 | 0.001 | 0.491 | 155 |
| HOMAB | PA | 0.01 | MT-TF | 5.0E−03 | 0.038 | 0.014 | 0.022 | 5 |
| HOMAB | PA | 0.01 | MT-TV | 6.0E−03 | 0.088 | 0.032 | 0.009 | 2 |
| HOMAB | PA | 0.05 | MT-RNR1 | 7.0E−03 | 0.002 | 0.001 | 1.323 | 119 |
| HOMAIR | EA | 0.05 | MT-TG | 7.0E−03 | 0.027 | 0.010 | 0.061 | 13 |
| BMI | EA | 0.05 | MT-ND3 | 7.0E−03 | 0.006 | 0.002 | 0.194 | 81 |
| HOMAB | PA | 0.05 | MT-TV | 8.0E−03 | 0.031 | 0.011 | 0.078 | 6 |
| BMI | EA | 0.05 | MT-TQ | 8.0E−03 | 0.018 | 0.007 | 0.064 | 12 |
| HOMAB | EA | 0.05 | MT-RNR1 | 9.0E−03 | 0.002 | 0.001 | 1.287 | 120 |
| HOMAB | EA | 0.05 | MT-TF | 9.0E−03 | 0.018 | 0.007 | 0.127 | 10 |
| HOMAB | EA | 0.05 | MT-TV | 9.0E−03 | 0.030 | 0.011 | 0.079 | 6 |
| HOMAB | EA | 0.01 | MT-RNR1 | 1.0E−02 | 0.003 | 0.001 | 0.292 | 61 |
Abbreviations and definitions: Trait, the trait used for a specific test; PA, Pan ancestry; EA, European ancestry; T, MAF-value threshold for selecting SNVs to be included in the gene-based-burden association test; Gene, gene name from mtDNA; p, p value from the gene-based burden test; the score test of the weighted sum of genotypes has the form of statistic, , where w is a weight for SNVj and Uj is the score statistic for SNVj; Beta and SE are result of regressing the trait on a weighted sum of genotypes; cMAFUsed, cumulative minor allele frequency; nSNVs, the number of SNVs used in the gene-based burden meta-analysis.
For the gene-based analysis, we used Burden tests which combine the contributions of rare genetic variants within a gene region. Such tests assume similar directionality and effect sizes for each variant.33 In contrast, the sequence kernel association test (SKAT) for unrelated or family-based studies is an efficient regression-based approach for rare genetic variant analysis.33, 37, 38 The meta-analyses were performed using seqMeta (see Web Resources). The gene-based Bonferroni p value was calculated as p ≤ 0.05/ (37 mtDNA genes) ≤ 1E−03.
Identification of MT-nDNA Candidate Genes
We established a list of 2,282 MT-nDNA candidate genes from three sources: (1) Mito Carta 2.0,6, 39 (2) Literature Lab (Acumenta Biotech), and (3) MitoMiner (4.0)7 (see also Supplemental Material and Methods, section 7). We used the two separate sets of human genes and mouse ortholog genes from MitoCarta. We used Literature Lab to perform a literature search and identified 36 terms based on MeSH mitochondria. From each of the 36 terms (Table S29), we selected only the upper quartile list of genes from a Log Probability Function - scoring distribution. Each term was tested for association in overlapping with genes in pathway analysis (Figure S15). We conditioned this list with only genes from human nomenclature and accepted only genes that had more than 15 abstracts cited per selected gene (see Supplemental Material and Methods section 7).
Using MitoMiner we identified additional MT-nDNA candidate genes. They were filtered with a MT-MitoMiner index ≥ 0.70, and by selecting the terms “Known mitochondrial” and “Predicted mitochondrial.” In MitoMiner, we kept only genes that were present in human nomenclature. MitoMiner included also the mitochondrial originated genes, which were later removed to keep our MT-nDNA list only of nuclear origin. Finally, three additional genes were added from a publication on MT-defects associated with β-cell dysfunction in a T2D mouse.40
MT-nDNA Candidate Genes Analysis
As a result of the above work, a list of 2,282 MT-nDNA candidate gene labels (Table S30) were used to identify any significant results from 20 GWAS papers full summary results for seven metabolic traits, representing 31 datasets (Tables 4, S4, and S6). For the adipose traits, the GWAS publication full summary results used were for BMI41, 42, 43, 44, 45, 46, 47, 48, 49, 50 and for WHR,43, 44, 47, 48, 49, 50, 51, 52 and the summary results data were retrieved from the GIANT Consortium repositories (see Web Resources). For glucose metabolism we used the summary results of glucose,53, 54, 55, 56 insulin,54, 55, 56, 57, 58, 59 HOMA-B,54 and HOMA-IR.54 These summary results data were retrieved from MAGIC Consortium archives (see Web Resources). For HbA1c we used two resources.60, 61 The published GWASs have large sample sizes, with a BMI study having a maximum of 339K individuals, WHR having 320K, glucose and insulin having 52K and 45K, respectively, HOMA-B and -IR having 46K, and HbA1c having 160K. Consequently, we obtained results for 109 MT-nDNA candidate genes, accompanied by 588 sentinel significant SNVs (one best per gene and trait combination, out of seven traits, Tables 4, S6, and S9).
Table 4.
GWAS Findings for Seven Traits (BMI, WHR, Glucose, Insulin, HOMA-B, HOMA-IR, HbA1c) for 109 MT-nDNA Candidate Genes
| No | SNV | Chrom | Position | Role | Gene | Full Gene Name (Cytogenetic Position) | BMI p | OBESITY p | WHR p | GLUC p | INS p | HOMA-B p | HOMAIR p | HbA1c p |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | rs622798 | 1 | 45549599 | upstream-2 kb | AKR1A1 | Aldo-Keto Reductase Family 1 Member A1 (1p34.1) | 8.56E−07 | – | – | – | – | – | – | – |
| 2 | rs1280316 | 1 | 66843725 | Intron | WDR78 | WD repeat domain 78 (1p31.3) | 6.81E−07 | – | – | – | – | – | – | – |
| 3 | rs1093013 | 1 | 75634658 | Intron | SLC44A5 | Solute carrier family 44 member 5 (1p31.1) | – | – | 4.86E−17 | – | – | – | – | – |
| 4 | rs6428792 | 1 | 119114244 | Intron | WARS2 | Tryptophanyl TRNA Synthetase 2, Mitochondrial (1p12) | – | – | 7.95E−18 | – | – | – | – | – |
| 5 | rs2301453 | 1 | 172389027 | Intron | DNM3 | Dynamin 3 (1q24.3) | – | – | 4.38E−17 | – | – | – | – | – |
| 6 | rs4844390 | 1 | 207761504 | Intron | CD46 | CD46 Molecule, Complement Regulatory Protein (1q32.2) | – | – | – | – | – | – | – | 6.90E−07 |
| 7 | rs11118296 | 1 | 219408638 | Intron | LYPLAL1 | Lysophospholipase Like 1 (1q41) | – | – | 3.00E−07 | – | – | – | – | – |
| 8 | rs6713865 | 2 | 23676937 | Intron | KLHL29 | Kelch like family member 29 (2p24.1) | – | – | – | – | – | – | – | 4.39E−13 |
| 9 | rs934778 | 2 | 25166355 | Intron | POMC | Proopiomelanocortin (2p23.3) | 7.15E−07 | – | – | – | – | – | – | |
| 10 | rs1107238 | 2 | 26235376 | Intron | HADHA | Hydroxyacyl-CoA Dehydrogenase/3-Ketoacyl-CoA Thiolase/Enoyl-CoA Hydratase (Trifunctional Protein), Alpha Subunit (2p23.3) | 8.86E−07 | – | – | – | – | – | – | |
| 11 | rs13404446 | 2 | 27296386 | intron | TRIM54 | Tripartite Motif Containing 54 (2p23.3) | – | – | – | 3.09E−09 | – | – | – | – |
| 12 | rs4665965 | 2 | 27313513 | intron | MPV17 | MPV17, Mitochondrial Inner Membrane Protein (2p23.3) | – | – | – | 1.83E−09 | – | – | – | – |
| 13 | rs3736594 | 2 | 27772914 | intron | MRPL33 | Mitochondrial Ribosomal Protein L33 (2p23.2) | – | – | – | 3.02E−13 | 4.67E−07 | – | – | – |
| 14 | rs4346434 | 2 | 43992607 | intron | LRPPRC | Leucine Rich Pentatricopeptide Repeat Containing (2p21) | – | – | 8.52E−08 | – | – | – | – | – |
| 15 | rs16843390 | 2 | 209655109 | intron | MAP2 | Microtubule Associated Protein 2 (2q34) | – | – | – | 3.32E−08 | – | – | – | – |
| 16 | rs715 | 2 | 210678331 | utr-3-prime | CPS1 | Carbamoyl-Phosphate Synthase 1 (2q34) | 5.81E−07 | – | – | – | – | – | – | – |
| 17 | rs933994 | 2 | 218785893 | intron | CYP27A1 | Cytochrome P450 Family 27 Subfamily A Member 1 (2q35) | 8.65E−07 | – | – | – | – | – | – | – |
| 18 | rs17036328 | 3 | 12348985 | intron | PPARG | Peroxisome Proliferator Activated Receptor Gamma (3p25.2) | 4.27E−07 | – | – | – | 3.59E−12 | – | – | – |
| 19 | rs3729931 | 3 | 12585017 | intron | RAF1 | Raf-1 Proto-Oncogene, Serine/Threonine Kinase (3p25.2) | – | – | 3.60E−10 | – | – | – | – | – |
| 20 | rs11715915 | 3 | 49417897 | nc-transcript | AMT | Aminomethyltransferase (3p21.31) | – | – | – | 4.90E−08 | – | – | – | – |
| 21 | rs12489828 | 3 | 52532998 | intron | NT5DC2 | 5′-Nucleotidase Domain Containing 2 (3p21.1) | – | – | 2.60E−10 | – | – | – | – | – |
| 22 | rs2365389 | 3 | 61250788 | intron | FHIT | Fragile Histidine Triad (3p14.2) | 3.75E−15 | – | – | – | – | – | – | – |
| 23 | rs332375 | 3 | 66377079 | intron | SLC25A26 | Solute Carrier Family 25 Member 26 (3p14.1) | – | – | 7.33E−07 | – | – | – | – | – |
| 24 | rs1735536 | 3 | 128377770 | intron | EEFSEC | Eukaryotic Elongation Factor, Selenocysteine-TRNA Specific (3q21.3) | 2.95E−10 | – | – | – | – | – | – | – |
| 25 | rs9844666 | 3 | 136255374 | intron | PCCB | Propionyl-CoA Carboxylase Beta Subunit (3q22.3) | 7.22E−07 | – | – | – | – | – | – | – |
| 26 | rs11924648 | 3 | 171000207 | intron | SLC2A2 | Solute Carrier Family 2 Member 2 (3q26.2) | – | – | – | 1.02E−17 | – | – | – | 4.05E−09 |
| 27 | rs10012946 | 4 | 6291623 | intron | WFS1 | Wolframin ER Transmembrane Glycoprotein (4p16.1) | – | – | – | 4.17E−07 | – | – | – | – |
| 28 | rs10518406 | 4 | 122841742 | intron | FGF2 | Fibroblast Growth Factor 2 (4q28.1) | 6.50E−07 | – | – | – | – | – | – | – |
| 29 | rs1458758 | 4 | 122914724 | intron | NUDT6 | Nudix Hydrolase 6 (4q28.1) | – | – | 6.22E−08 | – | – | – | – | – |
| 30 | rs303084 | 4 | 123145793 | intron | SPATA5 | Spermatogenesis associated 5 (4q28.1) | – | – | 3.40E−07 | – | – | – | – | – |
| 31 | rs12654264 | 5 | 75352778 | intron | HMGCR | 3-Hydroxy-3-Methylglutaryl-CoA Reductase (5q13.3) | 1.81E−08 | – | – | – | – | – | – | – |
| 32 | rs10478424 | 5 | 119453325 | intron | HSD17B4 | Hydroxysteroid 17-Beta Dehydrogenase 4 (5q23.1) | – | – | 1.40E−07 | – | – | – | – | – |
| 33 | rs2881156 | 5 | 135812973 | intron | SLC25A48 | Solute carrier family 25 member 48 (5q31.1) | – | 8.10E−07 | – | – | – | – | – | – |
| 34 | rs3828870 | 6 | 16743066 | intron | ATXN1 | Ataxin 1 (6p22.3) | – | – | – | – | – | – | – | 5.52E−07 |
| 35 | rs1800562 | 6 | 26092913 | intron | HFE | Hemochromatosis (6p22.2) | – | – | – | – | – | – | – | 4.67E−28 |
| 36 | rs1800629 | 6 | 31575254 | upstream-2KB | TNF | Tumor necrosis factor (6p21.33) | – | – | 7.30E−07 | – | – | – | – | – |
| 37 | rs6457796 | 6 | 34860776 | intron | UHRF1BP1 | UHRF1 binding protein 1 (6p21.31) | 1.15E−09 | – | – | – | – | – | – | – |
| 38 | rs10434 | 6 | 43785475 | utr-3-prime | VEGFA | Vascular endothelial growth factor A (6p21.1) | – | – | 8.80E−07 | – | – | – | – | – |
| 39 | rs1049354 | 6 | 88143732 | utr-3-prime | CNR1 | Cannabinoid receptor 1 (6q15) | 9.57E−07 | – | – | – | – | – | – | – |
| 40 | rs9400239 | 6 | 108656460 | intron | FOXO3 | Forkhead box O3 (6q21) | 1.61E−08 | – | – | – | – | – | – | – |
| 41 | rs1273733 | 6 | 121131419 | downstream-500B | TBC1D32 | TBC1 domain family member 32 (6q22.31) | 3.96E−12 | – | – | – | – | – | – | – |
| 42 | rs1049349 | 6 | 121449496 | utr-3-prime | GJA1 | Gap junction protein alpha 1; synonymous: CX43 (6q22.31) | 4.18E−15 | – | – | – | – | – | – | – |
| 43 | rs1293954 | 6 | 151669826 | intron | ESR1 | Estrogen receptor 1 (6q25.1-q25.2) | 4.41E−09 | – | – | – | – | – | – | – |
| 44 | rs1203576 | 7 | 40808233 | intron | SUGCT | C7orf10 (Succinyl-CoA:Glutarate-CoA Transferase, 7p14.1) | 1.48E−10 | – | – | – | – | – | – | – |
| 45 | rs2908289 | 7 | 44184343 | intron | GCK | Glucokinase (7p13) | – | – | – | 3.32E−88 | – | 6.00E−09 | – | 2.24E−19 |
| 46 | rs1088867 | 7 | 44705214 | intron | OGDH | Oxoglutarate dehydrogenase (7p13) | 7.90E−08 | – | – | – | – | – | – | – |
| 47 | rs16892421 | 8 | 106499705 | intron | OXR1 | Oxidation resistance 1 (8q23.1) | – | – | 6.94E−07 | – | – | – | – | – |
| 48 | rs7835803 | 8 | 120030213 | intron | DEPTOR | DEP domain containing MTOR interacting protein (8q24.12) | – | – | – | 4.97E−07 | – | – | – | – |
| 49 | rs2777795 | 9 | 104910084 | intron | ABCA1 | ATP binding cassette subfamily A member 1 (9q31.1) | – | – | 3.13E−08 | – | – | – | – | – |
| 50 | rs7023913 | 9 | 128255683 | downstream-500B | DNM1 | Dynamin 1 (9q34.11) | – | 7.30E−07 | – | – | – | – | – | – |
| 51 | rs3829109 | 9 | 136362314 | intron | DNLZ | DNL-Type Zinc Finger (9q34.3) | – | – | – | 1.13E−10 | – | – | – | – |
| 52 | rs1244497 | 10 | 7838019 | intron | TAF3 | TATA-box binding protein associated factor 3 (10p14) | 1.84E−11 | – | – | – | – | – | – | – |
| 53 | rs5030913 | 10 | 69246375 | intron | HKDC1 | Hexokinase domain containing 1 (10q22.1) | – | – | – | – | – | – | – | 3.56E−13 |
| 54 | rs4745982 | 10 | 69330087 | intron | HK1 | Hexokinase 1 (10q22.1) | – | – | – | – | – | – | – | 2.87E−65 |
| 55 | rs7899106 | 10 | 85651147 | intron | GRID1 | Glutamate Ionotropic Receptor Delta Type Subunit 1 (10q23.2) | 2.96E−08 | – | – | – | – | – | – | – |
| 56 | rs7917772 | 10 | 102727686 | intron | SFXN2 | Sideroflexin 2 (10q24.32) | – | – | 1.45E−09 | – | – | – | – | – |
| 57 | rs1004467 | 10 | 102834750 | intron | CYP17A1 | Cytochrome P450 Family 17 Subfamily A Member 1 (10q24.32) | 1.18E−07 | – | – | – | – | – | – | – |
| 58 | rs3740390 | 10 | 102878723 | intron | AS3MT | Arsenite Methyltransferase (10q24.32) | 4.82E−08 | – | – | – | – | – | – | – |
| 59 | rs4758633 | 11 | 219538 | intron | SIRT3 | Sirtuin 3 (11p15.5) | – | – | – | – | – | – | – | 3.44E−10 |
| 60 | rs757110 | 11 | 17396930 | missense | ABCC8 | ATP Binding Cassette Subfamily C Member 8 (11p15.1) | 4.23E−07 | – | – | – | – | – | – | – |
| 61 | rs10767664 | 11 | 27704439 | intron | BDNF | Brain Derived Neurotrophic Factor (11p14.1) | 5.53E−13 | – | – | – | – | – | – | – |
| 62 | rs11038913 | 11 | 46538180 | intron | AMBRA1 | Autophagy and beclin 1 regulator 1 (11p11.2) | – | – | – | 4.29E−08 | 4.91E−18 | – | – | – |
| 63 | rs11039149 | 11 | 47255124 | intron | NR1H3 | Nuclear Receptor Subfamily 1 Group H Member 3 (11p11.2) | – | – | – | 1.26E−12 | 4.13E−45 | – | – | – |
| 64 | rs7118178 | 11 | 47637583 | intron | MTCH2 | Mitochondrial carrier 2 (11p11.2) | 5.12E−08 | – | – | 3.84E−14 | 2.16E−29 | – | – | – |
| 65 | rs4246215 | 11 | 61796827 | utr-3-prime | FEN1 | Flap Structure-Specific Endonuclease 1 (11p12.2) | – | – | – | 4.46E−11 | – | – | – | – |
| 66 | rs174556 | 11 | 61813163 | intron | FADS1 | Fatty acid desaturase 1 (11q12.2) | – | – | – | 7.82E−18 | – | – | – | – |
| 67 | rs7943191 | 11 | 62561079 | intron | EEF1G | Eukaryotic translation elongation factor 1 gamma (11q12.3) | – | – | 4.36E−08 | – | – | – | – | – |
| 68 | rs11231150 | 11 | 62584330 | intron | TUT1 | Terminal uridylyl transferase 1, U6 snRNA-specific (11q12.3) | – | – | 5.20E−08 | – | – | – | – | – |
| 69 | rs1017639 | 11 | 68831066 | intron | CPT1A | Carnitine Palmitoyltransferase 1A (11q13.3) | 4.96E−10 | – | – | – | – | – | – | – |
| 70 | rs1296252 | 11 | 83273268 | intron | CCDC90B | Coiled-Coil Domain Containing 90B (11q13.3) | 7.60E−08 | – | – | – | – | – | – | – |
| 71 | rs2110073 | 12 | 6966719 | intron | PHB2 | Prohibitin 2 (12p13.31) | – | – | – | – | – | – | – | 4.44E−08 |
| 72 | rs7311050 | 12 | 7013532 | intron | LPCAT3 | Lysophosphatidylcholine Acyltransferase 3 (12p13.31) | – | – | – | – | – | – | – | 8.60E−08 |
| 73 | rs1049380 | 12 | 26336611 | downstream-500B | ITPR2 | Inositol 1,4,5-Trisphosphate Receptor Type 2 (12p11.23) | – | – | 1.42E−13 | – | – | – | – | – |
| 74 | rs2408955 | 12 | 48105348 | upstream-2KB | PFKM | Phosphofructokinase, muscle (12q13.11) | – | – | – | – | – | – | – | 1.42E−15 |
| 75 | rs35767 | 12 | 102481791 | missense | IGF1 | Insulin Like Growth Factor 1 (12q23.2) | – | – | – | – | 7.27E−08 | – | 7.57E−08 | – |
| 76 | rs4766578 | 12 | 111466567 | intron | ATXN2 | Ataxin 2 (12q24.12) | 2.85E−07 | – | – | – | – | – | – | 1.84E−07 |
| 77 | rs9581856 | 13 | 27451478 | upstream-2KB | MTIF3 | Mitochondrial Translational Initiation Factor 3 (13q12.2) | 1.03E−08 | – | – | – | – | – | – | – |
| 78 | rs1124607 | 13 | 27921083 | intron | PDX1 | Pancreatic And Duodenal Homeobox 1 (13q12.2) | – | – | – | 4.49E−07 | – | – | – | – |
| 79 | rs1325363 | 13 | 33192439 | intron | STARD13 | StAR related lipid transfer domain containing 13 (13q13.1-q13.2) | 7.25E−08 | – | – | – | – | – | – | – |
| 80 | rs1078892 | 13 | 40563883 | intron | FOXO1 | Forkhead Box O1 (13q14.11) | 5.11E−08 | – | – | – | – | – | – | – |
| 81 | rs7143963 | 14 | 102838088 | intron | TRAF3 | TNF Receptor Associated Factor 3 | 2.82E−07 | – | – | – | – | – | – | – |
| 82 | rs12908437 | 15 | 98744146 | intron | IGF1R | Insulin Like Growth Factor 1 Receptor | – | – | – | 6.32E−07 | – | – | – | – |
| 83 | rs740862 | 16 | 3639677 | intron | DNASE1 | Deoxyribonuclease 1 | 3.21E−07 | – | – | – | – | – | – | – |
| 84 | rs151181 | 16 | 28479196 | intron | CLN3 | CLN3, Battenin | 2.10E−07 | – | – | – | – | – | – | – |
| 85 | rs8055138 | 16 | 28880144 | intron | ATP2A1 | ATPase Sarcoplasmic/Endoplasmic Reticulum Ca2+ Transporting 1 | 8.17E−17 | – | – | – | – | – | – | – |
| 86 | rs749767 | 16 | 31113086 | downstream-500B | BCKDK | Branched Chain Ketoacid Dehydrogenase Kinase | 1.21E−09 | – | – | – | – | – | – | – |
| 87 | rs7186084 | 16 | 68782357 | intron | CDH1 | Cadherin 1 | – | – | – | – | – | – | – | 1.09E−07 |
| 88 | rs1847591 | 16 | 78908913 | intron | WWOX | WW domain containing oxidoreductase | – | – | 9.99E−07 | – | – | – | – | – |
| 89 | rs9904685 | 17 | 1352101 | intron | YWHAE | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein epsilon | 6.27E−07 | – | – | – | – | – | – | – |
| 90 | rs4646404 | 17 | 17516885 | intron | PEMT | Phosphatidylethanolamine N-Methyltransferase | – | – | 5.30E−11 | – | – | – | – | – |
| 91 | rs9914988 | 17 | 28856086 | intron | ERAL1 | Era like 12S mitochondrial rRNA chaperone 1 | – | – | – | – | – | – | – | 2.77E−11 |
| 92 | rs242559 | 17 | 45948522 | intron | MAPT | Microtubule Associated Protein Tau | – | – | – | 8.29E−07 | – | – | – | – |
| 93 | rs1319247 | 17 | 63106279 | intron | TANC2 | Etratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 | – | – | 6.02E−08 | – | – | – | – | – |
| 94 | rs12940622 | 17 | 80641771 | intron | RPTOR | Regulatory Associated Protein Of MTOR Complex 1 | 2.49E−09 | – | – | – | – | – | – | – |
| 95 | rs1044661 | 17 | 82943144 | intron | B3GNTL1/TBCD | UDP-GlcNAc:BetaGal Beta-1,3-N-Acetylglucosaminyltransferase Like 1/Tubulin Folding CofactorD | – | – | – | – | – | 1.74E−46 | ||
| 96 | rs1788785 | 18 | 23562376 | intron | NPC1 | NPC Intracellular Cholesterol Transporter 1 | 1.98E−08 | – | – | – | – | – | – | – |
| 97 | rs17066842 | 18 | 60373391 | upstream-2KB | MC4R | Melanocortin 4 Receptor | 6.40E−14 | – | – | – | – | – | – | – |
| 98 | rs12454712 | 18 | 63178651 | intron | BCL2 | BCL2, Apoptosis Regulator | – | – | 1.10E−09 | – | 1.39E−07 | – | – | – |
| 99 | rs757318 | 19 | 18709498 | intron | CRTC1 | CREB Regulated Transcription Coactivator 1 | 8.76E−09 | – | – | – | – | – | – | – |
| 100 | rs2075650 | 19 | 44892362 | intron | TOMM40 | Translocase Of Outer Mitochondrial Membrane 40 | 1.25E−08 | – | – | – | – | – | – | – |
| 101 | rs405509 | 19 | 44905579 | upstream-2 kb | APOE | Apolipoprotein E | 2.65E−07 | – | – | – | – | – | – | – |
| 102 | rs2281361 | 20 | 32140338 | intron | TM9SF4 | ransmembrane 9 superfamily member 4 | 9.78E−07 | – | – | – | – | – | – | – |
| 103 | rs878639 | 20 | 35306660 | intron | UQCC1 | Ubiquinol-Cytochrome C Reductase Complex Assembly Factor 1 | – | – | 1.50E−11 | – | – | – | – | – |
| 104 | rs2076574 | 20 | 41092733 | intron | TOP1 | DNA topoisomerase I | – | – | 4.11E−07 | – | – | – | – | – |
| 105 | rs5750373 | 22 | 37028990 | intron | MPST | Mercaptopyruvate Sulfurtransferase | – | – | – | – | – | – | – | 2.17E−07 |
| 106 | rs2284099 | 22 | 43155830 | intron | TSPO | Translocator Protein | – | – | 6.65E−07 | – | – | – | – | – |
| 107 | rs1050828 | 23 | 154536002 | missense | G6PD | Glucose-6-phosphate dehydrogenase | – | – | – | – | – | – | – | 8.23E−135 |
| 108 | rs1448032 | 23 | 155052530 | intron | FUNDC2 | FUN14 domain containing 2 | – | – | – | – | – | – | – | 5.92E−17 |
| 109 | rs5940514 | 23 | 155559972 | intron | TMLHE | Trimethyllysine hydroxylase, epsilon | – | – | – | – | – | – | – | 2.22E−19 |
p values in italics annotate SNVs that associate as significant with more than one trait.
To identify the 588 SNVs, a pre-specified selection process was followed. First, we downloaded the latest dbSNP of NCBI reference data (batch 150), and we assigned each SNV to a gene (pseudogenes excluded). The intergenic variants were assigned to the closest gene, up to half the distance between two genes. Then, we merged the 31 full-GWAS sets to the annotated dbSNP, thus each GWAS-SNP was assigned to a gene. Each summary result was merged with MT-nDNA gene labels (Table S30). The corresponding significant SNVs results, ones with the smallest p value, per gene and per trait, were accepted in the final list (p < 0.05/(2,282 × 7) = 3.13E−06). After this selection, we also performed an analysis based on 1000 Genomes to identify the number of independent SNVs within a gene.36, 62 The analysis produced a mean of independent SNVs per gene of 59, median of 38, and with a maximum of two outliers 542 (WWOX [MIM: 605131]) and 499 (FHIT [MIM: 601153]). If it was a gene-based test, then a conservative threshold p ≤ 0.05/(542 × 7 traits) ≤ 1.3E−05 could have been used, which is larger than the threshold we used, p ≤ 1E−06). Furthermore, to compare whether our MT-nDNA genes pass the genome-wide threshold of p ≤ 5E−08, generally used in GWAS publications, we merged our gene data with all possible reported SNVs from the GWAS-Catalog (accessed 01.27.2018, Table S12 and presented findings in the Results and Discussion paragraphs).
Annotation, Enrichment Analysis, and Gene Expression and Regulation
The mtDNA as well as MT-nDNA significant variants were annotated to NCBI dbSNP build 150 (HG38); genes and their protein biological functions were annotated to NCBI Entrez Gene, GeneCards and UniProtKB; enrichment analyses were performed with MetaCore and Literature Lab; pathways with KEGG and Reactome; gene expression and regulation were assessed using HaploReg, RegulomeDB, GTEx, and Human Protein Atlas; and protein interactions were assessed using STRING and NCBI summary of interactions from other databases (see Web Resources), with references to databases BIND, BioGRID, EcoCyc, HIV-1-human protein interaction data, and HPRD. Specifically, for GTEx gene expression eQTL analysis, the eSNVs were considered significant when the eSNV had a GTEx p < 1E−07 and was in high LD (r2 ≥ 0.80) with the best eSNV of the target gene. Depending on r2 ≥ 0.80 to 1, we called them similar to “lead” or “lead” eSNVs. Otherwise, if the LD r2 was < 0.02 to the target gene’s best eSNV, we called them “secondary” eSNVs (Table S8). Details of the resources and the corresponding references are provided in the Supplemental Material and Methods, sections 8 and 9. We have cited throughout the manuscript the corresponding gene MIM number from the Online Mendelian Inheritance in Man (see Web Resources).
Results
We evaluated the associations of mtDNA variants with seven key metabolic traits in meta-analyses of 45 cohorts (with up to N∼170,202). For details on the harmonization of the phenotypes and genotypes, see Material and Methods and Supplemental Material and Methods, sections 1–6. The Supplemental Study Descriptions and Table S1 with BMI mean and standard deviation values give a depiction of each contributing cohort in this study.
mtDNA Single-Variants Associations
Seven SNVs, two variants with average weighted MAF > 1% (Tables 1 and S2.a and Figures S1–S7) and five with MAF < 1% (Tables 1 and S2.b) displayed statistically significant evidence of association with six metabolic traits (Bonferroni threshold p ≤ 5E−04, see in Material and Methods: mtDNA Meta-Analyses). MT-8706 in MT-ATP6 (MIM: 516060) (see Web Resources), associated with WHR (with sample weighted average of cohorts’ MAF = 3.24%, p = 4.1E−04) and MT-16320 (rs62581338) of the D-loop (with sample weighted average of cohorts’ MAF = 1.58%, p = 7.6E−05) associated with fasting glucose. The five rare variants were MT-8896 (rs202120082, missense) in MT-ATP6 associated with WHR, MT-14124 in MT-ND5 (MIM: 516005) associated with fasting plasma insulin, MT-14272 in MT-ND6 (rs2853814 [MIM: 516006], missense) associated with BMI, MT-14353 in MT-ND6 associated with WHR, and MT-14584 in ND6 associated with BMI.
The evidence of association by cohort is reported in Tables S2.a, S2.b, and S3. Typically, the cohort with a large sample size displayed the strongest evidence of association. For example, the HCHS/SOL study contributed disproportionately to several mtDNA-trait associations (see in the Supplemental Study Descriptions, CHARGEmtDNA+ Study Description). Even further, for the MT-ATP6 (position 8706) association with WHR, the strongest association in HCHS/SOL was in those of Central/South American background (so more Native American ancestry as compared to African American ancestry, Tables S2.a and S3).
mtDNA Rare Variants Gene-Based Associations
The mitochondrial rare variants for gene-based analysis were mapped to the start and end positions of each gene from the NCBI Reference Sequence GenBank: NC_012920.1 (see Web Resources). The rare variant gene-based meta-analysis using SKAT did not yield any significant associations. In contrast, the Burden test yielded a significant association between rare variants in MT-ND3 and BMI (p = 1E−03, T < 0.05 including 82 SNVs). A forest plot representing the 82 SNVs and the overall MT-ND3-overall meta is shown in Figure S17. Several gene- HOMA-B, HbA1c, and BMI associations were suggestively significant employing both the SKAT and Burden approaches (MT-RNAs MT-TF [MIM: 590070], MT-TV [MIM: 590105], MT-TG [MIM: 590035], MT-TQ [MIM: 590030], and MT-RNR1 [MIM: 561000]; p ≤ 1E−02; Tables 2 and 3). A total of 131 (T5-test) and 123 (T1-test) low-frequency and/or rare variants contributed to T5-test and T1-test, respectively. We used two gene-based approaches, burden test and sequence kernel association test (SKAT), to evaluate the association between a gene and a phenotype. Burden tests access the cumulative effects of multiple variants by assuming that all variants have the same directionality in a genetic region. SKAT tests use a score-based variance component framework without assuming that all variants have the same directionality. Although the two methods are different, they all evaluate aggregate effects of multiple low-frequency and/or rare variants with a phenotype. A significant gene-test reflects an aggregate effect of multiple variants in the same area. If most variants in ND6 are not associated with the phenotype, the gene-based test might not be significant. Although statistical significance was not achieved, the association of MT-ND6 with BMI was suggestive in the burden (p = 0.06 [T5] and 0.04 [T1]) and SKAT (p = 0.15 [T5] and 0.08 [T1] tests).
Identification of MT-nDNA Candidate Variants Associated with Metabolic Traits
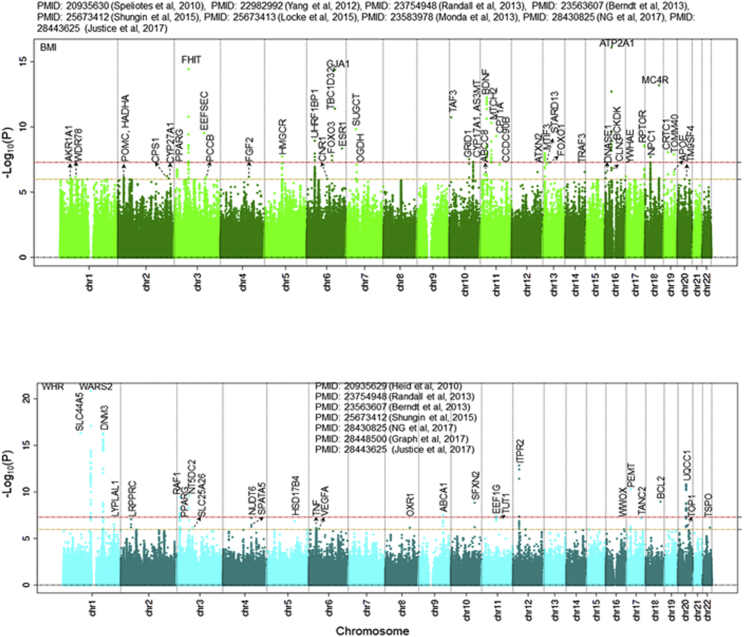
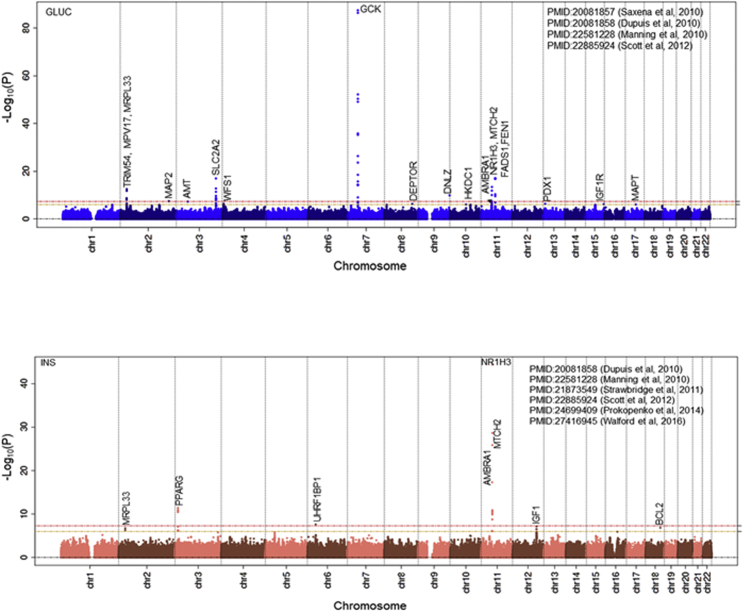
We identified 2,282 MT-nDNA candidate genes (see Material and Methods) and assessed their association from 20 GWASs with 31 datasets (Table S4). From the MT-nDNA candidate list, 109 genes reached statistical significance following correction for multiple testing (Bonferroni p < 1E−06) of which 46 were associated with BMI, 2 with extreme obesity, 26 with WHR, 18 with glucose, 7 with insulin, 1 with HOMA-B, 1 with HOMA-IR, and 20 with HbA1c, totaling 121 associations (Table 4). The use of additional SNVs belonging to the same 109 MT-nDNA genes, but now sourced from the GWAS catalog for the same 7 traits or any other traits, indicated that 84% (27% improvement) of MT-nDNA genes passed p ≤ 5E−08, while 16% remained at p ≤ 1E−06 without passing the p ≤ 5E−08 threshold. Of the MT-nDNA associations, GCK (MIM: 138079), MRPL33 (MIM: 610059), PPARG (MIM: 601487), SLC2A2 (MIM: 138160), AMBRA1 (MIM: 611359), NR1H3 (MIM: 602423), MTCH2 (MIM: 613221), IGF1 (MIM: 147440), and BCL2 (MIM: 151430) were associated with more than one trait (Figures 2, 3, S8, and S9). For example, the well-known GCK is a member of hexokinases that phosphorylates glucose to produce glucose-6-phosphate, the first step in most glucose metabolism pathways, and has pleiotropic effects (see Discussion).
Figure 2.
BMI and WHR Association Results with MT-nDNA Candidate Genes
Figure 3.
Glucose and Insulin Association Results with MT-nDNA Candidate Genes
Enrichment Analysis of MT-nDNA Candidate Genes
The 109 MT-nDNA selected genes are candidates for MT function based on their protein localization in mitochondria, as well as from mining the published literature (Table S5). We used Literature Lab and MetaCore software and the corresponding databases to perform enrichment analyses, which provided information for MT-nDNA gene-label relations with terms, pathways, diseases, gene ontology processes, and clustering (see Supplemental Material and Methods, section 8 and Figure S10). When comparing the 109 MT-nDNA candidate genes versus the remaining 2,173 (2,282 − 109), the 109 set showed enrichment (p = 1.7E−12) for “Signal transduction, Neuropeptide signaling pathways,” which included, among others, POMC (MIM: 176830) and MC4R (MIM: 155541), while no such enrichment was found for the 2,173 set. Out of 109 MT-nDNA candidate genes, 21 of the significantly associated genes were functionally related with cholesterol (Table S6 and Figure S11), 13 with glucose and insulin, and 5 with adipose/obesity. Of the 109 MT-nDNA genes, 13 associated with the “Type 2 diabetes”54, 61, 63 term (Table S7) while 18 were associated with “Cardiovascular disease”64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77 (Table S7). (For space limitation, a detailed enrichment analysis is provided in the Supplemental Material and Methods, section 8.) These findings demonstrate the importance of several MT-nDNA candidates to cardiometabolic outcomes.
eQTLs of MT-nDNA Candidate Variants
Several variants in or near MT-nDNA candidate genes were identified as expression QTL (eSNV) (Table S8). Based on RegulomeDB,78 three variants were the best in eSNVs features’ ranking. The first was rs242559, intronic to MAPT (MIM: 157140). Mutations in MAPT associate with lower mitochondrial nicotinamide adenine dinucleotide (NADH) levels,79 partially suppress complex I-driven respiration, and lower overall ATP production by oxidative phosphorylation, with cells relying on glycolysis to maintain ATP levels. The second, rs9897919, is a 3′ UTR variant for TBCD (MIM: 604649), tubulin folding cofactor D, and B3GNTL1 (17q25.3 [MIM: 615337]), a putative glycosyltransferase. The third, rs1788821, is intronic to NPC1 (MIM: 607623), which is an intracellular cholesterol transporter with a role in the egress of cholesterol from the endosomal/lysosomal compartments (Table S9).
The findings of RegulomeDB were reinforced by HaploReg (v.4.1).80 The MT-nDNA variants showed an enrichment in transcription regulation features. For instance, rs242559 of MAPT is localized within the promoter histone marks in skeletal muscle, at enhancer histone marks of 16 tissues, and at DNase marks of 4 tissues. In addition, the rs242559 polymorphism alters the protein binding site of GATA2, a transcription factor protein that binds in the promoter regions of target genes (Table S9).
To determine the eSNVs’ gene targets in specific tissues, we used GTEx (v.7.0)81, 82 with a summary in Table S8 and detailed in Table S10. The 42 unique eSNVs were assigned to 29 genes targeting regulation of 50 genes, distributed among 13 tissues (adipose, tibial artery, thyroid, skin, blood, brain, skeletal muscle, esophageal muscularis, fibroblast, liver, pancreas, testis, and tibial nerve). There were 28 unique lead and 18 unique secondary (LD r2 was < 0.02 to the target gene’s best eSNV) eSNVs identified (see Material and Methods). For example, rs2510344 of NPC1 regulates C18orf8 in skin and rs11663558 of NPC1 regulates its own NPC1 gene expressed highest in subcutaneous adipose tissue (GTEx data).
trans-eQTLs of MT-nDNA Candidate Variants
We combined the GWAS p value for the MT-nDNA SNVs with additional evidence from trans-gene expression regulation for a specific variant using GWAS3D83 (Figures S12.1–S12.3). The GWAS3D software selected 16 cell types, which included chromosomal looping data (5C or ChIA-PET or Hi-C) and important transcriptional marker data (H3K4me1, H3K27ac, DHSs, EP300, and CTCF).83 Among several trans-eQTLs, for example, rs2881156 (p = 8.1E−07) of SLC25A48 (5q31.1 [MIM: 616150], Figure S12.1) associated with obesity and trans-regulated expression of three genes: SAR1B (5q31.1 [MIM: 607690]) involved in protein transport from the endoplasmic reticulum to the Golgi (mutations in this gene are a cause of chylomicron retention disease [MIM: 246700]);84 TRPC7 (5q31.1), a regulator of intracellular calcium levels;85 and REEP2 (5q31.2 [MIM: 609347]), which enhances the function of sweet taste receptors86 and is about 80 times higher expressed in brain than in other tissues (GTEx data).
PPI Network
We analyzed 109 MT-nDNA proteins using the PPI network (see Web Resources)87, 88 to identify 4,132 interacting proteins. We present the 15 top genes (Table S11) with highest PageRank score for PPI,89, 90, 91 including the number of PPI, a short description of gene’s function from Gene Entrez of NCBI-db, associated trait(s), and association p value(s) (see also Supplemental Material and Methods, section 9). From the PPI analysis it is evident that a gene/protein hub (which is assumed important because of a relatively large number of interactions) is not necessary, the top-notch for association with a specific trait, as shown in the Discussion section.
Discussion
This large, comprehensive study of human mitochondrial and nuclear mitochondrial genetic variation in relation to seven key risk factors for metabolic disease provides important information toward a better understanding of the causes and mechanisms of these phenotypes. The use of a two-tiered approach—i.e., examining mtDNA variants for up to ∼170,202 individuals and nuclear MT-nDNA candidate gene polymorphisms for up to ∼339,000 individuals—facilitated the evaluation of the genetic underpinnings of seven metabolic traits reflecting adiposity/obesity and glucose-insulin metabolism and signaling. We identified two mtDNA SNVs (in MT-ATP6 and the D-loop) associated with WHR and fasting glucose, a burden of SNVs in a gene (MT-ND3) associated with BMI, and five rare SNVs of mtDNA (in MT-ATP6, MT-ND5, and MT-ND6) associated with BMI (2 of 3), with WHR (2 of 3), and with fasting insulin (1 of 3). Additionally, the MT-nDNA meta-analysis implicated significant associations with 62 of 109 protein coding candidate genes (p ≤ 5E−08), 29 additional passing the above threshold for the metabolic traits considered herein or any other traits querying the GWAS catalog (see Web Resources). Only 16% of MT-nDNA findings passed p ≤ 1E−06 but did not reach p ≤ 5E−08, and thus may be considered as new MT-nDNA contributions in association with seven metabolic traits (Tables 4 and S12). Enrichment analysis showed that 13 of MT-nDNA genes have also been identified as candidate genes for T2D risk and 18 for CVD risk. The eQTL analysis of MT-nDNA candidates revealed that approximately 27% of associations with seven metabolic traits also act as eSNVs in regulating expression of other cis-genes. In addition, trans-eQTL analysis yielded potential support for maternal and X chromosome epigenetic gene expression regulation. The PPI network analysis identified top PageRank-ed interacting genes from our significant MT-nDNA associations, but they did not necessarily represent the highest trait-specific associations. Overall, these findings indicate the important role of both the nuclear genome and mtDNA-related variants in energy production and, in turn, in the polygenic architecture of metabolic traits. Below we summarize and discuss our most salient study findings.
mtDNA and Metabolic Disease Associations
To date, nearly 290 genetic causes for rare disorders of mitochondrial energy generation have been identified.92 These rare mitochondrial genetic diseases commonly result in multiple clinical phenotypes with varying penetrance, likely due to differences in heteroplasmy.93 In our study, MT-ATP6 variants MT-8706 and MT-8896 (rs202120082), one with MAF > 1% and the other one rare, respectively, were both associated with WHR. MT-ATP6 is part of the proton-transporting portion of ATP synthase, contributing to its rotational mechanism.94, 95 rs202120082 (MT-8896) is a missense mutation (p.Ala124Thr) and conversion of the hydrophobic alanine to an uncharged, polar threonine at this site may have implications for ATP6’s ability to effectively pump protons and thus decrease production of ATP energy units.
MT-16320 (rs62581338; MAF > 1%), in the D-loop, which contains a few mtDNA replication origins and is possibly involved in accelerating mtDNA synthesis to satisfy developmental, physiological, or aging-related demands,96 displayed a significant association with glucose. MT-16320 is less than 1 kb from a major mtDNA transcription initiation site (IT) that overlaps with promoters (in the D-loop in H-strand, ITH1 at 561 bps and ITH2 at 630, and in L-strand at 407 bps (ITL1))97 and is 500 bps downstream of mitochondrial-encoded cytochrome b (MT-CYB [MIM: 516020], plus strand) and 2 kb upstream of MT-ND6 (minus strand).
Of the five rare SNVs, MT-14272 (rs2853814, missense), MT-14353, and MT-14584 of MT-ND6 were associated with adiposity-related traits. MT-ND6 is a core subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (complex I) and is part of the minimal assembly required for catalysis. Complex I functions in the transfer of electrons from NADH into the respiratory chain, while coupling the flow of electrons to the pumping of protons. We also demonstrated an association between insulin and rare variant MT-14124 in the closely related MT-ND5 gene. MT-ND5 is also a core subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (complex I). Future studies need to validate these rare variant mtDNA findings.
It is also important to note that we observed several rare gene-based suggestive associations between our metabolic traits (p ≤ 5E−02, Tables 2 and 3) and non-protein coding genes, primarily tRNAs. Mutations within tRNAs that impact translation would impact all of the mtDNA-encoded peptides and consequently would be expected to have significant effects on oxidative phosphorylation. The MT-ND3 gene displayed an aggregate of low-frequency variants associated with BMI. MT-ND3 is a subunit of the respiratory chain complex I that is part of the minimal assembly of core proteins required to catalyze NADH dehydrogenation and electron transfer to ubiquinone (coenzyme Q10). Interestingly, previous studies have demonstrated increased MT-ND3 gene expression associated with a higher histological severity of hepatic steatosis.98 Gene-based meta-analysis burden test, which employed inverse variance weighting to combine association results from 82 variants in MT-ND3 gene across cohorts, yielded a significant association between MT-ND3 and BMI (p = 1E−03, Figure S17). This figure illustrates that rare alleles of 82 SNVs contributing in the MT-ND3 meta-analysis have variable effects.
We used GTEx data for mtDNA genes as assembled by the Human Protein Atlas team (see Web Resources). They showed the highest expression of RNA for the MT-ATP6, MT-ND3, MT-ND5, and MT-ND6 significant mtDNA genes in the heart and brain, which represent the highest energy-demanding tissues as compared, for example, to adipose tissue, in a ratio of about 3:1 (Figure S13). The importance of MT-nDNA and mtDNA gene polymorphisms in the heart is supported by recent publications that have summarized the relation of MT to vascular function,99 as therapeutic targets in heart failure,100 and specific genes and pathways that relate to the heart.101, 102
MT-nDNA Candidate Genes Associations (Glycemic Traits)
Glucose sensing in β cells is largely controlled by the hexokinase proteins. Three of the genes encoding hexokinases, HK1 (MIM: 142600), HKDC1 (MIM: 617221), and GCK, were part of our MT-nDNA candidate list and were found to be significantly associated with glucose, HOMA-B, and/or HbA1c (Table 4). Hexokinases catalyze the conversion of glucose into glucose-6-phosphate. The phosphorylation of glucose directly couples extra-mitochondrial glycolysis to intra-mitochondrial oxidative phosphorylation. GCK, for example, with pleiotropic effects, produces an enzyme in the pancreas which plays a role in glucose-stimulated insulin secretion and affects glucose uptake and conversion to glycogen in the liver. The GCK GWAS-identified variant rs2908289 (Table 4) has been previously associated with glucose, HOMA-B, HbA1c,54, 60 and other variants in LD with it, more recently associated with BMI (rs4607517, r2 = 0.65, p = 8E−56),55 with T2D (rs1799884, r2 = 0.81, p = 5E−18),103 and with metabolic syndrome (rs3757840, r2 = 0.18, p = 4E−13).104 Mitochondrial metabolism, which drives the respiratory chain to produce ATP via oxidative phosphorylation, also contributes to glucose sensing, since disruption of mitochondrial oxidative metabolism blocks glucose-stimulated insulin secretion.40, 105, 106, 107 Our analysis shows that 13 additional MT-nDNA candidate genes are annotated with SNVs that associate with glucose/insulin metabolism (Figure S11). Diabetes mellitus has been associated with maternally inherited mutations in MT-TK (MIM: 590060), MT-TS2 (MIM: 590085), and MT-TE (MIM: 590025), where the molecular mechanisms involve impaired translation of mtDNA-encoded proteins.3 In addition, MT-nDNA candidate genes POLG (15q26.1 [MIM: 174763], DNA polymerase gamma); RRM2B (8q22.3 [MIM: 604712], a ribonucleotide reductase); OPA1 (3q29 [MIM: 605290], a nuclear-encoded mitochondrial protein with similarity to dynamin-related GTPases); and MPV17 (2p23.3 [MIM: 137960], a mitochondrial inner membrane protein) have been associated with diabetes mellitus, largely due to an impairment in mtDNA maintenance.108 In our study, rs1050828, a missense mutation of G6PD (Xq28 [MIM: 305900], 154536002 bps; p = 8.2E−135) was associated with HbA1c. G6PD catalyzes the rate-limiting step of oxidative pentose-phosphate pathways, a route for dissimilation of carbohydrates besides glycolysis. It contributes to the production of NADPH and pentose phosphatases for fatty acid and nucleic acid synthesis (Table S6 and Figure S14). G6PD plays an essential role in maintaining health by protecting against oxidative damage.109, 110 The same rs1050828 variant of G6PD has previously been reported as associated with HbA1c lowering in African Americans but not in other populations, leading to misdiagnosis/under-diagnosis of T2D.61 In the last century, a number of papers reported a maternal excess transmission of T2D,111, 112, 113 suggesting the possibility of an epigenetic transmission of diabetes mediated by the mother. To date, two genes, KLF14 (7q32.2 [MIM: 609393])114 and KCNQ1 (11p15.5-p15.4 [MIM: 607542]),115, 116 have been described as potential candidates that show parent-of-origin effects in association with T2D. However, based on our MT-nDNA list, these two genes are not MT-nDNA candidates. In our study, GWAS3D analysis identified G6PD as trans-regulated with the expression of MECP2 (Xq28 [MIM: 300005]) (Figure S12.3).117 MECP2 binds to methylated DNA and mediates transcriptional repression through interaction with histone deacetylase and the corepressor SIN3A (15q24.2 [MIM: 607776]) (Figure S14). Lai et al.118 reported meta-results that T2D-affected case subjects with G6PD deficiency had two times higher odds of developing diabetes than unaffected control subjects.119 Furthermore, G6PD protein has a weak score of 1 out of 5 to be found in MT. G6PD is mainly localized in the cytosol and also to the microtubule-organizing center and vesicles (Human Protein Atlas). G6PD protein has 69 interactions and was ranked 65th by the PageRank algorithm for the importance of PPI. These results taken together may suggest that G6PD contributes to the maternal epigenetics of T2D, through X chromosome inheritance and less through MT. Other candidates of MT-nDNA genes that may contribute to mitochondrial epigenetics120 are DNMT1 (19p13.2 [MIM: 126375], DNA methyltransferase 1), POLRMT (19p13.3 [MIM: 601778], RNA polymerase mitochondrial), TFB1M (6q25.3 [MIM: 607033], transcription factor B1, mitochondrial), TFB2M (1q44 [MIM: 607055], transcription factor B2, mitochondrial), and TFAM (10q21.1 [MIM: 600438], transcription factor A, mitochondrial), but showed no significant associations with existing data for the traits studied (for BMI: rs6926853, TFB1M, p = 1.3E−03;45 WHR: rs10465617, TFB2M, p = 3.1E−03;43 glucose: rs4804124, DNMT1, p = 7.3E−05;54 insulin: rs7253062, DNMT1, p = 4.4E−03;55 HOMA-B: rs12462004, DNMT1, p = 4.4E−03;54 HOMA-IR: rs892189, DNMT1, p = 7E−04;54 HbA1c: rs11006132, TFAM, p = 2.4E−0361).
MT-nDNA Candidate Genes Associations (Adipose Traits)
We found that a total of 26 of the 109 MT-nDNA candidate genes showed significant associations with WHR (Tables 4 and S6 and Figure S11). For example, WARS2 (1p12 [MIM: 604733]) encodes the MT tryptophanyl-tRNA synthase. We observed an intronic rs6428792 of WARS2 in association with WHR (p = 7.95E−18).50 Another SNV, rs10923724 in LD with rs6428792, r2 = 0.29, WHR p = 9E−25)52 upstream of WARS2 but closer to downstream of TBX15 (1p12 [MIM: 604127]), modifies WARS2 expression in skeletal muscle (p = 1.1E-36) and in adipose-subcutaneous tissues (p = 1E−s29) (GTEx data). In our PPI analysis of 109 MT-nDNA genes and 4,132 interactants, WARS2 showed two interactions and was ranked 2,028th based on the PageRank algorithm. Taken together, these findings (our study, GTEx, and GWAS) suggest that associations with WHR might be mediated also by differential expression of WARS2.121
There were 48 MT-nDNA candidates associating with BMI. For example, NPC1 (18q11.2, Tables 4 and S6) has been associated with early childhood onset and adult morbid obesity.122 NPC1 (a cholesterol transporter) is highly expressed in human white adipose tissue adipocytes with increased levels in obese individuals.123 Our lead SNV rs1788785 is an eSNV for NPC1 and is not in LD with the best NPC1 eSNV, supporting NPC1 as a candidate gene for obesity. Indeed, studies in mice have shown that a non-functioning NPC1 resulted in late-onset weight loss and less food intake.124 In humans, NPC1 is part of the cholesterol metabolism pathway (see Web Resources). Recently, NPC1 mutant cells were reported to have fragmented mitochondrial networks, increased respiration, alterations in the composition of the respiratory chain complex, and a substantial reduction in the cellular ATP level. Thus, a primary lysosomal defect in NPC1 mutant fibroblasts is accompanied by deregulation of the organization and function of the mitochondrial network.125 NPC1 was ranked 241st and had 13 interactions in the PPI network.
Other notable obesity-related variants/genes among MT-nDNA candidates include POMC (2p23.3, adrenocorticotropic peptide/hormone, p = 7.2E−07), which binds to melanocortin 2 receptor (MC2R, 18p11.21 [MIM: 607397]), stimulating release of cortisol, a steroid stress-response hormone. MC4R (18q21.32, p = 6.4E−14) was ranked the 3rd in BMI-effects compared to FTO (16q12.2 [MIM: 610966]), the top ranked for BMI effects in Speliotes et al.,41 Locke et al.,46 and Winkler et al.48 MC4R is another member of the melanocortin receptor family, which has a central role in energy homeostasis and somatic growth. MC4R is ranked the 82nd, with 12 PPI, when analyzed for the importance of PPI using the PageRank algorithm. CNR1 (6q15 [MIM: 114610], cannabinoid receptor, p = 9.6E−07) influences mitochondrial respiration. CRTC1 (19p13.11 [MIM: 607536], CREB regulated transcription coactivator 1, p = 8.8E−09) is a potent coactivator of PGC1a (4p15.2, officially known as PPARGC1A [MIM: 604517]), a transcriptional coactivator that regulates the genes involved in energy metabolism, and inducer of mitochondrial biogenesis.
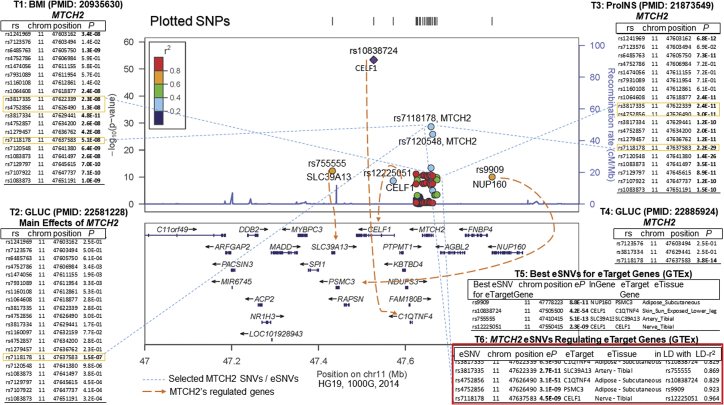
MTCH2 (11p11.2, mitochondrial carrier homolog 2, rs7118178, BMI p = 5.1E−08) was 9th in BMI effects compared to FTO in Speliotes et al.41 and 16th in Locke et al.46 MTCH2 is located in the inner membrane of MT, highly expressed in white adipose tissue and adipocytes, and is thought to play a regulatory role in adipocyte differentiation and biology.126 The MTCH2 GWAS-variant rs7118178 (Table 4) has been associated with BMI,46 glucose,36, 55 and pro-insulin58 (Figure 4, T1-4). MTCH2 has 87 protein interactions and ranked 14th when analyzed for the importance of PPI using PageRank algorithm. MTCH2 deficiency in mouse muscle has been shown to be beneficial, protecting mice from the obesogenic effect of high-fat diets, most likely the result of an increase in mitochondrial metabolism. MTCH2 is proposed as a repressor of muscle mitochondrial metabolism and size.127 Using both GWAS and eQTL information, three MTCH2 SNVs co-localize within a 15 kbp DNA region. Based on GTEx data, rs4752856 in MTCH2 (Figure 4, T6) regulates gene expression of C1QTNF4 (11p11.2 [MIM: 614911]) and PSMC3 (11p11.2 [MIM: 186852]) in subcutaneous adipose tissue. rs3817335, which is in high LD (r2 = 0.903) with rs4752856, regulates SLC39A13 (11p11.2 [MIM: 608735]) in tibial artery tissue, while rs7118178 (in less LD to two other SNVs, r2 = 0.083) regulates CELF1 (11p11.2 [MIM: 601074]) in tibial nerve tissue, and with results similar to the best eSNVs for the mentioned genes and in high LD with their eSNVs (Figure 4, T5). C1QTNF4 is reported as a potential cytokine that can induce the activation of both NFKB1 (MIM: 164011) and IL6/STAT3 (MIM: 147620/102582) signaling pathways128 and acts in the hypothalamus to modulate food intake and peripheral energy expenditure.129 PSMC3 is involved in ATP-dependent degradation of misfolded and damaged proteins and in removal of no longer required proteins. This protein is also involved in cell cycle progression, apoptosis, and DNA damage repair. A knockdown of PSMC3 in human immortalized fibroblasts increased cell proliferation.130 CELF1 has been associated with Alzheimer disease and obesity.131 Consequently, MTCH2 SNVs regulate gene expression of other cis-genes related to obesity, food intake, and/or cell number. Thus, MTCH2 effects on BMI may be more complex than previously accounted for.
Figure 4.
Selected SNVs of MTCH2 Significantly Associating with BMI, Glucose, Insulin, and a Few of Them Regulating the Expression (eSNVs) of at Least Four Genes: C1QTNF4, SLC39A13, PSMC3, and CELF1
First, we used LocusZoom142 for plotting regional information for MTCH2 selected SNVs, which have been reported by large GWAS publications (for BMI41 [T1], for glucose55 [T2], for proinsulin58 [T3], and another large analysis for glucose56 [T4]), for strength and extent of the association signal relative to genomic position, local linkage disequilibrium, and recombination patterns and the positions of genes in the region. Second, we used GTEx81 data to identify the SNVs of MTCH2 that are eSNVs by influencing the expression of other cis-target-genes (C1QTNF4, PSMC3, SLC39A13, CELF1) (T6). Third, we verified whether these eSNVs of MTCH2 (connected with blue dashed lines) are true expression regulators, by identifying the best eSNVs of the four target genes (T5, regulation effects shown with orange-dashed lines) and evaluating the LD r2 between the MTCH2 eSNV, e.g., rs4752856 (T6) and the best eSNV rs10838724 (T5) for the corresponding target gene (C1QTNF4), which was found to be 0.829 in adipose-subcutaneous tissue (T6). Fourth, the MTCH2 SNVs serving as eSNVs for the four cis-target-genes, shown in T6, colocalize in about 15K bps region. Thus, MTCH2 associates with BMI, glucose, and proinsulin, and among others influence the gene expression of other cis-genes (C1QTNF4, PSMC3, SLC39A13, CELF1), who in itself contribute also to individuals’ obesity risk (see Discussion).
Other Functions of MT-nDNA Candidate Genes
Mitochondrial inheritance has been previously associated with obesity, metabolic syndrome, insulin resistance, type 2 diabetes, and cardiovascular disease.3, 40, 132, 133, 134, 135 The complex genetics of MT affects processes of glucose, lipid, and amino acid catabolism, with ATP as the final energy product. By-products of these processes play an important role in signaling (e.g., ROS) and are used for epigenetic modifications (e.g., acetyl CoA). In our study, we would expect a preponderance of glycemic and lipid genes and proteins. Out of 109 MT-nDNA candidate genes, 21 of the significantly associated genes were functionally related with cholesterol (Table S6 and Figure S11), 16 with glucose and insulin, and 5 with adipose/obesity. The 109 MT-nDNA candidate genes have other functions too. For example, rs1044661 is located at 17q25.3, overlapping two genes that run in opposite strands: TBCD (involved in tubulin folding as cofactor D) and B3GNTL1 (a putative glucosyltransferase). This SNV was associated with HbA1c (p = 1.74E−46).61 Recently, Francis et al.136 hypothesized that the interaction between TBCD (17q25.3) and ARL2 (11q13.1 [MIM: 601175]) suggest that ARL2 serves as regulator of both mitochondrial fusion and microtubule dynamics. In 109 MT-nDNA candidates, WDR78 (1p31.3), TRIM54 (2p23.3 [MIM: 606474], regulating titin kinase [controlling elasticity in muscle] and microtubule-dependent signaling pathways in striated muscles), DNM3 (1q24.3 [MIM: 611445], involved in vesicular transport and with biased expression in brain), MAP2 (2q34 [MIM: 157130], involved in microtubule assembly and neurogenesis), GJA1 (6q22.31 [MIM: 121014], involved in the contraction of heart) and MAPT (17q21.31, expressed in nervous system) produce microtubule-associated proteins in interaction with MT (Table S6 and Figure S11). It should be noted that rs1050828, a missense mutation of G6PD residing on X chromosome, may impact HbA1c through non-glycemic factors rather than glucose metabolism.61 Additional functions of MT-nDNA candidate genes are described in the Supplemental Material and Methods, section 7.
Strengths and Limitations
Our study had many strengths. We used large sample sizes for studying mtDNA and MT-nDNA. At the mtDNA level we validated SNVs used per cohort and imputed based on Cosmopolitan MT-1000 Genomes. At the MT-nDNA level we used the strength of many consortia contributions for associations with seven traits. As a consequence, we performed comprehensive mtDNA and MT-nDNA analyses. The seven mtDNA variants significant associations showed no statistical significant deviations from the homogeneity/similarity of beta contributions before meta-pooling across studies as reported with Het-p in Table 1, as well as graphically represented in the Forest plot (Figure S16). For supporting MT-future studies, we have provided mtDNA updated Tables S15–S28 for arrays used in this study including MitoMap annotation. Also, we include Table S32 of predictions from the bioinformatics platforms that have been shown to have the highest sensitivity and specificity for the mtDNA variants.137 Our study also had some limitations. Rare mtDNA mutations are usually heteroplasmic point mutations and/or mtDNA lesions that typically result in a primary mitochondrial disease, which can manifest as a broad range of clinical outcomes. In this study we have interrogated the effect of inherited mtDNA variants, much commoner in the population than typical mtDNA mutations. Therefore, while there is a great deal of literature linking inherited mtDNA variants to disease, many of these studies have limited statistical power138 and several have never been independently replicated. So in many respects there is not a great deal of robust literature to call upon. The harmonization of mtDNA in each study was dependent on the number of mtDNA variants and their quality. Some variants were dropped before performing any imputation, because low-quality variants matched not only to the mtDNA genome, but also to nuclear genome (see mtDNA Variant Harmonization in Material and Methods). A number of quality-control filters were applied to mtDNA, which reduced individual sample size per marker or the number of useful markers. We did not address the association of significant markers by gender, because in our study sample sizes per mtDNA marker were variable and often small.
Perspective and Conclusion
We focused on associations of MT with adipose and glycemic traits, yet we acknowledge that there is still much to be learned from other traits, for example for triglycerides, high-density lipoprotein cholesterol, and C-reactive protein.46, 139, 140, 141 Notably, we identified 21 MT genes of lipid metabolism, although this was not the direct focus of our study. For example, TMLHE (Xq28 [MIM: 300777], trimethyllysine dioxygenase associated with HbA1c, p = 2.2E−19) is the first enzyme in the carnitine biosynthesis pathway. Carnitine play an essential role in the transport of activated fatty acids across the inner mitochondrial membrane and this gene is ubiquitous for its expression in heart. Hence, MT associations with other traits at the consortia-level remain to be explored. In conclusion, we identified common, rare SNVs, and a gene burden of genetic mtDNA variants, as well as 109 MT-nDNA genes associated with metabolic traits. Of the 109 MT-nDNA candidate genes, a subset pointed to associations with adipose, glucose metabolism, T2D, and CVD, and a subset of SNVs was inferred to contribute for differential regulatory genes expression. We documented in this study that the MT-candidates (SNVs/genes) have special contributions for functioning and energy homeostasis in adipose tissues and to glucose metabolism and insulin signaling.
Declaration of Interests
A.Y.C. is currently an employee of Merck & Co. C.T.-P. has received grants from Bayer pertaining to antithrombotic treatment of atrial fibrillation and cardiac valve replacements. D.O.M.-K. works as a part-time clinical research consultant for Metabolon, Inc. The remaining authors declare no competing financial interests.
Acknowledgments
A.E. Justice is supported by a K99/R00 from the National Institutes of Health, National Heart, Lung, and Blood Institute (NHLBI) (5K99HL130580-02 and 5R00HL130580-04). G. Hudson has received funding support by Wellcome Trust Centre for Mitochondrial Research, Grant Code G906919. J.I.R., X.G., Y.-D.I.C., K.D.T., Y.H., K.S., and J.Y. have received NIH support for research related to mitochondria. A.T.K., C. Liu, J.L.F., M. Graff, C.T.H., C.G., L.R.Y., M.F.F., D.E.A., D.I.C., K.Y., W.D.H., S.J.L., L.W., T.J.B., L.D.L.-G., T.I.A.S., G. Hudson, D.L., A.N.M., D.M.B., M.A.P., J.B.M., J.I.R., and K.E.N. were the writing group for this paper. M.A.P. and K.E.N. are from the CHARGE ADIPOSITY working group; J.I.R. and J.B.M. are from the CHARGE DIABETES working group.
Published: December 27, 2018
Footnotes
Supplemental Data include 17 figures, 32 tables, Supplemental Material and Methods, Supplemental Acknowledgments, and Supplemental Study Descriptions and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.12.001.
Contributor Information
Aldi T. Kraja, Email: aldikraja@wustl.edu.
Kari E. North, Email: kari_north@unc.edu.
Accession Numbers
Summary statistics for the mtDNA meta-analyses are available in the dbGaP repository for the CHARGE Consortium, phs000930.v8.p1.
Web Resources
1000 Genomes data (released April 2016), http://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/
GenBank (NCBI Reference Sequence: NC_012920.1), https://www.ncbi.nlm.nih.gov/nuccore/251831106
GIANT Consortium repositories, https://portals.broadinstitute.org/collaboration/giant/index.php/GIANT_consortium_data_files
GTEx (Gene Expression Portal), https://gtexportal.org
GWAS Catalog, http://www.ebi.ac.uk/gwas/
Human Mitochondrion Sequence, https://www.mitomap.org/MITOMAP/HumanMitoSeq
IMPUTE2, http://mathgen.stats.ox.ac.uk/impute/impute_v2.html
MAGIC consortium archives, https://www.magicinvestigators.org/downloads/
OMIM, http://www.omim.org/
Protein-Protein Interactions, ftp://ftp.ncbi.nih.gov/gene/GeneRIF/
seqMeta release notes, https://cran.r-project.org/web/packages/seqMeta/seqMeta.pdf
SHAPEIT2, https://mathgen.stats.ox.ac.uk/genetics_software/shapeit/shapeit.html#home
The Human Protein Atlas, http://www.proteinatlas.org/
Supplemental Data
References
- 1.Wang Z., Ying Z., Bosy-Westphal A., Zhang J., Schautz B., Later W., Heymsfield S.B., Müller M.J. Specific metabolic rates of major organs and tissues across adulthood: evaluation by mechanistic model of resting energy expenditure. Am. J. Clin. Nutr. 2010;92:1369–1377. doi: 10.3945/ajcn.2010.29885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ernster L., Schatz G. Mitochondria: a historical review. J. Cell Biol. 1981;91:227s–255s. doi: 10.1083/jcb.91.3.227s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chow J., Rahman J., Achermann J.C., Dattani M.T., Rahman S. Mitochondrial disease and endocrine dysfunction. Nat. Rev. Endocrinol. 2017;13:92–104. doi: 10.1038/nrendo.2016.151. [DOI] [PubMed] [Google Scholar]
- 4.Prasai K. Regulation of mitochondrial structure and function by protein import: A current review. Pathophysiology. 2017;24:107–122. doi: 10.1016/j.pathophys.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Lang B.F., Gray M.W., Burger G. Mitochondrial genome evolution and the origin of eukaryotes. Annu. Rev. Genet. 1999;33:351–397. doi: 10.1146/annurev.genet.33.1.351. [DOI] [PubMed] [Google Scholar]
- 6.Pagliarini D.J., Calvo S.E., Chang B., Sheth S.A., Vafai S.B., Ong S.E., Walford G.A., Sugiana C., Boneh A., Chen W.K. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith A.C., Robinson A.J. MitoMiner v3.1, an update on the mitochondrial proteomics database. Nucleic Acids Res. 2016;44(D1):D1258–D1261. doi: 10.1093/nar/gkv1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Febbo P.G., Mulligan M.G., Slonina D.A., Stegmaier K., Di Vizio D., Martinez P.R., Loda M., Taylor S.C. Literature Lab: a method of automated literature interrogation to infer biology from microarray analysis. BMC Genomics. 2007;8:461. doi: 10.1186/1471-2164-8-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haag-Liautard C., Coffey N., Houle D., Lynch M., Charlesworth B., Keightley P.D. Direct estimation of the mitochondrial DNA mutation rate in Drosophila melanogaster. PLoS Biol. 2008;6:e204. doi: 10.1371/journal.pbio.0060204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neiman M., Taylor D.R. The causes of mutation accumulation in mitochondrial genomes. Proc. Biol. Sci. 2009;276:1201–1209. doi: 10.1098/rspb.2008.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Craven L., Alston C.L., Taylor R.W., Turnbull D.M. Recent advances in mitochondrial disease. Annu. Rev. Genomics Hum. Genet. 2017;18:257–275. doi: 10.1146/annurev-genom-091416-035426. [DOI] [PubMed] [Google Scholar]
- 12.Schaefer A.M., Walker M., Turnbull D.M., Taylor R.W. Endocrine disorders in mitochondrial disease. Mol. Cell. Endocrinol. 2013;379:2–11. doi: 10.1016/j.mce.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malik A.N., Czajka A. Is mitochondrial DNA content a potential biomarker of mitochondrial dysfunction? Mitochondrion. 2013;13:481–492. doi: 10.1016/j.mito.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 14.Ashar F.N., Moes A., Moore A.Z., Grove M.L., Chaves P.H.M., Coresh J., Newman A.B., Matteini A.M., Bandeen-Roche K., Boerwinkle E. Association of mitochondrial DNA levels with frailty and all-cause mortality. J. Mol. Med. (Berl.) 2015;93:177–186. doi: 10.1007/s00109-014-1233-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mengel-From J., Thinggaard M., Dalgård C., Kyvik K.O., Christensen K., Christiansen L. Mitochondrial DNA copy number in peripheral blood cells declines with age and is associated with general health among elderly. Hum. Genet. 2014;133:1149–1159. doi: 10.1007/s00439-014-1458-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashar F.N., Zhang Y., Longchamps R.J., Lane J., Moes A., Grove M.L., Mychaleckyj J.C., Taylor K.D., Coresh J., Rotter J.I. Association of mitochondrial DNA copy number with cardiovascular disease. JAMA Cardiol. 2017;2:1247–1255. doi: 10.1001/jamacardio.2017.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y., Guallar E., Ashar F.N., Longchamps R.J., Castellani C.A., Lane J., Grove M.L., Coresh J., Sotoodehnia N., Ilkhanoff L. Association between mitochondrial DNA copy number and sudden cardiac death: findings from the Atherosclerosis Risk in Communities study (ARIC) Eur. Heart J. 2017;38:3443–3448. doi: 10.1093/eurheartj/ehx354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tin A., Grams M.E., Ashar F.N., Lane J.A., Rosenberg A.Z., Grove M.L., Boerwinkle E., Selvin E., Coresh J., Pankratz N., Arking D.E. Association between mitochondrial DNA copy number in peripheral blood and incident CKD in the Atherosclerosis Risk in Communities Study. J. Am. Soc. Nephrol. 2016;27:2467–2473. doi: 10.1681/ASN.2015060661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinonen S., Buzkova J., Muniandy M., Kaksonen R., Ollikainen M., Ismail K., Hakkarainen A., Lundbom J., Lundbom N., Vuolteenaho K. Impaired mitochondrial biogenesis in adipose tissue in acquired obesity. Diabetes. 2015;64:3135–3145. doi: 10.2337/db14-1937. [DOI] [PubMed] [Google Scholar]
- 20.De Pauw A., Tejerina S., Raes M., Keijer J., Arnould T. Mitochondrial (dys)function in adipocyte (de)differentiation and systemic metabolic alterations. Am. J. Pathol. 2009;175:927–939. doi: 10.2353/ajpath.2009.081155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altshuler-Keylin S., Kajimura S. Mitochondrial homeostasis in adipose tissue remodeling. Sci. Signal. 2017;10:10. doi: 10.1126/scisignal.aai9248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montgomery M.K., Turner N. Mitochondrial dysfunction and insulin resistance: an update. Endocr. Connect. 2015;4:R1–R15. doi: 10.1530/EC-14-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vernochet C., Damilano F., Mourier A., Bezy O., Mori M.A., Smyth G., Rosenzweig A., Larsson N.G., Kahn C.R. Adipose tissue mitochondrial dysfunction triggers a lipodystrophic syndrome with insulin resistance, hepatosteatosis, and cardiovascular complications. FASEB J. 2014;28:4408–4419. doi: 10.1096/fj.14-253971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiklund P., Zhang X., Pekkala S., Autio R., Kong L., Yang Y., Keinänen-Kiukaanniemi S., Alen M., Cheng S. Insulin resistance is associated with altered amino acid metabolism and adipose tissue dysfunction in normoglycemic women. Sci. Rep. 2016;6:24540. doi: 10.1038/srep24540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beasley T.M., Erickson S., Allison D.B. Rank-based inverse normal transformations are increasingly used, but are they merited? Behav. Genet. 2009;39:580–595. doi: 10.1007/s10519-009-9281-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delaneau O., Zagury J.F., Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 30.Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delaneau O., Marchini J., 1000 Genomes Project Consortium Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Nat. Commun. 2014;5:3934. doi: 10.1038/ncomms4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winkler T.W., Day F.R., Croteau-Chonka D.C., Wood A.R., Locke A.E., Mägi R., Ferreira T., Fall T., Graff M., Justice A.E., Genetic Investigation of Anthropometric Traits (GIANT) Consortium Quality control and conduct of genome-wide association meta-analyses. Nat. Protoc. 2014;9:1192–1212. doi: 10.1038/nprot.2014.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen H., Meigs J.B., Dupuis J. Sequence kernel association test for quantitative traits in family samples. Genet. Epidemiol. 2013;37:196–204. doi: 10.1002/gepi.21703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu C., Dupuis J., Larson M.G., Levy D. Association testing of the mitochondrial genome using pedigree data. Genet. Epidemiol. 2013;37:239–247. doi: 10.1002/gepi.21706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang H., Chanda P., Alonso A., Bader J.S., Arking D.E. Gene-based tests of association. PLoS Genet. 2011;7:e1002177. doi: 10.1371/journal.pgen.1002177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L., Lee S., Gim J., Qiao D., Cho M., Elston R.C., Silverman E.K., Won S. Family-based rare variant association analysis: A fast and efficient method of multivariate phenotype association analysis. Genet. Epidemiol. 2016;40:502–511. doi: 10.1002/gepi.21985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee S., Abecasis G.R., Boehnke M., Lin X. Rare-variant association analysis: study designs and statistical tests. Am. J. Hum. Genet. 2014;95:5–23. doi: 10.1016/j.ajhg.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calvo S.E., Clauser K.R., Mootha V.K. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016;44(D1):D1251–D1257. doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu H., Koshkin V., Allister E.M., Gyulkhandanyan A.V., Wheeler M.B. Molecular and metabolic evidence for mitochondrial defects associated with beta-cell dysfunction in a mouse model of type 2 diabetes. Diabetes. 2010;59:448–459. doi: 10.2337/db09-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Speliotes E.K., Willer C.J., Berndt S.I., Monda K.L., Thorleifsson G., Jackson A.U., Lango Allen H., Lindgren C.M., Luan J., Mägi R., MAGIC. Procardis Consortium Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang J., Loos R.J., Powell J.E., Medland S.E., Speliotes E.K., Chasman D.I., Rose L.M., Thorleifsson G., Steinthorsdottir V., Mägi R. FTO genotype is associated with phenotypic variability of body mass index. Nature. 2012;490:267–272. doi: 10.1038/nature11401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berndt S.I., Gustafsson S., Mägi R., Ganna A., Wheeler E., Feitosa M.F., Justice A.E., Monda K.L., Croteau-Chonka D.C., Day F.R. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat. Genet. 2013;45:501–512. doi: 10.1038/ng.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Randall J.C., Winkler T.W., Kutalik Z., Berndt S.I., Jackson A.U., Monda K.L., Kilpeläinen T.O., Esko T., Mägi R., Li S., DIAGRAM Consortium. MAGIC Investigators Sex-stratified genome-wide association studies including 270,000 individuals show sexual dimorphism in genetic loci for anthropometric traits. PLoS Genet. 2013;9:e1003500. doi: 10.1371/journal.pgen.1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Monda K.L., Chen G.K., Taylor K.C., Palmer C., Edwards T.L., Lange L.A., Ng M.C., Adeyemo A.A., Allison M.A., Bielak L.F., NABEC Consortium. UKBEC Consortium. BioBank Japan Project. AGEN Consortium A meta-analysis identifies new loci associated with body mass index in individuals of African ancestry. Nat. Genet. 2013;45:690–696. doi: 10.1038/ng.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Locke A.E., Kahali B., Berndt S.I., Justice A.E., Pers T.H., Day F.R., Powell C., Vedantam S., Buchkovich M.L., Yang J., LifeLines Cohort Study. ADIPOGen Consortium. AGEN-BMI Working Group. CARDIOGRAMplusC4D Consortium. CKDGen Consortium. GLGC. ICBP. MAGIC Investigators. MuTHER Consortium. MIGen Consortium. PAGE Consortium. ReproGen Consortium. GENIE Consortium. International Endogene Consortium Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shungin D., Winkler T.W., Croteau-Chonka D.C., Ferreira T., Locke A.E., Mägi R., Strawbridge R.J., Pers T.H., Fischer K., Justice A.E., ADIPOGen Consortium. CARDIOGRAMplusC4D Consortium. CKDGen Consortium. GEFOS Consortium. GENIE Consortium. GLGC. ICBP. International Endogene Consortium. LifeLines Cohort Study. MAGIC Investigators. MuTHER Consortium. PAGE Consortium. ReproGen Consortium New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winkler T.W., Justice A.E., Graff M., Barata L., Feitosa M.F., Chu S., Czajkowski J., Esko T., Fall T., Kilpeläinen T.O., CHARGE Consortium. DIAGRAM Consortium. GLGC Consortium. Global-BPGen Consortium. ICBP Consortium. MAGIC Consortium The influence of age and sex on genetic associations with adult body size and shape: a large-scale genome-wide interaction study. PLoS Genet. 2015;11:e1005378. doi: 10.1371/journal.pgen.1005378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ng M.C.Y., Graff M., Lu Y., Justice A.E., Mudgal P., Liu C.T., Young K., Yanek L.R., Feitosa M.F., Wojczynski M.K., Bone Mineral Density in Childhood Study (BMDCS) Group Discovery and fine-mapping of adiposity loci using high density imputation of genome-wide association studies in individuals of African ancestry: African Ancestry Anthropometry Genetics Consortium. PLoS Genet. 2017;13:e1006719. doi: 10.1371/journal.pgen.1006719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Justice A.E., Winkler T.W., Feitosa M.F., Graff M., Fisher V.A., Young K., Barata L., Deng X., Czajkowski J., Hadley D. Genome-wide meta-analysis of 241,258 adults accounting for smoking behaviour identifies novel loci for obesity traits. Nat. Commun. 2017;8:14977. doi: 10.1038/ncomms14977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heid I.M., Jackson A.U., Randall J.C., Winkler T.W., Qi L., Steinthorsdottir V., Thorleifsson G., Zillikens M.C., Speliotes E.K., Mägi R., MAGIC Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat. Genet. 2010;42:949–960. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Graff M., Scott R.A., Justice A.E., Young K.L., Feitosa M.F., Barata L., Winkler T.W., Chu A.Y., Mahajan A., Hadley D., CHARGE Consortium. EPIC-InterAct Consortium. PAGE Consortium Genome-wide physical activity interactions in adiposity - A meta-analysis of 200,452 adults. PLoS Genet. 2017;13:e1006528. doi: 10.1371/journal.pgen.1006528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saxena R., Hivert M.F., Langenberg C., Tanaka T., Pankow J.S., Vollenweider P., Lyssenko V., Bouatia-Naji N., Dupuis J., Jackson A.U., GIANT consortium. MAGIC investigators Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat. Genet. 2010;42:142–148. doi: 10.1038/ng.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dupuis J., Langenberg C., Prokopenko I., Saxena R., Soranzo N., Jackson A.U., Wheeler E., Glazer N.L., Bouatia-Naji N., Gloyn A.L., DIAGRAM Consortium. GIANT Consortium. Global BPgen Consortium. Anders Hamsten on behalf of Procardis Consortium. MAGIC investigators New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manning A.K., Hivert M.F., Scott R.A., Grimsby J.L., Bouatia-Naji N., Chen H., Rybin D., Liu C.T., Bielak L.F., Prokopenko I., DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium. Multiple Tissue Human Expression Resource (MUTHER) Consortium A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat. Genet. 2012;44:659–669. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scott R.A., Lagou V., Welch R.P., Wheeler E., Montasser M.E., Luan J., Mägi R., Strawbridge R.J., Rehnberg E., Gustafsson S., DIAbetes Genetics Replication and Meta-analysis (DIAGRAM) Consortium Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat. Genet. 2012;44:991–1005. doi: 10.1038/ng.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prokopenko I., Poon W., Mägi R., Prasad B R., Salehi S.A., Almgren P., Osmark P., Bouatia-Naji N., Wierup N., Fall T. A central role for GRB10 in regulation of islet function in man. PLoS Genet. 2014;10:e1004235. doi: 10.1371/journal.pgen.1004235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strawbridge R.J., Dupuis J., Prokopenko I., Barker A., Ahlqvist E., Rybin D., Petrie J.R., Travers M.E., Bouatia-Naji N., Dimas A.S., DIAGRAM Consortium. GIANT Consortium. MuTHER Consortium. CARDIoGRAM Consortium. C4D Consortium Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes. 2011;60:2624–2634. doi: 10.2337/db11-0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walford G.A., Gustafsson S., Rybin D., Stančáková A., Chen H., Liu C.T., Hong J., Jensen R.A., Rice K., Morris A.P. Genome-wide association study of the modified Stumvoll insulin sensitivity index identifies BCL2 and FAM19A2 as novel insulin sensitivity loci. Diabetes. 2016;65:3200–3211. doi: 10.2337/db16-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soranzo N., Sanna S., Wheeler E., Gieger C., Radke D., Dupuis J., Bouatia-Naji N., Langenberg C., Prokopenko I., Stolerman E., WTCCC Common variants at 10 genomic loci influence hemoglobin A1(C) levels via glycemic and nonglycemic pathways. Diabetes. 2010;59:3229–3239. doi: 10.2337/db10-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wheeler E., Leong A., Liu C.T., Hivert M.F., Strawbridge R.J., Podmore C., Li M., Yao J., Sim X., Hong J., EPIC-CVD Consortium. EPIC-InterAct Consortium. Lifelines Cohort Study Impact of common genetic determinants of Hemoglobin A1c on type 2 diabetes risk and diagnosis in ancestrally diverse populations: A transethnic genome-wide meta-analysis. PLoS Med. 2017;14:e1002383. doi: 10.1371/journal.pmed.1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chanda P., Huang H., Arking D.E., Bader J.S. Fast association tests for genes with FAST. PLoS ONE. 2013;8:e68585. doi: 10.1371/journal.pone.0068585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao W., Rasheed A., Tikkanen E., Lee J.J., Butterworth A.S., Howson J.M.M., Assimes T.L., Chowdhury R., Orho-Melander M., Damrauer S., CHD Exome+ Consortium. EPIC-CVD Consortium. EPIC-Interact Consortium. Michigan Biobank Identification of new susceptibility loci for type 2 diabetes and shared etiological pathways with coronary heart disease. Nat. Genet. 2017;49:1450–1457. doi: 10.1038/ng.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spracklen C.N., Chen P., Kim Y.J., Wang X., Cai H., Li S., Long J., Wu Y., Wang Y.X., Takeuchi F. Association analyses of East Asian individuals and trans-ancestry analyses with European individuals reveal new loci associated with cholesterol and triglyceride levels. Hum. Mol. Genet. 2017;26:1770–1784. doi: 10.1093/hmg/ddx062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nelson C.P., Goel A., Butterworth A.S., Kanoni S., Webb T.R., Marouli E., Zeng L., Ntalla I., Lai F.Y., Hopewell J.C., EPIC-CVD Consortium. CARDIoGRAMplusC4D. UK Biobank CardioMetabolic Consortium CHD working group Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat. Genet. 2017;49:1385–1391. doi: 10.1038/ng.3913. [DOI] [PubMed] [Google Scholar]
- 66.Eppinga R.N., Hagemeijer Y., Burgess S., Hinds D.A., Stefansson K., Gudbjartsson D.F., van Veldhuisen D.J., Munroe P.B., Verweij N., van der Harst P. Identification of genomic loci associated with resting heart rate and shared genetic predictors with all-cause mortality. Nat. Genet. 2016;48:1557–1563. doi: 10.1038/ng.3708. [DOI] [PubMed] [Google Scholar]
- 67.Warren H.R., Evangelou E., Cabrera C.P., Gao H., Ren M., Mifsud B., Ntalla I., Surendran P., Liu C., Cook J.P., International Consortium of Blood Pressure (ICBP) 1000G Analyses. BIOS Consortium. Lifelines Cohort Study. Understanding Society Scientific group. CHD Exome+ Consortium. ExomeBP Consortium. T2D-GENES Consortium. GoT2DGenes Consortium. Cohorts for Heart and Ageing Research in Genome Epidemiology (CHARGE) BP Exome Consortium. International Genomics of Blood Pressure (iGEN-BP) Consortium. UK Biobank CardioMetabolic Consortium BP working group Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat. Genet. 2017;49:403–415. doi: 10.1038/ng.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sabater-Lleal M., Huang J., Chasman D., Naitza S., Dehghan A., Johnson A.D., Teumer A., Reiner A.P., Folkersen L., Basu S., VTE Consortium. STROKE Consortium. Wellcome Trust Case Control Consortium 2 (WTCCC2) C4D Consortium. CARDIoGRAM Consortium Multiethnic meta-analysis of genome-wide association studies in >100 000 subjects identifies 23 fibrinogen-associated Loci but no strong evidence of a causal association between circulating fibrinogen and cardiovascular disease. Circulation. 2013;128:1310–1324. doi: 10.1161/CIRCULATIONAHA.113.002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Willer C.J., Schmidt E.M., Sengupta S., Peloso G.M., Gustafsson S., Kanoni S., Ganna A., Chen J., Buchkovich M.L., Mora S., Global Lipids Genetics Consortium Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mozaffarian D., Kabagambe E.K., Johnson C.O., Lemaitre R.N., Manichaikul A., Sun Q., Foy M., Wang L., Wiener H., Irvin M.R. Genetic loci associated with circulating phospholipid trans fatty acids: a meta-analysis of genome-wide association studies from the CHARGE Consortium. Am. J. Clin. Nutr. 2015;101:398–406. doi: 10.3945/ajcn.114.094557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yeo A., Li L., Warren L., Aponte J., Fraser D., King K., Johansson K., Barnes A., MacPhee C., Davies R. Pharmacogenetic meta-analysis of baseline risk factors, pharmacodynamic, efficacy and tolerability endpoints from two large global cardiovascular outcomes trials for darapladib. PLoS ONE. 2017;12:e0182115. doi: 10.1371/journal.pone.0182115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hinds D.A., Buil A., Ziemek D., Martinez-Perez A., Malik R., Folkersen L., Germain M., Mälarstig A., Brown A., Soria J.M., METASTROKE Consortium, INVENT Consortium Genome-wide association analysis of self-reported events in 6135 individuals and 252 827 controls identifies 8 loci associated with thrombosis. Hum. Mol. Genet. 2016;25:1867–1874. doi: 10.1093/hmg/ddw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ehret G.B., Munroe P.B., Rice K.M., Bochud M., Johnson A.D., Chasman D.I., Smith A.V., Tobin M.D., Verwoert G.C., Hwang S.J., International Consortium for Blood Pressure Genome-Wide Association Studies. CARDIoGRAM consortium. CKDGen Consortium. KidneyGen Consortium. EchoGen consortium. CHARGE-HF consortium Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van der Harst P., van Setten J., Verweij N., Vogler G., Franke L., Maurano M.T., Wang X., Mateo Leach I., Eijgelsheim M., Sotoodehnia N. 52 genetic loci influencing myocardial mass. J. Am. Coll. Cardiol. 2016;68:1435–1448. doi: 10.1016/j.jacc.2016.07.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pott J., Burkhardt R., Beutner F., Horn K., Teren A., Kirsten H., Holdt L.M., Schuler G., Teupser D., Loeffler M. Genome-wide meta-analysis identifies novel loci of plaque burden in carotid artery. Atherosclerosis. 2017;259:32–40. doi: 10.1016/j.atherosclerosis.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 76.Yasuno K., Bilguvar K., Bijlenga P., Low S.K., Krischek B., Auburger G., Simon M., Krex D., Arlier Z., Nayak N. Genome-wide association study of intracranial aneurysm identifies three new risk loci. Nat. Genet. 2010;42:420–425. doi: 10.1038/ng.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Aulchenko Y.S., Ripatti S., Lindqvist I., Boomsma D., Heid I.M., Pramstaller P.P., Penninx B.W., Janssens A.C., Wilson J.F., Spector T., ENGAGE Consortium Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat. Genet. 2009;41:47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Esteras N., Rohrer J.D., Hardy J., Wray S., Abramov A.Y. Mitochondrial hyperpolarization in iPSC-derived neurons from patients of FTDP-17 with 10+16 MAPT mutation leads to oxidative stress and neurodegeneration. Redox Biol. 2017;12:410–422. doi: 10.1016/j.redox.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ward L.D., Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Battle A., Brown C.D., Engelhardt B.E., Montgomery S.B., GTEx Consortium. Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group. Statistical Methods groups—Analysis Working Group. Enhancing GTEx (eGTEx) groups. NIH Common Fund. NIH/NCI. NIH/NHGRI. NIH/NIMH. NIH/NIDA. Biospecimen Collection Source Site—NDRI. Biospecimen Collection Source Site—RPCI. Biospecimen Core Resource—VARI. Brain Bank Repository—University of Miami Brain Endowment Bank. Leidos Biomedical—Project Management. ELSI Study. Genome Browser Data Integration &Visualization—EBI. Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz. Lead analysts. Laboratory, Data Analysis &Coordinating Center (LDACC) NIH program management. Biospecimen collection. Pathology. eQTL manuscript working group Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. [Google Scholar]
- 82.Saha A., Kim Y., Gewirtz A.D.H., Jo B., Gao C., McDowell I.C., Engelhardt B.E., Battle A., GTEx Consortium Co-expression networks reveal the tissue-specific regulation of transcription and splicing. Genome Res. 2017;27:1843–1858. doi: 10.1101/gr.216721.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li M.J., Wang L.Y., Xia Z., Sham P.C., Wang J. GWAS3D: Detecting human regulatory variants by integrative analysis of genome-wide associations, chromosome interactions and histone modifications. Nucleic Acids Res. 2013;41 doi: 10.1093/nar/gkt456. W150-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sané A.T., Seidman E., Peretti N., Kleme M.L., Delvin E., Deslandres C., Garofalo C., Spahis S., Levy E. Understanding chylomicron retention disease through Sar1b Gtpase gene disruption: insight from cell culture. Arterioscler. Thromb. Vasc. Biol. 2017;37:2243–2251. doi: 10.1161/ATVBAHA.117.310121. [DOI] [PubMed] [Google Scholar]
- 85.Yuasa K., Matsuda T., Tsuji A. Functional regulation of transient receptor potential canonical 7 by cGMP-dependent protein kinase Iα. Cell. Signal. 2011;23:1179–1187. doi: 10.1016/j.cellsig.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 86.Ilegems E., Iwatsuki K., Kokrashvili Z., Benard O., Ninomiya Y., Margolskee R.F. REEP2 enhances sweet receptor function by recruitment to lipid rafts. J. Neurosci. 2010;30:13774–13783. doi: 10.1523/JNEUROSCI.0091-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pattin K.A., Moore J.H. Role for protein-protein interaction databases in human genetics. Expert Rev. Proteomics. 2009;6:647–659. doi: 10.1586/epr.09.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Taniguchi K., Matsumura K., Kageyama S., Ii H., Ashihara E., Chano T., Kawauchi A., Yoshiki T., Nakata S. Prohibitin-2 is a novel regulator of p21WAF1/CIP1 induced by depletion of γ-glutamylcyclotransferase. Biochem. Biophys. Res. Commun. 2018;496:218–224. doi: 10.1016/j.bbrc.2018.01.029. [DOI] [PubMed] [Google Scholar]
- 90.Fimia G.M., Corazzari M., Antonioli M., Piacentini M. Ambra1 at the crossroad between autophagy and cell death. Oncogene. 2013;32:3311–3318. doi: 10.1038/onc.2012.455. [DOI] [PubMed] [Google Scholar]
- 91.Fimia G.M., Stoykova A., Romagnoli A., Giunta L., Di Bartolomeo S., Nardacci R., Corazzari M., Fuoco C., Ucar A., Schwartz P. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 92.Frazier A.E., Thorburn D.R., Compton A.G. Mitochondrial energy generation disorders: genes, mechanisms and clues to pathology. J. Biol. Chem. 2017 doi: 10.1074/jbc.R117.809194. jbc.R117.809194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Malik A.N., Parsade C.K., Ajaz S., Crosby-Nwaobi R., Gnudi L., Czajka A., Sivaprasad S. Altered circulating mitochondrial DNA and increased inflammation in patients with diabetic retinopathy. Diabetes Res. Clin. Pract. 2015;110:257–265. doi: 10.1016/j.diabres.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 94.Guo H., Bueler S.A., Rubinstein J.L. Atomic model for the dimeric FO region of mitochondrial ATP synthase. Science. 2017;358:936–940. doi: 10.1126/science.aao4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nakamoto R.K., Baylis Scanlon J.A., Al-Shawi M.K. The rotary mechanism of the ATP synthase. Arch. Biochem. Biophys. 2008;476:43–50. doi: 10.1016/j.abb.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fish J., Raule N., Attardi G. Discovery of a major D-loop replication origin reveals two modes of human mtDNA synthesis. Science. 2004;306:2098–2101. doi: 10.1126/science.1102077. [DOI] [PubMed] [Google Scholar]
- 97.Taanman J.W. The mitochondrial genome: structure, transcription, translation and replication. Biochim. Biophys. Acta. 1999;1410:103–123. doi: 10.1016/s0005-2728(98)00161-3. [DOI] [PubMed] [Google Scholar]
- 98.Wang H.N., Chen H.D., Chen K.Y., Xiao J.F., He K., Xiang G.A., Xie X. Highly expressed MT-ND3 positively associated with histological severity of hepatic steatosis. APMIS. 2014;122:443–451. doi: 10.1111/apm.12166. [DOI] [PubMed] [Google Scholar]
- 99.Fetterman J.L., Liu C., Mitchell G.F., Vasan R.S., Benjamin E.J., Vita J.A., Hamburg N.M., Levy D. Relations of mitochondrial genetic variants to measures of vascular function. Mitochondrion. 2018;40:51–57. doi: 10.1016/j.mito.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Brown D.A., Perry J.B., Allen M.E., Sabbah H.N., Stauffer B.L., Shaikh S.R., Cleland J.G., Colucci W.S., Butler J., Voors A.A. Expert consensus document: Mitochondrial function as a therapeutic target in heart failure. Nat. Rev. Cardiol. 2017;14:238–250. doi: 10.1038/nrcardio.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.McDermott-Roe C., Leleu M., Rowe G.C., Palygin O., Bukowy J.D., Kuo J., Rech M., Hermans-Beijnsberger S., Schaefer S., Adami E. Transcriptome-wide co-expression analysis identifies LRRC2 as a novel mediator of mitochondrial and cardiac function. PLoS ONE. 2017;12:e0170458. doi: 10.1371/journal.pone.0170458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee H.S., Park T. Pathway-driven approaches of interaction between oxidative balance and genetic polymorphism on metabolic syndrome. Oxid. Med. Cell. Longev. 2017;2017:6873197. doi: 10.1155/2017/6873197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Go M.J., Hwang J.Y., Kim Y.J., Hee Oh J., Kim Y.J., Heon Kwak S., Soo Park K., Lee J., Kim B.J., Han B.G. New susceptibility loci in MYL2, C12orf51 and OAS1 associated with 1-h plasma glucose as predisposing risk factors for type 2 diabetes in the Korean population. J. Hum. Genet. 2013;58:362–365. doi: 10.1038/jhg.2013.14. [DOI] [PubMed] [Google Scholar]
- 104.Kristiansson K., Perola M., Tikkanen E., Kettunen J., Surakka I., Havulinna A.S., Stancáková A., Barnes C., Widen E., Kajantie E. Genome-wide screen for metabolic syndrome susceptibility Loci reveals strong lipid gene contribution but no evidence for common genetic basis for clustering of metabolic syndrome traits. Circ Cardiovasc Genet. 2012;5:242–249. doi: 10.1161/CIRCGENETICS.111.961482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.MacDonald M.J., Fahien L.A. Insulin release in pancreatic islets by a glycolytic and a Krebs cycle intermediate: contrasting patterns of glyceraldehyde phosphate and succinate. Arch. Biochem. Biophys. 1990;279:104–108. doi: 10.1016/0003-9861(90)90468-e. [DOI] [PubMed] [Google Scholar]
- 106.MacDonald M.J., Fahien L.A., Brown L.J., Hasan N.M., Buss J.D., Kendrick M.A. Perspective: emerging evidence for signaling roles of mitochondrial anaplerotic products in insulin secretion. Am. J. Physiol. Endocrinol. Metab. 2005;288:E1–E15. doi: 10.1152/ajpendo.00218.2004. [DOI] [PubMed] [Google Scholar]
- 107.Dukes I.D., McIntyre M.S., Mertz R.J., Philipson L.H., Roe M.W., Spencer B., Worley J.F., 3rd Dependence on NADH produced during glycolysis for beta-cell glucose signaling. J. Biol. Chem. 1994;269:10979–10982. [PubMed] [Google Scholar]
- 108.El-Hattab A.W., Emrick L.T., Hsu J.W., Chanprasert S., Jahoor F., Scaglia F., Craigen W.J. Glucose metabolism derangements in adults with the MELAS m.3243A>G mutation. Mitochondrion. 2014;18:63–69. doi: 10.1016/j.mito.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nóbrega-Pereira S., Fernandez-Marcos P.J., Brioche T., Gomez-Cabrera M.C., Salvador-Pascual A., Flores J.M., Viña J., Serrano M. G6PD protects from oxidative damage and improves healthspan in mice. Nat. Commun. 2016;7:10894. doi: 10.1038/ncomms10894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cunningham A.D., Colavin A., Huang K.C., Mochly-Rosen D. Coupling between protein stability and catalytic activity determines pathogenicity of G6PD variants. Cell Rep. 2017;18:2592–2599. doi: 10.1016/j.celrep.2017.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dörner G., Mohnike A., Steindel E. On possible genetic and epigenetic modes of diabetes transmission. Endokrinologie. 1975;66:225–227. [PubMed] [Google Scholar]
- 112.Thomas F., Balkau B., Vauzelle-Kervroedan F., Papoz L., CODIAB-INSERM-ZENECA Study Group Maternal effect and familial aggregation in NIDDM. The CODIAB Study. Diabetes. 1994;43:63–67. doi: 10.2337/diab.43.1.63. [DOI] [PubMed] [Google Scholar]
- 113.Lin R.S., Lee W.C., Lee Y.T., Chou P., Fu C.C. Maternal role in type 2 diabetes mellitus: indirect evidence for a mitochondrial inheritance. Int. J. Epidemiol. 1994;23:886–890. doi: 10.1093/ije/23.5.886. [DOI] [PubMed] [Google Scholar]
- 114.Voight B.F., Scott L.J., Steinthorsdottir V., Morris A.P., Dina C., Welch R.P., Zeggini E., Huth C., Aulchenko Y.S., Thorleifsson G., MAGIC investigators. GIANT Consortium Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat. Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yasuda K., Miyake K., Horikawa Y., Hara K., Osawa H., Furuta H., Hirota Y., Mori H., Jonsson A., Sato Y. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat. Genet. 2008;40:1092–1097. doi: 10.1038/ng.207. [DOI] [PubMed] [Google Scholar]
- 116.Unoki H., Takahashi A., Kawaguchi T., Hara K., Horikoshi M., Andersen G., Ng D.P., Holmkvist J., Borch-Johnsen K., Jørgensen T. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat. Genet. 2008;40:1098–1102. doi: 10.1038/ng.208. [DOI] [PubMed] [Google Scholar]
- 117.Heard E., Rougeulle C., Arnaud D., Avner P., Allis C.D., Spector D.L. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell. 2001;107:727–738. doi: 10.1016/s0092-8674(01)00598-0. [DOI] [PubMed] [Google Scholar]
- 118.Lai Y.K., Lai N.M., Lee S.W. Glucose-6-phosphate dehydrogenase deficiency and risk of diabetes: a systematic review and meta-analysis. Ann. Hematol. 2017;96:839–845. doi: 10.1007/s00277-017-2945-6. [DOI] [PubMed] [Google Scholar]
- 119.Heymann A.D., Cohen Y., Chodick G. Glucose-6-phosphate dehydrogenase deficiency and type 2 diabetes. Diabetes Care. 2012;35:e58. doi: 10.2337/dc11-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.van der Wijst M.G., Rots M.G. Mitochondrial epigenetics: an overlooked layer of regulation? Trends Genet. 2015;31:353–356. doi: 10.1016/j.tig.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 121.Schleinitz D., Klöting N., Lindgren C.M., Breitfeld J., Dietrich A., Schön M.R., Lohmann T., Dreßler M., Stumvoll M., McCarthy M.I. Fat depot-specific mRNA expression of novel loci associated with waist-hip ratio. Int. J. Obes. 2014;38:120–125. doi: 10.1038/ijo.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Meyre D., Delplanque J., Chèvre J.C., Lecoeur C., Lobbens S., Gallina S., Durand E., Vatin V., Degraeve F., Proença C. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat. Genet. 2009;41:157–159. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- 123.Bambace C., Dahlman I., Arner P., Kulyté A. NPC1 in human white adipose tissue and obesity. BMC Endocr. Disord. 2013;13:5. doi: 10.1186/1472-6823-13-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jelinek D., Castillo J.J., Garver W.S. The C57BL/6J Niemann-Pick C1 mouse model with decreased gene dosage has impaired glucose tolerance independent of body weight. Gene. 2013;527:65–70. doi: 10.1016/j.gene.2013.05.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Woś M., Szczepanowska J., Pikuła S., Tylki-Szymańska A., Zabłocki K., Bandorowicz-Pikuła J. Mitochondrial dysfunction in fibroblasts derived from patients with Niemann-Pick type C disease. Arch. Biochem. Biophys. 2016;593:50–59. doi: 10.1016/j.abb.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 126.Bernhard F., Landgraf K., Klöting N., Berthold A., Büttner P., Friebe D., Kiess W., Kovacs P., Blüher M., Körner A. Functional relevance of genes implicated by obesity genome-wide association study signals for human adipocyte biology. Diabetologia. 2013;56:311–322. doi: 10.1007/s00125-012-2773-0. [DOI] [PubMed] [Google Scholar]
- 127.Buzaglo-Azriel L., Kuperman Y., Tsoory M., Zaltsman Y., Shachnai L., Zaidman S.L., Bassat E., Michailovici I., Sarver A., Tzahor E. Loss of muscle MTCH2 increases whole-body energy utilization and protects from diet-induced obesity. Cell Rep. 2016;14:1602–1610. doi: 10.1016/j.celrep.2016.01.046. [DOI] [PubMed] [Google Scholar]
- 128.Li Q., Wang L., Tan W., Peng Z., Luo Y., Zhang Y., Zhang G., Na D., Jin P., Shi T. Identification of C1qTNF-related protein 4 as a potential cytokine that stimulates the STAT3 and NF-κB pathways and promotes cell survival in human cancer cells. Cancer Lett. 2011;308:203–214. doi: 10.1016/j.canlet.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 129.Byerly M.S., Petersen P.S., Ramamurthy S., Seldin M.M., Lei X., Provost E., Wei Z., Ronnett G.V., Wong G.W. C1q/TNF-related protein 4 (CTRP4) is a unique secreted protein with two tandem C1q domains that functions in the hypothalamus to modulate food intake and body weight. J. Biol. Chem. 2014;289:4055–4069. doi: 10.1074/jbc.M113.506956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sepe M., Festa L., Tolino F., Bellucci L., Sisto L., Alfano D., Ragno P., Calabrò V., de Franciscis V., La Mantia G., Pollice A. A regulatory mechanism involving TBP-1/Tat-Binding Protein 1 and Akt/PKB in the control of cell proliferation. PLoS ONE. 2011;6:e22800. doi: 10.1371/journal.pone.0022800. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 131.Hinney A., Albayrak O., Antel J., Volckmar A.L., Sims R., Chapman J., Harold D., Gerrish A., Heid I.M., Winkler T.W., GERAD Consortium. IGAP Consortium. GIANT Consortium Genetic variation at the CELF1 (CUGBP, elav-like family member 1 gene) locus is genome-wide associated with Alzheimer’s disease and obesity. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2014;165B:283–293. doi: 10.1002/ajmg.b.32234. [DOI] [PubMed] [Google Scholar]
- 132.Ridler C. Obesity: Inheritance via mitochondria. Nat. Rev. Endocrinol. 2016;12:497. doi: 10.1038/nrendo.2016.108. [DOI] [PubMed] [Google Scholar]
- 133.Semenkovich C.F. We know more than we can tell about diabetes and vascular disease: The 2016 Edwin Bierman Award Lecture. Diabetes. 2017;66:1735–1741. doi: 10.2337/db17-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Szendroedi J., Phielix E., Roden M. The role of mitochondria in insulin resistance and type 2 diabetes mellitus. Nat. Rev. Endocrinol. 2011;8:92–103. doi: 10.1038/nrendo.2011.138. [DOI] [PubMed] [Google Scholar]
- 135.West A.P., Shadel G.S. Mitochondrial DNA in innate immune responses and inflammatory pathology. Nat. Rev. Immunol. 2017;17:363–375. doi: 10.1038/nri.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Francis J.W., Turn R.E., Newman L.E., Schiavon C., Kahn R.A. Higher order signaling: ARL2 as regulator of both mitochondrial fusion and microtubule dynamics allows integration of 2 essential cell functions. Small GTPases. 2016;7:188–196. doi: 10.1080/21541248.2016.1211069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Castellana S., Fusilli C., Mazzoccoli G., Biagini T., Capocefalo D., Carella M., Vescovi A.L., Mazza T. High-confidence assessment of functional impact of human mitochondrial non-synonymous genome variations by APOGEE. PLoS Comput. Biol. 2017;13:e1005628. doi: 10.1371/journal.pcbi.1005628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Samuels D.C., Carothers A.D., Horton R., Chinnery P.F. The power to detect disease associations with mitochondrial DNA haplogroups. Am. J. Hum. Genet. 2006;78:713–720. doi: 10.1086/502682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Guo Y., Lanktree M.B., Taylor K.C., Hakonarson H., Lange L.A., Keating B.J., IBC 50K SNP array BMI Consortium Gene-centric meta-analyses of 108 912 individuals confirm known body mass index loci and reveal three novel signals. Hum. Mol. Genet. 2013;22:184–201. doi: 10.1093/hmg/dds396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Middelberg R.P., Ferreira M.A., Henders A.K., Heath A.C., Madden P.A., Montgomery G.W., Martin N.G., Whitfield J.B. Genetic variants in LPL, OASL and TOMM40/APOE-C1-C2-C4 genes are associated with multiple cardiovascular-related traits. BMC Med. Genet. 2011;12:123. doi: 10.1186/1471-2350-12-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kraja A.T., Chasman D.I., North K.E., Reiner A.P., Yanek L.R., Kilpeläinen T.O., Smith J.A., Dehghan A., Dupuis J., Johnson A.D., Cross Consortia Pleiotropy Group. Cohorts for Heart and. Aging Research in Genetic Epidemiology. Genetic Investigation of Anthropometric Traits Consortium. Global Lipids Genetics Consortium. Meta-Analyses of Glucose. Insulin-related traits Consortium. Global BPgen Consortium. ADIPOGen Consortium. Women’s Genome Health Study. Howard University Family Study Pleiotropic genes for metabolic syndrome and inflammation. Mol. Genet. Metab. 2014;112:317–338. doi: 10.1016/j.ymgme.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.