Small lakes are an important source of greenhouse gases in the boreal zone. These lakes are severely impacted by the winter season, when ice and snow cover obstruct gas exchange between the lake and the atmosphere and diminish light availability in the water column. Currently, climate change is resulting in reduced spring snow cover. A short-term removal of the snow from the ice stimulated algal primary producers and subsequently heterotrophic bacteria. Concurrently, the relative abundance of methanotrophic bacteria decreased and methane concentrations increased. Our results increase the general knowledge of microbial life under ice and, specifically, the understanding of the potential impact of climate change on boreal lakes.
KEYWORDS: climate change, greenhouse gas, lakes, methane, methanotrophs, microorganisms, primary production, snow cover
ABSTRACT
Climate change scenarios anticipate decreased spring snow cover in boreal and subarctic regions. Forest lakes are abundant in these regions and substantial contributors of methane emissions. To investigate the effect of reduced snow cover, we experimentally removed snow from an anoxic frozen lake. We observed that the removal of snow increased light penetration through the ice, increasing water temperature and modifying microbial composition in the different depths. Chlorophyll a and b concentrations increased in the upper water column, suggesting activation of algal primary producers. At the same time, Chlorobiaceae, one of the key photosynthetic bacterial families in anoxic lakes, shifted to lower depths. Moreover, a decrease in the relative abundance of methanotrophs within the bacterial family Methylococcaceae was detected, concurrent with an increase in methane concentration in the water column. These results indicate that decreased snow cover impacts both primary production and methane production and/or consumption, which may ultimately lead to increased methane emissions after spring ice off.
IMPORTANCE Small lakes are an important source of greenhouse gases in the boreal zone. These lakes are severely impacted by the winter season, when ice and snow cover obstruct gas exchange between the lake and the atmosphere and diminish light availability in the water column. Currently, climate change is resulting in reduced spring snow cover. A short-term removal of the snow from the ice stimulated algal primary producers and subsequently heterotrophic bacteria. Concurrently, the relative abundance of methanotrophic bacteria decreased and methane concentrations increased. Our results increase the general knowledge of microbial life under ice and, specifically, the understanding of the potential impact of climate change on boreal lakes.
INTRODUCTION
Small forest lakes are a typical feature of boreal and subarctic regions (1). These small water bodies with high organic loads are hot spots in the carbon cycle and one of the most prominent environmental sources of greenhouse gas emissions in these regions (2, 3). The microorganisms inhabiting such lakes are the main drivers of these biogeochemical processes (4). Microbial community structure and functioning are strongly impacted by environmental conditions and seasonality. Winter conditions have a particularly strong impact on life in the lake, as ice and snow cover isolate the lake water column from the surrounding environment (5).
One of the most striking impacts of ice and snow cover is the impairment of light availability in the lake. Food webs in most lakes are based on primary production by algae, which provide substrates and oxygen for lake bacteria and serve as important food sources for zooplankton. In wintertime, decreased light availability curtails photosynthesis beneath the ice (6–8), while ice cover also inhibits oxygen transfer from the atmosphere, both contributing to lower under-ice oxygen concentrations. Moreover, aerobic organisms consume residual oxygen in the water beneath the ice (9), leading to a hypoxic-to-anoxic gradient from the lake surface to bottom waters. Anoxic conditions facilitate anaerobic processes, such as methanogenesis, and decrease methanotrophic capacity (10). As a result, methane accumulates under ice and is emitted during ice-break in the spring (11, 12). At the same time, some organisms might benefit from winter conditions, as previously shown for taxa within the bacterial phylum Verrucomicrobia (13). To date, however, studies that address winter conditions are lagging behind open water season research (14).
Currently, climate change is resulting in altered seasonal patterns in subarctic regions (15) and patterns of snow cover worldwide (16). In the Northern Hemisphere, these changes include sudden extreme conditions, as seen in northern Scandinavia during the 2007/08 winter, when snow cover loss occurred abruptly due to a warming event (17). Long-term decreases in spring snow cover have also been observed (16). As snow cover is a major determinant of the amount of light under ice in lakes (5, 18), changes in its thickness and distribution may directly or indirectly affect microbial communities and their activities. For example, cell division among diatoms adapted to low-light conditions—such as those under snow-covered ice—is inhibited when exposed to higher light levels (18). In addition, algal blooms under ice have been shown to be controlled by snow cover (19). Since algae are the sole source of oxygen under ice, a fluctuation in snow cover likely has an impact on algal processes, on other organisms that interact directly and indirectly with algae, and on interactions between nonalgal organisms. We conducted a snow removal experiment on a frozen, oxygen-stratified lake to address the impact of snow cover variation on conditions in the water column and on microbial organisms, with emphasis on methane cyclers and primary producers.
We expected that removal of snow cover would deepen light penetration (5, 18) and hypothesized that this would stimulate primary production. Further, we hypothesized that higher photosynthetic activity would improve oxygen availability and impact the aerobic bacteria in general and specifically increase methanotrophic activity.
RESULTS
Our sampling scheme included six time points and six depths (0.65, 1.00, 1.35, 1.85, 2.35, and 0.5 m above the sediment surface at each location—either 2.55 or 2.85 m) measured from the top surface of the ice layer). We sampled a vertical profile of the lake on three occasions before the snow removal and three times after snow removal, with 1 day between each sampling, except for the last sampling that was 2 days after the previous sampling. Thus, the total duration of the experiment was 2 weeks. The snow depth on the frozen lake was 18 to 21 cm through the experiment, and ice thickness was approximately 50 cm, consisting of mainly black ice with a few centimeters of white ice at the top. Prior to the snow removal, the lake had a shallow oxic layer (epilimnion), with a steep oxygen depletion layer from 0.6 to 0.7 m, where the oxygen concentration was under 0.5 mg liter−1 (Fig. 1).
FIG 1.
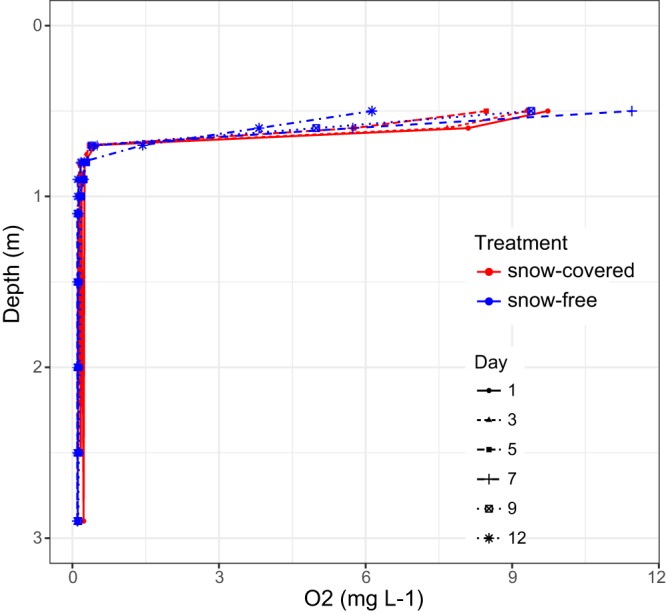
Oxygen concentration (in milligrams per liter) under the ice during the experiment.
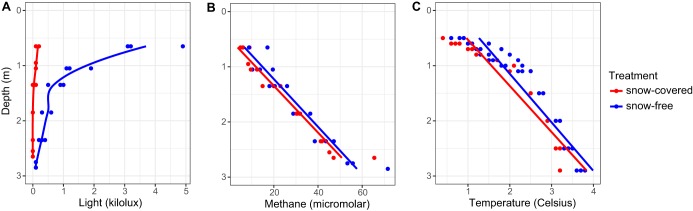
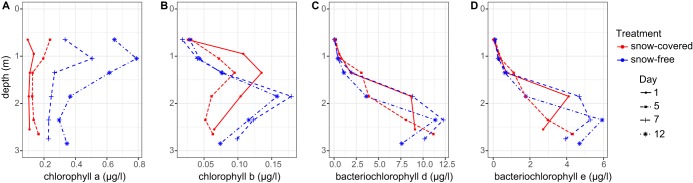
We observed a significant light intensity increase at all depths of the lake after the snow removal, although the extent of the change decreased with increasing depth (Fig. 2 and Table 1). The water temperature also increased throughout the water column after the snow was removed. Moreover, chlorophyll a concentration increased in the three upper layers (0.65 to 1.35 m), whereas chlorophyll b increased in the three bottom layers (1.85 to 2.85 m). Furthermore, the concentration of bacteriochlorophyll d and e appeared to increase at a depth of 2.35 m (Fig. 3). On the basis of these results, we presume that light had a direct effect on the phytoplankton, increasing their chlorophyll production, and likely affecting their photosynthetic activity and oxygen production. After the snow removal, an increase in oxygen concentration in the layer directly under ice was observed, which rapidly decreased over time (Fig. 1). However, differences in oxygen concentration were not statistically significant (Table 1).
FIG 2.
Light intensity (A), methane concentration (B), and temperature (C) in the lake water column during the experiment.
TABLE 1.
Impact of treatment and depth on environmental and chemical factors and on the bacterial communitya
| Factor or bacterial familyb | Between depths |
Within depths |
||
|---|---|---|---|---|
| Treatment | Depth | Treatment | Treatment × depth | |
| Light | 0.012 | <0.001 | 0.025 | 0.169 |
| Temperature | 0.555 | 0.005 | 0.003 | 0.547 |
| Oxygen | 0.309 | 0.012 | 0.346 | 0.014 |
| Methane | 0.012 | <0.001 | 0.023 | 0.169 |
| Methylococcaceae | ||||
| DNA | 0.292 | 0.033 | 0.009 | 0.121 |
| RNA | 0.094 | 0.012 | 0.277 | 0.259 |
| Comamonadaceae | ||||
| DNA | 0.364 | 0.067 | 0.005 | 0.114 |
| RNA | 0.113 | 0.023 | <0.001 | 0.970 |
| Flavobacteriaceae | ||||
| DNA | 0.040 | 0.004 | 0.653 | 0.995 |
| RNA | 0.240 | 0.073 | 0.615 | 0.093 |
| Chlorobiaceae | ||||
| DNA | 0.181 | 0.034 | 0.680 | 0.321 |
| RNA | 0.084 | 0.014 | 0.635 | 0.021 |
The results of repeated measures two-way ANOVA for testing the impact of treatment and depth to light intensity, temperature, concentration of oxygen and methane, and the relative abundance of dominant taxonomic bacterial groups in the lake water column. Significant results (P < 0.05) are shown in bold type, while marginal significant results (P < 0.1) are shown in italic type.
The dominant bacterial group or family and whether the bacterial community analysis was based on DNA or RNA are shown.
FIG 3.
Concentrations of chlorophyll a (A), chlorophyll b (B), bacteriochlorophyll d (C), and bacteriochlorophyll e (D) in the lake water column during the experiment. The concentrations are shown in micrograms per liter.
As expected, prior to snow removal, methane concentrations increased with depth. However, in contrast to our hypothesis, methane concentrations were significantly higher after the snow removal (Fig. 2 and Table 1). There was a depth-related decrease in phosphate concentrations at 0.65, 1.35, 1.85, and 2.35 m after the treatment, but none of the nutrient concentrations changed significantly during the experiment (see Table S1 in the supplemental material).
Concentrations (and standard deviations) of nitrate, nitrite, ammonium, phosphate, and sulfate in Lake Lomtjärn during the experiment. Download Table S1, DOCX file, 0.1 MB (56.4KB, docx) .
Copyright © 2019 Garcia et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Overall, the 11 most abundant operational taxonomic units (OTUs) each contained at least 1% of the sequences. Together, these 11 OTUs accounted for 32% of the sequences. The most abundant OTU belonged to the Chlorobiaceae family, while the second and third most abundant OTUs belonged to the Methylococcaceae family. Snow removal had a significant effect on the total community composition at the DNA level but not at the RNA level (permutational analysis of variance [PERMANOVA], pDNA = 0. 0.008, pRNA = 0. 073). Depth was an important driver of community composition (PERMANOVA, pDNA < 0.001, pRNA < 0.001). The major microbial groups in water at a depth of 0.65 m were Betaproteobacteria (26% before and 32% after snow removal in relative abundance of DNA-based bacterial community composition; 30% before and 46% after snow removal in RNA-based community) and Gammaproteobacteria (23% before and 7% after snow removal in DNA; 37% before and 18% after snow removal in RNA) (see Fig. S1 in the supplemental material). Most of the gammaproteobacterial organisms observed in the water column belonged to the family Methylococcaceae, which consists solely of methane-consuming bacteria.
Composition of the bacterial community according to the proportions of 16S rRNA genes in the DNA (top panel) and RNA (bottom panel) from the surface of the lake (depth 1) to the bottom (depth 6). Download FIG S1, PDF file, 0.01 MB (13.5KB, pdf) .
Copyright © 2019 Garcia et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
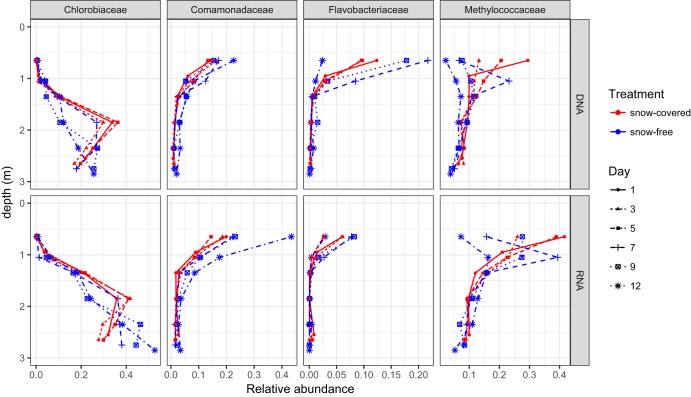
Changes in both DNA and RNA were observed for most of the groups that exhibited the most substantial alterations posttreatment. Among the most abundant taxonomic groups, we observed an increase in the relative abundance of the Comamonadaceae and Flavobacteriaceae families after the snow removal, which were dominant groups in the uppermost layer. Concurrently, the abundance of Methylococcaceae decreased following the experimental treatment (Fig. 4 and Table 1). Finally, the maximum abundance of Chlorobi, a dominant microbial group in the anoxic compartment, appeared to have shifted to deeper layers (Fig. 4).
FIG 4.
Relative abundance of phototrophic Chlorobiaceae, heterotrophic bacteria (Comamonadaceae and Flavobacteriaceae), and methanotrophic Methylococcaceae in the lake water column during the experiment.
DISCUSSION
Our results show that the impact of decreased snow cover on the lake microbiome and metabolism is a complex interaction between biotic and abiotic lake characteristics. Increases in chlorophyll concentrations throughout the water column confirmed our first hypothesis, which postulated that the increase of light following the snow removal would stimulate the primary producers. However, our second hypothesis was verified only partially, as the increase in oxygen concentration that we observed did not persist. Nevertheless, we detected changes in the relative abundances of heterotrophic and methanotrophic bacteria following snow cover removal. Finally, we reject our third hypothesis, which is the anticipated increase of methanotrophic activity with decreased snow cover.
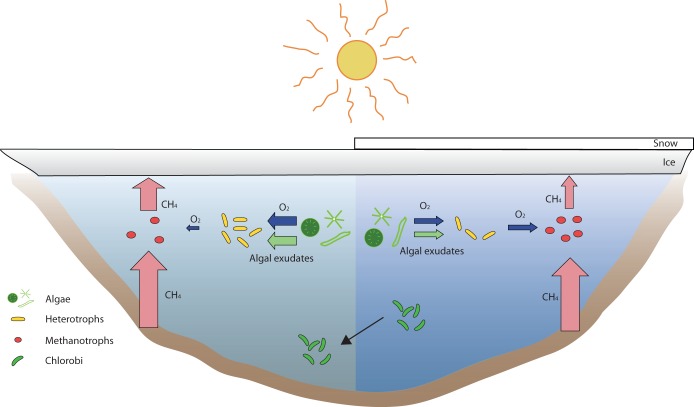
Here, the results show that the algal community was stimulated, producing substrates that likely enhance the activities of aerobic, heterotrophic bacteria within the families Comamonadaceae and Flavobacteriaceae (Fig. 5). Members of these families are aerobic heterotrophs capable of thriving on increased availability of oxygen and organic compounds originating from increased activity of the primary producers (20–22). The immediate oxygen drawdown may be due to decomposition of algal exudates and other alga-derived compounds by heterotrophic bacteria (Fig. 5). In accordance with this assumption, bacterial abundance almost doubled in the upper layer of the lake, concurrent with the oxygen utilization (see Table S2 in the supplemental material). At the same time, the relative abundance of methanotrophic bacteria decreased (Fig. 5). Many methanotrophs are known to have low growth rates (23); hence, heterotrophs potentially outgrew the methanotrophs due to increased availability of algal substrates and oxygen. Other explanations for the decrease of methanotrophs could be low phosphate concentration, which has been linked to impaired methanotrophic activity (24). Increased algal activity in the lake after snow removal may have resulted in phosphate depletion, impeding the growth of methanotrophs. Alternatively, increased primary production could have contributed more substrate to methanogenesis in surface sediments, likely increasing the concentration of methane in the water column through diffusion (12). Decreased methanotrophy and increased methanogenesis are consistent with the increased methane concentrations after the snow removal. However, the specific reasons for the reduced relative abundances of methanotrophs, as well as the impact of a longer period of snow-free ice cover on methanotrophic communities, require further investigation.
FIG 5.
A conceptual figure visualizing the dynamics of algal primary producers, heterotrophic bacteria, methanotrophic bacteria, and bacterial phototrophs (Chlorobi). The right side illustrates the conditions found in the lake with ice and snow cover. The left side illustrates how the conditions changed after the removal of snow, such as increase in light and increased methane throughout the water column. In the first depth sampled, methanotrophs decreased after snow removal, while heterotrophs increased. Moreover, the Chlorobi populations shifted to lower depths in the water column.
Bacterial abundance during the experiment (number of bacteria per milliliter). Download Table S2, DOCX file, 0.04 MB (46.9KB, docx) .
Copyright © 2019 Garcia et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Several experimental features should be taken into consideration in interpreting our results. One important factor to consider is the short duration of our experiment, which makes our results mainly applicable to short-term effects of snow depletion. However, as under-ice algal growth rates can be comparable to those of blooming cyanobacteria (25, 26), and bacterial communities beneath ice are capable of swiftly responding to environmental change (27), we were able to detect shifts in communities related to decreased snow cover within the narrow experimental window. The extent and nature of community shifts in response to changed conditions on a longer time frame must be investigated through other experimental approaches. Another important factor to consider is that we estimated the photosynthetic activity using chlorophyll concentrations, which is an indirect way to measure primary production. However, chlorophyll measurements are widely used as an estimator of photosynthetic activity. In the ocean, for example, chlorophyll concentrations reliably reflect the photosynthetic potential of primary producers (28, 29). Moreover, the presence of bacterial photosynthetic community members was detected not only by bacteriochlorophyll but also by DNA and RNA amplicon sequencing (i.e., Chlorobi; Fig. 3 and 4), suggesting active photosynthesis in the ice-covered lake.
Chlorobi is a common community member in boreal lakes (30–32), such as our study lake, and may contribute substantially to inorganic carbon assimilation in this environment (30). Because of the abundance of bacteriochlorophylls in their antenna complexes, some of these green sulfur bacteria are able to grow at extremely low light levels (1 to 10 nmol photons m−2 s−1), under which no other types of chlorophototrophs can grow (33, 34). Decrease in the abundance of Chlorobi after snow removal at a depth of 1.85 m was likely due to light intensity increase, making conditions more favorable to organisms that use chlorophyll a and b (Fig. 1). Moreover, an increase of Chlorobi—in DNA- and RNA-based relative abundances—was observed in deeper layers of the lake (Fig. 4). This finding, coupled with the increase in bacteriochlorophyll d and e around 2.35 m, suggests that decreased snow cover alters the taxonomical composition of the primary producers of the lake, pushing the optimal conditions for Chlorobi to lower depths of the lake. Changes in primary producers may have implications for lake carbon balance, owing to likely shifts in the efficiency of carbon dioxide uptake and microbial interactions; however, further studies are needed to verify these hypothesized mechanisms.
For the bacterial community in general, depth was an important driver of community composition, likely explained by decreasing oxygen concentrations down the water column. A decrease in oxygen typically cooccurs with a shift in redox potential, both of which are key factors structuring bacterial communities (35, 36). The dominant community members in the lake were the same as previously reported for similar lakes (30–32, 37), suggesting that our results likely apply to a large portion of boreal lakes.
Conclusions.
Our results suggest that decreased snow cover can be expected to impact total lake metabolism and potentially increase methane emission after ice-break. Climate change may additionally lead to shortened ice cover period with earlier ice-off (14), the implications of which have yet to be investigated in boreal lakes. A shorter ice cover period would likely coincide with changes in snow dynamics, and the end results will highly depend on the response of microbial communities. Our results are representative of short-term impacts of decreased snow; the consequences of this phenomenon across other boreal lakes and over longer time scales must be investigated in future studies. In any case, our observations strongly suggest that decreased snow in ice-covered lakes has the immediate potential to increase methane concentrations in the water column, which could lead to increased methane emissions to the atmosphere once the ice melts.
MATERIALS AND METHODS
The experiment was conducted in March 2016 on Lake Lomtjärnan (63°20'56.9"N 14°27'28.3"E), a small forest lake located in Krokom, Sweden. The surface area of the lake is about 1 ha, and the maximum depth is 3.5 m. The lake is located on a mire surrounded by a coniferous forest. The lake has characteristics shared by millions of lakes in the boreal region, such as decreasing oxygen concentration and light intensity from the surface to the bottom and a nutrient and temperature gradient (see Table S1 in the supplemental material). The lake is covered with ice during the winter months, approximately from November to April. The experiment consisted of two parts: first, the lake was monitored while there was still snow cover on the ice surface, and second, the impact of snow cover removal—from an area of approximately 400 m2 above the deepest part of the lake—was observed. Snow was removed manually using snow shovels on day 6 of the monitoring. Under-ice water was sampled by drilling holes through the ice at various locations to ensure that the sampled parameters and microbial communities were as close to an unaltered state as possible, beyond the changes potentially imposed by the experimental snow removal. After sampling, each hole was filled and covered with snow to limit oxygen and light penetration to the water column.
Light intensity and temperature were measured with 18 HOBO loggers (HOBO Pendant temperature/light 64K data logger, Onset Computer Corporation, USA). On the first day of the experiment, loggers were placed under the ice, measuring parameters from the bottom of the ice to the bottom of the lake every 0.1 to 0.75 m; the loggers were maintained at the same place throughout the duration of the experiment. Laterally, the sensors were approximately 50 cm from the hole in the ice. The light values are presented as daily averages for the time between sunrise and sunset (approximately 10 AM to 3 PM).
During each sampling occasion, samples were taken to measure chemical parameters (chlorophyll a and b, nitrite, nitrate, phosphate, sulfate, ammonia, fluoride, chloride, oxygen, methane, and carbon dioxide) and DNA- and RNA-based community analyses. Oxygen concentration was measured with YSI 55 combined temperature and oxygen probe (Yellow Springs Instruments, Yellow Springs, OH, USA). Nutrients were measured by standard methods.
For DNA and RNA, samples were taken with Sterivex filters (Millipore, Billerica, MA, USA). Filtration for RNA was limited to 15 min, whereas filtration for DNA continued until the filter was clogged. Samples were taken from five different depths on day 1, as well as from six different depths on days 3 and 5 (before the snow removal), and days 7, 9, and 12 (after the snow removal).
Chlorophyll pigments were extracted on ice, in tubes containing 2 ml of 90% acetone, using an ultrasonic bath; these extractions occurred overnight at −20°C. Samples were then centrifuged (3,000 × 10 min at 4°C) and filtered (0.45-μm syringe filters). Extracts were analyzed using HPLC on an Agilent 1100 Series HPLC system (Agilent Technologies, Waldbronn, Germany) fitted with three RP-18e Chromolith Performance columns (100 by 4.6 mm) (Merck, Darmstadt, Germany) connected in series. The flow rate was 1.4 ml/min, and the gradient program used is described in detail elsewhere (38). The column temperature was 25°C, and the injection volume was 100 μl (70 μl sample plus 30 μl 0.5 M ammonium acetate). Absorbance was measured with a diode array detector between 300 to 800 nm (resolution 2 nm and slit with 4 nm). Chlorophyll a and b were identified and quantified using standard solutions (DHI Laboratory Products, Hoersholm, Denmark); bacteriochlorophylls were separately identified using previously published chromatograms, spectra, and extinction coefficients (39–43). Methane concentration was analyzed as described previously (44), except that room air was used instead of nitrogen for the headspace. The methane concentration was also analyzed from room air and subtracted from the final gas concentrations.
DNA and RNA were coextracted using a phenol-chloroform method (45) with modifications (30). RNA was then transcribed into cDNA as previously described (46) using RevertAid H Minus First Strand cDNA synthesis kit (Thermo Scientific). Thereafter, RNA and DNA samples were amplified for bacterial 16S rRNA genes using primers 341r and 805f (47). The PCR conditions were set as previously described (48). The samples were then pooled in equimolar amounts and sequenced with Illumina MiSeq at Science for Life Laboratory (Uppsala, Sweden). The resulting 2.5 million sequences were processed using mothur (49) as described elsewhere (50), except that operational taxonomic unit (OTU) clustering was done using abundance-based greedy clustering.
The effects of snow removal and water depth were tested with two-way repeated-measures analyses of variance (RM-ANOVA) for the environmental parameters and by two-way nested permutational ANOVA (PERMANOVA) with 9,999 permutations for the community composition data. Univariate data were log transformed to fulfill the assumptions of RM-ANOVA. All statistical analyses were done in R version 3.4.3 (51). Packages phyloseq, vegan, and ggplot2 were used (52–54).
Accession number(s).
Raw sequences have been submitted to the European Nucleotide Archive (ENA) under accession numbers ERS2597919 to ERS2597988.
ACKNOWLEDGMENTS
This work was supported by grants from the Olsson-Borgh Foundation (grant to S.P. and S.L.G.), the Academy of Finland (grant 265902 to S.P.), and the Royal Swedish Academy of Sciences (grant BS2017-0044). The computations were performed on resources provided by SNIC through the Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under project SNIC 2017/1-616. For the sequencing, the support from Science for Life Laboratory is acknowledged.
We thank the shoveling team, mainly Zoltan Török, Philipp Baur, and Jussi Peura. We thank J. P. Balmonte for editing the manuscript.
REFERENCES
- 1.Sobek S, Soderback B, Karlsson S, Andersson E, Brunberg AK. 2006. A carbon budget of a small humic lake: an example of the importance of lakes for organic matter cycling in boreal catchments. Ambio 35:469–475. doi: 10.1579/0044-7447(2006)35[469:ACBOAS]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 2.Algesten G, Sobek S, Bergstrom AK, Agren A, Tranvik LJ, Jansson M. 2004. Role of lakes for organic carbon cycling in the boreal zone. Global Change Biol 10:141–147. doi: 10.1111/j.1365-2486.2003.00721.x. [DOI] [Google Scholar]
- 3.Tranvik LJ, Downing JA, Cotner JB, Loiselle SA, Striegl RG, Ballatore TJ, Dillon P, Finlay K, Fortino K, Knoll LB, Kortelainen PL, Kutser T, Larsen S, Laurion I, Leech DM, McCallister SL, McKnight DM, Melack JM, Overholt E, Porter JA, Prairie Y, Renwick WH, Roland F, Sherman BS, Schindler DW, Sobek S, Tremblay A, Vanni MJ, Verschoor AM, von Wachenfeldt E, Weyhenmeyer GA. 2009. Lakes and reservoirs as regulators of carbon cycling and climate. Limnol Oceanogr 54:2298–2314. doi: 10.4319/lo.2009.54.6_part_2.2298. [DOI] [Google Scholar]
- 4.Newton RJ, Jones SE, Eiler A, McMahon KD, Bertilsson S. 2011. A guide to the natural history of freshwater lake bacteria. Microbiol Mol Biol Rev 75:14–49. doi: 10.1128/MMBR.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greenbank J. 1945. Limnological conditions in ice-covered lakes, especially as related to winter-kill of fish. Ecol Monogr 15:343–392. doi: 10.2307/1948427. [DOI] [Google Scholar]
- 6.White DM, Clilverd HM, Tidwell AC, Little L, Lilly MR, Chambers M, Reichardt D. 2008. A tool for modeling the winter oxygen depletion rate in arctic lakes. J Am Water Resour Assoc 44:293–304. doi: 10.1111/j.1752-1688.2007.00162.x. [DOI] [Google Scholar]
- 7.Clilverd H, White D, Lilly M. 2009. Chemical and physical controls on the oxygen regime of ice-covered Arctic lakes and reservoirs. J Am Water Resour Assoc 45:500–511. doi: 10.1111/j.1752-1688.2009.00305.x. [DOI] [Google Scholar]
- 8.Hampton SE, Galloway AWE, Powers SM, Ozersky T, Woo KH, Batt RD, Labou SG, O'Reilly CM, Sharma S, Lottig NR, Stanley EH, North RL, Stockwell JD, Adrian R, Weyhenmeyer GA, Arvola L, Baulch HM, Bertani I, Bowman LL, Carey CC, Catalan J, Colom-Montero W, Domine LM, Felip M, Granados I, Gries C, Grossart H-P, Haberman J, Haldna M, Hayden B, Higgins SN, Jolley JC, Kahilainen KK, Kaup E, Kehoe MJ, MacIntyre S, Mackay AW, Mariash HL, McKay RM, Nixdorf B, Nõges P, Nõges T, Palmer M, Pierson DC, Post DM, Pruett MJ, Rautio M, Read JS, Roberts SL, Rücker J, Sadro S, et al. 2017. Ecology under lake ice. Ecol Lett 20:98–111. doi: 10.1111/ele.12699. [DOI] [PubMed] [Google Scholar]
- 9.Bellido JL, Tulonen T, Kankaala P, Ojala A. 2009. CO2 and CH4 fluxes during spring and autumn mixing periods in a boreal lake (Pääjarvi, southern Finland). J Geophys Res Biogeosci 114:G04007. doi: 10.1029/2009JG000923. [DOI] [Google Scholar]
- 10.Dunfield P, Knowles R, Dumont R, Moore TR. 1993. Methane production and consumption in temperate and sub-Arctic peat soils: response to temperature and pH. Soil Biol Biochem 25:321–326. doi: 10.1016/0038-0717(93)90130-4. [DOI] [Google Scholar]
- 11.Smith LK, Lewis WM. 1992. Seasonality of methane emissions from five lakes and associated wetlands of the Colorado Rockies. Global Biogeochem Cycles 6:323–338. doi: 10.1029/92GB02016. [DOI] [Google Scholar]
- 12.Sepulveda-Jauregui A, Anthony KMW, Martinez-Cruz K, Greene S, Thalasso F. 2015. Methane and carbon dioxide emissions from 40 lakes along a north-south latitudinal transect in Alaska. Biogeosciences 12:3197–3223. doi: 10.5194/bg-12-3197-2015. [DOI] [Google Scholar]
- 13.Tran P, Ramachandran A, Khawasik O, Beisner BE, Rautio M, Huot Y, Walsh DA. 2018. Microbial life under ice: metagenome diversity and in situ activity of Verrucomicrobia in seasonally ice-covered lakes. Environ Microbiol 20:2568–2584. doi: 10.1111/1462-2920.14283. [DOI] [PubMed] [Google Scholar]
- 14.Hampton SE, Moore MV, Ozersky T, Stanley EH, Polashenski CM, Galloway AWE. 2015. Heating up a cold subject: prospects for under-ice plankton research in lakes. J Plankton Res 37:277–284. doi: 10.1093/plankt/fbv002. [DOI] [Google Scholar]
- 15.Callaghan TV, Bergholm F, Christensen TR, Jonasson C, Kokfelt U, Johansson M. 2010. A new climate era in the sub-Arctic: accelerating climate changes and multiple impacts. Geophys Res Lett 37:L14705. doi: 10.1029/2009GL042064. [DOI] [Google Scholar]
- 16.Brown RD, Mote PW. 2009. The response of Northern Hemisphere snow cover to a changing climate. J Climate 22:2124–2145. doi: 10.1175/2008JCLI2665.1. [DOI] [Google Scholar]
- 17.Bokhorst SF, Bjerke JW, Tommervik H, Callaghan TV, Phoenix GK. 2009. Winter warming events damage sub-Arctic vegetation: consistent evidence from an experimental manipulation and a natural event. J Ecol 97:1408–1415. doi: 10.1111/j.1365-2745.2009.01554.x. [DOI] [Google Scholar]
- 18.Jewson DH, Granin NG, Zhdanov AA, Gnatovsky RY. 2009. Effect of snow depth on under-ice irradiance and growth of Aulacoseira baicalensis in Lake Baikal. Aquat Ecol 43:673–679. doi: 10.1007/s10452-009-9267-2. [DOI] [Google Scholar]
- 19.Welch HE, Bergmann MA. 1989. Seasonal development of ice algae and its prediction from environmental factors near Resolute, N.W.T., Canada. Can J Fish Aquat Sci 46:1793–1804. doi: 10.1139/f89-227. [DOI] [Google Scholar]
- 20.Hahn MW, Kasalicky V, Jezbera J, Brandt U, Jezberova J, Simek K. 2010. Limnohabitans curvus gen. nov., sp nov., a planktonic bacterium isolated from a freshwater lake. Int J Syst Evol Microbiol 60:1358–1365. doi: 10.1099/ijs.0.013292-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willems A, De Ley J, Gillis M, Kersters K. 1991. Comamonadaceae, a new family encompassing the Acidovorans rRNA complex, including Variovorax paradoxus gen. nov., comb. nov. for Alcaligenes paradoxus (Davis 1969). Int J Syst Bacteriol 41:445–450. doi: 10.1099/00207713-41-3-445. [DOI] [Google Scholar]
- 22.O'Sullivan LA, Rinna J, Humphreys G, Weightman AJ, Fry JC. 2006. Culturable phylogenetic diversity of the phylum 'Bacteroidetes' from river epilithon and coastal water and description of novel members of the family Flavobacteriaceae: Epilithonimonas tenax gen. nov., sp. nov. and Persicivirga xylanidelens gen. nov., sp. nov. Int J Syst Evol Microbiol 56:169–180. doi: 10.1099/ijs.0.63941-0. [DOI] [PubMed] [Google Scholar]
- 23.Krueger M, Wolters H, Gehre M, Joye SB, Richnow HH. 2008. Tracing the slow growth of anaerobic methane-oxidizing communities by N-15-labelling techniques. FEMS Microbiol Ecol 63:401–411. doi: 10.1111/j.1574-6941.2007.00431.x. [DOI] [PubMed] [Google Scholar]
- 24.Denfeld BA, Ricao Canelhas M, Weyhenmeyer GA, Bertilsson S, Eiler A, Bastviken D. 2016. Constraints on methane oxidation in ice-covered boreal lakes. J Geophys Res Biogeosci 121:1924–1933. doi: 10.1002/2016JG003382. [DOI] [Google Scholar]
- 25.Arrigo KR, Perovich DK, Pickart RS, Brown ZW, van Dijken GL, Lowry KE, Mills MM, Palmer MA, Balch WM, Bahr F, Bates NR, Benitez-Nelson C, Bowler B, Brownlee E, Ehn JK, Frey KE, Garley R, Laney SR, Lubelczyk L, Mathis J, Matsuoka A, Mitchell BG, Moore GWK, Ortega-Retuerta E, Pal S, Polashenski CM, Reynolds RA, Schieber B, Sosik HM, Stephens M, Swift JH. 2012. Massive phytoplankton blooms under Arctic sea ice. Science 336:1408–1408. doi: 10.1126/science.1215065. [DOI] [PubMed] [Google Scholar]
- 26.Robarts RD, Zohary T. 1987. Temperature effects on photosynthetic capacity, respiration, and growth rates of bloom-forming cyanobacteria. New Zeal J Mar Freshw Res 21:391–399. doi: 10.1080/00288330.1987.9516235. [DOI] [Google Scholar]
- 27.Monier A, Findlay HS, Charvet S, Lovejoy C. 2014. Late winter under ice pelagic microbial communities in the high Arctic Ocean and the impact of short-term exposure to elevated CO2 levels. Front Microbiol 5:490. doi: 10.3389/fmicb.2014.00490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Antoine D, Andre JM, Morel A. 1996. Oceanic primary production. 2. Estimation at global scale from satellite (coastal zone color scanner) chlorophyll. Global Biogeochem Cycles 10:57–69. doi: 10.1029/95GB02832. [DOI] [Google Scholar]
- 29.Moore JK, Abbott MR. 2000. Phytoplankton chlorophyll distributions and primary production in the Southern Ocean. J Geophys Res 105:28709–28722. doi: 10.1029/1999JC000043. [DOI] [Google Scholar]
- 30.Taipale S, Jones RI, Tiirola M. 2009. Vertical diversity of bacteria in an oxygen-stratified humic lake, evaluated using DNA and phospholipid analyses. Aquat Microb Ecol 55:1–16. doi: 10.3354/ame01277. [DOI] [Google Scholar]
- 31.Karhunen J, Arvola L, Peura S, Tiirola M. 2013. Green sulphur bacteria as a component of the photosynthetic plankton community in small dimictic humic lakes with an anoxic hypolimnion. Aquat Microb Ecol 68:267–272. doi: 10.3354/ame01620. [DOI] [Google Scholar]
- 32.Peura S, Eiler A, Bertilsson S, Nykanen H, Tiirola M, Jones RI. 2012. Distinct and diverse anaerobic bacterial communities in boreal lakes dominated by candidate division OD1. ISME J 6:1640–1652. doi: 10.1038/ismej.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beatty JT, Overmann J, Lince MT, Manske AK, Lang AS, Blankenship RE, Van Dover CL, Martinson TA, Plumley FG. 2005. An obligately photosynthetic bacterial anaerobe from a deep-sea hydrothermal vent. Proc Natl Acad Sci U S A 102:9306–9310. doi: 10.1073/pnas.0503674102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olson JM. 1998. Chlorophyll organization and function in green photosynthetic bacteria. Photochem Photobiol 67:61–75. doi: 10.1111/j.1751-1097.1998.tb05166.x. [DOI] [Google Scholar]
- 35.Garcia SL, Salka I, Grossart H-P, Warnecke F. 2013. Depth-discrete profiles of bacterial communities reveal pronounced spatio-temporal dynamics related to lake stratification. Environ Microbiol Rep 5:549–555. doi: 10.1111/1758-2229.12044. [DOI] [PubMed] [Google Scholar]
- 36.Peura S, Sinclair L, Bertilsson S, Eiler A. 2015. Metagenomic insights into strategies of aerobic and anaerobic carbon and nitrogen transformation in boreal lakes. Sci Rep 5:12102. doi: 10.1038/srep12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Linz AM, Crary BC, Shade A, Owens S, Gilbert JA, Knight R, McMahon KD. 2017. Bacterial community composition and dynamics spanning five years in freshwater bog lakes. mSphere 2:e00169-17. doi: 10.1128/mSphere.00169-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Airs RL, Atkinson JE, Keely BJ. 2001. Development and application of a high resolution liquid chromatographic method for the analysis of complex pigment distributions. J Chromatogr A 917:167–177. doi: 10.1016/S0021-9673(01)00663-X. [DOI] [PubMed] [Google Scholar]
- 39.Borrego CM, Garcia-Gil LJ, Vila X, Cristina XP, Figueras JB, Abella CA. 1997. Distribution of bacteriochlorophyll homologs in natural populations of brown-colored phototrophic sulfur bacteria. FEMS Microbiol Ecol 24:301–309. doi: 10.1111/j.1574-6941.1997.tb00447.x. [DOI] [Google Scholar]
- 40.Borrego CM, Garcia-Gil J, Cristina XP, Vila X, Abella CA. 1998. Occurrence of new bacteriochlorophyll d forms in natural populations of green photosynthetic sulfur bacteria. FEMS Microbiol Ecol 26:257–267. doi: 10.1111/j.1574-6941.1998.tb00510.x. [DOI] [Google Scholar]
- 41.Picazo A, Rochera C, Vicente E, Miracle MR, Camacho A. 2013. Spectrophotometric methods for the determination of photosynthetic pigments in stratified lakes: a critical analysis based on comparisons with HPLC determinations in a model lake. Limnetica 32:139–157. [Google Scholar]
- 42.Hurley JP, Watras CJ. 1991. Identification of bacteriochlorophylls in lakes via reverse-phase HPLC. Limnol Oceanogr 36:307–315. doi: 10.4319/lo.1991.36.2.0307. [DOI] [Google Scholar]
- 43.Borrego CM, Arellano JB, Abella CA, Gillbro T, Garcia-Gil J. 1999. The molar extinction coefficient of bacteriochlorophyll e and the pigment stoichiometry in Chlorobium phaeobacteroides. Photosynth Res 60:257–264. doi: 10.1023/A:1006230820007. [DOI] [Google Scholar]
- 44.Kankaala P, Taipale S, Nykänen H, Jones RI. 2007. Oxidation, efflux, and isotopic fractionation of methane during autumnal turnover in a polyhumic, boreal lake. J Geophys Res Biogeosci 112:G02003. doi: 10.1029/2006JG000336. [DOI] [Google Scholar]
- 45.Griffiths RI, Whiteley AS, O'Donnell AG, Bailey MJ. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl Environ Microbiol 66:5488–5491. doi: 10.1128/AEM.66.12.5488-5491.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szekely AJ, Berga M, Langenheder S. 2013. Mechanisms determining the fate of dispersed bacterial communities in new environments. ISME J 7:61–71. doi: 10.1038/ismej.2012.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herlemann DPR, Labrenz M, Jurgens K, Bertilsson S, Waniek JJ, Andersson AF. 2011. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J 5:1571–1579. doi: 10.1038/ismej.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sinclair L, Osman OA, Bertilsson S, Eiler A. 2015. Microbial community composition and diversity via 16S rRNA gene amplicons: evaluating the Illumina platform. PLoS One 10:e0116955. doi: 10.1371/journal.pone.0116955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.R Core Team. 2014. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/.
- 52.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dixon P. 2003. VEGAN, a package of R functions for community ecology. J Veg Sci 14:927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
- 54.Ginestet C. 2011. ggplot2: elegant graphics for data analysis. J R Stat Soc Ser A Stat Soc 174:245. doi: 10.1111/j.1467-985X.2010.00676_9.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Concentrations (and standard deviations) of nitrate, nitrite, ammonium, phosphate, and sulfate in Lake Lomtjärn during the experiment. Download Table S1, DOCX file, 0.1 MB (56.4KB, docx) .
Copyright © 2019 Garcia et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Composition of the bacterial community according to the proportions of 16S rRNA genes in the DNA (top panel) and RNA (bottom panel) from the surface of the lake (depth 1) to the bottom (depth 6). Download FIG S1, PDF file, 0.01 MB (13.5KB, pdf) .
Copyright © 2019 Garcia et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacterial abundance during the experiment (number of bacteria per milliliter). Download Table S2, DOCX file, 0.04 MB (46.9KB, docx) .
Copyright © 2019 Garcia et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.