Abstract
The pathogenesis of infectious diseases depends on the interaction of host and pathogen. In Plasmodium falciparum malaria, host and parasite processes can be assessed by dual RNA-sequencing of blood from infected patients. Here we performed dual transcriptome analyses on samples from 46 malaria-infected Gambian children to reveal mechanisms driving the systemic pathophysiology of severe malaria. Integrating these transcriptomic data with estimates of parasite load and detailed clinical information allowed consideration of potentially confounding effects due to differing leukocyte proportions in blood, parasite developmental stage, and whole-body pathogen load. We report hundreds of human and parasite genes differentially expressed between severe and uncomplicated malaria, with distinct profiles associated with coma, hyperlactatemia, and thrombocytopenia. High expression of neutrophil granule-related genes was consistently associated with all severe malaria phenotypes. We observed severity-associated variation in the expression of parasite genes which determine cytoadhesion to vascular endothelium, rigidity of infected erythrocytes, and parasite growth rate. Up to 99% of human differential gene expression in severe malaria was driven by differences in parasite load, whereas parasite gene expression showed little association with parasite load. Co-expression analyses revealed interactions between human and P. falciparum, with prominent co-regulation of translation genes in severe malaria between host and parasite. Multivariate analyses suggested that increased expression of granulopoiesis and interferon-γ related genes, together with inadequate suppression of type-1 interferon signalling, best explained severity of infection. These findings provide a framework for understanding the contributions of host and parasite to the pathogenesis of severe malaria and identifying targets for adjunctive therapy.
Introduction
Most studies of infectious disease pathogenesis focus on either host or pathogen, despite the fact that outcome is determined by their interaction. Dual RNA-sequencing has been developed as a method for transcriptomic assessment of such interactions (1, 2), although it has not been widely applied to study systemic infection in humans. In malaria, the pathogenic stage of the parasite is restricted to blood where important interactions between parasites and host leukocytes can be assessed by dual RNA-sequencing (3). The blood is also the conduit for systemic responses to infection, and gene expression in blood will reflect the inflammatory and metabolic milieu to which leukocytes and parasites are exposed.
Plasmodium falciparum malaria is one of the most important infectious diseases affecting humans (3). Most malaria deaths occur in children, in whom three major syndromes are associated with increased risk of death and distinguish severe malaria from uncomplicated malaria: 1. cerebral malaria (manifesting as coma), 2. hyperlactatemia / acidosis (often manifesting as deep breathing), and 3. severe anemia (3–6). These are usually accompanied by the laboratory finding of low platelet count (thrombocytopenia) (7). Severe malaria syndromes can occur in isolation or in overlapping combinations (4), and mortality is highest when cerebral malaria and hyperlactatemia / acidosis coexist (4). Severe malaria is most likely when there is a high parasite load (8–11) and numerous accompanying pathophysiological derangements have been described (4, 5, 12), broadly arising from inflammation, vascular endothelial dysfunction and parasite sequestration (accumulation of parasitized erythrocytes in small blood vessels which obstruct blood flow) (4, 13). These mechanisms interact with one another, so defining their individual contributions to specific features of severe malaria is challenging (13, 14).
The host immune response is a major determinant of outcome in rodent models of severe malaria (15, 16) and it has long been supposed that an excessive host response may also contribute to some forms of human severe malaria (17, 18). A similar concept exists to explain severity in other infections such as bacterial sepsis, Ebola and Respiratory Syncytial Virus (19–21), however direct evidence is often lacking and the confounding effect of pathogen load on the magnitude of the host response is rarely quantified. Controlled human infection models have provided insights into the relationship between pathogen load and early immune responses to infection (22) but cannot be extended to investigate severe disease. To better understand the pathogenesis of severe infection, a systemic, integrated view of host-pathogen interaction accounting for variation in pathogen load is required.
The feasibility of host and parasite dual RNA-sequencing in malaria has been demonstrated in individuals with uncomplicated malaria (23). Here, we extend the application of dual RNA-sequencing to infectious disease pathogenesis by integrating gene expression analysis with detailed clinical and laboratory data that characterise the systemic pathophysiology of severe malaria. We further refine the analysis by accounting for three major confounders which may vary within and between severity groups. This allows us to characterise human and parasite differential gene expression between severe and uncomplicated malaria and the role of parasite load in determining the host response. These data provide a unique insight into host-pathogen interactions associated with severity of infection in humans and reveal new perspectives on the likely pathogenic mechanisms of human severe malaria.
Results
Dual RNA-sequencing and adjustment for cellular heterogeneity
We performed dual RNA-sequencing on whole blood, collected prior to treatment, from 46 Gambian children with P. falciparum uncomplicated malaria (n = 21) and severe malaria (n = 25) (Table 1). These children were recruited from a region with relatively low malaria transmission and consistent with this epidemiology, (4) the severe malaria group contained children with cerebral malaria, hyperlactatemia, or a combination thereof, but no cases of severe anemia. After exclusion of parasite var, rifin, stevor (14) and other highly polymorphic regions for which reference genome-based mapping is not possible, we obtained medians of 26.6 million human uniquely mapped reads from each subject (26.6 million severe malaria, 26.7 million uncomplicated malaria, Mann-Whitney P = 0.913) and 9.61 million parasite uniquely mapped reads (10.3 million severe malaria, 5.03 million uncomplicated malaria, Mann-Whitney P = 0.346) (Fig 1A). We detected expression of 12253 human and 3880 parasite genes. Commensurate with the high parasitemias seen in these children (Table 1), parasite read depth was considerably greater than in a previous study of Indonesians with uncomplicated malaria (23).
Table 1. Characteristics of study subjects (n = 46).
CM, cerebral malaria; CH, cerebral malaria plus hyperlactatemia; HL, hyperlactatemia (CM, CH, and HL are all subgroups of severe malaria); UM, uncomplicated malaria; PfHRP2, P. falciparum histidine rich protein 2; Hb, hemoglobin concentration; WBC, white blood cell count. Data are median (IQR), superscripts indicate the number of subjects with data for each variable if less than the total; P for Kruskall-Wallis test comparing all groups (degrees of freedom = 3) except for sex where P is for Fisher’s exact test. Leukocyte population numbers and proportions measured by clinical hematology analyser.
| CM (n = 5) | CH (n = 12) | HL (n = 8) | UM (n = 21) | P | |
|---|---|---|---|---|---|
| Age (years) | 4.3 (4.2-4.8) | 4.9 (3.6-5.7) | 5.0 (3.8-8.3) | 6.0 (4.0-9.0) | 0.51 |
| Male (%) | 3 (60%) | 5 (42%) | 7 (88%) | 13 (62%) | 0.24 |
| Parasitemia (%) | 8.3 (5.3-9.0)4 | 12.6 (9.4-19.0) | 9.6 (1.8-12.2) | 5.1 (3.8-7.0) | 0.008 |
|
Parasites (x105 /uL) |
2.3 (1.7-3.1) 3 | 3.5 (2.7-8.4) 11 | 2.8 (0.7-5.0) | 2.3 (1.6-3.2) | 0.062 |
| Clones | 2 (1.5-2.5) 4 | 2 (1-2) 9 | 1 (1-2) 5 | 2 (1-2) 15 | 0.67 |
|
PfHRP2 (ng/mL) |
202 (93-528) 4 | 763 (374-1750) | 470 (164-2214) | 163 (128-227) | 0.004 |
| Duration of illness (days) | 2.0 (1.7-3.0) | 2.0 (2.0-2.5) | 2.0 (2.0-3.5) | 2.7 (2.0-3.0) | 0.22 |
| Hb (g/dL) | 9.7 (7.4-10.4) | 9.3 (7.8-11.5) 11 | 9.1 (7.4-11.0) | 10.8 (9.9-12.1) | 0.12 |
| WBC (x109/L) | 9.8 (8.2-12.9) 4 | 8.8 (6.4-9.4) 11 | 15.3 (7.9-16.8) 7 | 9.5 (7.7-11.8) | 0.41 |
| Platelets (x109/L) | 41 (40-82) 4 | 36 (23-65) 11 | 59 (33-132) | 122 (96-132) | 0.013 |
|
Lymphocytes (x109/L) |
2.7 (2.1-3.6)4 | 2.9 (2.4-3.6)11 | 3.1 (1.8-5.2)7 | 2.4 (1.4-3.1)20 | 0.57 |
| Lymphocyte (%) | 29.8 (20.6-37.3) 4 |
37.8 (29.9-49.9)11 |
22.3 (14.7-37.3)7 |
23.9 (16.0-33.5)20 |
0.087 |
|
Neutrophils (x109/L) |
6.4 (4.0-8.7)4 | 4.0 (2.9-4.3)10 | 6.5 (5.8-10.4)7 | 7.0 (5.3-7.7)20 | 0.045 |
|
Neutrophil (%) |
55.1 (49.0-69.6) 4 |
48.3 (39.6-56.2) 10 |
61.5 (55.6-74.9) 7 |
68.0 (59.9-79.6)20 |
0.016 |
|
Monocytes (x109/L) |
0.6 (0.6-0.7)4 | 0.6 (0.5-0.9)10 | 0.8 (0.6-1.3)7 | 0.7 (0.4-0.9)20 | 0.58 |
|
Monocyte (%) |
7.1 (6.0-7.7)4 |
7.8 (6.8-8.6)10 |
6.6 (5.1-7.8) 7 |
6.7 (4.8-7.3)20 |
0.12 |
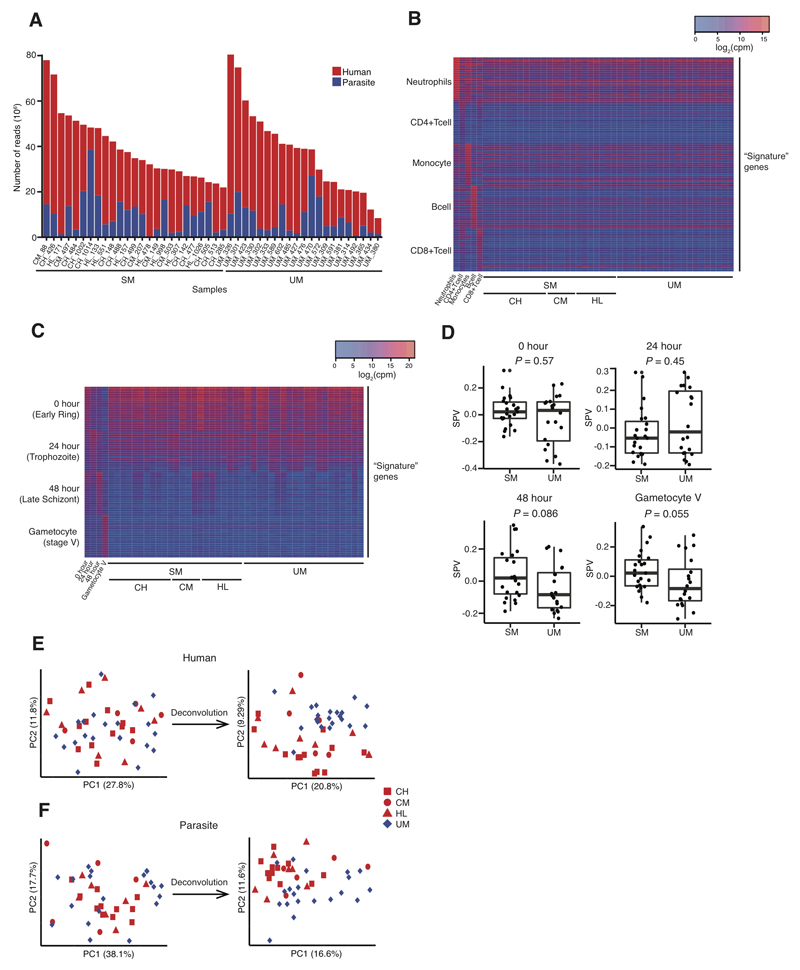
Figure 1. Whole blood dual RNA-sequencing and deconvolution.
(A) Uniquely mapped reads from human (red) and P. falciparum (blue) from subjects with severe (SM, n = 25) and uncomplicated malaria (UM, n = 21). (B,C) Heatmaps showing signature gene expression for different leukocyte (B) and parasite developmental stage (C) populations and their relative intensity in individual subjects with SM, including different SM phenotypes (CH, cerebral malaria plus hyperlactatemia; CM, cerebral malaria; HL, hyperlactatemia), and UM. (D) Surrogate proportion variables for parasite developmental stages compared between severe malaria and uncomplicated malaria using the Mann-Whitney test (bold line, box and whiskers indicate median, interquartile range and up to 1.5-times interquartile range from the lower and upper ends of the box respectively). (E,F) Principal component plots showing the effect of deconvolution on the segregation of subjects with UM and SM, adjusting human (E) and parasite (F) gene expression for differences in proportions of leukocytes or parasite developmental stages respectively. Analyses of human gene expression (B, E): SM, n = 25; UM, n = 21. Analyses of parasite gene expression (C, D,F): SM, n = 23; UM, n = 20.
Systemic infection provokes changes in blood leukocyte subpopulations which can dominate changes in gene expression and confound their interpretation (24). Amongst our study subjects there were significant differences between clinical groups in the proportions of neutrophils (Kruskal-Wallis P = 0.01) and neutrophil counts (Kruskal-Wallis P = 0.05) in blood (Table 1). We performed gene signature-based deconvolution (25) to estimate heterogeneity in the contribution of the major leukocyte subpopulations to the RNA in our samples (Fig 1B, fig S1). Parasite gene expression in vivo is also dominated by the mixture of parasite developmental stages at the time of sampling because there is phasic variation in gene expression (26) and total RNA content increases during the intraerythrocytic developmental cycle (27). Therefore we also applied the deconvolution approach with reference gene signatures derived from highly synchronous parasite cultures (26, 28) to identify the contribution of parasites at different developmental stages (Fig 1C). As the method was developed and validated for distinct human cell types, we confirmed the effectiveness of estimation of parasite developmental stage mixture by comparison with previously proposed stage-specific marker genes (29), and by assessment of performance in synthetic datasets of known composition (fig S2). We compared relative contributions of parasite developmental stages between severe and uncomplicated malaria samples and observed a trend towards greater contributions from late stage asexual parasites and gametocytes in children with severe malaria (Fig 1D), consistent with previous reports (30).
In order to remove the confounding effect of heterogeneity in leukocyte and parasite mixtures we adjusted gene expression values for the proportions of detected cell types, essentially allowing us to compare gene expression as if all subjects had the same leukocyte and parasite population compositions. Adjustment for heterogeneity in the mixture of leukocytes and parasite developmental stages improved segregation of severe and uncomplicated malaria cases (Fig 1E, F; multivariate ANOVA for human gene expression P = 0.0013 and P = 0.00012, and for parasite gene expression P = 0.0049 and P = 0.00019, before and after adjustment, respectively). Therefore we used adjusted gene expression values for all subsequent analyses.
Gene expression associated with severe malaria and parasite load
To identify differentially expressed genes between clinical groups, and to identify gene expression associated with continuous variable severity features, we used a generalised linear model approach incorporating leukocyte populations or parasite stage as covariates, and we considered a false discovery rate (FDR)-adjusted P < 0.05 as significant. Considering all subjects, there were 770 human significantly differentially expressed genes between severe and uncomplicated malaria (Fig 2A, table S1). Genes more highly expressed in severe malaria versus uncomplicated malaria most notably included MMP8 (matrix metallopeptidase 8), OLFM4 (olfactomedin 4), DEFA3 (defensin A3), and ELANE (neutrophil elastase), all encoding neutrophil granule proteins (31). Given our adjustment for cellular heterogeneity, these results likely reflect a true increase in transcription of these genes rather than a greater proportion of neutrophils in the blood, and differences remained when absolute neutrophil counts were multiplied by expression (Fig 2B). We performed Gene Ontology (GO) analyses to better understand the biological functions of the differentially expressed genes (Fig 2C) and identified enrichment of co-translational protein targeting, cell motility and immune response functions (table S2). We used Ingenuity Pathway Analysis (Qiagen Bioinformatics) to predict upstream regulators of the differentially expressed genes, and colony stimulating factor 3 (CSF3, also known as granulocyte colony stimulating factor, GCSF), Fas cell surface death receptor, and Prostaglandin E receptor 2 signalling were amongst the most over-represented (table S3). We repeated similar analyses to identify and interpret genes differentially expressed between uncomplicated malaria and different clinical phenotypes of severe malaria (fig S3, tables S1&S2), which revealed many consistent associations but also some notable differences. For example, the number of differentially expressed genes substantially increased when comparing the subgroup of subjects with cerebral malaria plus hyperlactatemia (the most severe phenotype, n = 12) vs uncomplicated malaria (n=21), possibly reflecting their greater severity and homogeneity of disease (table S1, figure S3). The most highly expressed genes in these patients relative to uncomplicated malaria included genes for neutrophil granules and heat shock proteins.
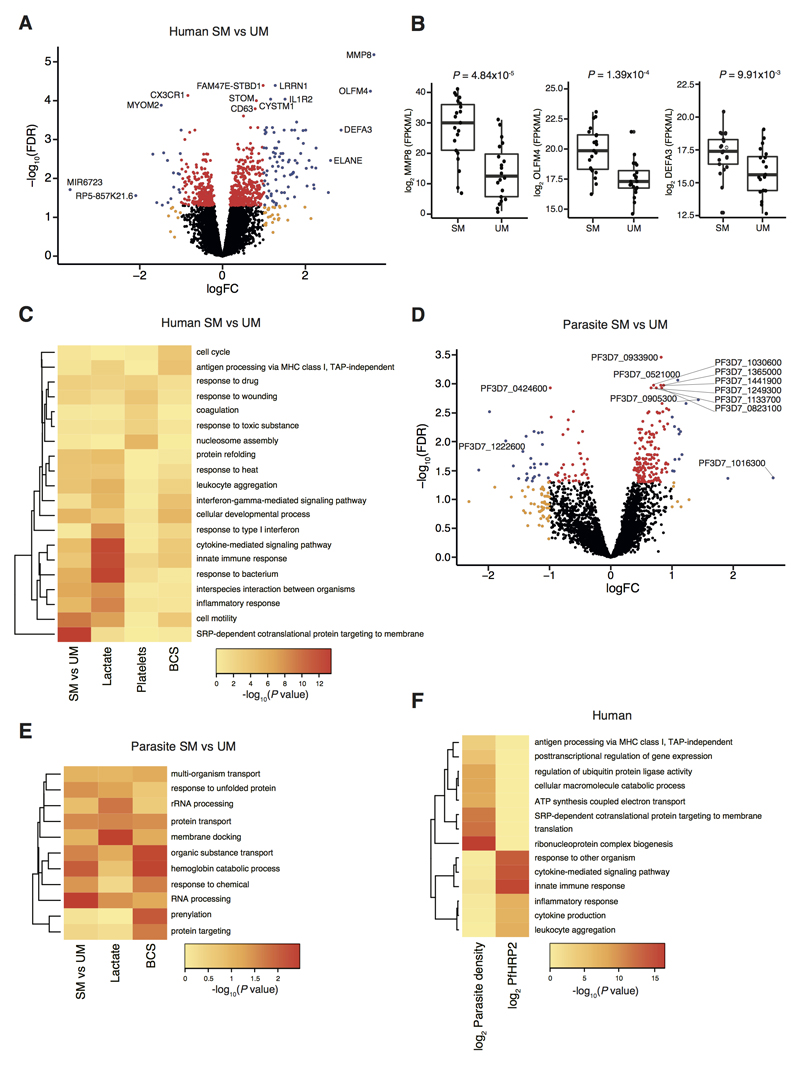
Figure 2. Association of gene expression with features of severe malaria and parasite load.
(A) Volcano plot showing extent and significance of up- or down- regulation of human gene expression in severe malaria (SM) compared with uncomplicated malaria (UM) (red and blue, P <0.05 after Benjamini-Hochberg adjustment for false discovery rate [FDR]; orange and blue, absolute log2-fold change (FC) in expression > 1; SM n = 25, UM n = 21). (B) Comparison of selected neutrophil-related gene expression multiplied by absolute neutrophil count in blood between SM (n = 21) and UM (n = 20) (bold line, box and whiskers indicate median, interquartile range and up to 1.5-times interquartile range from the lower and upper ends of the box respectively, P for Mann-Whitney test). (C) Heatmap comparing enrichment of gene ontology terms for human genes significantly differentially expressed between severe malaria and uncomplicated malaria or significantly associated with blood lactate, platelet count or BCS. (D) P. falciparum differential gene expression in severe malaria compared to uncomplicated malaria (colour coding as in (A); SM n = 23, UM n = 20). (E) Heatmap comparing enrichment of gene ontology terms for parasite genes significantly differentially expressed between severe malaria and uncomplicated malaria and or significantly associated with blood lactate or BCS. (F) Heatmap comparing gene ontology terms for human genes significantly associated with log parasite density and log PfHRP2.
In order to gain a greater insight into specific pathophysiological processes, we examined the quantitative association of gene expression with clinical and laboratory parameters (table 1) which characterise specific aspects of severe malaria pathophysiology: consciousness level (using the Blantyre Coma Scale, BCS), blood lactate concentration, platelet count and hemoglobin concentration (table S4). 738 genes were significantly associated with BCS level (using ordinal regression with FDR P < 0.05, table S4) and decreasing consciousness level (lower BCS) was associated with both higher expression of genes involved in the cell cycle and lower expression of genes involved in MHC class I antigen presentation and interferon-γ (IFN-γ) signalling (Fig 2C, table S2). Predicted upstream regulators included estrogen receptor 1 and transglutaminase 2 (table S3). 1012 human genes were associated with lactate concentration (table S4), amongst which immune response pathways were again prominent, but a negative association between lactate and type 1 IFN signalling was particularly notable (Fig 2C, table S2), and the most strongly predicted upstream regulators were IFN-γ, IFN-α, and TNF (table S3). 178 genes were associated with platelet count (table S4) and the most enriched pathways differed considerably from those in the preceding analyses (Fig 2C, table S2), with nucleosome assembly (predominantly histone genes), coagulation, and response to wounding genes all negatively correlated, and the most strongly predicted upstream regulators being IL13, RB transcriptional corepressor 1, and IL1RN (table S3). No human genes correlated with hemoglobin concentration. Taken together, these results identify common transcriptional features of severe malaria but also implicate distinct mechanisms underlying the different pathophysiological processes which can occur in severe malaria.
There were 236 parasite genes differentially expressed between severe and uncomplicated malaria (Fig 2D, table S5). The parasite gene with highest expression in severe relative to uncomplicated malaria was PF3D7_1016300, a gene which encodes a glycophorin binding protein (GBP) expressed in the cytoplasm of infected erythrocytes and which influences adhesion and rigidity of the red cell (32, 33). The most down-regulated parasite gene (with known function) in severe relative to uncomplicated malaria was PF3D7_1222600 which encodes the AP2 domain transcription factor AP2-G. This protein controls the balance between gametocytogenesis and asexual replication, and knockout of the orthologue in P. berghei ANKA enhances the in vivo asexual parasite growth rate (34, 35). As with human gene expression, there were more differentially expressed genes in the comparison of uncomplicated malaria vs. the cerebral malaria plus hyperlactatemia group (fig S3, table S5). Here the most differentially expressed parasite genes included PF3D7_0202000 (knob-associated histidine rich protein), PF3D7_1016300 (GBP), PF3D7_0201900 (erythrocyte membrane protein 3), and PF3D7_0424600 (PHIST-b protein), all of which encode proteins that interact with the erythrocyte cytoskeleton to influence cytoadhesion and deformability of the infected erythrocyte, making them plausible determinants of severity (33). Parasite genes differentially expressed in severe compared to uncomplicated malaria were enriched in specific biological functions including RNA processing, protein transport, and hemoglobin catabolism (Fig 2E, table S6).
445 parasite genes were significantly associated with BCS level (using ordinal regression with FDR P < 0.05), and those most significantly associated with lower BCS were PF3D7_0919800 (TLD domain-containing protein), PF3D7_1133700 (FHA domain-containing protein), and PF3D7_1408200 (AP2 domain transcription factor AP2-G2), the latter two being important determinants of asexual parasite growth rate (35, 36) (table S7). The most enriched functions associated with BCS included transport, hemoglobin catabolism and prenylation (Fig 2D). 100 parasite genes associated with lactate (table S7). The most significant (FDR P = 2.4x10-6) was PF3D7_0201900 (encoding erythrocyte membrane protein 3, EMP3), consistent with infected erythrocyte rigidity and cytoadhesion (32, 37, 38) being important determinants of microvascular obstruction and hyperlactatemia (39). Pathway enrichments amongst these genes differed from those most associated with BCS and included membrane docking and rRNA processing (Fig 2D and table S6). Few parasite genes were associated with hemoglobin concentration or platelet count (table S7). Taken together, these findings indicate that different patterns of parasite gene expression are associated with, and may therefore contribute to, specific aspects of host pathophysiology.
In order to establish whether changes in parasite gene expression might be cause or consequence of severe malaria, we tested the effect of hyperlactatemia on parasite gene expression in vitro. 61 genes were differentially expressed between lactate supplemented (n = 4) and control (n = 5) early ring-stage parasite cultures, particularly enriched in genes associated with transcription and RNA processing (tables S5 and S6). Two of the genes most highly induced by lactate supplementation were PF3D7_1016300 (GBP) and PF3D7_0202000 (knob-associated histidine rich protein), genes which were also highly expressed in the cerebral malaria plus hyperlactatemia phenotype. This suggests lactate may influence the virulence phenotype of parasites, consistent with a recent report that Plasmodium can sense and respond to the host metabolic environment (40).
Previous studies have shown a correlation between the expression levels of host genes and circulating parasitemia (41, 42). Parasite load differed between our subjects with severe and uncomplicated malaria (Table 1) and we were interested to determine the extent to which this explained the differences in whole blood gene expression. Peripheral blood parasite quantification (parasite density) underestimates the total number of parasites in the body because of sequestration of parasites in the microvasculature (13, 14). The soluble parasite protein, P. falciparum histidine rich protein 2 (PfHRP2), has been used as a plasma biomarker of total parasite load (circulating plus sequestered parasites) and is more strongly associated with severity (8, 9, 11) and death (8, 9). We examined the association of host and parasite gene expression with both circulating parasite density and PfHRP2 (restricting comparisons to subjects with data for both). We found 1886 human genes significantly (FDR P < 0.05) correlated with log parasite density and 616 significantly correlated with log PfHRP2 (102 common to both), whilst only 2 and 10 parasite genes were significant in the corresponding analyses (none common to both) (tables S4 and S7). Human genes correlated with log parasite density were particularly enriched in pathways related to translation (especially exported proteins), oxidative phosphorylation, and antigen presentation (Fig 2F, table S2), with predicted upstream regulation by RPTOR independent companion of MTOR complex 2 (RICTOR), hepatocyte nuclear factor 4 alpha (HNF4A) and X-box binding protein 1(XBP1); table S3). Genes correlated with log PfHRP2 were enriched in innate immune response functions (Fig 2F, table S2), and the most strongly predicted upstream regulators were interferon-γ (IFN-γ), transglutaminase 2, and IFN-α2 (table S3). These findings suggest that the nature of the systemic host response is associated with the localisation of parasites.
We next asked to what extent the differences in gene expression between severe and uncomplicated malaria phenotypes were dependent on parasite load. Restricting analyses to subjects with both parasite density and PfHRP2 measurements, the number of human genes differentially expressed in severe vs uncomplicated malaria remained almost unchanged after adjustment for parasite density but was reduced by 98.6% after adjustment for PfHRP2, whilst parasite differentially expressed genes changed much less after either of the same adjustments (Table 2, tables S1 and S5). Findings were similar when adjusting for parasite load in comparisons of each of the severe malaria subtypes vs uncomplicated malaria (tables S1 and S5). Repeating this analysis on an independent dataset of human microarray gene expression in Malawian children with cerebral malaria (43) revealed 994 differentially expressed genes (FDR P < 0.05) between children with (n = 55) and without (n = 17) malaria-associated retinopathy (table S1), with 608 (61%) genes found to be differentially expressed after adjustment for parasite density, and none differentially expressed after adjustment for PfHRP2.
Table 2. Numbers of differentially expressed genes before and after adjustment for parasite load.
*Only subjects with complete data for every parameter are included. Number of genes associated with severity category or laboratory marker of severity before and after adjustment for parasite load (% of number in unadjusted analysis where applicable). BCS, Blantyre coma scale; SM, severe malaria; UM, uncomplicated malaria.
| Human genes | Parasite genes | |||||||
|---|---|---|---|---|---|---|---|---|
| n* | Unadjusted | Log parasite density | Log PfHRP2 | n* | Unadjusted | Log parasite density | Log PfHRP2 | |
| SM vs UM | 43 | 907 | 914 (101%) |
13 (1.4%) |
41 | 516 | 562 (109%) |
329 (64%) |
| BCS | 43 | 738 | 491 (67%) |
12 (1.6%) |
41 | 445 | 340 (76%) |
148 (33%) |
| Lactate | 40 | 1012 | 526 (52%) |
51 (5.0%) | 38 | 100 | 109 (109%) |
47 (47%) |
| Platelets | 43 | 178 | 66 (37%) |
46 (25%) | 41 | 1 | 1 (100%) |
1 (100%) |
| Hemoglobin | 43 | 0 | 0 | 0 | 41 | 6 | 5 (83%) |
4 (67%) |
Taken together these findings suggest that total body parasite load, as represented by PfHRP2, is a dominant determinant of host gene expression in malaria, particularly of inflammatory and immune response genes, and differences in total body parasite load drive the majority of the human gene expression differences between severe and uncomplicated malaria. However, if genes remain associated with severity after adjustment for parasite load, this may indicate intrinsic variation in the host response which determines susceptibility to severe disease. In our dataset, only 13 genes remained significant (FDR P < 0.05) after adjustment for PfHRP2 (Table 2, table S1). Of particular interest amongst these, MMP8 encodes the metallopeptidase MMP8 (also known as collagenase 1) which causes endothelial barrier damage in several infection models (44, 45); AZI2 encodes 5-azacytidine induced 2 (also known as NF-Kappa-B-Activating Kinase-Associated Protein 1, NAP1) (46), a regulator of the type 1 interferon response, a pathway which is known to control severity of disease in rodent malaria models (47); whilst CX3CR1 encodes the receptor for fractalkine (a biomarker of cerebral malaria in humans (48)), expressed on subset of monocytes which are particularly efficient at killing malaria parasites (49), and controls the trafficking of monocytes during inflammation (50).
Parasite load was also a major driver of the associations between human gene expression and BCS level, lactate, and platelet count, although platelet count-associated genes were relatively less dependent on parasite load (Table 2). The few remaining genes significantly associated with lactate after adjustment for PfHRP2 included PKM (encoding the glycolytic enzyme pyruvate kinase M) and GYS1 (encoding the glycogenic enzyme glycogen synthase 1) (table S4), suggesting hyperlactatemia is partly associated with parasite load-independent variation in control of host glucose metabolism.
Co-expression networks of host and parasite genes
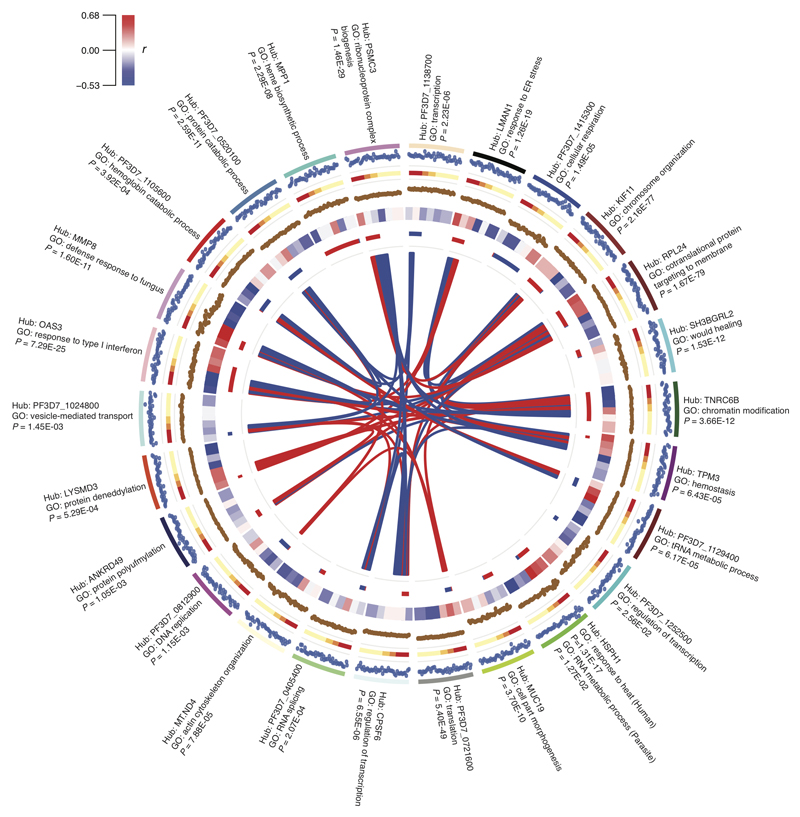
The expression of genes with common functional roles is often correlated and can be identified through co-expression network analysis (51). We applied this methodology to paired host and parasite gene expression data from each individual (without prior adjustment for parasite load) to identify co-expressed groups of genes from either or both species. We term these groups of genes “modules”, and we named each module according to the “hub gene” which has the greatest connectivity to other genes within the module. First we analysed all subjects together, generating a network with 26 modules (Fig 3 and table S8): 10 modules contained exclusively human genes, 5 exclusively parasite genes, and 11 both human and parasite genes (most of these highly skewed to a single species). Only the HSPH1 (heat shock protein family H [Hsp110] member 1) module contained more than 10 genes from both human and parasite, and was strongly enriched in human heat shock response and parasite RNA metabolism genes. All modules showed significant (P < 0.05) GO enrichments, regardless of host or parasite origin. The composite expression of genes within a module can be described by a module eigengene value (51, 52) and there were associations between module eigengene values and malaria severity, parasite load, consciousness level, and other laboratory parameters (Figure 3). Some host-dominated and parasite-dominated modules were also highly correlated with each other, most notably the RPL24 (ribosomal protein L24) module (highly enriched in translation pathways) strongly correlated with the functionally similar PF3D7_0721600 (putative 40S ribosomal protein S5) parasite module. We excluded multi-mapping reads as an explanation for this, and suggest that this indicates co-regulation of conserved host and parasite translation machinery. Furthermore, most of these genes were also differentially expressed between severe and uncomplicated malaria.
Figure 3. Interspecies gene expression modules and their associations with severity.
Circos plot showing gene expression modules obtained from whole genome correlation network analysis using expression of all human and parasite genes from each subject (severe malaria, n = 22; uncomplicated malaria, n = 19) as the input. From outside to inside: labels, hub gene and most enriched GO term (with P-value) for each module; track 1, module eigengene value for each subject; track 2, clinical phenotype (Red, CH; Orange, CM; Green, HL; Yellow, uncomplicated malaria); track 3, hub gene expression (log CPM) for each subject; track 4, heatmap for correlation with laboratory measurements (clockwise, blocks: log parasite density, log PfHRP2, lactate, platelets, hemoglobin, BCS; colour intensity represents correlation coefficient as shown in colour key); track 5, module size and composition (length proportional to number of genes in module; red, human genes; blue, parasite genes); polygons connect modules with significant (FDR P < 0.01) Pearson correlation between eigengene values (width proportional to -log10 FDR P-value; red=positive correlation, blue=negative correlation).
Association of co-expression modules with severity
Co-expression network modules can be used as units of analysis, affording considerable dimension reduction for whole-genome expression data. We used module eigengene values and parasite load (with which many modules were correlated, Fig 3) in linear regression models to determine the best within-sample predictors of severity, starting with all significant (P < 0.01) univariate associations and proceeding by backward selection (table S9). The resulting multivariate model combined MMP8, OAS1 (2'-5'-oligoadenylate synthetase 1) and LYSMD3 (LysM, putative peptidoglycan-binding, domain containing 3) module eigengenes, but not parasite load. In fact, these modules represent distinct aspects of the immune response (table S8): the MMP8 module, highly enriched in defence response genes with predicted upstream regulators CEBPA (CCAAT/enhancer binding protein alpha, a myeloid transcription factor) and CSF3, likely reflects granulopoiesis (31); the OAS1 module is highly enriched for type-1 IFN response genes; and the small LYSMD3 module, with limited GO enrichment, contains a functional network around IFN-γ (figure S4). The direction of association of the OAS1 module with severity changed from negative in univariate analysis to positive in the multivariate analysis, suggesting that inadequate suppression of the type-1 IFN response in conjunction with upregulation of granulopoiesis and IFN-γ signalling may contribute to pathogenesis.
Differential co-expression in severe malaria
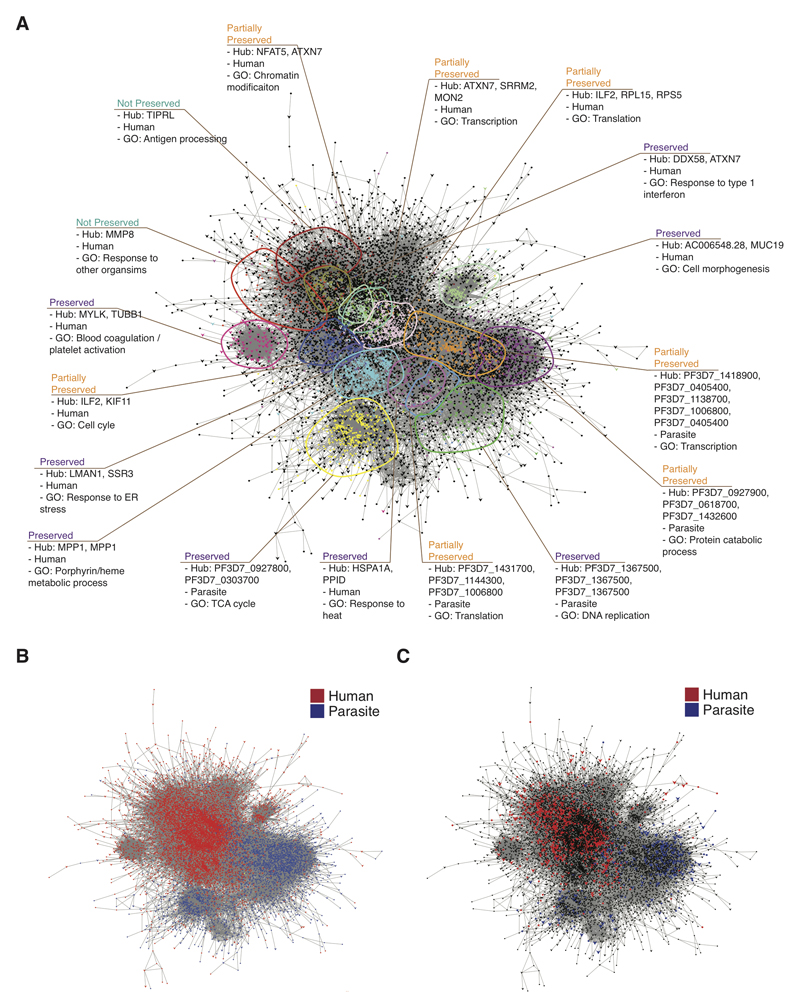
Considering all subjects together in the generation of co-expression networks maximises power to detect consistently co-regulated genes but may not identify sets of genes where co-regulation is altered by severity. For this reason we also created separate co-expression networks for uncomplicated and severe malaria and compared modules to identify differential co-expression (Fig 4, table S10). Eight modules showed substantial preservation between networks, seven were partially preserved, and two were unique to severe malaria (Figure 4A). Partial preservation was common amongst modules comprised predominantly from human or parasite genes (Figure 4A,B), and module preservation was not dependent on the proportion of module genes differentially expressed between severe and uncomplicated malaria (Figure 4A,C). A MMP8 module (exclusively human genes, many encoding neutrophil granule and phagosome components) was uniquely present in severe malaria subjects, and 38% of its member genes were differentially expressed in the comparison between severe malaria and uncomplicated malaria (table S10). The module was enriched in host defence functions and predicted to be regulated by CEBPA, CSF3 and TNF (table S10). These findings strongly suggest the MMP8 module represents emergency granulopoiesis (31) and mark this as a specific feature of severe malaria. The TIPRL (TOR Signaling Pathway Regulator) module (99.2% human genes) was also unique to severe malaria but contained very few (1.3%) differentially expressed genes, had limited GO enrichment, and the most strongly predicted upstream regulator was the transcription factor HNF4A (table S10). Both TIPRL and HNF4A have regulatory roles in metabolic, inflammatory and apoptosis signal pathways, so the minimal change in expression of this module may represent an aberrant response in severe malaria (53, 54). Amongst the partially preserved modules we observed that host and parasite translation pathways were more tightly co-regulated in severe than uncomplicated malaria, with genes being distributed across fewer modules in severe malaria (Fig 4A, table S10). This once again suggests that there is an interaction between these processes that is associated with severity.
Figure 4. Severity-associated differential co-expression within the interspecies gene expression network.
(A-C) Cytoscape visualisation of merged co-expression networks derived separately from severe malaria (n = 22) and uncomplicated malaria (n = 19). Networks were merged such that genes found in both sub-networks (represented as arrow-shaped, larger-sized nodes) are connected to genes found in only one sub-network (represented as circular-shaped and smaller-sized nodes). (A) Genes and gene clusters are coloured and annotated by module, species, most enriched gene ontology terms, and conservation between sub-networks. Preserved, module pairs from severe malaria and uncomplicated malaria sub-networks overlap with each otherbut no other modules; partially preserved, module clusters in one sub-networkoverlap with two or more modules in the other sub-network; unique, gene clustering only found in one sub-network. Genes in black do not belong to any characterized module. (B) Identical network layout with genes coloured by species (red, human; blue, P. falciparum). (C) Identical network layout with genes coloured by whether they are significantly differentially expressed in severe malaria vs uncomplicated malaria (red, human; blue, P. falciparum; black, not differentially expressed).
Interaction of parasite and host gene expression accounting for parasite load
We sought to integrate pathogen load into analysis of interaction between host and parasite gene expression. To reduce dimensionality we generated human-only gene expression modules from all subjects (table S11), identified those modules significantly (P < 0.05) associated with severity, and then identified parasite genes with significant (FDR P < 0.05) pairwise associations with these modules in a linear model accounting for log PfHRP2. The human modules associated with severity were similar to those identified in preceding analyses (Table 3). The most significantly associated parasite genes and the most enriched parasite GO terms were those involved in RNA processing and translation (Table 3, table S11), suggesting that these processes in the parasite drive multiple aspects of the host transcriptional response independent of their effect on parasite load.
Table 3. Parasite genes correlated with human gene co-expression modules after adjustment for parasite load.
+/- indicates number of parasite genes positively / negatively correlated with each human module eigengene value. All “top genes” in the table are positively correlated with the module eigengene value.
| Human Module | Correlated parasite genes after adjustment for parasite load | |||||
|---|---|---|---|---|---|---|
| Hub gene | Top GO term |
n (+/-) |
Top genes | FDR P | Top GO terms | P |
| Receptor Transporter Protein 4 (RTP4) | GO:0034340 response to type I interferon | 2 (2/0) |
PF3D7_0827500 (apicoplast ribosomal protein L21 precursor) | 0.014 | GO:0006412 translation | 0.018 |
| PF3D7_0111800 (eukaryotic translation initiation factor 4E) | 0.035 | |||||
| Trinucleotide Repeat Containing 6B (TNRC6B) | GO:0016569 chromatin modification | 97 (29/ 68) | PF3D7_1119200 (unknown function) | 0.00012 | GO:0008380 RNA splicing | 4.15 x10-6 |
| PF3D7_1309100 (60S ribosomal protein L24) | 0.00027 | GO:0006396 RNA processing | 3.88 x10-5 | |||
| PF3D7_0825500 (protein KRI1) | 0.00036 | |||||
| Heat shock protein family H (Hsp110) member 1 (HSPH1) | GO:0006457 protein folding | 21 (17/ 4) | PF3D7_0933100 (unknown function) | 0.0071 | GO:0000338 protein deneddylation | 0.0038 |
| PF3D7_1118400 (haloacid dehalogenase-like hydrolase) | 0.0079 | |||||
| PF3D7_0521800 (AFG1-like ATPase) | 0.01 | |||||
| Matrix metallopeptidase 8 (MMP8) | GO:0009617 response to bacterium | 18 (16/ 2) | PF3D7_1356200 (mitochondrial import inner membrane translocase subunit TIM23) | 0.0045 | GO:0019219 regulation of nucleobase-containing compound metabolic process | 0.0065 |
| PF3D7_1119100 (tRNA m(1)G methyltransferase) | 0.0071 | |||||
| PF3D7_0823100 (RWD domain-containing protein) | 0.0073 | |||||
| Peptidylprolyl isomerase B (PPIB) | GO:0034976 response to endoplasmic reticulum stress | 24 (24/ 0) | PF3D7_1420300 (DNL-type zinc finger protein) | 0.0022 | GO:0006364 rRNA processing | 0.0058 |
| PF3D7_0821200 (unknown function) | 0.0057 | |||||
| PF3D7_1119200 (unknown function) | 0.0057 | |||||
| Ribosomal protein L24 (RPL24) | GO:0006614 SRP-dependent co-translational protein targeting to membrane | 93 (78/ 15) | PF3D7_0530600 (XAP-5 DNA binding protein) | 5.3 x10-6 | GO:0006396 RNA processing | 6.92 x10-4 |
| PF3D7_1309100 (60S ribosomal protein L24) | 5.3 x10-6 | GO:0008380 RNA splicing | 1.10 x10-3 | |||
| PF3D7_0821200 (unknown function) | 3.3 x10-5 | |||||
Neutrophil-related proteins in plasma
The most differentially expressed genes in comparisons between severe and uncomplicated malaria encode neutrophil granule proteins (figure 2A). Relationships between transcription, translation, storage and release of granule proteins are expected to be complex, but we sought evidence of correlation between gene expression and circulating protein concentrations. In subjects with residual stored plasma we found significant correlations between gene expression and plasma concentrations of defensin A3 (P = 0.0049, rho =0.47, n = 34) and elastase (P = 0.045, rho = 0.35, n = 34) (figure S5). MMP8 expression was significantly correlated with plasma concentrations in subjects with severe malaria (P = 0.02, rho = 0.59, n = 15), but not in uncomplicated malaria (P = 0.37, rho = -0.21, n = 20) or all subjects combined (P = 0.88, rho = -0.026, n = 35). Upstream regulator analyses described earlier suggested that GCSF (CSF3) was a major regulator of genes in the MMP8 module. We found that plasma GCSF concentrations significantly correlated with the eigengene values (table S8) for this module (P = 0.0030, rho = 0.64, n = 19). We tested whether neutrophil degranulation occurred in response to parasite material by stimulating healthy donor blood cells with P. falciparum schizont lysate, and detected increases in MMP8 release, reaching similar concentrations to those observed in plasma during malaria (figure S5).
Discussion
We used dual RNA-sequencing to identify simultaneous host and parasite gene expression and their systemic interactions associated with severity of P. falciparum malaria in humans. Whilst gene expression is only one of the many biological processes involved, our findings add to the argument for an integrated understanding of infectious diseases and make a strong case that neither host nor pathogen should be studied in isolation when possible.
We have identified many associations between gene expression and features of severity, providing plausible insight into the pathogenesis of severe malaria (figure S6). One of our most striking findings was the overriding effect of parasite load on differences in human gene expression between severe and uncomplicated malaria. Previous studies have examined the association between human gene expression and circulating parasitemia (23, 41, 42), but we found that estimation of total body (both circulating and sequestered) parasite load was necessary to appreciate the full effect on host response. Our findings imply that the host response in severe malaria is not excessive per se, but rather that it is an appropriate host response to an excessive pathogen load. This has important implications for malaria research and likely for other infectious disease, immunology, and pathogenesis research in humans. Without accounting for pathogen load, associations between host factors (such as genetic variants or comorbidities) and severity of infection may be misinterpreted. Unfortunately, total body pathogen load is much harder to measure in other infections in humans where pathogens are not restricted to the blood (55).We found that specific sets of host and parasite genes were associated with different pathophysiological consequences of malaria, although our power to detect associations was limited in the smallest subgroup analyses. Distinct sets of host genes were associated with BCS, lactate concentration and platelet count. Hyperlactatemia in malaria is often ascribed to anaerobic metabolism arising from microvascular obstruction by adherent and rigid parasitized erythrocytes (4, 39). These properties are partly determined by the expression of particular members of the var, rif and stevor gene families (14), which we did not include in our analysis because their extreme degree of polymorphism prevents reference genome-based quantification. Despite this limitation we still found associations between lactate concentration and severity and in vivo variation in the expression of other parasite genes known to modify the biophysical properties of the infected erythrocyte. Some variation in parasite gene expression may have a genetic basis (56) but our in vitro data suggests that it may also occur as a response to the host environment.
The human genes most correlated with lactate were immune response-related, suggesting that inflammation, and perhaps its effect on glycolysis, may be involved (57, 58). If lactate production is associated with the strength of the host response then the changes in expression of parasite genes in response to lactate might favour sequestration and evasion of innate immune cells as a parasite survival strategy. Cerebral malaria in humans is usually ascribed to parasite sequestration in the cerebral microvasculature, but the association between BCS and antigen presentation via MHC class I, interferon-γ, and type-1 interferon signalling, would be consistent with the localisation of these immunopathological mechanisms away from the blood and into the brain microvasculature as seen in rodent experimental cerebral malaria (15, 16, 59). Thromobocytopenia is almost invariable in malaria and its mechanism is poorly understood (7). Our findings implicate the well-recognised activation of endothelial surfaces and coagulation pathways in malaria (7) as a cause, but also lead us to suspect a role for histone-induced thrombocytopenia (60, 61).
Many of our results converge on a putative role for neutrophils in severe malaria. Different analytical approaches repeatedly identified the association of genes encoding neutrophil granules (such as MMP8, OLFM4, DEFA3, ELANE) and upstream regulators of granulopoiesis (CSF3 and CEBPA) with severe outcomes. Granule proteins are enriched in immature neutrophils (31, 62) which are mobilised from the bone marrow to the circulation in malaria (63, 64), and there is plentiful evidence that neutrophil degranulation occurs in severe malaria (43, 65). Release of neutrophil granule proteins can be highly damaging to host tissues (62), and increased production and release of these proteins could contribute to many of the pathological features of severe malaria. Both neutrophil granule proteins and histones are released into the circulation during production of neutrophil extracellular traps (NETs) (62, 66), a phenomenon which has been described in malaria (64). It is noteworthy that similar neutrophil-related signatures are not reported in the whole blood transcriptomes of rodent models which have been examined to date (42, 67), creating a challenge for experimental validation. However, neutrophil depletion has been shown to prevent experimental cerebral malaria (68), and whilst this is not a viable therapeutic option in humans, pharmacological inhibitors of specific neutrophil functions such as NETosis are being evaluated in other diseases (66).
We observed a relationship between type-1 IFN responses and severity of malaria, which may help to tie together data from previous observations in humans and animal models. In a small study, expression of type-1 IFN response genes in blood from uncomplicated malaria was higher than in severe malaria, leading to the suggestion that this may be protective against developing severe malaria (69). However, we found that type-1 IFN response genes were negatively correlated with parasite load, suggesting down-regulation with increasing parasite load (and severity) is a more likely explanation. Our multivariate analyses using gene expression modules to explain severity suggested that insufficient suppression of type-1 IFN signalling was in fact associated with severity. This would be more consistent with results in mouse malaria models where genetic or antibody-mediated ablation of type-1 IFN signalling improves outcome (70–73).
Very few parasite genes correlated with parasite load at the time of clinical presentation. This may be a consequence of the dynamic nature of parasite load, which is determined by parasite growth rate, the number of replication cycles in the host (duration of infection), and a reciprocal interaction with the constraining host response. Thus lower expression of the gene encoding ApiAP2-G in severe malaria may increase the asexual parasite growth rate (34) and make severe malaria more likely, without this gene exhibiting any correlation with parasite load. It may also seem paradoxical that this gene is down-regulated in severe malaria given that gametocytes are usually more common in severe malaria (30). Development of mature gametocytes takes 10-12 days, for much of which they are not in the systemic circulation (74). We speculate that the mature gametocytes detected at the time of clinical presentation may reflect preferential gametocytogenesis in early infection and perhaps a subsequent reduction in ApiAP2-G that promotes enhanced asexual replication and severe disease.
We noted that both human and parasite translation pathways were associated with severe malaria, and these pathways showed the strongest evidence of interaction between species, with co-regulation appearing tighter in more severe disease. Increased translation is important for production of host defence effector proteins (75) and parasite proteins which enable survival (76) and it is feasible that these processes drive each other. This raises the intriguing question of whether addition of a translation inhibiting anti-malarial such as mefloquine (77) to standard artesunate treatment may have added benefit in severe malaria.
The differences in human and parasite gene expression between severe and uncomplicated malaria were much clearer after adjusting for heterogeneity of leukocyte population and parasite developmental stage. Although the importance of accounting for such variation is well recognised (24), it is rarely done in infectious disease transcriptomic studies. Several studies have used alternative methods to account for parasite developmental stage distribution and have shown that this has a major impact on observed associations between parasite gene expression and clinical phenotype (78, 79).
Whilst we cannot establish causation from an observational study such as this, our findings should be launch points for future work assessing the implicated mechanisms and their potential as targets for adjunctive therapies. The identification of multiple and sometimes distinct host and parasite mechanisms associated with differing aspects of pathophysiology potentially bodes ill for adjunctive therapies, which might need to have multiple targets and perhaps be personalized to differing severe malaria manifestations. However the common association of neutrophil granule protein genes with all severe malaria manifestations suggests that targeting neutrophil function may be a therapeutic strategy in the future.
Materials and Methods
Detailed Supplementary Methods are available on-line
Study design
The primary aim of the study was to analyse differential human and parasite gene expression between children with severe malaria and uncomplicated malaria and to determine association of gene expression with parasite load. Secondary aims were 1) to analyse differences in gene expression associated with different severe malaria syndromes and with continuous variable markers of pathophysiology, 2) to evaluate co-expression of host and parasite genes, and 3) to evaluate differential co-expression associated with severe malaria. Sample size was determined pragmatically based on the availability of suitable samples with necessary clinical and laboratory data. We aimed to achieve close to 30 million mapped human reads and 5 million mapped parasite reads per sample and calculated the likely number of reads required to achieve this based on the percentage parasitemia in each subject and the likely amount of RNA per ring-stage parasite (27). From available RNA samples we aimed to have roughly equal numbers of severe and uncomplicated malaria cases and within the severe malaria cases we aimed to have eight subjects with each phenotype. After assessment of RNA quantity and quality some samples were unsuitable for RNA-sequencing which resulted in the final composition of groups being slightly unbalanced.
Subjects and samples
Gambian children (under 16 years old) with P. falciparum malaria were recruited as previously described (11, 80, 81). Informed consent was obtained and the study was approved by the Gambian Government / MRC Laboratories Joint Ethics Committee (SCCs 670, 1077, 1143, 1178, 1179, 1180, 1207 and L2013.07V2). Malaria was defined by fever and >5000 asexual parasites/μL of blood. Cerebral malaria was defined as BCS of 1 or 2, or a BCS of 3 if the motor response was 1, not due to other causes (11, 80). Hyperlactatemia / acidosis was defined as blood lactate concentration > 5mmol/L (11). Subjects meeting both criteria were described as having cerebral malaria plus hyperlactatemia / acidosis. Blood was collected at the time of presentation to the clinic, prior to any treatment (80).
For RNA-sequencing, severe and uncomplicated malaria groups were matched as closely as possible by age and gender (Supplementary Dataset 1). For uncomplicated malaria samples we aimed to include an equal number with parasitemia above and below 5%.
RNA-sequencing
RNA was collected and extracted as described previously (80). 1μg of total RNA was used with the ScriptSeq v2 RNA-sequencing library preparation kit (Illumina) and ribosmal RNA (rRNA) and globin messenger RNA (mRNA) were depleted using the Globin-Zero Gold kit (Epicentre). Strand-specific libraries were sequenced using the 2x100 bp protocol with an Illumina HiSeq 2500.
Genomes and RNA annotations
Reference genomes were obtained for human (hg38) (82) and P. falciparum (release 24) (83), and gene annotations were obtained from GENCODE (release 22) (84) and PlasmoDB (release 24) (83).
Statistical analysis
Characteristics were compared between subject groups using the Kruskal-Wallis or Mann-Whitney tests for continuous data and Fisher’s exact test for categorical data. The Wilcoxon matched pairs test was used to compare paired data. Correlations were performed using Spearman correlation or Pearson correlation (when data was normally distributed). To determine genome-wide gene expression associated with variables of interest, P-values were calculated for individual genes as described in the Supplementary Methods, and a false-discovery rate adjusted P < 0.05 was considered significant. Fisher’s exact test was used to identify gene set enrichments. Logistic regression was used to determine associations between severity and module eigengene values.
Supplementary Material
One sentence summary.
Host and parasite RNA-sequencing is combined with parasite load estimates to reveal mechanisms associated with human severe malaria.
Acknowledgements
We are grateful to the study subjects, staff at MRCG, Jammeh Foundation for Peace Hospital, and Brikama Health Centre; Konrad Paszkiewicz and staff at Exeter Sequencing Service at the University of Exeter; Johanna Daily for providing subject level data for GSE72058.
Funding: This work was funded by the UK Medical Research Council (MRC) and the UK Department for International Development (DFID) under the MRC/DFID Concordat agreement and is also part of the EDCTP2 programme supported by the European Union (MR/L006529/1 to AJC)), the European Research Council (AdG-2011-294428 to DJC and LBS), MRC core funding of the MRC Gambia Unit (MRCG), and the Wellcome Trust (098051 to AJC); Exeter Sequencing Service is supported by Medical Research Council Clinical Infrastructure award (MR/M008924/1), Wellcome Trust Institutional Strategic Support Fund (WT097835MF), Wellcome Trust Multi User Equipment Award (WT101650MA) and BBSRC LOLA award (BB/K003240/1).
Footnotes
This manuscript has been accepted for publication in Science Translational Medicine. This version has not undergone final editing. Please refer to the complete version of record at www.sciencetranslationalmedicine.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior written permission of AAAS.
Author contributions: AJC, ML and DJC conceived the study; HJL, TDO and AG performed formal analysis; AG, MW and AJC performed laboratory analyses; MW, DJC, DN and LBS provided samples; DJC, TDO and LBS provided methodology; AJC and HJL wrote the original draft; all authors contributed to review and editing of the manuscript; LJC, DJC, ML, TDO and AJC provided supervision; AJC obtained funding.
Competing interests: The authors declare no competing financial interests.
Data availability: RNA-seq data from human samples have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-6413. The accession number for in vitro parasite RNA-seq is E-MTAB-6573. Source data for Table 1 are provided with the paper as Supplementary Dataset 1.
References and notes
- 1.Westermann AJ, Barquist L, Vogel J. Resolving host-pathogen interactions by dual RNA-seq. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006033. e1006033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Westermann AJ, Gorski SA, Vogel J. Dual RNA-seq of pathogen and host. Nat Rev Microbiol. 2012;10:618–630. doi: 10.1038/nrmicro2852. [DOI] [PubMed] [Google Scholar]
- 3.Phillips MA, Burrows JN, Manyando C, van Huijsduijnen RH, Van Voorhis WC, Wells TNC. Malaria. Nat Rev Dis Primers. 2017;3 doi: 10.1038/nrdp.2017.50. 17050. [DOI] [PubMed] [Google Scholar]
- 4.Cunnington AJ, Walther M, Riley EM. Piecing together the puzzle of severe malaria. Sci Transl Med. 2013;5:211ps218. doi: 10.1126/scitranslmed.3007432. [DOI] [PubMed] [Google Scholar]
- 5.Wassmer SC, Grau GE. Severe malaria: what's new on the pathogenesis front? Int J Parasitol. 2017;47:145–152. doi: 10.1016/j.ijpara.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marsh K, Forster D, Waruiru C, Mwangi I, Winstanley M, Marsh V, Newton C, Winstanley P, Warn P, Peshu N, et al. Indicators of life-threatening malaria in African children. N Engl J Med. 1995;332:1399–1404. doi: 10.1056/NEJM199505253322102. [DOI] [PubMed] [Google Scholar]
- 7.O'Sullivan JM, Preston RJ, O'Regan N, O'Donnell JS. Emerging roles for hemostatic dysfunction in malaria pathogenesis. Blood. 2016;127:2281–2288. doi: 10.1182/blood-2015-11-636464. [DOI] [PubMed] [Google Scholar]
- 8.Dondorp AM, Desakorn V, Pongtavornpinyo W, Sahassananda D, Silamut K, Chotivanich K, Newton PN, Pitisuttithum P, Smithyman AM, White NJ, Day NP. Estimation of the total parasite biomass in acute falciparum malaria from plasma PfHRP2. PLoS Med. 2005;2:e204. doi: 10.1371/journal.pmed.0020204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hendriksen IC, Mwanga-Amumpaire J, von Seidlein L, Mtove G, White LJ, Olaosebikan R, Lee SJ, Tshefu AK, Woodrow C, Amos B, Karema C, et al. Diagnosing severe falciparum malaria in parasitaemic African children: a prospective evaluation of plasma PfHRP2 measurement. PLoS Med. 2012;9:e1001297. doi: 10.1371/journal.pmed.1001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hendriksen IC, White LJ, Veenemans J, Mtove G, Woodrow C, Amos B, Saiwaew S, Gesase S, Nadjm B, Silamut K, Joseph S, et al. Defining falciparum-malaria-attributable severe febrile illness in moderate-to-high transmission settings on the basis of plasma PfHRP2 concentration. J Infect Dis. 2013;207:351–361. doi: 10.1093/infdis/jis675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cunnington AJ, Bretscher MT, Nogaro SI, Riley EM, Walther M. Comparison of parasite sequestration in uncomplicated and severe childhood Plasmodium falciparum malaria. J Infect. 2013;67:220–230. doi: 10.1016/j.jinf.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haldar K, Murphy SC, Milner DA, Taylor TE. Malaria: mechanisms of erythrocytic infection and pathological correlates of severe disease. Annu Rev Pathol. 2007;2:217–249. doi: 10.1146/annurev.pathol.2.010506.091913. [DOI] [PubMed] [Google Scholar]
- 13.Cunnington AJ, Riley EM, Walther M. Stuck in a rut? Reconsidering the role of parasite sequestration in severe malaria syndromes. Trends Parasitol. 2013;29:585–592. doi: 10.1016/j.pt.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wahlgren M, Goel S, Akhouri RR. Variant surface antigens of Plasmodium falciparum and their roles in severe malaria. Nat Rev Microbiol. 2017;15:479–491. doi: 10.1038/nrmicro.2017.47. [DOI] [PubMed] [Google Scholar]
- 15.Dunst J, Kamena F, Matuschewski K. Cytokines and Chemokines in Cerebral Malaria Pathogenesis. Front Cell Infect Microbiol. 2017;7:324. doi: 10.3389/fcimb.2017.00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howland SW, Claser C, Poh CM, Gun SY, Renia L. Pathogenic CD8+ T cells in experimental cerebral malaria. Semin Immunopathol. 2015;37:221–231. doi: 10.1007/s00281-015-0476-6. [DOI] [PubMed] [Google Scholar]
- 17.Higgins SJ, Kain KC, Liles WC. Immunopathogenesis of falciparum malaria: implications for adjunctive therapy in the management of severe and cerebral malaria. Expert Rev Anti Infect Ther. 2011;9:803–819. doi: 10.1586/eri.11.96. [DOI] [PubMed] [Google Scholar]
- 18.Gazzinelli RT, Kalantari P, Fitzgerald KA, Golenbock DT. Innate sensing of malaria parasites. Nat Rev Immunol. 2014;14:744–757. doi: 10.1038/nri3742. [DOI] [PubMed] [Google Scholar]
- 19.van der Poll T, van de Veerdonk FL, Scicluna BP, Netea MG. The immunopathology of sepsis and potential therapeutic targets. Nat Rev Immunol. 2017;17:407–420. doi: 10.1038/nri.2017.36. [DOI] [PubMed] [Google Scholar]
- 20.Prescott JB, Marzi A, Safronetz D, Robertson SJ, Feldmann H, Best SM. Immunobiology of Ebola and Lassa virus infections. Nat Rev Immunol. 2017;17:195–207. doi: 10.1038/nri.2016.138. [DOI] [PubMed] [Google Scholar]
- 21.Openshaw PJM, Chiu C, Culley FJ, Johansson C. Protective and harmful immunity to RSV Infection. Annu Rev Immunol. 2017;35:501–532. doi: 10.1146/annurev-immunol-051116-052206. [DOI] [PubMed] [Google Scholar]
- 22.Scholzen A, Sauerwein RW. Immune activation and induction of memory: lessons learned from controlled human malaria infection with Plasmodium falciparum. Parasitology. 2016;143:224–235. doi: 10.1017/S0031182015000761. [DOI] [PubMed] [Google Scholar]
- 23.Yamagishi J, Natori A, Tolba ME, Mongan AE, Sugimoto C, Katayama T, Kawashima S, Makalowski W, Maeda R, Eshita Y, Tuda J, et al. Interactive transcriptome analysis of malaria patients and infecting Plasmodium falciparum. Genome Res. 2014;24:1433–1444. doi: 10.1101/gr.158980.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen-Orr SS, Gaujoux R. Computational deconvolution: extracting cell type-specific information from heterogeneous samples. Curr Opin Immunol. 2013;25:571–578. doi: 10.1016/j.coi.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chikina M, Zaslavsky E, Sealfon SC. CellCODE: a robust latent variable approach to differential expression analysis for heterogeneous cell populations. Bioinformatics. 2015;31:1584–1591. doi: 10.1093/bioinformatics/btv015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otto TD, Wilinski D, Assefa S, Keane TM, Sarry LR, Bohme U, Lemieux J, Barrell B, Pain A, Berriman M, Newbold C, et al. New insights into the blood-stage transcriptome of Plasmodium falciparum using RNA-Seq. Mol Microbiol. 2010;76:12–24. doi: 10.1111/j.1365-2958.2009.07026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoeijmakers WA, Bartfai R, Stunnenberg HG. Transcriptome analysis using RNA-Seq. Methods Mol Biol. 2013;923:221–239. doi: 10.1007/978-1-62703-026-7_15. [DOI] [PubMed] [Google Scholar]
- 28.Lopez-Barragan MJ, Lemieux J, Quinones M, Williamson KC, Molina-Cruz A, Cui K, Barillas-Mury C, Zhao K, Su XZ. Directional gene expression and antisense transcripts in sexual and asexual stages of Plasmodium falciparum. BMC Genomics. 2011;12:587. doi: 10.1186/1471-2164-12-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joice R, Narasimhan V, Montgomery J, Sidhu AB, Oh K, Meyer E, Pierre-Louis W, Seydel K, Milner D, Williamson K, Wiegand R, et al. Inferring developmental stage composition from gene expression in human malaria. PLoS Comput Biol. 2013;9:e1003392. doi: 10.1371/journal.pcbi.1003392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nacher M, Singhasivanon P, Silachamroon U, Treeprasertsuk S, Tosukhowong T, Vannaphan S, Gay F, Mazier D, Looareesuwan S. Decreased hemoglobin concentrations, hyperparasitemia, and severe malaria are associated with increased Plasmodium falciparum gametocyte carriage. J Parasitol. 2002;88:97–101. doi: 10.1645/0022-3395(2002)088[0097:DHCHAS]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 31.Cowland JB, Borregaard N. Granulopoiesis and granules of human neutrophils. Immunol Rev. 2016;273:11–28. doi: 10.1111/imr.12440. [DOI] [PubMed] [Google Scholar]
- 32.Maier AG, Rug M, O'Neill MT, Brown M, Chakravorty S, Szestak T, Chesson J, Wu Y, Hughes K, Coppel RL, Newbold C, et al. Exported proteins required for virulence and rigidity of Plasmodium falciparum-infected human erythrocytes. Cell. 2008;134:48–61. doi: 10.1016/j.cell.2008.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Koning-Ward TF, Dixon MW, Tilley L, Gilson PR. Plasmodium species: master renovators of their host cells. Nat Rev Microbiol. 2016;14:494–507. doi: 10.1038/nrmicro.2016.79. [DOI] [PubMed] [Google Scholar]
- 34.Sinha A, Hughes KR, Modrzynska KK, Otto TD, Pfander C, Dickens NJ, Religa AA, Bushell E, Graham AL, Cameron R, Kafsack BFC, et al. A cascade of DNA-binding proteins for sexual commitment and development in Plasmodium. Nature. 2014;507:253–257. doi: 10.1038/nature12970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Modrzynska K, Pfander C, Chappell L, Yu L, Suarez C, Dundas K, Gomes AR, Goulding D, Rayner JC, Choudhary J, Billker O. A knockout screen of ApiAP2 genes reveals networks of interacting transcriptional regulators controlling the Plasmodium life cycle. Cell Host Microbe. 2017;21:11–22. doi: 10.1016/j.chom.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balu B, Singh N, Maher SP, Adams JH. A genetic screen for attenuated growth identifies genes crucial for intraerythrocytic development of Plasmodium falciparum. PLoS One. 2010;5:e13282. doi: 10.1371/journal.pone.0013282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glenister FK, Coppel RL, Cowman AF, Mohandas N, Cooke BM. Contribution of parasite proteins to altered mechanical properties of malaria-infected red blood cells. Blood. 2002;99:1060–1063. doi: 10.1182/blood.v99.3.1060. [DOI] [PubMed] [Google Scholar]
- 38.Cooke BM, Glenister FK, Mohandas N, Coppel RL. Assignment of functional roles to parasite proteins in malaria-infected red blood cells by competitive flow-based adhesion assay. Br J Haematol. 2002;117:203–211. doi: 10.1046/j.1365-2141.2002.03404.x. [DOI] [PubMed] [Google Scholar]
- 39.Ishioka H, Ghose A, Charunwatthana P, Maude R, Plewes K, Kingston H, Intharabut B, Woodrow C, Chotivanich K, Sayeed AA, Hasan MU, et al. Sequestration and red cell deformability as determinants of hyperlactatemia in Falciparum malaria. J Infect Dis. 2016;213:788–793. doi: 10.1093/infdis/jiv502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mancio-Silva L, Slavic K, Grilo Ruivo MT, Grosso AR, Modrzynska KK, Vera IM, Sales-Dias J, Gomes AR, MacPherson CR, Crozet P, Adamo M, et al. Nutrient sensing modulates malaria parasite virulence. Nature. 2017;547:213–216. doi: 10.1038/nature23009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Griffiths MJ, Shafi MJ, Popper SJ, Hemingway CA, Kortok MM, Wathen A, Rockett KA, Mott R, Levin M, Newton CR, Marsh K, et al. Genomewide analysis of the host response to malaria in Kenyan children. J Infect Dis. 2005;191:1599–1611. doi: 10.1086/429297. [DOI] [PubMed] [Google Scholar]
- 42.Idaghdour Y, Quinlan J, Goulet JP, Berghout J, Gbeha E, Bruat V, de Malliard T, Grenier JC, Gomez S, Gros P, Rahimy MC, et al. Evidence for additive and interaction effects of host genotype and infection in malaria. Proc Natl Acad Sci U S A. 2012;109:16786–16793. doi: 10.1073/pnas.1204945109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feintuch CM, Saidi A, Seydel K, Chen G, Goldman-Yassen A, Mita-Mendoza NK, Kim RS, Frenette PS, Taylor T, Daily JP. Activated Neutrophils Are Associated with Pediatric Cerebral Malaria Vasculopathy in Malawian Children. MBio. 2016;7:e01300–01315. doi: 10.1128/mBio.01300-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vandenbroucke RE, Dejonckheere E, Van Lint P, Demeestere D, Van Wonterghem E, Vanlaere I, Puimege L, Van Hauwermeiren F, De Rycke R, Mc Guire C, Campestre C, et al. Matrix metalloprotease 8-dependent extracellular matrix cleavage at the blood-CSF barrier contributes to lethality during systemic inflammatory diseases. J Neurosci. 2012;32:9805–9816. doi: 10.1523/JNEUROSCI.0967-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schubert-Unkmeir A, Konrad C, Slanina H, Czapek F, Hebling S, Frosch M. Neisseria meningitidis induces brain microvascular endothelial cell detachment from the matrix and cleavage of occludin: a role for MMP-8. PLoS Pathog. 2010;6:e1000874. doi: 10.1371/journal.ppat.1000874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sasai M, Oshiumi H, Matsumoto M, Inoue N, Fujita F, Nakanishi M, Seya T. Cutting Edge: NF-kappaB-activating kinase-associated protein 1 participates in TLR3/Toll-IL-1 homology domain-containing adapter molecule-1-mediated IFN regulatory factor 3 activation. J Immunol. 2005;174:27–30. doi: 10.4049/jimmunol.174.1.27. [DOI] [PubMed] [Google Scholar]
- 47.Mooney JP, Wassmer SC, Hafalla JC. Type I interferon in malaria: a balancing act. Trends Parasitol. 2017;33:257–260. doi: 10.1016/j.pt.2016.12.010. [DOI] [PubMed] [Google Scholar]
- 48.Tahar R, Albergaria C, Zeghidour N, Ngane VF, Basco LK, Roussilhon C. Plasma levels of eight different mediators and their potential as biomarkers of various clinical malaria conditions in African children. Malar J. 2016;15:337. doi: 10.1186/s12936-016-1378-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chimma P, Roussilhon C, Sratongno P, Ruangveerayuth R, Pattanapanyasat K, Perignon JL, Roberts DJ, Druilhe P. A distinct peripheral blood monocyte phenotype is associated with parasite inhibitory activity in acute uncomplicated Plasmodium falciparum malaria. PLoS Pathog. 2009;5:e1000631. doi: 10.1371/journal.ppat.1000631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hamon P, Loyher PL, Baudesson de Chanville C, Licata F, Combadiere C, Boissonnas A. CX3CR1-dependent endothelial margination modulates Ly6C(high) monocyte systemic deployment upon inflammation in mice. Blood. 2017;129:1296–1307. doi: 10.1182/blood-2016-08-732164. [DOI] [PubMed] [Google Scholar]
- 51.Fuller TF, Ghazalpour A, Aten JE, Drake TA, Lusis AJ, Horvath S. Weighted gene coexpression network analysis strategies applied to mouse weight. Mamm Genome. 2007;18:463–472. doi: 10.1007/s00335-007-9043-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakashima A, Tanimura-Ito K, Oshiro N, Eguchi S, Miyamoto T, Momonami A, Kamada S, Yonezawa K, Kikkawa U. A positive role of mammalian Tip41-like protein, TIPRL, in the amino-acid dependent mTORC1-signaling pathway through interaction with PP2A. FEBS Lett. 2013;587:2924–2929. doi: 10.1016/j.febslet.2013.07.027. [DOI] [PubMed] [Google Scholar]
- 54.Marcil V, Seidman E, Sinnett D, Boudreau F, Gendron FP, Beaulieu JF, Menard D, Precourt LP, Amre D, Levy E. Modification in oxidative stress, inflammation, and lipoprotein assembly in response to hepatocyte nuclear factor 4alpha knockdown in intestinal epithelial cells. J Biol Chem. 2010;285:40448–40460. doi: 10.1074/jbc.M110.155358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cunnington AJ. The importance of pathogen load. PLoS Pathog. 2015;11:e1004563. doi: 10.1371/journal.ppat.1004563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mackinnon MJ, Li J, Mok S, Kortok MM, Marsh K, Preiser PR, Bozdech Z. Comparative transcriptional and genomic analysis of Plasmodium falciparum field isolates. PLoS Pathog. 2009;5:e1000644. doi: 10.1371/journal.ppat.1000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Norata GD, Caligiuri G, Chavakis T, Matarese G, Netea MG, Nicoletti A, O'Neill LA, Marelli-Berg FM. The cellular and molecular basis of translational immunometabolism. Immunity. 2015;43:421–434. doi: 10.1016/j.immuni.2015.08.023. [DOI] [PubMed] [Google Scholar]
- 58.O'Neill LA, Pearce EJ. Immunometabolism governs dendritic cell and macrophage function. J Exp Med. 2016;213:15–23. doi: 10.1084/jem.20151570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hunt NH, Ball HJ, Hansen AM, Khaw LT, Guo J, Bakmiwewa S, Mitchell AJ, Combes V, Grau GE. Cerebral malaria: gamma-interferon redux. Front Cell Infect Microbiol. 2014;4:113. doi: 10.3389/fcimb.2014.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fuchs TA, Bhandari AA, Wagner DD. Histones induce rapid and profound thrombocytopenia in mice. Blood. 2011;118:3708–3714. doi: 10.1182/blood-2011-01-332676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alhamdi Y, Abrams ST, Lane S, Wang G, Toh CH. Histone-associated thrombocytopenia in patients who are critically ill. JAMA. 2016;315:817–819. doi: 10.1001/jama.2016.0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Amulic B, Cazalet C, Hayes GL, Metzler KD, Zychlinsky A. Neutrophil function: from mechanisms to disease. Annu Rev Immunol. 2012;30:459–489. doi: 10.1146/annurev-immunol-020711-074942. [DOI] [PubMed] [Google Scholar]
- 63.Berens-Riha N, Kroidl I, Schunk M, Alberer M, Beissner M, Pritsch M, Kroidl A, Froschl G, Hanus I, Bretzel G, von Sonnenburg F, et al. Evidence for significant influence of host immunity on changes in differential blood count during malaria. Malar J. 2014;13:155. doi: 10.1186/1475-2875-13-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baker VS, Imade GE, Molta NB, Tawde P, Pam SD, Obadofin MO, Sagay SA, Egah DZ, Iya D, Afolabi BB, Baker M, et al. Cytokine-associated neutrophil extracellular traps and antinuclear antibodies in Plasmodium falciparum infected children under six years of age. Malar J. 2008;7:41. doi: 10.1186/1475-2875-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dietmann A, Helbok R, Lackner P, Issifou S, Lell B, Matsiegui PB, Reindl M, Schmutzhard E, Kremsner PG. Matrix metalloproteinases and their tissue inhibitors (TIMPs) in Plasmodium falciparum malaria: serum levels of TIMP-1 are associated with disease severity. J Infect Dis. 2008;197:1614–1620. doi: 10.1086/587943. [DOI] [PubMed] [Google Scholar]
- 66.Jorch SK, Kubes P. An emerging role for neutrophil extracellular traps in noninfectious disease. Nat Med. 2017;23:279–287. doi: 10.1038/nm.4294. [DOI] [PubMed] [Google Scholar]
- 67.Oakley MS, Anantharaman V, Venancio TM, Zheng H, Mahajan B, Majam V, McCutchan TF, Myers TG, Aravind L, Kumar S. Molecular correlates of experimental cerebral malaria detectable in whole blood. Infect Immun. 2011;79:1244–1253. doi: 10.1128/IAI.00964-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Porcherie A, Mathieu C, Peronet R, Schneider E, Claver J, Commere PH, Kiefer-Biasizzo H, Karasuyama H, Milon G, Dy M, Kinet JP, et al. Critical role of the neutrophil-associated high-affinity receptor for IgE in the pathogenesis of experimental cerebral malaria. J Exp Med. 2011;208:2225–2236. doi: 10.1084/jem.20110845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krupka M, Seydel K, Feintuch CM, Yee K, Kim R, Lin CY, Calder RB, Petersen C, Taylor T, Daily J. Mild Plasmodium falciparum malaria following an episode of severe malaria is associated with induction of the interferon pathway in Malawian children. Infect Immun. 2012;80:1150–1155. doi: 10.1128/IAI.06008-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haque A, Best SE, Ammerdorffer A, Desbarrieres L, de Oca MM, Amante FH, de Labastida Rivera F, Hertzog P, Boyle GM, Hill GR, Engwerda CR. Type I interferons suppress CD4(+) T-cell-dependent parasite control during blood-stage Plasmodium infection. Eur J Immunol. 2011;41:2688–2698. doi: 10.1002/eji.201141539. [DOI] [PubMed] [Google Scholar]
- 71.Haque A, Best SE, Montes de Oca M, James KR, Ammerdorffer A, Edwards CL, de Labastida Rivera F, Amante FH, Bunn PT, Sheel M, Sebina I, et al. Type I IFN signaling in CD8-DCs impairs Th1-dependent malaria immunity. J Clin Invest. 2014;124:2483–2496. doi: 10.1172/JCI70698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sharma S, DeOliveira RB, Kalantari P, Parroche P, Goutagny N, Jiang Z, Chan J, Bartholomeu DC, Lauw F, Hall JP, Barber GN, et al. Innate immune recognition of an AT-rich stem-loop DNA motif in the Plasmodium falciparum genome. Immunity. 2011;35:194–207. doi: 10.1016/j.immuni.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zander RA, Guthmiller JJ, Graham AC, Pope RL, Burke BE, Carr DJ, Butler NS. Type I interferons induce T regulatory 1 responses and restrict humoral immunity during experimental malaria. PLoS Pathog. 2016;12:e1005945. doi: 10.1371/journal.ppat.1005945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Josling GA, Llinas M. Sexual development in Plasmodium parasites: knowing when it's time to commit. Nat Rev Microbiol. 2015;13:573–587. doi: 10.1038/nrmicro3519. [DOI] [PubMed] [Google Scholar]
- 75.Arguello RJ, Rodriguez Rodrigues C, Gatti E, Pierre P. Protein synthesis regulation, a pillar of strength for innate immunity? Curr Opin Immunol. 2015;32:28–35. doi: 10.1016/j.coi.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 76.Vembar SS, Droll D, Scherf A. Translational regulation in blood stages of the malaria parasite Plasmodium spp.: systems-wide studies pave the way. Wiley Interdiscip Rev RNA. 2016;7:772–792. doi: 10.1002/wrna.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wong W, Bai XC, Sleebs BE, Triglia T, Brown A, Thompson JK, Jackson KE, Hanssen E, Marapana DS, Fernandez IS, Ralph SA, et al. Mefloquine targets the Plasmodium falciparum 80S ribosome to inhibit protein synthesis. Nat Microbiol. 2017;2 doi: 10.1038/nmicrobiol.2017.31. 17031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lemieux JE, Gomez-Escobar N, Feller A, Carret C, Amambua-Ngwa A, Pinches R, Day F, Kyes SA, Conway DJ, Holmes CC, Newbold CI. Statistical estimation of cell-cycle progression and lineage commitment in Plasmodium falciparum reveals a homogeneous pattern of transcription in ex vivo culture. Proc Natl Acad Sci U S A. 2009;106:7559–7564. doi: 10.1073/pnas.0811829106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pelle KG, Oh K, Buchholz K, Narasimhan V, Joice R, Milner DA, Brancucci NM, Ma S, Voss TS, Ketman K, Seydel KB, et al. Transcriptional profiling defines dynamics of parasite tissue sequestration during malaria infection. Genome Med. 2015;7:19. doi: 10.1186/s13073-015-0133-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Walther M, Jeffries D, Finney OC, Njie M, Ebonyi A, Deininger S, Lawrence E, Ngwa-Amambua A, Jayasooriya S, Cheeseman IH, Gomez-Escobar N, et al. Distinct roles for FOXP3 and FOXP3 CD4 T cells in regulating cellular immunity to uncomplicated and severe Plasmodium falciparum malaria. PLoS Pathog. 2009;5:e1000364. doi: 10.1371/journal.ppat.1000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Walther M, De Caul A, Aka P, Njie M, Amambua-Ngwa A, Walther B, Predazzi IM, Cunnington A, Deininger S, Takem EN, Ebonyi A, et al. HMOX1 gene promoter alleles and high HO-1 levels are associated with severe malaria in Gambian children. PLoS Pathog. 2012;8:e1002579. doi: 10.1371/journal.ppat.1002579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Casper J, Zweig AS, Villarreal C, Tyner C, Speir ML, Rosenbloom KR, Raney BJ, Lee CM, Lee BT, Karolchik D, Hinrichs AS, et al. The UCSC Genome Browser database: 2018 update. Nucleic Acids Res. 2018;46:D762–D769. doi: 10.1093/nar/gkx1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Aurrecoechea C, Brestelli J, Brunk BP, Dommer J, Fischer S, Gajria B, Gao X, Gingle A, Grant G, Harb OS, Heiges M, et al. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37:D539–D543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.DeLuca DS, Levin JZ, Sivachenko A, Fennell T, Nazaire MD, Williams C, Reich M, Winckler W, Getz G. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28:1530–1532. doi: 10.1093/bioinformatics/bts196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tarr SJ, Díaz-Ingelmo O, Stewart LB, Hocking SE, Murray L, Duffy CW, Otto TD, Chappell L, Rayner JC, Awandare GA, Conway DJ. Plasmodium falciparum mature schizont transcriptome variation among clinical isolates and laboratory-adapted clones. bioRxiv. doi: 10.1101/329532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Abbas AR, Baldwin D, Ma Y, Ouyang W, Gurney A, Martin F, Fong S, van Lookeren Campagne M, Godowski P, Williams PM, Chan AC, et al. Immune response in silico (IRIS): immune-specific genes identified from a compendium of microarray expression data. Genes Immun. 2005;6:319–331. doi: 10.1038/sj.gene.6364173. [DOI] [PubMed] [Google Scholar]
- 91.Benjamini Y, Hochberg Y. Controlling false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 92.Supek F, Bosnjak M, Skunca N, Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hu Y, Yan C, Hsu CH, Chen QR, Niu K, Komatsoulis GA, Meerzaman D. OmicCircos: a simple-to-use R package for the circular visualization of multidimensional omics data. Cancer Inform. 2014;13:13–20. doi: 10.4137/CIN.S13495. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.