Abstract
Chorioamnionitis is an intrauterine infection involving inflammation of the chorion, amnion and placenta. It leads to a fetal systemic inflammatory response that can alter the transcription of neonatal immune genes. We have previously shown that neonatal monocytes gain the activating histone tail modification H3K4me3 at promoters of immunologically important genes as development progresses from preterm neonate to adult. In this study, we applied ChIP-seq and RNA-seq to evaluate the impact of chorioamnionitis on the neonatal monocyte H3K4me3 histone modification landscape over the course of fetal and neonatal immune system development. Chorioamnionitis exposure in neonatal monocytes resulted in a net increase in total monocyte H3K4me3, primarily in introns and intergenic regions. Immune gene expression was decreased in chorioamnionitis-exposed monocytes, with the majority of enriched transcripts falling into pathways that are not linked to the immune system. Over half of all neonatal monocyte H3K4me3 peaks, independent of their location, were associated with active gene transcription. Overall, chorioamnionitis exposure resulted in global remodeling of the neonatal monocyte H3K4me3 landscape and changes in the expression of known immune genes. These changes resulted in a less robust inflammatory response upon exposure to a secondary challenge, which may explain why chorioamnionitis-exposed neonates have an increased risk of sepsis.
Keywords: chorioamnionitis, histone modification, monocyte, transcription, epigenetics
Graphical Abstract
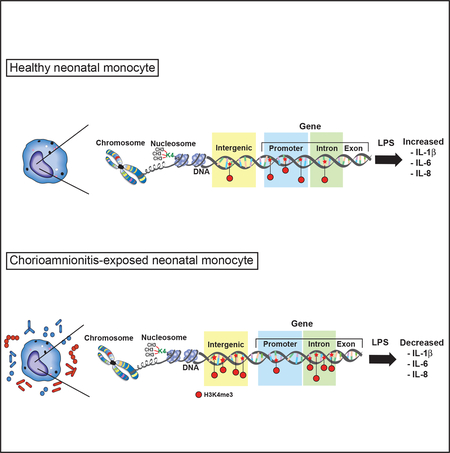
Exposure to chorioamnionitis, a common intrauterine infection, alters the neonatal monocyte histone modification landscape. Specifically, the activating histone modification mark H3K4me3 is removed from promoters and instead enriched in introns and intergenic regions. This results in reduced expression of immune pathway genes, leading to a decreased inflammatory response when a secondary immune stimulus is encountered.
Introduction
Chorioamnionitis is a condition involving infection and inflammation of the chorion, amnion and placenta and is the most common cause of preterm birth, although it complicates up to 4% of term births as well[1, 2]. Chorioamnionitis exposure can lead to a fetal systemic inflammatory response that alters the developing immune system and places exposed infants at a higher risk of developing early onset sepsis[3–5]. It is unclear if this sepsis risk is due to a common infectious agent leading to both conditions or as a result of remodeling of the neonatal immune system that leaves exposed infants vulnerable to infections. Recent studies demonstrate that exposure to histologic chorioamnionitis alters the neonatal immune transcriptosome, with activation of innate immune pathways[6, 7]. Chorioamnionitis exposure can also have long-term consequences for exposed infants, including neurodevelopment impairment, cerebral palsy and the development of immune-mediated diseases later in life, including wheezing and asthma[8–11]. This highlights the importance of perinatal exposures on the developing immune system, which may result in life-long alterations in immune responses.
Monocytes are first responders of the innate immune system, sensing pathogens and initiating an inflammatory cascade that results in the clearance of microbes[12]. Neonatal monocytes demonstrate major deficiencies in chemotaxis, microbial killing and pro-inflammatory cytokine expression, all of which contribute to a heightened risk of infection early in the neonatal period[13–15]. Epigenetics refers to modifications of DNA structure and chromatin accessibility that alter gene expression without affecting the underlying genetic code. Many changes to DNA structure occur as the result of histone tail modifications, which bind to DNA and package it into chromatin. The addition of three methyl groups to lysine (K) 4 on histone (H) 3 (H3K4me3) near gene promoters allows for active gene transcription[16]. We have previously shown that neonatal monocytes gain H3K4me3 at promoter sites of immunologically important genes, including IL1B, TNF, and CCR2, as development progresses from preterm neonate to adult[17]. This developmental change in the monocyte histone modification landscape was found to contribute to neonatal-specific immune responses that leave them vulnerable to infections. It is currently unknown how exposures in the perinatal period, including chorioamnionitis, alter the normal developmental progression of the histone modification landscape in the neonatal immune system, and what impact this has on monocyte transcription and function.
In this study we utilized chromatin immunoprecipitation followed by massively parallel DNA sequencing (ChIP-seq) to determine the impact of chorioamnionitis exposure on the neonatal monocyte H3K4me3 histone modification landscape as development progressed from extremely preterm to term neonate. We then performed whole transcriptome shotgun sequencing (RNA-seq) on unstimulated and lipopolysaccharide (LPS) stimulated monocytes from term healthy and chorioamnionitis-exposed neonates to evaluate the impact of chorioamnionitis exposure on monocyte transcription at baseline and upon a secondary inflammatory stimulus. These experiments revealed that chorioamnionitis exposure remodeled the neonatal monocyte H3K4me3 landscape and altered gene transcription, with decreased expression of known immune genes. Additionally, H3K4me3 presence near a known gene, not just at promoter sites, predicted mRNA expression over 50% of the time.
Results
Chorioamnionitis exposure altered the abundance and location of neonatal monocyte H3K4me3
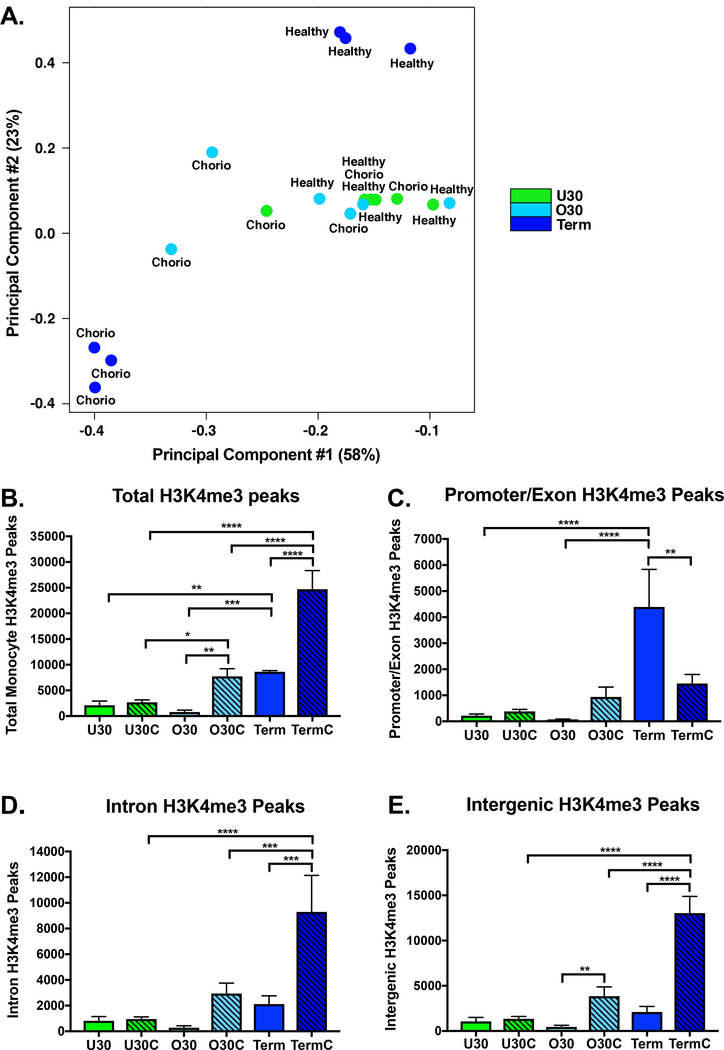
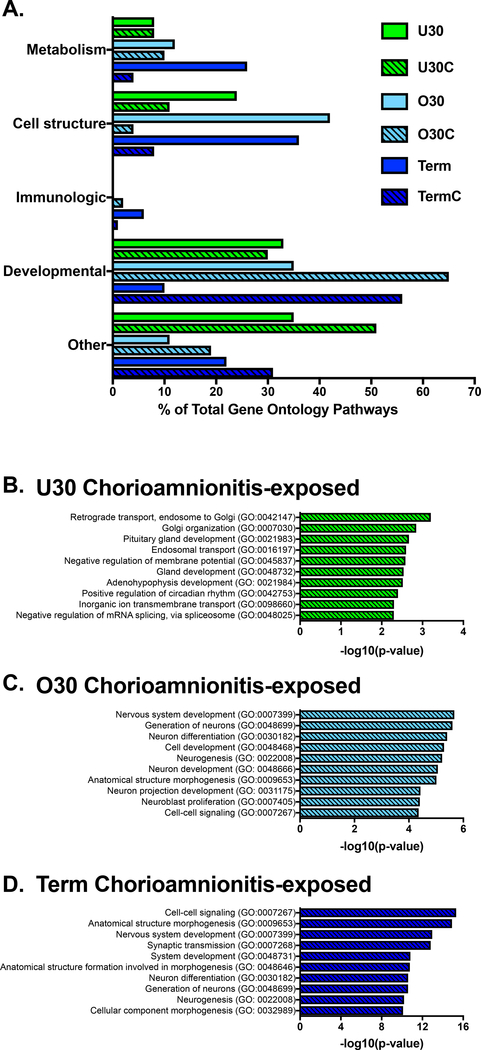
To determine the influence of chorioamnionitis on the monocyte H3K4me3 landscape we performed ChIP-seq on preterm and term monocytes with and without exposure to histopathologic chorioamnionitis. We evaluated the global distribution of H3K4me3 in umbilical cord blood purified CD14+ monocytes from six different experimental groups: under 30-week extremely preterm healthy infants (U30), under 30-week extremely preterm chorioamnionitis-exposed infants (U30C), over 30-week preterm healthy infants (O30), over 30-week preterm chorioamnionitis-exposed infants (O30C), term healthy infants (Term) and term chorioamnionitis-exposed infants (TermC). Table 1 compares patient characteristics between the healthy and chorioamnionitis-exposed infants in each gestational age group. Overall, the groups for each age and exposure group were well matched for gestational age, birth weight, sex, maternal smoking, gestational hypertension and gestational diabetes. The only difference noted was that infants in the O30C and Term groups were more likely to be born by vaginal delivery than those in the comparison groups. Fig. 1 summarizes differences in H3K4me3 abundance and location between these different experimental groups. A principal component analysis of monocyte H3K4me3 normalized read counts reveals that preterm healthy and chorioamnionitis-exposed monocytes cluster together along principal component 2, with some differences between preterm healthy and chorioamnionitis-exposed monocytes on principal component 1 (Fig. 1A). The term healthy sample replicates cluster together and the term chorioamnionitis-exposed sample replicates cluster together but are distinct from each other in both principal components 1 and 2 (Fig. 1A). We have previously reported an increase in total monocyte H3K4me3 peaks as development progresses from preterm to term neonate[17]. In addition to this developmentally related increase in monocyte H3K4me3, chorioamnionitis exposure in preterm and term monocytes significantly increases the total number of H3K4me3 monocyte peaks (Fig. 1B). To obtain a broad overview of H3K4me3 distribution, the human genome was divided into four distinct categories according to the UCSC Genome Browser: promoters, introns, exons and intergenic regions[18]. Chorioamnionitis exposure had little to no impact on the genomic location of H3K4me3 peaks in extremely preterm monocytes (Fig. 1C-E). Chorioamnionitis exposure resulted in decreased promoter/exon peaks in term monocytes (Fig. 1C). In contrast, exposure to chorioamnionitis resulted in increased intergenic H3K4me3 peaks in preterm and term monocytes with increased intron peaks in term monocytes (Fig. 1D-E). All gestational age and exposure groups had H3K4me3 enriched in gene ontology pathways necessary for basic cell survival and function, including cellular metabolism, cell signaling and maintenance of cell structure (Fig. 2A). Chorioamnionitis exposure had minimal impact on the gene ontology pathways enriched for H3K4me3 in extremely preterm monocytes. In contrast, chorioamnionitis exposure in preterm and term monocytes was associated with increased H3K4me3 enrichment in gene ontology pathways associated with development, with heavy enrichment in pathways important in nervous system development (Fig. 2A). The top gene ontology pathways enriched for H3K4me3 in healthy extremely preterm, preterm and term neonatal monocytes was previously published[17], and the top 10 H3K4me3 enriched gene ontology pathways in chorioamnionitis-exposed neonates are shown in Fig. 2B-D. Immunologically relevant gene ontology pathways enriched for H3K4me3 in chorioamnionitis-exposed monocytes are detailed in Table 2.
Table 1:
ChIP-seq and RNA-seq patient characteristics
| Characteristic | ChIP-seq U30a | ChIP-seq O30b | ||||
|---|---|---|---|---|---|---|
| U30a | U30Ca,d | p-value | O30b | O30Cb,d | p-value | |
| Total pooled samples (n) | 9 | 9 | 9 | 9 | ||
| Gestational age (weeks) | 27.79 ± 1.98 | 28.16 ± 1.57 | p=0.67 | 32.54 ± 1.29 | 33.86 ± 2.43 | p=0.17 |
| Birth weight (grams) | 1055 ± 414 | 1177 ± 205 | p=0.44 | 1887 ± 5225 | 2264 ± 509 | p=0.14 |
| Males | 5 (56%) | 4 (44%) | p=0.64 | 4 (44%) | 6 (67%) | p=0.34 |
| Vaginal delivery | 1 (11%) | 4 (44%) | p=0.11 | 1 (11%) | 8 (89%) | p=0.001* |
| Maternal smoking | 0 (0%) | 1 (11%) | p=0.3 | 0 (0%) | 2 (22%) | p=0.13 |
| Gestational hypertension | 2 (22%) | 0 (0%) | p=0.13 | 3 (33%) | 1 (11%) | p=0.26 |
| Gestational diabetes | 0 (0%) | 0 (0%) | p>0.99 | 0 (0%) | 0 (0%) | p>0.99 |
| Prenatal steroids | 9 (100%) | 9 (100%) | p>0.99 | 8 (89%) | 6 (67%) | p=0.26 |
| ChIP-seq Termc | RNA-seq Termc | |||||
| Termc | TermCc,d | p-value | Termc | TermCc,d | p-value | |
| Total pooled samples (n) | 9 | 9 | 9Ue 9Lf |
6Ue 9Lf |
||
| Gestational age (weeks) | 39.9 ± 1.47 | 40.25 ± 1.08 | p=0.57 | 38.65 ± 1.29 38.65 ± 1.29 |
39.29 ± 1.31 39.84 ± 1.34 |
p=0.37 p=0.07 |
| Birth weight (grams) | 3779 ± 556 | 3544 ± 366 | p=0.31 | 3283 ± 623 3283 ± 623 |
3259 ± 693 3354 ± 580 |
p=0.94 p=0.81 |
| Males | 6 (67%) | 6 (67%) | p>0.99 | 3 (33%) 3 (33%) |
3 (50%) 4 (44%) |
p=0.52 p=0.63 |
| Vaginal delivery | 8 (89%) | 4 (44%) | p=0.05 | 4 (44%) 4 (44%) |
3 (50%) 4 (44%) |
p=0.83 p>0.99 |
| Maternal smoking | 0 (0%) | 0 (0%) | p>0.99 | 0 (0%) 0 (0%) |
0 (0%) 0 (0%) |
p>0.99 p>0.99 |
| Gestational hypertension | 0 (0%) | 0 (0%) | p>0.99 | 0 (0%) 0 (0%) |
0 (0%) 1 (11%) |
p>0.99 p=0.3 |
| Gestational diabetes | 0 (0%) | 1 (11%) | p=0.3 | 0 (0%) 0 (0%) |
0 (0%) 1 (11%) |
p>0.99 p=0.3 |
| Prenatal steroids | 0 (0%) | 0 (0%) | p>0.99 | 0 (0%) 0 (0%) |
0 (0%) 0 (0%) |
p>0.99 p>0.99 |
U30 = under 30 weeks gestation,
O30 = 30–36 weeks gestation,
Term = term gestation,
C=chorioamnionitis exposure,
U=unstimulated,
L=LPS stimulated. The student’s t-test was used to test for differences between quantitative data and the Chi-square test was used to test for differences between categorical data.
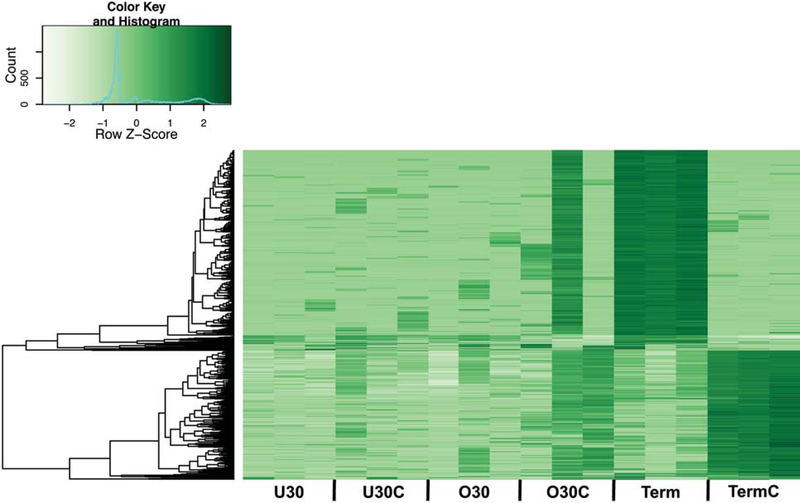
Figure 1: Chorioamnionitis exposure increased term neonatal monocyte H3K4me3 with a preferential gain in introns and intergenic regions.
A) Principal component analysis of neonatal monocyte H3K4me3 normalized read counts. All healthy and chorioamnionitis-exposed preterm monocytes clustered together along principal component 2, with some separation of a few of the chorioamnionitis-exposed monocytes along principal component 1. Term healthy and chorioamnionitis-exposed monocytes had distinct clustering along both principal components 1 and 2, with separation from each other and the preterm monocytes. Chorio=chorioamnionitis exposure. B) Total monocyte H3K4me3 peaks by gestational age and chorioamnionitis exposure. C) Promoter and exon monocyte H3K4me3 peaks by gestational age and chorioamnionitis exposure. Promoter locations were defined as 1 kb upstream or downstream from transcriptional start sites. D) Intron monocyte H3K4me3 peaks by gestational age and chorioamnionitis exposure. E) Intergenic monocyte H3K4me3 peaks by gestational age and chorioamnionitis exposure. U30(n=3)=under 30 weeks gestation non-chorioamnionitis exposed neonates, U30C(n=3)=under 30 weeks gestation chorioamnionitis exposed neonates, O30(n=3)=30–36 weeks gestation non-chorioamnionitis exposed neonates, O30C(n=3)=30–36 weeks gestation chorioamnionitis exposed neonates, Term(n=3)=37+ weeks gestation non-chorioamnionitis exposed neonates, TermC(n=3)=37+ weeks gestation chorioamnionitis exposed neonates. Box represents mean, error bars are SD. Differences between groups were assessed using the Kruskal-Wallis test with multiple comparisons. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Figure 2: Chorioamnionitis stimulated placement of H3K4me3 peaks in gene ontology pathways related to development.
A) Biological gene ontology pathways enriched for H3K4me3 in neonatal monocytes at different gestational ages. The top 10 biological gene ontology pathways enriched for H3K4me3 in chorioamnionitis exposed B) extremely preterm monocytes, C) preterm monocytes and D) term monocytes. U30(n=3)=under 30 weeks gestation non-chorioamnionitis exposed neonates, U30C(n=3)=under 30 weeks gestation chorioamnionitis exposed neonates, O30(n=3)=30–36 weeks gestation non-chorioamnionitis exposed neonates, O30C(n=3)=30–36 weeks gestation chorioamnionitis exposed neonates, Term(n=3)=37+ weeks gestation non-chorioamnionitis exposed neonates, TermC(n=3)=37+ weeks gestation chorioamnionitis exposed neonates.
Table 2:
Immunologically relevant gene ontology pathways enriched for H3K4me3 in neonatal monocytes
| Group | GO Term | Description | Number of genes | p-value |
|---|---|---|---|---|
| U30Ca,d | ||||
| GO:0050829 | Defense response to Gram-negative bacterium | 2 | 0.019 | |
| GO:0006959 | Humoral immune response | 4 | 0.024 | |
| GO:0002437 | Inflammatory response to antigenic stimulus | 2 | 0.025 | |
| GO:0042742 | Defense response to bacterium | 4 | 0.036 | |
| GO:0098542 | Defense response to other organism | 6 | 0.037 | |
| O30Cb,d | ||||
| GO:0071559 | Response to transforming growth factor beta | 27 | 0.011 | |
| GO:0050918 | Positive chemotaxis | 9 | 0.018 | |
| GO:0006935 | Chemotaxis | 68 | 0.019 | |
| GO:0071634 | Regulation of transforming growth factor beta production | 5 | 0.023 | |
| GO:0033081 | Regulation of T cell differentiation in thymus | 5 | 0.027 | |
| GO:0048291 | Isotype switching to IgG isotypes | 3 | 0.036 | |
| GO:0038123 | Toll-like receptor TLR1:TLR2 signaling pathway | 10 | 0.043 | |
| TermCc,d | ||||
| GO:0048291 | Isotype switching to IgG isotypes | 8 | 0.0007 | |
| GO:0006935 | Chemotaxis | 221 | 0.001 | |
| GO:0002824 | Positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 30 | 0.003 | |
| GO:0045830 | Positive regulation of isotype switching | 9 | 0.004 | |
| GO:0030217 | T cell differentiation | 68 | 0.006 | |
| GO:0002714 | Positive regulation of B cell mediated immunity | 13 | 0.006 | |
| GO:0002708 | Positive regulation of lymphocyte mediated immunity | 29 | 0.007 | |
| GO:0045581 | Negative regulation of T cell differentiation | 14 | 0.007 | |
| GO:0033081 | Regulation of T cell differentiation in thymus | 12 | 0.008 | |
| GO:0030098 | Lymphocyte differentiation | 94 | 0.011 | |
| GO:0002819 | Regulation of adaptive immune response | 42 | 0.012 | |
| GO:0002705 | Positive regulation of leukocyte mediated immunity | 33 | 0.012 | |
| GO:0043030 | Regulation of macrophage activation | 12 | 0.013 | |
| GO:0045954 | Positive regulation of natural killer cell mediated cytotoxicity | 9 | 0.02 | |
| GO:0046639 | Negative regulation of alpha-beta T cell differentiation | 7 | 0.021 | |
| GO:0046636 | Negative regulation of alpha-beta T cell activation | 10 | 0.022 | |
| GO:0032634 | Interleukin-5 production | 8 | 0.026 | |
| GO:0045063 | T-helper 1 cell differentiation | 8 | 0.026 | |
| GO:0043032 | Positive regulation of macrophage activation | 6 | 0.028 | |
| GO:0070664 | Negative regulation of leukocyte proliferation | 23 | 0.033 | |
| GO:0002889 | Regulation of immunoglobulin mediated immune response | 16 | 0.038 | |
| GO:0050868 | Negative regulation of T cell activation | 28 | 0.041 | |
| GO:0050851 | Antigen receptor-mediated signaling pathway | 49 | 0.041 | |
| GO:0002717 | Positive regulation of natural killer cell mediated immunity | 9 | 0.044 | |
| GO:0032700 | Negative regulation of interleukin-17 production | 6 | 0.047 | |
| GO:2000515 | Negative regulation of CD4-positive, alpha-beta T cell activation | 6 | 0.047 | |
| GO:0043368 | Positive T cell selection | 11 | 0.047 | |
| GO:0050856 | Regulation of T cell receptor signaling pathway | 11 | 0.047 | |
| GO:0050853 | B cell receptor signaling pathway | 16 | 0.047 | |
| GO:0002706 | Regulation of lymphocyte mediated immunity | 36 | 0.048 | |
| GO:0002696 | Positive regulation of leukocyte activation | 83 | 0.048 |
U30 = under 30 weeks gestation,
O30 = 30–36 weeks gestation,
Term = term gestation,
C=chorioamnionitis exposure
Chorioamnionitis exposure resulted in removal and deposition of H3K4me3 in monocytes at all gestational ages
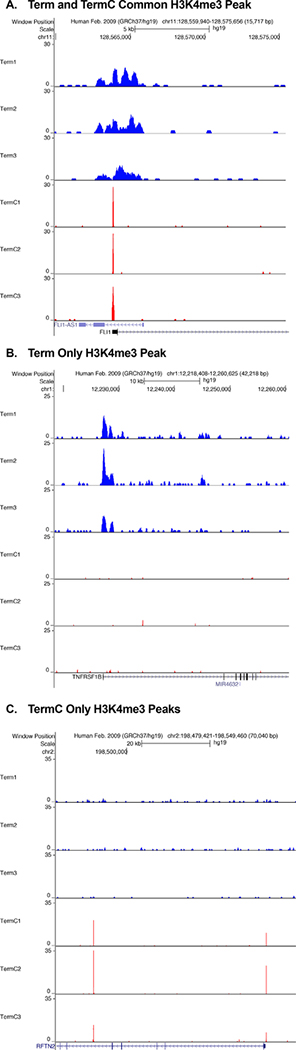
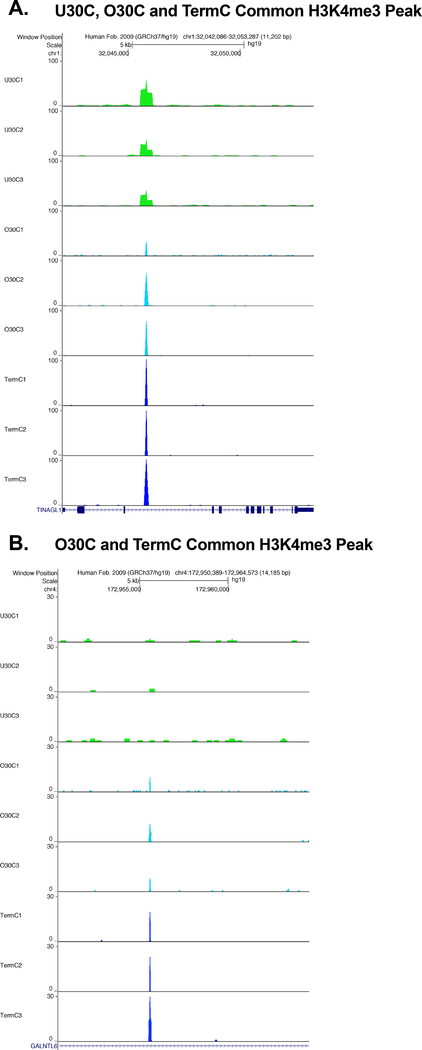
We then derived consensus H3K4me3 peaksets for each gestational age and exposure group, and peaks needed to be present in two out of three replicates to be included. The H3K4me3 consensus peaks present in each gestational age and exposure group are outlined in Table 3. Next, we compared H3K4me3 peak locations (occupancy) between gestation and exposure groups. At all gestations there were unique H3K4me3 peaks present in both the healthy and chorioamnionitis-exposed monocytes, with varying amounts of shared H3K4me3 peak locations depending on the gestation (Fig. 3A-C). When the chorioamnionitis-exposed monocytes were compared between gestations, there were some common H3K4me3 peaks shared by all groups, with the largest overlap between the preterm (O30) and term groups (Fig. 3D). In the extremely preterm neonates there were no differences in average H3K4me3 peak width between exposure groups. In contrast, in the preterm and term populations the chorioamnionitis-exposed monocyte H3K4me3 peaks were narrower than those in the healthy monocytes (Table 4). Representative unique and common H3K4me3 peaks for the term population are shown in Fig. 4. Representative common H3K4me3 peaks for the chorioamnionitis-exposed monocytes are shown in Fig. 5. A binding affinity analysis was then performed to determine differentially bound H3K4me3 peaks between gestational age and exposure groups. A binding affinity heatmap demonstrated that term healthy monocytes and term chorioamnionitis-exposed monocytes had distinctly different H3K4me3 binding affinity patterns, while preterm healthy and chorioamnionitis-exposed monocyte H3K4me3 binding affinity was more varied (Fig. 6). An overview of differentially bound H3K4me3 sites between different gestational age and exposure groups is presented in Table 5 with a more comprehensive list in Supplemental Data. Representative differentially bound H3K4me3 peaks between term chorioamnionitis-exposed and unexposed monocytes are shown in Fig. 7. Gene ontology pathways enriched for differentially bound H3K4me3 between gestational age and exposure groups are detailed in Table 6. The most striking differences in gene ontology pathways enriched for differentially bound H3K4me3 were between term chorioamnionitis-exposed and unexposed monocytes. Term unexposed monocytes had increased H3K4me3 in a broad array of gene pathways, including protein catabolism, transcriptional regulation, apoptosis, response to virus, intracellular signaling and autophagy (Table 6). In contrast, although chorioamnionitis exposure resulted in an overall increase in monocyte H3K4me3 peaks, term chorioamnionitis-exposed monocytes had very few gene pathways enriched for H3K4me3, suggesting that chorioamnionitis stimulated H3K4me3 deposition in a more random and poorly regulated manner (Table 6).
Table 3:
Number and location of monocyte consensus H3K4me3 peaks by gestational age and chorioamnionitis exposure
| Peak Location | U30a | U30Ca,d | O30b | O30Cb,d | Termc | TermCc,d |
|---|---|---|---|---|---|---|
| Total Consensus Peaks | 496 | 432 | 374 | 4254 | 4250 | 21,457 |
| Promoter Peaks | 4 | 6 | 2 | 26 | 292 | 92 |
| Exon Peaks | 10 | 7 | 6 | 63 | 217 | 429 |
| Intron Peaks | 65 | 67 | 57 | 757 | 646 | 3436 |
| Intergenic Peaks | 417 | 352 | 309 | 3408 | 3095 | 17,500 |
U30 = under 30 weeks gestation,
O30 = 30–36 weeks gestation,
Term = term gestation,
C=chorioamnionitis exposure
Figure 3: There were varying degrees of shared and unique monocyte H3K4me3 peaks in healthy and chorioamnionitis-exposed monocytes based on gestational age.
Venn diagrams demonstrating unique and common H3K4me3 peaks between healthy and chorioamnionitis-exposed A) extremely preterm monocytes, B) preterm monocytes and C) term monocytes. D) Venn diagram demonstrating common and unique H3K4me3 peaks between chorioamnionitis-exposed monocytes at different gestations. U30=under 30 weeks gestation, O30=30–36 weeks gestation, Term=37+ weeks gestation, Healthy=no chorioamnionitis exposure, Chorio=chorioamnionitis exposure.
Table 4:
Neonatal monocyte H3K4me3 peak width
| Age Group | Healthy | Chorioamnionitis-exposed | p-value |
|---|---|---|---|
| U30a | 270±240 bp | 383±255 bp | 0.999 |
| O30b | 307±179 bp | 142±124 bp | <0.0001 |
| Termc | 474±364 bp | 148±79 bp | <0.0001 |
U30 = under 30 weeks gestation,
O30 = 30–36 weeks gestation,
Term = term gestation, values are mean±SD, p-values calculated using Mann-Whitney test
Figure 4: Representative common and unique H3K4me3 peaks in term healthy and chorioamnionitis-exposed monocytes.
A) Term chorioamnionitis-exposed and non-exposed monocytes had a common H3K4me3 peak at the promoter site of FLI1 (Fli-1 proto-oncogene, ETS transcription factor). B) Term healthy monocytes had a unique H3K4me3 peak at the promoter site of TNFRSF1B (TNF receptor superfamily member 1B) that was not present in chorioamnionitis-exposed monocytes. C) Term chorioamnionitis-exposed monocytes had multiple unique H3K4me3 peaks inside gene RFTN2 (Raftlin family member 2) that were not present in unexposed monocytes. These visual representations were obtained using the UCSC genome browser at http://genome.ucsc.edu
Figure 5: Representative common H3K4me3 peaks in chorioamnionitis-exposed monocytes at different gestations.
A) Chorioamnionitis-exposed monocytes at all gestational ages shared a common H3K4me3 peak inside the gene TINAGL1 (Tubulointerstitial nephritis antigen like 1). B) preterm and term chorioamnionitis-exposed monocytes shared a common H3K4me3 peak inside the gene GALNTL6 (Polypeptide N-acetylgalactosaminyltransferase like 6) that was not present in extremely preterm chorioamnionitis-exposed monocytes. These visual representations were obtained using the UCSC genome browser at http://genome.ucsc.edu
Figure 6: Term healthy and term chorioamnionitis exposed monocytes had distinctly different H3K4me3 binding affinities.
Heatmap demonstrating affinities for differentially bound H3K4me3 peaks between extremely preterm, preterm and term chorioamnionitis-exposed and unexposed monocytes. The binding affinity of differentially bound H3K4me3 in extremely preterm and preterm healthy and chorioamnionitis-exposed monocytes varied both within gestational age and exposure groups. In contrast, all the replicates within term healthy and term chorioamnionitis-exposed monocytes had similar differentially bound H3K4me3 sites, which were very different between exposure groups. U30(n=3)=under 30 weeks gestation non-chorioamnionitis-exposed neonates, U30C(n=3)=under 30 weeks gestation chorioamnionitis-exposed neonates, O30(n=3)=30–36 weeks gestation non-chorioamnionitis-exposed neonates, O30C(n=3)=30–36 weeks gestation chorioamnionitis-exposed neonates, Term(n=3)=37+ weeks gestation non-chorioamnionitis-exposed neonates, TermC(n=3)=37+ weeks gestation chorioamnionitis-exposed neonates.
Table 5:
Differentially bound H3K4me3 peaks
| Group 1 | Group 2 | Total Differentially Bound H3K4me3 Peaks | H3K4me3 Peaks Increased in Group 1 | H3K4me3 Peaks Increased in Group 2 |
|---|---|---|---|---|
| U30a | U30Ca,d | 1 | 0 | 1 |
| O30b | O30Cb,d | 68 | 37 | 31 |
| Termc | TermCc,d | 9633 | 4387 | 5246 |
| U30Ca,d | O30Cb,d | 60 | 48 | 12 |
| U30Ca,d | TermCc,d | 6280 | 1141 | 5139 |
| O30Cb,d | TermCc,d | 4600 | 2975 | 1625 |
U30 = under 30 weeks gestation,
O30 = 30–36 weeks gestation,
Term = term gestation,
C=chorioamnionitis exposure
Figure 7: Representative differentially bound H3K4me3 peaks in term healthy and chorioamnionitis-exposed monocytes.
A) Term healthy monocytes had increased H3K4me3 binding affinity at the promoter site of IL6ST (Interleukin 6 signal transducer) when compared to term chorioamnionitis-exposed monocytes. B) Term chorioamnionitis-exposed monocytes had increased H3K4me3 binding affinity inside gene CD84 (Leukocyte differentiation antigen CD84) when compared to term unexposed monocytes. These visual representations were obtained using the UCSC genome browser at http://genome.ucsc.edu. Differential binding was determined and p-values were calculated using the DiffBind package.
Table 6:
Gene ontology biological pathways enriched for differentially bound H3K4me3
| Groups | Increased in Group | GO Term | Term | DBe Genes in Pathway | p-value | FDRf |
|---|---|---|---|---|---|---|
| U30a vs U30Ca,d | U30 | None | None | None | None | None |
| U30a vs U30Ca,d | U30C | None | None | None | None | None |
| O30b vs O30Cb,d | O30 | None | None | None | None | None |
| O30b vs O30Cb,d | O30C | None | None | None | None | None |
| Termc vs TermCc,d | Term | 0000209 | Protein polyubiquitination | 77 | 3.24E-15 | 6.22E-12 |
| Term | 0043161 | Proteasome-mediated ubiquitin-dependent protein catabolic process | 78 | 4.61E-13 | 8.91E-12 | |
| Term | 0006468 | Protein phosphorylation | 131 | 2.47E-10 | 4.77E-7 | |
| Term | 0045893 | Positive regulation of transcription, DNA-templated | 143 | 4.63E-10 | 8.94E-7 | |
| Term | 0006357 | Regulation of transcription from RNA polymerase II promoter | 124 | 3.24E-9 | 6.26E-6 | |
| Term | 0006915 | Apoptotic process | 150 | 6.28E-9 | 1.21E-5 | |
| Term | 0016567 | Protein ubiquitination | 103 | 2.56E-8 | 4.95E-5 | |
| Term | 0016032 | Viral process | 88 | 8.42E-8 | 1.63E-4 | |
| Term | 0098609 | Cell-cell adhesion | 81 | 1.42E-7 | 2.74E-4 | |
| Term | 0002223 | Stimulatory C-type lectin receptor signaling pathway | 40 | 4.86E-7 | 9.39E-4 | |
| Term | 0018105 | Peptidyl-serine phosphorylation | 45 | 5.37E-7 | 0.001 | |
| Term | 0048010 | Vascular endothelial growth factor receptor signaling pathway | 31 | 5.9E-7 | 0.0011 | |
| Term | 0045892 | Negative regulation of transcription, DNA-templated | 127 | 9.56E-7 | 0.0018 | |
| Term | 0030036 | Actin cytoskeleton organization | 45 | 1.83E-6 | 0.0035 | |
| Term | 0015031 | Protein transport | 104 | 2.01E-6 | 0.0039 | |
| Term | 0050852 | T cell receptor signaling pathway | 49 | 2.53E-6 | 0.0049 | |
| Term | 0042147 | Retrograde transport, endosome to Golgi | 29 | 2.61E-6 | 0.005 | |
| Term | 0007050 | Cell cycle arrest | 47 | 3.39E-6 | 0.0065 | |
| Term | 0006914 | Autophagy | 45 | 3.64E-6 | 0.007 | |
| Term | 0016569 | Covalent chromatin modification | 40 | 4.04E-6 | 0.0078 | |
| Term | 0007049 | Cell cycle | 64 | 5.31E-6 | 0.01 | |
| Term | 0006897 | Endocytosis | 46 | 5.43E-6 | 0.01 | |
| Term | 0038095 | Fc-epsilon receptor signaling pathway | 55 | 6E-6 | 0.012 | |
| Term | 0070936 | Protein K48-linked ubiquitination | 22 | 7.68E-6 | 0.015 | |
| Term | 0046777 | Protein autophosphorylation | 52 | 2.15E-5 | 0.042 | |
| Term | 0016241 | Regulation of macroautophagy | 20 | 3.73E-5 | 0.072 | |
| Term | 0043065 | Positive regulation of apoptotic process | 79 | 3.75E-5 | 0.072 | |
| Term | 0051301 | Cell division | 89 | 4.76E-5 | 0.092 | |
| Term | 0034976 | Response to endoplasmic reticulum stress | 28 | 5.11E-5 | 0.099 | |
| Termc vs TermCc,d | TermC | 0045944 | Positive regulation of transcription from RNA polymerase II promoter | 128 | 7.2E-8 | 1.35E-4 |
| TermC | 0061337 | Cardiac conduction | 16 | 1.39E-6 | 0.0026 | |
| TermC | 0007267 | Cell-cell signaling | 43 | 7.61E-6 | 0.014 | |
| U30Ca,d vs O30Cb,d | U30C | None | None | None | None | None |
| U30Ca,d vs O30Cb,d | O30C | None | None | None | None | None |
| U30Ca,d vs TermCc,d | U30C | 0000122 | Negative regulation of transcription from RNA polymerase II promoter | 51 | 4E-6 | 0.007 |
| TermC | 0045944 | Positive regulation of transcription from RNA polymerase II promoter | 119 | 4.92E-6 | 0.009 | |
| O30Cb,d vs TermCc,d | O30C | 0000209 | Protein polyubiquitination | 59 | 4.02E-13 | 7.62E-10 |
| O30C | 0006351 | Transcription, DNA-templated | 321 | 1.69E-11 | 3.21E-8 | |
| O30C | 0043161 | Proteasome-mediated ubiquitin-dependent protein catabolic process | 58 | 1.14E-10 | 2.16E-7 | |
| O30C | 0006357 | Regulation of transcription from RNA polymerase II promoter | 94 | 6.17E-9 | 1.17E-5 | |
| O30C | 0016032 | Viral process | 70 | 1.39E-8 | 2.64E-5 | |
| O30C | 0098609 | Cell-cell adhesion | 65 | 1.82E-8 | 3.45E-5 | |
| O30C | 0006915 | Apoptotic process | 111 | 3.26E-8 | 6.18E-5 | |
| O30C | 0006468 | Protein phosphorylation | 94 | 3.41E-8 | 6.46E-5 | |
| O30C | 0002223 | Stimulatory C-type lectin receptor signaling pathway | 34 | 4.87E-8 | 9.23E-5 | |
| O30C | 0070936 | Protein K48-linked ubiquitination | 21 | 7.97E-8 | 1.51E-4 | |
| O30C | 0045944 | Positive regulation of transcription from RNA polymerase II promoter | 164 | 1.04E-6 | 0.002 | |
| O30C | 0038095 | Fc-epsilon receptor signaling pathway | 44 | 2.02E-6 | 0.0038 | |
| O30C | 0050852 | T cell receptor signaling pathway | 38 | 4.37E-6 | 0.008 | |
| O30C | 0000122 | Negative regulation of transcription from RNA polymerase II promoter | 124 | 6.06E-6 | 0.011 | |
| O30C | 0007049 | Cell cycle | 49 | 7.43E-6 | 0.014 | |
| O30C | 0042147 | Retrograde transport, endosome to Golgi | 22 | 2.24E-5 | 0.042 | |
| O30C | 0007050 | Cell cycle arrest | 35 | 2.39E-5 | 0.045 | |
| O30C | 0030036 | Actin cytoskeleton organization | 33 | 2.64E-5 | 0.05 | |
| O30C | 0051403 | Stress-activated MAPK cascade | 11 | 4.61E-5 | 0.087 | |
| O30Cb,d vs TermCc,d | TermC | None | None | None | None | None |
U30 = under 30 weeks gestation,
O30 = 30–36 weeks gestation,
Term = term gestation,
C=chorioamnionitis exposure,
DB=differentially bound,
FDR=false discovery rate
Chorioamnionitis exposure changed neonatal monocyte transcription both in unstimulated and LPS stimulated conditions
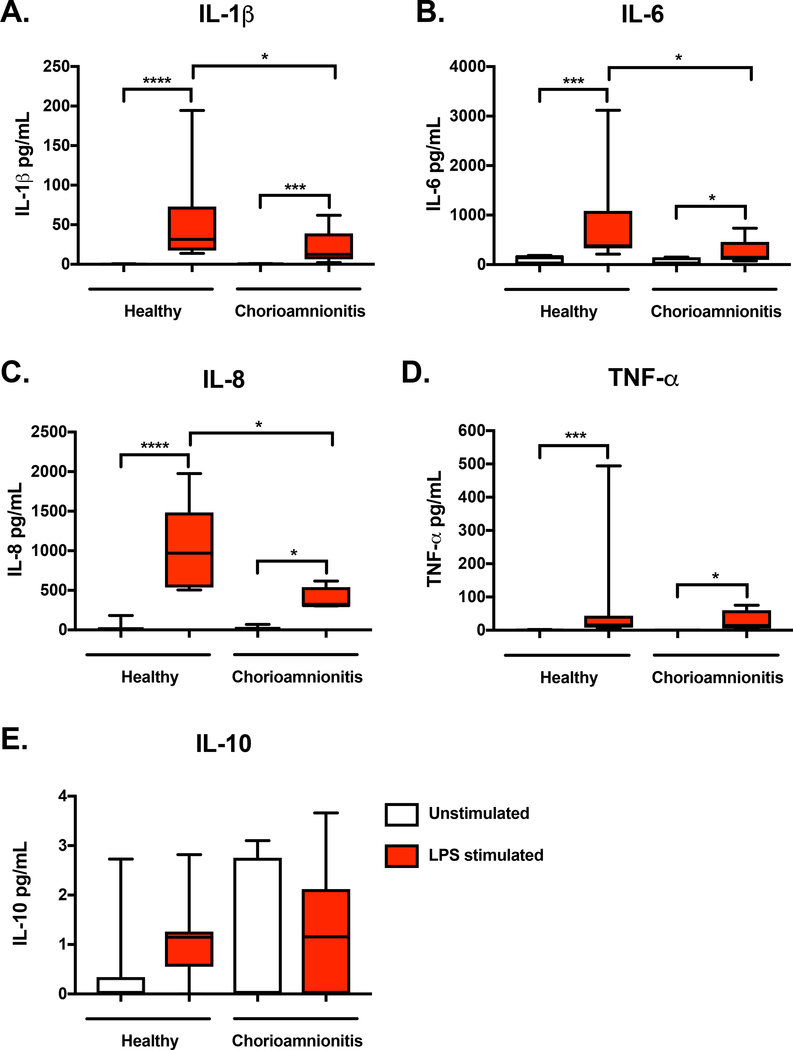
In order to determine the impact of this chorioamnionitis induced global monocyte H3K4me3 landscape remodeling on gene transcription, we performed RNA-seq on a separate cohort of infants. Given the low H3K4me3 peak abundance in preterm monocytes, we chose to focus solely on term monocytes. Table 1 compares the patient characteristics between RNA-seq groups. The healthy and chorioamnionitis-exposed groups were well matched for gestational age, birth weight, sex, mode of delivery, maternal smoking, gestational hypertension and gestational diabetes. RNA-seq was performed on umbilical cord blood purified CD14+ monocytes from four different experimental groups: term healthy unstimulated monocytes (U), term healthy LPS stimulated monocytes (L), term chorioamnionitis-exposed unstimulated monocytes (CU) and term chorioamnionitis-exposed LPS stimulated monocytes (CL). RNA-seq of unstimulated and LPS stimulated monocytes was performed to determine if chorioamnionitis exposure impacted gene transcription in a resting state and if it altered gene transcription upon a secondary pathogenic stimulus. A principal component analysis of normalized transcript read counts revealed that the unstimulated chorioamnionitis-exposed and unexposed monocytes clustered together, the healthy LPS stimulated monocytes clustered together, but the chorioamnionitis-exposed LPS stimulated monocytes displayed significantly more variance (Fig. 8A). A pearson’s correlation heatmap of normalized read counts of differentially expressed transcripts revealed that the differentially expressed transcripts were very consistent between replicates, with separation between exposure groups (Fig. 8B-C). An overview of the differentially expressed transcripts between exposure and treatment groups is presented in Table 7 with a more detailed list in Supplemental Data. As expected, LPS stimulation of non-chorioamnionitis exposed monocytes resulted in increased mRNA expression in gene ontology pathways related to cytokine activity and the inflammatory response (Table 8). Of note, there was increased expression of the immune related genes IL6, IFIT2, IFI44 and CSF3 after LPS stimulation in the unexposed monocytes, which is consistent with previously published data[19]. In contrast, LPS stimulation of chorioamnionitis-exposed monocytes did not result in enriched transcript expression in any gene ontology pathways, although there was increased expression of the immune related genes IL6 and CSF3 (Table 8). This suggests that chorioamnionitis exposure results in a dysregulated monocyte inflammatory response when a secondary pathogen was encountered. Gene ontology pathways enriched for differentially expressed transcripts between chorioamnionitis-exposed and unexposed monocytes primarily involved oxygen and bicarbonate transport, with a large induction in hemoglobin associated genes in the chorioamnionitis-exposed monocytes, both in unstimulated and LPS stimulated conditions (Table 8). Network analysis of differentially expressed transcripts in either LPS stimulated healthy or chorioamnionitis-exposed monocytes showed significant overlap in KEGG pathways with gene enrichment in both groups, including response to infection and TGF-beta signaling (Figure 9A-B). Given the lack of gene enrichment in distinct immunologic pathways between chorioamnionitis-exposed and healthy monocytes, we sought to determine if there was a difference in monocyte function upon a secondary inflammatory exposure. To assess this, we measured inflammatory cytokine expression 24 hours after LPS stimulation. Both healthy and chorioamnionitis-exposed term monocytes had significantly increased expression of the pro-inflammatory cytokines IL-1β, IL-6, IL-8 and TNF-α compared to the unstimulated controls (Fig 10A-D). There was very little IL-10 detected in either the unstimulated or LPS stimulated samples (Fig. 10E). There was decreased expression of the pro-inflammatory cytokines IL-1β, IL-6 and IL-8 in chorioamnionitis-exposed term monocytes when compared to term healthy monocytes (Fig. 10A-C). This indicates that while there are no clear differences in transcript expression in distinct inflammatory pathways when monocytes with previous chorioamnionitis exposure encounter a secondary pathogen, there is a difference in monocyte function, particularly pro-inflammatory cytokine expression.
Figure 8: Chorioamnionitis exposure altered global term neonatal monocyte gene expression both in unstimulated and LPS stimulated conditions.
A) Principal component analysis of RNA-seq normalized read counts using ggplot2 (http://ggplot.tidyverse.org). B) Clustered pearson correlation of differentially expressed mRNA transcripts using normalized read counts revealed that unstimulated healthy and chorioamnionitis-exposed term neonatal monocytes had distinct transcript expression profiles. U(n=3)=term healthy unstimulated monocytes, CU(n=2)=term chorioamnionitis-exposed unstimulated monocytes. C) Clustered pearson correlation of differentially expressed mRNA transcripts based on normalized read counts showed that LPS stimulated healthy and chorioamnionitis-exposed monocytes had distinct patterns of RNA expression. L(n=3)=term healthy LPS stimulated monocytes, CL(n=3)=term chorioamnionitis-exposed LPS stimulated monocytes. Differentially expressed mRNA transcripts were determined using the edgeR package.
Table 7:
Differentially expressed mRNA transcripts
| Group 1 | Group 2 | Total Differentially Expressed Transcripts | Transcripts Increased in Group 1 | Transcripts Increased in Group 2 |
|---|---|---|---|---|
| Ua | Lb | 516 | 266 | 250 |
| CUc | CLd | 304 | 223 | 81 |
| Ua | CUc | 135 | 23 | 112 |
| Lb | CLd | 594 | 263 | 331 |
U=term healthy unstimulated,
L=term healthy LPS stimulated,
CU=term chorioamnionitis-exposed unstimulated,
CL=term chorioamnionitis-exposed LPS stimulated
Table 8:
Gene ontology pathways enriched for differentially expressed transcripts
| Groups | Increased in Group | GO Term | Term | Ontology | DEe Genes in Pathway | p-value | Benjamini |
|---|---|---|---|---|---|---|---|
| Ua vs Lb | |||||||
| U | 0003676 | Nucleic acid binding | MF | 29 | 8.3E-6 | 2.8E-3 | |
| U | 0046872 | Metal ion binding | MF | 45 | 3.4E-5 | 5.6E-3 | |
| L | 0005615 | Extracellular space | CC | 32 | 1.3E-5 | 2.6E-03 | |
| L | 0005125 | Cytokine activity | MF | 10 | 6.5E-5 | 2.0E-02 | |
| L | 0006954 | Inflammatory response | BP | 15 | 3.7E-5 | 4E-2 | |
| CUc vs CLd | |||||||
| U | None | None | None | None | None | None | |
| L | None | None | None | None | None | None | |
| Ua vs CUc | |||||||
| U | None | None | None | None | None | None | |
| CU | 0015671 | Oxygen transport | BP | 6 | 7.1E-9 | 3.3E-6 | |
| CU | 0005344 | Oxygen transporter activity | MF | 6 | 3.8E-9 | 6E-7 | |
| CU | 0015701 | Bicarbonate transport | BP | 7 | 6.9E-8 | 1.6E-05 | |
| Lb vs CLd | |||||||
| L | None | None | None | None | None | None | |
| CL | 0005344 | Oxygen transporter activity | MF | 8 | 3.9E-10 | 1.4E-07 | |
| CL | 0015671 | Oxygen transport | BP | 8 | 7.5E-10 | 8.6E-07 | |
| CL | 0019825 | Oxygen binding | MF | 9 | 3.4E-7 | 6.2E-05 | |
| CL | 0005506 | Iron ion binding | MF | 11 | 8.1E-5 | 9.7E-03 |
U=term healthy unstimulated monocytes,
L=term healthy LPS stimulated monocytes,
U=term chorioamnionitis-exposed unstimulated monocytes,
CL=term chorioamnionitis-exposed LPS stimulated monocytes,
DE=differentially expressed
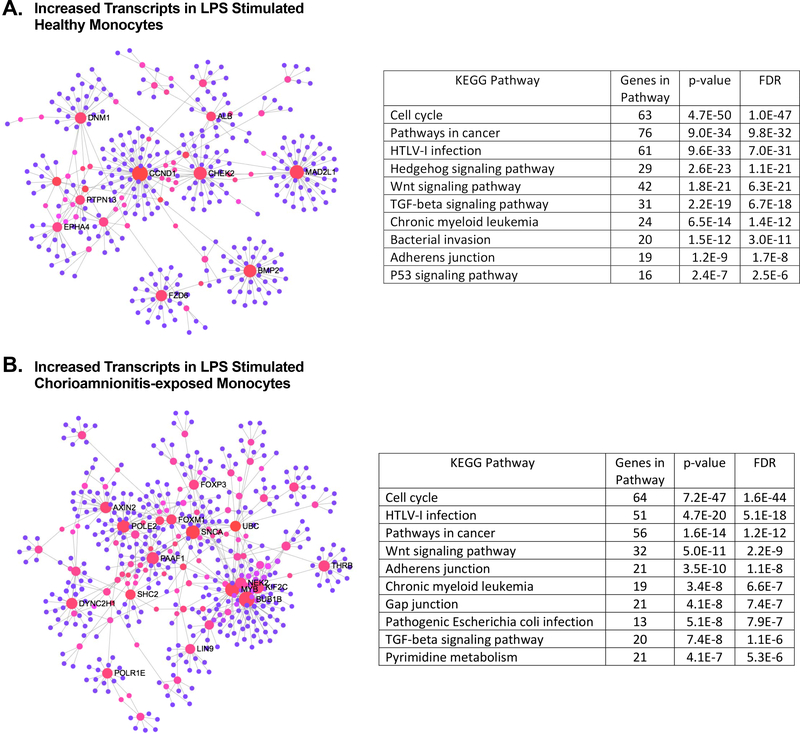
Figure 9: There is significant overlap in KEGG pathways containing enriched differentially expressed transcripts between LPS stimulated healthy and chorioamnionitis-exposed term neonatal monocytes.
A) Network of differentially expressed transcripts with increased expression in LPS stimulated term healthy monocytes. The top 10 KEGG pathways with enriched transcripts are presented. B) Network of differentially expressed transcripts with increased expression in LPS stimulated term chorioamnionitis-exposed monocytes. The top 10 KEGG pathways containing enriched transcripts are detailed. Network visualization was performed and p-values were derived using NetworkAnalyst.ca.
Figure 10: Term chorioamnionitis-exposed monocytes exhibit decreased pro-inflammatory cytokine expression when a secondary inflammatory stimulus is encountered.
Protein expression of A) IL-1β, B) IL-6, C) IL-8, D) TNF-α and E) IL-10 were measured in unstimulated and LPS stimulated conditions (100 ng/mL) after 24 hours in culture. Box represents 25th–75th percentile range, whiskers represent minimum to maximum values, line represents median. n=10 healthy term monocytes, n=9 chorioamnionitis-exposed term monocytes, *p<0.05, ***p<0.001, ****p<0.0001. Differences between groups were assessed using the Kruskal-Wallis test with multiple comparisons.
H3K4me3 peak presence in neonatal monocytes predicted RNA expression
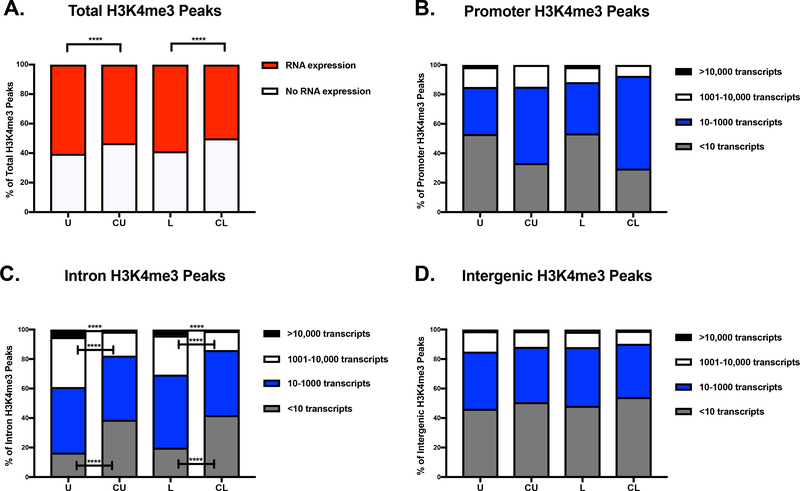
We then took consensus H3K4me3 peaks that annotated to known genes and compared them to normalized transcript read counts to evaluate if the presence and location of H3K4me3 peaks around a gene was associated with transcript expression. RNA expression was considered present if the normalized read count was greater than 10 using the edgeR package[20]. In healthy term monocytes, the presence of an H3K4me3 peak within or around a known gene was associated with RNA expression 60.4% of the time in unstimulated cells and 58.7% of the time in LPS stimulated cells. In chorioamnionitis-exposed term monocytes the presence of an H3K4me3 peak within or around a known gene was associated with significantly less RNA expression, only 53.2% of the time in unstimulated cells and 50% of the time in LPS stimulated cells (Fig. 11A). Next, we evaluated if the ability of H3K4me3 peaks to predict gene expression was dependent on its location, as H3K4me3 is known to be primarily located at promoter regions of actively transcribed genes[21]. Promoter H3K4me3 peaks in the healthy monocytes were associated with RNA expression less than 50% of the time, while promoter H3K4me3 peaks in the chorioamnionitis-exposed monocytes were associated with RNA expression over 50% of the time (Fig. 11B). Of note, there were few promoter H3K4me3 consensus peaks in the chorioamnionitis-exposed monocytes (92), with a significantly higher number in the healthy monocytes (292), making it difficult to draw any conclusions from these findings. In contrast, there were significantly more intron H3K4me3 peaks in the chorioamnionitis-exposed monocytes that were not associated with gene expression. Furthermore, significantly more intron H3K4me3 peaks in the healthy monocytes were associated with high level RNA expression (Fig. 11C). Interestingly, intergenic H3K4me3 peaks, which could be as distant as 800 kb from the gene of interest, predicted RNA expression approximately 50% of the time in all exposure and treatment groups (Fig. 11D). Overall, the presence of H3K4me3 near a gene predicted RNA expression approximately 50% of the time, independent of the actual location of the H3K4me3. Next, we compared differentially bound H3K4me3 peaks with differentially expressed mRNA transcripts between exposure and treatment groups (Table 9 and Table 10). Overall, approximately 10% of differentially expressed transcripts were associated with differentially bound H3K4me3 peaks. Conversely, less than 2% of differentially bound H3K4me3 peaks were associated with differentially expressed mRNA transcripts.
Figure 11: H3K4me3 peak presence near a known gene in neonatal monocytes was a good predictor of active gene transcription.
A) Total consensus H3K4me3 peaks that annotate to known genes in healthy and chorioamnionitis-exposed term neonatal monocytes compared to known annotated mRNA transcripts. mRNA expression was defined as a normalized read count of >10. Consensus H3K4me3 peaks that annotated to known genes compared to quantitative normalized mRNA transcript levels at B) promoter sites, C) introns and D) intergenic sites. U(n=3)=term healthy unstimulated monocytes, CU(n=2)=term chorioamnionitis-exposed unstimulated monocytes, L(n=3)=term healthy LPS stimulated monocytes, CL(n=3)=term chorioamnionitis-exposed LPS stimulated monocytes. Chi-square test was used to evaluate differences in proportions between groups, ****p<0.0001.
Table 9:
Comparison of differentially bound H3K4me3 and differentially expressed mRNA transcripts
| Increased Differentially Bound H3K4me3 | Increased Differential mRNA Transcripts | Comparison Group | Total Non-duplicated Differentially Bound H3K4me3 | Total Differential mRNA Transcripts | Shared Differential H3K4me3 and mRNA Transcripts |
|---|---|---|---|---|---|
| Healthya | Uc | CUe | 3386 | 23 | 1 |
| Healthya | Ld | CLf | 3386 | 263 | 23 |
| Choriob | CUe | Uc | 1930 | 112 | 9 |
| Choriob | CLf | Ld | 1930 | 331 | 31 |
Healthy=term healthy monocytes,
Chorio=term chorioamnionitis-exposed monocytes,
U=term healthy unstimulated,
L=term healthy LPS stimulated,
CU=term chorioamnionitis-exposed unstimulated,
CL=term chorioamnionitis-exposed LPS stimulated
Table 10:
Locations of shared differentially bound H3K4me3 and differentially expressed mRNA
| Increased Differentially Bound H3K4me3 | Increased Differential mRNA Transcripts | Comparison Group | Gene | H3K4me3 Peak Location | Differentially Bound H3K4me3 p-value | Differentially Expressed mRNA Transcript p-value |
|---|---|---|---|---|---|---|
| Healthya | Uc | CUe | STXBP5-AS1 | Promoter | 2.58E-21 | 4E-4 |
| Healthya | Ld | CLf | ITGB3BP | Promoter | 3.07E-31 | 0.0019 |
| TPGS1 | Promoter | 2.28E-29 | 6.5E-4 | |||
| TACSTD2 | Inside gene | 1.06E-26 | 0.0016 | |||
| GPSM2 | Inside gene | 2.19E-24 | 0.0013 | |||
| LRR1 | Promoter | 8.4E-24 | 0.0025 | |||
| RFNG | Promoter | 1.6E-23 | 0.0035 | |||
| PIGZ | Upstream | 2.59E-23 | 2.5E-4 | |||
| PGM2 | Inside gene | 2.76E-23 | 0.001 | |||
| RGS9BP | Promoter | 2.87E-23 | 2E-4 | |||
| FRS3 | Upstream | 3.24E-19 | 2E-4 | |||
| NAA50 | Inside gene | 6.06E-19 | 0.0034 | |||
| ZNF747 | Promoter | 2.15E-18 | 5.2E-4 | |||
| IMPACT | Inside gene | 3.95E-18 | 0.0034 | |||
| FKBP9 | Promoter | 2.21E-17 | 4.3E-5 | |||
| PRRG4 | Promoter | 4.34E-17 | 1.3E-5 | |||
| NDC1 | Inside gene | 4.63E-17 | 0.0028 | |||
| PKIB | Upstream | 6.33E-17 | 1.2E-5 | |||
| RALGAPA2 | Promoter | 8.71E-16 | 9.9E-5 | |||
| B4GALT6 | Upstream | 6.67E-15 | 0.0041 | |||
| SNX33 | Promoter | 6.68E-15 | 2.3E-4 | |||
| KCTD3 | Promoter | 4.24E-13 | 2.5E-4 | |||
| TTC12 | Promoter | 5.72E-12 | 3E-5 | |||
| MAD2L1 | Promoter | 1.63E-11 | 7.2E-5 | |||
| Choriob | CUe | Uc | RASEF | Downstream | 1.1E-6 | 1.4E-4 |
| LRRC70 | Downstream | 9.4E-6 | 5.7E-4 | |||
| KLHDC9 | Upstream | 1.3E-4 | 8.7E-5 | |||
| CFAP46 | Inside gene | 9E-4 | 1.6E-4 | |||
| KEL | Upstream | 0.0017 | 1.6E-4 | |||
| SARDH | Upstream | 0.0025 | 3.7E-5 | |||
| ZNF608 | Downstream | 0.0026 | 3.6E-5 | |||
| CLEC1B | Upstream | 0.0045 | 1.7E-4 | |||
| MAP9 | Upstream | 0.015 | 2.6E-4 | |||
| Choriob | CLf | Ld | LRGUK | Inside gene | 4.1E-9 | 0.0017 |
| DNAJC5B | Upstream | 3.6E-8 | 4E-4 | |||
| HIBCH | Upstream | 2E-6 | 0.0042 | |||
| FOXP3 | Inside gene | 3.1E-6 | 5.5E-4 | |||
| ALAS2 | Inside gene | 2.7E-5 | 0.002 | |||
| MECOM | Inside gene | 2.8E-5 | 6.9E-4 | |||
| ARV1 | Inside gene | 3.9E-5 | 0.0014 | |||
| KLHDC9 | Upstream | 1.3E-4 | 3.9E-5 | |||
| MRPL45P2 | Downstream | 2E-4 | 1.8E-4 | |||
| TRUB1 | Upstream | 2.2E-4 | 7E-5 | |||
| CACNA2D2 | Inside gene | 2.3E-4 | 5.3E-4 | |||
| EXTL2 | Downstream | 4.8E-4 | 0.0031 | |||
| LGR5 | Upstream | 8.9E-4 | 0.0014 | |||
| NMNAT2 | Inside gene | 0.0011 | 0.0022 | |||
| MIR652 | Downstream | 0.0013 | 8.7E-4 | |||
| DMBX1 | Upstream | 0.0025 | 1.3E-4 | |||
| DAPK2 | Inside gene | 0.0033 | 5.5E-5 | |||
| RASGRF1 | Inside gene | 0.0035 | 4.1E-5 | |||
| CEP44 | Upstream | 0.0036 | 0.0015 | |||
| CETN3 | Downstream | 0.0043 | 0.0023 | |||
| KCNIP4 | Inside gene | 0.005 | 1.4E-4 | |||
| CMKLR1 | Upstream | 0.0055 | 2.4E-5 | |||
| MDGA1 | Promoter | 0.01 | 0.0023 | |||
| TTYH1 | Upstream | 0.01 | 0.0022 | |||
| DYNC2H1 | Inside gene | 0.014 | 6.8E-8 | |||
| AXIN2 | Downstream | 0.014 | 2.9E-4 | |||
| ZNF385C | Upstream | 0.022 | 0.0011 | |||
| NCAPH | Downstream | 0.024 | 1E-5 | |||
| DET1 | Upstream | 0.035 | 0.0024 | |||
| VIL1 | Upstream | 0.035 | 0.0038 | |||
| ACADSB | Downstream | 0.036 | 2.2E-4 |
Healthy=term healthy monocytes,
Chorio=term chorioamnionitis-exposed monocytes,
U=term healthy unstimulated,
L=term healthy LPS stimulated,
CU=term chorioamnionitis-exposed unstimulated,
CL=term chorioamnionitis-exposed LPS stimulated
Discussion
Although chorioamnionitis is a frequent complication encountered around the time of delivery, there are few studies in humans that have investigated its effect on neonatal immune cell development and function. In this study we demonstrated that exposure to chorioamnionitis resulted in global remodeling of the monocyte H3K4me3 landscape, with both removal and deposition of H3K4me3 in extremely preterm, preterm and term neonates. In preterm and term neonates this remodeling resulted in an overall gain in monocyte H3K4me3, which was primarily occurring in introns and intergenic regions. We also demonstrated that chorioamnionitis exposure altered global term neonatal monocyte transcription, both at baseline and after a secondary pathogenic stimulus. Furthermore, monocyte H3K4me3 peaks located near known genes was associated with mRNA expression over 50% of the time, with better correlation noted in non-chorioamnionitis-exposed monocytes. However, differentially bound H3K4me3 peaks between healthy and chorioamnionitis-exposed monocytes were a poor predictor of differentially expressed mRNA.
We have previously demonstrated that neonatal monocytes gain H3K4me3 as development progresses from extremely preterm neonate to adult. We have also shown that the majority of H3K4me3 peaks in neonatal monocytes are located in intergenic regions while they are located in promoter and exon regions in adult monocytes[17]. Here we demonstrate that chorioamnionitis exposure resulted in a global remodeling of the neonatal monocyte H3K4me3 landscape, with both removal and deposition of H3K4me3 that resulted in an overall gain of H3K4me3 in preterm and term monocytes. Interestingly, this H3K4me3 gain was primarily in introns and intergenic regions, so although the total number of H3K4me3 peaks in term chorioamnionitis-exposed monocytes approximates that of adult monocytes, the location of the peaks and global H3K4me3 landscape were strikingly different. This demonstrates that the gain of H3K4me3 that occurs over the course of development from neonate to adult can be disrupted by a pathogenic inflammatory exposure in the neonatal period. We suspect that developmentally related H3K4me3 removal and deposition occur in a very nuanced manner, requiring a series of appropriately timed specific stimuli for this process to occur effectively. It is clear that exposure to chorioamnionitis disrupts this process, but the long-term implications of this are unknown.
The H3K4me3 peaks in preterm and term healthy monocytes are broader than the peaks in the chorioamnionitis-exposed monocytes of the same gestation. Broad H3K4me3 peaks, typically defined as greater than 1.5 kb, are associated with enhanced gene expression[22, 23]. In this study there is a significantly larger correlation between H3K4me3 peak presence and mRNA expression in term healthy monocytes (58–60% of H3K4me3 peaks are associated with mRNA transcript expression) when compared to term chorioamnionitis-exposed monocytes (50–53% of H3K4me3 peaks are associated with mRNA transcript expression). It is possible that the difference in H3K4me3 peak width between exposure groups is contributing to this difference in mRNA expression. H3K4me3 is an activating histone tail modification that is primarily believed to be located at promoter and 5’-coding regions of actively transcribed genes[21, 24]. It is interesting that upon chorioamnionitis exposure H3K4me3 is removed from promoter regions and deposited in introns and intergenic regions, where is does not have a well-described function. It is also surprising that H3K4me3 peaks predicted gene expression greater than 50% of the time regardless of its location. This demonstrates that H3K4me3 is not only associated with active gene transcription when it is located at promoter regions, but can also direct active gene transcription of nearby genes when located in introns and intergenic regions as well. It has been postulated that some 3’ H3K4me3 peaks have a role in antisense transcription[25] and recent studies have demonstrated that non-promoter H3K4me3 can act as transcriptional enhancers in T-cells and cancer cell lines[26, 27]. Therefore, it is plausible that non-promoter monocyte H3K4me3 peaks can influence the expression of genes located far distances away, in part due to the three-dimensional structure and extensive folding of cellular DNA. It is unclear why differential H3K4me3 binding between exposure groups did not predict differential mRNA expression. It may be that the presence or absence of H3K4me3 is more important in influencing gene expression than smaller differences in abundance or affinity that differential binding algorithms highlight. Additionally, the ChIP-seq and RNA-seq experiments were performed on different patients, which may have contributed to this poor correlation. It is also likely that there are factors in addition to H3K4me3 influencing mRNA expression that were not accounted for in this study.
To date, studies evaluating the impact of chorioamnionitis exposure on human neonatal immune function have focused primarily on preterm neonates. It is true that chorioamnionitis is more prevalent in preterm births, but it still complicates up to 4% of term deliveries[2, 28]. Studies focusing on neonatal adaptive immunity have shown that chorioamnionitis exposure amplifies Th17 and Th1 type responses in preterm infants[29, 30]. A study focusing on both innate and adaptive immunity employed a broad transcriptomic approach, using a microarray to evaluate mRNA expression in whole blood of preterm neonates with and without chorioamnionitis exposure[6]. While this approach revealed that chorioamnionitis exposure altered gene expression in both innate and adaptive immune pathways, the approach made it impossible to determine which cells were responsible for these differences. A more recent study performed RNA-seq on purified cord blood monocytes from very preterm neonates (<32 weeks) with and without chorioamnionitis exposure both before and after stimulation with the most common etiology of neonatal late onset sepsis, Staphylococcus epidermidis[7]. This study found that chorioamnionitis exposure in preterm infants is associated with a hypo-responsive transcriptional profile, which may alter a preterm infants’ risk of sepsis. While chorioamnionitis in term pregnancies is a less common and more heterogeneous condition than chorioamnionitis in preterm pregnancies, it can still result in a significant perinatal inflammatory exposure that has the potential to impact neonatal immune function. We have provided evidence here that chorioamnionitis exposure in term pregnancies resulted in altered monocyte gene transcription, with less induction of genes involved in the inflammatory cascade, consistent with a hypo-responsive transcriptional profile. This translated to decreased pro-inflammatory cytokine expression when monocytes with previous chorioamnionitis exposure encountered a secondary inflammatory stimulus. Chorioamnionitis exposure was also noted to upregulate genes vital to oxygen and gas exchange, including numerous hemoglobin-associated transcripts. Suboptimal tissue gas exchange in hemoglobinopathies is thought to result in compensatory hemoglobin synthesis in different cell lineages, including monocytes[31]. Inflammation associated immune cell activation causes increased oxygen consumption and cellular respiration, so it is possible that the chorioamnionitis-exposed monocytes are upregulating hemoglobin-associated transcripts in an attempt to compensate for this[32].
Both term and preterm fetuses exposed to chorioamnionitis can mount a systemic inflammatory response, resulting in elevated fetal cortisol levels and fetal immune cell activation with increased expression of the cytokines and chemokines IL-1β, IL-6, TNF-α, G-CSF and CXCL1[33–37]. There is also an association between chorioamnionitis exposure and the development of early onset neonatal sepsis that persists even after adjusting for confounding factors such as birth weight and gestational age[38–41]. This association has been attributed to either a common infectious agent leading to both conditions or as a result of immune system reprogramming that results in increased susceptibility to infection in the early neonatal period. A growing body of evidence points to immune system reprogramming, including the fact that maternal antibiotic administration does not impact the increased early onset sepsis risk associated with chorioamnionitis exposure[42, 43]. Our study also supports the immune system reprogramming hypothesis, and demonstrates that exposure to chorioamnionitis results in alteration in the neonatal monocyte transcriptosome, both at baseline and with a more pronounced difference upon a secondary pathogenic stimulus. LPS was chosen as the secondary pathogenic stimulus as gram-negative organisms, primarily Escherichia coli, are some of the leading causes of early onset neonatal sepsis and are one of the earliest pathogenic exposures a neonate is likely to encounter[44]. It is currently unclear how long this chorioamnionitis induced immune system reprogramming lasts, as studies are conflicting about whether chorioamnionitis exposure is protective against or increases susceptibility to neonatal late onset sepsis, which occurs beyond the first 72 hours of life[38, 45, 46]. Little is known about the impact of chorioamnionitis exposure on the susceptibility to other bacterial or viral infections in the neonatal, infant and childhood periods, although further studies specifically addressing this question would be informative.
We realize that this study has several limitations. All of the patients included in this study were recruited from a single center, which could have introduced bias into the results. We attempted to control for interpatient variability and genetic diversity by pooling patient samples and using separate patient groups for the ChIP-seq and RNA-seq experiments, but this approach may have masked the identification of important differences between individual patients. The purity of the CD14+ isolation was 92%, which could have resulted in non-monocyte population transcripts being included in the RNA-seq analysis. We do not believe this significantly impacted the main conclusions of the study as the non-monocyte cell populations included in the mononuclear cell layer after Ficoll separation are primarily lymphocytes, which will have little to no response to LPS stimulation. While there was significant induction of hemoglobin-associated transcripts in chorioamnionitis-exposed monocytes, we believe that red blood cell contamination of our samples is unlikely to be the cause given the purity of our magnetic bead separation and the absence of red blood cells upon microscopic examination of the samples after the separation. We performed ChIP-seq and RNA-seq on total CD14+ monocytes rather than monocyte subsets due to limitations in available cell numbers and a recent paper showing no significant difference in monocyte subsets between premature infants with and without chorioamnionitis exposure[7]. We acknowledge that distinct monocyte subsets, including classical (CD14++CD16-), intermediate (CD14++CD16+) and nonclassical (CD14+CD16+) monocytes exhibit unique gene expression profiles and that this may influence the interpretation of our results[47]. Additionally, the inclusion of 2–3 replicates per age/exposure group (n=6–9 total patients) in the ChIP-seq and RNA-seq experiments is low, although this number of replicates has been shown to be sufficient for site discovery in next generation sequencing experiments by the ENCODE consortium[48]. There was a difference in the proportion of infants born by vaginal delivery between healthy preterm and chorioamnionitis-exposed preterm infants. It is possible that mode of delivery rather than chorioamnionitis exposure may have contributed to important differences in monocyte H3K4me3 between these groups as mode of delivery is known to influence neonatal immune function[49]. Additionally, we were unable to control for all potential confounding variables (maternal and neonatal antimicrobial exposure, etc.), which should be taken into account when interpreting our results. We also did not account for other factors regulating mRNA expression, including additional histone modifications (H3K4me1, H3K27me3, etc.), DNA methylation and microRNA expression.
Conclusion
This study reveals that chorioamnionitis exposure remodeled the neonatal monocyte H3K4me3 landscape, with age-inappropriate H3K4me3 patterning. Chorioamnionitis exposure also altered term neonatal monocyte transcription, with a less robust inflammatory response upon a secondary pathogenic exposure. Additionally, H3K4me3 peak presence near a gene was predictive of mRNA expression, irrespective of the genomic location of the H3K4me3. Together these finding demonstrate that a pathogenic inflammatory exposure in the neonatal period has a profound effect on monocyte transcription and transcriptional regulation that may result in long-term immune dysfunction.
Materials and Methods
Participants
The local Institutional Review Board approved this research protocol and written informed consent was obtained from the parents of all neonatal participants. The study methods conformed to the standards set by the Declaration of Helsinki. Enrolled infants were born at the University of Michigan Medical Center and were classified as term (gestational age 37 weeks or greater), preterm (gestational age 30–36 weeks) or extremely preterm (gestational age under 30 weeks). There is no consensus on what gestational age cutoffs should be used when studying the preterm immune system, and many different cutoff values have been published[17, 50, 51]. We chose the above gestational age cutoffs because as we have published them previously, and believe they provide an immunologically meaningful distinction between groups[17]. Chorioamnionitis was diagnosed upon histopathological examination of the placenta as previously described[52]. Healthy infants had no evidence of inflammation on histopathological examination of the placenta. We enrolled a total of 28 healthy term infants, 27 chorioamnionitis-exposed term infants, 9 healthy preterm infants, 9 chorioamnionitis-exposed preterm infants, 9 healthy extremely preterm infants, and 9 chorioamnionitis-exposed extremely preterm infants.
Blood
Umbilical cord blood was collected from enrolled infants immediately after the delivery of the placenta. The umbilical cord blood was stored in the University of Michigan blood bank at 4oC and the samples were retrieved from the blood bank within 5 days of collection as previously described[17]. The blood samples were diluted 1:2 with sterile 0.9% saline and Ficoll-Isopaque density gradient centrifugation was used to isolate mononuclear cells. Mononuclear cells underwent CD14+ magnetic bead isolation according to the manufacturer’s instructions (Miltenyi Biotec, Auburn, CA, USA, 130–050-201). The average purity of the monocytes was 92% by flow cytometry using anti-human CD14 (BioLegend, San Diego, CA, USA, clone HCD14, PerCP/cy5.5). Isolated CD14+ cells were stored in recovery cell culture freezing media (Life Technologies, Carlsbad, CA, USA) in cryovials at −80oC until use. Monocyte viability post freezing was 70–75% by flow cytometry using DAPI (Molecular probes, Eugene, OR).
High Throughput ChIP-Sequencing
ChIP-seq was performed on purified CD14+ monocytes from healthy and chorioamnionitis-exposed term infants, preterm infants, and extremely preterm infants as previously described[17]. Each age group contained 3 biological replicates. In order to obtain approximately 5×105 cells per immunoprecipitation, each neonatal sample replicate contained 3 pooled samples (Table 11). The ultrasonication was performed on wet ice using the Branson Digital Sonifier 450 for 240 seconds at 40% amplitude with 0.7 seconds ‘on’ and 1.3 seconds ‘off’. The immunoprecipitation was performed with 4 micrograms of H3K4me3 antibody (Abcam, Cambridge, MA, USA, ab8580). After library preparation, the samples were pooled in equimolar concentrations and single-end ChIP-seq (50 nt) was performed on an Illumina HiSeq 2500 machine, resulting in 20–100 million reads/sample. The quality of the samples was assessed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and ChIPQC[53]. All samples passed quality control measures, which are outlined in Table 12.
Table 11:
ChIP-seq patient sample composition
| Group | Replicate | Age (weeks gestation) and Sex | # of Cells used in ChIP |
|---|---|---|---|
| U301 | 1 | 24M, 24.86F, 29.14M | 8.8×105 |
| U302 | 2 | 28.14F, 29.14F, 29.29M | 8.5×105 |
| U303 | 3 | 28.14M, 28.14F, 29.29M | 8.2×105 |
| U30C1 | 1 | 29.71M, 28F, 29M | 8.5×105 |
| U30C2 | 2 | 29.71M, 28.43M, 28.71F | 2.2×106 |
| U30C3 | 3 | 25.86F, 25.29F, 28.71F | 7×105 |
| O301 | 1 | 30.71M, 31.43F, 31.43F | 7.0×105 |
| O302 | 2 | 32.43M, 33.43M, 34.29F | 5.5×105 |
| O303 | 3 | 31.71M, 33.43F, 34F | 6.0×105 |
| O30C1 | 1 | 34.43M, 34M, 34.71F | 1.4×106 |
| O30C2 | 2 | 30M, 36.71F, 35.57M | 1×106 |
| O30C3 | 3 | 30.86M, 31.86F, 36.57M | 4.8×105 |
| Term1 | 1 | 37.14F, 38M, 41M | 5.0×105 |
| Term2 | 2 | 39.86F, 41M, 41.14M | 5.5×105 |
| Term3 | 3 | 39.43M, 40.43F, 41.14M | 6.0×105 |
| TermC1 | 1 | 40.14M, 41.29F, 40.86M | 3.6×105 |
| TermC2 | 2 | 37.86M, 39.71M, 40.71F | 4.7×105 |
| TermC3 | 3 | 39.86F, 41.43M, 40.43M | 3.9×105 |
U30 = under 30 weeks gestation, O30 = 30–36 weeks gestation, Term = term gestation, C=chorioamnionitis exposure, M=male, F=female
Table 12:
H3K4me3 ChIP-seq quality control data
| Sample | Sequencing Depth (reads) | Read Quality (Phred Scorea) | Uniquely Mapping Reads | Reads Mapped (%) | SSDb | FRIPc |
|---|---|---|---|---|---|---|
| U301 | 57,276,495 | 39 | 72,464 | 100 | 14.3 | 22.8 |
| U302 | 49,342,352 | 39 | 72,665 | 100 | 11.6 | 18.7 |
| U303 | 29,802,440 | 39 | 291,617 | 100 | 4.07 | 5.89 |
| U30C1 | 70,437,662 | 38.4 | 396,500 | 100 | 6.71 | 7.9 |
| U30C2 | 50,711,822 | 38.3 | 134,943 | 100 | 8.75 | 12.5 |
| U30C3 | 43,468,108 | 38.3 | 379,625 | 100 | 5.02 | 7.14 |
| O301 | 39,347,378 | 39 | 87,324 | 100 | 7.98 | 12 |
| O302 | 62,638,230 | 39 | 310,426 | 100 | 6.87 | 7.01 |
| O303 | 44,318,249 | 39 | 215,790 | 100 | 5.52 | 6.55 |
| O30C1 | 49,479,468 | 38.4 | 983,885 | 100 | 4.85 | 5.22 |
| O30C2 | 103,309,230 | 36.6 | 208,965 | 100 | 3.66 | 6.46 |
| O30C3 | 56,772,587 | 36.6 | 111,612 | 100 | 3.08 | 6.85 |
| Term1 | 71,263,489 | 39 | 252,327 | 100 | 12.8 | 14.8 |
| Term2 | 72,201,221 | 39 | 251,160 | 100 | 11.6 | 14 |
| Term3 | 60,628,889 | 39 | 145,300 | 100 | 14.2 | 18.9 |
| TermC1 | 72,842,732 | 36.4 | 96,237 | 100 | 6.37 | 5.29 |
| TermC2 | 82,604,593 | 36.4 | 104,332 | 100 | 12.2 | 10.6 |
| TermC3 | 79,096,520 | 36.6 | 126,490 | 100 | 4.83 | 5.81 |
Phred scores greater than 30 are considered excellent, with a base call accuracy of over 99.9%.
SSD is the standard deviation of signal pile-up along the genome normalized to the total number of reads; values greater than 2 indicate very good chromatin immunoprecipitation enrichment.
FRIP is the percentage of reads that overlap peaks; values greater than 5 indicate successful chromatin immunoprecipitation enrichment.
ChIP-seq Mapping and Data Analysis
Scythe (https://github.com/vsbuffalo/scythe) was used to trim adapter sequences from the raw reads. The reads were then aligned to the Homo sapiens genome assembly (Build 37, hg19) with Bowtie2 version 2.0.0 using default parameters[54]. The results were normalized to total read count and library size. MACS2 version 2.1.1.20160309 was used to call peaks by comparing the immunoprecipitated samples to input samples[55]. DiffBind was used to derived consensus peaksets from the biological replicates, and peaks needed to be present in two out of three replicates to be included[56]. The consensus peaksets were used for downstream data analysis. The EdgeR package was used to identify differentially bound sites[20]. The false discovery rate for differentially bound sites was set to <0.1[57, 58]. Peaks were annotated and Gene Ontology terms were identified using ChIPpeakAnno[59]. The UCSC Genome Browser was used for data visualization[18]. Venneuler version 1.1–1 was used construct proportional venn diagrams (http://www.rforge.net/venneuler/). The datasets supporting the results are available in the GEO repositories GSE81957 and GSE111873.
Cell culture
Purified CD14+ monocytes were washed twice in sterile RPMI after thawing. The cells were plated at a concentration of 5×104 cells/well in 96-well polystyrene cell culture plates in 100 μl of RPMI media containing 400 mM/L L-glutamine, 10% adult human serum, 1% penicillin/streptomycin, 1% sodium pyruvate and 1% nonessential amino acids. The cells were either left unstimulated or were stimulated with 100 ng/mL lipopolysaccharide (Sigma Aldrich, St. Louis, MO, USA, purified from E. coli 055:B5) and were incubated at 37oC with 5% CO2. Cells were collected for RNA-seq preparation 6 hours after stimulation and for measurement of protein concentration 24 hours after stimulation. Protein levels of the cytokines IL-1β, IL-6, IL-8, TNF-α and IL-10 were measured from cell culture supernatants using a bead-based multiplex assay (Bio-Rad, Hercules, CA, USA).
RNA-seq
RNA-seq was performed on umbilical cord blood purified CD14+ monocytes from healthy and chorioamnionitis-exposed term neonates in both unstimulated and LPS stimulated conditions. Each exposure/stimulation group contained 3 replicates, except the chorioamnionitis-exposed unstimulated group, which contained 2 replicates as one did not pass quality control. Each replicate contained 3 pooled samples (Table 13). RNA was isolated using an RNeasy micro kit with an on column DNAse digestion following the manufacturer’s directions (Qiagen, Germantown, MD, USA). cDNA was synthesized and libraries were prepared using the SMARTer stranded total RNA pico kit following the manufacturer’s instructions (Takara, Mountain View, CA, USA). The samples were pooled in equimolar concentrations and paired-end sequencing (50 nt) was performed on an Illumina HiSeq 2500 machine, resulting in 10–20 million reads/sample. The quality of the samples was assessed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). All samples passed quality control measures, except for replicate 3 of the unstimulated chorioamnionitis-exposed group, which was not sequenced (Table 14).
Table 13:
RNA-seq patient sample composition
| Group | Replicate | Age (weeks gestation) and Sex | # of Cells used in RNA-seq |
|---|---|---|---|
| U1 | 1 | 37F, 39.57M, 37.86F | 1.7×105 |
| U2 | 2 | 40.43F, 38.86M, 39.14F | 1.8×105 |
| U3 | 3 | 37.86F, 40.14M, 37F | 1.6×105 |
| L1 | 1 | 37F, 39.57M, 37.86F | 1.7×105 |
| L2 | 2 | 40.43F, 38.86M, 39.14F | 1.8×105 |
| L3 | 3 | 37.86F, 40.14M, 37F | 1.6×105 |
| CU1 | 1 | 38.14F, 41F, 40.43M | 1.3×105 |
| CU2 | 2 | 39.86F, 38.14M, 38.14M | 1.6×105 |
| CU3 | 3 | 41.14F, 41.14M, 40.57F | 1.7×105 |
| CL1 | 1 | 38.14F, 41F, 40.43M | 1.3×105 |
| CL2 | 2 | 39.86F, 38.14M, 38.14M | 1.6×105 |
| CL3 | 3 | 41.14F, 41.14M, 40.57F | 1.7×105 |
U=healthy unstimulated monocytes, L=healthy LPS stimulated monocytes, CU=chorioamnionitis-exposed unstimulated monocytes, CL=chorioamnionitis-exposed LPS stimulated monocytes, M=male, F=female
Table 14:
RNA-seq quality control data
| Sample | Sequencing Depth (reads) | Read Quality (Phred Scorea) | RNA Concentration (nanograms) | RINb |
|---|---|---|---|---|
| U1 | 13,325,665 | 35.8 | 26.4 | 8.9 |
| U2 | 14,003,586 | 35.6 | 43.5 | 9.2 |
| U3 | 16,590,521 | 35.6 | 13.1 | 1.1 |
| L1 | 14,090634 | 35.8 | 11.3 | 8.6 |
| L2 | 16,437,185 | 35.8 | 36.9 | 8.7 |
| L3 | 14,342,866 | 35.8 | 5.7 | 8.6 |
| CU1 | 14,797,385 | 35.8 | 46.5 | 9.0 |
| CU2 | 14,179,302 | 35.8 | 45.9 | 6.8 |
| CU3 | Not sequenced | Not sequenced | <5 | <1 |
| CL1 | 15,336,848 | 35.7 | 13 | 6.5 |
| CL2 | 11,726,579 | 35.7 | 27.4 | 8.8 |
| CL3 | 14,963,332 | 35.6 | 16.1 | 8.2 |
Phred scores greater than 30 are considered excellent, with a base call accuracy of over 99.9%.
RNA integrity number representing the degree of RNA degradation; values below 7 indicate some degree of degradation. While a RIN value of greater than 7 is recommended to proceed with RNA-seq, accurate transcript quantification can be provided by degraded RNA samples[65].
RNA-seq data analysis
Adapter sequences were trimmed from raw reads using Trimmomatic[60]. Reads were aligned to the Homo sapiens genome assembly (Build 37, hg19) with HISAT2 version 2.1.0 using the paired-end option[61]. Subread version 1.6.0 was used to align reads, quantify transcript abundance and assign reads to genomic features[62]. Reads were normalized to total read counts and differentially expressed transcripts were identified using the edgeR package[20]. The false discovery rate for differentially expressed transcripts was set to <0.1[57, 58]. Genes were annotated using MyGene[63]. Pathway analysis was performed using the edgeR package. Network analysis was performed using NetworkAnalyst[64]. The datasets supporting the results are available in the GEO repository GSE111927.
Statistical analysis
Prism 6 was used for basic data analysis. The differences between groups were evaluated with the student’s t-test for parametric quantitative data, the Mann-Whitney test for nonparametric quantitative data, ANOVA for multiple comparisons of parametric data and Chi-square test to evaluate relationships between categorical variables. Values of p<0.05 were considered to be significant.
Supplementary Material
Acknowledgements
This study was funded by the NIH Child Health Research Center Grant 5K12HD028820–22 and the University of Michigan Institute for Clinical and Health Research Grant UL1TR000433.
Abbreviations:
- H3K4me3
trimethylation of lysine 4 of histone 3
- ChIP-seq
chromatin immunoprecipitation followed by massively parallel DNA sequencing
- RNA-seq
whole transcriptome shotgun sequencing
- LPS
lipopolysaccharide
- U30
under 30 week extremely preterm healthy infants
- U30C
under 30 week extremely preterm chorioamnionitis-exposed infants
- O30
over 30 week preterm healthy infants
- O30C
over 30 week preterm chorioamnionitis-exposed infants
- Term
term healthy infants
- TermC
term chorioamnionitis-exposed infants
- U
term healthy unstimulated monocytes
- L
term healthy LPS stimulated monocytes
- CU
term chorioamnionitis-exposed unstimulated monocytes
- CL
term chorioamnionitis-exposed LPS stimulated monocytes
Footnotes
References
- 1.Goldenberg RL, Culhane JF, Iams JD & Romero R (2008) Epidemiology and causes of preterm birth, Lancet (London, England). 371, 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gibbs RS & Duff P (1991) Progress in pathogenesis and management of clinical intraamniotic infection, American journal of obstetrics and gynecology. 164, 1317–26. [DOI] [PubMed] [Google Scholar]
- 3.Leviton A, Hecht JL, Allred EN, Yamamoto H, Fichorova RN & Dammann O (2011) Persistence after birth of systemic inflammation associated with umbilical cord inflammation, Journal of reproductive immunology. 90, 235–43. [DOI] [PubMed] [Google Scholar]
- 4.Schuchat A, Zywicki SS, Dinsmoor MJ, Mercer B, Romaguera J, O’Sullivan MJ, Patel D, Peters MT, Stoll B & Levine OS (2000) Risk factors and opportunities for prevention of early-onset neonatal sepsis: a multicenter case-control study, Pediatrics. 105, 21–6. [DOI] [PubMed] [Google Scholar]
- 5.Schrag SJ, Hadler JL, Arnold KE, Martell-Cleary P, Reingold A & Schuchat A (2006) Risk factors for invasive, early-onset Escherichia coli infections in the era of widespread intrapartum antibiotic use, Pediatrics. 118, 570–6. [DOI] [PubMed] [Google Scholar]
- 6.Weitkamp JH, Guthrie SO, Wong HR, Moldawer LL, Baker HV & Wynn JL (2016) Histological chorioamnionitis shapes the neonatal transcriptomic immune response, Early human development. 98, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Jong E, Hancock DG, Wells C, Richmond P, Simmer K, Burgner D, Strunk T & Currie AJ (2018) Exposure to chorioamnionitis alters the monocyte transcriptional response to the neonatal pathogen Staphylococcus epidermidis, Immunology and cell biology. [DOI] [PubMed] [Google Scholar]
- 8.Andrews WW, Goldenberg RL, Faye-Petersen O, Cliver S, Goepfert AR & Hauth JC (2006) The Alabama Preterm Birth study: polymorphonuclear and mononuclear cell placental infiltrations, other markers of inflammation, and outcomes in 23- to 32-week preterm newborn infants, American journal of obstetrics and gynecology. 195, 803–8. [DOI] [PubMed] [Google Scholar]
- 9.O’Shea TM, Shah B, Allred EN, Fichorova RN, Kuban KC, Dammann O & Leviton A (2013) Inflammation-initiating illnesses, inflammation-related proteins, and cognitive impairment in extremely preterm infants, Brain, behavior, and immunity. 29, 104–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nasef N, Shabaan AE, Schurr P, Iaboni D, Choudhury J, Church P & Dunn MS (2013) Effect of clinical and histological chorioamnionitis on the outcome of preterm infants, American journal of perinatology. 30, 59–68. [DOI] [PubMed] [Google Scholar]
- 11.Kumar R, Yu Y, Story RE, Pongracic JA, Gupta R, Pearson C, Ortiz K, Bauchner HC & Wang X (2008) Prematurity, chorioamnionitis, and the development of recurrent wheezing: a prospective birth cohort study, The Journal of allergy and clinical immunology. 121, 878–84.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murray PJ & Wynn TA (2011) Protective and pathogenic functions of macrophage subsets, Nature reviews Immunology. 11, 723–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tatad AM, Nesin M, Peoples J, Cheung S, Lin H, Sison C, Perlman J & Cunningham-Rundles S (2008) Cytokine expression in response to bacterial antigens in preterm and term infant cord blood monocytes, Neonatology. 94, 8–15. [DOI] [PubMed] [Google Scholar]
- 14.Strunk T, Prosser A, Levy O, Philbin V, Simmer K, Doherty D, Charles A, Richmond P, Burgner D & Currie A (2012) Responsiveness of human monocytes to the commensal bacterium Staphylococcus epidermidis develops late in gestation, Pediatric research. 72, 10–8. [DOI] [PubMed] [Google Scholar]
- 15.Marodi L, Kaposzta R & Nemes E (2000) Survival of group B streptococcus type III in mononuclear phagocytes: differential regulation of bacterial killing in cord macrophages by human recombinant gamma interferon and granulocyte-macrophage colony-stimulating factor, Infection and immunity. 68, 2167–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruthenburg AJ, Allis CD & Wysocka J (2007) Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark, Molecular cell. 25, 15–30. [DOI] [PubMed] [Google Scholar]
- 17.Bermick JR, Lambrecht NJ, denDekker AD, Kunkel SL, Lukacs NW, Hogaboam CM & Schaller MA (2016) Neonatal monocytes exhibit a unique histone modification landscape, Clinical epigenetics. 8, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM & Haussler D (2002) The human genome browser at UCSC, Genome research. 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lissner MM, Thomas BJ, Wee K, Tong AJ, Kollmann TR & Smale ST (2015) Age-Related Gene Expression Differences in Monocytes from Human Neonates, Young Adults, and Older Adults, PloS one. 10, e0132061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson MD, McCarthy DJ & Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data, Bioinformatics (Oxford, England). 26, 139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian Y, Jia Z, Wang J, Huang Z, Tang J, Zheng Y, Tang Y, Wang Q, Tian Z, Yang D, Zhang Y, Fu X, Song J, Liu S, van Velkinburgh JC, Wu Y & Ni B (2011) Global mapping of H3K4me1 and H3K4me3 reveals the chromatin state-based cell type-specific gene regulation in human Treg cells, PloS one. 6, e27770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen K, Chen Z, Wu D, Zhang L, Lin X, Su J, Rodriguez B, Xi Y, Xia Z, Chen X, Shi X, Wang Q & Li W (2015) Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor-suppressor genes, Nature genetics. 47, 1149–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benayoun BA, Pollina EA, Ucar D, Mahmoudi S, Karra K, Wong ED, Devarajan K, Daugherty AC, Kundaje AB, Mancini E, Hitz BC, Gupta R, Rando TA, Baker JC, Snyder MP, Cherry JM & Brunet A (2014) H3K4me3 breadth is linked to cell identity and transcriptional consistency, Cell. 158, 673–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karlic R, Chung HR, Lasserre J, Vlahovicek K & Vingron M (2010) Histone modification levels are predictive for gene expression, Proceedings of the National Academy of Sciences of the United States of America. 107, 2926–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui P, Liu W, Zhao Y, Lin Q, Ding F, Xin C, Geng J, Song S, Sun F, Hu S & Yu J (2012) The association between H3K4me3 and antisense transcription, Genomics, proteomics & bioinformatics. 10, 74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao F, Fang Y, Tan HK, Goh Y, Choy JYH, Koh BTH, Hao Tan J, Bertin N, Ramadass A, Hunter E, Green J, Salter M, Akoulitchev A, Wang W, Chng WJ, Tenen DG & Fullwood MJ (2017) Super-Enhancers and Broad H3K4me3 Domains Form Complex Gene Regulatory Circuits Involving Chromatin Interactions, Scientific reports. 7, 2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russ BE, Olshansky M, Li J, Nguyen MLT, Gearing LJ, Nguyen THO, Olson MR, McQuilton HA, Nussing S, Khoury G, Purcell DFJ, Hertzog PJ, Rao S & Turner SJ (2017) Regulation of H3K4me3 at Transcriptional Enhancers Characterizes Acquisition of Virus-Specific CD8(+) T Cell-Lineage-Specific Function, Cell reports. 21, 3624–3636. [DOI] [PubMed] [Google Scholar]
- 28.Onderdonk AB, Hecht JL, McElrath TF, Delaney ML, Allred EN & Leviton A (2008) Colonization of second-trimester placenta parenchyma, American journal of obstetrics and gynecology. 199, 52.e1–52.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rito DC, Viehl LT, Buchanan PM, Haridas S & Koenig JM (2017) Augmented Th17-type immune responses in preterm neonates exposed to histologic chorioamnionitis, Pediatric research. 81, 639–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matta P, Sherrod SD, Marasco CC, Moore DJ, McLean JA & Weitkamp JH (2017) In Utero Exposure to Histological Chorioamnionitis Primes the Exometabolomic Profiles of Preterm CD4(+) T Lymphocytes, Journal of immunology (Baltimore, Md : 1950). 199, 3074–3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altinoz MA, Ozcan EM, Ince B & Guloksuz S (2016) Hemoglobins as new players in multiple sclerosis: metabolic and immune aspects, Metabolic brain disease. 31, 983–92. [DOI] [PubMed] [Google Scholar]
- 32.Schmid D, Burmester GR, Tripmacher R, Kuhnke A & Buttgereit F (2000) Bioenergetics of human peripheral blood mononuclear cell metabolism in quiescent, activated, and glucocorticoid-treated states, Bioscience reports. 20, 289–302. [DOI] [PubMed] [Google Scholar]
- 33.Yoon BH, Romero R, Jun JK, Maymon E, Gomez R, Mazor M & Park JS (1998) An increase in fetal plasma cortisol but not dehydroepiandrosterone sulfate is followed by the onset of preterm labor in patients with preterm premature rupture of the membranes, American journal of obstetrics and gynecology. 179, 1107–14. [DOI] [PubMed] [Google Scholar]
- 34.Gomez R, Romero R, Ghezzi F, Yoon BH, Mazor M & Berry SM (1998) The fetal inflammatory response syndrome, American journal of obstetrics and gynecology. 179, 194–202. [DOI] [PubMed] [Google Scholar]
- 35.Nakamura T, Hatanaka D, Kusakari M, Kashima K, Takizawa Y, Takahashi H, Yoshioka T, Takahashi N & Kamohara T (2018) Neonatal Leukemoid Reaction with Fetal Inflammatory Response Syndrome Is Associated with Elevated Serum Granulocyte Colony Stimulating Factor and Interleukin-6, The Tohoku journal of experimental medicine. 244, 145–149. [DOI] [PubMed] [Google Scholar]
- 36.Bergeron J, Gerges N, Guiraut C, Grbic D, Allard MJ, Fortier LC, Vaillancourt C & Sebire G (2016) Activation of the IL-1beta/CXCL1/MMP-10 axis in chorioamnionitis induced by inactivated Group B Streptococcus, Placenta. 47, 116–123. [DOI] [PubMed] [Google Scholar]
- 37.Kunze M, Klar M, Morfeld CA, Thorns B, Schild RL, Markfeld-Erol F, Rasenack R, Proempeler H, Hentschel R & Schaefer WR (2016) Cytokines in noninvasively obtained amniotic fluid as predictors of fetal inflammatory response syndrome, American journal of obstetrics and gynecology. 215, 96.e1–8. [DOI] [PubMed] [Google Scholar]
- 38.Garcia-Munoz Rodrigo F, Galan Henriquez G, Figueras Aloy J & Garcia-Alix Perez A (2014) Outcomes of very-low-birth-weight infants exposed to maternal clinical chorioamnionitis: a multicentre study, Neonatology. 106, 229–34. [DOI] [PubMed] [Google Scholar]
- 39.Dempsey E, Chen MF, Kokottis T, Vallerand D & Usher R (2005) Outcome of neonates less than 30 weeks gestation with histologic chorioamnionitis, American journal of perinatology. 22, 155–9. [DOI] [PubMed] [Google Scholar]
- 40.Soraisham AS, Singhal N, McMillan DD, Sauve RS & Lee SK (2009) A multicenter study on the clinical outcome of chorioamnionitis in preterm infants, American journal of obstetrics and gynecology. 200, 372.e1–6. [DOI] [PubMed] [Google Scholar]
- 41.Botet F, Figueras J, Carbonell-Estrany X, Arca G & The Castrillo Study G (2010) Effect of maternal clinical chorioamnionitis on neonatal morbidity in very-low birthweight infants: a case-control study, Journal of perinatal medicine. 38, 269–73. [DOI] [PubMed] [Google Scholar]
- 42.Garcia-Munoz Rodrigo F, Galan Henriquez GM & Ospina CG (2014) Morbidity and mortality among very-low-birth-weight infants born to mothers with clinical chorioamnionitis, Pediatrics and neonatology. 55, 381–6. [DOI] [PubMed] [Google Scholar]
- 43.Ofman G, Vasco N & Cantey JB (2016) Risk of Early-Onset Sepsis following Preterm, Prolonged Rupture of Membranes with or without Chorioamnionitis, American journal of perinatology. 33, 339–42. [DOI] [PubMed] [Google Scholar]
- 44.Weston EJ, Pondo T, Lewis MM, Martell-Cleary P, Morin C, Jewell B, Daily P, Apostol M, Petit S, Farley M, Lynfield R, Reingold A, Hansen NI, Stoll BJ, Shane AL, Zell E & Schrag SJ (2011) The burden of invasive early-onset neonatal sepsis in the United States, 2005–2008, The Pediatric infectious disease journal. 30, 937–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strunk T, Doherty D, Jacques A, Simmer K, Richmond P, Kohan R, Charles A & Burgner D (2012) Histologic chorioamnionitis is associated with reduced risk of late-onset sepsis in preterm infants, Pediatrics. 129, e134–41. [DOI] [PubMed] [Google Scholar]
- 46.Puri K, Taft DH, Ambalavanan N, Schibler KR, Morrow AL & Kallapur SG (2016) Association of Chorioamnionitis with Aberrant Neonatal Gut Colonization and Adverse Clinical Outcomes, PloS one. 11, e0162734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wong KL, Tai JJ, Wong WC, Han H, Sem X, Yeap WH, Kourilsky P & Wong SC (2011) Gene expression profiling reveals the defining features of the classical, intermediate, and nonclassical human monocyte subsets, Blood. 118, e16–31. [DOI] [PubMed] [Google Scholar]
- 48.Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, Chen Y, DeSalvo G, Epstein C, Fisher-Aylor KI, Euskirchen G, Gerstein M, Gertz J, Hartemink AJ, Hoffman MM, Iyer VR, Jung YL, Karmakar S, Kellis M, Kharchenko PV, Li Q, Liu T, Liu XS, Ma L, Milosavljevic A, Myers RM, Park PJ, Pazin MJ, Perry MD, Raha D, Reddy TE, Rozowsky J, Shoresh N, Sidow A, Slattery M, Stamatoyannopoulos JA, Tolstorukov MY, White KP, Xi S, Farnham PJ, Lieb JD, Wold BJ & Snyder M (2012) ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia, Genome research. 22, 1813–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, Bjorksten B, Engstrand L & Andersson AF (2014) Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section, Gut. 63, 559–66. [DOI] [PubMed] [Google Scholar]
- 50.Varvarigou AA, Thomas I, Rodi M, Economou I, Mantagos S & Mouzaki A (2012) Respiratory distress syndrome (RDS) in premature infants is underscored by the magnitude of Th1 cytokine polarization, Cytokine. 58, 355–60. [DOI] [PubMed] [Google Scholar]
- 51.Zasada M, Kwinta P, Durlak W, Bik-Multanowski M, Madetko-Talowska A & Pietrzyk JJ (2014) Development and maturation of the immune system in preterm neonates: results from a whole genome expression study, BioMed research international. 2014, 498318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Redline RW, Faye-Petersen O, Heller D, Qureshi F, Savell V & Vogler C (2003) Amniotic infection syndrome: nosology and reproducibility of placental reaction patterns, Pediatric and developmental pathology : the official journal of the Society for Pediatric Pathology and the Paediatric Pathology Society. 6, 435–48. [DOI] [PubMed] [Google Scholar]
- 53.Carroll TS, Liang Z, Salama R, Stark R & de Santiago I (2014) Impact of artifact removal on ChIP quality metrics in ChIP-seq and ChIP-exo data, Frontiers in genetics. 5, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ourthiague DR, Birnbaum H, Ortenlof N, Vargas JD, Wollman R & Hoffmann A (2015) Limited specificity of IRF3 and ISGF3 in the transcriptional innate-immune response to double-stranded RNA, Journal of leukocyte biology. 98, 119–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu L, Zinkgraf M, Petzold HE, Beers EP, Filkov V & Groover A (2015) The Populus ARBORKNOX1 homeodomain transcription factor regulates woody growth through binding to evolutionarily conserved target genes of diverse function, The New phytologist. 205, 682–94. [DOI] [PubMed] [Google Scholar]
- 56.Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, Ali S, Chin SF, Palmieri C, Caldas C & Carroll JS (2012) Differential oestrogen receptor binding is associated with clinical outcome in breast cancer, Nature. 481, 389–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu R, Holik AZ, Su S, Jansz N, Chen K, Leong HS, Blewitt ME, Asselin-Labat ML, Smyth GK & Ritchie ME (2015) Why weight? Modelling sample and observational level variability improves power in RNA-seq analyses, Nucleic acids research. 43, e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jenkins NT, Padilla J, Thorne PK, Martin JS, Rector RS, Davis JW & Laughlin MH (2014) Transcriptome-wide RNA sequencing analysis of rat skeletal muscle feed arteries. I. Impact of obesity, Journal of applied physiology (Bethesda, Md : 1985). 116, 1017–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu LJ, Gazin C, Lawson ND, Pages H, Lin SM, Lapointe DS & Green MR (2010) ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data, BMC bioinformatics. 11, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bolger AM, Lohse M & Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data, Bioinformatics (Oxford, England). 30, 2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim D, Langmead B & Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements, Nature methods. 12, 357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liao Y, Smyth GK & Shi W (2013) The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote, Nucleic acids research. 41, e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu C, Macleod I & Su AI (2013) BioGPS and MyGene.info: organizing online, gene-centric information, Nucleic acids research. 41, D561–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xia J, Gill EE & Hancock RE (2015) NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data, Nature protocols. 10, 823–44. [DOI] [PubMed] [Google Scholar]
- 65.Gallego Romero I, Pai AA, Tung J & Gilad Y (2014) RNA-seq: impact of RNA degradation on transcript quantification, BMC biology. 12, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.