Abstract
Many animal species present sex differences. Sex-associated genes (SAGs), which have female-biased or male-biased expression, have major influences on the remarkable sex differences in important traits such as growth, reproduction, disease resistance and behaviors. However, the SAGs resulting in the vast majority of phenotypic sex differences are still unknown. To provide a useful resource for the functional study of SAGs, we manually curated public RNA-seq datasets with paired female and male biological replicates from the same condition and systematically re-analyzed the datasets using standardized methods. We identified 27,793 female-biased SAGs and 64,043 male-biased SAGs from 2,828 samples of 21 species, including human, chimpanzee, macaque, mouse, rat, cow, horse, chicken, zebrafish, seven fly species and five worm species. All these data were cataloged into SAGD, a user-friendly database of SAGs (http://bioinfo.life.hust.edu.cn/SAGD) where users can browse SAGs by gene, species, drug and dataset. In SAGD, the expression, annotation, targeting drugs, homologs, ontology and related RNA-seq datasets of SAGs are provided to help researchers to explore their functions and potential applications in agriculture and human health.
INTRODUCTION
Sexually reproducing animals usually demonstrate remarkable differences between females and males in morphological, physiological and behavioral phenotypes (1,2). Such differences are caused by the large number of sex-associated genes (SAGs), whose expressions vary between females and males (3,4). The study of SAGs is important not only for understanding gene regulation and evolution, but also for their application to animal reproduction and pest control (3–7). Moreover, the increasing evidence indicates that SAGs are a key factor affecting the risk of developing all kinds of diseases including neurodegenerative diseases, cardiovascular diseases and cancers, and they have been linked to precision medicine (8–10).
With the advent of RNA-seq technologies, it becomes possible to accurately quantify expression differences between males and females on a genome-wide scale. Numerous studies have been performed by RNA-seq to identify SAGs and the results revealed that a large fraction of genes are SAGs (11–14). Based on the samples, experimental and statistical methods used, up to 95% of genes may be identified as SAGs (12,15–17). However, there is a lack of a comprehensive database characterizing all the SAGs derived from RNA-seq data of the sequenced animal genomes through the same pipeline.
To date, there has been only one database about SAGs called Sebida (18), which collected SAGs from microarray data of three insect species (Drosophila melanogaster, Drosophila simulans and Anopheles gambiae). It was established in 2006 and has not been updated in recent years. Some central repositories of gene expression (e.g. Expression Atlas (19), GEO Profiles (20)) cover comprehensive expression profiles including those from RNA-seq datasets with sex variables. However, these repositories are not designed exclusively for comparing the expressions between paired biological replicates under the same condition, thus they are not suitable for SAG study.
To make gene expression comparisons between sexes across species possible, we presented SAGD (sex-associated gene database) integrating data from 2,828 RNA-seq samples to compare male versus female gene expression in 21 sequenced genomes. Users can compare the expression changes of SAGs in different species, tissues, and developmental stages, and can screen out candidate genes. This database will be a valuable resource for researchers and clinicians to conduct studies and practice on the function of SAGs.
MATERIALS AND METHODS
Extraction of metadata information from public resources of RNA-seq samples
We extracted metadata information of RNA-seq samples by integrating databases Expression Atlas (https://www.ebi.ac.uk/gxa/) (19), NCBI Short Read Archive (SRA, http://www.ncbi.nlm.nih.gov/sra/), and NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) (21).
We first extracted metadata information from Expression Atlas (19) with well-curated sample information. We downloaded the gzipped tar archive of all Expression Atlas analysis results (4 April 2018) and extracted condensed-sdrf.tsv files for all the assays. Sample feature information including organism, organism part, sex and age was organized into a matrix with species, tissue, sex, and stage as columns.
We then extracted metadata information from SRA containing descriptive information of sample attributes in forms of free text which is difficult to parse. We queried SRA projects with sex and gonad, and we curated species, tissue, sex, and stage features out of various attributes of SRA samples. The curated information was organized into one matrix.
As a complement, we queried ‘sex’ for series in GEO (20) on 14 July 2018, and refined the 2,545 search results with study type ‘Expression profiling by high throughput sequencing’. We examined the series, and manually curated feature values of series samples into another matrix.
Subsequent selection of RNA-seq samples
We incorporated the three matrices mentioned above, and uniformed nomenclature of the feature values. Then, we grouped the samples by the combination information of project, species, tissue, and stage. Subsequently, we extracted sample library information from SRA by the Bioconductor SRAdb (1.42.2) (21) along with its sqlite database (modified on 8 June 2018). To select all potential RNA-Seq runs consisting of raw reads of sequenced RNAs, we maintained SRA runs with ‘TRANSCRIPTOMIC’ library source, ‘ILLUMINA’ platform, and ‘RNA-Seq’ library strategy. Then, we selected RNA-Seq runs from species with sequenced genomes in Ensembl or Ensembl Metazoa (22) for the purpose of gene expression analysis based on annotation.
Next, we retained groups with both female and male biological replicates for differential expression analysis setting sex as a major difference variable. We selected only the group with most biological replicates for further analysis when there was more than one group in a species/tissue/stage combination. Up to 20 biological replicates for each sex from each group were randomly picked. In total, 15,718 SRA runs of potential RNA-Seq data for sequenced genomes were maintained for further selection.
Analysis of RNA-seq data
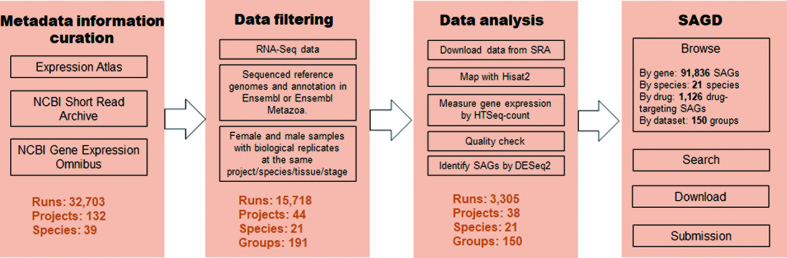
All selected groups of raw RNA-seq datasets were processed through the same pipeline (Figure 1). The raw RNA-seq data were downloaded from SRA, and mapped to corresponding reference genomes by HISAT2 (version 2.0.5) (23) under the guidance of gene annotation from Ensembl (release 92) or Ensembl Metazoa (Release 40) (22). HTSeq-count (version 0.9.1) (24) was employed to quantify the reads uniquely aligned to each gene so that one read would not be assigned to several paralogs. Read counts were merged by sample so that technical replicates would be integrated. We then normalized read counts and identified differentially expressed genes between female and male samples in each group by DESeq2 (version 1.20.0) (25). We also normalized read counts into the FPKM values (Fragments Per Kilobase of transcript per Million mapped reads). We defined genes with padj <0.05 and |log2 (M/F ratio)| ≥2 in each group as SAGs.
Figure 1.
Overall design of SAGD. SAGD curated metadata information from Expression Atlas, SRA and GEO, and selected groups of RNA-seq datasets with female and male biological replicates with the same project/species/tissue/stage combination. All RNA-seq raw data was processed using a standard pipeline. SAGD includes ‘Browse’, ‘Search’, ‘Download’ and ‘Submission’.
For quality control, we measured the replicability among biological replicates. We defined a standard sample for each sex/project/species/tissue/stage combination as the median value of normalized read counts of samples in the combination (15), and calculated the Spearman correlation coefficients of normalized read counts between each sample and its corresponding standard sample. Samples with a correlation over 0.8 were defined as qualified (15).
Database implementation
The SAGD database was built with the Flask open source framework (http://flask.pocoo.org/). All data were integrated into MongoDB (version 3.2.11). The web interface was designed and implemented using AngularJS (version 1.6.9) and was improved with some AngularJS libraries and several JavaScript libraries for a more useful interface. Our website was tested with several popular web browsers and Google Chrome was recommended.
RESULTS
Data summary
In total, we identified 27,793 female-biased SAGs and 64,043 male-biased SAGs in 21 species by curating high-throughput datasets of 2,828 samples from 38 projects (Table 1). There were more male-biased genes than female-biased genes in 117 of all the 150 groups (Table 1, Supplementary Table S1). In XX/XY sex chromosome systems, 121 of the 142 groups showed a higher percent of female-biased genes on the X chromosome than male-biased genes (Supplementary Table S1). Among SAGs, there were 4,871 female-biased human SAGs (8.3% human genes) and 17,223 male-biased human SAGs (29.5% human genes) derived from 1,800 samples covering 4 developmental stages and 60 tissues in 16 projects (Table 1). Combining the drug information, we found that 1,126 SAGs were drug targets and thus they might be associated with sex difference of drug response.
Table 1.
Statistics of RNA-seq datasets and SAGs in each species
| SAG_F4 | SAG_M5 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Project | Sample | Tissue | Stage | Group | #2 | %3 | # | % | |
| Bos taurus | 1 | 39 | 4 | 1 | 4 | 45 | 0.2 | 43 | 0.2 |
| Caenorhabditis brenneri | 1 | 6 | 1 | 1 | 1 | 3,583 | 10.8 | 5,465 | 16.4 |
| Caenorhabditis elegans | 2 | 28 | 2 | 2 | 3 | 1,249 | 2.7 | 3,408 | 7.3 |
| Caenorhabditis japonica | 1 | 6 | 1 | 1 | 1 | 1,855 | 5.7 | 3,384 | 10.4 |
| Caenorhabditis remanei | 1 | 6 | 1 | 1 | 1 | 2,799 | 8.5 | 4,485 | 13.6 |
| Danio rerio | 1 | 4 | 1 | 1 | 1 | 3 | 0.0 | 14 | 0.0 |
| Drosophila ananassae | 1 | 4 | 1 | 1 | 1 | 295 | 1.9 | 2,406 | 15.2 |
| Drosophila melanogaster | 3 | 160 | 9 | 1 | 9 | 2,906 | 16.4 | 7,425 | 41.9 |
| Drosophila mojavensis | 1 | 4 | 1 | 1 | 1 | 1,001 | 6.8 | 2,433 | 16.6 |
| Drosophila pseudoobscura | 2 | 12 | 3 | 1 | 3 | 2,991 | 17.6 | 5,866 | 34.6 |
| Drosophila simulans | 2 | 11 | 2 | 1 | 2 | 930 | 6.0 | 2,800 | 18.2 |
| Drosophila virilis | 1 | 4 | 1 | 1 | 1 | 417 | 2.8 | 2,370 | 15.7 |
| Drosophila yakuba | 1 | 4 | 1 | 1 | 1 | 595 | 3.7 | 2,526 | 15.5 |
| Equus caballus | 1 | 24 | 2 | 1 | 2 | 10 | 0.0 | 20 | 0.1 |
| Gallus gallus | 1 | 87 | 9 | 1 | 9 | 2,349 | 9.4 | 469 | 1.9 |
| Homo sapiens | 16 | 1,800 | 60 | 4 | 66 | 4,871 | 8.3 | 17,223 | 29.5 |
| Macaca mulatta | 1 | 12 | 1 | 1 | 1 | 26 | 0.1 | 131 | 0.4 |
| Mus musculus | 7 | 276 | 20 | 3 | 20 | 606 | 1.1 | 1,092 | 2.0 |
| Pan troglodytes | 1 | 12 | 1 | 1 | 1 | 68 | 0.2 | 125 | 0.4 |
| Pristionchus pacificus | 1 | 6 | 1 | 1 | 1 | 973 | 3.3 | 1,898 | 6.4 |
| Rattus norvegicus | 3 | 323 | 12 | 2 | 21 | 221 | 0.7 | 460 | 1.4 |
| Total1 | 38 | 2,828 | 93 | 6 | 150 | 27,793 | 4.6 | 64,043 | 10.6 |
Notes: 1Duplicates were removed before summing up.
2Number of the SAGs.
3Percent of the genes in the genome.
4Female-biased genes.
5Male-biased genes.
To explore the conservation of sex bias within and among species, we compared sex bias of SAGs in adult somatic tissues among different human groups, as well as the groups between human and other species. The comparison revealed that 2.4–38.9% SAGs shared the same sex bias among different human groups (Supplementary Table S2), whereas only 0.2–9.2% SAGs shared the same sex bias between human species and other species (Supplementary Table S3). Multiple SAGs were found to be human-specific. For example, the gene LTF was female-biased in the adult liver of human, while unbiased in other species. It was reported to affect endometriosis (26), and is the drug target of NIMESULIDE for the treatment of excessive uterine bleeding during menstruation. The low conservation of sex bias across species could result from the varied sample size and experimental methods among groups. Alternatively, it might suggest that sex bias depends on species, and thus researchers need to be cautious when using animal models to study sex differences in drug response.
Browse and search of the database
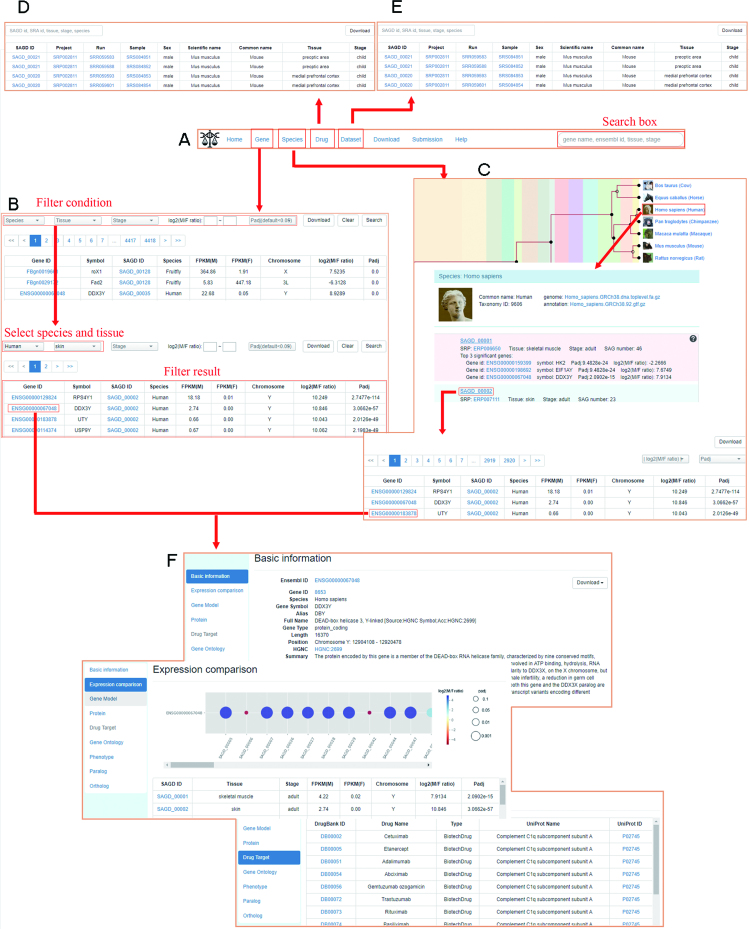
We designed a user-friendly webpage for the database. A quick search box was provided on the top navigation bar to search by keywords (i.e. gene symbol, ensemble ID, tissue and stage). Users can also browse SAGs of multiple species by gene, species, dataset and drug (Figure 2A).
Figure 2.
An overview of SAGD. (A) The homepage of SAGD. (B) Browse by gene. (C) Browse by species. The species tree was plotted by TimeTree (www.timetree.org) (29) with modifications. (D) Browse by drug. (E) Browse by dataset. (F) Information of each SAG.
On the webpage of gene, users can browse and search SAGs by species, tissue and developmental stage, and can refine the results with the range of sex bias (log2 (M/F ratio)) and difference significance (padj) (Figure 2B). For example, if users want to browse SAGs in human liver, they only need to select ‘Human’ and ‘liver’ in the drop-down menus of ‘Species’ and ‘Tissue’ on the top left, and click the ‘search’ button on the top right. The searching results will be exhibited in a table that contains padj, FPKM of each sex, and log2(M/F ratio) for each gene (Figure 2B). Users can start a new search after clicking the ‘clear’ button (Figure 2B).
On the webpage of species, users can view the phylogeny of 21 species covered by SAGD, and can browse SAGs in each dataset of the selected species (Figure 2C). The phylogeny is presented as a species tree with time scale. Users can select a species of interest and browse all its groups (Figure 2C). Group information contains project, tissue, stage, SAG number and top 3 significant genes. We colored the groups based on the log2 (M/F ratio) of the most significant gene for visualization. Users can browse all the genes in their interested group via the links of SAGD ID and find corresponding group datasets (Figure 2C).
On the webpage of drug, users can browse and search the SAG-targeting drugs by keywords Gene ID, DrugBank ID, Drug Name and Drug Type (Figure 2D).
On the webpage of dataset, users can browse all the RNA-seq datasets, and search by keywords SRA accession, species, tissue, stage and sex to find their interested datasets (Figure 2E).
All the four browse methods guide users to gene information pages, on which we integrated basic gene information from Ensembl BioMarts (27), expression comparison information from our RNA-seq data analysis, and drug target information from DrugBank (28). Sex-biased gene expressions across groups were shown in bubble plots (Figure 2F).
Downloads
All the search results can be downloaded as CSV files for customized analysis by clicking the Download button on the top right of almost all pages. Alternatively, SAGD offers users the RNA-seq data analysis results of each group in CSV files on the Download page.
Data submission
Users can submit relevant data by sending us a data information table via email. Currently, SAGD only accepts open access RNA-seq data from SRA for species with reference genomes and annotations from Ensembl or Ensembl Metazoa. The submitted data would be added to SAGD after curation and analysis as described in the section of Materials and Methods.
DISCUSSION
SAGD aims to provide users a comprehensive resource for SAGs by curating available high-quality raw RNA-seq datasets through the same pipeline. Multiple efforts were made to ensure the validity of this database. For example, (i) We curated metadata information including project, species, sex, tissue and developmental stage for the datasets from multiple sources. Manual inspection was conducted to ensure correctness and comprehensiveness. (ii) We only used datasets from the same project, species, tissue and developmental stage for SAG identification so as to ensure sex to be the major difference variable. (iii) We selected the groups with the most and at least two biological replicates for each condition, and performed quality control to ensure good replicability among biological replicates. (iv) We provided customized, instead of fixed, filters including sex bias and statistical significance level so that users could define their own SAGs. However, the determination of SAGs is a complicated issue encompassing a vast number of assumptions and hypotheses. Users should be cautious that if a gene is apparently sex-biased or not sex-biased in this database, the level of evidence should be examined carefully.
SUMMARY AND FUTURE DIRECTIONS
With the rapid accumulation of RNA-seq data, it is worthwhile to explore the function of SAGs by curating RNA-seq data from multiple species. SAGD facilitates users to explore their interested SAGs across projects, species, tissues and stages through customized browsing options.
The comparative analysis of SAGs within and across species requires comparable group pairs under the same condition. For such analysis, we will update SAGD regularly by adding more SAGs when additional RNA-seq datasets and reference genomes become available. SAGD will also provide more experimentally supported data as a solid resource for the studies of sex differences and comparative genomics.
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge Xiao-Shu Chen from Sun Yat-sen University for the assistance of data curation. We thank the contributors for providing the RNA-seq datasets. We thank Ze-Sheng Zhang from Hohai University for the assistance of web page design. We are also grateful to our users and all members in our lab for their valuable suggestions and comments. This work was conducted under dbGaP-approved protocol 16932 (accession phs000424.v7.p2).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Huazhong Agricultural University Scientific & Technological Self-innovation Foundation [2016RC011]; National Natural Science Foundation of China [31701259 and 31871305 to Z.X.C, 31822030 and 31771458 to A.Y.G]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Bachtrog D., Mank J.E., Peichel C.L., Kirkpatrick M., Otto S.P., Ashman T.L., Hahn M.W., Kitano J., Mayrose I., Ming R. et al. Sex determination: why so many ways of doing it. PLoS Biol. 2014; 12:e1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tree of Sex, C Tree of sex: a database of sexual systems. Scientific Data. 2014; 1:140015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ellegren H., Parsch J.. The evolution of sex-biased genes and sex-biased gene expression. Nat. Rev. Genet. 2007; 8:689–698. [DOI] [PubMed] [Google Scholar]
- 4. Grath S., Parsch J.. Sex-Biased gene expression. Annu. Rev. Genet. 2016; 50:29–44. [DOI] [PubMed] [Google Scholar]
- 5. Parsch J., Ellegren H.. The evolutionary causes and consequences of sex-biased gene expression. Nat. Rev. Genet. 2013; 14:83–87. [DOI] [PubMed] [Google Scholar]
- 6. Hall A.B., Basu S., Jiang X., Qi Y., Timoshevskiy V.A., Biedler J.K., Sharakhova M.V., Elahi R., Anderson M.A., Chen X.G. et al. SEX DETERMINATION. A male-determining factor in the mosquito Aedes aegypti. Science. 2015; 348:1268–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Graves J.A. Evolution of vertebrate sex chromosomes and dosage compensation. Nat. Rev. Genet. 2016; 17:33–46. [DOI] [PubMed] [Google Scholar]
- 8. Gilks W.P., Abbott J.K., Morrow E.H.. Sex differences in disease genetics: evidence, evolution, and detection. Trends Genet. 2014; 30:453–463. [DOI] [PubMed] [Google Scholar]
- 9. Yuan Y., Liu L., Chen H., Wang Y., Xu Y., Mao H., Li J., Mills G.B., Shu Y., Li L. et al. Comprehensive characterization of molecular differences in cancer between male and female patients. Cancer Cell. 2016; 29:711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Morrow E.H. The evolution of sex differences in disease. Biol. Sex Differ. 2015; 6:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen Z.X., Sturgill D., Qu J., Jiang H., Park S., Boley N., Suzuki A.M., Fletcher A.R., Plachetzki D.C., FitzGerald P.C. et al. Comparative validation of the D. melanogaster modENCODE transcriptome annotation. Genome Res. 2014; 24:1209–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Meisel R.P., Malone J.H., Clark A.G.. Disentangling the relationship between sex-biased gene expression and X-linkage. Genome Res. 2012; 22:1255–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim B., Suo B., Emmons S.W.. Gene function prediction based on developmental transcriptomes of the two sexes in C. elegans. Cell Rep. 2016; 17:917–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Blekhman R., Marioni J.C., Zumbo P., Stephens M., Gilad Y.. Sex-specific and lineage-specific alternative splicing in primates. Genome Res. 2010; 20:180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lin Y., Golovnina K., Chen Z.X., Lee H.N., Negron Y.L., Sultana H., Oliver B., Harbison S.T.. Comparison of normalization and differential expression analyses using RNA-Seq data from 726 individual Drosophila melanogaster. BMC Genomics. 2016; 17:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lin Y., Chen Z.X., Oliver B., Harbison S.T.. Microenvironmental gene expression plasticity among individual drosophila melanogaster. G3. 2016; 6:4197–4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Assis R., Zhou Q., Bachtrog D.. Sex-biased transcriptome evolution in Drosophila. Genome Biol. Evol. 2012; 4:1189–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gnad F., Parsch J.. Sebida: a database for the functional and evolutionary analysis of genes with sex-biased expression. Bioinformatics. 2006; 22:2577–2579. [DOI] [PubMed] [Google Scholar]
- 19. Papatheodorou I., Fonseca N.A., Keays M., Tang Y.A., Barrera E., Bazant W., Burke M., Fullgrabe A., Fuentes A.M., George N. et al. Expression Atlas: gene and protein expression across multiple studies and organisms. Nucleic Acids Res. 2018; 46:D246–D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M. et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013; 41:D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhu Y., Stephens R.M., Meltzer P.S., Davis S.R.. SRAdb: query and use public next-generation sequencing data from within R. BMC Bioinformatics. 2013; 14:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kersey P.J., Allen J.E., Allot A., Barba M., Boddu S., Bolt B.J., Carvalho-Silva D., Christensen M., Davis P., Grabmueller C. et al. Ensembl Genomes 2018: an integrated omics infrastructure for non-vertebrate species. Nucleic Acids Res. 2018; 46:D802–D808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim D., Langmead B., Salzberg S.L.. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015; 12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Anders S., Pyl P.T., Huber W.. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Polak G., Wertel I., Tarkowski R., Morawska D., Kotarski J.. Decreased lactoferrin levels in peritoneal fluid of women with minimal endometriosis. Eur. J. Obstet. Gynecol. Reprod. Biol. 2007; 131:93–96. [DOI] [PubMed] [Google Scholar]
- 27. Kinsella R.J., Kahari A., Haider S., Zamora J., Proctor G., Spudich G., Almeida-King J., Staines D., Derwent P., Kerhornou A. et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database. 2011; 2011:bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z. et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018; 46:D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kumar S., Stecher G., Suleski M., Hedges S.B.. TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 2017; 34:1812–1819. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.