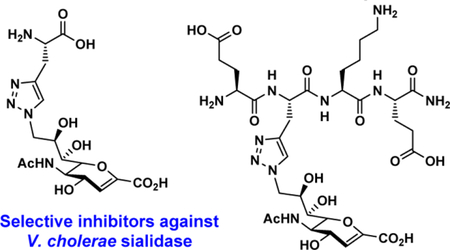
Abstract
Sialidases or neuraminidases are enzymes that catalyze the cleavage of terminal sialic acids from oligosaccharides and glycoconjugates. They play important roles in bacterial and viral infection and have been attractive targets for drug development. Structure-based drug design has led to potent inhibitors against neuraminidases of influenza A viruses that have been used successfully as approved therapeutics. However, selective and effective inhibitors against bacterial and human sialidases are still being actively pursued. Guided by crystal structural analysis, several derivatives of 2-deoxy-2,3-didehydro-N-acetylneuraminic acid (Neu5Ac2en or DANA) were designed and synthesized as triazole-linked transition state analogs. Inhibition studies revealed that glycopeptide analog E-(TriazoleNeu5Ac2en)-AKE and compound (TriazoleNeu5Ac2en)-A were selective inhibitors against Vibrio cholerae sialidase, while glycopeptide analog (TriazoleNeu5Ac2en)-AdE selectively inhibited Vibrio cholerae and A. ureafaciens sialidases.
Keywords: Carbohydrate, Glycopeptide, Neuraminidase, Sialidase, Sialidase inhibitor
Graphical Abstract
1. Introduction
Sialic acids are negatively charged monosaccharides with a nine-carbon backbone and are often found as the terminal residues of cell-surface glycoconjugates involved in many important cellular events.1 Sialidases or neuraminidases (EC 3.2.1.18) are exoglycosidases that catalyze the cleavage of terminal sialic acids from oligosaccharides and glycoconjugates. They have been found in many organisms including bacteria, viruses, fungi, and mammals.2 All sialidases share a common catalytic domain of a canonical six-bladed β -propeller fold despite variation on the sources, lengths, and protein sequences.2–4 Based on protein sequence similarities, most viral neuraminidases are grouped in glycoside hydrolase family 34 (GH34) and GH83 in the Carbohydrate Active Enzyme (CAZy) database (www.CAZy.org).3–6 For bacterial and eukaryotic exosialidases and trans-sialidases, most are in CAZy GH33 family except for SiaHI from a Gram-negative oral anaerobe Tannerella forsythia (GH NC), 7, 8 a GH1 family enzyme with both β -glucuronidase and sialidase activities, as well as multifunctional bacterial sialyltransferases with sialidase and trans-sialidase functions in CAZy glycosyltransferase GT42 and GT80 families.9–11 Some sialidases have one or more carbohydrate-binding modules (e.g. CBM32, CBM40) and other domains in addition to their catalytic domain.12–15
Sialidases are known virulence factors for some microorganisms.16–18 For example, sialidases secreted by epidemic strains of Vibrio cholerae play an important role in infection and are potential drug targets against cholera.19 Streptococcus pneumoniae is a Gram-positive bacterium and a human opportunistic pathogen capable of causing respiratory- tract infections, pneumonia, otitis media, bacteremia, sepsis, and meningitis.20–22 Three sialidases (SpNanA, SpNanB, and SpNanC) have been identified and characterized from Streptococcus pneumoniae strains. 23–25 Among these, both SpNanA and SpNanB have been found to be essential for bacterial infection26 and are considered as valid drug targets.27
Rational design of sialidase inhibitors based on protein crystal structures has been implemented successfully for identifying effective influenza A virus neuraminidase inhibitors as anti- influenza drugs including Zanamivir (Relenza, GlaxoSmithKline),16 Oseltamivir (Tamiflu, Gilead/Roche), and more recently Peramivir (Rapivab, BioCryst).28–31 Development of selective inhibitors against bacterial sialidases lags behind despite that a number of crystal structures are available and the mechanisms of actions are known.32, 33
A sialidase transition state analog, 2-deoxy-2,3-didehydro-N- acetylneuraminic acid (Neu5Ac2en (1) or DANA, 1, Figure 1),34 is a nonspecific inhibitor against numerous sialidases from influenza viruses, bacteria, and human. In this study, by comparing the crystal structures of SpNanA catalytic domain and its complex with Neu5Ac2en and 9-azido-9-deoxy-Neu5Ac2en (Neu5Ac9N32en, 2 ), several triazole-linked Neu5Ac2en-derived structures were designed and synthesized as potential inhibitors against bacterial sialidases. Selective inhibitors against Vibrio cholerae sialidase have been identified which can serve as probes to investigate its roles in bacterial infection.
Figure 1.
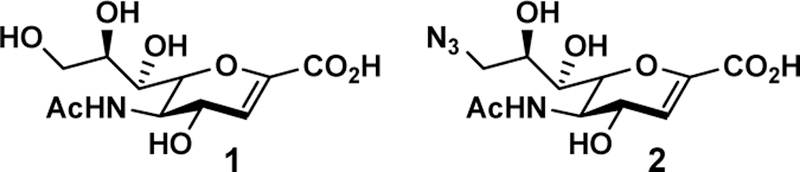
Structures of Neu5Ac2en (1) and Neu5Ac9N32en (2).
2. Results and discussion
2.1. Structure of SpNanA and design of sialidase inhibitors
We have shown previously that modifications of Neu5Ac2en at C9 and C5 can improve the selectivity of its inhibition against some sialidases.35 Installation of an azido group at C9 of Neu5Ac2en also provides a chemical handle for further modification to improve the selectivity for sialidase inhibitors.36 We obtained a novel crystal structure of the catalytic domain of SpNanA in complex with 9-azido-9-deoxy-Neu5Ac2en (Neu5Ac9N32en, 2 , Figure 1) (PDB accession code 5KKY) and compared this structure to its complex with Neu5Ac2en.34 The Neu5Ac9N32en binds to SpNanA in a similar way as Neu5Ac2en in other known structures,18, 37 but surprisingly the SpNanA is a homodimer with Neu5Ac9N32en in two different conformations in the active sites of two different subunits (Figure 2).
Figure 2.
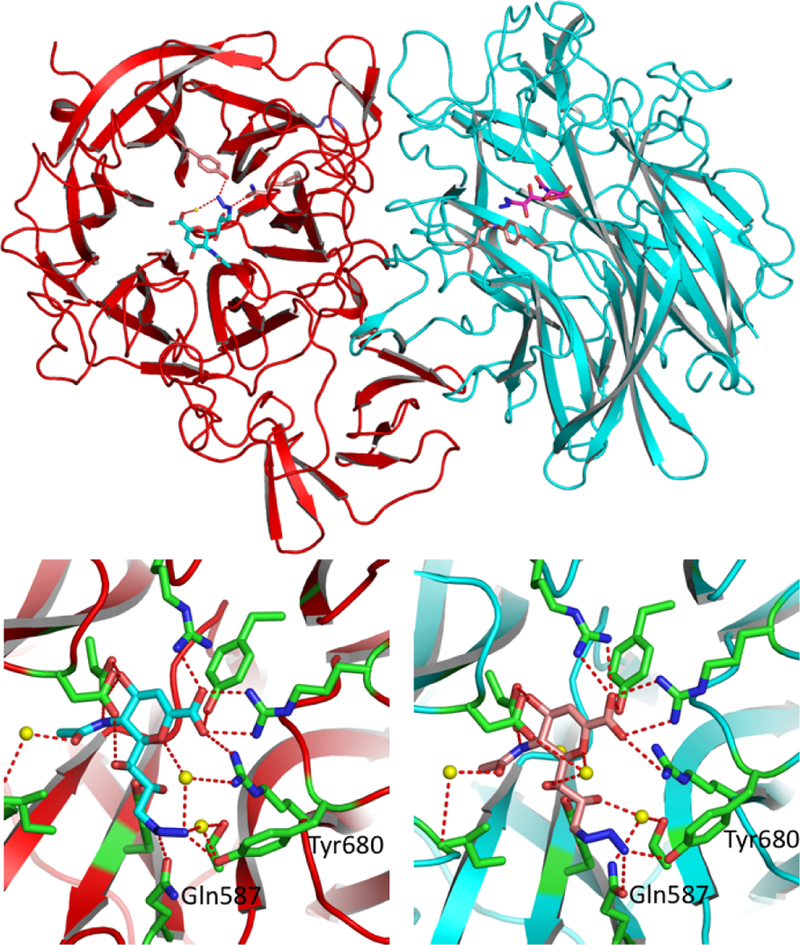
The novel complex structure between SpNanA and Neu5Ac9N32en (2). There are two subunits. A (red) and B (blue) in an asymmetric unit shown in the up panel. The bound Neu5Ac9N32en molecules in subunits A and B are shown in light blue and pink sticks, respectively. The close up views of Neu5Ac9N32en binding sites in subunits A (left) and B (right) are shown in the lower panel. The side chains of amino acid residues involving in the hydrogen bond interactions with Neu5Ac9N32en are shown in green sticks. Potential hydrogen bond interactions between the Neu5Ac9N32en and the protein are shown in red dashed lines.
The 9-azide group on Neu5Ac9N32en forms hydrogen bonds to the side chain of Tyr680 and Gln587 in the active sites of both subunits in the asymmetric unit. Interestingly, in subunit B, the 9- azide group on Neu5AcN32en forms an extra hydrogen bond to the main-chain oxygen of Gly692 from another adjacent subunit due to crystal packing (not shown in Figure 2). Furthermore, the peptide segment around Gly692 (Glu691-Gly692-Lys693- Glu694) is nicely patched onto the inhibitor. As a result, the Neu5Ac9N32en binding site is covered by a peptide segment Glu691-Gly692-Lys693-Glu694 (Figure 3). The proximity of “Glu691-Gly692-Lys693-Glu694” of the neighboring subunit with Neu5Ac9N32en” indicates the potential favorable interaction of peptides with the protein surface of the active site. Linkage of peptide with Neu5Ac9N32en.
Figure 3.
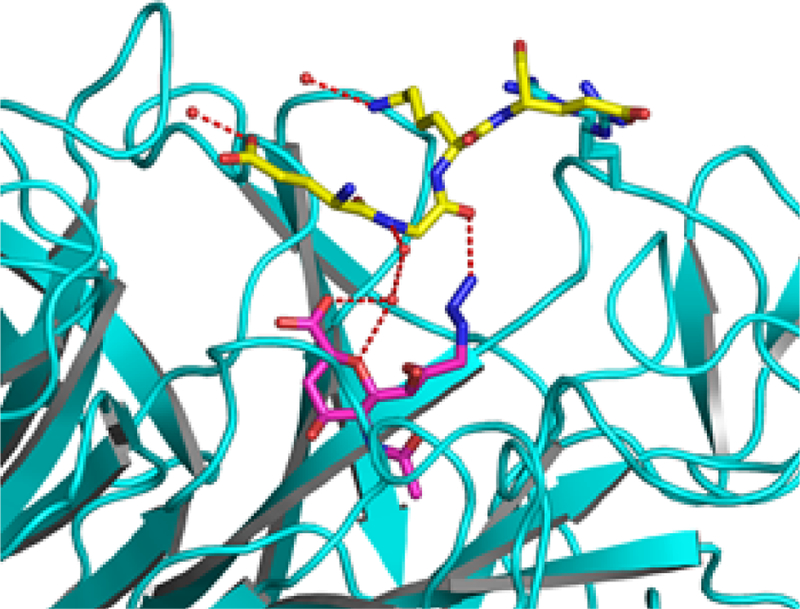
Crystal packing contact of Neu5Ac9N32en (2) in the active site of the B subunit of SpNanA with the adjacent subunit. The inhibitor (pink sticks) is hydrogen-bond (shown in red dashed lines) interacting with the peptide segment (yellow sticks) Glu691-Gly692-Lys693-Glu694 from the adjacent symmetric-related subunit A (different from the subunit A in the same asymmetric unit shown in Figure 2).
Guided by the crystal structure of SpNanA complexed with Neu5Ac9N32en as well as the virtual docking of SpNanA with different peptide-modified Neu5Ac9N32en, we hypothesized that a peptide-modified Neu5Ac9N32en would result in a selective inhibitor against SpNanA. A convenient design would be linking the peptide and Neu5Ac9N32en via a Cu(I)-catalyzed azide and alkyne cycloaddition (CuAAC) reaction.38
2.2. Synthesis of glycopeptide sialidase inhibitors
To test the hypothesis, three triazole-linked C9-modified Neu5Ac2en analogs were designed. These were synthesized by clicking Neu5Ac9N32en (2) to derivatives of an amino acid L-alanine (3), a dipeptide of an L-alanine and a D-glutamate (4), or a tetrapeptide of L-glutamate-L-alanine-L-lysine-L-glutamate (5) with a propargyl group replacing the side chain of the L-alaninee residue. As shown in Scheme 1, compound 3 was synthesized by removal of the fluorenylmethyloxycarbonyl (Fmoc) group from commercially available Fmoc-protected propargyl glycine. Propargyl-modified peptides 4–5 were produced using a standard solid phase peptide synthesis (SPPS) process with a Fmoc- protected strategy (Scheme 1). Glycoconjugate inhibitors including monoamino acid-containing glycoconjugate (TriazoleNeu5Ac2en)-A (6), dipeptide-containing glycoconjugate (TriazoleNeu5Ac2en)-AdE (7), and tetrapeptide- containing glycoconjugate E-(TriazoleNeu5Ac2en)-AKE (8) were then readily obtained by CuAAC with in situ generation of Cu(I) from CuSO4·5H2O and sodium ascorbate (Scheme 2).39
Scheme 1.
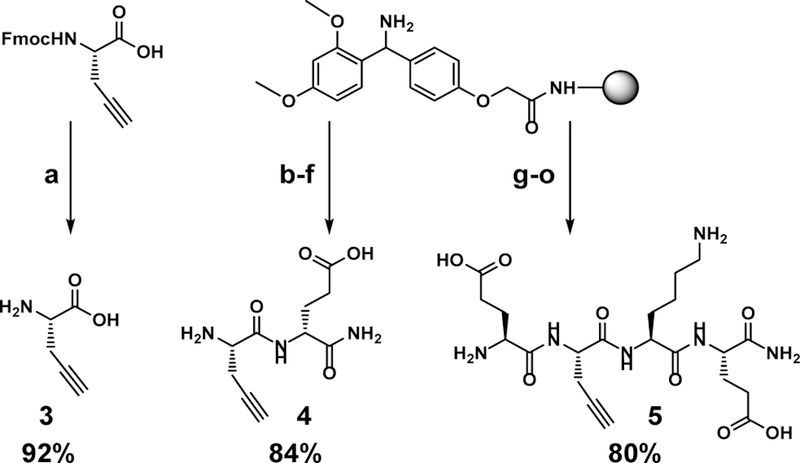
Synthesis of propargyl-containing amino acid (3) or peptides 4–5. (a) piperidine, DMF; (b) Fmoc-D-Glu, COMU, DIEA, DMF; (c) piperidine, DMF; (d) Fmoc-L-propargyl-Gly, COMU, DIEA, DMF; (e) piperidine, DMF; (f) TFA: H2O: TIPS = 95:2.5:2.5; (g) Fmoc-L-Glu, COMU, DIEA, DMF; (h) piperidine, DMF; (i) Fmoc-L-Lys, COMU, DIEA, DMF; (j) piperidine, DMF; (k) Fmoc-L-propargyl-Gly, COMU, DIEA, DMF; (l) piperidine, DMF; (m) Fmoc-L-Glu, COMU, DIEA, DMF; (n) piperidine, DMF; (o) TFA: H2O: TIPS = 95: 2.5: 2.5.
Scheme 2.
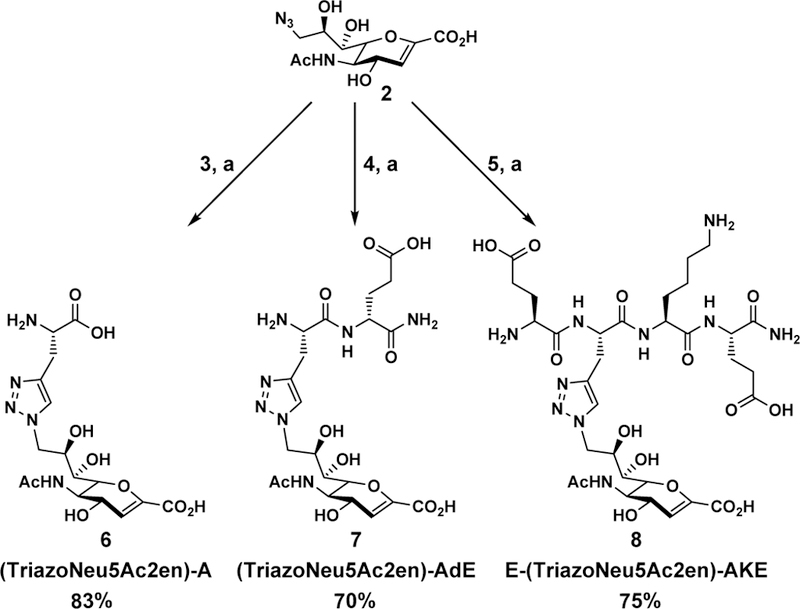
Synthesis of glycoconjugates 6–8 from Neu5Ac9N32en (2). (a) CuSO4·5H2O, sodium ascorbate, H2O.
2.3. Inhibition studies
Inhibition studies were performed using a microtiter-plate- based colorimetric assay.41 Neu5Acα2–3GalβpNP was used as the substrate in the presence of a β-galactosidase which was responsible to cleave the GalβpNP released by the sialidase of interest to form p NP whose reading at A405 nm at pH higher than 9.5 was corresponding to the activity of the sialidase.
Several enzymes were tested. These included a recombinant human cytosolic sialidase NEU2 (hNEU2),41 and a panel of bacterial sialidases such as commercially available sialidases from V. cholerae , C. perfringens , and A. ureafaciens , as well as recombinant sialidases SpNanA, SpNanB, and SpNanC,23, 40 Bifidobacterium longum subsp. infantis ATCC15697 sialidase 2 (BiNanH2),42 and Pasteurella multocida multifunctional sialyltransferase 1 with α2–3-sialidase activity (PmST1).9
Initially, each compound was used in a concentration of 1 mM (Table 1) and 0.1 mM (Table 2) to obtain percentage inhibition values to identify potential inhibitor candidates for further analysis. As shown in Table 2, the presence of 0.1 mM of Neu5Ac2en (1) led to more than 50% inhibition for all sialidases tested except for SpNanB, SpNanC, and PmST1. This was consistent with previous results,35, 43 indicating that Neu5Ac2en (1) is a general inhibitor against most hydrolytic sialidases. Its 9- azido-9-deoxy-modification in Neu5Ac9N32en (2) was well tolerated by sialidases that are susceptible for Neu5Ac2en inhibition except for hNEU2 and BiNanH2. In comparison, conjugating of Neu5Ac9N32en (2) with a propargyl amino acid, dipeptide, or tetrapeptide enhanced the selectivity of the inhibitor significantly. Compounds (TriazoleNeu5Ac2en)-A (6) and E- (TriazoleNeu5Ac2en)-AKE (8) retained the inhibitory activity against V. cholerae sialidase but not other sialidases tested. Quite interestingly, (TriazoleNeu5Ac2en)-AdE (7) with a D-glutamate residue in the dipeptide was better tolerated by some Neu5Ac9N32en (2)-sensitive sialidases including SpNanA and A. ureafaciens sialidase in addition to V. cholerae sialidase.
Table 1.
Percentage inhibition with 1.0 mM of inhibitor using Neu5Acα2–3GalβpNP as the sialidase substrate.
| Sialidases | Neu5Ac2en (1) | Neu5Ac9N32en (2) | (TriazoleNeu5Ac2en)-A (6) | (TriazoleNeu5Ac2en)-AdE (7) | E-(TriazoleNeu5Ac2en)-AKE (8) |
|---|---|---|---|---|---|
| SpNanA | 99.9 ± 1.8 | 98.8 ± 0.9 | 84.4 ± 0.2 | 94.7 ± 1.2 | 51.5 ± 2.1 |
| SpNanB | 31.9 ± 1.5 | 35.2 ± 1.4 | 35.5 ± 0.2 | 39.0 ± 0.6 | 25.3 ± 0.8 |
| SpNanC | 23.7 ± 3.5 | 39.2 ± 2.4 | 47.8 ± 2.2 | 29.7 ± 1.6 | 9.8 ± 0.7 |
| V. cholerae | 94.5 ± 1.9 | 95.6 ± 0.7 | 93.2 ± 0.2 | 93.5 ± 0.2 | 91.6 ± 2.0 |
| A. ureafaciens | 99.0 ± 0.0 | 99.5 ± 0.3 | 85.6 ± 3.1 | 96.2 ± 0.0 | 79.8 ± 0.0 |
| C. perfringens | 92.6 ± 0.5 | 89.3 ± 1.9 | 69.7 ± 0.7 | 84.6 ± 0.0 | 56.5 ± 1.9 |
| BiNanH2 | 91.1± 1.0 | 86.7 ± 1.0 | 14.7 ± 2.1 | 76.4 ± 1.2 | 13.8 ± 3.2 |
| PmST1 | 9.3 ± 2.8 | 14.9 ± 2.5 | 9.1 ± 3.3 | 6.0 ± 1.5 | 1.1 ± 1.3 |
| hNEU2 | 95.5 ± 0.4 | 74.2 ± 0.6 | 39.1 ± 1.1 | 57.6 ± 3.2 | 8.6 ± 1.8 |
Table 2.
Percentage inhibition with 0.1 mM of inhibitor using Neu5Acα2–3GalβpNP as the sialidase substrate.
| Sialidases | Neu5Ac2en (1) | Neu5Ac9N32en (2) | (TriazoleNeu5Ac2en)-A (6) | (TriazoleNeu5Ac2en)-AdE (7) | E-(TriazoleNeu5Ac2en)-AKE (8) |
|---|---|---|---|---|---|
| SpNanA | 94.9 ± 2.2 | 78.6 ± 0.8 | 42.9 ± 2.4 | 61.9 ± 1.5 | 4.5 ± 2.1 |
| SpNanB | 2.7 ± 0.3 | 6.6 ± 2.1 | 8.8 ± 1.2 | 12.6 ± 1.8 | 9.7 ± 0.8 |
| SpNanC | 6.3 ± 1.6 | 13.4 ± 2.2 | 15.3 ± 2.1 | 13.5 ± 1.8 | 14.0 ± 3.0 |
| V. cholerae | 88.2 ± 0.6 | 86.0 ± 0.4 | 82.0 ± 3.0 | 85.8 ± 1.1 | 72.5 ± 0.9 |
| A. ureafaciens | 95.8 ± 0.6 | 99.2 ± 0.4 | 42.1 ± 3.5 | 95.7 ± 1.6 | 29.9 ± 1.3 |
| C. perfringens | 72.1 ± 0.0 | 57.1 ± 0.1 | 17.6 ± 1.3 | 48.9 ± 0.6 | 10.8 ± 2.1 |
| BiNanH2 | 66.7 ± 1.7 | 49.4 ± 1.6 | 0.0 ± 0.3 | 3.3 ± 1.0 | 0.1 ± 5.0 |
| PmST1 | 2.1 ± 0.8 | 1.7 ± 4.5 | 2.8 ± 0.2 | 0.1 ± 5.0 | 0.6 ± 2.5 |
| hNEU2 | 74.9 ± 0.0 | 27.2 ± 1.2 | 12.2 ± 2.5 | 17.7 ± 0.8 | 0.2 ± 0.7 |
As shown in Table 3, IC50 values obtained for potential inhibitors confirmed that monoamino acid-conjugate (TriazoleNeu5Ac2en)-A (6) and tetrapeptide-conjugate E- (TriazoleNeu5Ac2en)-AKE (8) were selective inhibitors against V. cholerae sialidase with an IC50 value of 13.5 ± 0.5 μM and 28.9 ± 3.2 μM, respectively, which were close to that of Neu5Ac9N32en (2) (20 ± 1 μM). Dipeptide-conjugate (TriazoleNeu5Ac2en)-AdE (7) was also a selective inhibitor against V. cholerae and A. ureafaciens sialidases.
Table 3.
IC50 values (μM) of potential inhibitors against bacterial and human sialidases using Νeu5Αcα2–3GalβpNP as a substrate.
| Sialidases | Neu5Ac2en (1) | Neu5Ac9N32en (2) | (TriazoleNeu5Ac2en)-A (6) | (TriazoleNeu5Ac2en)-AdE (7) | E-(TriazoleNeu5Ac2en)-AKE (8) |
|---|---|---|---|---|---|
| SpNanA | 10.0 ± 0.2b | 44.3 ± 1.8 | 100 – 1000 | 65.6 ± 4.8 | 100 – 1000 |
| SpNanB | > 1000 | > 1000 | > 1000 | > 1000 | > 1000 |
| SpNanC | > 1000 | > 1000 | > 1000 | > 1000 | > 1000 |
| V. cholerae | 8.6 ± 1.0a | 20 ± 1a | 13.5 ± 0.5 | 10.9 ± 0.8 | 28.9 ± 3.2 |
| A. ureafaciens | 8.1 ± 0.3b | 0.72 ± 0.02 | 100 – 1000 | 1.87 ± 0.07 | 100 – 1000 |
| C. perfringens | 20 ± 1a | 59 ± 5a | 100 – 1000 | 100 – 1000 | 100 – 1000 |
| BiNanH2 | 40 ± 2.0b | 100 – 1000 | > 1000 | 100 – 1000 | > 1000 |
| PmST1 | > 1000 | > 1000 | > 1000 | > 1000 | > 1000 |
| hNEU2 | 18 ± 1a | 100 – 1000 | > 1000 | 100 – 1000 | > 1000 |
2.4. Docking studies
Molecular docking studies were performed to better understand the inhibitory effects of compounds 6 and 7 on V. cholerae sialidase (Figure 4). Flexible docking of compound 8 was unsuccessful due to its size and flexibility. The binding of the core Neu5Ac2en structure in 2 , 6 , and 7 was unchanged relative to co-crystallized 1 (PDB accession number 1w0o, Figure 4A). Edge-to-face pi-pi interactions between the triazole and F638 (Figure 4C–4D) were predicted for 6 and 7 , while the azide of 2 was predicted to fit underneath F638 (Figure 4B). No hydrogen bonding contacts were predicted between the peptide and the enzyme, although intramolecular hydrogen bonds between the carboxyl groups of compound 7 (Figure 4D) were predicted. These results indicate that the additional binding interaction between the triazole moiety and the protein positions the peptide chain in compounds 6 and 7 away from the active site.
Figure 4.
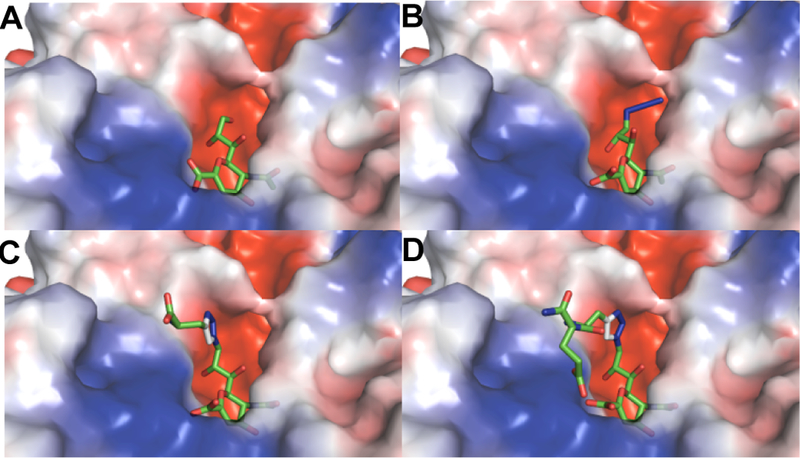
Docking results presenting semi-transparent electrostatic potential surface (red is negative and blue is positive) of V. cholerae sialidase structure (PDB accession number 1w0o) with A , co-crystallized Neu5Ac2en (1); as well as docked B, 2; C, 6; and D, 7.
The alignment of the sequences of the sialidases used in this study (Figure S1) shows that only C. perfringens sialidase contains a residue homologous to V. cholerae F638. This residue is Y248, and although it is aromatic and potentially capable of pi stacking with the triazole moiety of the inhibitors, the two sialidases share only 26% sequence identity. It is therefore not surprising that the inhibitors do not interact with the two enzymes in the same manner.
3. Conclusions
Based on analysis of crystal structures of SpNanA in the presence of Neu5Ac2en and Neu5Ac9N32en, respectively, several triazole-linked C9-modified Neu5Ac2en analogs were synthesized and their inhibitory activities were tested. Although the peptide-conjugates obtained were not selective inhibitors against SpNanA, monoamino acid-conjugate (TriazoleNeu5Ac2en)-A (6) and tetrapeptide-conjugate E-(TriazoleNeu5Ac2en)-AKE (8) were shown to be selective inhibitors against V. cholerae sialidase, and dipeptide-conjugate (TriazoleNeu5Ac2en)-AdE (7) was a selective inhibitor against V. cholerae and A. ureafaciens sialidases. This indicates that conjugating a peptide to a proper position of sialidase transition state analog via a triazole linker is a suitable strategy to obtain sialidase inhibitors with improved selectivity.
4. Experimental section
4.1. Materials
All chemicals were obtained from commercial suppliers and used without further purification. 1H NMR (400 or 800 MHz) and 13C NMR (400 or 800 MHz) spectra were recorded on a Bruker Avance-400 Spectrometer (400 MHz for 1H, 100 MHz for 13C) or a Avance-800 Spectrometer (800 MHz for 1H, 200 MHz for 13C). High resolution electrospray ionization (ESI) mass spectra were obtained using a Thermo Electron LTQ-Orbitrap Hybrid MS at the Mass Spectrometry Facility in the University of California, Davis. Column chromatography was performed using RediSep Rf silica columns or an ODS-SM column (51 g, 50 μ m, 120 Å, Yamazen) on the CombiFlash® Rf 200i system. HPLC purification was carried out using Shimadzu SCL-10AVP with reverse-phase C18 column. Analytical thin-layer chromatography was performed on silica gel plates 60 GF254 (Sorbent technologies) using anisaldehyde stain for detection. Fmoc protected propargyl glycine and D-glutamic acid (Chem-Impex), Rink amide resin ChemMatrix® (pcas BioMatrix Inc.), Fmoc- protected L-glutamic acid and L-lysine ( AAPPTec), DIEA (Acros), COMU (NovaBiochem). Neu5Ac2en and Neu5Ac9N32en were synthesized as reported previously.35 Sialidases from V. cholerae , and A. ureafaciens were purchased from Prozyme. Sialidases from C. perfringens (CpNanI) was purchased from Sigma Aldrich. SpNanA and SpNanB,23 hNEU2,41 Bifidobacterium longum subsp. infantis ATCC15697 sialidase 2 (BiNanH2),42 and PmST19 were expressed and purified as described previously.
4.2. Protein crystallization
The crystals of SpNanA-Neu5Ac9N32en complex were grown by the sitting-drop, vapor-diffusion method. Before crystallization, the purified protein (~5 mg/mL) was incubated with 5 mM of Neu5Ac9N32en for 30 min. Screening of crystallization conditions was performed using sitting-drop vapor diffusion in 96-well plates (Hampton Research) at 291 K by mixing 2 μ L of the protein solution with 2 μ L of the reagent solution from the sparse matrix Crystal Screens 1 and 2, and Index Screen (Hampton Research). The best crystals were grown from a reservoir solution containing Tris-HCl (pH 8.5, 100 mM), lithium sulfate (0.2 M), 25% PEG3350. Crystals were plate- shaped and took 2–3 days to reach a maximal length of 0.05 mm.
4.3. X-ray diffraction data collection and structure determination
Crystals were transferred from the crystallization plate to a well solution supplemented with 25% glycerol and then frozen directly by liquid nitrogen. Data were collected at NIH at 100 K using Rigaku Raxis IV X-ray generator and CCD detector. Data processing and scaling were performed with HKL2000.44 The structure was solved by molecular replacement using Phaser45 and SpNanA structure (PDB: 2VVZ)34 as a search model. The model was built with Coot46 and refined with Phenix.47 Final R and Rfree values were 15.4% and 20.7%, respectively. The final refined coordinates for SpNanA bound with Neu5Ac9N32en and its structure factor have been deposited in RCSB Protein Data Bank with accession code 5KKY.
4.4. Synthesis of propargyl glycine (3)
Fmoc-protected propargyl glycine (200 mg) was dissolved in 20 mL of piperidine/DMF solution (1:4). Reaction was stirred for 30 minutes at room temperature. The volume was reduced by rotary evaporation and 25 mL of cold ether was added to precipitate the amino acid. The suspended mixture was centrifuged at 4000 rpm at 4 °C for 10 minutes and washed twice with cold ether (10 mL × 2). The amino acid was loaded onto a C18 cartridge for flash purification (ACN/H2O). 1H NMR (D2O, 400 MHz): δ 3.89 (t, 1H, J = 5.4 Hz), 2.87–2.81 (m, 2H,), 2.52 (t, 1H, J =2.6Hz).13CNMR (D2O, 101 MHz):δ 172.7, 77.5, 73.4, 52.8, 20.4. HRMS (ESI) calculated (M-H) 112.0404, found 112.0415.
4.5. General procedures for synthesizing compounds 4 and 5
Peptides 4 and 5 were synthesized using standard SPPS on Rink Amide high yield resin (loading = 0.45 mmol/g). The beads were swollen in DMF for 1 hr prior to coupling. Each peptide bond formation was done with 3 eq. of the Fmoc protected amino acid, 3 eq. of COMU coupling reagent, and 6 eq. of DIEA in DMF. Reaction was mixed constantly at room temperature for 30–60 minutes and monitored by Kaiser test. Beads were washed with DMF (3×) MeOH (3×) DMF (3×) after coupling. The N- terminus Fmoc deprotection was done in 1:4 piperidine:DMF mixed constantly for 30 minutes at room temperature. Beads were washed with DMF (6×) after deprotection. The peptide was cleaved from the bead using the cleavage cocktail TFA:H2O:TIPS = 95:2.5:2.5 by volume. The TFA was evaporated and cold diethyl ether was added to precipitate peptide. The solution was centrifuged at 4000 rpm at 4 °C for 10 minutes and decanted (3× in cold ether). The peptide was dissolved in water for reverse-phase HPLC purification (ACN/H2O).
(propargyl)-AdE (4)
1H NMR (D2O 600 MHz): δ 4.41 (dd, 1H, J = 9.6, 4.8 Hz), 4.21 (t, 1H, J = 6.0 Hz) 2.92 (m, 2H), 2.60 (app. d, 1H), 2.41–2.50 (m, 2H), 2.12–2.18 (m, 1H), 1.94–2.01 (m, 1H) 13C NMR (D2O, 150 MHz): δ 178.1, 175.6, 168.3, 76.1, 74.3, 53.0, 51.4, 30.9, 26.6, 20.9, HRMS (ESI) calculated (M+H) 242.1135, found 242.1151.
E-(propargyl)-AKE (5)
1H NMR (D2O 400 MHz): 4.56 (t, J = 6.5 Hz, 1H), 4.40 – 4.30 (m, 2H), 4.14 (t, J = 6.5 Hz, 1H), 3.00 (t, J = 7.6 Hz, 2H), 2.76 (dd, J = 6.6, 2.7 Hz, 2H), 2.65 – 2.37 (m, 5H), 2.29 – 1.60 (m, 8H), 1.55 – 1.30 (m, 2H). 13C NMR (101 MHz, D2O) δ 177.41, 177.08, 175.65, 173.35, 171.29, 169.27, 78.81, 72.69, 53.62, 52.74, 52.34, 52.20, 39.13, 30.28, 30.19, 29.85, 26.25, 26.21(2C), 21.93, 21.04. HRMS (ESI) calculated (M - H) 497.2365, found 497.2336.
4.6. General procedures for synthesizing sialidase inhibitors 6–8
The synthesis of inhibitors 6–8 was done using a 1:1 molar ratio (0.03–0.1 mmol) of Neu5Ac9N32en and the propargyl- modified peptide brought to a 200 mM solution in H2O. Sodium ascorbate (10 mol%) was added followed by 2 mol% of copper(II) sulfate. The mixture was stirred at room temperature overnight. The reaction was concentrated and purified using SEC with Bio-Gel P2 before reverse-phase HPLC purification (ACN/H2O).
(TriazoleNeu5Ac2en)-A (6).
1H NMR (D2O, 800 MHz): δ 7.91 (s, 1H), 5.68 (d, 1H, J = 2.2 Hz), 4.79 (m, 1H), 4.50 (dd, 1H, J = 14.3 and 7.9 Hz), 4.45 (d, 1H, J = 9.1 Hz), 4.27–4.30 (m, 1H), 4.19 (d, 1H, J = 10.9 Hz), 4.02–4.05 (m, 1H), 3.98–4.02 (m, 1H), 3.49 (d, 1H, J = 3.49 Hz), 3.32–3.35 (m,1H), 3.25–3.28 (m, 1H) 2.04 (s, 3H). 13C NMR (D2O, 200 MHz): δ 175.1, 173.5, 169.8, 148.1, 142.0, 125.7, 107.9, 75.4, 69.5, 68.5, 67.7, 54.7, 53.7, 50.2, 26.7, 22.3. HRMS (ESI) calculated (M-H) 428.1423, found 428.1412.
(TriazoleNeu5Ac2en)-AdE (7).
1H NMR (D2O, 800 MHz): δ 7.93 (s, 1H), 5.68 (d, 1H, J = 1.8 Hz), 4.81 (d, 1H, J = 2.7 Hz), 4.51 (dd, 1H, J = 14.4, 7.8 Hz), 4.46 (dd, 1H, J = 8.9, 2.3 Hz), 4.28–4.30 (m, 1H), 4.17–4.20 (m, 2H), 4.03–4.07 (m, 1H), 4.00 (t, 1H, J = 6.9 Hz), 3.48 (d, 1H, J = 8.8Hz), 3.17–3.23 (m, 2H) 2.14–2.18 (m, 1H) 2.08–2.13(m, 1H), 1.96 (s, 3H), 1.96–2.01 (m, 1H), 1.84–1.91 (m, 1H); 13C NMR (D2O, 200 MHz): δ 181.1, 176.3 (2C), 174.7, 169.5, 147.9, 141.9, 125.4, 107.6, 74.9, 69.2, 68.3, 67.5, 53.8, 53.6, 53.4, 49.8, 33.5, 28.7, 27.6, 22.0, HRMS (ESI) calculated (M - H) 556.2009, found 556.1983.
E-(TriazoleNeu5Ac2en)-AKE (8).
1H NMR (D2O, 800 MHz): 7.88 (s, 1H), 6.03 (s, 1H), 4.69 (s, 1H), 4.47 (m, 2H), 4.28 (m, 3H), 4.06 (m, 2H), 3.53 (d, J = 9.4 Hz, 1H), 3.21 (m, 2H), 2.96 (t, J = 7.8 Hz, 2H), 2.49 (q, J = 9.1, 8.6 Hz, 4H), 2.19 – 1.91 (m, 7H), 1.82 – 1.54 (m, 5H), 1.37 (s, 3H). 13C NMR (201 MHz, D2O) δ 176.88, 176.08, 175.64, 174.80, 173.24, 171.56, 168.92, 166.89, 145.08, 142.02, 125.14, 110.62, 75.38, 69.15, 68.48, 66.92, 53.52, 53.48, 53.30, 52.64, 52.01, 49.64, 39.02, 30.27, 29.93, 28.98, 27.03, 26.16, 25.92, 25.81, 21.94, 21.80. HRMS (ESI) calculated (M - H) 813.3384, found 813.3343.
4.7. Inhibition assay
Percentage Inhibition assays were carried out in duplicate sets in a 384-well plate. All reactions were final volume of 20 μ L containing Neu5Acα2–3GalβpNP (0.3 mM) and β-galactosidase (12 μg) with inhibitor concentration of 0.1 mM or 1.0 mM. The assay conditions for various sialidases were as follows: A . ureafacienssialidase (0.5 mU), NaOAc buffer (100 mM, pH 5.5); C. perfringenssialidase (NanI, 1.3 mU), MES buffer (100 mM, pH 5.0); V. cholera sialidase (0.6 mU), NaCl (150 mM), CaCl2 (10 mM), NaOAc buffer (100 mM, pH 5.5); SpNanA (0.75 ng), NaOAc buffer (100 mM, pH 6.0); SpNanB (3 ng), NaOAc buffer (100 mM, pH 6.0); SpNanC (10 ng), MES buffer (100 mM, pH 6.5); PmST1 (0.2 μ g), NaOAc buffer (100 mM, pH 5.5), CMP (0.4 mM); hNEU2 (0.6 μ g), MES buffer (100 mM, pH 5.0); BiNanH2 (4 ng), NaOAc buffer (100 mM, pH 5.0). The reactions were carried out for 30 minutes and quenched with CAPS buffer (N-cyclohexyl-3-aminopropane sulfonic acid, 40 μ L, 0.5 M, pH 10.5). The amount of p-nitrophenolate formed was determined by measuring A405 nm of the reaction mixtures using a microplate reader.
Inhibition assays for obtaining IC50 values were carried out in duplicates in a 384-well plate similarly as described above except that twelve different concentrations of inhibitors in the range of 0 to 10 mM were used (varied from 10 mM, 5 mM, 2.5 mM, 1 mM, 0.5 mM, 0.1 mM, 50 μM, 25 μM, 10 μM, 5 μM, 2.5 μM, 1 μM, 0.5 μM, 0.1 μM, 50 nM, 10 nM, 5 nM, and 0 nM). IC50 values were obtained by fitting the average values to get the concentration-response plots using software Grafit 5.0.
4.8. Docking studies
Three dimensional coordinate files for inhibitors 6 and 7 were generated using Open Babel,48 while inhibitor 2 was generated using PyMOL from Neu5Ac2en (1) in V. cholerae sialidase crystal structure (PDB 1w0o). Docking was performed using Autodock Vina49 and the docking results were analyzed in PyMOL.50
Supplementary Material
Acknowledgements
This work was partially supported by United States National Institutes of Health grants R01AI130684 and R43AI108115. Bruker Avance-800 NMR spectrometer was funded by United States National Science Foundation grant DBIO-722538.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Mann MC, Thomson RJ, Dyason JC, McAtamney S, Itzstein MV. Modelling, synthesis and biological evaluation of novel glucuronide- based probes of Vibrio cholerae sialidase. Bioorg Med Chem 2006;14: 1518–1537. [DOI] [PubMed] [Google Scholar]
- 2.Taylor G Sialidases: structures, biological significance and therapeutic potential. Curr Opin Struct Biol 1996;6:830–837. [DOI] [PubMed] [Google Scholar]
- 3.Henrissat B A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem 1991;280:309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henrissat B, Bairoch A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem 1993;293:781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henrissat B, Bairoch A. Updating the sequence-based classification of glycosyl hydrolases. Biochem 1996;316:695–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies G, Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure 1995;3:853–859. [DOI] [PubMed] [Google Scholar]
- 7.Ishikura H, Arakawa S, Nakajima T, Tsuchida N, Ishikawa I. Cloning of the Tannerella forsythensis (Bacteroides forsythus) siaHI gene and purification of the sialidase enzyme. J Med Microbiol 2003;52:1101– 1107. [DOI] [PubMed] [Google Scholar]
- 8.Honma K, Mishima E, Sharma A. Role of Tannerella forsythia NanH sialidase in epithelial cell attachment. Infect immun 2011;79:393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu H, Chokhawala H, Karpel R, et al. A multifunctional Pasteurella multocida sialyltransferase: a powerful tool for the synthesis of sialoside libraries. J Am Chem Soc 2005;127:17618–17619. [DOI] [PubMed] [Google Scholar]
- 10.Cheng J, Yu H, Lau K, et al. Multifunctionality of Campylobacter jejuni sialyltransferase CstII: characterization of GD3/GT3 oligosaccharide synthase, GD3 oligosaccharide sialidase, and trans-sialidase activities. Glycobiology 2008;18:686–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng J, Huang S, Yu H, Li Y, Lau K, Chen X. Trans-sialidase activity of Photobacterium damsela alpha2,6-sialyltransferase and its application in the synthesis of sialosides. Glycobiology 2010;20:260–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crennell S, Garman E, Laver G, Vimr E, Taylor G. Crystal structure of Vibrio cholerae neuraminidase reveals dual lectin-like domains in addition to the catalytic domain. Structure 1994;2:535–544. [DOI] [PubMed] [Google Scholar]
- 13.Newstead SL, Watson JN, Bennet AJ, Taylor G. Galactose recognition by the carbohydrate-binding module of a bacterial sialidase. Acta Crystallogr D Biol Crystallogr 2005;61:1483–1491. [DOI] [PubMed] [Google Scholar]
- 14.Boraston AB, Ficko-Blean E, Healey M. Carbohydrate recognition by a large sialidase toxin from Clostridium perfringens . Biochemistry 2007;46:11352–11360. [DOI] [PubMed] [Google Scholar]
- 15.Park KH, Kim MG, Ahn HJ, et al. Structural and biochemical characterization of the broad substrate specificity of Bacteroides thetaiotaomicron commensal sialidase. Biochim Biophys Acta 2013;1834:1510–1519. [DOI] [PubMed] [Google Scholar]
- 16.von Itzstein M, Wu W- Y, Kok GB, et al. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature 1993;363:418–423. [DOI] [PubMed] [Google Scholar]
- 17.Paton JC, Andrew PW, Boulnois GJ, Mitchell TJ. Molecular analysis of the pathogenicity of Streptococcus Pneumoniae : The role of Pneumococcal proteins. Annu Rev Microbiol 1993;47:89–115. [DOI] [PubMed] [Google Scholar]
- 18.Gut H, Xu G, Taylor GL, Walsh MA. Structural basis for Streptococcus pneumoniae NanA inhibition by influenza antivirals zanamivir and oseltamivir carboxylate. J Mol Biol 2011;409:496–503. [DOI] [PubMed] [Google Scholar]
- 19.Jermyn WS, Boyd EF. Characterization of a novel Vibrio pathogenicity island (VPI-2) encoding neuraminidase (nanH) among toxigenic Vibrio cholerae isolates. Microbiology 2002;148:3681–3693. [DOI] [PubMed] [Google Scholar]
- 20.Bogaert D, de Groot R, Hermans PWM. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect Dis 2004;4: 144–154. [DOI] [PubMed] [Google Scholar]
- 21.Gray BM, Converse GM, Dillon HC. Epidemiologic Studies of Streptococcus pneumoniae in Infants: Acquisition, Carriage, and Infection during the First 24 Months of Life. J Infect Dis 1980;142:923– 933. [DOI] [PubMed] [Google Scholar]
- 22.Shenoy AT, Orihuela CJ. Anatomical site-specific contributions of pneumococcal virulence determinants. Pneumonia 2016;8:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tasnima N, Yu H, Li Y, Santra A, Chen X. Chemoenzymatic synthesis of para-nitrophenol (pNP)-tagged alpha2–8-sialosides and high-throughput substrate specificity studies of alpha2–8-sialidases. Org Biomol Chem 2016;15:160–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Owen CD, Lukacik P, Potter JA, Sleator O, Taylor GL, Walsh MA. Streptococcus pneumoniae NanC: structural insights into the specificity and mechanism of a sialidase that produces a sialidase inhibitor. J Biol Chem 2015;290: 27736–27748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu G, Kiefel MJ, Wilson JC, Andrew PW, Oggioni MR, Taylor GL. Three Streptococcus pneumoniae sialidases: Three different products. J Am Chem Soc 2011;133:1718–1721. [DOI] [PubMed] [Google Scholar]
- 26.Manco S, Hernon F, Yesilkaya H, Paton JC, Andrew PW, Kadioglu A. pneumococcal neuraminidases A and B both have essential roles during infection of the respiratory tract and sepsis. Infect Immun 2006;74:4014– 4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brear P, Telford J, Taylor GL, Westwood NJ. Synthesis and structural characterisation of selective non-carbohydrate-based inhibitors of bacterial sialidases. ChemBioChem 2012;13:2374–2383. [DOI] [PubMed] [Google Scholar]
- 28.Kim CU, Lew W, Williams MA, et al. Influenza neuraminidase inhibitors possessing a novel hydrophobic interaction in the enzyme active site: design, synthesis, and structural analysis of carbocyclic sialic acid analogues with potent anti-influenza activity. J Am Chem Soc 1997;119:681–690. [DOI] [PubMed] [Google Scholar]
- 29.von Itzstein M The war against influenza: discovery and development of sialidase inhibitors. Nat Rev Drug Discov 2007;6: 967–974. [DOI] [PubMed] [Google Scholar]
- 30.Laborda P, Wang SY, Voglmeir J. Influenza neuraminidase inhibitors: synthetic approaches, derivatives and biological activity. Molecules 2016;21:1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Babu YS, Chand P, Bantia S, et al. Discovery of a novel, highly potent, orally active, and selective influenza neuraminidase inhibitor through structure-based drug design. J Med Chem 2000;43:3482–3486. [DOI] [PubMed] [Google Scholar]
- 32.Buschiazzo A, Amaya MaF, Cremona MaL, Frasch AC, Alzari PM. The crystal structure and mode of action of trans-sialidase, a key enzyme in Trypanosoma cruzi Pathogenesis. Mol Cell 2002;10:757–768. [DOI] [PubMed] [Google Scholar]
- 33.Amaya MaF, Watts AG, Damager I, et al. Structural insights into the catalytic mechanism of Trypanosoma cruzi trans-sialidase. Structure 2004;12:775–784. [DOI] [PubMed] [Google Scholar]
- 34.Xu G, Li X, Andrew PW, Taylor GL. Structure of the catalytic domain of Streptococcus pneumoniae sialidase NanA. Acta Crystallogr Sect F, Struct Biol Cryst Commun 2008;64:772–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khedri Z, Li Y, Cao H, et al. Synthesis of selective inhibitors against V. cholerae sialidase and human cytosolic sialidase NEU2. Org Biomol Chem 2012;10:6112–6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Albohy A, Zhang Y, Smutova V, Pshezhetsky AV, Cairo CW. Identification of selective nanomolar inhibitors of the human neuraminidase, NEU4. ACS Med Chem Lett 2013;4:532–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsiao Y- S, Parker D, Ratner AJ, Prince A, Tong L. Crystal structures of respiratory pathogen neuraminidases. Biochem Biophys Res Commun 2009;380:467–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tiwari VK, Mishra BB, Mishra KB, Mishra N, Singh AS, Chen X. Cu-catalyzed click reaction in carbohydrate chemistry. Chem Rev 2016;116:3086–3240. [DOI] [PubMed] [Google Scholar]
- 39.Chan TR, Hilgraf R, Sharpless KB, Fokin VV. Polytriazoles as copper(I)- stabilizing ligands in catalysis. Org Lett 2004;6:2853–2855. [DOI] [PubMed] [Google Scholar]
- 40.Xiao A, Li Y, Li X, et al. Sialidase-catalyzed one-pot multienzyme (OPME) synthesis of sialidase transition-state analogue inhibitors. ACS Catal 2018;8:43–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y, Cao H, Yu H, et al. Identifying selective inhibitors against the human cytosolic sialidase NEU2 by substrate specificity studies. Mol Biosys 2011;7:1060–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sela DA, Li Y, Lerno L, et al. An infant-associated bacterial commensal utilizes breast milk sialyloligosaccharides. J Biol Chem 2011;286: 11909–11918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiao A, Slack TJ, Li Y, et al. Streptococcus pneumoniae sialidase SpNanB-catalyzed one-pot multienzyme (OPME) synthesis of 2,7- anhydro-sialic acids as selective sialidase inhibitors. J Org Chem 2018; doi: 10.1021/acs.joc.8b01519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode: Academic Press; 1997. [DOI] [PubMed] [Google Scholar]
- 45.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cystallogr 2007;40: 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D, Biol Crystallogr 2004;60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 47.Adams PD, Afonine PV, Bunkoczi G, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D, Biol Crystallogr 2010;66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open babel: an open chemical toolbox. J Cheminform 2011;3:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010;31:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. The PyMOL Molecular Graphics System, V. S., LLC.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


