New therapeutic approaches are needed against Mycobacterium abscessus, a respiratory mycobacterial pathogen that evades efforts to successfully treat infected patients. Clofazimine and bedaquiline, two drugs used for the treatment of multidrug-resistant tuberculosis, are being considered as alternatives for the treatment of lung diseases caused by M. abscessus.
KEYWORDS: EMSA, MmpL, Mycobacterium abscessus, TetR regulator, bedaquiline, clofazimine, drug resistance mechanisms, efflux pumps
ABSTRACT
New therapeutic approaches are needed against Mycobacterium abscessus, a respiratory mycobacterial pathogen that evades efforts to successfully treat infected patients. Clofazimine and bedaquiline, two drugs used for the treatment of multidrug-resistant tuberculosis, are being considered as alternatives for the treatment of lung diseases caused by M. abscessus. With the aim to understand the mechanism of action of these agents in M. abscessus, we sought herein to determine the means by which M. abscessus can develop resistance. Spontaneous resistant strains selected on clofazimine, followed by whole-genome sequencing, identified mutations in MAB_2299c, encoding a putative TetR transcriptional regulator. Unexpectedly, mutants with these mutations were also cross-resistant to bedaquiline. MAB_2299c was found to bind to its target DNA, located upstream of the divergently oriented MAB_2300-MAB_2301 gene cluster, encoding MmpS/MmpL membrane proteins. Point mutations or deletion of MAB_2299c was associated with the concomitant upregulation of the mmpS and mmpL transcripts and accounted for this cross-resistance. Strikingly, deletion of MAB_2300 and MAB_2301 in the MAB_2299c mutant strain restored susceptibility to bedaquiline and clofazimine. Overall, these results expand our knowledge with respect to the regulatory mechanisms of the MmpL family of proteins and a novel mechanism of drug resistance in this difficult-to-treat respiratory mycobacterial pathogen. Therefore, MAB_2299c may represent an important marker of resistance to be considered in the treatment of M. abscessus diseases with clofazimine and bedaquiline in clinical settings.
INTRODUCTION
Mycobacterium abscessus is an emerging nontuberculous mycobacterium (NTM) commonly associated with contaminated traumatic skin wounds or postsurgical soft tissue infections (1). Among the rapid growers, M. abscessus also represents the most frequently isolated species in cystic fibrosis (CF) patients, with a prevalence of 3% to 6% in this population (2), or in patients with other underlying lung disorders, such as non-CF bronchiectasis and chronic obstructive pulmonary disease (COPD), resulting in nodular and cavitary granulomas and persistent lung infection (3–6). CF patients with chronic M. abscessus infection have higher rates of lung function decline than those with no NTM infections (7). Recent surveys have identified M. abscessus to be a major threat in many CF centers worldwide (5), and this alarming situation is worsened by epidemiological studies documenting the transmission of dominant circulating M. abscessus clones that have spread globally between hospitals (8). In addition, lung infections caused by M. abscessus remain extremely difficult to treat, mainly because of its natural resistance to most currently available antibiotics (9). The prognosis of pulmonary infections is poor, particularly in the context of CF, with a cure rate of 30% to 50%, in spite of lengthy courses of antibiotics often complemented by surgery (10). In contrast to many other NTM infections, antibiotherapy against M. abscessus often fails, leading to lasting sputum culture positivity, and no antibiotic regimen reliably cures these infections (3, 11–14).
There is an important medical need to discover and develop more efficient and safer treatments to fight against M. abscessus. The proposed strategy for fueling a drug pipeline can rely on (i) de novo drug discovery to identify new pharmacophores and targets, (ii) repurposing of known drugs as new treatments of M. abscessus infections, or (iii) the repositioning of antibiotics that act against pharmacologically validated targets but that have been developed for the treatment of other infectious diseases (15). Among the repositioned drugs over which there is increasing interest, clofazimine (CFZ) and bedaquiline (BDQ) are currently being evaluated in clinical trials for their activities against M. abscessus pulmonary infections.
The riminophenazine clofazimine (CFZ) is a lipophilic agent with cationic amphiphilic properties used as an antileprosy drug and currently repurposed as an antituberculosis (anti-TB) drug (16). CFZ also has unique characteristics, such as a slow metabolic elimination, preferential accumulation inside macrophages, and a low incidence of drug resistance (16). In M. tuberculosis, CFZ acts as a prodrug which is reduced by the NADH dehydrogenase (Ndh2) and, upon spontaneous reoxidation by O2, releases reactive oxygen species (ROS) (17), explaining why high levels of resistance to CFZ are rare. Due to the recent widespread emergence of M. abscessus, there has been a renewed interest in the repurposing of CFZ for the treatment of M. abscessus infections. In vitro studies reported the efficacy of CFZ in multidrug regimens for the treatment of M. abscessus infections, in which it showed synergistic activity with amikacin (18) or tigecycline (19). CFZ was also found to prevent the regrowth of M. abscessus exposed to clarithromycin and amikacin (13). Although CFZ has increasingly been used in the treatment of lung diseases in clinical practice (10, 20), only limited data on its effectiveness are available. In one study, CFZ was found to be safe, reasonably tolerated, and active when given orally for the treatment of NTM infections, including M. abscessus infections, and was proposed to be an alternative drug for the treatment of NTM diseases (21). In another study, CFZ-containing regimens also showed improved treatment outcomes in patients with pulmonary diseases due to M. abscessus (22).
BDQ is a diarylquinoline antibiotic that has been approved by the Food and Drug Administration and the European Medicines Agency for the treatment of multidrug-resistant tuberculosis (MDR-TB) (23). BDQ acts by targeting the c subunit of the essential FoF1 ATP synthase (24–27), and studies have also proposed that it may inhibit the ATP synthase via another mechanism involving the ε subunit of the enzyme, in addition to binding to its c subunit (28, 29). BDQ exhibits very low MICs against various NTM, including M. abscessus clinical isolates from CF and non-CF patients (27, 30, 31), but despite being an excellent growth inhibitor at low doses, it lacks bactericidal activity against M. abscessus (27). Studies in immunocompromised mouse models led to conflictual conclusions, reporting, on the one hand, a benefit of BDQ in reducing bacterial loads in gamma interferon knockout mice (32) and, on the other hand, no decrease in the bacillary loads in the lungs or an inability to prevent death in nude mice (33). However, BDQ is highly efficient in reducing pathophysiological signs, such as abscesses and cords in M. abscessus-infected zebrafish, and in protecting zebrafish larvae from death (27). The mode of action of BDQ in M. abscessus relies on rapid ATP depletion and was demonstrated by genetically transferring single point mutations into atpE, which conferred high levels of resistance to the drug (27). Preliminary results of studies using BDQ as salvage therapy for pulmonary infections with M. avium or M. abscessus suggested that BDQ has clinical activity but its efficacy appears to be relatively moderate, as suggested by a low sputum culture conversion rate (34). Therefore, the clinical utility of BDQ against M. abscessus infections requires more studies.
To further delineate the mechanism of action of CFZ and BDQ in M. abscessus, we sought to determine how M. abscessus develops resistance to these agents. Herein, mutations were identified in a new TetR regulator, MAB_2299c. These mutations were associated with low levels of resistance to both CFZ and BDQ. Genetic and biochemical analyses were applied to determine the specificity of this regulatory system in M. abscessus and to describe the contribution of a key residue important in driving the DNA-binding activity of MAB_2299c to its operator. The results expand our knowledge with respect to the regulatory mechanisms of the MmpL family of efflux pumps and on a novel mechanism of drug resistance in M. abscessus.
RESULTS
Mutations in MAB_2299c confer coresistance to CFZ and BDQ.
With the aim of identifying the mechanism of resistance to CFZ in M. abscessus, 6 spontaneous CFZ-resistant mutants were first reared in passages of the reference M. abscessus ATCC 19977 S strain in liquid medium containing increasing concentrations of CFZ and then isolated on solid 7H11 medium supplemented with oleic acid-albumin-dextrose-catalase (OADC) (7H11OADC medium). All 6 resistors exhibited low resistance levels (MIC, 8 µg/ml) compared to the resistance level of the parental strain (MIC, 4 µg/ml) (Table 1). Spontaneous resistant mutants arose at a frequency of ≈2 × 10−7. Whole-genome sequencing identified mutations in the MAB_2299c locus in all 6 mutants, which were subsequently confirmed by PCR amplification and Sanger sequencing. This approach identified single nucleotide polymorphisms (SNP) in mutants CFZ-R1 (L40W replacement), CFZ-R4 (stop codon), CFZ-R7 (L151P replacement), and CFZ-R9 (G215S replacement), as well as a single nucleotide deletion in CFZ-R3 or a single nucleotide insertion in CFZ-R6 leading to frameshift mutations (Table 1). These observations converge to a prominent role of MAB_2299c in the CFZ resistance phenotype. Interestingly, all 6 CFZ-resistant mutants were also coresistant to BDQ with MIC levels of 2 µg/ml, corresponding to a 4-fold increased MIC compared to that for the parental strain (Table 1). In contrast, all mutants remained susceptible to amikacin (AMK). These results imply that mutations in MAB_2299c confer cross-resistance to CFZ and BDQ but not to AMK, pointing out a unique resistance mechanism.
TABLE 1.
Drug susceptibility profiles and genotypes of 6 spontaneous CFZ-resistant M. abscessus strainsa
| Strain | MIC99 (µg/ml) |
Mutation in MAB_2299c |
|||
|---|---|---|---|---|---|
| CFZ | BDQ | AMK | SNP/indel | Amino acid change | |
| Wild type | 4 | 0.5 | 8 | ||
| CFZ-R1 | 8 | 2 | 8 | T119G | L40W |
| CFZ-R3 | 8 | 2 | 8 | C276del | P92fs |
| CFZ-R4 | 8 | 2 | 8 | G541T | E181stop |
| CFZ-R6 | 8 | 2 | 8 | ins318A | D106fs |
| CFZ-R7 | 8 | 2 | 8 | T452C | L151P |
| CFZ-R9 | 8 | 2 | 8 | G643A | G215S |
The mutants were derived from the ATCC 199177 S parental strain and selected in the presence of CFZ. MIC99 values were determined on Middlebrook 7H10 agar. Single nucleotide polymorphisms (SNP) and/or indels were identified in MAB_2299c. The corresponding amino acid changes are also indicated. CFZ, clofazimine; BDQ, bedaquiline; AMK, amikacin; del, deletion; ins, insertion; fs, frameshift.
MAB_2299c encodes a TetR repressor controlling expression of a specific MmpS/MmpL pair.
Sequence alignments and BLAST analyses indicated that MAB_2299c encodes a putative TetR transcriptional regulator. TetR family members possess a conserved N-terminal helix-turn-helix (HTH) DNA-binding domain and a C-terminal ligand regulatory domain and are commonly associated with antibiotic resistance by regulating expression of genes coding for multidrug resistance efflux pumps (35). Interestingly, the two gene pairs (MAB_2300-MAB_2301 and MAB_2302-MAB_2303) adjacent to MAB_2299c and transcribed in the opposite direction code for MmpS (MAB_2300 and MAB_2302) and MmpL (MAB_2301 and MAB_2303) integral membrane proteins (Fig. 1). MmpL proteins are part of the superfamily of resistance, nodulation, and division (RND) transporters and were reported to act as efflux pumps for azoles, CFZ, and BDQ in M. tuberculosis (36, 37) and for thiacetazone analogues in M. abscessus (38, 39).
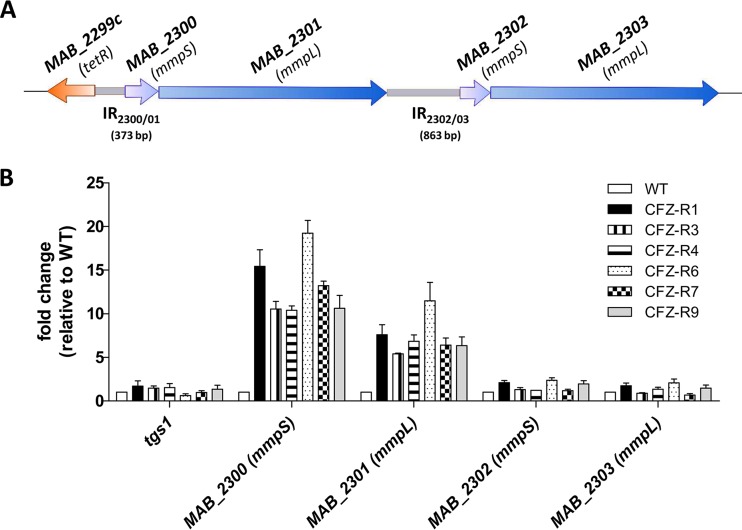
FIG 1.
MAB_2299c regulates expression of the MAB_2300-MAB_2301 locus. (A) Schematic representation of the genetic environment of MAB_2299c and its two adjacent mmpS-mmpL gene pairs (MAB_2300-MAB_2301 and MAB_2302-MAB_2303). The sizes and positions of the two intergenic regions (IR2300/01 and IR2302/03) are indicated. (B) Transcriptional expression of the MAB_2300-MAB_2303 genes in the parental M. abscessus strain and in 6 spontaneous CFZ-resistant mutants harboring mutations in MAB_2299c. The results are expressed as the fold change in mRNA levels between the mutant strains and the parental strain. Error bars indicate the standard deviation. Relative gene expression was calculated using the 2−ΔΔCT threshold cycle (CT) method with PCR efficiency correction. The data are representative of those from three independent experiments.
To explore whether the TetR regulator MAB_2299c controls the expression of the neighboring mmpS and mmpL genes, quantitative reverse transcription-PCR (qRT-PCR) was first performed both in the parental M. abscessus S strain and in the 6 CFZ- and BDQ-resistant derivatives harboring the various MAB_2299c alleles. The results clearly showed the induction of both the MAB_2300 and the MAB_2301 transcripts in all 6 mutants (Fig. 1B). The expression levels of tgs1, which encodes the triacylglycerol synthase, involved in the synthesis and accumulation of triglycerides in M. abscessus (40), and which was included as an unrelated gene control, were found to remain unchanged in the various strains tested (Fig. 1B). In contrast, no induction of the expression levels of MAB_2302 and MAB_2303 transcripts was observed in the different mutant strains (Fig. 1B). This suggests that MAB_2299c represses expression of only the MAB_2300-MAB_2301 (mmpS-mmpL) pair.
MAB_2299c binds to a palindromic sequence upstream of MAB_2300.
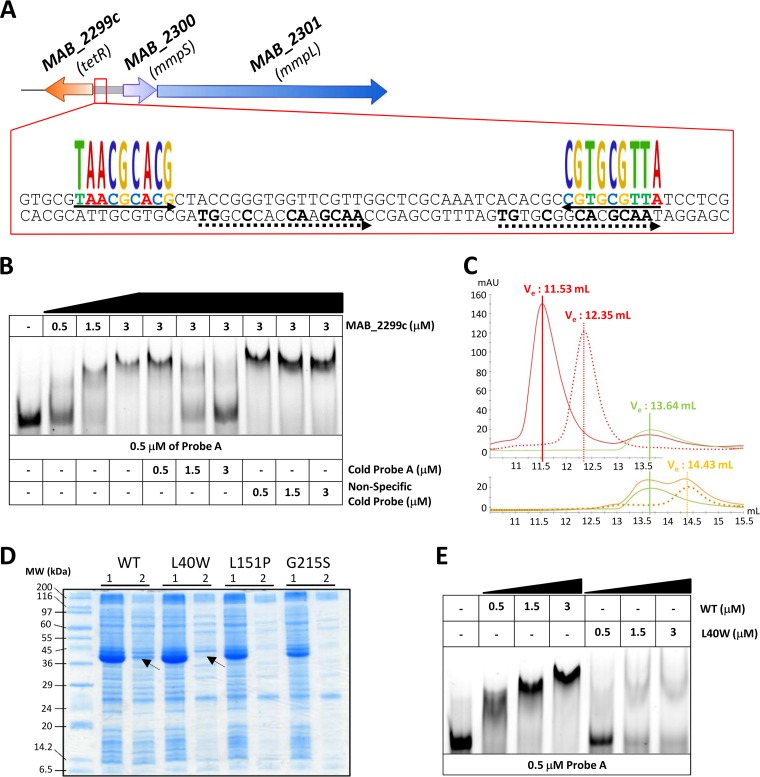
Using MEME, a motif-based sequence analysis tool (41), a 64-bp DNA segment within the 373-bp intergenic region between MAB_2299c and MAB_2300 (IR2300/01) harboring two palindromic sequences as well as two degenerated repeat motifs was identified (Fig. 2A). To test whether this 64-bp fragment represents a DNA-binding site for MAB_2299c, electrophoretic mobility shift assays (EMSA) were performed using increasing concentrations of purified MAB_2299c in the presence of the corresponding labeled fragment (probe A; Fig. 2B). Under these conditions, a DNA-protein complex was detected. The specificity of binding was demonstrated in a competition assay by adding increasing concentrations of the corresponding unlabeled DNA fragment (cold probe), which led to a dose-dependent decrease in DNA-protein complex formation (Fig. 2B). Moreover, the shift was maintained in the presence of an excess of a nonrelated probe, further indicating that a specific protein-DNA complex was formed only when the TetR regulator was incubated with DNA containing its specific target. These results were also confirmed by size exclusion chromatography (SEC), where the MAB_2299c-probe A complex eluted at 11.53 ml and could be readily separated from the protein alone (which eluted at 13.64 ml) or the DNA target (which eluted at 12.35 ml) (Fig. 2C, top). In contrast, no protein-DNA complex was eluted when MAB_2299c was incubated with the DNA target from MAB_4384, a previously characterized TetR regulator in M. abscessus (38, 39) (Fig. 2C, bottom), further highlighting the specificity of the MAB_2299c/IR2300/01 interaction. In addition, the pronounced shift impairment using mutated derivatives of probe A lacking either the palindromic sequence (ΔPalin) or the degenerated double repeats (ΔDR), generated by replacing the original sequences with random nucleotides, suggests that both the palindrome and the double repeats are required for the optimal binding of MAB_2299c to its operator (see Fig. S1 in the supplemental material).
FIG 2.
Binding activity of MAB_2299c to the intergenic region upstream of MAB_2300-MAB_2301. (A) Schematic representation of the 64-bp DNA operator identified within IR2300/01 corresponding to the sequence of probe A. The oligonucleotides recognized by MAB_2299c are composed of two palindromes (underlined by arrows) and two degenerated double repeats (underlined by dashed arrows). (B) EMSA and competition assays using probe A and purified MAB_2299c. Gel shifts were revealed by fluorescence emission using a 5′ fluorescein-labeled probe A. (C) Gel filtration profiles of free probe A, free MAB_2299c, and the TetR-DNA complex. Probe A (red dotted line) and MAB_2299c (green line) were isolated individually by size exclusion chromatography and displayed elution volumes (Ve) of 12.35 ml and 13.64 ml, respectively. When mixed together, a stable MAB_2299c-probe A complex (red line) with an elution volume of 11.53 ml was observed. The MAB_4384-specific palindromic sequence (orange dotted line) eluted at 14.43 ml. However, when mixed together with MAB_2299c, no protein-DNA complex was formed and both the DNA and protein were eluted separately (orange line) as two distinct peaks, with the elution volumes corresponding exactly to those for the MAB_4384 palindrome and MAB_2299c protein alone. mAU, milli-absorbance units. (D) Expression of the various MAB_2299c variants in E. coli. Lanes 1, total crude extract; lanes 2, clarified/soluble extract. The theoretical molecular mass of MAB_2299c-6His-TrxA is 41,420 Da. MW, molecular weight. (E) Impaired DNA-binding activity of the MAB_2299c L40W mutant, as shown by EMSA using either the soluble MAB_2299c (WT) or MAB_2299c (L40W) protein. Gel shifts were revealed by the fluorescence emission thanks to fluorescein-labeled probe A.
Screening of the entire 863-bp IR2302/03 located upstream of the second mmpS-mmpL pair (Fig. 1A) using MEME failed to identify inverted repeats that looked similar to the ones found in IR2300/01, suggesting that the regulator is unlikely to recognize a DNA-binding sequence in the upstream region of MAB_2302-MAB_2303. To confirm this hypothesis, EMSAs were done by incubating increasing concentrations of MAB_2299c with 3 overlapping probes covering the entire IR2302/03 region (Fig. S2A). Even at the highest protein concentration tested, no protein-DNA complexes were observed with either of the three probes (Fig. S2B). These results are in agreement with the qRT-PCR results (Fig. 1B) and support the view that MAB_2299c is unable to bind to IR2302/03 and to regulate the expression of this second mmpS-mmpL pair.
Overall, these results confirm the strict DNA sequence requirements for the optimal binding of MAB_2299c to IR2300/01.
Leu40 is critical for optimal DNA-binding activity of MAB_2299c.
To get insights into the mechanisms by which the different point mutations in CFZ-R1, CFZ-R7, and CFZ-R9 confer resistance to CFZ and BDQ, the three corresponding MAB_2299c alleles were cloned into pET32a and introduced into E. coli for expression. Figure 2D shows that although the 3 mutated proteins were highly expressed, only a very small fraction of the L40W protein was found in the soluble fraction, whereas the vast majority of the L151P and G215S mutants remained insoluble. Despite many attempts and by using large E. coli cultures, we failed to obtain enough soluble L151P and G215S proteins for subsequent purification but succeeded in generating the L40W derivative in a soluble form. Due to its localization within the N-terminal domain of the TetR regulator, known to participate in the DNA-binding activity of the protein (35), we addressed whether the L40W substitution conferring resistance to CFZ and BDQ affects the DNA-binding activity of MAB_2299c. EMSAs were therefore performed using purified MAB_2299c (L40W) and probe A. Compared to the shift profile with wild-type (WT) MAB_2299c, the formation of the DNA-protein complex was severely impaired in the presence of the mutated protein, even at high protein concentrations (Fig. 2E).
Overall, these results support the importance of Leu40 in the DNA-binding capacity of MAB_2299c and explain the reduced ability of the mutants to bind to the operator region, in agreement with the derepression of MAB_2300-MAB_2301 transcription in the CFZ-R1 M. abscessus strain carrying the L40W substitution (Fig. 1B).
An unmarked deletion of MAB_2299c leads to upregulation of the MAB_2300-MAB_2301 efflux pump and coresistance to CFZ and BDQ.
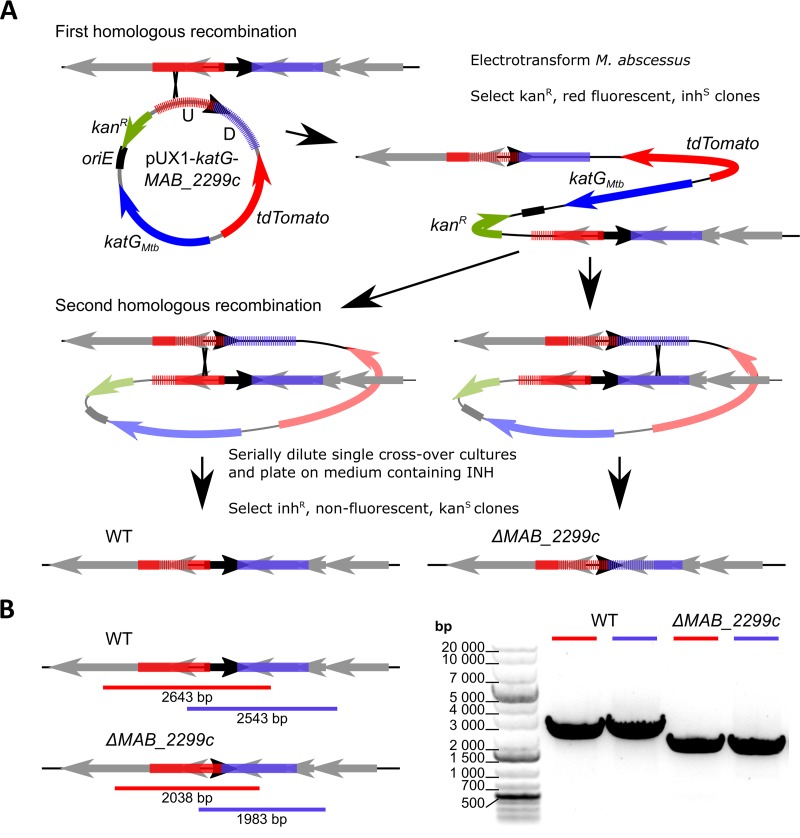
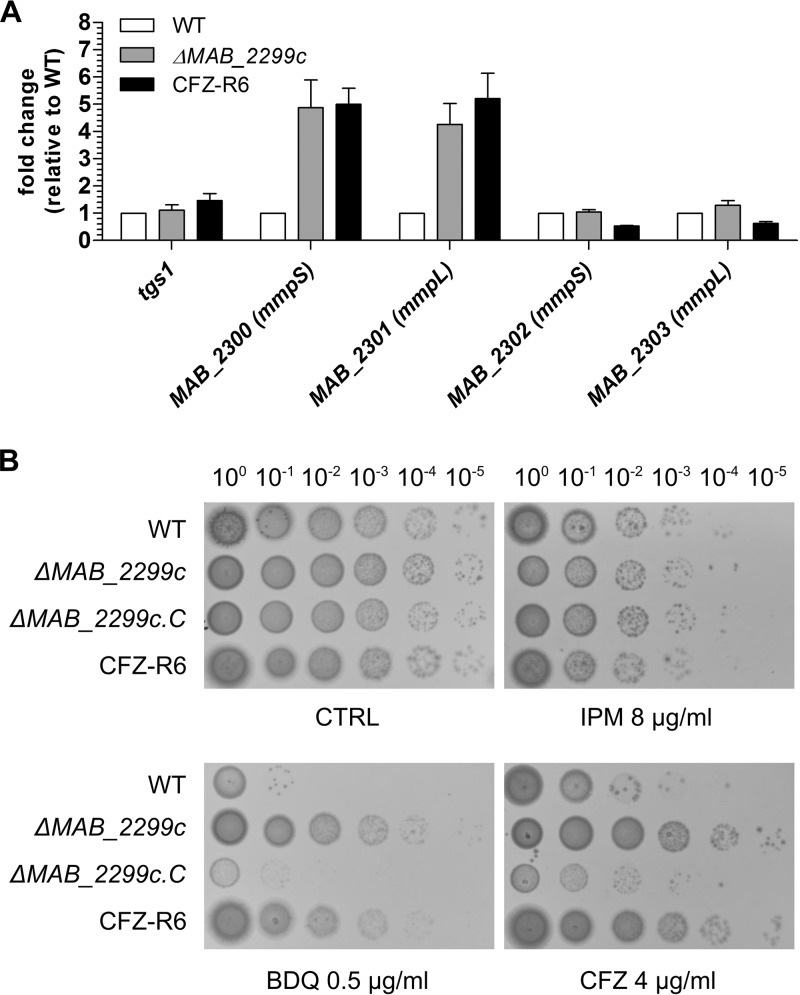
Because point mutations or premature stop codons in the MAB_2299c mutants did not prevent the altered proteins from interacting with other protein partners/DNA sequences and eventually generating aberrant phenotypes, we further confirmed the contribution of MAB_2299c in the profile of susceptibility/resistance to CFZ and BDQ by generating an M. abscessus deletion mutant. To achieve this aim, we first improved a flexible suicide vector that we previously created, pUX1 (43), by cloning into it the M. tuberculosis katG, which was recently demonstrated as an efficient counterselectable marker in M. abscessus in the presence of isoniazid (INH) (42). Through the positive selection afforded by kanamycin resistance and tdTomato markers and the negative selection afforded by katG, this vector, pUX1-katG, could be used in a two-step homologous recombination procedure to generate scarless unmarked deletion mutants in M. abscessus. The different steps of this method leading to an unmarked MAB_2299c deletion are depicted in Fig. 3A. As a first step, sequences of approximately 1 kb directly upstream and downstream of a 560-bp internal fragment of the 666-bp MAB_2299c open reading frame were cloned adjacently into pUX1-katG. After transformation of M. abscessus with pUX1-katG-MAB_2299c, colonies that had undergone a first homologous recombination between the plasmid and the bacterial chromosome either up- or downstream of the MAB_2299c gene were easily identified by their red fluorescence against a large background of colonies spontaneously resistant to the selective antibiotic. After a single passage in liquid culture without kanamycin, the MAB_2299c single-crossover clones were serially diluted and the dilutions were plated on INH-containing solid medium. Clones that were INH resistant, nonfluorescent, and kanamycin sensitive arose at an approximate frequency of 10−4 to 10−3 CFU/ml. These clones were selected and subsequently genotyped to confirm the correct unmarked deletion of MAB_2299c by PCR/sequencing analysis (Fig. 3B). No observable changes in the colony morphology or in the in vitro growth rate were noticed in this unmarked deletion mutant, subsequently designated ΔMAB_2299c (Fig. S3). ΔMAB_2299c was then subjected to qRT-PCR analysis. The results in Fig. 4A clearly show a 5-fold increase in the transcription level of MAB_2300 and MAB_2301, but not that of MAB_2302 and MAB_2303. Interestingly, these induction levels were comparable to those found in CFZ-R6. In agreement with the qRT-PCR results performed using the various CFZ spontaneous mutants, the MAB_2299c gene deletion did not affect expression of the MAB_2302-MAB_2303 pair. As expected, the CFZ and BDQ susceptibility pattern of the ΔMAB_2299c mutant was comparable to that of CFZ-R6 (Fig. 4B). Importantly, both the ΔMAB_2299c and CFZ-R6 mutants displayed enhanced growth on agar supplemented with 0.5 µg/ml BDQ or 4 µg/ml CFZ (Fig. 4B, bottom), whereas, under the same conditions, the growth of the parental M. abscessus strain was severely inhibited. That this resistance phenotype was restricted to CFZ and BDQ only was supported by the similar growth and susceptibility to imipenem (IPM) of all strains. As anticipated, complementation of the ΔMAB_2299c mutant by introducing a pMV261-MAB_2299c construct (leading to ΔMAB_2299c.C) fully restored susceptibility to both CFZ and BDQ (Fig. 4B). In contrast, the susceptibility profile of the ΔMAB_2299c mutant to anti-TB front-line drugs (INH, ethambutol, or rifampin) was comparable to that of the WT and complemented strains (Table S3). Together, these results confirm the importance of MAB_2299c in resistance to CFZ and BDQ.
FIG 3.
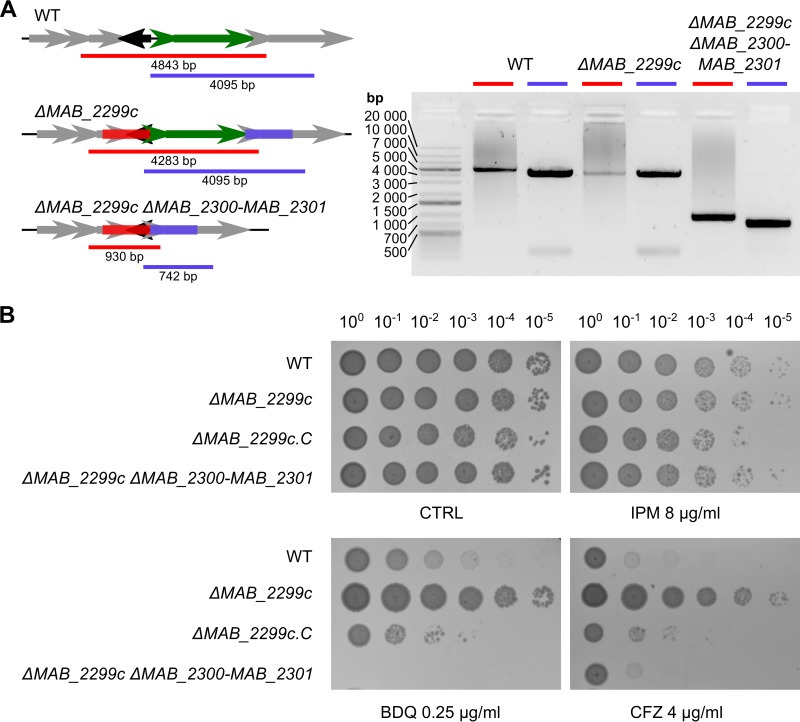
Generation of an unmarked MAB_2299c deletion mutant in M. abscessus. (A) Line drawing illustrating the general protocol followed to ascertain the ΔMAB_2299c mutant. katGMtb, M. tuberculosis katG. (B) (Left) Line drawing of the genomic context in the M. abscessus WT and ΔMAB_2299c mutant that also illustrates the PCR strategy followed to confirm the deletion of MAB_2299c. Red and blue bars, the PCR products obtained during the screening of potential mutant clones; black arrow, MAB_2299c. (Right) A gel confirming the ΔMAB_2299c genotype. The red and blue bars correspond to the amplicons obtained in the left panel. These amplicons were subjected to sequencing analysis to confirm the correct deletion of the MAB_2299c gene.
FIG 4.
Deletion of MAB_2299c confers resistance to both CFZ and BDQ. (A) Results of a qRT-PCR experiment showing the induction levels of the mmpS-mmpL (MAB_2300-MAB_2301) pair but not MAB_2302-MAB_2303 when MAB_2299c is deleted from the M. abscessus genome. (B) Profile of the susceptibility or resistance of M. abscessus WT, M. abscessus ΔMAB_2299c, and the complemented derivative of ΔMAB_2299c (ΔMAB_2299c.C) to BDQ, CFZ, or IPM. Five microliters of 10-fold serially diluted bacterial suspensions of exponentially growing cultures was spotted on Middlebrook 7H10 plates in the absence (control [CTRL]) or presence of drugs at the indicated concentrations. The plates were incubated at 37°C for 4 days.
Deletion of MAB_2300-MAB_2301 in the ΔMAB_2299c mutant restores susceptibility to CFZ and BDQ.
To address whether resistance to CFZ and BDQ is directly mediated by an MmpS/MmpL-dependent efflux machinery which is induced in ΔMAB_2299c, both MAB_2300 and MAB_2301 were deleted in the ΔMAB_2299c mutant. The simultaneous knockout of MAB_2300 and MAB_2301 in the unmarked MAB_2299c deletion mutant was achieved using a strategy similar to the one used to construct the ΔMAB_2299c mutant. The ΔMAB_2299c ΔMAB_2300-MAB_2301 genotype of this triple-knockout mutant was confirmed by PCR, as illustrated in Fig. 5A. Importantly, whereas the susceptibility of the triple mutant to CFZ was reverted to WT levels, the MIC of BDQ for this mutant was even lower than the MIC observed for the WT strain (Fig. 5B and Table S3). This indicates that abolition of the MAB_2300-MAB_2301 efflux pump system rendered the ΔMAB_2299c mutant sensitive to both drugs. As expected, the triple mutant’s sensitivity to IPM remained similar to that of the ΔMAB_2299 progenitor (Fig. 5B and Table S3).
FIG 5.
Deletion of MAB_2300 and MAB_2301 in ΔMAB_2299c restores sensitivity to CFZ and BDQ. (A) Line drawing (left) of the MAB_2299c-MAB_2301 genomic context in the M. abscessus WT, ΔMAB_2299c, and ΔMAB_2299c ΔMAB_2300-MAB_2301 strains. Also illustrated is the PCR strategy followed to confirm the deletion of MAB_2300 and MAB_2301. Black arrow, MAB_2299c; small and large green arrows, MAB_2300 and MAB_2301, respectively. (B) Profile of susceptibility of the ΔMAB_2299c ΔMAB_2300-MAB_2301 triple-knockout strain to BDQ, CFZ, or IPM compared to the other strains reared in this study. Five microliters of 10-fold serially diluted bacterial suspensions obtained from exponentially growing cultures was spotted on Middlebrook 7H10 plates in the absence (control [CTRL]) or presence of drugs at the indicated concentrations. The plates were incubated at 37°C for 4 days.
Overall, these results suggest that both CFZ and BDQ are specific substrates of the MAB_2301 (MmpL) efflux pump and that the sole deletion of this MmpS/MmpL system is sufficient to reverse the CFZ and BDQ resistance phenotype of the ΔMAB_2299c mutant.
DISCUSSION
M. abscessus has recently emerged as one of the most clinically relevant NTM (8, 14) and accounts for more than 80% of all pulmonary infections caused by rapidly growing mycobacteria (44). It poses a serious threat, particularly in patients suffering from lung disorders, such as CF or bronchiectasis (14, 44). Occasional fatalities due to M. abscessus infections (44, 45) can be a result of the extreme difficulty with the treatment of patients as a result of the intrinsic and broad antibiotic resistance of this bacterium conferred by an impermeable cell envelope, the presence of drug-modifying enzymes and a large abundance of efflux pumps (3, 11, 46). Therefore, to reach an unmet medical need in the treatment of M. abscessus diseases, the repositioning of the anti-TB drugs CFZ and BDQ has recently been evaluated in retrospective case series, although data related to their mode of action and/or resistance mechanisms in M. abscessus remain scarce.
In this study, whole-genome sequencing of in vitro-selected mutants showing cross-resistance to CFZ and BDQ identified mutations in MAB_2299c that were subsequently confirmed by PCR/sequencing. MAB_2299c belongs to the TetR family of transcriptional regulators, representing the most abundant family of regulators in mycobacteria (47). In line with the general observation that 60% of TetR regulators are divergently oriented with the genes that they control, MAB_2299c was found in the opposite orientation relative to that of its MAB_2300 and MAB_2301 target genes. That the different point mutations identified (L40W, L151P, and G215S) were associated with similar fold increases in the upregulation of MAB_2300-MAB_2301 in the mutants carrying premature stop codons suggested a loss of DNA-binding activity, resulting in derepression of MAB_2300-MAB_2301 gene expression. This hypothesis was confirmed by EMSA, which clearly demonstrated an impaired DNA-binding activity of the L40W mutant, with Leu40 being located in the N-terminal domain, which carries the DNA-binding activity in TetR regulators (35). The L151P and G215S mutated proteins, albeit being expressed in high yields in E. coli, remained largely insoluble, suggesting that the L151 and G215 residues play a role in the folding and/or stability of the MAB_2299c multimer. Therefore, substitution of these important residues is very likely to affect the overall structure of the regulator and, consequently, its biological function. During preparation of the manuscript, an independent study identified mutations in several genes, including MAB_2299c, in CFZ-resistant M. abscessus strains selected in vitro (48). Whereas most mutations conferred a loss of function caused by indels or stop codons, three amino acid replacements differing from ours were found, namely, C110Y, H173R, and A214S. Interestingly, the mutation at position A214 is located next to G215, found in our study, further emphasizing an important role of this region in the activity/structure of the regulator. However, despite the lack of any mechanistic data regarding the involvement of these mutations in the function of MAB_2299c and in MmpS/MmpL expression and a potential link with BDQ resistance, the study by Chen et al. (48) strongly supports our findings regarding the implication that MAB_2299c is a major determinant of resistance to CFZ and BDQ. Our results are also reminiscent of those of previous work describing a similar cross-resistance to both BDQ and CFZ identified by a whole-genome sequencing comparison of initial and relapse isolates of Mycobacterium intracellulare which occurred during a trial of BDQ as salvage therapy for M. intracellulare lung disease (49). Mutations in MmpT5, another TetR member controlling expression of the adjacent mmpS5-mmpL5 drug efflux operon, was identified as the cause for this cross-resistance. Susceptibility testing indicated that mmpT5 mutations are associated with 2- to 8-fold increases in MICs for BDQ and CFZ (49).
MAB_2300-MAB_2301 encodes an MmpS/MmpL efflux pump system which is separated by an intergenic region from MAB_2302-MAB_2303, encoding a second putative efflux pump system. Whether MAB_2299c controls the expression of the first, the second, or both MmpS/MmpL pairs was addressed by qRT-PCR. Our results clearly indicate that only MAB_2300-MAB_2301 is under the control of MAB_2299c, and this specificity of regulation was further confirmed by functional complementation experiments with a MAB_2299c deletion mutant. Restoration of the CFZ and BDQ susceptibility profile to WT levels was achieved following ectopic expression of MAB_2299c in the deletion mutant. Strikingly, deletion of the MAB_2300-MAB_2301 locus in ΔMAB_2299c abrogated the resistance to both drugs, thus suggesting that the MmpS/MmpL machinery encoded by MAB_2300-MAB_2301 acts as a multisubstrate efflux pump that is responsible for the drug resistance phenotype in M. abscessus. Similarly, studies in M. tuberculosis showed resistance to azoles, CFZ, and BDQ involving a wide set of mutations in Rv0678, encoding a transcriptional regulator from the MarR family, causing overexpression of the MmpS5/MmpL5 efflux pump (36, 37, 50–52). In addition, the level of resistance to both drugs in the M. tuberculosis Rv0678 mutants was similar to that found in the M. abscessus MAB_2299c mutants. To explain why the MAB_2300/MAB_2301 pair participates in BDQ and CFZ extrusion while the MAB_4383c/MAB_4382c pair that we previously identified to be the closest ortholog in M. abscessus of MmpS5/MmpL5, rather, excludes thiacetazone derivatives and not BDQ and CFZ (38), we performed multiple-sequence alignments and subsequent sequence identity determination of the nucleotide and protein sequences of the 17 MmpS/MmpL pairs encoded by the M. abscessus genome (see Table S4 in the supplemental material). The identity scores at both the nucleotide and protein levels supported our previous observations that MAB_4383c/MAB_4382c is the closest orthologous pair to MmpS5/MmpL5 from M. tuberculosis. Interestingly, considering the protein sequence identity, MAB_2301 is the second closest ortholog to MmpL5 from M. tuberculosis after MAB_4382c, explaining the functional similarity in CFZ and BDQ export. However, a full explanation of the differences in substrate specificity that exist between MmpS5/MmpL5 orthologs/paralogs will rely on the elucidation of the high-resolution three-dimensional structures of MmpS5/MmpL5 alone and, more pertinently, in complex with their substrates. Given the very high relatedness between MAB_2300/MAB_2301 and other MmpS-MmpL pairs in M. abscessus (Table S4), one cannot exclude the possibility that other MmpS-MmpL efflux pumps mediate cross-resistance to CFZ and BDQ. This can be investigated thanks to the new suicide vector pUX1-katG, described in this study, which allows the easy and rapid generation of scarless genetic alterations in the M. abscessus chromosome, facilitated by the presence of the brightly red fluorescent tdTomato positive selectable marker and the KatG counterselectable marker.
In M. tuberculosis, efflux inhibition using efflux pump inhibitors such as verapamil or reserpine decreases the MICs of BDQ and CFZ in vitro (52, 53). Whether these inhibitors would also potentiate the effect of CFZ and/or BDQ in resistant M. abscessus strains remains to be established. However, in both organisms, the resistance levels were low and were about 4-fold greater than those of their parental strains. This may be linked to the low induction level of the MAB_2300-MAB_2301 gene cluster under derepressed conditions (in the MAB_2299c mutants or deletion strain). This also contrasts with the very high induction level reported previously for mmpS5-mmpL5 in mutants or a deletion strain of MAB_4384, which also accounted for very high levels of resistance to thiacetazone analogues (38, 39). Overall, these observations, combined with the fact that M. abscessus possesses a very large mmpL repertoire (54) and also a high abundance of TetR transcriptional regulators, allow us to speculate that similar drug resistance mechanisms are employed by this pathogen to express its natural pattern of resistance to many more antimicrobial agents.
In summary, this study adds new functional and mechanistic insights into the TetR-dependent regulation mechanisms responsible for cross-resistance to CFZ and BDQ in M. abscessus, which may have important clinical implications. Future studies should help to elucidate whether the emergence of MAB_2299c variants occurs during BDQ or CFZ treatment in patients with M. abscessus lung disease.
MATERIALS AND METHODS
Strains, growth conditions, and reagents.
All M. abscessus strains used in this study are listed in Table S1 in the supplemental material. The strains were grown in Middlebrook 7H9 broth (BD Difco) supplemented with 0.05% Tween 80 (Sigma-Aldrich) and 10% oleic acid, albumin, dextrose, catalase (OADC enrichment; BD Difco) (7H9T/OADC) at 30°C (unless otherwise stated) or in Sauton’s medium in the presence of antibiotics, when required. On plates, colonies were selected on Middlebrook 7H10 or 7H11 agar (BD Difco) supplemented with 10% OADC enrichment (7H10OADC or 7H11OADC, respectively) or on LB agar. All drugs were purchased from Sigma-Aldrich.
Drug susceptibility testing.
MICs were determined on cation-adjusted Mueller-Hinton broth using the microdilution method or on Middlebrook 7H10OADC agar plates. The MIC99 was defined as the minimal drug concentration required to inhibit 99% growth and was recorded by counting the colonies obtained after 3 to 4 days of incubation at 30°C. All experiments were done on three independent occasions. The frequency of spontaneous resistance was determined by counting the number of resistant colonies growing on 7H10OADC and 7H11OADC agar supplemented with 8 µg/ml CFZ after plating 5 × 107 CFU.
Generating clones resistant to clofazimine.
Resistance to CFZ was induced by subjecting an M. abscessus ATCC 19977 susceptible clone to sub-MICs of the antibiotic. The bacteria were grown in 7H9 supplemented with albumin-dextrose-catalase (ADC; 10%) and glycerol (0.04%) containing sub-MICs of CFZ for 5 days. The bacteria were then spun in a centrifuge at 3,000 × g and washed with phosphate-buffered saline. Bacteria that grew at the highest concentration of CFZ were used to inoculate new cultures containing increasing concentrations of CFZ. The bacteria were then grown in 7H9 supplemented with ADC (10%) and glycerol (0.04%) without CFZ for 5 days and streaked on 7H11 agar plates supplemented with OADC (10%) and glycerol (0.04%) containing CFZ. Single colonies isolated from these plates were subjected to MIC determinations and DNA extraction and sequencing. All growth was done at 37°C.
Whole-genome sequencing and target identification.
DNA from six spontaneous resistant mutants and one susceptible isolate was sequenced on an Illumina MiSeq platform, producing 150-bp paired-end reads. The raw reads were subsequently mapped to the M. abscessus reference genome using the BWA-MEM algorithm (55). Variants were called using the SAMtools (v.1.2) and BCFtools (v.1.2) packages and the parameters described previously (56, 57). Single nucleotide polymorphisms (SNP) were identified in MAB_2299c, and confirmation of these mutations was done by PCR amplification and sequencing.
DNA constructs.
All constructs used in this study are listed in Table S1. For expression of MAB_2299c in E. coli, MAB_2299c was PCR amplified from pure genomic DNA of M. abscessus CIP104536T using primers MAB_2299c_Fw and MAB_2299c_Rv (Table S2) and Phusion polymerase (Thermo Fisher Scientific). The allele carrying the L40W mutation was amplified from strain CFZ-R1 using the same primers. The PCR products were then cloned into pET32a that had been cut with KpnI and HindIII, allowing introduction of thioredoxin and polyhistidine tags at the N terminus of the protein. A tobacco etch virus (TEV) cleavage site was also incorporated before the 1st amino acid of the protein to remove the N-terminal tags from the rest of the protein. To complement the M. abscessus ΔMAB_2299c mutant, MAB_2299c was PCR amplified using the Compl_MAB_2299c primers (Table S2), digested with EcoRI, and ligated into pMV261 that had been digested with MscI and EcoRI, allowing the constitutive overexpression of MAB_2299c under the control of the hsp60 promoter, thus yielding pMV261-MAB_2299c (Table S1).
Quantitative real-time PCR.
Isolation of RNA, reverse transcription, and qRT-PCR were done as reported earlier (38), using the primers listed in Table S2.
Expression and purification of MAB_2299c variants.
The E. coli BL21(DE3) strain (New England Biolabs) containing the pRARE2 vector was transformed with the pET32a constructs containing either the wild-type (WT) or the mutated MAB_2299c gene harboring the L40W mutation. Protein expression was done in lysogeny broth (LB) medium containing 200 µg/ml ampicillin and 30 µg/ml chloramphenicol. When an optical density at 600 nm of 0.6 to 1 was reached, the cultures underwent a 30-min cold shock in icy water prior to protein synthesis induction with 1 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG). Cultures were then grown for 20 h at 16°C prior to centrifugation (6,000 × g, 4°C, 20 min). Bacterial pellets were resuspended in lysis buffer (50 mM Tris-HCl, pH 8, 200 mM NaCl, 20 mM imidazole, 1 mM benzamidine, 5 mM β-mercaptoethanol), and the cells were disrupted by sonication before the lysates were clarified by an additional centrifugation (16,000 × g, 4°C, 45 min). Proteins were then purified by a first ion metal affinity chromatography (IMAC) step (Ni-nitrilotriacetic acid Sepharose; GE Healthcare Life Sciences). To remove the two N-terminal tags (thioredoxin, His tag), the eluted proteins were mixed in a 1:50 ratio with the TEV protease and dialyzed overnight at 4°C against 50 mM Tris-HCl, pH 8, 200 mM NaCl, and 5 mM β-mercaptoethanol. The dialyzed proteins were subjected to a second IMAC step before being further dialyzed against a buffer consisting of 50 mM Tris-HCl, pH 8, and 5 mM β-mercaptoethanol. The proteins were then purified by cation exchange chromatography (HiTrap SP Sepharose Fast Flow; GE Healthcare Life Sciences) and eluted in the same buffer using a linear NaCl gradient. Size exclusion chromatography (SEC; ENrich SEC 650; Bio-Rad) was then performed using an elution buffer containing 50 mM Tris-HCl, pH 8, 200 mM NaCl, and 5 mM β-mercaptoethanol to generate highly pure proteins.
EMSA.
Using the MEME suite 4.20.0 online tool, a DNA operator of 64 nucleotides containing two perfect palindromes and a degenerated double repeat sequence was identified within the intergenic region between MAB_2299c and MAB_2300. Thus, the 64-bp double-stranded sequence (probe A) was labeled at the 5′ end with fluorescein (Sigma-Aldrich) and incubated for 1 h at room temperature in 1× Tris base-acetic acid-EDTA (TAE) buffer with increasing amounts of MAB_2299c or MAB_2299c (L40W). After 30 min of electrophoresis in 1× TAE buffer at 100 V, gel shifts were revealed by fluorescence using an Amersham Imager 600 imager (GE Healthcare Life Sciences). This native DNA operator as well as mutated probes are listed in Table S2. The 373-bp intergenic region located between MAB_2299c and MAB_2300 and the three overlapping probes covering the entire intergenic region between MAB_2301 and MAB_2302 (Table S2) were amplified using a similar strategy and subjected to electrophoretic mobility shift assays (EMSA) using purified MAB_2299c.
Isolation of the MAB_2299c-probe A complex by SEC.
Protein-DNA complex formation was assessed by high-performance liquid chromatography (HPLC) using an Akta Pure 25M chromatography system (GE Healthcare Life Sciences) on an ENrich SEC 650 column (Bio-Rad). The DNA alone, the protein alone, and the protein-DNA mixture were eluted at 4°C at a flow rate of 0.4 ml/min in a buffer containing 50 mM Tris-HCl, pH 8, 200 mM NaCl, 5 mM β-mercaptoethanol. Probe A (60 µg) and MAB_2299c (186 µg) were independently loaded onto the column. A mixture of 60 µg probe A and 186 µg MAB_2299c was coincubated for 1 h at room temperature and loaded onto the column. As a negative control, 21 µg of a 45-bp DNA operator targeted by the TetR regulator MAB_4384 was coincubated with 186 µg MAB_2299c and loaded onto the column.
Generation of MAB_2299c and MAB_2300-MAB_2301 recombination plasmids.
To generate pUX1-katG, used to perform gene disruption by two-step homologous recombination, the M. tuberculosis katG gene was first PCR amplified using the overlap extension mutagenesis approach (58) to introduce a synonymous mutation in the gene, removing an NheI restriction site. Briefly, two separate PCR mixtures containing either the primer set katG_outer_Fw and katG_inner_Rev or the primer set katG_inner_Fw and katG_outer_Rev, purified genomic DNA from M. tuberculosis, and Phusion polymerase (Thermo Fisher Scientific) were set up. The PCR products were purified and added (10 ng each) to a new Phusion polymerase PCR mixture without any primers or genomic DNA. This mixture was then subjected to two cycles of 95°C for 30 s, 55°C for 2 min, and 72°C for 3 min before the outer primers were added and 25 more cycles of 95°C for 10 s, 55°C for 30 s, and 72°C for 2 min were performed. The final PCR product was AvrII digested and ligated to AvrII-XmnI-linearized pUX1 to produce pUX1-katG. To generate pUX1-katG-MAB_2299c, the same overlap extension approach with outer and inner primers was first used to generate a single fused PCR amplicon containing 1-kb up- and downstream sequences of MAB_2299c so that it effectively carried a 560-bp deletion in the 666-bp MAB_2299c open reading frame. This PCR product was NheI digested and ligated to NheI-XmnI-linearized pUX1-katG to produce pUX1-katG-MAB_2299c. The pUX1-katG-MAB_2300-MAB_2301 plasmid used to make a ΔMAB_2299c ΔMAB_2300-MAB_2301 triple-knockout mutant was made using the same approach.
Unmarked deletions of MAB_2299c and MAB_2300-MAB_2301 in M. abscessus.
The generation of an unmarked MAB_2299c deletion in M. abscessus relied on two separate homologous recombination events. First, highly electrocompetent M. abscessus was transformed with pUX1-katG-MAB_2299c, as previously described (43). Kanamycin-resistant (growing at 200 µg/ml), red fluorescent colonies were selected, and the broth was cultured in the presence of kanamycin and subjected to PCR screening using either the primer set MAB_2299c_U_scrn_Fw and MAB_2299c_U_scrn_Rev or the primer set MAB_2299c_D_scrn_Fw and MAB_2299c_D_scrn_Rev (Table S2). Based on the PCR product sizes obtained using the two primer sets, clones that had undergone a single homologous recombination event in either the upstream or the downstream sequence flanking MAB_2299c were identified. Positive clones were washed to remove the antibiotics, cultured for 4 h in the absence of antibiotic, and then 10-fold serially diluted, and the dilutions were plated on 7H10 agar plates containing 50 µg/ml isoniazid (INH). INH-resistant, nonfluorescent colonies were restreaked on plates without selective antibiotics to obtain single colonies that were used to inoculate the broth cultures. These cultures were confirmed to be kanamycin sensitive and used to extract and purify genomic DNA, which was subsequently used to perform the same PCR screening described above to identify clones that had undergone a second homologous recombination event effectively deleting 85% of the MAB_2299c open reading frame from the M. abscessus genome. The PCR products were sequenced, and special attention was paid in the subsequent analyses of the sequencing data to verify the correct junctions between cloned sequences and chromosomal sequences to exclude the possibility of spurious PCR amplification products. The ΔMAB_2299c ΔMAB_2300-MAB_2301 triple-knockout mutant was reared in a similar way, with the only difference being that the ΔMAB_2299c strain was the progenitor strain transformed with pUX1-katG-MAB_2300-MAB_2301.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Fondation pour la Recherche Médicale (FRM) (grant number DEQ20150331719 to L.K.; grant number ECO20160736031 to M.R.); the InfectioPôle Sud, which funded the Ph.D. fellowship of A.V.G.; the Wellcome Trust (098051, 107032AIA); and the Cystic Fibrosis Trust (to D.R.-R., I.E., J.P., and R.A.F.).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We have no conflict of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01316-18.
REFERENCES
- 1.Singh M, Dugdale CM, Solomon IH, Huang A, Montgomery MW, Pomahac B, Yawetz S, Maguire JH, Talbot SG. 2016. Rapid-growing mycobacteria infections in medical tourists: our experience and literature review. Aesthet Surg J 36:NP246–NP253. doi: 10.1093/asj/sjw047. [DOI] [PubMed] [Google Scholar]
- 2.Roux A-L, Catherinot E, Ripoll F, Soismier N, Macheras E, Ravilly S, Bellis G, Vibet M-A, Le Roux E, Lemonnier L, Gutierrez C, Vincent V, Fauroux B, Rottman M, Guillemot D, Gaillard J-L, Herrmann J-L, for the OMA Group. 2009. Multicenter study of prevalence of nontuberculous mycobacteria in patients with cystic fibrosis in France. J Clin Microbiol 47:4124–4128. doi: 10.1128/JCM.01257-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Medjahed H, Gaillard J-L, Reyrat J-M. 2010. Mycobacterium abscessus: a new player in the mycobacterial field. Trends Microbiol 18:117–123. doi: 10.1016/j.tim.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 4.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, Iseman M, Olivier K, Ruoss S, von Reyn CF, Wallace RJ, Winthrop K, ATS Mycobacterial Diseases Subcommittee, American Thoracic Society, Infectious Diseases Society of America. 2007. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med 175:367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- 5.Floto RA, Olivier KN, Saiman L, Daley CL, Herrmann J-L, Nick JA, Noone PG, Bilton D, Corris P, Gibson RL, Hempstead SE, Koetz K, Sabadosa KA, Sermet-Gaudelus I, Smyth AR, van Ingen J, Wallace RJ, Winthrop KL, Marshall BC, Haworth CS. 2016. US Cystic Fibrosis Foundation and European Cystic Fibrosis Society consensus recommendations for the management of non-tuberculous mycobacteria in individuals with cystic fibrosis: executive summary. Thorax 71:88–90. doi: 10.1136/thoraxjnl-2015-207983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Catherinot E, Roux A-L, Macheras E, Hubert D, Matmar M, Dannhoffer L, Chinet T, Morand P, Poyart C, Heym B, Rottman M, Gaillard J-L, Herrmann J-L. 2009. Acute respiratory failure involving an R variant of Mycobacterium abscessus. J Clin Microbiol 47:271–274. doi: 10.1128/JCM.01478-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esther CR, Esserman DA, Gilligan P, Kerr A, Noone PG. 2010. Chronic Mycobacterium abscessus infection and lung function decline in cystic fibrosis. J Cyst Fibros 9:117–123. doi: 10.1016/j.jcf.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryant JM, Grogono DM, Rodriguez-Rincon D, Everall I, Brown KP, Moreno P, Verma D, Hill E, Drijkoningen J, Gilligan P, Esther CR, Noone PG, Giddings O, Bell SC, Thomson R, Wainwright CE, Coulter C, Pandey S, Wood ME, Stockwell RE, Ramsay KA, Sherrard LJ, Kidd TJ, Jabbour N, Johnson GR, Knibbs LD, Morawska L, Sly PD, Jones A, Bilton D, Laurenson I, Ruddy M, Bourke S, Bowler IC, Chapman SJ, Clayton A, Cullen M, Daniels T, Dempsey O, Denton M, Desai M, Drew RJ, Edenborough F, Evans J, Folb J, Humphrey H, Isalska B, Jensen-Fangel S, Jönsson B, Jones AM, et al. 2016. Emergence and spread of a human-transmissible multidrug-resistant nontuberculous mycobacterium. Science 354:751–757. doi: 10.1126/science.aaf8156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown-Elliott BA, Nash KA, Wallace RJ. 2012. Antimicrobial susceptibility testing, drug resistance mechanisms, and therapy of infections with nontuberculous mycobacteria. Clin Microbiol Rev 25:545–582. doi: 10.1128/CMR.05030-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jarand J, Levin A, Zhang L, Huitt G, Mitchell JD, Daley CL. 2011. Clinical and microbiologic outcomes in patients receiving treatment for Mycobacterium abscessus pulmonary disease. Clin Infect Dis 52:565–571. doi: 10.1093/cid/ciq237. [DOI] [PubMed] [Google Scholar]
- 11.Nessar R, Cambau E, Reyrat JM, Murray A, Gicquel B. 2012. Mycobacterium abscessus: a new antibiotic nightmare. J Antimicrob Chemother 67:810–818. doi: 10.1093/jac/dkr578. [DOI] [PubMed] [Google Scholar]
- 12.Maurer FP, Bruderer VL, Ritter C, Castelberg C, Bloemberg GV, Böttger EC. 2014. Lack of antimicrobial bactericidal activity in Mycobacterium abscessus. Antimicrob Agents Chemother 58:3828–3836. doi: 10.1128/AAC.02448-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferro BE, Srivastava S, Deshpande D, Pasipanodya JG, van Soolingen D, Mouton JW, van Ingen J, Gumbo T. 2016. Failure of the amikacin, cefoxitin, and clarithromycin combination regimen for treating pulmonary Mycobacterium abscessus infection. Antimicrob Agents Chemother 60:6374–6376. doi: 10.1128/AAC.00990-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffith DE. 2014. Mycobacterium abscessus subsp abscessus lung disease: “trouble ahead, trouble behind….” F1000Prime Rep 6:107. doi: 10.12703/P6-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu M-L, Aziz DB, Dartois V, Dick T. 2018. NTM drug discovery: status, gaps and the way forward. Drug Discov Today 23:1502–1519. doi: 10.1016/j.drudis.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reddy VM, O’Sullivan JF, Gangadharam PR. 1999. Antimycobacterial activities of riminophenazines. J Antimicrob Chemother 43:615–623. doi: 10.1093/jac/43.5.615. [DOI] [PubMed] [Google Scholar]
- 17.Yano T, Kassovska-Bratinova S, Teh JS, Winkler J, Sullivan K, Isaacs A, Schechter NM, Rubin H. 2011. Reduction of clofazimine by mycobacterial type 2 NADH:quinone oxidoreductase: a pathway for the generation of bactericidal levels of reactive oxygen species. J Biol Chem 286:10276–10287. doi: 10.1074/jbc.M110.200501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen G-H, Wu B-D, Hu S-T, Lin C-F, Wu K-M, Chen J-H. 2010. High efficacy of clofazimine and its synergistic effect with amikacin against rapidly growing mycobacteria. Int J Antimicrob Agents 35:400–404. doi: 10.1016/j.ijantimicag.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 19.Singh S, Bouzinbi N, Chaturvedi V, Godreuil S, Kremer L. 2014. In vitro evaluation of a new drug combination against clinical isolates belonging to the Mycobacterium abscessus complex. Clin Microbiol Infect 20:O1124–O1127. doi: 10.1111/1469-0691.12780. [DOI] [PubMed] [Google Scholar]
- 20.Czaja CA, Levin AR, Cox CW, Vargas D, Daley CL, Cott GR. 2016. Improvement in quality of life after therapy for Mycobacterium abscessus group lung infection. A prospective cohort study. Ann Am Thorac Soc 13:40–48. doi: 10.1513/AnnalsATS.201508-529OC. [DOI] [PubMed] [Google Scholar]
- 21.Martiniano SL, Wagner BD, Levin A, Nick JA, Sagel SD, Daley CL. 2017. Safety and effectiveness of clofazimine for primary and refractory nontuberculous mycobacterial infection. Chest 152:800–809. doi: 10.1016/j.chest.2017.04.175. [DOI] [PubMed] [Google Scholar]
- 22.Yang B, Jhun BW, Moon SM, Lee H, Park HY, Jeon K, Kim DH, Kim S-Y, Shin SJ, Daley CL, Koh W-J. 2017. Clofazimine-containing regimen for the treatment of Mycobacterium abscessus lung disease. Antimicrob Agents Chemother 61:e02052-16. doi: 10.1128/AAC.02052-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matteelli A, Carvalho AC, Dooley KE, Kritski A. 2010. TMC207: the first compound of a new class of potent anti-tuberculosis drugs. Future Microbiol 5:849–858. doi: 10.2217/fmb.10.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andries K, Verhasselt P, Guillemont J, Göhlmann HWH, Neefs J-M, Winkler H, Van Gestel J, Timmerman P, Zhu M, Lee E, Williams P, de Chaffoy D, Huitric E, Hoffner S, Cambau E, Truffot PC, Lounis N, Jarlier V. 2005. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307:223–227. doi: 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- 25.Koul A, Dendouga N, Vergauwen K, Molenberghs B, Vranckx L, Willebrords R, Ristic Z, Lill H, Dorange I, Guillemont J, Bald D, Andries K. 2007. Diarylquinolines target subunit c of mycobacterial ATP synthase. Nat Chem Biol 3:323–324. doi: 10.1038/nchembio884. [DOI] [PubMed] [Google Scholar]
- 26.Haagsma AC, Podasca I, Koul A, Andries K, Guillemont J, Lill H, Bald D. 2011. Probing the interaction of the diarylquinoline TMC207 with its target mycobacterial ATP synthase. PLoS One 6:e23575. doi: 10.1371/journal.pone.0023575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dupont C, Viljoen A, Thomas S, Roquet-Banères F, Herrmann J-L, Pethe K, Kremer L. 2017. Bedaquiline inhibits the ATP synthase in Mycobacterium abscessus and is effective in infected zebrafish. Antimicrob Agents Chemother 61:e01225-17. doi: 10.1128/AAC.01225-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biukovic G, Basak S, Manimekalai MSS, Rishikesan S, Roessle M, Dick T, Rao SPS, Hunke C, Grüber G. 2013. Variations of subunit {varepsilon} of the Mycobacterium tuberculosis F1Fo ATP synthase and a novel model for mechanism of action of the tuberculosis drug TMC207. Antimicrob Agents Chemother 57:168–176. doi: 10.1128/AAC.01039-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kundu S, Biukovic G, Grüber G, Dick T. 2016. Bedaquiline targets the ε subunit of mycobacterial F-ATP synthase. Antimicrob Agents Chemother 60:6977–6979. doi: 10.1128/AAC.01291-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pang Y, Zheng H, Tan Y, Song Y, Zhao Y. 2017. In vitro activity of bedaquiline against nontuberculous mycobacteria in China. Antimicrob Agents Chemother 61:e02627-16. doi: 10.1128/AAC.02627-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vesenbeckh S, Schönfeld N, Roth A, Bettermann G, Krieger D, Bauer TT, Rüssmann H, Mauch H. 2017. Bedaquiline as a potential agent in the treatment of Mycobacterium abscessus infections. Eur Respir J 49:1700083. doi: 10.1183/13993003.00083-2017. [DOI] [PubMed] [Google Scholar]
- 32.Obregón-Henao A, Arnett KA, Henao-Tamayo M, Massoudi L, Creissen E, Andries K, Lenaerts AJ, Ordway DJ. 2015. Susceptibility of Mycobacterium abscessus to antimycobacterial drugs in preclinical models. Antimicrob Agents Chemother 59:6904–6912. doi: 10.1128/AAC.00459-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lerat I, Cambau E, Roth Dit Bettoni R, Gaillard J-L, Jarlier V, Truffot C, Veziris N. 2014. In vivo evaluation of antibiotic activity against Mycobacterium abscessus. J Infect Dis 209:905–912. doi: 10.1093/infdis/jit614. [DOI] [PubMed] [Google Scholar]
- 34.Philley JV, Wallace RJ, Benwill JL, Taskar V, Brown-Elliott BA, Thakkar F, Aksamit TR, Griffith DE. 2015. Preliminary results of bedaquiline as salvage therapy for patients with nontuberculous mycobacterial lung disease. Chest 148:499–506. doi: 10.1378/chest.14-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cuthbertson L, Nodwell JR. 2013. The TetR family of regulators. Microbiol Mol Biol Rev 77:440–475. doi: 10.1128/MMBR.00018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milano A, Pasca MR, Provvedi R, Lucarelli AP, Manina G, Ribeiro ALDJL, Manganelli R, Riccardi G. 2009. Azole resistance in Mycobacterium tuberculosis is mediated by the MmpS5-MmpL5 efflux system. Tuberculosis (Edinb) 89:84–90. doi: 10.1016/j.tube.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 37.Hartkoorn RC, Uplekar S, Cole ST. 2014. Cross-resistance between clofazimine and bedaquiline through upregulation of MmpL5 in Mycobacterium tuberculosis. Antimicrob Agents Chemother 58:2979–2981. doi: 10.1128/AAC.00037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Halloum I, Viljoen A, Khanna V, Craig D, Bouchier C, Brosch R, Coxon G, Kremer L. 2017. Resistance to thiacetazone derivatives active against Mycobacterium abscessus involves mutations in the MmpL5 transcriptional repressor MAB_4384. Antimicrob Agents Chemother 61:e02509-16. doi: 10.1128/AAC.02509-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richard M, Gutiérrez AV, Viljoen AJ, Ghigo E, Blaise M, Kremer L. 2018. Mechanistic and structural insights into the unique TetR-dependent regulation of a drug efflux pump in Mycobacterium abscessus. Front Microbiol 9:649. doi: 10.3389/fmicb.2018.00649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Viljoen A, Blaise M, de Chastellier C, Kremer L. 2016. MAB_3551c encodes the primary triacylglycerol synthase involved in lipid accumulation in Mycobacterium abscessus. Mol Microbiol 102:611–627. doi: 10.1111/mmi.13482. [DOI] [PubMed] [Google Scholar]
- 41.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rominski A, Selchow P, Becker K, Brülle JK, Dal Molin M, Sander P. 2017. Elucidation of Mycobacterium abscessus aminoglycoside and capreomycin resistance by targeted deletion of three putative resistance genes. J Antimicrob Chemother 72:2191–2200. doi: 10.1093/jac/dkx125. [DOI] [PubMed] [Google Scholar]
- 43.Viljoen A, Gutiérrez AV, Dupont C, Ghigo E, Kremer L. 2018. A simple and rapid gene disruption strategy in Mycobacterium abscessus: on the design and application of glycopeptidolipid mutants. Front Cell Infect Microbiol 8:69. doi: 10.3389/fcimb.2018.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Griffith DE, Girard WM, Wallace RJ. 1993. Clinical features of pulmonary disease caused by rapidly growing mycobacteria. An analysis of 154 patients. Am Rev Respir Dis 147:1271–1278. doi: 10.1164/ajrccm/147.5.1271. [DOI] [PubMed] [Google Scholar]
- 45.Sermet-Gaudelus I, Le Bourgeois M, Pierre-Audigier C, Offredo C, Guillemot D, Halley S, Akoua-Koffi C, Vincent V, Sivadon-Tardy V, Ferroni A, Berche P, Scheinmann P, Lenoir G, Gaillard J-L. 2003. Mycobacterium abscessus and children with cystic fibrosis. Emerg Infect Dis 9:1587–1591. doi: 10.3201/eid0912.020774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ripoll F, Pasek S, Schenowitz C, Dossat C, Barbe V, Rottman M, Macheras E, Heym B, Herrmann J-L, Daffé M, Brosch R, Risler J-L, Gaillard J-L. 2009. Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus. PLoS One 4:e5660. doi: 10.1371/journal.pone.0005660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Balhana RJC, Singla A, Sikder MH, Withers M, Kendall SL. 2015. Global analyses of TetR family transcriptional regulators in mycobacteria indicates conservation across species and diversity in regulated functions. BMC Genomics 16:479. doi: 10.1186/s12864-015-1696-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen Y, Chen J, Zhang S, Shi W, Zhang W, Zhu M, Zhang Y. 2018. Novel mutations associated with clofazimine resistance in Mycobacterium abscessus. Antimicrob Agents Chemother 62:e00544-18. doi: 10.1128/AAC.00544-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alexander DC, Vasireddy R, Vasireddy S, Philley JV, Brown-Elliott BA, Perry BJ, Griffith DE, Benwill JL, Cameron ADS, Wallace RJ. 2017. Emergence of mmpT5 variants during bedaquiline treatment of Mycobacterium intracellulare lung disease. J Clin Microbiol 55:574–584. doi: 10.1128/JCM.02087-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang S, Chen J, Cui P, Shi W, Zhang W, Zhang Y. 2015. Identification of novel mutations associated with clofazimine resistance in Mycobacterium tuberculosis. J Antimicrob Chemother 70:2507–2510. doi: 10.1093/jac/dkv150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Somoskovi A, Bruderer V, Hömke R, Bloemberg GV, Böttger EC. 2015. A mutation associated with clofazimine and bedaquiline cross-resistance in MDR-TB following bedaquiline treatment. Eur Respir J 45:554–557. doi: 10.1183/09031936.00142914. [DOI] [PubMed] [Google Scholar]
- 52.Andries K, Villellas C, Coeck N, Thys K, Gevers T, Vranckx L, Lounis N, de Jong BC, Koul A. 2014. Acquired resistance of Mycobacterium tuberculosis to bedaquiline. PLoS One 9:e102135. doi: 10.1371/journal.pone.0102135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gupta S, Cohen KA, Winglee K, Maiga M, Diarra B, Bishai WR. 2014. Efflux inhibition with verapamil potentiates bedaquiline in Mycobacterium tuberculosis. Antimicrob Agents Chemother 58:574–576. doi: 10.1128/AAC.01462-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Viljoen A, Dubois V, Girard-Misguich F, Blaise M, Herrmann J-L, Kremer L. 2017. The diverse family of MmpL transporters in mycobacteria: from regulation to antimicrobial developments. Mol Microbiol 104:889–904. doi: 10.1111/mmi.13675. [DOI] [PubMed] [Google Scholar]
- 55.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv arXiv:1303-3997v2 [q-bio.GN]. [Google Scholar]
- 56.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harris SR, Feil EJ, Holden MTG, Quail MA, Nickerson EK, Chantratita N, Gardete S, Tavares A, Day N, Lindsay JA, Edgeworth JD, de Lencastre H, Parkhill J, Peacock SJ, Bentley SD. 2010. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aiyar A, Xiang Y, Leis J. 1996. Site-directed mutagenesis using overlap extension PCR. Methods Mol Biol 57:177–191. doi: 10.1385/0-89603-332-5:177. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.