Abstract
Clinical data acquired over the last decade on non-small cell lung cancer (NSCLC) treatment with small molecular weight Epidermal Growth Factor Receptor (EGFR) inhibitors have shown significant influence of EGFR point mutations and in-frame deletions on clinical efficacy. Identification of small molecules capable of inhibiting the clinically relevant EGFR mutant forms is desirable, and novel chemical scaffolds might provide knowledge regarding selectivity among EGFR forms and shed light on new strategies to overcome current clinical limitations. Design, synthesis, docking studies and in vitro evaluation of N-(3-(3-phenylureido)quinoxalin-6-yl) acrylamide derivatives (7a-m) against EGFR mutant forms are described. Compounds 7h and 7l were biochemically active in the nanomolar range against EGFRwt and EGFRL858R. Molecular docking and reaction enthalpy calculations have shown the influence of the combination of reversible and covalent binding modes with EGFR on the inhibitory activity. The inhibitory profile of 7h against a panel of patient-derived tumor cell lines was established, demonstrating selective growth inhibition of EGFR related cells at 10 μM among a panel of 30 cell lines derived from colon, melanoma, breast, bladder, kidney, prostate, pancreas and ovary tumors.
Introduction
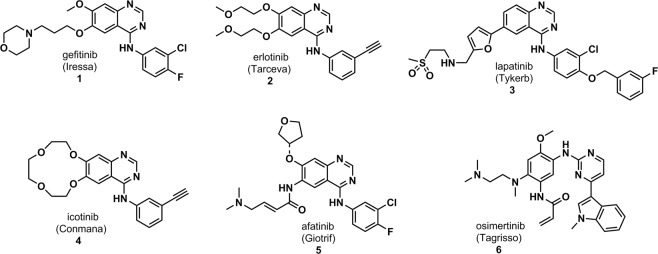
Over the last years there has been a growing interest for the identification of protein kinase inhibitors to treat several disorders1,2. Tyrosine kinase inhibitors (TKIs) drug discovery projects have emerged as an attractive field once approved drugs have been considered as efficient pharmacological tools mainly in cancer treatment3,4. To date, more than 35 small molecular weight protein kinase inhibitors have been approved for clinical use targeting a small number of protein kinases5,6. Protein kinases are druggable targets and their inhibition induces apoptosis in tumor cell lines presenting the oncogenic addiction phenomenon7,8. This feature assures the protein kinase inhibitors’ drugs-selective cytotoxicity to tumor cells overexpressing the targeted protein and sparing non-tumor cells. Therefore, targeted cancer therapy is more tolerable to adverse effects than classic chemotherapy9. Yet, common adverse effects observed in targeted therapy are related to “on-target” inhibition of wild type EGFR10,11. Molecular modifications on chemical structures might dramatically change not only the potency of TKIs over their on-target12–14, but also affect their selectivity towards other non-related protein kinase (off-targets)15,16. This effect can be clearly observed in Epidermal Growth Factor Receptor inhibitors (EGFRi) drugs gefitinib (1), erlotinib (2), lapatinib (3), icotinib (4) afatinib (5), osimertinib (6) (Fig. 1).
Figure 1.
EGFR inhibitor drugs.
Among the kinome, the Epidermal Growth Factor Receptor (EGFR) is one of the most studied protein kinases and its relationship with cancer has been known since the 1980’s17. EGFR is a receptor tyrosine kinase, a member of ErbB family together with HER-2, HER-3 and HER-4, related to cellular proliferation, cellular differentiation and cell survival18,19. As a receptor, EGFR has an extracellular domain, a transmembrane segment and an intracellular kinase domain20. Ligands such as EGF, TGF-alpha, epiregulin and others, bind to the extracellular domain. The EGFR activation process is initiated by homo- or heterodimerization with ErbB family members and corresponding signaling pathways, mainly PI3K/AKT and MAPK, are triggered. Deregulated activity of EGFR is related to aggressive tumors with poor prognosis21,22.
Clinical data acquired over the last decade, especially on non-small cell lung cancer (NSCLC) treatment with small molecular weight EGFR inhibitors, have shown significant influence of EGFR point mutations and in-frame deletions on clinical efficacy23,24. Activating EGFR point mutation L858R and exon19 in-frame deletion mutations are related to clinical response to erlotinib, gefitinib and afatinib25,26. These mutations comprise more than 80% of driver mutations found in EGFR driven NSCLC27,28. After one year of treatment, initially responsive patients showed a resistance to erlotinib or gefitinib-based tumor treatment associated to a secondary point mutation at the gatekeeper residue T790M29,30. Kinetic studies showed ATP and reversible EGFR inhibitors to have different affinities for EGFR wild-type and mutant forms31. ATP has a higher affinity to its catalytic binding site in EGFR-wt and EGFR harboring T790M mutation and, therefore, reversible inhibitors show lower potency, shorter residence time and lack of clinical efficacy in these forms. On the other hand, ATP has a lower affinity to EGFR L858R, and first generation EGFRi beat ATP with regard to its binding pocket, showing longer residence time and better clinical response.
The discovery of new TKI capable of inhibiting the clinically relevant EGFR mutant forms is desirable, and novel chemical scaffolds might provide knowledge regarding selectivity among EGFR forms and shed light on new strategies to overcome current clinical limitations. In this context, this work describes the design, synthesis and in vitro evaluation of new acrylamide-quinoxaline derivatives as a novel scaffold for EGFR inhibition.
Results and Discussion
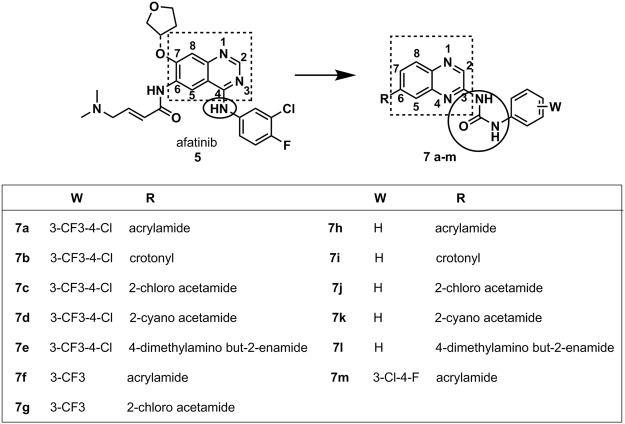
Molecular design of quinoxaline EGFR inhibitors
The molecular design conception was based on the bioisosteric replacement of the quinazoline aromatic ring by a quinoxaline scaffold32, maintaining sp2 nitrogen atoms for hydrogen bond interactions to the hinge region33. Subsequently, the aniline moiety was replaced by a urea subunit. Aiming to explore an eventual covalent interaction with EGFR cysteine 797 residue34, different electrophilic subunits were introduced to the position analogous to afatinib (4), allowing the design of compounds 7a-m (Fig. 2). The election of the covalent reactive groups was based on previous works describing EGFR inhibition towards reversible and irreversible covalent bond with cysteine residues35–38. Additionally, chemical reactivity studies and promiscuity profiles of the covalent reactive groups were also considered39,40.
Figure 2.
Molecular conception of quinoxaline urea derivatives 7a-m designed as EGFR covalent inhibitors.
Chemistry
Synthesis of the derivatives 7a-m was performed through the synthetic methodology depicted in Fig. 3, employing 7-nitroquinoxaline-2-amine (8) as key intermediate. A simple multi-gram procedure to obtain 8 was developed, using the non-expensive and readily available o-phenylendiamine as starting material41. Substituted phenylureas (9a-d) were obtained by the reaction of a 7-nitroquinoxinoxaline-2-amine (8) derivative with isocyanates in dry toluene under reflux in moderate yields (58–82%)42. The nitro group was reduced to the corresponding aniline (10a-d) using tin (II) chloride dihydrate under reflux in ethanol with 37–91% yields43. In the final step, the anilines 10a-d reacted with the previously selected acyl chlorides in dry THF in the presence of DIPEA as organic base to furnish the desired compounds 7a-m after flash chromatography purification step44. Crotonoyl chloride, 2-cyanoacetyl chloride and (E)-4-(dimethylamino) but-2-enoyl chloride were prepared using their corresponding commercial available carboxylic acids in a procedure previous to the acylation reaction using oxalyl chloride in dichloromethane and catalytic DMF45.
Figure 3.
Reagents and conditions: (a) substituted phenyl isocyanates, dry toluene, reflux, 2–4 h, 58–82%; (b) SnCl2.2H2O, ethanol, reflux, overnight, 37–91%; (c) acyl chlorides, DIPEA, dry THF, 0 °C, 2–3 h, 12–70%.
EGFR wt and mutant forms inhibition
EGFR-wt and mutant forms biochemical inhibitory activities for 7a-m are depicted in Table 1. Enzymatic in vitro determination showed that para-substituted compounds 7a-e and 7m have weak or no inhibitory activity on EGFR forms. Moreover, meta-trifluoromethyl-substituted derivatives (7f, 7g) showed weak activity against EGFRwt and EGFRL858R and no inhibitory activity against EGFRL858R/T790M. These results indicate that substitutions at the phenyl ring of the urea moiety are deleterious to EGFR inhibitory activity. Biochemical data also suggest that the replacement of the aniline quinazoline scaffold, common in the structure of well-known EGFR inhibitors (e.g. afatinib, gefitinib), with quinoxaline phenylurea results in compounds sensitive to steric constraints at the binding site, probably associated with the augmentation of the distance between the heteroaromatic ring’s nitrogen, which is able to interact with the hinge region, and the urea phenyl ring. However, non-substituted phenylurea derivatives 7h and 7l, bearing as covalent reactive groups the acrylamide and 4-dimethylamino but-2-enamide moieties, respectively, displayed notorious EGFR inhibitory activity in the low nanomolar range. Compound 7h, was identified as the most potent inhibitor of EGFRwt (IC50 = 25 ± 4 nM) and EGFRL858R (IC50 = 18 ± 3 nM). Despite that, weak inhibition of EGFRL858R/T790M was observed. Compound 7l was able to inhibit all EGFR forms (IC50 = 101 ± 12 nM for EGFRwt, IC50 = 32 ± 14 nM for EGFRL858R and IC50 = 132 ± 49 nM for EGFRL858R/T790M) in 2–3 digits nanomolar range. As the covalent inhibition process relies on two steps46, an initial reversible ligand-receptor interaction followed by a covalent reaction of a nucleophile (EGFR C797 residue) with an electrophile (covalent reactive group), potency differences for EGFR inhibition observed between compounds 7h-l (phenyl substituted) are expected. The presence of the tertiary amine group at the covalent reactive group can not only provide a better solubility profile, but can also act as an “in situ” catalyst during the covalent bond formation with cysteine residue35,39. Therefore, it might be responsible for the apparent potency in EGFR inhibition, including EGFRL858R/T790M, exhibited by compound 7l. Compounds harboring 2-chloro acetamide as covalent reactive group were able to inhibit EGFR forms, even when the urea moiety was substituted with 3-CF3 and 4-Cl groups (compounds 7c, 7g). This data reinforces that compounds with highly reactive warheads might inhibit protein kinase activity in a non-specific manner, making this warhead an improper subunit to design selective TKIs.
Table 1.
EGFR (EGFRwt, EGFRL858R and EGFRL858R/T790M) IC50 values (nM ± S.D./n ≥ 3) determined for compounds 7a-m.
| IC50 (nM) ± S.D. | |||
|---|---|---|---|
| compounds | EGFRwt | EGFRL858R | EGFRL858R/T790M |
| gefitinib | <1 | <1 | 185 ± 98 |
| afatinib | <1 | <1 | <1 |
| osimertinib | 1 ± 0.6 | <1 | <1 |
| (7a) | >10000 | >10000 | >10000 |
| (7b) | >10000 | >10000 | >10000 |
| (7c) | >10000 | 3237 ± 1257 | 2710 ± 412 |
| (7d) | >10000 | >10000 | 9867 ± 265 |
| (7e) | >10000 | >10000 | 6139 ± 3469 |
| (7f) | 2499 ± 735 | 2163 ± 1623 | >10000 |
| (7g) | 179 ± 61 | 2099 ± 505 | 9829 ± 297 |
| (7h) | 25 ± 4 | 18 ± 3 | 1682 ± 1506 |
| (7i) | >10000 | 5577 ± 4163 | >10000 |
| (7j) | 123 ± 134 | 270 ± 202 | 823 ± 613 |
| (7k) | 3117 ± 787 | 445 ± 105 | 6293 ± 2789 |
| (7l) | 101 ± 12 | 32 ± 14 | 132 ± 49 |
| (7m) | >10000 | >10000 | >10000 |
According to our data, compound 7h, which has the simplest structure in the series, showed the highest inhibitory activity on EGFRwt and EGFRL858R forms. In comparison to the data obtained for compounds 7a, 7f and 7m, these data endorse the deleterious contribution of substituents at the phenylurea moiety; and point out that further molecular modifications are needed for more efficient EGFR mutant forms’ inhibitors’ identification.
Molecular docking
The experimental data are indicative that the presence of a meta or para substituent at the phenyl group was deleterious for the EGFR inhibition, so attempts to elucidate the binding mode with the enzyme were only implemented with the non-substituted compounds 7h-7l, by means of molecular docking with GOLD 5.4 in the afatinib-containing wt-EGFR structure (PDB code: 4G5J).
Compounds 7h, 7i and 7l have Michael acceptor groups, whereas compounds 7j and 7k have chloride and cyanide at the α-carbon to the carbonyl, respectively, which can act as leaving groups, so that a covalent bond can be possibly formed with the Cys797A sulfur atom by all compounds.
Initially, simple and covalent docking of the three Micheal acceptor inhibitors were performed to identify possible binding modes that could help in the explanation of the loss of activity of compound 7i compared to the two other compounds.
The ChemPLP fitness function presented the best performance both in simple (RMSD equal to 2.81 Å) and covalent redocking studies (2.50 Å) based on the 4G5J [51] crystallographic structure. Simple docking studies confirmed the hypothesis that covalent ligands firstly form noncovalent adducts in the ATP binding site before the covalent bond is formed.
It was observed that all compounds have the same binding mode before the covalent bond is formed (Figs S1 and S2, supplementary material).
Covalent docking studies were performed at the electrophilic α-carbon of the carbonyl subunit (compounds 7j and 7k) and at the β-carbon of the enone subunit (7h, 7i and 7l).
Although molecular docking programs are effective in producing ligand-enzyme interaction geometries, the respective scores do not match the experimental activity data so well. For this reason, for compounds 7j and 7k the generated enzyme-inhibitor complexes (Fig. S3, supplementary material) were then used as input geometries for the calculation with the semi-empirical method PM7 [50] of the reaction enthalpies, which play a significant role in the enzyme-inhibitor complex stability. The results were analyzed from the point of view of the relative reaction enthalpies for the formation of a ligand-enzyme adduct, obtained by the nucleophilic substitution of the cysteine residue (Cys797) at the α-carbon of carbonyl subunit (Fig. 4A). As can be seen in Table 2, the reaction enthalpy for the formation of the enzyme-inhibitor complex of 7j is much more favorable than that of 7k, in qualitative accordance with the greater activity of the former.
Figure 4.
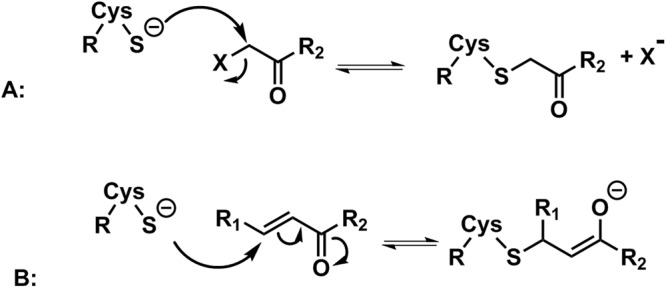
Cysteine (Cys797) residue attack scheme at the electrophilic carbon of the α-carbon of carbonyl subunit (A) and the enone subunit (B) of the quinoxaline urea derivatives.
Table 2.
Calculated enzyme-inhibitor reaction relative enthalpies (kcal/mol) according to the reaction depicted in Fig. 6 (PM7 method, dielectric constant = 78.4).
| Ligand | Relative Enthalpy |
|---|---|
| According to scheme 2A | |
| 7j | 0 |
| 7k | 61.46 |
| According to scheme 2B | |
| 7h | 0 |
| 7i | 22.56 |
| 7l | 2.61 |
In the same way for compounds 7h, 7i and 7l, with Michael acceptor groups, the calculated enthalpies generated from the enzyme-inhibitor complexes (Fig. S4, supplementary material) were analyzed. The relative reaction enthalpies for the formation of the ligand-enzyme adducts were obtained from the nucleophilic attack of the cysteine residue (Cys797) at the electrophilic carbon of the enone subunit (Fig. 4B). The Michael complex formed from compound 7h is slightly more stable than the complex generated from compound 7l and both are much more stable than the one produced from compound 7i (Table 2). Once again, the reaction enthalpy data are in qualitative accordance with the experimental results: the lower stability of the enzyme-inhibitor complex from compound 7i may explain its lower activity, since the unfavorable reaction enthalpy would contribute to reduce the formation of the enzyme-inhibitor complex in comparison with compounds 7l and 7h.
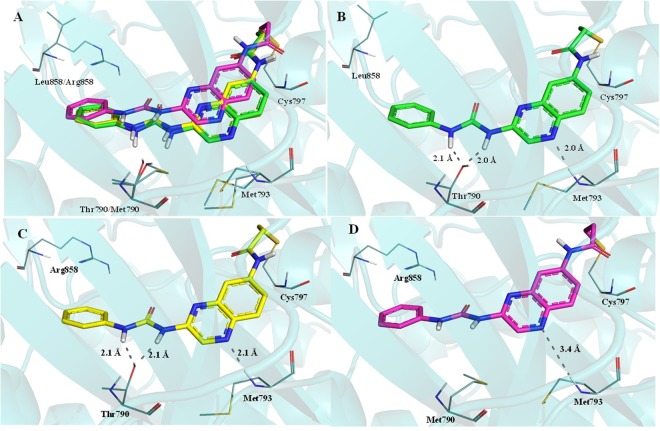
Additionally, covalent docking studies with the quinoxaline ureas 7h and 7l in the ATP binding site of EGFRwt, EGFRL858R and EGFRL858R/T790M were performed in an attempt to provide a structural rationale for the differences observed in their activities against these enzymes. Strong binding interactions in EGFRwt and EGFRL858R were observed for compound 7h, involving the NH group of the hinge’s Met793 and the OH group of Thr790 (Fig. 5). On the other hand, no interaction with Met790 was observed for compound 7h in the ATP binding site of EGFR double mutant form (i.e. EGFRL858R/T790M), although a weak interaction has been observed with Met793, probably due to the absence of the Thr790 hydroxyl group and the steric hindrance of the Met790 side chain (Fig. 5). These results can explain the equipotency of 7h in inhibiting EGFRwt and EGFRL858R, and its lower activity against EGFRL858R/T790M.
Figure 5.
Compound 7h interaction profile in the ATP binding site of EGFRwt, EGFRL858R and EGFRL858R/T790M (A). Interaction profile of compound 7h in EGFRwt (carbon atoms in green, B), in EGFRL858R (carbon atoms in yellow, C) and in EGFRL858R/T790M (carbon atoms in magenta, D); Dashed gray lines: Hydrogen bonds.
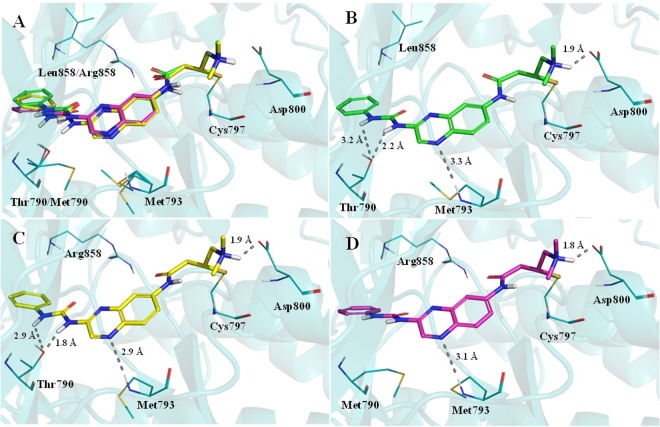
As depicted in Fig. 6, covalent docking showed that urea 7l binds properly and similarly in EGFRwt, EGFRL858R and EGFRL858R/T790M. The quinoxaline’s nitrogen makes a similar interaction with the Met793, and the quaternary amine establishes ionic interactions with Asp800. The urea subunit was able to interact with Thr790 hydroxyl group only in the EGFRwt and EGFRL858R forms (Fig. 6). The covalent adducts formed between Cys797 and compound 7l, for all the EGFR forms, are presented in Fig. 6.
Figure 6.
Compound 7l interaction profile in the ATP binding site of EGFRwt, EGFRL858R and EGFRL858R/T790M (A). Interaction profile of compound 7l in EGFRwt (carbon atoms in green, B), in EGFRL858R (carbon atoms in yellow, C) and in EGFRL858R/T790M (carbon atoms in magenta, D). Dashed gray lines: Hydrogen bonds.
Cellular panel screening
Patient-derived tumor cells proved to be an important approach to improve predictability of effectiveness during future drug development clinical phases47,48. Based on this premise, the inhibitory activity of compounds 7a-m towards a patient-derived tumor cell panel49 was evaluated, using a single screening concentration of 10 μM. Despite its nanomolar potency against the three EGFR forms, compound 7l did not inhibit the Oncotest® tumor cell lines’ growth in a significant manner. These results suggest permeation issues for compound 7l. In fact, it is worth mentioning that, although the data observed in a cellular assay have no direct correlation to EGFR enzymatic tests, this phenomenon is not unusual and is related to the nature of the assays itself. In enzymatic assays there is no influence of membrane permeability issues, while in cell viability assays compounds must penetrate the cell membrane and resist the cellular environment for long enough for the ligand-receptor interaction to occur and to trigger the biological activity50. The most potent EGFRwt and EGFRL858R inhibitor, 7h, showed important cytotoxic activity. It displayed a moderate growth inhibition effect (55–77%) upon sensible (GXF 251L, CXF DiFi and LXFA PC9) and first generation EGFR inhibitor resistant (LXFL 529L and LXFA NCI-H1975) patient-derived tumor cell lines (Table 3).
Table 3.
Cytotoxic activity on a patient-derived (PD) tumor cell line panel determined for compound 7h at 10 μM. Results are expressed by cellular growth inhibition percentage related to negative control.
| Tumor type | PD tumor cell line | % inhibition | Tumor type | PD tumor cell line | % inhibition |
|---|---|---|---|---|---|
| Stomach | GXF 251Lb | 55% | Breast | MAXF 401NL | 72% |
| GXA MKN45 | 8% | MAXF MDA231 | 67% | ||
| Colon | CXF DIFIb | 77% | MAXF MCF7 | 76% | |
| CXF Colo205 | 56% | Bladder | BXF 1218L | 84% | |
| CXF SW620 | 82% | BXF T24 | 76% | ||
| CXF RKO | 82% | Kidney | RXF 486L | 86% | |
| CXF HCT116 | 92% | RXF 786-O | 80% | ||
| Lung | LXFA PC9b | 75% | Glioblastoma | CNXF A172 | 86% |
| LXFA NCI-H1975c | 68% | Prostate | PRXF DU145 | 91% | |
| LXFL H460 | 33% | Uterus | UXF 1138L | 91% | |
| LXFL 529La | 74% | Liver | LIXF 575L | 40% | |
| LXFA 629L | 76% | Ovary | OVXF 899L | 87% | |
| Melanoma | MEXF 276L | 88% | Pancreas | PAXF 546L | 58% |
| MEXF 1737l | 15% | PAXF 1657l | 89% | ||
| MEXF 1539L | 93% | Sarcoma | SXF1301L | 89% |
aEGFR overexpressed, bEGFRdel-exon19 expressed, cEGFRwt expressed.
Conclusions
A novel and original scaffold was identified for the wild-type and clinically relevant mutant forms of EGFR inhibition. Among the compounds herein described, 7h is a potent inhibitor of EGFRwt and E.GFRL858R in the nanomolar range and showed cellular cytotoxic activity (>80% at 10 µM) on Oncotest® patient-derived tumor cell lines, while being selective for EGFR related cells. Molecular docking and reaction enthalpy calculations have shown the influence of the combination of reversible and covalent binding modes with EGFR on the inhibitory activity.
Methods
Synthesis and characterization of compounds
All commercially available reagents and solvents were used without further purification, except compound 7-nitroquinoxalin-2-amine (8) synthetized as previously described41. 1H and 13C nuclear magnetic resonance (NMR) and DEPT135 spectra were determined in DMSO-d6 or pyridine-d5 solutions using a Bruker AC-200 or a Bruker Avance 400 spectrometer. NOESY 1D experiments were determined using a Varian Unity-300 spectrometer. The chemical shifts are indicated in parts per million (δ) from solvent residual peaks and the coupling constant values (J) are indicated in Hz. Signal multiplicities are represented by: s (singlet), d (doublet), dd (double doublet), t (triplet), dt (double triplet), m (multiplet) and br (broad signal). Infrared spectra were obtained using a Thermo Scientific Nicolet’s Avatar iS10 spectrometer equipped with smart endurance diamond ATR unit for direct measurements. Mass spectra were obtained from a TLC-MS interface CAMAG in negative mode and from a Hewlett Packard HP 5973 mass selective detector (70 eV). Melting points (m.p.) were determined using a MP70 Mettler Toledo and are uncorrected. The purity of compounds was determined by HPLC (Merck Hitachi L- 6200 intelligent pump, Merck Hitachi AS-2000 auto sampler, Merck Hitachi L-4250 UV vis detector) using a ZORBAX Eclipse XDB C8 column (5 mm), employing a gradient of 0.01 M KH2PO4 (pH 2.3) and methanol as solvent system with a flow rate of 1.5 mL/min and detection at 254 nm.
General methodology for the synthesis of 1-(7-nitroquinoxalin-2-yl)-3-phenylureas (9a-d)
To a round bottom flask containing a suspension of 7-nitroquinoxalin-2-amine (0.5 g, 2.5 mmol) in dry toluene, one equivalent (2.5 mmol) of substituted phenylisocyanate was added. Suspension was kept in refluxing conditions for 2–4 hours. After reactant consumption, at room temperature, solids were filtered and washed with toluene to obtain salmon color solids.
1-(7-nitroquinoxalin-2-yl)-3-(4-chloro-3-(trifluormethyl)phenyl)urea (9a)
Compound 9a was synthetized via condensation of 8 with 4-chloro-3-(trifluoromethyl)phenyl isocyanate resulting in a salmon powder with 82% yield. Melting point (m.p.) was 266–269 °C. 1H NMR (200 MHz, DMSO-d6) δ (ppm): 10.90 (1H, s), 10.75 (1H, s), 9.14 (1H, s), 8.91 (1H, d, J = 4 Hz), 8.32 (1H, dd), 8.30 (1H, d, J = 4 Hz,), 8.19 (1H, d, J = 9 Hz), 7.91 (1H, dd), 7.70 (1H, d, J = 9 Hz). 1H NMR (200 MHz, piridine-d5) δ (ppm): 11.82 (1H, s), 9.29 (1H, s), 8.91 (1H, d, J = 2 Hz), 8.70 (1H, d, J = 2 Hz), 8.40 (1H, dd, J = 2, 10 Hz), 8.26 (1H, d, J = 8 Hz), 8.00 (1H, d, J = 8 Hz), 7.57 (1H, d, J = 8 Hz), 4.98 (br, exchange with D2O). 13C NMR (50 MHz, piridine-d5) δ (ppm): 153.0, 150.8, 149.3, 143.9, 142.0, 139.2, 139.0, 136.3, 132.8, 131.5, 129.2, 128.6, 124.3, 123.7, 121.4, 120.0–119.7. IR (ATR: cm−1): 2971, 1702, 1589, 1538, 1347, 737. MS: ESI-: m/z 410.2 [M-1]-; 412.2 [M + 2-1]-.
1-(7-nitroquinoxalin-2-yl)-3-(3-(trifluormethyl)phenyl)urea (9b)
Compound 9b was synthetized via condensation of 8 with 3-(trifluoromethyl)phenyl isocyanate resulting in a salmon powder with 65% yield. m.p. was 250–252 °C. 1H NMR (200 MHz, DMSO-d6) δ (ppm): 10.84 (1H, s), 10.71 (1H, s), 9.17 (1H, s), 8.90 (1H, d, J = 4 Hz), 8.33 (1H, dd), 8.20 (2H, m), 7,85 (1H, d, J = 7 Hz), 7.61 (1H, t, J = 8 Hz), 7.45 (1H, d, J = 8 Hz). 13C NMR (50 MHz, DMSO-d6) δ (ppm): 151.7, 148.7, 148.1, 143.1, 143.0, 140.8, 139.1, 138.5, 130.4, 130.0, 129.3, 123.8, 122.9, 120.6, 119.8, 115.7. IR (ATR: cm−1): 3072, 2968, 1695, 1620, 1545, 1348, 728. MS: ESI-: m/z 376.2 [M-1]-.
1-(7-nitroquinoxalin-2-yl)-3-phenylurea (9c)
Compound 9c was synthetized via condensation of 8 with phenyl isocyanate resulting in a salmon powder with 67% yield. m.p. was 253–255 °C. 1H NMR (200 MHz, DMSO-d6) δ (ppm): 10.63 (1H, s), 10.59 (1H, s), 9.16 (1H, s), 8.83 (1H, d, J = 2 Hz), 8.35 (1H, dd), 8.21 (1H, d, J = 8 Hz), 7.68 (2H, d, J = 8 Hz), 7.38 (2H, t, J = 8 Hz), 7.11 (1H, t, J = 8 Hz). 13C NMR and DEPT 135 (50 MHz, DMSO-d6) δ (ppm): 151.5, 148.8, 148.1, 143.2, 140.7, 138.5, 138.2, 130.5, 128.9, 123.5, 122.7, 120.5, 119.8. IR (ATR: cm−1): 3069, 2974, 1690, 1617, 1538, 1343, 737. MS: ESI-: m/z 308.2 [M-1]-.
1-(3-chloro-4-fluorophenyl)-3-(7-nitroquinoxalin-2-yl)urea (9d)
Compound 9d was synthetized via condensation of 8 with 3-chloro-4-fluorophenyl isocyanate resulting in a salmon powder with 68% yield. m.p. was 253–256 °C.1H NMR (200 MHz, DMSO-d6) δ(ppm): 10.81 (1H, s), 10.73 (1H, s), 9.10 (1H, s), 8.95 (1H, d, J = 4 Hz), 8.35 (1H, dd, J = 2 and 9 Hz), 8.20 (1H, d, J = 9 Hz), 8.02 (1H, dd, J = 2 and 7 Hz), 7.69–7.61 (1H, m), 7.43 (1H, t, J = 9 Hz). 13C NMR (50 MHz, DMSO-d6) δ (ppm): 151.7, 148.6, 148.1, 143.2, 140.7, 138.4, 135.4, 135.4, 130.4, 122.9, 121.5, 120.7, 120.6, 119.5, 119.1, 117.2, 116.7. IR (ATR: cm−1): 2986, 1694, 1612, 1542, 1345, 728. MS: ESI-: m/z 360.2 [M-1]-; 362.2 [M + 2–1].
General methodology for the synthesis of 1-(7-aminoquinoxalin-2-yl)-3-phenylureas (10a-d)
In a round bottom flask, derivatives 9a-d and five equivalent of tin (II) chloride dihydrate (SnCl2.2H2O) in ethanol were refluxed for 17 hours. After the nitro group reduction to an aniline group, the solvent was evaporated under reduced pressure, sodium carbonate solution was used to adjust pH to 7, ethyl acetate was added for product extraction by the organic phase, further dried with anhydrous sodium sulfate, filtered and reduced to furnish anilines as yellow solids.
1-(7-aminoquinoxalin-2-yl)-3-(4-chloro-3-(trifluormethyl)phenyl) urea (10a)
Compound 10a was synthetized via reduction of 9a with SnCl2.2H2O resulting in a yellow powder with 91% yield and the m.p. was >300 °C. 1H NMR (200 MHz, DMSO-d6) δ (ppm): 11.40 (1H, s), 10.21 (1H, s), 8.51 (1H, s), 8.24 (1H, d, J = 2 Hz), 7.76–7.72 (2H, m), 7.63 (1H, d, J = 9 Hz), 7.03 (1H, dd), 6.82 (1H, d, J = 2 Hz), 6.07 (2H, br). 13C NMR and DEPT 135 (100 MHz, DMSO-d6) δ (ppm): 152.1, 151.4, 147.2, 141.1, 138.2, 136.8, 132.4, 132.3, 132.1, 129.4, 129.4, 127.9, 124.1, 123.6, 121.4, 119.0, 103.9. IR (ATR: cm−1): 3219, 2980, 1691, 1621, 1573, 745. MS: ESI-: m/z 380.2 [M-1]-; 382.2 [M + 2–1]-.
1-(7-aminoquinoxalin-2-yl)-3-(3-(trifluormethyl)phenyl)urea (10b)
Compound 10b was synthetized via reduction of 9b with SnCl2.2H2O resulting in a yellow powder with 82% yield and the m.p. was 257–261 °C. 1H NMR (200 MHz, DMSO-d6) δ (ppm): 11.43 (1H, s), 10.18 (1H, s), 8.51 (1H, s), 8.14 (1H, s), 7.69–7.61 (3H, m), 7.43 (1H, d, J = 8 Hz), 7.03 (1H, dd, J = 2 and 9 Hz), 6.83 (1H, d, J = 2 Hz), 6.06 (2H, br). 13C NMR (100 MHz, DMSO-d6) δ (ppm): 152.2, 151.4, 147.3, 141.1, 139.6, 132.3, 130.1, 129.7, 129.4, 128.9, 122.8, 119.2, 118.9, 115.0, 104.1, 103.8. IR (ATR: cm−1): 3361, 3222, 2986, 1692, 1621, 1567, 746. MS: ESI-: m/z 347.2 [M-1]-.
1-(7-aminoquinoxalin-2-yl)-3-phenylurea (10c)
Compound 10c was synthetized via reduction of 9c with SnCl2.2H2O resulting in a yellow powder with 61% yield and the m.p. was 243–247 °C. 1H NMR (200 MHz, DMSO-d6) δ (ppm): 11.35 (1H, s), 10.14 (1H, s), 8.54 (1H, s), 7.64–7.57 (3H, m), 7.35 (1H, t, J = 8 Hz), 7.06 (1H, t, J = 8 Hz), 7.03 (1H, dd, J = 2 and 8 Hz), 6.81 (1H, d, J = 2 Hz), 6.04 (1H, br). 13C NMR and DEPT 135 (50 MHz, DMSO-d6) δ (ppm): 152.1, 151.4, 147.6, 141.2, 138.8, 132.5, 132.2, 129.4, 129.0, 122.9, 118.9, 118.6 (C6), 103.8. IR (ATR: cm−1): 3463, 3339, 3217, 2984, 1687, 1627, 1562, 750. MS: ESI-: m/z 278.2 [M-1].
1-(7-aminoquinoxalin-2-yl)-3-(3-chloro-4-fluorophenyl) urea (10d)
Compound 10d was synthetized via reduction of 9d with SnCl2.2H2O resulting in a yellow powder with 37% yield. 1H NMR (200 MHz, DMSO-d6) δ (ppm): 11.36 (1H, s), 10.18 (1H, s), 8.46 (1H, s), 7.92 (1H, dd, J = 2 and 7 Hz), 7.62 (1H, d, J = 9 Hz), 7.48–7.41 (2H, m), 7.05 (1H, dd, J = 2 and 9 Hz), 6.83 (1H, d, J = 2 Hz), 6.05 (2H, br). IR (ATR: cm−1): 3461, 3333, 3217, 2981, 1693, 1622, 1564, 739. MS: ESI-: m/z 330.2 [M-1]-; 332.2 [M + 2–1]-.
General methodology for the synthesis of N-(3-(3-phenylureido)quinoxalin-6-yl) amide derivatives
Anilines 10a-d were solubilized in dry THF and 1.1 equivalent of N,N-diisopropylethylamine (DIPEA) was added in a round bottom flask under argon atmosphere. Then, corresponding acyl chloride (1 equivalent) was dropwise added into the mixture. The reaction mixture was kept in ice cold bath until the total conversion of reagents to products. Isolation and purification of compounds were performed by flash chromatography (mobile phases: dichloromethane: methanol 4–8%; dichloromethane: ethyl acetate 10–90%; ethyl acetate: methanol: ammonia in methanol 9:0.8:0.2 or ethyl acetate: methanol 10%).
N-(3-(3-(4-chloro-3-(trifluoromethyl)phenyl)ureido)quinoxalin-6-yl) acrylamide (7a)
Compound 7a was synthetized via condensation of 10a with acryloyl chloride resulting in a white powder with 70% yield after flash chromatography (mobile phase: DCM:MeOH 4–8%) and the m.p. was 272–274 °C. 1H NMR (200 MHz, DMSO-d6) δ (ppm): 10.88 (1H, s), 10.60 (1H, s), 10.37 (1H, s), 8.97 (1H, s), 8.38 (1H, d, J = 2 Hz), 8.23 (1H, s), 7.95 (1H, d, J = 10 Hz), 7.81 (1H, dd, J = 2 and 9 Hz), 7.72 (2H, s), 6.46 (1H, dd, J = 10 and 17 Hz), 6.35 (1H, dd, J = 2 and 17 Hz), 5.85 (1H, dd, J = 2 and 9 Hz). 13C NMR (50 MHz, DMSO-d6) δ (ppm): 163.7, 151.9, 147.6, 140.9, 139.8, 138.1, 137.5, 135.4, 132.2, 131.5, 129.2, 127.9–126.6, 124.2, 123.8, 120.7, 120.0, 117.8, 114.2. IR (ATR: cm−1): 2924, 1687, 1584, 1130, 1032. MS: ESI-: m/z 434.2 [M-1]-; 436.2 [M + 2–1]-. Purity (HPLC at 254 nm; R.T.): 100%; 8.37 minutes.
(E)-N-(3-(3-(4-chloro-3-(trifluoromethyl)phenyl)ureido)quinoxalin-6-yl)but-2-enamide (7b)
Compound 7b was synthetized via condensation of 10a with crotonyl chloride resulting in a salmon powder with 37% yield after flash chromatography (mobile phase: DCM:MeOH 4–8%) and the m.p. was 277–280 °C. 1H NMR (200 MHz, DMSO-d6) δ (ppm): 10.90 (1H, s), 10.40 (1H, s), 10.34 (1H, s), 8.95 (1H, s), 8.35 (1H, d, J = 2 Hz), 8.22 (1H, s), 7.93 (1H, d, J = 10 Hz), 7.78 (1H, dd, J = 2 and 10 Hz), 7.72 (2H, s), 6.88 (1H, dd, J = 7 and 15 Hz), 6.19 (1H, dd, J = 2 and 15 Hz), 1.90 (3H, dd, J = 1,5 and 7 Hz). 13C NMR (50 MHz, DMSO-d6) δ (ppm): 164.0, 151.9, 147.6, 141.2, 141.0, 139.8, 138.1, 137.3, 135.2, 132.2, 129.1, 127.2–126.6, 125.7, 124.1, 123.8, 120.7, 117.8, 113.9, 17.6. IR (ATR: cm−1): 3266, 2971, 1695, 1675, 1641, 1618, 1582, 1124, 1031. MS: ESI-: m/z 448.1 [M-1]-; 450.1 [M + 2–1]-. Purity (HPLC at 254 nm; R.T.): 100.0%; 8.75 minutes.
2-chloro-N-(3-(3-(4-chloro-3-(trifluoromethyl)phenyl)ureido)quinoxalin-6-yl)acetamide (7c)
Compound 7c was synthetized via condensation of 10a with chloroacethyl chloride resulting in a pearl powder with 38% yield after flash chromatography (mobile phase: DCM: AcOEt 10–90%) and the m.p. was 253–255 °C. 1H NMR (200 MHz, DMSO-d6) δ (ppm): 10.88 (1H, s), 10.76 (1H, s), 10.39 (1H, s), 8.99 (1H, s), 8.29 (1H, d, J = 2 Hz), 8.21 (1H, s), 7.97 (1H, d, J = 8 Hz), 7.75 (3H, m), 4.36 (2H, s). 13C NMR (50 MHz, DMSO-d6) δ (ppm): 165.3, 151.9, 147.7, 140.3, 139.7, 138.1, 137.8, 135.4, 132.2, 129.3, 127.2–126.6, 124.2, 123.8, 120.6, 117.9, 114.3, 43.6. IR (ATR: cm−1): 3335, 2962, 1687, 1615, 1584, 1130,1030. MS: ESI-: m/z 456.0 [M-1]-; 458.0 [M + 2–1]-. Purity (HPLC at 254 nm; R.T.): 97.0%; 8.60 minutes.
N-(3-(3-(4-chloro-3-(trifluoromethyl)phenyl)ureido)quinoxalin-6-yl)-2-cyanoacetamide (7d)
Compound 7d was synthetized via condensation of 10a with 2-cyanoacethyl chloride resulting in a pearl powder with 17% yield after flash chromatography (mobile phase: DCM:AcOEt 10–90%) and the m.p. was 243–246 °C. 1H NMR (200 MHz, DMSO-d6) δ (ppm): 10.85 (1H, s), 10.76 (1H, s), 10.38 (1H, s), 8.99 (1H, s), 8.23 (2H, m), 7.95 (1H, d, J = 9 Hz), 7.70 (3H, m), 4.02 (2H, s). 13C NMR (50 MHz, DMSO-d6) δ (ppm): 161.9, 151.9, 147.7, 140.2, 139.7, 138.1, 137.9, 135.4, 132.2, 129.4, 127.2–126.6, 124.1, 123.8, 120.4, 120.0, 117.9, 115.7, 114.3, 27.1. IR (ATR: cm−1): 3336, 2971, 2264, 1694, 1596, 1127, 1032. MS: ESI-: m/z 447.0 [M-1]-; 449.0 [M + 2–1]-. Purity (HPLC at 254 nm; R.T.): 97.4%; 7.64 minutes.
(E)-N-(3-(3-(4-chloro-3-(trifluoromethyl)phenyl)ureido)quinoxalin-6-yl)-4-(dimethylamino)but-2-enamide (7e)
Compound 7e was synthetized via condensation of 10a with 4-(dimethylamino)but-2-enamide chloride resulting in a pearl powder with 23% yield after flash chromatography (mobile phase: AcOEt:MeOH:NH3 in MeOH 9:0.8:0.2) and the m.p. was 217–220 °C. 1H NMR (200 MHz, DMSO-d6) δ (ppm): 10.88 (1H, s), 10.50 (1H, s), 10.34 (1H, s), 8.96 (1H, s), 8.37 (1H, d, J = 2 Hz), 8.22 (1H, s), 7.94 (1H, d, J = 9 Hz), 7.78 (1H, dd, J = 2 and 9 Hz), 7.22 (1H, s), 6.89–8.76 (1H, dt, J = 6 and 16 Hz), 6.34 (1H, d, J = 16 Hz), 3.09 (2H, d, J = 6 Hz), 2.20 (6H, s). 13C NMR (150 MHz, DMSO-d6) δ (ppm): 163.1, 151.9, 147.6, 140.8, 140.7, 139.9, 138.2, 137.5, 135.4, 132.2, 129.2, 127.0–126.9, 125.5, 124.0, 123.7, 121.8, 120.7, 117.7, 114.2, 58.1, 43.4 MS: ESI-: m/z 491.3 [M-1]-; 493.,3 [M + 2–1]-. Purity (HPLC at 254 nm; R.T.): 100.0%; 5.90 minutes.
N-(3-(3-(3-(trifluoromethyl)phenyl)ureido)quinoxalin-6-yl)acrylamide (7f)
Compound 7f was synthetized via condensation of 10b with acryloyl chloride resulting in a pearl powder with 41% yield after flash chromatography (mobile phase: DCM: AcOEt 10–90%) and the m.p. was 268–271 °C. 1H NMR (200 MHz, DMSO-d6) δ (ppm): 10.89 (1H, s), 10.61 (1H, s), 10.34 (1H, s), 8.98 (1H, s), 8.38 (1H, d, J = 2 Hz), 8.15 (1H, s), 7.96 (1H, d, J = 9 Hz), 7.83 (1H, dd, J = 2 and 9 Hz), 7.79–7.62 (2H, m), 7.44 (1H, d, J = 8 Hz), 6.50 (1H, dd, J = 9 and 17 Hz), 6.35 (1H, dd, J = 2 and 17 Hz), 5.85 (1H, dd, J = 2 and 9 Hz). 13C NMR (50 MHz, DMSO-d6) δ (ppm): 163.7, 152.0, 147.7, 140.9, 139.8, 139.4, 137.6, 135.3, 131.6, 130.2, 129.4, 129.2, 127.9, 122.9, 121.4, 120.7, 117.8, 115.1, 114.2. IR (ATR: cm−1): 3271, 2976, 1687, 1666, 1625, 1577, 1120. MS: ESI-: m/z 400.3 [M-1]-. Purity (HPLC at 254 nm; R.T.): 100%; 7.78 minutes.
2-chloro-N-(3-(3-(3-(trifluoromethyl)phenyl)ureido)quinoxalin-6-yl)acetamide (7g)
Compound 7g was synthetized via condensation of 10b with chloroacethyl chloride resulting in a white powder with 45% yield after flash chromatography (mobile phase: DCM:AcOEt 10–90%) and the m.p. was 260–263 °C. 1H NMR (200 MHz, DMSO-d6) δ (ppm): 10.89 (1H, s), 10.76 (1H, s), 10.36 (1H, s), 8.99 (1H, s), 8.28 (1H, d, J = 2 Hz), 8.14 (1H, s), 7.97 (1H, d, J = 9 Hz), 7.75 (1H, dd, J = 2 and 9 Hz), 7.67–7.57) (2H, m), 7.44 (1H, d, J = 7 Hz), 4.36 (2H, s). 13C NMR (50 MHz, DMSO-d6) δ (ppm): 165.3, 151.9, 147.8, 140.3, 139.7, 139.4, 137.8, 135.4, 130.2, 130.0, 129.4, 129.3, 122.8, 120.5, 115.2, 114.3, 105.7, 43.64. IR (ATR: cm−1): 3305, 2970, 1684, 1666, 1621, 1575, 1120.MS: ESI-: m/z 422.1 [M-1]-; 423.9 [M + 2–1]-. Purity (HPLC at 254 nm; R.T.): 97,5%; 8.07 minutes.
N-(3-(3-phenylureido)quinoxalin-6-yl)acrylamide (7h)
Compound 7h was synthetized via condensation of 10c with acryloyl chloride resulting in a white powder with 38% yield after flash chromatography (mobile phase: DCM: AcOEt 10–90%) and the m.p. was 280–283 °C. 1H NMR (200 MHz, DMSO-d6) δ (ppm): 10.85 (1H, s), 10.60 (1H, s), 10.27 (1H, s), 8.91 (1H, s), 8.36 (1H, d, J = 2 Hz), 7.95 (1H, d, J = 8 Hz), 7.82 (1H, dd, J = 2 and 8 Hz), 7.58 (2H, d, J = 8 Hz), 7.38 (2H, t, J = 8 Hz), 7.09 (1H, t, J = 8 Hz), 6.59–6.46 (1H, dd, J = 10 and 17 Hz), 6.40–6.30 (1H, dd, J = 2 and 16 Hz), 5.88–5.82 (1H, dd, J = 2 and 10 Hz). 13C NMR and DEPT135 (50 MHz, DMSO-d6) δ (ppm): 163.7, 151.8, 147.9, 140.9, 139.6, 138.5, 137.7, 135.1, 131.5, 129.2, 129.1, 127.9, 123.2, 120.4, 119.1, 114.0. IR (ATR: cm−1): 3266, 3032, 2977, 1686, 1662, 1621, 1585. MS: ESI-: m/z 332.2 [M-1]-. Purity (HPLC at 254 nm; R.T.): 95.5%; 6.13 minutes.
(E)-N-(3-(3-phenylureido)quinoxalin-6-yl)but-2-enamide (7i)
Compound 7i was synthetized via condensation of 10c with crotonyl chloride resulting in a white powder with 29% yield after flash chromatography (mobile phase: DCM: AcOEt 10–90%) and the m.p. was 273–276 °C. 1H NMR (200 MHz, DMSO-d6) δ (ppm): 10.86 (1H, s), 10.40 (1H, s), 10.25 (1H, s), 8.89 (1H, s), 8.34 (1H, d, J = 2 Hz), 7.93 (1H, d, J = 10 Hz), 7.79 (1H, dd, J = 2 and 9 Hz), 7.58 (2H, d, J = 7 Hz), 7.38 (2H, t, J = 8 Hz), 7.09 (1H, t, J = 7 Hz), 6.99–6.81 (1H, m), 6.20 (1H, d, J = 16 Hz), 1.90 (3H, d, J = 6 Hz). 13C NMR (50 MHz, DMSO-d6) δ (ppm): 164.0, 151.8, 147.9, 141.2, 141.1, 139.7, 138.5, 137.5, 135.0, 129.1, 129.1, 125.7, 123.2, 120.4, 119.1, 113.8, 17.7. IR (ATR: cm−1): 3274, 2974, 1693, 1673, 1636, 1586. MS: ESI-: m/z 346.3 [M-1]-. Purity (HPLC at 254 nm; R.T.): 98.5%; 6.58 minutes.
2-chloro-N-(3-(3-phenylureido)quinoxalin-6-yl)acetamide (7j)
Compound 7j was synthetized via condensation of 10c with 2-chloroacethyl chloride resulting in a white powder with 36% yield after flash chromatography (mobile phase: DCM:AcOEt 10–90%) and the m.p. was 249–252 °C. 1H NMR (200 MHz, DMSO-d6) δ (ppm): 10.83 (1H, s), 10.76 (1H, s), 10.29 (1H, s), 8.93 (1H, s), 8.26 (1H, s), 7.96 (1H, d, J = 8 Hz), 7.76 (1H, d, J = 8 Hz), 7.58 (2H, d, J = 8 Hz), 7.38 (2H, t, J = 7 Hz), 7.09 (1H, t, J = 7 Hz), 4.36 (2H, s). 13C NMR and DEPT 135 (50 MHz, DMSO-d6) δ (ppm): 165.3, 151.8, 148.0, 140.3, 139.6, 138.5, 137.9, 135.2, 129.3, 129.1, 123.2, 120.2, 119.1, 114.2, 43.7. IR (ATR: cm−1): 3267, 2980, 1681, 1622, 1585. MS: ESI-: m/z 354.1 [M-1]-; 356.1 [M + 2-1]-. Purity (HPLC at 254 nm; R.T.): 96.4%; 6.12 minutes.
2-cyano-N-(3-(3-phenylureido)quinoxalin-6-yl)acetamide (7k)
Compound 7k was synthetized via condensation of 10c with 2-cyanoacethyl chloride resulting in a white powder with 12% yield after flash chromatography (mobile phase: DCM:AcOEt 10–90%) and the m.p. was 238–241 °C. 1H NMR (200 MHz, DMSO-d6) δ (ppm): 10.79 (1H, s), 10.76 (1H, s), 10.26 (1H, s), 8.94 (1H, s), 8.20 (1H, s), 7.96 (1H, d, J = 8 Hz), 7.70 (1H, d, J = 8 Hz), 7.58 (2H, d, J = 8 Hz), 7.38 (2H, t, J = 8 Hz), 7.09 (1H, t, J = 6 Hz), 4.01 (2H, s). 13C NMR (50 MHz, DMSO-d6) δ (ppm): 161.9, 151.8, 148.0, 140.2, 139.6, 138.5, 138.0, 135.2, 129.4, 129.1, 123.2, 120.1, 119.1, 115.7, 114.1, 27.1. MS: ESI-: m/z 345.3 [M-1]-. Purity (HPLC at 254 nm; R.T.): 98.1%; 5.05 minutes.
(E)-4-(dimethylamino)-N-(3-(3-phenylureido)quinoxalin-6-yl)but-2-enamide (7l)
Compound 7l was synthetized via condensation of 10c with 4-(dimethylamino)but-2-enamide chloride resulting in a white powder with 18% yield after flash chromatography (mobile phase: AcOEt:MeOH:NH3 in MeOH 9:0.8:0.2) and the m.p. was 222–225 °C. 1H NMR (200 MHz, DMSO-d6) δ (ppm): 10.86 (1H, s), 10.51 (1H, s), 10.26 (1H, s), 8.90 (1H, s), 8.36 (1H, d, J = 2 Hz), 7.93 (1H d, J = 9 Hz), 7.80 (1H, dd, J = 2 and 8 Hz), 7.58 (2H, d, J = 7 Hz), 7.38 (2H, t, J = 7 Hz), 7.09 (1H, t, J = 6 Hz), 6.90–6.77 (1H, m), 6.34 (1H, d, J = 14 Hz), 3.09 (2H, d, J = 6 Hz), 2.20 (6H, s). 13C NMR and DEPT 135 (50 MHz, DMSO-d6)δ(ppm): 163.8, 151.8, 147.9, 142.6, 141.1, 139.7, 138.5, 137.6, 135.1, 129.2, 129.1, 125.6, 123.2, 120.4, 119.1, 113.9, 59.8, 45.2. IR (ATR: cm−1): 3228, 2976, 1687, 1636, 1586. MS: ESI-: m/z 389.4 [M-1]-. Purity (HPLC at 254 nm; R.T.): 95.5%; 3.16 minutes.
N-(3-(3-(3-chloro-4-fluorophenyl)ureido)quinoxalin-6-yl) acrylamide (7m)
Compound 7m was synthetized via condensation of 10d with acryloyl chloride resulting in a white powder with 24% yield after flash chromatography (mobile phase: DCM: AcOEt 10–90%) and the m.p. was 259–262 °C. 1H NMR (200 MHz, DMSO-d6) δ (ppm): 10.83 (1H, s), 10.59 (1H, s), 10.34 (1H, s), 8.91 (1H, s), 8.35 (1H, d, J = 2 Hz), 7.92 (2H, s e dd, J = 8 Hz), 7.82 (1H, dd, J = 2 and 9 Hz), 7.44 (1H, d, J = 8 Hz), 6.52 (1H, dd, J = 10 and 17 Hz), 6.34 (1H, dd, J = 17 and 2 Hz), 5.85 (1H, dd, J = 2.5 and 10 Hz). 13C NMR (50 MHz, DMSO-d6) δ (ppm): 163.7, 151.9, 147.7, 140.9, 139.6, 137.6, 135.8, 135.7, 135.2, 131.5, 129.2, 127.9, 120.7, 120.6, 119.6, 119.3, 117.3, 116.9, 114.2. IR (ATR: cm−1): 3281, 2981, 1686, 1667, 1614, 1572. MS: ESI-: m/z 384.3 [M-1]-; 386.3 [M + 2-1]-. Purity (HPLC at 254 nm; R.T.): 100%; 7.68 minutes.
Biochemical assay (TR-FRET)
IC50 determinations for EGFR and its mutants (Carna Biosciences, lot13CBS-0005K for EGFRwt; Carna, lot13CBS-0537B for EGFR-L858R and Carna, lot12CBS-0765B for EGFR-L858R/T790M) were performed with the HTRF KinEASE-TK assay from Cisbio according to the manufacturer’s instructions. Briefly, the amount of EGFR in each reaction well was set to 0.60 ng EGFR wild-type (0.67 nM), 0.10 ng EGFR L858R (0.11 nM) or 0.07 ng EGFR T790M/L858R (0.08 nM), respectively. An artificial substrate peptide (TK-substrate from Cisbio) was phosphorylated by EGFR. After completion of the reaction (reaction times: 25 min for wild-type, 15 min for L858R, 20 min for T790M/L858R), the reaction was stopped by addition of a buffer containing EDTA as well as, an anti-phosphotyrosine antibody labeled with europium cryptate and streptavidin labeled with the fluorophore XL665. FRET between europium cryptate and XL665 was measured after an additional hour of incubation to quantify the phosphorylation of the substrate peptide. ATP concentrations were set at their respective Km-values (9.5 μM for EGFR-wt, 9 μM for EGFR-L858R and 4 μM for EGFR-L858R/T790M) while a substrate concentration of 1 μM, 225 nM and 200 nM, respectively, was used. Kinase and inhibitor were preincubated for 30 min (EGFR-wt) and 1 h (EGFR-L858R and EGFR-L858R/T790M) before the reaction was started by addition of ATP and substrate peptide. An EnVision multimode plate reader (Perkin Elmer) was used to measure the fluorescence of the samples at 620 nm (Eu-labeled antibody) and 665 nm (XL665 labeled streptavidin) 50 μs after excitation at 320 nm. The quotient of both intensities for reactions made with eight different inhibitor concentrations was then analyzed using the Quattro Software Suite for IC50-determination. Each reaction was performed in duplicate, and at least three independent determinations of each IC50 were made.
Molecular docking
All the compounds were constructed with Spartan’16 (Key ID: 713413641076066525). A Monte Carlo conformational search was performed with the molecular mechanics method MMFF (Merck Molecular Force Field) and the geometry of the lowest energy conformer of each compound was re-optimized with the semi-empirical method PM651. For compounds 7e and 7l, the amino group was also considered in the protonated form.
The EGFRwt crystallographic structure available in the Protein Data Bank with code 4G5J (resolution: 2.8 Å. in complex with Afatinib)52 was used for docking runs with the GOLD 5.4 program (Validation code: 44d6-05f1-f186-5a7f-190c-7fec-7eff-bb85-9e39). The default fitness score function ChemPLP53 was evaluated for re-docking of the co-crystallized ligand (Afatinib). Crystallographic water molecules were removed during the docking runs, and the binding site was determined within 6 Å around the ligand (Afatinib) in complex with 4G5J. The covalent docking mode was used with the protein link at Cys797 with the ligand link atom at the α or β carbon to the carbonyl group, depending on the ligand structure.
Docking runs were performed in triplicate and the poses presenting the best scores were analyzed to identify potential interactions with amino acid residues of the enzyme’s binding site. The amino acids with more favorable interactions were selected and a semi-rigid docking was performed, which allowed for flexibility of the side chains of Glu762A, Met766A, Leu788A, Thr790A or Met790A, Met793A, Thr854A and Asp855A residues for EGFRwt, EGFRL858R and EGFRL858R/T790M.
The reaction enthalpy for the ligands/enzyme complexes (∆Hr) was calculated from enthalpies for formation (∆Hf) calculated with the semi-empirical PM7 method using the MOPAC2016 program (Stewart Computational Chemistry), according to the following equation:
where L is the ligand; X, leaving group; E, the enzyme; and E − L, the covalently bonded complex.
To reduce the computational cost, only the amino acid residues located within 12 Å from Met793 were used and the calculations were performed with the solvent continuum model using the water dielectric constant, 78.451.
Electronic supplementary material
Acknowledgements
The authors would like to thank Heiner Fiebig and Gerhard Kelter from Oncotest GmbH for determination of cellular inhibition data, INCT-INOFAR (BR573.564/2008-6 and E-26/170.020/2008), CAPES/PSDE (BEX 4188/14-4), ICEPHA-Grants, DFG LA 1453/2-1.
Author Contributions
D.N.A., C.M.R.S., D.R., E.J.B., S.L., L.M.L. conceived and designed the experiment D.N.A., J.L., H.H.F., E.M.B.S. performed the experiments; D.N.A., J.L., H.H.F., E.M.B.S., C.M.R.S., S.L., L.M.L. analyzed the data; E.M.B.S., E.J.B., L.M.L., D.R., S.L. contributed reagents/materials/analysis tools; D.N.A., J.L., H.H.F., C.M.R.S., S.L., L.M.L. wrote the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Stefan Laufer, Email: stefan.laufer@uni-tuebingen.de.
Lidia Moreira Lima, Email: lidialima@icb.ufrj.br.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-36846-7.
References
- 1.Cohen P, Alessi DR. Kinase drug discovery–what’s next in the field? ACS Chem. Biol. 2013;8:96–104. doi: 10.1021/cb300610s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Müller, S., Chaikuad, A., Gray, N. S. & Knapp, S. The ins and outs of selective kinase inhibitor development. Nat. Chem. Biol. 11, 818–821 (2015). [DOI] [PubMed]
- 3.Knapp S, Sundström M. Recently targeted kinases and their inhibitors - The path to clinical trials. Curr. Opin. Pharmacol. 2014;17:58–63. doi: 10.1016/j.coph.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 4.Fabbro D. 25 Years of Small Molecular Weight Kinase Inhibitors: Potentials and Limitations. Mol. Pharmacol. 2015;10:766–775. doi: 10.1124/mol.114.095489. [DOI] [PubMed] [Google Scholar]
- 5.Ferguson, F. M. & Gray, N. S. Kinase inhibitors: the road ahead. Nat. Rev. Drug Discov. 17, 353–377 (2018). [DOI] [PubMed]
- 6.Wu P, Nielsen TE, Clausen MH. Small-molecule kinase inhibitors: An analysis of FDA-approved drugs. Drug Discovery Today. 2016;21:5–10. doi: 10.1016/j.drudis.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 7.Weinstein IB, Joe AK. Mechanisms of Disease: Oncogene addiction - A rationale for molecular targeting in cancer therapy. Nat. Clin. Pract. Oncol. 2006;3:448–457. doi: 10.1038/ncponc0558. [DOI] [PubMed] [Google Scholar]
- 8.Baselga J. Targeting tyrosine kinases in cancer: the second wave. Science. 2006;312:1175–1178. doi: 10.1126/science.1125951. [DOI] [PubMed] [Google Scholar]
- 9.Fedorov O, Müller S, Knapp S. The (un)targeted cancer kinome. Nat. Chem. Biol. 2010;6:166–169. doi: 10.1038/nchembio.297. [DOI] [PubMed] [Google Scholar]
- 10.Peters JU. Polypharmacology - Foe or friend? J. Med. Chem. 2013;56:8955–8971. doi: 10.1021/jm400856t. [DOI] [PubMed] [Google Scholar]
- 11.Morphy R. Selectively nonselective kinase inhibition: striking the right balance. J. Med. Chem. 2010;53:1413–1437. doi: 10.1021/jm901132v. [DOI] [PubMed] [Google Scholar]
- 12.Rewcastle GW, et al. Tyrosine kinase inhibitors. 5. Synthesis and structure-activity relationships for 4-[(phenylmethyl) amino]-and 4-(phenylamino) quinazolines as potent adenosine 5′-triphosphate binding site inhibitors of the tyrosine kinase domain of the epidermal growth f. J. Med. Chem. 1995;38:3482–3487. doi: 10.1021/jm00018a008. [DOI] [PubMed] [Google Scholar]
- 13.Bridges AJ, et al. Tyrosine kinase inhibitors. 8. An unusually steep structure-activity relationship for analogues of 4-(3-bromoanilino)-6,7-dimethoxyquinazoline (PD 153035), a potent inhibitor of the epidermal growth factor receptor. J. Med. Chem. 1996;39:267–276. doi: 10.1021/jm9503613. [DOI] [PubMed] [Google Scholar]
- 14.Tsou HR, et al. 6-Substituted-4-(3-bromophenylamino)quinazolines as putative irreversible inhibitors of the epidermal growth factor receptor (EGFR) and human epidermal growth factor receptor (HER-2) tyrosine kinases with enhanced antitumor activity. J. Med. Chem. 2001;44:2719–2734. doi: 10.1021/jm0005555. [DOI] [PubMed] [Google Scholar]
- 15.Davis MI, et al. Comprehensive analysis of kinase inhibitor selectivity. Nat. Biotechnol. 2011;29:1046–1051. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
- 16.Karaman MW, et al. A quantitative analysis of kinase inhibitor selectivity. Nat. Biotechnol. 2008;26:127–32. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 17.Yarden Y, Pines G. The ERBB network: at last, cancer therapy meets systems biology. Nat. Rev. Cancer. 2012;12:553–563. doi: 10.1038/nrc3309. [DOI] [PubMed] [Google Scholar]
- 18.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat. Rev. Cancer. 2005;5:341–54. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 19.Wieduwilt MJ, Moasser MM. The epidermal growth factor receptor family: Biology driving targeted therapeutics. Cell. Mol. Life Sci. 2008;65:1566–1584. doi: 10.1007/s00018-008-7440-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tebbutt N, Pedersen MW, Johns TG. Targeting the ERBB family in cancer: couples therapy. Nat. Rev. Cancer. 2013;13:663–673. doi: 10.1038/nrc3559. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H, et al. ErbB receptors: From oncogenes to targeted cancer therapies. Journal of Clinical Investigation. 2007;117:2051–2058. doi: 10.1172/JCI32278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hynes NE, MacDonald G. ErbB receptors and signaling pathways in cancer. Current Opinion in Cell Biology. 2009;21:177–184. doi: 10.1016/j.ceb.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 23.Ohashi, K., Maruvka, Y. E., Michor, F. & Pao, W. Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor – Resistant Disease. J. Clin. Oncol. 31 (2013). [DOI] [PMC free article] [PubMed]
- 24.Chong CR, Jänne PA. The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat. Med. 2013;19:1389–400. doi: 10.1038/nm.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zou B, et al. Deciphering mechanisms of acquired T790M mutation after EGFR inhibitors for NSCLC by computational simulations. Sci. Rep. 2017;7:1–13. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paez JG. EGFR Mutations in Lung Cancer: Correlation with Clinical Response to Gefitinib Therapy. Science (80-.). 2004;304:1497–1501. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 27.Sharma SV, Bell DW, Settleman JE, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat. Rev. Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 28.Lynch TJ, et al. Activating Mutations in the Epidermal Growth Factor Receptor Underlying Responsiveness of Non–Small-Cell Lung Cancer to Gefitinib. N. Engl. J. Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 29.Ma C, Wei S, Song Y. T790M and acquired resistance of EGFR TKI: A literature review of clinical reports. Journal of Thoracic Disease. 2011;3:10–18. doi: 10.3978/j.issn.2072-1439.2010.12.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oxnard GR, et al. New strategies in overcoming acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in lung cancer. Clinical Cancer Research. 2011;17:5530–5537. doi: 10.1158/1078-0432.CCR-10-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yun C, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc. Natl. Acad. Sci. USA. 2008;105:2070–5. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lima L, Barreiro E. Bioisosterism: A Useful Strategy for Molecular Modification and Drug Design. Curr. Med. Chem. 2005;12:23–49. doi: 10.2174/0929867053363540. [DOI] [PubMed] [Google Scholar]
- 33.Yun CH, et al. Structures of Lung Cancer-Derived EGFR Mutants and Inhibitor Complexes: Mechanism of Activation and Insights into Differential Inhibitor Sensitivity. Cancer Cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fry DW, et al. Specific, irreversible inactivation of the epidermal growth factor receptor and erbB2, by a new class of tyrosine kinase inhibitor. Proc. Natl. Acad. Sci. USA. 1998;95:12022–7. doi: 10.1073/pnas.95.20.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carmi C, et al. Epidermal growth factor receptor irreversible inhibitors: chemical exploration of the cysteine-trap portion. Mini Rev. Med. Chem. 2011;11:1019–30. doi: 10.2174/138955711797247725. [DOI] [PubMed] [Google Scholar]
- 36.Potashman MH, Duggan ME. Covalent modifiers: An orthogonal approach to drug design. J. Med. Chem. 2009;52:1231–1246. doi: 10.1021/jm8008597. [DOI] [PubMed] [Google Scholar]
- 37.Liu Q, et al. Developing irreversible inhibitors of the protein kinase cysteinome. Chem. Biol. 2013;20:146–159. doi: 10.1016/j.chembiol.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalgutkar AS, Dalvie DK. Drug discovery for a new generation of covalent drugs. Expert Opin. Drug Discov. 2012;7:561–581. doi: 10.1517/17460441.2012.688744. [DOI] [PubMed] [Google Scholar]
- 39.Flanagan ME, et al. Chemical and computational methods for the characterization of covalent reactive groups for the prospective design of irreversible inhibitors. J. Med. Chem. 2014;57:10072–10079. doi: 10.1021/jm501412a. [DOI] [PubMed] [Google Scholar]
- 40.Jöst C, Nitsche C, Scholz T, Roux L, Klein CD. Promiscuity and selectivity in covalent enzyme inhibition: A systematic study of electrophilic fragments. J. Med. Chem. 2014;57:7590–7599. doi: 10.1021/jm5006918. [DOI] [PubMed] [Google Scholar]
- 41.Do Amaral DN, Sá Alves FR, Barreiro EJ, Laufer SA, Lima LM. Multi-gram Preparation of 7-Nitroquinoxalin-2-amine. J. Braz. Chem. Soc. 2017;28:1874–1878. [Google Scholar]
- 42.Kubo K, et al. Novel potent orally active selective VEGFR-2 tyrosine kinase inhibitors: synthesis, structure-activity relationships, and antitumor activities of N-phenyl-N′-{4-(4-quinolyloxy)phenyl}ureas. J. Med. Chem. 2005;48:1359–1366. doi: 10.1021/jm030427r. [DOI] [PubMed] [Google Scholar]
- 43.Hui X, et al. Synthesis and antiprotozoal activity of some new synthetic substituted quinoxalines. Bioorganic Med. Chem. Lett. 2006;16:815–820. doi: 10.1016/j.bmcl.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 44.Ward RA, et al. Structure and Reactivity Based Development of Covalent Inhibitors of the Activating and Gatekeeper Mutant Forms of the Epidermal Growth Factor Receptor (EGFR) J. Med. Chem. 2013;56:7025–7048. doi: 10.1021/jm400822z. [DOI] [PubMed] [Google Scholar]
- 45.Kort ME, et al. Discovery and biological evaluation of 5-aryl-2-furfuramides, potent and selective blockers of the Nav1.8 sodium channel with efficacy in models of neuropathic and inflammatory pain. J. Med. Chem. 2008;51:407–416. doi: 10.1021/jm070637u. [DOI] [PubMed] [Google Scholar]
- 46.Singh J, Petter RC, Baillie Ta, Whitty A. The resurgence of covalent drugs. Nat. Rev. Drug Discov. 2011;10:307–317. doi: 10.1038/nrd3410. [DOI] [PubMed] [Google Scholar]
- 47.Gao H, et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat. Med. 2015;21:1318–1325. doi: 10.1038/nm.3954. [DOI] [PubMed] [Google Scholar]
- 48.Edwards AM, et al. Preclinical target validation using patient-derived cells. Nat. Rev. Drug Discov. 2015;14:149–150. doi: 10.1038/nrd4565. [DOI] [PubMed] [Google Scholar]
- 49.Günther M, Juchum M, Kelter G, Fiebig H, Laufer S. Lung Cancer: EGFR Inhibitors with Low Nanomolar Activity against a Therapy-Resistant L858R/T790M/C797S Mutant. Angew. Chemie - Int. Ed. 2016;55:10890–10894. doi: 10.1002/anie.201603736. [DOI] [PubMed] [Google Scholar]
- 50.Knight ZA, Shokat KM. Features of selective kinase inhibitors. Chem. Biol. 2005;12:621–637. doi: 10.1016/j.chembiol.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 51.Stewart JJP. Optimization of parameters for semiempirical methods VI: More modifications to the NDDO approximations and re-optimization of parameters. J. Mol. Model. 2013;19:1–32. doi: 10.1007/s00894-012-1667-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Solca F, et al. Target binding properties and cellular activity of afatinib (BIBW 2992), an irreversible ErbB family blocker. J. Pharmacol. Exp. Ther. 2012;343:342–50. doi: 10.1124/jpet.112.197756. [DOI] [PubMed] [Google Scholar]
- 53.Korb O, Stützle T, Exner TE. Empirical scoring functions for advanced Protein-Ligand docking with PLANTS. J. Chem. Inf. Model. 2009;49:84–96. doi: 10.1021/ci800298z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.