Poor-quality medicines undermine the treatment of infectious diseases, such as tuberculosis, which require months of treatment with rifampin and other drugs. Rifampin resistance is a critical concern for tuberculosis treatment.
KEYWORDS: antimicrobial resistance, Escherichia coli, Mycobacterium smegmatis, rifampin, rpoB, substandard medicines, tuberculosis
ABSTRACT
Poor-quality medicines undermine the treatment of infectious diseases, such as tuberculosis, which require months of treatment with rifampin and other drugs. Rifampin resistance is a critical concern for tuberculosis treatment. While subtherapeutic doses of medicine are known to select for antibiotic resistance, the effect of drug degradation products on the evolution of resistance is unknown. Here, we demonstrate that substandard drugs that contain degraded active pharmaceutical ingredients select for gene alterations that confer resistance to standard drugs. We generated drug-resistant Escherichia coli and Mycobacterium smegmatis strains by serially culturing bacteria in the presence of the rifampin degradation product rifampin quinone. We conducted Sanger sequencing to identify mutations in rifampin-resistant populations. Strains resistant to rifampin quinone developed cross-resistance to the standard drug rifampin, with some populations showing no growth inhibition at maximum concentrations of rifampin. Sequencing of the rifampin quinone-treated strains indicated that they acquired mutations in the DNA-dependent RNA polymerase B subunit. These mutations were localized in the rifampin resistance-determining region (RRDR), consistent with other reports of rifampin-resistant E. coli and mycobacteria. Rifampin quinone-treated mycobacteria also had cross-resistance to other rifamycin class drugs, including rifabutin and rifapentine. Our results strongly suggest that substandard drugs not only hinder individual patient outcomes but also restrict future treatment options by actively contributing to the development of resistance to standard medicines.
INTRODUCTION
The ability to treat infectious diseases, such as tuberculosis, is an ongoing global health issue. Rifampin is a broad-spectrum rifamycin-derived antibiotic that is the basis of antituberculosis monotherapy and combination treatment regimens (1). Of the 10 million new tuberculosis cases in 2016, 600,000 were rifampin resistant, necessitating the use of second line treatments with increased toxicity (2, 3). Rifampin may also be used as a prophylaxis against staphylococcal and meningococcal infections and has efficacy against a broad range of pathogens, including Escherichia coli and pseudomonas. Clinically, resistance may arise during periods of poor adherence and pharmacokinetic variability and due to inappropriate treatments (4). In vitro studies demonstrate that subinhibitory doses of drugs may select for antibiotic-resistant organisms (5).
The drug target of rifampin is the rpoB subunit of the DNA-dependent RNA polymerase (6). Resistance to rifampin predominately arises due to mutations in the rpoB gene (7), resulting in a decreased affinity of rifampin to its binding site (8). Three noncontiguous regions of the rpoB gene have been recognized as resistance clusters due to the high frequency of mutations at these sites in strains of drug-resistant pathogens (6). Single amino acid changes in the resistance clusters may confer a high degree of resistance.
The phenomenon of cross-resistance occurs when bacteria gain resistance to an antibiotic they have not been exposed to after gaining resistance to another antibiotic (9). Cross-resistance is common among antibiotics from similar classes; for example, Oz et al. reported cross-resistance between 3 DNA gyrase inhibitors in E. coli independently cultured under a single drug condition (10). Therefore, bacteria may be expected to acquire resistance to a standard antibiotic after exposure to a structurally similar drug degradation product.
In addition to challenges associated with incidence, transmission, and adherence, poor-quality medicines also undermine the treatment of infectious diseases (11). Substandard medicines vary from standard drugs due to poor formulations associated with incorrect dose or bioavailability or due to postmanufacturing issues, such as drug expiry and improper storage conditions (12, 13). Antimicrobial agents are among the most prevalent drugs to be counterfeit or substandard (12).
Drug degradation products may partially or fully replace the standard active pharmaceutical ingredient, resulting in a diminished dose to patients (14). Such underdosing regimens are associated with the evolution of antimicrobial resistance (15). Therefore, substandard drugs are hypothesized to be a contributory factor in the worldwide trend toward antibiotic resistance. However, whether bacterial exposure to drug degradation products may select for resistance to standard antibiotics remains poorly understood.
With regard to tuberculosis treatments, poorly synthesized or improperly stored rifampin may contain impurities and drug degradation products (16). Rifampin’s main degradation product occurs from nonenzymatic autoxidation to form rifampin quinone (17); for comparison between the two structures, see Fig. S1 in the supplemental material. 3-formyl rifampin and 1-amino 4-methylpiperazine form by the hydrolysis of rifampin in acidic conditions (18). The presence of rifampin quinone in rifampin-containing tablets is a marker of poor quality (19). Rifampin quinone may cause immunosuppression in animal models (20) and may underlie rifampin-associated adverse drug interactions (21, 22).
While there has been a general discussion linking resistance to quality, a direct link between drug degradation products and resistance has not been observed in a model system. Here, as a model to test the impact of substandard drugs on resistance development, we examined rifampin resistance arising from subtherapeutic concentrations and a drug degradation product in two disparate bacterial species, E. coli and M. smegmatis, the model organism for the study of Mycobacterium tuberculosis. Here, we demonstrate that bacteria evolve resistance against both rifampin and rifampin quinone. Alarmingly, we found that bacteria that are resistant against the drug degradation product rifampin quinone were also resistant to clinically relevant rifamycins, including rifampin, rifabutin, and rifapentine. Our results strongly suggest that substandard drugs actively compound the worldwide antibiotic resistance problem by selecting for the evolution of resistance to standard drugs.
RESULTS
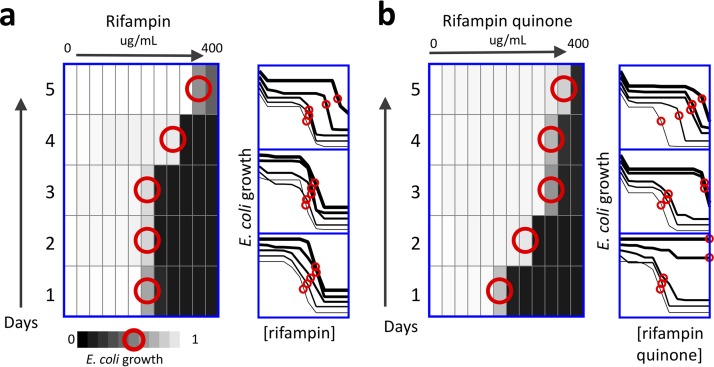
We developed an in vitro model to examine the role of substandard drugs in the acquisition of rifampin resistance by growing bacteria in increasing concentrations of rifampin (RIF) or rifampin quinone (RFQ). We first studied E. coli, a Gram-negative bacterium with a short doubling time, and evaluated its dose response to RIF or RFQ in 1-day cycles. The MIC values in E. coli are ∼25 µg/ml for RIF and 12.5 µg/ml for RFQ; the 50% inhibitory concentration (IC50) values are approximately one half the MIC for each compound. E. coli evolved resistance to RIF or RFQ in three biological replicates. After only five cycles of selection, we observed resistance in all the strains, ranging from 2-fold to 14-fold for RIF and 32-fold to 64-fold for RFQ (Fig. 1). We labeled these strains as RIF-res 1 to 3 and RFQ-res 1 to 3 and prepared glycerol stocks for further experimentation. These stocks were grown in cultures without selective pressure in drug-free medium prior to follow-up studies.
FIG 1.
Selection of resistance in E. coli exposed to rifampin or the drug degradation product rifampin quinone. E. coli were cultured in a range of concentrations of either RIF (a) or the drug degradation product RFQ (b) with 2-fold increments of doses. Each day, bacteria were selected from the concentration that inhibited growth by approximately 50% (IC50; red circles), diluted in fresh media, and aliquoted to a fresh range of drugs. All experiments were conducted in triplicate, with each heatmap corresponding to the top dose-response curves. The right shift in dose-response curves over time demonstrates that E. coli acquire up to a 14-fold increase in IC50 after exposure to rifampin (a) and a 32-fold increase in MIC after exposure to rifampin quinone (b).
RIF-resistant E. coli strains retained their increased MIC to RIF, confirming that these strains acquired stable resistance (Fig. 2a). Next, we studied whether RFQ-resistant strains were resistant to rifampin. To answer this question, we cultured RFQ-res strains in 2-fold increasing concentrations of RIF (Fig. 2b). RFQ-exposed E. coli showed up to a 64-fold increase in the rifampin MIC compared with solvent-treated controls, despite no previous exposure to the standard drug. The level of RIF resistance was also higher in RFQ-res than RIF-res strains. This result indicates that substandard antibiotics may confer resistance to standard antibiotics.
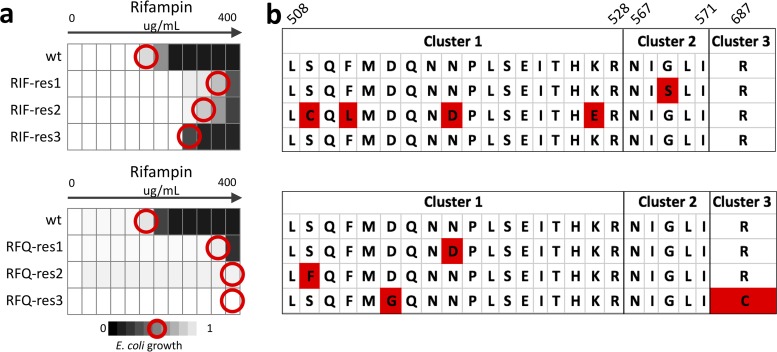
FIG 2.
E. coli exposed to rifampin or the drug degradation product rifampin quinone show similar patterns of rifampin resistance and genetic changes. E. coli resistant to either RIF (RIF-res, a) or the drug degradation product RFQ (RFQ-res; b) over 5 days were assessed for stable increase in RIF MIC compared with solvent-treated controls (wild type [WT]). RFQ-res populations showed cross-resistance to rifampin, with up to a 64-fold increase in IC50 (red circles). Each population was assessed for genetic changes in the RRDR of the rpoB gene (c). The majority of populations that acquired resistance to either RIF or RFQ acquired nonsynonymous mutations in the RRDR clusters of the rpoB gene. These mutations are consistent with previous reports of rifampin resistance due to rpoB mutations.
We next evaluated all three RIF-res and three RFQ-res populations for mutations in the rifampin resistance-determining region (RRDR) of the rpoB gene. Two of the RIF-res strains had nonsynonymous mutations in the RRDR, and the third strain did not have any detectable mutations in the RRDR (Fig. 2c). Similarly, all of the RFQ-res strains had mutations in the RRDR, explaining their resistance to RIF. The cluster I mutation N518D was found in both RIF-res and RFQ-res populations and has previously been associated with RIF-resistant tuberculosis (23). RFQ-res populations also had S512F and D516G mutations that have been previously reported in RIF-resistant E. coli and M. tuberculosis samples (S512F [24]; D516G [25, 26]). These results suggest that substandard antibiotics may cause the selection of resistance to standard antibiotics via well-defined mutations that confer resistance to the standard drug.
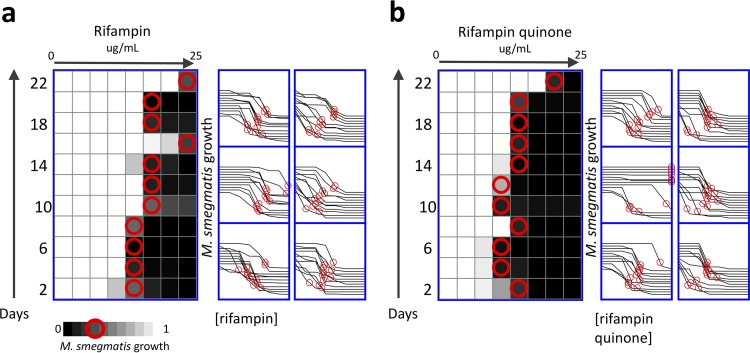
The long (∼1 day) doubling time of Mycobacterium tuberculosis makes evolution experiments difficult to conduct. Therefore, we used the tuberculosis model organism M. smegmatis, a mycobacterium species with a shorter doubling time (2 h), for our experiments. M. smegmatis cells were ∼10-fold more sensitive to RIF and RFQ compared with E. coli. The MIC was approximately 3 µg/ml for both RIF and RFQ in M. smegmatis, and the 75% inhibitory concentration (IC75) was approximately 1.5 µg/ml for both compounds. Using the experimental setup described above, we evaluated M. smegmatis for the selection of RIF resistance due to substandard drugs (Methods). We selected six independent M. smegmatis strains resistant to RIF (named RIF-res1 to 6) and six strains resistant to RFQ (named RFQ-res1 to 6). Bacteria were selected from the concentration that is closest to the IC75, diluted in fresh medium and aliquoted to a fresh range of drugs (Fig. 3), every 48 hours for 11 cycles. After 22 days of serial passages, M. smegmatis exposed to rifampin quinone drugs evolved an increase in MIC compared with solvent-treated controls (t test, P = 0.03), with population RFQ-res2 demonstrating up to a 13-fold increase in MIC (Fig. 3). We prepared glycerol stocks of all strains and used these stocks to grow cultures in drug-free media for further experimentation in M. smegmatis.
FIG 3.
Selection of resistance in Mycobacterium smegmatis exposed to rifampin and the drug degradation product rifampin quinone. M. smegmatis bacteria were cultured in a range of concentrations of either rifampin (a) or the drug degradation product rifampin quinone (b), with 2-fold increments of concentrations (n = 6). These selections represent independent experiments conducted in parallel. Every 48 h, bacteria were selected from the concentration at approximately IC75 (red circles), diluted in fresh media, and aliquoted to a fresh range of drugs. All experiments were conducted with 6 biological replicates, with each heatmap corresponding to the upper left dose-response curves. The right shift in dose-response curves over time demonstrates that M. smegmatis acquired up to a 10-fold increase in IC75 after exposure to rifampin (a) and a 13-fold increase in IC75 after exposure to rifampin quinone (b).
As expected, RIF-exposed M. smegmatis strains maintained resistance to RIF after culture in selection-free media, with an MIC increase of 4 to 64-fold compared with the parental population. We next evaluated if RFQ-exposed M. smegmatis strains demonstrated cross-resistance to RIF. We observed strong RIF resistance in four of these six strains. The RFQ-res2 population acquired a 128-fold increase in MIC, despite being only exposed to RFQ.
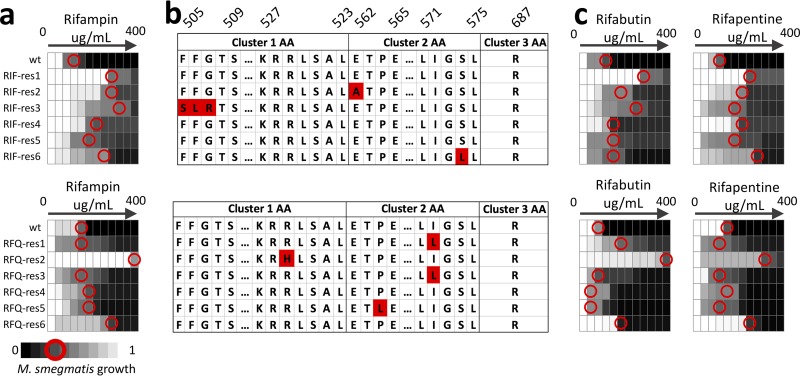
We assessed each population for genetic changes in the RRDR of the rpoB gene. Three RIF-res populations and four RFQ-res populations acquired nonsynonymous mutations in the RRDR (Fig. 4b). R529H (27), I572L (28), and P564L (29) mutations from RFQ-res M. smegmatis were previously reported in RIF-resistant E. coli and mycobacteria. Therefore, we conclude that RFQ exposure may select for genetic variants that are RIF resistant in M. smegmatis.
FIG 4.
Mycobacterium smegmatis exposed to rifampin and the drug degradation product rifampin quinone show similar patterns of rifampin resistance and genetic changes. M. smegmatis resistant to either rifampin (RIF-res) or the drug degradation product rifampin quinone (RFQ-res) over 22 days was assessed for stable increase in RIF MIC, compared with solvent-treated controls (WT; a). All plots present dose response with 2-fold increments of drug concentration. RFQ-res populations showed cross-resistance to RIF with up to 1a 28-fold increase in IC75 (red circles). Each population was assessed for genetic changes in the RRDR of the rpoB gene (b). Amino acid sequences of the RRDR clusters 1, 2, and 3 are displayed, with amino acids numbered using the E. coli mapping notation. The displayed RRDR was truncated to include regions with mutations in the evolved populations. The majority of populations that acquired resistance to either RIF or RFQ acquired nonsynonymous mutations in the RRDR of the rpoB gene. These mutations were consistent with previous reports of rifampin resistance in M. tuberculosis and E. coli. RIF- and RFQ-resistant mycobacteria also showed cross-resistance to other rifamycin drugs, rifabutin and rifapentine (c). Two RFQ-res populations (RFQ-res1 and 3) that did not have resistance to RIF showed cross-resistance to rifabutin.
Cross-resistance is not uncommon between drugs that have structural similarity. RIF is one such compound, belonging to the rifamycin class of antibiotics with closely related drugs rifabutin (RFB) and rifapentine (RFP). RIF-res and RFQ-res populations were further tested for cross-resistance to RFB and RFP (Fig. 4c). Each of the RIF-resistant strains was resistant to both RFB and RFP. The RFB IC75 was increased by 2-fold in half of the RIF-RES populations and >2-fold in the remainder. Half of the six RFQ-res populations were at least 2-fold resistant to both RFB and RFP, despite never having been exposed to either of the compounds. Strain RFQ-res2, which had very high resistance to RIF, also had high resistance to RFB (500-fold) and RFP (30-fold). These observations support the idea that strains exposed to substandard drugs may acquire resistance to standard drugs with similar molecular structures.
DISCUSSION
According to the FDA and the World Health Organization, 10% to 25% of drugs worldwide are substandard, the majority of which are antimicrobial agents (11, 30–32). Up to one third of rifampin-containing drugs failed quality testing depending on world region (33–35). Despite these considerations, there has been no systematic study exploring the proximal and distal negative outcomes associated with substandard antimicrobial drugs. Apart from affecting the proximal treatment success of individual patients receiving subtherapeutic doses of medicine, it has been conjectured that substandard drugs affect treatment success in the future by selecting for the evolution of resistance to standard drugs (36).
Our study demonstrates that substandard drugs promote drug resistance through exposure to an agent similar to the standard drug. Bacterial strains rapidly evolved resistance to the drug degradation product RFQ, and these strains were resistant to the standard drug RIF and two similar antibiotics, RFB and RFP, despite never being exposed to any of these drugs. One of the RFQ-treated mycobacterial populations became highly resistant to rifampin, with only some growth inhibition at 100 times the wild-type MIC. Gene analysis indicated that resistance to the substandard drug RFQ was often associated with at least 1 mutation in the RRDR, explaining the convergent evolution in RIF and RFQ conditions. Previous clinical studies have found the rates of double and multiple mutations in the rpoB gene of rifampin-resistant Mycobacteria tuberculosis vary dramatically by geographic region (10% to 75% of tested isolates) (37, 38). Interestingly, the two RFQ-res mycobacteria strains that did not acquire resistance to rifampin (RFQ-res1 and RFQ-res3) had cross-resistance to rifabutin. These strains had the exact same I572L mutation in the rpoB gene. Varying patterns of cross-resistance between rifampin, rifabutin, and rifapentine have previously been demonstrated in Mycobacterium tuberculosis with different RRDR mutations (26).
Rifampin-resistant tuberculosis is classically thought to arise through factors such as transmission, poor treatment adherence, immune status, and poor absorption (39). Substandard drugs contribute to these factors by hindering efforts to control disease and undermining patient trust in the medical system (4).
While the results of our study are promising and identify new areas of inquiry to investigate the role of poor-quality and degraded drugs in selecting resistance, we recognize that there are a number of limitations of our study. It is possible that there were mutations outside the region that we sequenced in the rpoB gene or that mutations arose in another gene. There may also be heteroresistance caused by a mixture of subpopulations (40). While E. coli and M. smegmatis are both widely used model systems, neither are the causative agent of tuberculosis. The methodology and proof-of-concept described in our study can serve as a basis for more extensive studies on Mycobacterium tuberculosis. Host-pathogen interactions also influence resistance outcomes. Investigating these interactions through the lens of poor quality drugs in animal models is beyond the scope of our study but nonetheless is an important future direction to get a comprehensive understanding of resistance in vivo. The extent to which poor-quality drugs affect resistance will also depend on patient behavior, socioeconomics, and the extent of degradation of the drugs. Our study does not include these public health factors but nonetheless provides evidence that drug degradation can act as a strong driver for resistance.
Despite these limitations, we believe that our study demonstrates a direct and previously unexplored link between rifampin drug quality and the selection of antimicrobial resistance in two disparate bacterial species. It remains to be seen how these observations will translate to other substandard drugs. However, our study provides a proof-of principle strongly suggesting that substandard antibiotics affect not only the current treatment success but also the future treatments by selecting for mutations that confer resistance to standard antibiotics.
MATERIALS AND METHODS
Experimental conditions.
The strains include wild-type MG1655 Escherichia coli and MC2 155 Mycobacterium smegmatis, cultured at 37°C in LB medium or Middlebrook 7H9 with ADC supplement plus 0.2% glycerol, respectively. Bacteria were grown in 2-fold increments of RIF or RFQ (Sigma) (41), with the maximum concentration at 400 µg/ml, (400, 200, 100, 50, 25, etc., through ∼0.4 µg/ml) and a final solvent concentration of 2% dimethyl sulfoxide (DMSO). Bacterial growth was monitored by endpoint optical density (OD600), with growth normalized to the drug-free condition. Different inhibitory concentration (IC) levels were used for the different bacteria due to variation in the slopes of the dose-response curves for each species. IC50 and IC75 were defined as the concentration of drug required to inhibit growth by 50% or 75%, respectively. On the first day of experiments, this corresponded to 12.5 µg/ml for rifampin, 6.25 µg/ml for rifampin quinone in E. coli, and approximately 1.5 µg/ml for both compounds in M. smegmatis, respectively. We selected the bacteria in the well closest to IC50 (E. coli) or IC75 (M. smegmatis) to seed new bacterial cultures on the same concentration series of RIF or RFQ after ∼22 h for E. coli and ∼48 h for M. smegmatis. Bacteria serially passaged in media plus 2% DMSO for the duration of the experiments served as the control groups. E. coli was cultured over 5 days, and M. smegmatis was cultured over 22 days. All data presented are biological replicates with n = 3 for E. coli and n = 6 for M. smegmatis. Resistance was defined as a greater than 2-fold increase in IC level.
Genotyping of RNA polymerase B rifampin-resistance clusters.
Sequences were compared to the reference rpoB genes for E. coli (NCBI gene identity [ID], 948488) and M. smegmatis (NCBI gene ID, 4535217), using ApE plasmid-editing software to identify mutations. Amino acid sequences of the RRDR were numbered using the E. coli mapping notation throughout. RIF-res and RFQ-res populations were streaked onto drug-free LB agar plates and incubated at 37°C overnight to isolate colonies for sequencing. The RRDR of the rpoB gene was assessed by Sanger sequencing (Quintara Biosciences) using the following primers: E. coli-rpoB-FWD (5′-TCTCTGGGCGATCTGGATAC-3′), E. coli-rpoB-REV (5′-CAACAGCACGTTCCATACCA-3′), M. smegmatis-rpoB-FWD (5′-GCTGATCCAGAACCAGATCC-3′), and M. smegmatis-rpoB-REV (5′-GATGACACCGGTCTTGTCG-3′).
Supplementary Material
ACKNOWLEDGMENTS
Z.B.W. is supported by NIGMS Training Program in Biomolecular Pharmacology (grant no. T32GM008541). M.H.Z. is supported by US Pharmacopeia: Developing Superior Screening Technology for Medicines Quality Control in Low Resource Countries.
We thank Eric J. Rubin for providing M. smegmatis. We also thank Murat Cokol for help with the figures and Atena Shemirani for assistance with maintaining cell cultures.
Z.B.W. and M.H.Z. designed the study; Z.B.W. conducted the experiments and analysis; and Z.B.W. and M.H.Z. wrote the paper.
We declare no competing financial interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01243-18.
REFERENCES
- 1.Horsburgh CR Jr, Barry CE III, Lange C. 2015. Treatment of tuberculosis. N Engl J Med 373:2149–2160. doi: 10.1056/NEJMra1413919. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. 2017. Global tuberculosis report 2017. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 3.van den Boogaard J, Kibiki GS, Kisanga ER, Boeree MJ, Aarnoutse RE. 2009. New drugs against tuberculosis: problems, progress, and evaluation of agents in clinical development. Antimicrob Agents Chemother 53:849–862. doi: 10.1128/AAC.00749-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnold A, Cooke GS, Kon OM, Dedicoat M, Lipman M, Loyse A, Butcher PD, Ster IC, Harrison TS. 2017. Drug resistant TB: UK multicentre study (DRUMS): treatment, management and outcomes in London and West Midlands 2008–2014. J Infect 74:260–271. doi: 10.1016/j.jinf.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 5.Kohanski MA, DePristo MA, Collins JJ. 2010. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol Cell 37:311–320. doi: 10.1016/j.molcel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, Darst SA. 2001. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104:901–912. doi: 10.1016/S0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- 7.Farhat MR, Shapiro BJ, Kieser KJ, Sultana R, Jacobson KR, Victor TC, Warren RM, Streicher EM, Calver A, Sloutsky A, Kaur D, Posey JE, Plikaytis B, Oggioni MR, Gardy JL, Johnston JC, Rodrigues M, Tang PK, Kato-Maeda M, Borowsky ML, Muddukrishna B, Kreiswirth BN, Kurepina N, Galagan J, Gagneux S, Birren B, Rubin EJ, Lander ES, Sabeti PC, Murray M. 2013. Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nat Genet 45:1183–1189. doi: 10.1038/ng.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pang Y, Lu J, Wang Y, Song Y, Wang S, Zhao Y. 2013. Study of the rifampin monoresistance mechanism in Mycobacterium tuberculosis. Antimicrob Agents Chemother 57:893–900. doi: 10.1128/AAC.01024-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams DL, Spring L, Collins L, Miller LP, Heifets LB, Gangadharam PR, Gillis TP. 1998. Contribution of rpoB mutations to development of rifamycin cross-resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 42:1853–1857. doi: 10.1128/AAC.42.7.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oz T, Guvenek A, Yildiz S, Karaboga E, Tamer YT, Mumcuyan N, Ozan VB, Senturk GH, Cokol M, Yeh P, Toprak E. 2014. Strength of selection pressure is an important parameter contributing to the complexity of antibiotic resistance evolution. Mol Biol Evol 31:2387–2401. doi: 10.1093/molbev/msu191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pincock S. 2003. WHO tries to tackle problem of counterfeit medicines in Asia. BMJ 327:1126. doi: 10.1136/bmj.327.7424.1126-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnston A, Holt DW. 2014. Substandard drugs: a potential crisis for public health. Br J Clin Pharmacol 78:218–243. doi: 10.1111/bcp.12298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Crevel R, Nelwan RH, Borst F, Sahiratmadja E, Cox J, van der Meij W, de Graaff M, Alisjahbana B, de Lange WC, Burger D. 2004. Bioavailability of rifampicin in Indonesian subjects: a comparison of different local drug manufacturers. Int J Tuber Lung Dis 8:500–503. [PubMed] [Google Scholar]
- 14.Hall Z, Allan EL, van Schalkwyk DA, van Wyk A, Kaur H. 2016. Degradation of artemisinin-based combination therapies under tropical conditions. Am J Trop Med Hyg 94:993–1001. doi: 10.4269/ajtmh.15-0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caminero JA. 2008. Likelihood of generating MDR-TB and XDR-TB under adequate National Tuberculosis Control Programme implementation. Int J Tuber Lung Dis 12:869–877. [PubMed] [Google Scholar]
- 16.Mohan B, Sharda N, Singh S. 2003. Evaluation of the recently reported USP gradient HPLC method for analysis of anti-tuberculosis drugs for its ability to resolve degradation products of rifampicin. J Pharm Biomed Anal 31:607–612. doi: 10.1016/S0731-7085(02)00715-X. [DOI] [PubMed] [Google Scholar]
- 17.Bolt HM, Remmer H. 1976. Implication of rifampicin-quinone in the irreversible binding of rifampicin to macromolecules. Xenobiotica 6:21–32. doi: 10.3109/00498257609151608. [DOI] [PubMed] [Google Scholar]
- 18.Prankerd RJ, Walters JM, Parnes JH. 1992. Kinetics for degradation of rifampicin, an azomethine-containing drug which exhibits reversible hydrolysis in acidic solutions. Int J Pharm 78:59–67. doi: 10.1016/0378-5173(92)90355-6. [DOI] [Google Scholar]
- 19.World Health Organization. 2007. Rifampicin, isoniazid and pyrazinamide dispersible tablets. WHO Drug Inf 21:232. [Google Scholar]
- 20.Konrad P, Stenberg P. 1988. Rifampicin quinone is an immunosuppressant, but not rifampicin itself. Clin Immunol Immunopathol 46:162–166. doi: 10.1016/0090-1229(88)90017-7. [DOI] [PubMed] [Google Scholar]
- 21.Piriou A, Jacqueson A, Warnet JM, Claude JR. 1983. Enzyme induction with high doses of rifampicin in Wistar rats. Toxicol Lett 17:301–306. doi: 10.1016/0378-4274(83)90242-4. [DOI] [PubMed] [Google Scholar]
- 22.Shi F, Li X, Pan H, Ding L. 2017. NQO1 and CYP450 reductase decrease the systemic exposure of rifampicin-quinone and mediate its redox cycle in rats. J Pharm Biomed Anal 132:17–23. doi: 10.1016/j.jpba.2016.09.040. [DOI] [PubMed] [Google Scholar]
- 23.Andres S, Hillemann D, Rusch-Gerdes S, Richter E. 2014. Occurrence of rpoB mutations in isoniazid-resistant but rifampin-susceptible Mycobacterium tuberculosis isolates from Germany. Antimicrob Agents Chemother 58:590–592. doi: 10.1128/AAC.01752-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durão P, Gülereşi D, Proença J, Gordo I. 2016. Enhanced survival of rifampin- and streptomycin-resistant Escherichia coli inside macrophages. Antimicrob Agents Chemother 60:4324–4332. doi: 10.1128/AAC.00624-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan Y, Hu Z, Zhao Y, Cai X, Luo C, Zou C, Liu X. 2012. The beginning of the rpoB gene in addition to the rifampin resistance determination region might be needed for identifying rifampin/rifabutin cross-resistance in multidrug-resistant Mycobacterium tuberculosis isolates from southern China. J Clin Microbiol 50:81–85. doi: 10.1128/JCM.05092-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jamieson FB, Guthrie JL, Neemuchwala A, Lastovetska O, Melano RG, Mehaffy C. 2014. Profiling of rpoB mutations and MICs for rifampin and rifabutin in Mycobacterium tuberculosis. J Clin Microbiol 52:2157–2162. doi: 10.1128/JCM.00691-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin DJ, Gross CA. 1988. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol 202:45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- 28.McCammon MT, Gillette JS, Thomas DP, Ramaswamy SV, Graviss EA, Kreiswirth BN, Vijg J, Quitugua TN. 2005. Detection of rpoB mutations associated with rifampin resistance in Mycobacterium tuberculosis using denaturing gradient gel electrophoresis. Antimicrob Agents Chemother 49:2200–2209. doi: 10.1128/AAC.49.6.2200-2209.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hauck Y, Fabre M, Vergnaud G, Soler C, Pourcel C. 2009. Comparison of two commercial assays for the characterization of rpoB mutations in Mycobacterium tuberculosis and description of new mutations conferring weak resistance to rifampicin. J Antimicrob Chemother 64:259–262. doi: 10.1093/jac/dkp204. [DOI] [PubMed] [Google Scholar]
- 30.Nayyar GML, Breman JG, Newton PN, Herrington J. 2012. Poor-quality antimalarial drugs in southeast Asia and sub-Saharan Africa. Lancet Infect Dis 12:488–496. doi: 10.1016/S1473-3099(12)70064-6. [DOI] [PubMed] [Google Scholar]
- 31.World Health Organization. 2017. WHO global surveillance and monitoring system for substandard and falsified medical products. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 32.Zaman MH. 2018. Bitter Pills: the global war on counterfeit drugs. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 33.Kenyon TA, Kenyon AS, Kgarebe BV, Mothibedi D, Binkin NJ, Layloff TP. 1999. Detection of substandard fixed-dose combination tuberculosis drugs using thin-layer chromatography. Int J Tuber Lung Dis 3:347–350. [PubMed] [Google Scholar]
- 34.Bate R, Jensen P, Hess K, Mooney L, Milligan J. 2013. Substandard and falsified anti-tuberculosis drugs: a preliminary field analysis. Int J Tuber Lung Dis 17:308–311. doi: 10.5588/ijtld.12.0355. [DOI] [PubMed] [Google Scholar]
- 35.Taylor RB, Shakoor O, Behrens RH, Everard M, Low AS, Wangboonskul J, Reid RG, Kolawole JA. 2001. Pharmacopoeial quality of drugs supplied by Nigerian pharmacies. Lancet 357:1933–1936. doi: 10.1016/S0140-6736(00)05065-0. [DOI] [PubMed] [Google Scholar]
- 36.Newton PN, Green MD, Fernandez FM. 2010. Impact of poor-quality medicines in the “developing” world. Trends Pharmacol Sci 31:99–101. doi: 10.1016/j.tips.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bahrmand AR, Titov LP, Tasbiti AH, Yari S, Graviss EA. 2009. High-level rifampin resistance correlates with multiple mutations in the rpoB gene of pulmonary tuberculosis isolates from the Afghanistan border of Iran. J Clin Microbiol 47:2744–2750. doi: 10.1128/JCM.r00548-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jing W, Pang Y, Zong Z, Wang J, Guo R, Huo F, Jiang G, Ma Y, Huang H, Chu N. 2017. Rifabutin resistance associated with double mutations in rpoB gene in Mycobacterium tuberculosis isolates. Front Microbiol 8:1768. doi: 10.3389/fmicb.2017.01768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koch A, Mizrahi V, Warner DF. 2014. The impact of drug resistance on Mycobacterium tuberculosis physiology: what can we learn from rifampicin? Emerg Microbes Infect 3:e17. doi: 10.1038/emi.2014.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heep M, Brandstatter B, Rieger U, Lehn N, Richter E, Rusch-Gerdes S, Niemann S. 2001. Frequency of rpoB mutations inside and outside the cluster I region in rifampin-resistant clinical Mycobacterium tuberculosis isolates. J Clin Microbiol 39:107–110. doi: 10.1128/JCM.39.1.107-110.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, Han L, He J, He S, Shoemaker BA, Wang J, Yu B, Zhang J, Bryant SH. 2016. PubChem substance and compound databases. Nucleic Acids Res 44:D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.