Chronic hepatitis B virus (HBV) infection is a major cause of advanced liver disease and hepatocellular carcinoma (HCC) world-wide (1). Current antiviral treatments control HBV replication and reduce progressive liver disease. However, viral cure is rarely achieved and patients still remain at risk for HCC (1). The pathogenesis of HBV-induced HCC is thought to be multifactorial with both direct and indirect mechanisms (2): HBV-related HCCs can also arise in non-cirrhotic livers, supporting the notion that HBV plays a direct role in liver transformation by triggering both common and etiology specific oncogenic pathways in addition to stimulating the host immune response and driving liver chronic necro-inflammation. Moreover, certain viral genotypes and HBV variants have been proposed to increase HCC risk (3): Epidemiological studies suggest that HBV genotypes C, D and F carry a higher lifetime risk of HCC development than genotype A and B (1). In the United States, Alaska Native (AN) individuals are characterized by the highest HBV seroprevalence, with a high incidence of HCC. HBV genotype F and its subtype 1 (F1) is highly prevalent in AN individuals with HCC and has therefore suggested to predispose to HCC (4,5). Among naturally occurring viral variants, HBV core promoter and precore/core mutants have been shown to be associated with enhanced HCC risk (6). Mutations in the basal core promoter (BCP) region enhance viral replication by creating a binding site for hepatocyte nuclear factor (HNF) protein leading to increased transcription of viral RNA (7) or enhanced viral encapsidation (8). The double mutation A1762T/G1764A is the most common mutation found in the BCP. In the pre-core region (PC), the predominant mutation is G1896A, which modulates of HBeAg synthesis at the translational level by introducing a stop codon (6). However, there are only few functional studies investigating the functional relationship between genetic alterations and disease biology in in vivo model systems. A better understanding of the functional relationship between HBV variants and their impact on liver disease and carcinogenesis is important not only for the understanding of their mechanism of action but also has potential impact for determining HCC risk and patient management.
In this issue of Hepatology, a collaborative research effort between the teams of Dr. Brian J. McMahon, Anchorage and Prof. Yasuhito Tanaka, Nagoya, Japan has addressed this important question by identifying genetic variants associated with HCC and investigating their phenotype in cell culture and animal models (9). Hayashi and colleagues first analyzed the clinical and virological significance of genotype F1b in long-term serial samples from 20 HCC patients with HBV infection. The authors showed that all isolates were of genotype F1b. Core T1938C (V13A) and A2051C (N51H) mutations had accumulated together with BCP and PC mutations, suggesting a functional association between these variants and hepatocarcinogenesis (9). The N-terminal part of core protein is crucial for self-assembly, while the C-terminal part is essential for nucleocapsid formation. Interestingly, T1938C and A2051C mutations are located in the capsid assembly domain of the core protein. Using an expression cloning approach combined with site-directed mutagenesis, the authors observed that the core A2051C mutation enhances HBV replication by stabilizing core protein dimerization putatively facilitating nucleocapsid formation. Next, to address the structure-function relevance of the variants selected in HCC in vivo they used human liver chimeric mice - a state-of-the-art animal model for the study of HBV infection in engrafted human hepatocytes in the mouse liver. Chimeric mice were inoculated with wild-type virus or recombinant virus containing the BCP A1762T/G1764A mutations, the PC G1896A mutation or the combination BCP/PC/2051. Interestingly, the variants exhibited higher levels of replication and protein expression than the wild-type strain as indicated by enhanced levels of HBV DNA and HBsAg in the serum of infected mice.
To better understand the disease biology associated with the mutations, the authors investigated the functional consequences of viral infection on the liver transcriptome using microarray. Comparative analyses between wild-type and mutant strains showed that the mutations resulted in the upregulation of the expression of genes associated with cell proliferation and hepatocarcinogenesis: these included MYC (c-myc avian myelocytomatosis viral oncogene homolog), GAB2 (Grb2-Associated Binding Protein 2), BDKRB2 (bradykinin receptor B2), FST (follistatin) and MAP3K8 (mitogen-activated protein). Furthermore, the authors observed that the BCP/PC/2051 mutations were associated with a modulated expression of liver inflammation and fibrosis-related genes such as C-X-C chemokine receptor type 7, endothelin 1, insulin-like growth factor binding protein 3 and intercellular adhesion molecule 1 (Figure 1).
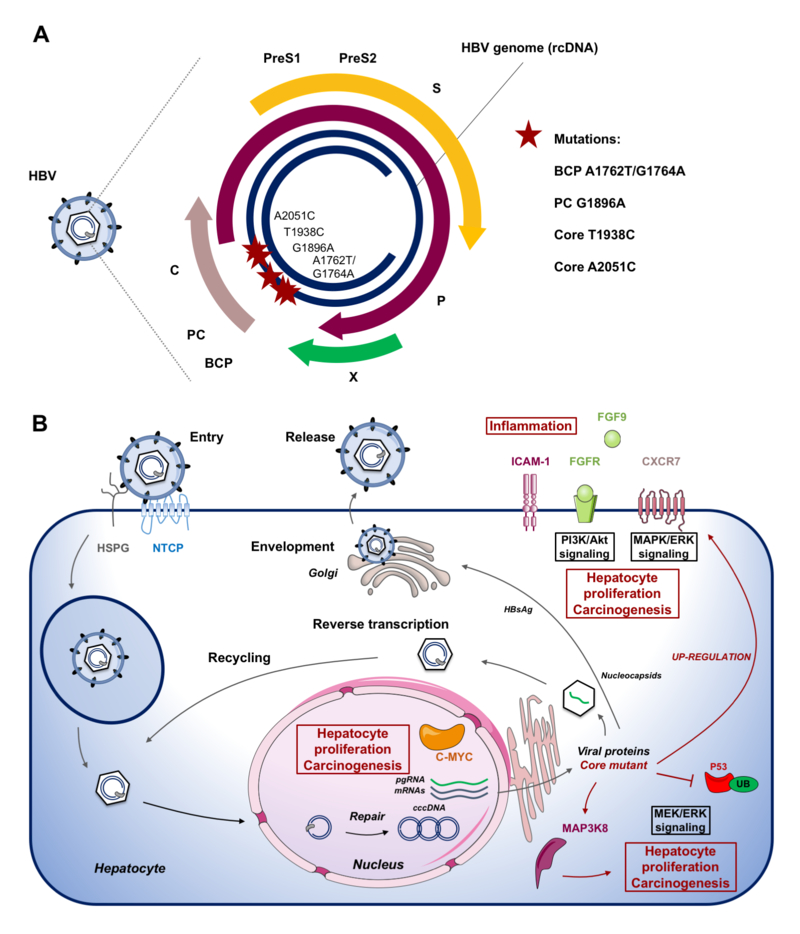
Figure 1: HBV core mutations associated with fibrosis and hepatocarcinogenesis.
A. Schematic representation of the HBV genome with clinical mutations. An accumulation of Core T1938C and A2051C mutations together with BCP (A1762T/G1764A) and PC (G1896A) mutations were observed in Alaskan native HCC patients (9). The mutations, leading to an increased viral replication, are indicated by stars on the genome. B. Effects of HBV core mutants on the HBV life cycle and host cell pathways in liver chimeric mice as observed by Hayashi et al. (9). Following viral entry mediated by HSPG and NTCP, the HBV genome is released into the nucleus where rcDNA is converted into cccDNA. cccDNA serves as template for viral transcripts including pgRNA, which is encapsidated and reverse transcribed into de novo HBV genomic DNA before assembly and release of the viral particle. Hayashi et al. demonstrated that core mutations enhance HBV replication in genotype F1b. In human liver chimeric mice infection with mutant virus resulted in up-regulation/activation of pathways potentially driving fibrosis and carcinogenesis. Abbreviations: BCP, basal core promotor; c-MYC, c-myc avian myelocytomatosis viral oncogene homolog; cccDNA, covalently closed circular DNA; CXCR7, C-X-C chemokine receptor type 7; FGF9, fibroblast growth factor 9; FGFR, fibroblast growth factor receptor; ICAM-1, intercellular adhesion molecule 1; MAP3K8, mitogen-activated protein; NTCP, NA+-taurocholate costransporting polypeptide; P, polymerase; HSPG, heparan sulfate proteoglycan; PC, core promotor; pgRNA, pregenome RNA; PreS, PreS region; S, surface protein; X, X protein.
Finally, the authors studied the liver disease biology associated with this variant using detailed histopathological analyses of the humanized part of the livers. Importantly, infections with the core variant appeared to be associated with fibrosis in the liver of HBV-infected chimeric mice. The authors also observed an enhanced inflammation, although the meaning of this finding remains to be determined since the immune system of these mice is heavily disturbed by the severe combined immunodeficiency (SCID) phenotype. Additional in vivo studies using recombinant viruses containing BCP/PC mutations with and without the A2051C or the T1938C mutation revealed that the phenotype of fibrosis appeared to be predominantly caused by the A2051C mutation.
Collectively, these data provide strong evidence that this variant may have a causal role for HCC observed in the AN patients. One potential limitation of the study is that the variant infection did not result in HCC in the mouse model. However, the absence of hepatocarcinogenesis is a known feature of this animal model, which is most likely due the absence of a functional immune system with lack of inflammatory pro-carcinogenic antiviral immune responses. Alternatively, the duration of the experiment was not long enough to observe hepatocarcinogenesis. Collectively, this study provides an important advancement in the field by showing convincing evidence of a robust structural and functional relationship between a patient-derived core variant and liver disease biology. In particular, the observation of variant-associated liver fibrosis is an important finding since fibrosis stage has been shown to be a key predictor and driver of hepatocarcinogenesis in various etiologies (10).
What are the clinical implications of these findings and the next studies further advancing the findings of the authors? First, it would be of interest to study the prevalence of the identified core mutation in a larger Alaskan cohort as well as cohorts infected with other HBV genotypes. Further studies may also address the question whether certain genotypes, such as genotype F, predispose to the mutation by genotype-specific sequences. Finally, it would be of interest to study whether or not the mutants are associated with human HCC that occurs in the absence of liver cirrhosis. These studies would give important insights of the impact and prevalence of the mutation the association with HCC risk in general. It may be of interest to assess whether the detection of this variant could serve as an additional predictor of HCC risk triggering e.g. more intense surveillance of patients harboring this mutation. Second, a follow-up study on the molecular virus-host interactions of this variant may potentially provide an opportunity to uncover novel targets for chemopreventive approaches for HCC.
By providing a convincing structure-function relationship of the core mutant with liver fibrosis in a state-of-the-art animal model, the study significantly advances our understanding of the role of variants in the pathogenesis of liver disease and HCC and will stimulate further research on the role of viral factors in hepatocarcinogenesis.
Acknowledgements.
The authors acknowledge support by the European Union (ERC-2014-AdG-671231-HEPCIR, EU-H2020–667273- HEPCAR), ANRS (15/1099), ARC (IHU201301187), ANR (Laboratoire d’Excellence HEPSYS ANR-10-LAB-28 and Infect-ERA hepBccc) and the National Institutes of Health (NIAID U19AI123862–01, NIAID R03AI131066, NCI R21CA209940).
Footnotes
Conflict of interest. The authors declare no conflict of interest.
Comment on: “A novel association between core mutations in hepatitis B virus genotype F1b and hepatocellular carcinoma in Alaskan Native People” by Sanae Hayashi, Anis Khan, Brenna C. Simons, Chriss Homan, Takeshi Matsui, Kenji Ogawa, Keigo Kawashima, Shuko Murakami, Satoru Takahashi, Masanori Isogawa, Kazuho Ikeo, Masashi Mizokami, Brian J. McMahon, Yasuhito Tanaka. Hepatology 2018, in press.
References
- 1.Trépo C, Chan HLY, Lok A. Hepatitis B virus infection. Lancet. 2014;384:2053–2063. [DOI] [PubMed] [Google Scholar]
- 2.Levrero M, Zucman-Rossi J. Mechanisms of HBV-induced hepatocellular carcinoma. J. Hepatol 2016;64:S84–S101. [DOI] [PubMed] [Google Scholar]
- 3.Lin C-L, Kao J-H. Natural history of acute and chronic hepatitis B: The role of HBV genotypes and mutants. Best Pract Res Clin Gastroenterol 2017;31:249–255. [DOI] [PubMed] [Google Scholar]
- 4.Gounder PP, Bulkow LR, Snowball M, Negus S, Spradling PR, McMahon BJ. Hepatocellular carcinoma risk in Alaska native children and young adults with hepatitis B virus: retrospective cohort analysis. J. Pediatr 2016;178:206–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ching LK, Gounder PP, Bulkow L, Spradling PR, Bruce MG, Negus S, et al. Incidence of hepatocellular carcinoma according to hepatitis B virus genotype in Alaska Native people. Liver Int 2016;36:1507–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin C-L, Kao J-H. Hepatitis B virus genotypes and variants. Cold Spring Harb Perspect Med 2015;5:a021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Günther S, Piwon N, Iwanska A, Schilling R, Meisel H, Will H. Type, prevalence, and significance of core promoter/enhancer II mutations in hepatitis B viruses from immunosuppressed patients with severe liver disease. J. Virol 1996;70:8318–8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baumert TF, Rogers SA, Hasegawa K, Liang TJ. Two core promotor mutations identified in a hepatitis B virus strain associated with fulminant hepatitis result in enhanced viral replication. J. Clin. Invest 1996;98:2268–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayashi S, Khan A, Simons BC, Homan C, Matsui T, Ogawa K, et al. A novel association between core mutations in hepatitis B virus genotype F1b and hepatocellular carcinoma in Alaskan Native People. Hepatology 2018, in press [DOI] [PubMed] [Google Scholar]
- 10.Hagström H, Nasr P, Ekstedt M, Hammar U, Stål P, Hultcrantz R, et al. Fibrosis stage but not NASH predicts mortality and time to development of severe liver disease in biopsy-proven NAFLD. J. Hepatol 2017;67:1265–1273. [DOI] [PubMed] [Google Scholar]