Abstract
Approximately 1 million women smoke during pregnancy despite evidence demonstrating serious juvenile and/or adult diseases being linked to early-life exposure to cigarette smoke. Susceptibility could be determined by factors in previous generations, that is, prenatal or “maternal” exposures to toxins. Prenatal exposure to airborne pollutants such as mainstream cigarette smoke has been shown to induce early-life insults (i.e., gene changes) in Offspring that serve as biomarkers for disease later in life. In this investigation, we have evaluated genome-wide changes in the lungs of mouse Dams and their juvenile Offspring exposed prenatally to mainstream cigarette smoke. An additional lung model was tested alongside the murine model, as a means to find an alternative in vitro, human tissue-based replacement for the use of animals in medical research. Our toxicogenomic and bio-informatic results indicated that in utero exposure altered the genetic patterns of the fetus, which could put them at greater risk for developing a range of chronic illnesses in later life. The genes altered in the in vitro, cell culture model were reflected in the murine model of prenatal exposure to mainstream cigarette smoke. The use of alternative in vitro models derived from human medical waste tissues could be viable options to achieve human endpoint data and conduct research that meets the remits for scientists to undertake the 3Rs practices.
Keywords: in vitro, in vivo, mainstream cigarette smoke, prenatal exposure, toxicogenomics
Introduction
Lung cancer was estimated to have accounted for over 14% of cancer cases in the United States during 2014 and >27% of cancer-related mortalities. Indeed, nearly 70% of incidences of lung cancer are predicted to end in mortality.1 Despite the high incidence and mortality rates, lung cancer has historically received a disproportionately low share of funding for research. The U.S. National Institute of Health calculated the U.S. spent <5% of all cancer research funding on dedicated lung cancer research.2 The imbalance of research funding in the United States is indicative of a wider global trend, which is often attributed to the wider social stigma that lung cancer is a direct result of smoking and the consequence of their conscious transgressions. Indeed, the complex mixture of over 4000 cigarette smoke compounds, of which many either direct or second-hand, are known to have direct links to, cancer, cell irritation, and death.3–5 Yet, despite this, around 25% of cases of lung cancer are not being directly linked to smoking.6 Second-hand cigarette smoke, air pollution, inhalation of carcinogens, and hereditary genes are all known to increase risk of developing lung cancer.7 These also contribute to the multiple respiratory diseases burdening health worldwide.8 Chronic obstructive pulmonary disease (COPD), respiratory tract infections, pneumonia, and asthma, all contribute to increasing financial cost and strain on medical care with COPD prevalence as high as 9% in some U.S. states.9
The lack of progress in lung cancer therapeutics combined with international goals for replacement, reduction, and refinement of animal testing10,11 is a driving need to shift toward alternative models.12,13 These alternative models have historically been limited to monolayer cultures of specific cell lines, which fail to account for the intricacy and real-world variability offered through animal testing.14 However, a complication associated with in vivo testing arises from intra-subject variation of immune responses, which is often activated by foreign object particulates or pathogens.15 Use of in vitro testing can, as such, simplify understanding of pathological pathways to the route mechanism and is particularly useful in multistimuli studies.
Only in recent years have there been advances in the growth of complex tissues capable of providing the intermediary between simple in vitro cell monolayers and complex in vivo animal models.16 Laboratory-grown models reduce the burden on animal testing, allowing multiple cell-type interactions to be examined without the more confounding aspects resulting from the presence of systemic systems and the immune response. Furthermore, the ability to test multiple cell-type responses using human tissues, negating the reliance on use of alternative species as models, provides more ethically and biologically relevant research into the carcinogenic and damaging effects of inhaled toxicants such as cigarette smoke and air pollution.17 MatTek’s EpiAirway™ is a multicellular, differentiated model of the human bronchial epithelium derived from healthy human primary tracheobronchial cells. The model is aimed at replicating the epithelial tissue of the human respiratory tract.18,19
This study was aimed at confirming, through transcriptomics, the suitability of a laboratory-grown human lung model (EpiAirway) as an alternative model to study the genetic effects on gene regulation and associated pathways caused by tobacco smoke inhalation.20–22 The study also aimed to look at the downstream implications on Offspring carried during exposure. The EpiAirway model and pregnant female mice were exposed to mainstream cigarette smoke (MCS) or filtered air before lung tissue RNA extraction, and gene regulation was assessed by the Agilent Single Colour Microarray.23
Materials and Methods
In vitro cell culture exposures
The EpiAirway cell cultures were transported from MatTek in the United States in a 24-well plate format. The cells were equilibrated at 37°C with 4.5% CO2 for 24 hours following manufacturer’s guidelines. Culture preconditioning, acute exposure (24 hours) of the EpiAirway lung tissue (ELT) to MCS, was carried out at the air–liquid interface (ALI) as per the methods by Sexton et al.,21 respectively. Each insert had a surface area of 1 cm2 and was apically dosed.
Animal exposures
MCS was generated through burning 3R4F reference filtered cigarettes (Kentucky Tobacco Research and Development Centre, Lexington, KY) on an automated CS generation system (Baumgartner-Jaeger CSM 2070; CH Technologies, Inc., Westwood, NJ) and both Dam and EpiAirway lung tissue cultures were exposed as described in Ng et al.3 Smoke was drawn from the cigarettes under ISO standard conditions (35 mL puff drawn over 2 seconds every 1 minute) and diluted in filtered air using an RM20s smoke engine (Borgwaldt Technik GmbH, Hamburg, Germany). Exposure was designed to be equivalent to an adult human smoking ~10 cigarettes/day for 18 consecutive days. For our cigarette smoke animal studies, we exposed 4 h/day running a continuous 15 mg/m3 concentration in the chamber. B6C6F1 mice (Jackson Laboratory, Bar Harbor, ME) were acclimatized and mated as described in Ng et al.3 Pregnant females (Dams) were exposed to MCS (or filtered air) by whole body inhalation 4 h/day for 5 days/week during gestation until parturition (18 days) and sacrificed postexposure, and the Dams’ and the Offspring’ lungs (n = 3 replicates each) were extracted. EpiAirway lung tissue (ELT; n = 3 replicates) was exposed to the same level of MCS (or filtered air) and for the same duration. Diluted smoke (1/50 smoke:air v/v) was continually delivered to exposure chambers (UK patent number WO 03/100417 A1) containing the culture inserts for a period of 1 hour. In the absence of cells, the total deposition of particulates on the base of the cell culture inserts was determined to be 1.84 μg/cm2. We generated 1 mg/m3 for the cell culture studies. Lungs were preserved in RNAlater (Qiagen) for genomic analysis at Cardiff University (Wales).
Sample preparation
Total RNA was extracted and purified from Dams and ELTs using the RNeasy Mini Kit (Qiagen, United Kingdom). Purity and integrity of extracted RNA was assessed using an Agilent 2100 BioAnalyzer (Agilent Technologies, Palo Alto, CA). Five hundred ng of total RNA from each of the lung tissue samples and the Universal Human/mouse Reference RNA (Stratagene, La Jolla, CA) were used for amplification of RNA and labeled with cyanine Cy3 using the Agilent’s Low RNA Input Linear Amplification Kit (Agilent Technologies) according to the manufacturer’s instructions. Labeled samples and reference cRNAs were purified using RNeasy mini spin columns (Qiagen, United Kingdom) and eluted in 30 μL of nuclease-free water. After amplification and labeling, cRNA quantity and Cy dye incorporation were determined using a Nanodrop ND.1000 UV-VIS-Spectrophotometer version 3.2.1 (Agilent Technologies). For each hybridization, 1 μg Cy3-labeled cRNA was mixed, fragmented, and hybridized at 65°C for 17 hours onto Agilent Whole human/mouse genome 4 × 44 K 60mer Oligo Microarrays. Labeled cRNA from three different ELT, Dams, or Offspring tissues were each hybridized to the arrays. After washing, microarrays were scanned using an Agilent Array scanner (G2505C) (Agilent Technologies) and the images were analyzed. Reproducibility and reliability of each single microarray were assessed using Quality Control report data. Data were extracted using Agilent feature extraction software (version 9.5.3) and the GE2-v5_95_Feb07 protocol. In addition, genes with either uniformly low expression or low expression variation across the experiments were eliminated.
Analyzing gene expression
Microarray data were analyzed using GeneSpring (version 13.0) to highlight differentially expressed genes. Samples were grouped by exposure to MCS and filtered air for the Dams, Offspring, and the ELT, normalizing the arrays to the 75th percentile. Quality control on each data set was performed to minimize false detection rate (FDR). A moderated t-test used a cutoff p-value of 0.05 and minimum 1.4-fold change without FDR. These values were chosen as Dalman et al.24 concluded that lower fold change cutoff produces more significant results in gene ontology (GO). Upregulated and downregulated genes, which satisfied these criteria, underwent gene enrichment analysis. FDR was accounted for through the use of Gene enrichment of DAVID’s Benjamini–Hochberg score for corrected p-value, rather than through the use of Bonferroni-style approaches at earlier stages, which, while reducing false positives, often simultaneously exclude true positives. GO terms and significant pathways for the upregulated and downregulated genes were identified through the use of DAVID (version 6.7),25,26 and the strength of association was assessed through p-value and Benjamini score, where gene ontology was deconstructed by biological process (BP) using REVIGO27 with an allowed similarity of 0.7 and visualized using Cytoscape (version 3.2.1).28,29
Results
Following normalization and quality control, 27,758, 26,571, and 25,250 out of 44,000 features were retained for the ELT, Dams, and Offspring, respectively. Following a moderated t-test with p-value 0.05 cutoff and minimum 1.4-fold change, 716 (500 upregulated and 216 downregulated) genes in the ELT, 437 (283 upregulated and 154 downregulated) genes in the Dams and 9825 (5208 upregulated and 4617 downregulated) genes in the Offspring were identified as significantly altered, comparing exposure of MCS and filtered air (Fig. 1). The top 10 differentially (up and down) expressed genes for the ELT, Dams, and Offspring were identified and listed in the Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/aivt).
FIG. 1.
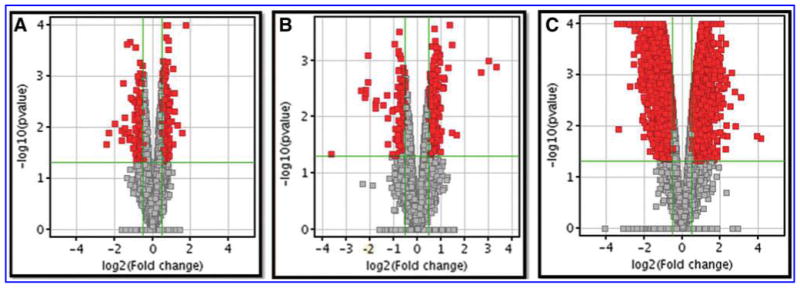
Selection of differentially expressed genes of ELT (A) Dam lung tissue (B) and Offspring lung tissue (C). Volcano plot of differentially expressed genes between ELT and Dam and Offspring tissues exposed to MCS and filtered air. The vertical lines correspond to 1.4-fold up and down expression and the horizontal line represents a p-value of 0.05. ELT, EpiAirway lung tissue.
Of the top upregulated genes in ELT, many have direct functions in regulation and cell adhesion [c-fos], cell division [cdc20b], and matrix proteins [matn1], while the top ELT downregulated genes have links to cell binding [fn1], calcium/zinc ion binding in proteolysis [mmp12], and calcium ion binding in protease inhibition [spock1]. The top Dam-upregulated genes, most have direct functions in immunity (e.g., Ighg–Immunoglobulin heavy constant-γ), but also includes killer cell lectin-like receptors [klra17] and Mediterranean fever [mefv], and the top Dam-downregulated genes have links to fat regulation and cell life regulation [retn], regulation of lipid biosynthetic process [thrsp], and mucus production [muc5b]. The top Offspring-upregulated genes have functions in histocompatibility, [h2ab1], hemoglobin [hbb-bt], and immunity [ly6d]) and the top Offspring-downregulated genes have links to chloride ion channels [BEST1], tight junctions [tjp2], and GTPases [agap1].
The genes that saw the greatest fold change in expression indicate large gene network pathways and therefore, gene ontology was performed to see what the global trends in gene regulation of the cell were involved. Upregulated and downregulated genes underwent GO analysis through DAVID and the top 10 enriched terms for the genes associated with BP, cellular component (CC), and molecular function (MF) were identified (Table 1).
Table 1.
Top 10 Enrichment Terms Extracted from DAVID
| Term | p | Term | p | Term | p | |||
|---|---|---|---|---|---|---|---|---|
| ELT BP | Microtubule-based movement | 8.60E-09 | Dams BP | Immune response | 6.70E-19 | Offspring BP | Cellular process | 1.23E-91 |
| Microtubule-based process | 2.31E-06 | Immune system process | 9.00E-16 | Cellular metabolic process | 1.30E-52 | |||
| Cellular developmental process | 7.30E-06 | Response to external stimulus | 4.09E-09 | Cellular metabolic process | 1.86E-43 | |||
| Ciliary or flagellar motility | 1.17E-05 | Response to stimulus | 7.60E-09 | Metabolic process | 2.31E-41 | |||
| Ectoderm development | 1.94E-05 | Nuclear division | 9.73E-09 | Macromolecule metabolic process | 1.82E-35 | |||
| Developmental process | 2.70E-05 | Mitosis | 9.73E-09 | Primary metabolic process | 9.05E-35 | |||
| Multicellular organismal process | 5.30E-05 | M phase of mitotic cell cycle | 1.35E-08 | Cellular protein metabolic process | 8.79E-34 | |||
| Cell differentiation | 9.25E-05 | Organelle fission | 1.72E-08 | Gene expression | 1.64E-25 | |||
| Epidermis development | 1.16E-04 | Cell division | 3.80E-08 | Protein metabolic process | 5.00E-23 | |||
| Tissue development | 2.23E-04 | Chemotaxis | 7.76E-08 | CC organization | 1.53E-21 | |||
| ELT CC | Cytoskeleton | 1.36E-14 | Dams CC | Cell surface | 4.25E-10 | Offspring CC | Intracellular | 3.90E-135 |
| Cilium | 3.87E-11 | External side of plasma membrane | 1.37E-09 | Intracellular part | 4.56E-124 | |||
| Axoneme | 1.47E-10 | Extracellular space | 7.40E-08 | Cytoplasm | 5.02E-96 | |||
| Cell projection | 2.06E-10 | Chromosome, centromeric region | 9.11E-07 | Organelle | 8.86E-83 | |||
| Cytoskeletal part | 2.08E-10 | Extracellular region part | 2.18E-06 | Intracellular organelle | 1.40E-82 | |||
| Microtubule | 6.81E-10 | Condensed chromosome, centromeric region | 1.31E-05 | Membrane-bounded organelle | 7.70E-59 | |||
| Microtubule cytoskeleton | 6.29E-09 | Condensed chromosome kinetochore | 4.88E-05 | Intracellular membrane-bounded organelle | 1.33E-58 | |||
| Microtubule associated complex | 2.32E-08 | Extracellular region | 5.70E-05 | Cytoplasmic part | 1.25E-52 | |||
| Cell projection part | 6.26E-08 | Chromosomal part | 8.49E-05 | Intracellular organelle part | 8.85E-38 | |||
| Dynein complex | 8.15E-08 | Kinetochore | 1.35E-04 | Organelle part | 7.09E-37 | |||
| ELT MF | Microtubule motor activity | 4.25E-09 | Dams MF | Sugar binding | 2.08E-11 | Offspring MF | Binding | 1.21E-65 |
| Structural molecule activity | 2.94E-07 | Carbohydrate binding | 2.15E-10 | Protein binding | 8.57E-42 | |||
| Motor activity | 3.72E-07 | Chemokine activity | 5.32E-06 | Structural constituent of ribosome | 2.72E-26 | |||
| Metalloendopeptidase activity | 4.56E-05 | Chemokine receptor binding | 6.39E-06 | Cytoskeletal protein binding | 2.27E-16 | |||
| Metallopeptidase activity | 1.60E-04 | Cytokine activity | 6.58E-06 | Catalytic activity | 1.04E-14 | |||
| Calcium ion binding | 2.67E-04 | Receptor binding | 6.38E-05 | Purine ribonucleotide binding | 3.90E-14 | |||
| Structural constituent of cytoskeleton | 4.27E-04 | Sh3/Sh2 adaptor activity | 2.26E-04 | Ribonucleotide binding | 3.90E-14 | |||
| Nucleoside-triphosphatase activity | 0.003537 | Signal transducer activity | 2.34E-04 | Purine nucleotide binding | 1.13E-13 | |||
| Extracellular matrix structural constituent | 0.004979 | Molecular transducer activity | 2.34E-04 | Actin binding | 1.25E-13 | |||
| Pyrophosphatase activity | 0.006094 | Protein binding | 2.89E-04 | Nucleotide binding | 2.24E-13 |
BP, biological process; CC, cellular component; ELT, EpiAirway lung tissue; MF, molecular function.
DAVID ranks associated genes with a GO term to provide an enrichment score, with highly enriched terms having a greater number of associated genes. Many of the top 10 BPs and MFs and all the CCs for ELT relate to cytoskeletal genes and cell development. The Dams had BPs relating to cell cycle and additionally significant immune response alterations. The CCs identified were mostly involved with the chromosomal organization, while MFs had highly enriched terms in receptor binding and transmission. This suggested that ELT was primarily affected in cellular organization, while the Dams were primarily responding to external stimuli, above a cellular organization response. To investigate similarities in response, common GO terms between ELT and the Dams were collated. Offspring had many GO terms associated with metabolic processes, organelle structure, and protein binding.
To assess the similarities between ELT as a model for replacement of the Dams, common processes for all GO terms for ELT and Dams were identified (Table 2). The common terms can largely be linked to the processes of cellular adhesion and response to a stimulus. This indicated that the Dam immune response was masking similar mechanical pathway changes with the ELT. The global KEGG pathways identified through DAVID display the complexity of the Dams when compared to the ELT (Table 3). ELTs highlight 5 pathways, including extracellular matrix (ECM)-receptor interactions and pathways in cancer and cardiac stress. Dam tissue had a large range of 19 disease pathways altered that included immunity, cell cycle regulation, diabetes, and cell adhesion. The Offspring, however, showed a vast network of 61 disease and cancer pathways, which included Alzheimer’s and Parkinson’s, chronic myeloid leukemia, colorectal cancer, and Type II diabetes.
Table 2.
Common Gene Ontology Terms Found Between EpiAirway Lung Tissue and Dams
| GO term | Description | ELT P-value | Dams P-value |
|---|---|---|---|
| GO:0044421 | Extracellular region part | 3.41E-06 | 2.18E-06 |
| GO:0005576 | Extracellular region | 8.30E-05 | 5.70E-05 |
| GO:0005615 | Extracellular space | 6.59E-04 | 7.40E-08 |
| GO:0005488 | Binding | 2.16E-02 | 1.70E-03 |
| GO:0044459 | Plasma membrane part | 2.87E-02 | 9.75E-03 |
| GO:0016043 | CC organization | 3.42E-02 | 5.07E-03 |
| GO:0042060 | Wound healing | 3.68E-02 | 5.18E-02 |
| GO:0042221 | Response to chemical stimulus | 3.89E-02 | 3.02E-02 |
| GO:0009605 | Response to external stimulus | 4.10E-02 | 4.09E-09 |
| GO:0007155 | Cell adhesion | 4.10E-02 | 4.67E-02 |
| GO:0022610 | Biological adhesion | 4.17E-02 | 4.77E-02 |
| GO:0016485 | Protein processing | 4.88E-02 | 7.93E-02 |
| GO:0040011 | Locomotion | 4.88E-02 | 9.39E-04 |
| GO:0042127 | Regulation of cell proliferation | 5.01E-02 | 6.26E-02 |
| GO:0032879 | Regulation of localization | 5.34E-02 | 4.75E-05 |
| GO:0051604 | Protein maturation | 6.81E-02 | 9.50E-02 |
| GO:0030674 | Protein binding, bridging | 7.96E-02 | 3.08E-03 |
| GO:0045834 | Positive regulation of lipid metabolic process | 9.35E-02 | 6.88E-02 |
GO, gene ontology.
Table 3.
Differentially Regulated Gene Interaction Identified in Kegg Pathways
| Kegg term | Gene count | –LogP | Kegg term | Gene count | –LogP |
|---|---|---|---|---|---|
| ELT | |||||
| Focal adhesion | 12 | 2.78 | ARVC | 5 | 1.22 |
| ECM-receptor interaction | 8 | 2.94 | Small cell lung cancer | 5 | 1.10 |
| p53 signaling pathway | 6 | 1.99 | Oocyte meiosis | 5 | 1.07 |
| Calcium signaling pathway | 8 | 1.26 | 7 | 1.79 | |
| Dams | |||||
| Natural killer cell mediated cytotoxicity | 20 | 11.81 | Systemic lupus erythematosus | ||
| Graft vs. host disease | 13 | 9.06 | ECM-receptor interaction | 6 | 1.62 |
| Cytokine–cytokine receptor interaction | 21 | 7.33 | NOD-like receptor signaling pathway | 5 | 1.47 |
| Hematopoietic cell lineage | 9 | 3.63 | Antigen processing and presentation | 6 | 1.47 |
| Chemokine signaling pathway | 12 | 3.04 | p53 signaling pathway | 5 | 1.32 |
| Allograft rejection | 7 | 3.03 | Autoimmune thyroid disease | 5 | 1.27 |
| Type I diabetes mellitus | 7 | 2.84 | Oocyte meiosis | 6 | 1.11 |
| Circadian rhythm | 4 | 2.72 | T cell receptor signaling pathway | 6 | 1.07 |
| Toll-like receptor signaling pathway | 8 | 2.47 | CAMs | 7 | 1.07 |
| Cytosolic DNA-sensing pathway | 6 | 2.35 | |||
| Offspring | |||||
| Focal adhesion | 110 | 11.70 | Purine metabolism | 64 | 1.94 |
| Huntington’s disease | 99 | 9.65 | Nucleotide excision repair | 22 | 1.91 |
| ECM-receptor interaction | 49 | 6.31 | Glioma | 30 | 1.89 |
| Oxidative phosphorylation | 68 | 5.96 | Nonsmall cell lung cancer | 26 | 1.83 |
| Adherens junction | 44 | 5.37 | RNA polymerase | 15 | 1.68 |
| Alzheimer’s disease | 86 | 5.09 | Colorectal cancer | 37 | 1.60 |
| Ubiquitin mediated proteolysis | 68 | 5.08 | Propanoate metabolism | 16 | 1.60 |
| Regulation of actin cytoskeleton | 99 | 5.00 | SNARE interactions in vesicular transport | 19 | 1.55 |
| Axon guidance | 64 | 4.40 | mTOR signaling pathway | 25 | 1.54 |
| Endocytosis | 91 | 4.36 | Type II diabetes mellitus | 23 | 1.50 |
| Parkinson’s disease | 64 | 4.15 | Leukocyte transendothelial migration | 48 | 1.45 |
| Proteasome | 28 | 3.73 | B cell receptor signaling pathway | 34 | 1.41 |
| Viral myocarditis | 47 | 3.59 | Graft-vs.-host disease | 26 | 1.41 |
| Valine, leucine and isoleucine degradation | 27 | 3.46 | p53 signaling pathway | 30 | 1.41 |
| Small cell lung cancer | 43 | 3.44 | Long-term potentiation | 30 | 1.32 |
| Fc gamma R-mediated phagocytosis | 48 | 3.40 | Hypertrophic cardiomyopathy | 35 | 1.32 |
| Spliceosome | 58 | 3.39 | ErbB signaling pathway | 36 | 1.31 |
| Insulin signaling pathway | 62 | 3.03 | CAMs | 59 | 1.26 |
| Pancreatic cancer | 36 | 2.80 | Glycosylphosphatidylinositol-anchor biosynthesis | 13 | 1.22 |
| Renal cell carcinoma | 35 | 2.73 | |||
| Pyrimidine metabolism | 45 | 2.71 | Lysine degradation | 19 | 1.21 |
| Vascular smooth muscle contraction | 53 | 2.45 | Gap junction | 35 | 1.18 |
| Glutathione metabolism | 27 | 2.42 | Dilated cardiomyopathy | 37 | 1.16 |
| RNA degradation | 30 | 2.37 | (ARVC) | 31 | 1.16 |
| Tight junction | 58 | 2.33 | Aldosterone-regulated sodium reabsorption | 19 | 1.11 |
| Pathways in cancer | 124 | 2.30 | Natural killer cell mediated cytotoxicity | 47 | 1.10 |
| Prostate cancer | 41 | 2.23 | Cell cycle | 49 | 1.09 |
| Phosphatidylinositol signaling system | 35 | 2.13 | Fatty acid biosynthesis | 5 | 1.09 |
| Neurotrophin signaling pathway | 55 | 2.06 | GnRH signaling pathway | 38 | 1.04 |
| Chronic myeloid leukemia | 35 | 2.02 | Systemic lupus erythematosus | 40 | 1.03 |
| Apoptosis | 39 | 2.00 | |||
CAMs, cell adhesion molecules.
The links between BP, CC, and MF GO term processes were assessed utilizing REVIGO and visualized in Cytoscape. ELT BPs showed several clusters of GO terms (Fig. 2). The large group of GO terms has been mapped by color and listed with their associated GO terms. One group classified under high density lipid (HDL) particle remodeling included many cellular organization processes and were heavily interlinked with cellular development processes. Response to inorganic substance formed many intragroup links, but connected to HDL particle remodeling through a single node process of intracellular signal transduction. The majority of ELT CCs are interlinked cytoskeletal processes and extracellular structures, and basolateral plasma membrane, while MFs had interlinks between molecular binding, structural activity, and kinase/transferase activity (Supplementary Table S2).
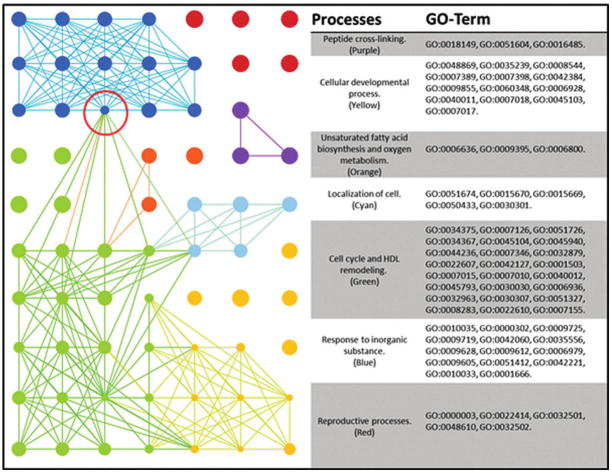
FIG. 2.
ELT-BP interaction map. GO terms processed through REVIGO are visualized through Cytoscape. Node sizes are correlated to the “uniqueness” value determined by REVIGO, where smaller nodes share more similarity with neighboring GO terms. The red circle indicates bottleneck between the “Response to inorganic substance’s cluster and the larger and more heavily interlinked ‘Cell cycle and high density lipid remodeling’.” BP, biological process; GO, gene ontology.
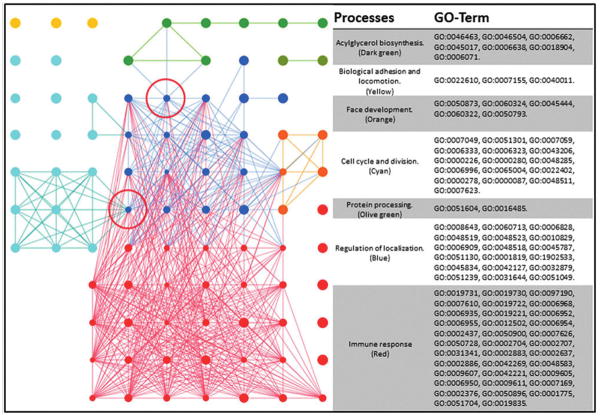
The Dams BP-associated REVIGO groupings showed a far more complex mapping (Fig. 3). The large group of GO terms has been mapped by color and listed with their associated GO terms. There is a vast and heavily interconnected cluster of immune processes, which also form multiple interactions with regulation of localization. Smaller clusters of nuclear division and acylglycerol biosynthesis connect through to this massive cluster through single nodes of positive regulation of CC organization and lipid metabolism. There was a small CC interconnection with processes relating to the cell surface, extracellular space, and protein/DNA interaction. MF interconnections between cytokine activity, protein activity, and binding was also identified (Supplementary Table S3).
FIG. 3.
Dams-BP interaction map. GO terms processed through REVIGO are visualized through Cytoscape. Node sizes are correlated to the “uniqueness” value determined by REVIGO, where smaller nodes share more similarity with neighboring GO terms. The red circle indicates bottleneck between the “cell cycle and division” cluster and the larger and more heavily interlinked “immune system.” A bottle neck also connects “Acylglycerol biosynthesis” pathways with “Regulation of localization.”
Large similarities between the ELT and Dams exist in cell cycle regulation and localization. While the ELT sees alteration to processes associated with response to inorganic substances on a cellular level, the Dams have a heavy immune response as a whole. The influence of this immune response can distract from the cellular mechanistic responses and is outlined in greater detail in the discussion.
The Offspring REVIGO map of associated GO terms displayed a vast and highly interconnected network, both intraprocesses and interprocesses (Fig. 4). The large group of GO terms has been mapped by color and listed with their associated GO terms. Translation and regulation of GTPases had the largest networks of GO terms in the BP map, while there were also a large number of regulatory changes in localization, cell–substrate adhesion and tube development, and antigen processing. There was a large network of GO terms associated with the mitochondrion cell projection and the basement membrane for the CC map, and cytoskeletal binding, motor activity, ubiquitin-protein transferase activity, and zinc iron binding in the MF map (Supplementary Table S4).
FIG. 4.
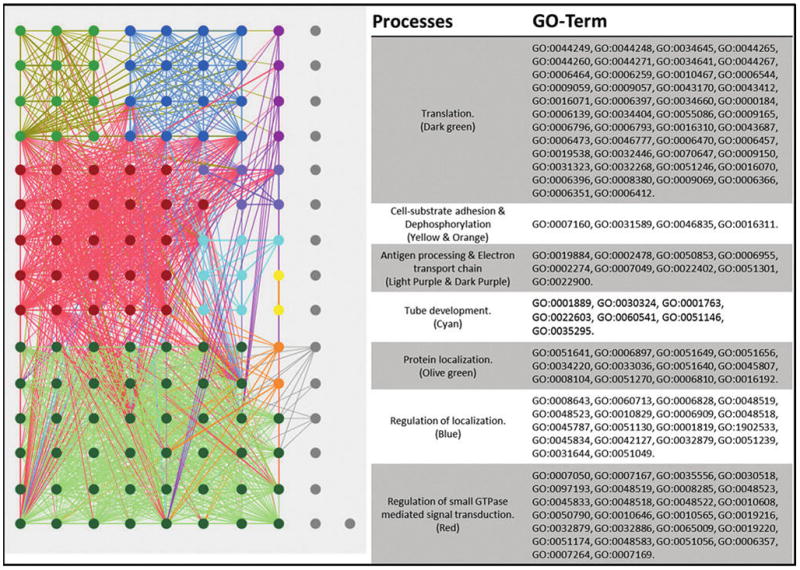
Offspring-BP interaction map. GO terms processed through REVIGO are visualized through Cytoscape. Node sizes are correlated to the “uniqueness” value determined by REVIGO, where smaller nodes share more similarity with neighboring GO terms. The highly interconnecting map does not have any bottlenecks in process interaction as seen in the ELT and Dam maps. The nodes in gray show multiple smaller unlinked GO terms.
Discussion
Despite the high incidence and mortality that accompany lung cancer and other pulmonary diseases, research funding remains disproportionately low. The additional pressures for refinement, reduction, and replacement of animal testing is a driving need for alternative models that can more accurately represent human in vivo responses. This pilot study aimed at determining if the ELT (EpiAirway) could provide this alternative without the obfuscating presence of an immune system. It was hoped the ELT would provide a reductionist view, useful for understanding initial, site of impact, and mechanics of the cellular interactions.
Global transcriptomic pathway analysis allows analysis between individuals and also cross-species comparison. Requiring higher fold changes has often been used to filter out normal fluctuations in gene regulation. This accompanied with early FDR compensation can exclude many relevant genes. Indeed, some genes only require a minimal fluctuation in their regulation to have a profound effect and are highly regulated to avoid fluctuations.29 Filtering genes with a lower threshold for fold change and allowing significance to be assessed in gene enrichment processes provide a more informative and reliable picture of global cellular response.24
Comparatively, fewer differentially expressed genes met the criteria for gene enrichment in the ELT model than in the Dams. The top differentially regulated genes and their functions were initially identified. In the ELT, many of the upregulated genes had functions linked to cell cycle, while there was downregulation of cell adhesion and calcium homeostasis regulation. In the Dams, there was heavy upregulation of immunoglobulins and immunity receptors and downregulation of fat regulation, detection, and signaling pathways.
These differences were observed (Table 1) for the “immune” versus the “mechanical” damage in the top 10 enriched terms, where the top 10 in the Dams included immune response and response to external stimulus, but the ELT is cellular structure. However, there were similarities, suggesting the same mechanical issues are occurring, but they are hidden behind the overwhelming immune response. For example, common GO terms included extra-CCs (i.e., regions and spaces) and response to chemicals and external stimuli (Table 2). We also observed common Kegg pathways, such as the P53 signaling and ECM interaction pathways (Table 3). More pathways were exacerbated in Dams, again linked to the exceptional immune response, but cell cycle and cell adhesion responses were common. The interlinking of these pathways demonstrated not only the interconnectivity of stress responses and the comparative size of the immune response but also common stress responses. Histologically, these broad similarities were observed, such as the loss of tight junctions, cytoskeleton differences, and inflammatory responses associated with the exacerbated immune response.
With regard to the additional analyses of the Offspring gene changes, although this research focus was the ELT versus Dam model comparison, the heavy alterations (i.e., 9825 differentially expressed genes) nonetheless provide an interesting data set. For example, GO terms included those associated largely with translation, regulation of small GTPase-mediated signal transduction, and regulation of localization (Fig. 4). Fetal development is a time when many genes are being turned on and off and the impacts of chemical exposures might well explain the significant number that was observed. Many of the known later life impacts following prenatal exposures to CS have been identified (Table 3), such as diabetes, Alzheimer’s, and multiple cancer pathways.5
Often times, animal models do not recapitulate what is observed epidemiologically when it comes to cigarette smoke-induced carcinogenesis. However, a study by Hutt et al.30 used the B6C3F1 mouse strain (the same used in this study) and found that lifetime exposure of female mice to MCS to 250 mg PM/m3 for 6 h/day, 5 days/week, induced an increased rate of focal alveolar hyperplasia, pulmonary adenomas, papillomas, and adenocarcinomas versus unexposed control mice, those exposed to MCS, had 10-fold increase in hyperplastic lesions, 4.6-fold increase in adenomas, 7.25-fold increase in adenocarcinomas, and 5-fold increase in metastatic pulmonary adenocarcinoma. The selection of the mouse strain, that is, the B6C3F1 hybrid strain, has been used in carcinogenesis assays by many researchers, as well as the National Toxicology Program (USA) due to its lung tumor response to certain chemicals like cigarette smoke and chemicals present in cigarette smoke. In addition, as reviewed by Pandiri,31 “Meta-analysis of transcriptomic alterations in human and mouse lung tumors revealed significant similarities in lung cancer pathways in both species.32–34 These data indicate that mouse lung tumors are similar to human adenocarcinomas at the morphologic and molecular levels and that mouse lung tumors are relevant in evaluating carcinogenic hazards associated with environmental exposures.”
In conclusion, the gene changes observed in the in vitro, three-dimensional cell culture model of the human bronchial epithelium mirrored the responses detected in the mouse model of prenatal exposure to MCS. The ELT model could be utilized as the first step (i.e., before using animal models) to screening aerosolized compounds such as combustion-derived air pollution,35 environmental tobacco smoke,20–22 diesel exhaust,36 coal fly ash particles,37 and shipping emissions.38 The benefits of using alternative in vivo-like in vitro ALI models of the human lung are self-evident. The ELT model is both cost- and time-effective for toxicity testing of aerosolized and soluble compounds given that cell culture consumables are highly affordable and permit rapid analyses, in comparison to animal models that are expensive due to costs of the animals and their maintenance, which could last for years, versus days and/or weeks for in vitro cell culturing practices.39 Finally, when considering the contentious ethical issues surrounding the use of animals for medical research, when using alternative systems like MatTek’s EpiAirway platform, there is an immediate impact for the 3Rs and human endpoint data are acquired, negating the need to extrapolate data from animals into effects in man.
Supplementary Material
Acknowledgments
Studies were supported in part by NYU NIEHS Center Grant ES000260.
Footnotes
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.National Institutes of Health, U.S. (NIH) [last accessed Apr. 23, 2015];Estimates of funding for various research condition and disease categories (RCDC) NIH research portfolio online reporting tools. 2015 http://report.nih.gov/categorical_spending.aspx.
- 3.Ng SP, Silverstone AE, Lai Z, et al. Effects of prenatal exposure to cigarette smoke on offspring tumour susceptibility and associated immune mechanisms. Toxicol Sci. 2006;89:135–144. doi: 10.1093/toxsci/kfj006. [DOI] [PubMed] [Google Scholar]
- 4.Faux SP, Tai T, Throne D, et al. The role of oxidative stress in the biological responses of lung epithelial cells to cigarette smoke. Biomakers. 2009;14(Suppl 1):90–96. doi: 10.1080/13547500902965047. [DOI] [PubMed] [Google Scholar]
- 5.Doherty SP, Grabowski J, Hoffman C, et al. Early life insult from cigarette smoke may be predictive of chronic diseases later in life. Biomarkers. 2009;14(Suppl 1):97–101. doi: 10.1080/13547500902965898. [DOI] [PubMed] [Google Scholar]
- 6.Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers—A different disease. Nature Rev Cancer. 2007;7:778–790. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- 7.Samet JM, Avila-Tang E, Boffetta P, et al. Lung cancer in never smokers: Clinical epidemiology and environmental risk factors. Clin Cancer Res. 2009;15:5626–5645. doi: 10.1158/1078-0432.CCR-09-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bousquet J, Khaltaev N, Cruz A. Global Surveillance, Prevention and Control of Chronic Respiratory Diseases. Geneva: World Health Organization; 2007. [Google Scholar]
- 9.Centre for Disease Control and Prevention, U.S. (CDC) [last accessed Aug. 4, 2015];Chronic obstructive pulmonary disease (COPD)–Data and Statistics. 2014 www.cdc.gov/copd/data.htm.
- 10.Animal Procedures Committee APC. Review of Cost-Benefit Assessment in the Use of Animals in Research June 2003. Home Office publication, Communication Directorate; London: 2003. [Google Scholar]
- 11.Russell WMS, Burch RL. The Principles of Humane Experimental Technique. Methuen; London: 1959. [Google Scholar]
- 12.BéruBé K, Gibson C, Job C, et al. Human lung tissue engineering: A critical tool for safer medicines. Cell Tissue Bank. 2011;12:11–13. doi: 10.1007/s10561-010-9204-6. [DOI] [PubMed] [Google Scholar]
- 13.BéruBé KA. Medical waste tissues—Breathing life back into respiratory research. Altern Lab Anim. 2013;41:429–434. doi: 10.1177/026119291304100604. [DOI] [PubMed] [Google Scholar]
- 14.Kroll A, Pillukat MH, Hahn D, et al. Current in vitro methods in nanoparticle risk assessment: Limitations and challenges. Eur J Pharm Biopharm. 2009;72:370–377. doi: 10.1016/j.ejpb.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 15.Cressler CE, Nelson WA, Day T, et al. Disentangling the interaction among host resources, the immune system and pathogens. Ecol Lett. 2014;17:284–293. doi: 10.1111/ele.12229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bredenkamp N, Ulyanchenko S, O’Neill KE, et al. An organized and functional thymus generated from FOXN1-reprogrammed fibroblasts. Nature Cell biol. 2014;16:902–908. doi: 10.1038/ncb3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adam M, Schikowski T, Carsin AE, et al. Adult lung function and long-term air pollution exposure. ESCAPE: A multicentre cohort study and meta-analysis. Eur Respir J. 2015;45:38–50. doi: 10.1183/09031936.00130014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.BéruBé K, Prytherch Z, Job C, et al. Human primary bronchial lung cell constructs: The new respiratory models. Toxicology. 2009;278:311–318. doi: 10.1016/j.tox.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 19.BéruBé K, Pitt A, Hayden, Patrick, et al. Filter-well technology for advanced three-dimensional cell culture: Perspectives for respiratory research. Altern Lab Anim. 2010;38(Suppl 1):49–65. doi: 10.1177/026119291003801S04. [DOI] [PubMed] [Google Scholar]
- 20.Balharry D, Sexton KJ, BéruBé KA. An in vitro approach to assess the toxicity of inhaled tobacco smoke components: Nicotine, cadmium, formaldehyde and urethane. Toxicology. 2008;244:66–76. doi: 10.1016/j.tox.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 21.Sexton K, Balharry D, BéruBé KA. Genomic biomarkers of pulmonary exposure to tobacco smoke components. Pharmacogenet Genomics. 2008;18:853–860. doi: 10.1097/FPC.0b013e328307bddf. [DOI] [PubMed] [Google Scholar]
- 22.Sexton K, Balharry D, Brennan P, et al. Proteomic profiling of human respiratory epithelia by iTRAQ reveals biomarkers of exposure and harm by tobacco smoke components. Biomarkers. 2011;16:567–576. doi: 10.3109/1354750X.2011.608855. [DOI] [PubMed] [Google Scholar]
- 23.Agilent. [last accessed April 23, 2015];One-color microarray-based gene expression analysis low input quick Amp Labeling Protocol Version 6.9.1. 2015 www.agilent.com/cs/library/usermanuals/Public/G4140-90040_GeneExpression_OneColor_6.9.pdf.
- 24.Dalman MR, Deeter A, Nimishakavi G, et al. Fold change and p-value cutoffs significantly alter microarray interpretations. BMC Bioinformatics. 2011;13:S11. doi: 10.1186/1471-2105-13-S2-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 26.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Supek F, Bosnjak M, Skunca N, et al. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shannon P, Markiel A, Ozier O, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raser JM, O’shea EK. Noise in gene expression: Origins, consequences and control. Science. 2005;39:2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hutt JA, Vuillemenot BR, Barr EB, et al. Life-span inhalation exposure to mainstream cigarette smoke induces lung cancer in B6C3F1 mice through genetic and epigenetic pathways. Carcinogenesis. 2005;26:1999–2009. doi: 10.1093/carcin/bgi150. [DOI] [PubMed] [Google Scholar]
- 31.Pandiri A. Comparative pathobiology of environmentally induced lung cancers in humans and rodents. Toxicol Pathol. 2015;43:107–114. doi: 10.1177/0192623314556516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stearman RS, Dwyer-Nield L, Zerbe L, et al. Analysis of orthologous gene expression between human pulmonary adenocarcinoma and a carcinogen-induced murine model. Am J Pathol. 2005;167:1763–1775. doi: 10.1016/S0002-9440(10)61257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonner AE, Lemon WJ, Devereux TR, et al. Molecular profiling of mouse lung tumors: Association with tumor progression, lung development, and human lung adenocarcinomas. Oncogene. 2004;23:1166–1176. doi: 10.1038/sj.onc.1207234. [DOI] [PubMed] [Google Scholar]
- 34.Pandiri AR, Sills RC, Ziglioli V, et al. Differential transcriptomic analysis of spontaneous lung tumors in B6C3F1 mice: Comparison to human non-small cell lung cancer. Toxicol Pathol. 2012;40:1141–1159. doi: 10.1177/0192623312447543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price H, Jones T, BéruBé K. Resolution of the mediators of in vitro oxidative reactivity in size-segregated fractions that may be masked in the urban PM10 cocktail. Sci Total Environ. 2014;485:588–595. doi: 10.1016/j.scitotenv.2014.03.056. [DOI] [PubMed] [Google Scholar]
- 36.Hoogendoorn B, BéruBé K, Gregory C, et al. Gene and protein responses of human lung tissue explants exposed to ambient particulate matter of different sizes. Inhal Toxicol. 2012;24:966–975. doi: 10.3109/08958378.2012.742600. [DOI] [PubMed] [Google Scholar]
- 37.Lawson M. MRes Thesis. Wales, UK: Cardiff School of Biosciences, Cardiff University; 2015. The Respiratory Toxicity of Artic Coal Fly Ash. [Google Scholar]
- 38.Oeder S, et al. Particulate matter from both heavy fuel oil and diesel fuel shipping emissions show strong biological effects on human lung cells at realistic and comparable in vitro exposure conditions. PLoS One. 2015;10:e0126536. doi: 10.1371/journal.pone.0126536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prytherch ZC, BéruBé KA. A normal and biotransforming model of the human bronchial epithelium for the toxicity testing of aerosols and solubilised substances. Altern Lab Anim. 2014;42:377–381. doi: 10.1177/026119291404200607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.