Abstract
Inflammatory bowel diseases (IBDs) are chronic inflammatory disorders affecting the gastrointestinal tract. The incidence of IBD is increasing, with more cases occurring in developed countries. Multiple factors such as genetics, environmental changes, gut microbiota, and immune abnormalities have been associated with development of IBD. In recent years, it has become increasingly apparent that epigenetic modifications of chromatin and the manner in which chromatin is organized in the nucleus are additionally important elements that can influence responses induced by the factors described above, and may therefore contribute to the onset and pathogenesis of IBD. Epigenetics and chromatin organization regulate diverse functions that include maintenance of homeostasis in the intestinal epithelium, the development and differentiation of immune cells, and modulation of responses generated by the immune system to defend against potential pathogens. Furthermore, changes in epigenetic chromatin marks and in chromatin organization have now been linked to differential gene expression in IBD patient cells. Although direct evidence for a role of histone modifications in IBD is currently very limited, in this review, we summarize the links between various epigenetic modifications, the proteins that catalyze or recognize these modifications, and the development or progression of IBD in human and experimental IBD. We also discuss how epigenetics influence the organization of DNA contacts to regulate gene expression and the implications this may have for diagnosing and treating IBD.
Keywords: epigenetics, chromatin, IBD, enhancers, regulatory elements, DNA loops, histone modification, condensin, CTCF, cohesin, Crohn's disease, ulcerative colitis
INTRODUCTION
Inflammatory bowel diseases (IBD), including Crohn’s disease (CD) and ulcerative colitis (UC), are gastrointestinal disorders that lead to chronic and relapsing intestinal injury. These diseases have unknown etiology, with several factors such as environment, genetics, diet, and changes in microbiome composition playing important roles in the disease development and pathogenesis.1–4 Approximately 1.6 million Americans suffer from inflammatory bowel disease.5 Crohn’s disease is characterized by inflammation occurring at any location along the gastrointestinal canal but is most commonly localized to the terminal ileum and the colon. Crohn’s disease manifestations include transmural and patchy lesions, fibrotic strictures,6 and creeping fat.7 Ulcerative colitis is restricted to the colon and rectum, involves continuous lesions, and affects the mucosal layer of the bowel.8
The genetics of IBD are complex, and genome-wide association studies have identified over 200 IBD susceptibility loci.9–11 However, many of these polymorphisms are located in noncoding regions of the genome and are hypothesized to contain transcriptional regulatory elements.11, 12 The activity of these regulatory elements and the genes whose expression they control are influenced by epigenetic modifications.11, 12 Epigenetic changes can be heritable and primarily involve chemical modifications to DNA (DNA methylation) without changing the underlying DNA sequence, or modifications to histone proteins which are components of the nucleosomes around which DNA wraps. These modifications can influence expression of genes, leading to either activation or repression of transcription. In addition to epigenetic modifications, specific ATP-dependent remodeling complexes act to position nucleosomes, help compact chromatin, and/or promote long-distance interactions between different segments of DNA.13–16 Disruptions in the above processes can lead to disturbances in gene expression, promoting inflammation, and potentially contributing to IBD pathogenesis.
In this review, we will focus on epigenetic modifications hypothesized to play important roles in the pathogenesis of IBD, and we will also discuss several links between protein complexes involved in chromatin remodeling and IBD pathogenesis. We will not discuss the role played by DNA methyltransferases and demethylases in IBD, and we refer the reader to excellent, recent reviews on these subjects.17–19
HISTONE MODIFICATION AND CHROMATIN ORGANIZATION REGULATE GENE EXPRESSION
Within the nucleus of a cell, the minimal unit of chromatin is the nucleosome, which is comprised of 147 bp of DNA wrapped 1.7 times around a core of 8 histone proteins.20 This core histone octamer comprises a hetero-tetramer of histones H3 and H4, surrounded by 2 histone H2A and H2B heterodimers (Fig. 1). Nucleosomes are separated from each other by 10 to 60 bp of linker DNA, eventually forming a 10 nm diameter chromatin fiber. Compaction of chromatin into 100 to 400 nm fibers occurs during early mitosis, resulting in condensed chromosomes. The N- and C-terminal tails of the core histones project out from the nucleosome and are responsible for mediating higher-order folding of chromatin.21 Different amino acids on the histone tails, namely lysine, arginine, serine, and threonine, are targeted by enzymes for post-translational modifications (PTMs) (Fig. 1), which then influence whether a gene is accessible for binding by transcription factors and the RNA Polymerase II machinery. In its more compacted state, chromatin is less accessible by these factors, and therefore, gene expression will generally be suppressed. Conversely, relaxed chromatin is more accessible, and genes will generally be transcriptionally active.21 Several different classes of histone-modifying enzymes exist to modify lysines; these modifications include acetylation and deacetylation, methylation and demethylation, phosphorylation, ubiquitination, sumoylation, crotonylation, ADP-ribosylation, and deamination.21, 22
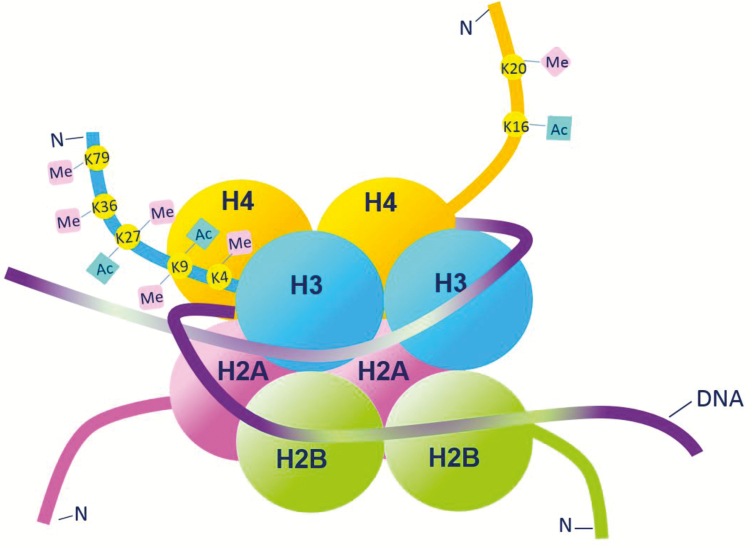
FIGURE 1.
The nucleosome is the basic packaging unit of chromatin. The nucleosome is composed of an octamer of histone proteins, comprising a hetero-tetramer of histones H3 and H4, surrounded by 2 hetero-dimers of histones H2A and H2B. DNA is wrapped approximately 1.7 times around the histone core. Lysine (K) residues present on the N-terminal tails of histones undergo several post-translational modifications such as addition of acetyl (Ac) and methyl (Me) groups. These histone modifications contribute to the epigenetic regulation of gene transcription.
Gene expression is controlled by a variety of DNA regulatory elements that include promoters, enhancers, insulators, and silencers. Various chromatin-organizing complexes, including CTCF (CCCTC-binding factor), cohesin, and condensins, bind to the regulatory elements, associate with DNA, and/or facilitate the generation of DNA loops at these locations.23–33 Through formation of loops, DNA regulatory elements can be positioned into close proximity with specific gene promoters, thus activating or repressing transcription, depending on the nature of the regulatory element (Fig. 2A-B). For example, loop formation can position an enhancer element, which will activate gene expression, next to a gene promoter, causing the transcription factors and chromatin-modifying proteins that are bound to the enhancer to activate transcription of the gene (Fig. 2A). Conversely, a repressive regulatory element positioned in close proximity to a gene promoter can direct repressive DNA and histone modifications to be made within the loop, thus repressing gene expression (Fig. 2B).
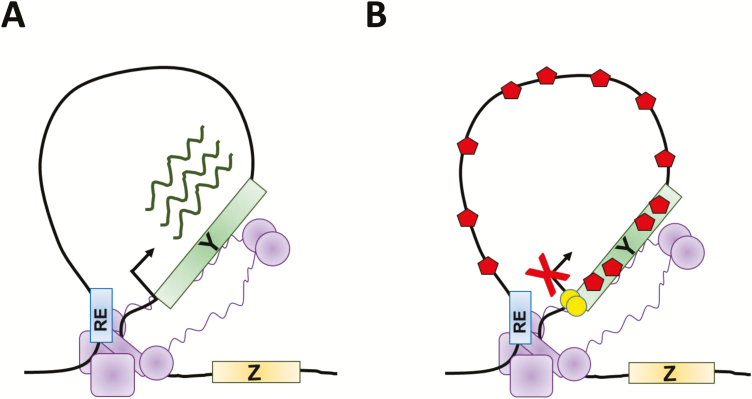
FIGURE 2.
The 3-dimensional organization of chromatin plays an important role in regulating gene expression. A–B, Illustrations of different ways in which the formation of DNA loops mediated by genome organizing complexes (purple) can regulate gene expression. In A, a regulatory element (RE) that activates gene expression, such as an enhancer, interacts with the promoter of gene Y which, following binding of transcription factors, results in enhancement of transcription. In B, the RE confers a repressive activity, possibly by directing DNA methylation (yellow circles) of gene Y’s promoter, or by facilitating binding of chromatin remodeling proteins that promote repressive histone modifications (red hexagons). This represses transcription of gene Y.
HISTONE MODIFICATIONS AND CHROMATIN ORGANIZERS INFLUENCE DEVELOPMENT AND FUNCTION OF CELLS IN THE INTESTINE
The mammalian intestine is comprised of a mucosal layer, a submucosal layer, a muscularis layer, and a serosal layer. The mucosal layer contains epithelial cells, cells in the lamina propria, and cells of the muscularis mucosa. The intestinal epithelium acts as the first line of defense against both pathobionts and invasive species present within the gut microbiome. Defects in the development and differentiation of epithelial cells can lead to defects in this barrier function and may contribute to IBD development and progression.34–37 The intestinal lamina propria is comprised of a variety of immune cell populations that mediate a balance between response to pathogens and tolerance to host antigens. This is facilitated by cells of the innate immune system [mainly neutrophils, natural killer (NK) cells, dendritic cells, macrophages, and innate lymphoid cells] and adaptive immune system (B, T, and NKT cells). Although both innate and adaptive immune cells infiltrate the intestinal mucosa during IBD, the proper development, differentiation, and activation of T cells seems to be especially important for maintaining intestinal homeostasis.38, 39 Interestingly, histone modifications and chromatin-organizing proteins contribute to the development and function of both the intestinal epithelium and T cells, as discussed later on.
Intestinal Epithelium
The intestinal epithelium, a single layer of columnar cells lining the intestinal lumen, is organized into crypt-villus units, with crypts forming invaginations in the underlying mesenchymal tissue and villi projecting into the lumen. The pluripotent stem cells of the intestine, also referred to as crypt base columnar cells or CBCs, reside at the bottom of the crypt and divide frequently to give rise to transit-amplifying (TA) cells, which migrate upwards in the crypt and eventually differentiate into absorptive or secretory lineages. The absorptive cells or enterocytes are involved in nutrient absorption, whereas the secretory cells include mucus-secreting goblet cells, hormone-producing enteroendocrine cells, mechanosensory Tuft cells, and Paneth cells that produce antimicrobial peptides.40 All of the differentiated cell types except the Paneth cells are located in the upper crypts or the villi; Paneth cells migrate towards the crypt bottoms and are located near the CBCs.
The identity of intestinal cells is defined by specific signals that act to activate or repress transcription of certain genes; this is mainly facilitated by coordination of interactions between transcription factors, promoters, and distal regulatory elements in loops (Fig. 2). However, cells also need to maintain a certain level of plasticity in their transcription so they can respond to various environmental cues and preserve their homeostatic state. One group of proteins that is instrumental in defining repressed states of gene expression via epigenetic modifications, while simultaneously maintaining transcriptional plasticity, are the Polycomb proteins.41 The Polycomb machinery, which comprises Polycomb repressive complexes (PRC) 1 and 2, maintains a level of transcriptional repression in intestinal epithelial stem cells by facilitating histone modifications such as trimethylation of H3K27 by PRC2, resulting in a more compact chromatin state (Fig. 3). Experiments in mice suggest that in the TA zone within the crypts, PRC2 adds H3K27me3 marks to repress the expression of terminal differentiation markers such as sucrase-isomaltase (SI), and might function to help TA cells maintain their proliferative state.42 Polycomb repressive complex 1 is essential for maintaining the pool of LGR5+ intestinal epithelial stem cells; loss of PRC1 leads to a loss of stem cell identity, causing them to exit their niche prior to expression of differentiation markers. Importantly, loss of PRC2 leads to impairment in the regenerative abilities of the stem cells following extensive damage induced by irradiation.41, 43 This observation was confirmed in human intestinal epithelial cells as well; PRC2 depletion in colon carcinoma cells and in human, primary, intestinal crypt cells resulted in increased differentiation of stem cells into the absorptive lineage, along with increased expression of SI protein. Therefore, in the TA compartment, Polycomb machinery might facilitate proliferation while inhibiting terminal differentiation.42 This is important in regard to IBD because the catalytic histone methyltransferase subunit of PRC2, Enhancer of Zeste Homolog 2 (EZH2), is downregulated in IBD patients compared with non-IBD patients.44 Additionally, intestinal epithelial cell–specific knockdown of EZH2 in mice led to increased intestinal inflammation following treatments with DSS (dextran sodium sulfate) or trinitrobenzenesulfonic acid (TNBS), compared with wild-type mice subjected to similar treatments.44 Conversely, overexpression of EZH2 in intestinal epithelia of mice conferred protection from inflammation after treatments with DSS, indicating the importance of Polycomb complexes in intestinal regeneration after epithelial damage.44
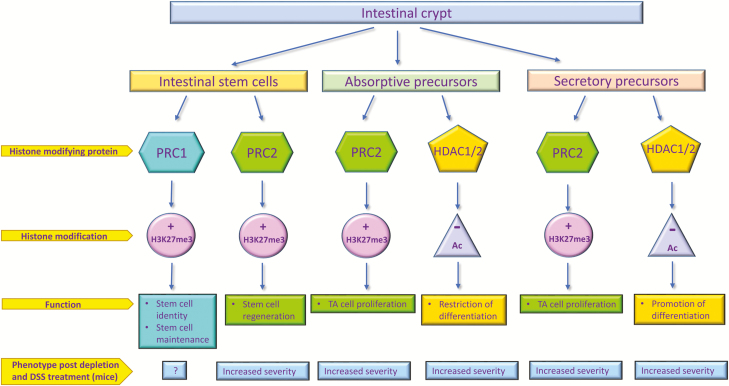
FIGURE 3.
Histone modifications help to maintain homeostasis in the intestinal epithelium. Histone methylation induced by Polycomb repressive complexes are responsible for intestinal stem cell identity and regeneration following epithelial damage, as well as for proliferation of absorptive and secretory precursors in the transit amplifying (TA) compartments within the intestinal crypt. Histone acetylation and deacetylation play important roles in restricting differentiation of absorptive precursors while promoting differentiation of secretory cells. PRC, Polycomb repressive complex; HDAC, Histone deacetylase; TA, transit-amplifying.
Acetylation/de-acetylation is important for regulating proliferation and differentiation of both absorptive and secretory precursor cells in the intestinal epithelium (Fig. 3). Promoters of genes encoding terminal differentiation markers like SI are hyperacetylated during absorptive cell differentiation. The histone deacetylase (HDAC) inhibitors, which promote acetylation by blocking removal of acetyl groups from lysines, phenocopy this effect both in human and murine intestinal epithelial cells.45 Furthermore, depletion of HDAC1 and HDAC2 in the mouse intestinal epithelium, early in development, promotes development of absorptive lineages and represses secretory cell development. Depletion in adults, however, leads to a loss of crypt cells, as demonstrated by an inability of these cells to proliferate.45, 46 Intestinal epithelial–specific dual depletion of HDAC 1 and 2 increases the severity of DSS-induced colitis47; depletion of only HDAC2 surprisingly leads to a lower severity of colitis after DSS,47 whereas depletion of only HDAC1 results in a colitis phenotype similar to wild type mice post treatment with DSS.48 The HDAC2-deficient mice expressing one Hdac1 allele exhibit normal intestinal architecture but are more susceptible to DSS-induced colitis, whereas HDAC1-deficient mice expressing one Hdac2 allele show disruptions to intestinal architecture in addition to increased susceptibility to DSS.48 This suggests that although HDAC1 may be more important for maintaining proper intestinal architecture, appropriate levels of both HDAC1 and HDAC2 are crucial for recovery from intestinal injury.
Chromosome organizers also contribute to the regulation of epithelial differentiation and barrier function. Loss of CTCF-binding sites at a specific imprinted locus in mice resulted in decreased intestinal epithelial cell differentiation.49 While the effects of condensin depletion on the development and differentiation of epithelial cells are currently unknown, new roles were recently discovered for condensins in the regulation of innate immunity within the intestinal epithelium. Depletion of condensin II subunit, CAP-D3, in human intestinal epithelial cells infected with enteropathogenic bacteria, including Salmonella typhimurium, diminished the ability of cells to clear bacteria.50 Moreover, CAP-D3 was found to repress the transient upregulation of several genes encoding proteins that act to block autophagy. In CAP-D3-depleted cells, these proteins accumulate early in the infection, thus providing a growth advantage to the bacteria.50 These studies also found that colonic epithelial cells from patients with active UC exhibit diminished levels of CAP-D3. Given that prior enteric infections are a risk factor and can predispose towards the development of IBD,51–54 it is possible that the chromatin-organizing and transcriptional regulatory functions of CAP-D3/Condensin II may play a role in preventing IBD development and/or pathogenesis.
T Cells
The CD4+ T effector cells, particularly the Th1, Th2, and Th17 cell subsets, are important for defense against pathogens, while CD4+ regulatory T cells (Tregs) help to control activity and proliferation of effector T cells. Inflammatory bowel disease is commonly associated with hyperactivity of effector CD4+ T cells and/or defects in immune tolerance mediated by defects in Treg development or Treg immunosuppressive function.55
The JMJD3 (Jumonji Domain-containing protein 3) histone demethylase has been shown to regulate T cell differentiation and gene transcription in T cells.56–58 JMJD3 demethylates H3K27me2/3, a histone mark that is typically associated with repression of transcription. Loss of JMJD3 in mice promotes differentiation of Th2 and Th17 cells while inhibiting Treg and Th1 differentiation (Fig. 4A). Therefore, it is not surprising that introduction of CD4+ T cells from JMJD3-ablated mice into a Th1-dependent colitis mouse model leads to milder colitis59 (Fig. 4D). CD4+ T cells from JMJD3-deficient mice also show higher levels of H3K27 methylation (me2 and me3), consistent with loss of JMJD3-mediated derepression of gene expression at specific genes such as Cd44, Ccnd2, Ifng, Irf4, Ccr2 and Fosl2, which could be important targets for preventing colitis.59 However, JMJD3 has also been shown to inhibit Th17 differentiation but still acted to protect mice against experimental colitis, suggesting that absence of JMJD3 might contribute to an overall reduction in inflammation via suppression of pro-inflammatory T cell responses.60
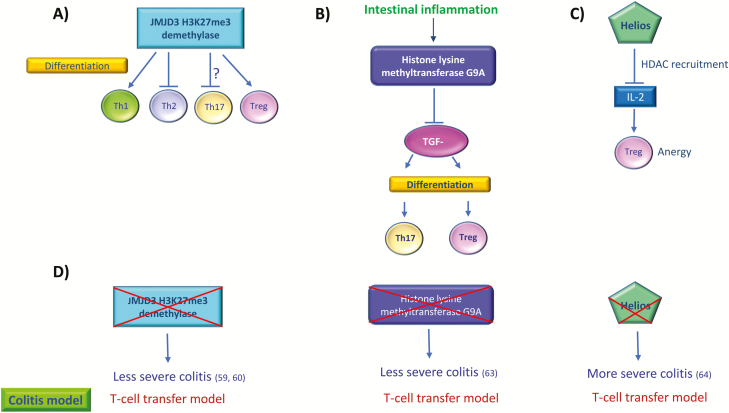
FIGURE 4.
T cell differentiation and function within the intestinal lamina propria are regulated by histone modifications. A, JMJD3 H3K27me3 demethylase modulates the differentiation of T-cell subsets. The “?” sign refers to a conflicting study which shows that JMJD3 promotes Th17 differentiation (60). B, During intestinal inflammation, histone lysine methyltransferase G9A restricts TGF-beta-mediated differentiation of Th17 and Treg cells. C, Transcription factor Helios recruits histone deacetylases to suppress IL-2 expression in Tregs, leading to Treg anergy. D, Loss of mechanisms that regulate histone modifications can impact the severity of colitis. Loss of JMJD3 caused less severe colitis in mice, whereas depletion of G9A protects against severe colitis. Loss of Helios leads to increased IL-2 expression in Tregs and loss of anergy, in addition to more severe colitis in mice. Supporting references are in brackets. JMJD3-Jumonji domain-containing protein 3.
Transforming growth factor (TGF)-β, interleukin (IL)-6, IL-21, and IL-23 are the major cytokines involved in Th17 cell differentiation.61 One study showed that during intestinal inflammation, TGF-β can convert commensal bacterial antigen-producing Th1 cells into IL-17+ Th17 cells, thereby producing pathogenic IL-17+, interferon-gamma (IFN-γ+), CD4+ T cells that result in different levels of severity of colitis.62 The histone lysine methyltransferase G9A produced by T cells is required for a response to intestinal inflammation, and causes methylation at H3K9 at the promoters of Il17a, Il17f, Rorc and Foxp3, which restricts TGF-β1 mediated Th17 and Treg differentiation (Fig. 4B).63 Loss of G9A increased responses to TGF-β1 by increasing chromatin accessibility in naïve T cells, increasing Th17 and Treg differentiation, and thus protecting from T cell transfer colitis63 (Fig. 4D). Pharmacological inhibition of G9A in T cells caused a similar effect, leading to increased differentiation into Th17 and Treg lineages. Therefore, targeting of the H3K9me2 mark either by G9A inhibition, or by introducing demethylases that can remove the H3K9me2 methylation marks could help to manipulate Th cell subset populations, offering a potential therapy toward the treatment of IBD.
Transcription factor-mediated recruitment of histone modifiers can also affect T cell function. Helios is an Ikaros family transcription factor expressed at high levels in Treg cells. Helios normally represses Il2 expression in Tregs by recruiting HDACs to the Il2 promoter, leading to histone deacetylation and an anergic phenotype (Fig. 4C). Upon stimulation with antibodies against CD3 and CD28, Helios-depleted Treg cells exhibit increased IL-2 expression and decreased forkhead box P3 (Foxp3) binding to the Il2 promoter.64 Transfer of Helios depleted Tregs into Rag1-/- mice, where IBD is induced by injection of CD4+CD25--naïve T cells, leads to increased weight loss and increased severity of colitis (Fig. 4D) as compared with control groups, thus underscoring the importance of Helios-mediated HDAC activity in the regulation of Tregs and their contribution to intestinal inflammation.
In addition to regulating the expression of genes involved in innate immunity, the genome organizing functions of CTCF and condensins may also be important for T cell development and activation. Conditional depletion of CTCF in double positive thymocytes does not impede peripheral T cell development, but does impair activation and proliferation following stimulation in vitro.65 Similarly, T cell activation is also regulated by condensins; activation of naïve T cells in response to IL-2 and TCR engagement induces changes in higher order chromatin structure via CAP-H2 of the condensin II complex. Mutation of CAP-H2 in mice, therefore, resulted in aberrant chromatin condensation during T cell development and failure to maintain a quiescent state.
INTESTINAL CYTOKINE PRODUCTION IS REGULATED BY HISTONE MODIFICATIONS AND CHROMATIN ORGANIZERS
A balance between pro-inflammatory and anti-inflammatory cytokines is essential for the maintenance of intestinal homeostasis. Various cytokines, including IFN-γ, tumor necrosis factor-alpha (TNF-α), IL-12, IL-23, IL-17, IL-13, IL-5, and IL-9, have been shown to be significantly upregulated in intestinal tissues from IBD patients and IBD mouse models.66, 67 In wild type mice, ten-eleven translocation enzyme 2 (Tet2) recruits HDAC2 to repress pro-inflammatory cytokine IL-6 expression.68 Loss of Tet2 in mice leads to an exacerbated inflammatory response with higher levels of IL-6 production compared with controls in mice injected intraperitoneally with lipopolysaccharide (LPS); Tet2-deficient mice also display an increased susceptibility to DSS-induced colitis. IκBζ is an Il6 specific transcription factor that recruits Tet2 to the Il6 promoter during late stages of response to LPS, suggesting a role for Tet2 in resolution of inflammation via IL-6.68
Interestingly, chemical inhibition of histone modifications at the Il6 promoter also ameliorates colitis symptoms. Atractylodin, a drug which inhibits the H3K9me3 demethylase KDM4A (lysine specific demethylase 4A), induced H3K9 trimethylation in fibroblasts stimulated with TNF-α, which impaired binding between NF-κB and the Il6 promoter, leading to decreased intestinal inflammation. Other KDM4A inhibitors have also demonstrated similar effects on IL-6 and colitis symptoms, suggesting that KDM4A could be a novel target to resolve inflammation during colitis.69
Interleukin-10 (IL-10), secreted by CD4+ Th2 cells, macrophages, and dendritic cells, is a major anti-inflammatory cytokine. It modulates the immune response by suppressing effector activities of Th1, Th17, NK cells, and macrophages, and depletion of IL-10 in mice leads to colitis.70 The lysine acetyltransferase KAT2B activates IL-10 through promoter acetylation.71 Interestingly, KAT2B is significantly downregulated in the inflamed colonic mucosa of CD and UC patients compared with healthy controls and noninflamed mucosa from IBD cases.71
The receptor for TGF-β, TGFβRII, induces signaling which leads to polarization of macrophages towards an anti-inflammatory M2 phenotype.72 Expression of a defective TGFβRII in murine macrophages leads to enhanced susceptibility to colitis following DSS treatment. The zinc finger protein KLF10 (TGF–β inducible early gene 1) epigenetically regulates TGFβRII transcription in macrophages by promoter binding and recruitment of histone acetyl transferase P300/CBP associated factor (PCAF) to induce H3 acetylation.73 When KLF10 is depleted, colonic macrophages exhibit lower TGFβRII and IL-10 expression levels, leading to a pro-inflammatory phenotype. Transfer of KLF10 depleted macrophages to WT mice also increases the severity of DSS-induced colitis, suggesting that transcription factor KLF10 is an important regulator of epigenetics and macrophage anti-inflammatory function in the intestine.73
The global de-regulation of histone acetylation can also have severe consequences for cytokine production and intestinal homeostasis. SIRTuins are histone deacetylases known to play roles in metabolic regulation and aging.74 SIRT2 regulates cytokine production in macrophages and influences bone marrow–derived macrophage (BMDM) development.75 Before the DSS challenge, both wild-type and SIRT2 KO mice had similar intestinal permeability and cytokine production.75 After the DSS challenge, SIRT2 KO mice had higher intestinal permeability and higher plasma levels of pro-inflammatory cytokines such as TNF-α and IL-1β than wild-type mice treated with DSS.75 It should also be mentioned that SIRT2 is capable of deacetylating a major regulator of cytokine production, NF-κB; SIRT2 knockout mice show hyperacetylation of the p65 subunit of NF-κB in macrophages, leading to an inflammatory phenotype and impaired resistance against DSS colitis.75
The formation of contacts between regulatory elements and gene promoters at cytokine loci is essential for controlling cytokine gene expression in various cell types. TNF-α, lymphotoxin-α (LT-α), and lymphotoxin-β are pro-inflammatory cytokines whose aberrant expression is linked to several diseases including IBD.76–80 The genes encoding these cytokines are clustered within the human genome. Excitingly, CTCF was shown to bind to this locus and, following TNF-α stimulation, to regulate looping of repressive regulatory elements (insulators), allowing for expression of TNF-α and LT-α.81 Cohesin and CTCF were found to bind at the IFNγ locus in primary CD4+ T cells at both the gene promoter, and at a suspected enhancer region within the gene.82 Experiments showed that activated Th1 cells possessing increased amounts of CTCF and cohesin at these binding sites within the IFNγ locus physically associate with one another through CTCF/cohesin-dependent DNA looping, which then activates IFNγ expression.82 Interestingly, a similar situation involving DNA looping at cytokine loci occurs within Th2 cells. Following Th2 differentiation, the expression of IL4, IL5, and IL13 is increased.81 The Signal Transducer and Activator of Transcription 6 (STAT6) and GATA binding protein 3 (GATA3) transcription factors are essential for establishment and/or maintenance of this gene expression program,83 and recently, Th2 cells from CTCF deficient mice were shown to exhibit significantly reduced levels of IL4, IL5, and IL13 but normal levels of IFNγ. These data suggest that the ability of CTCF and cohesin to facilitate DNA looping is very important for regulating the cytokine expression profiles of differentiated T cell subsets and that the formation of loops occurs in a cell-type specific manner, possibly involving regulation by other proteins, as well.
HISTONE READERS AS THERAPEUTIC TARGETS FOR IBD
The recognition of histone modifications by specific proteins (referred to as histone readers) can lead to activation of various signaling cascades and immune responses, thus making these histone readers attractive therapeutic targets. Bromodomain family (BRD) proteins, BRD2, BRD3, BRD4, and BRDT, bind acetylated lysines on histone H4 or other proteins to regulate transcription. Bromodomain family members can also regulate RNA polymerase II activity.84 These functions of BRD proteins have been shown to lead to activation of pro-inflammatory cytokines via interactions with NF-κB in macrophages.85 Interestingly, BRD proteins have been shown to interact with condensin proteins86 and with CTCF,87 making them exciting candidates to potentially target the functions of genome organizing proteins and/or target aberrant DNA organization.
Currently, several BET (bromodomain and extra-terminal domain) inhibitors are undergoing clinical trials for treatment of various diseases.88 These inhibitors possess functions ranging from induction of apoptosis and inhibition of tumor growth to downregulation of pro-inflammatory cytokines and transcription factors.88 These particular functions also make them excellent therapeutic candidates for disorders such as IBD and several autoimmune diseases. Recently, BRD4 was found to regulate expression of genes important for lineage-specific progression during naïve T cell differentiation into Th1, Th2, Th17, and Treg cells.89 Specific inhibition and uncoupling of BRD4 from key Th17 genes by BET inhibitor MS402 induced Th17 differentiation inhibition without affecting Th1, Th2, or Treg differentiation.90 Excitingly, MS402 also prevents and reduces the severity of T cell transfer mediated colitis in mice by inhibiting pro-inflammatory Th17 differentiation.90 These findings suggest that MS402 may be a potential therapeutic for inflammatory disorders like IBD, multiple sclerosis, and rheumatoid arthritis that are linked to an increase in Th17 cell development.
Another BRD4 inhibitor, I-BET151, attenuates the maturation and costimulatory potential of dendritic cells in both humans and mice. Treatment with I-BET151 reduces maturation marker and pro-inflammatory cytokine expression in dendritic cells and inhibits mature dendritic cell–mediated, antigen-dependent proliferation of naïve CD4+ T cells.91 Dendritic cells treated with I-BET151 remain immature, and this led to increased generation of immunosuppressive Foxp3+ Tregs in vitro. Disappointingly, mice (Rag1-/- mice transferred with CD45RBhigh sorted T cells) experiencing chronic colitis due to impaired Treg function displayed only modest improvement in disease symptoms when treated with I-BET151.91 One explanation could be timing; I-BET151 administration in these mice occurred after colitis symptoms appeared and after priming of naïve T cells by dendritic cells had already occurred. Since earlier experiments negatively implicated I-BET151 in dendritic cell maturation and T cell activation, treatment with I-BET151 at an earlier stage could lead to higher Treg levels by immature dendritic cells, which might make a difference in ameliorating colitis.
In addition to BRD family proteins, other bromodomain-containing proteins have been implicated in IBD. The bromodomain protein SP140 (which also possesses PHD and SAND domains) is highly expressed in immune cells, reads methylated histones, and binds DNA. Single nucleotide polymorphisms (SNPs) within Sp140 are associated with CD,92 in addition to multiple sclerosis and B cell chronic lymphocytic leukemia. Crohn’s disease–associated Sp140 SNPs cause defects in alternative splicing, generating a truncated isoform and reduced transcript levels.93 Deficiency in SP140 in macrophages leads to their ineffective maturation through deregulated transcriptional programs, leading to downregulation of several LPS-induced cytokines, chemokines, and transcription factors including TNF, IL-6, CXCL10, and NF-κB2. Defective innate immune responses can be a driving force for intestinal dysbiosis and inflammation; consequently, peripheral blood mononuclear cells (PBMCs) of CD patients carrying Sp140 SNPs had dampened responses to bacterial and viral ligands. Similarly, Sp140 depletion in mice led to increased severity of DSS-induced colitis, suggesting that SP140 is critical for intestinal homeostasis. In terms of therapy, SP140 levels could potentially be used as a marker for responsiveness to anti-TNF therapies; lower SP140 levels in CD patients correlate with lower TNF levels due to defects in innate immune responses, and this consequently leads to better responses to anti-TNF therapy.93
CHANGES TO THE 3-DIMENSIONAL ORGANIZATION OF CHROMATIN-ENCOMPASSING IBD-ASSOCIATED SNPS MAY INFLUENCE DISEASE PATHOGENESIS
Genome-wide association studies (GWAS) have vastly increased our knowledge of loci associated with common diseases. However, these studies have their limitations because they do not provide information on disease mechanisms associated with loci or directly identify causal variants associated with disease due to linkage disequilibrium between variants.94 Genome-wide association studies of IBD have identified over 200 susceptibility loci, with the majority of the SNPs being located in noncoding regions of the genome.9–11 Some of these SNPs are assumed to be markers for variants within nearby protein-coding genes. However, as discussed above, noncoding regions are highly associated with the presence of DNA regulatory elements (REs), including enhancers which can be looped to facilitate long distance contacts with gene promoters and modulate expression of protein-coding genes.12, 95, 96 The SNPs identified by GWAS might, therefore, contribute to IBD pathogenesis by altering the functions of these REs and, consequently, the IBD-specific, protein-coding genes they regulate.97 In fact, 92 of the known IBD-associated SNPs were recently shown to localize to potential REs, as evidenced by the presence of H3K27ac marks.12 The genes that are regulated by these IBD-associated REs should, therefore, also be considered to be IBD candidate genes. Analysis of the 3-dimensional landscape of chromatin interactions via 3C-derived techniques, such as chromosome conformation capture-on-chip-seq (4C-seq), identifies physical interactions between REs and downstream regulatory genes.97, 98 4C-seq was performed in several different cell types to interrogate the interactions that occurred at the 92 IBD-associated loci containing REs.97 These studies identified 902 novel IBD candidate genes, many of which were reported to be involved in IBD pathogenesis but had not been identified through traditional GWAS. We analyzed selected IBD-associated SNPs identified by Jostins et al9 using a web-based tool called Capture Hi-C plotter (CHiCP), which helps to visualize interactions between gene promoters and distal regulatory elements such as enhancers based on promoter capture Hi-C datasets.99 This analysis revealed several novel, potential interactions between gene promoters and DNA REs within these loci (Table 1). For example, c-Fos, a leucine zipper containing proto-oncogene, heterodimerizes with c-Jun and forms the AP-1 complex that can bind DNA at promoters or enhancers of target genes important in cell proliferation, apoptosis, and differentiation, leading to their expression.100 The expression of c-Fos is induced early during inflammation by pro-inflammatory cytokines such as TNF-α and IL-6.101 The IBD-specific SNP, rs4899554, is associated with the promoter region of c-Fos, suggesting that it might regulate its expression and subsequently impact Fos-mediated, downstream pathways. The CCAAT/enhancer binding protein beta (C/EBPB), a leucine zipper containing transcription factor, is activated by various inflammatory stimuli including LPS and IL-17 and can regulate expression of a number of inflammatory genes including Il6, Il4, Il5, and TNFA.102 The IBD-associated SNP, rs913678, interacts with the CEBPB promoter. The promoter of IFNGR1 was associated with IBD GWAS SNP, rs6920220; IFNGR1 encodes the receptor for IFN-γ, a pro-inflammatory cytokine that is highly important in the regulation of antitumor and antiviral immunity.103 The IBD GWAS SNP, rs12654812, associates with the promoter of DDX41, which functions in innate immunity by recognizing foreign DNA and promoting signaling through the TBK1-STING-IRF3 cascade to activate type I interferons.104, 105 Finally, AKNA, an AT-hook transcription factor which mediates inflammation, development, and B cell differentiation106 may be regulated by contacts with the IBD GWAS SNP, rs4246905. More evidence that aberrant genome organization may contribute to IBD pathogenesis comes from where CTCF was shown to be instrumental in the recruitment of JunD and PU.1 transcription factors to a CTCF binding site containing an IBD-associated single nucleotide polymorphism. The gene closest to this region is ZFP36L1, which has anti-inflammatory activity, and CTCF recruitment of transcription factors near this gene could alter its expression, which could have significance in IBD pathogenesis.94 Additionally, elegant studies performed on colon tissue from CD and non-IBD patients revealed the existence of 2 clinically relevant CD subtypes based on their chromatin accessibility and gene expression profiles.107 These experiments showed that potential enhancers, which are typically active in normal ileum, were accessible and active in CD patient colon tissue, and this correlated with a more ileal-like gene expression pattern. Further studies will be necessary to identify the proteins and histone modifications that facilitate these and other interactions between regulatory elements and promoters of genes involved in regulating intestinal cellular homeostasis and inflammation, and to elucidate the downstream effects that loss of these contacts have on IBD pathogenesis.
Table 1.
Candidate Genes Linked To IBD-Associated SNPs, Identified Via The Capture Hi-C Plotter (CHiCP) (https://www.chicp.org/). aConfirmed by 4C-seq in (97)
| Chr | Position (Mb) | SNP | Interacting Gene Promoters | Cell Types With Significant Interactions |
|---|---|---|---|---|
| 5 | 176.79 | rs12654812 | MXD3 | monocytes, erythroblasts, endothelial precursors, naive B, naive CD4, neutrophils, total B, total CD4 activated and nonactivated, total CD4 MF, total CD8 |
| PRR7 | monocytes, naive CD8, total CD8, total B, total CD4 nonactivated | |||
| DDX41 | monocytes, naive B, total B, naive CD8, total CD8, naive CD4, total CD4 nonactivated, total CD4 MF | |||
| DOK3 | monocytes, naive CD8, total CD8, total B | |||
| DBN1 | monocytes | |||
| LMAN2 | endothelial precursors, M0, M1, M2 macrophages, erythroblasts, megakaryocytes, naive B, total B, naive CD4, total CD4 activated and nonactivated, total CD4 MF, naive CD8, total CD8 | |||
| RGS14 | endothelial precursors, M0, M1, M2 macrophages, erythroblasts, megakaryocytes, naive B, total B, naive CD4, total CD4 activated and nonactivated, total CD4 MF, naive CD8, total CD8 | |||
| SLC34A1 | erythroblasts, M1 macrophages, megakaryocytes, total CD4 activated | |||
| 6 | 138.00 | rs6920220 | TNFAIP3 | monocytes, M1, M2 macrophages, naive CD8, total CD8, total CD4 activated |
| IFNGR1 a | monocytes | |||
| 9 | 117.55 | rs4246905 | AKNA | monocytes, endothelial precursors, M0, M1, M2 macrophages, naive B, neutrophils, total B, total CD4 activated, total CD4 MF, total CD8 |
| TNFSF8 | endothelial precursors, naive CD4, total CD4 activated | |||
| 14 | 75.70 | rs4899554 | FOS | monocytes, M1, M2 macrophages, naive B, neutrophils, total CD4 activated |
| NEK9 | monocytes, erythroblasts, M1 macrophages, neutrophils, naive CD4 | |||
| TMED10 | M2 macrophages, naive B, total B, naive CD8, total CD8, total CD4 activated, total CD4 MF | |||
| 20 | 48.95 | rs913678 | CEBPB | monocytes, M2 macrophages, neutrophils |
Future Directions
IBD is a complex disease with multiple etiological factors playing a role in its development and pathogenesis. Along with a hereditary component, environmental factors such as diet, prior enteric infections, and the microbiome are highly involved in disease development. Epigenetic modifications have an important influence on the development of the intestinal epithelium and lamina propria immune cells, both of which are critical in the proper functioning and maintenance of the intestine as a whole. Epigenetic changes can also alter levels of cytokines in the intestinal epithelial milieu, influencing the levels of inflammation. Thus, efforts should be increased to study the role of epigenetic modifications in diseases including IBD. However, there are several obstacles to consider when studying epigenetics and disease. First, the presence of an epigenetic modification does not guarantee, by itself, that transcription of the associated gene will be affected. Several factors such as the presence of transcription factors and long-range interactions between promoter-enhancer elements can also govern expression of a gene, and it is important to consider these compounding factors when analyzing results between different individuals with the same disease. Second, correlating specific epigenetic modifications with disease activity is often difficult, and large scale, high-throughput studies are therefore necessary to assess the penetrance of epigenetic modifications and to be confident of their association with a given disease. Many labs are now using a technique called Assay for Transposase Accessible Chromatin-using sequencing (ATAC-seq) to identify the global changes to chromatin accessibility in patient samples.108 Third, many diseased states are typically associated with multiple epigenetic modifications that might contribute to the disease, making it hard to understand which modification is rate-limiting in helping to drive disease development. Lastly, epigenetic modifications can be tissue and/or cell-type specific, and ideally, drugs targeting these modifications should, therefore, be administered in a tissue or cell type–specific manner to induce the desired effects in the tissue/cells of interest, without causing off-target effects in other tissues in the body.
There are various ways in which current knowledge about epigenetics can be effectively harnessed to improve both diagnoses and treatment outcomes for IBD. Several candidate drugs that can modulate enzymes, including those that are involved in DNA methylation, histone acetylation/methylation, and bromodomain activity such as Vorinostat, curcumin, and JQ1, are undergoing clinical trials for use in resolving inflammation in several diseases, including IBD.109, 110 However, many of these drugs have broad activity, affecting several enzymes in their target class, leading to serious side effects.109, 110 There is, therefore, a need for drugs that target these enzymes with higher specificity. In addition to the use of manufactured compounds as potential therapies for chronic inflammation, bacterial metabolites such as butyrate111 and plant-based compounds such as phenethyl isothiocyanate found in cruciferous vegetables112 can in turn have several effects on chromatin via modulation of epigenetic marks. These naturally derived components have shown promise for reducing inflammation associated with IBD and promoting healing of the intestinal epithelium.111, 112
Finally, the identification of novel biomarkers for UC and CD using epigenetic profiling methods could improve the early and accurate diagnosis of these diseases, allowing appropriate lines of treatment to be pursued. Recently, very elegant studies were performed using biopsies from 94 IBD patients, classified as having active or inactive disease, and non-IBD patient colons to profile gene expression of epigenetic marks and identify regulatory elements that could be controlling gene expression.113 These studies performed a technique called CAGE (cap analysis of gene expression), which provides a comprehensive map of genome-wide transcription start sites (TSS) and promoters of genes by sequencing the 5’-end of capped RNAs and detects cell-specific enhancers that actively transcribe enhancer RNA.114 Genes upregulated in IBD patients vs controls were enriched for functions in the inflammatory response and cytokine production, along with colon specific functions such as extracellular matrix remodeling, antimicrobial peptide secretion, and barrier integrity—important in IBD pathogenesis. Downregulated genes in IBD patients vs controls were associated with functions that include steroid/drug processing, cell cycle and growth, maintaining fluid balance in the colon, and xenobiotic response. Actively upregulated genes in CD vs UC were linked to steroid, lipid, and lipoprotein metabolism, whereas genes upregulated specifically in UC vs CD were involved in barrier function. The CAGE study indicated that the inflammatory responses in UC and CD are similar; differences are mostly seen in genes important for gut and gut epithelial functions. Based on TSS enrichment data, CAGE also indicated that both epithelial cells and immune cells had functions in IBD pathogenesis. Excitingly, the analyses identified TSS that acted as biomarkers to distinguish between UC, CD, and non-IBD cases with approximately 85% accuracy. This study represents an excellent example of how information on gene expression, epigenetics, and chromatin interactions can be harnessed to improve IBD patient health. Future studies interrogating the roles of the identified biomarkers and/or the controlling regulatory elements in IBD mouse models could shed light on whether they are also important for IBD pathogenesis.
GLOSSARY OF TERMS
Chromatin – Macromolecular complex of DNA, proteins and RNA that make up the genetic material in the nucleus of cells. DNA is packaged into the nucleus with the assistance of histones and other proteins.
Histones – Highly basic proteins that bind tightly to acidic DNA and compact it into nucleosomes.
Nucleosome – Minimal repeating unit of eukaryotic chromatin, consisting of 147 base pairs of DNA wrapped around a histone octamer. Presents as “beads on a string” under electron microscopy.
Heterochromatin – Tightly packaged form of chromatin, making DNA less accessible to transcription factors and other DNA-binding proteins. Two types-constitutive and facultative heterochromatin, exist within the nucleus. Associated with repression of gene expression.
Euchromatin – Less tightly packaged form of chromatin, making DNA more accessible to DNA-binding proteins. Associated with activation of gene expression.
Chromatin remodeling – ATP-dependent process of re-structuring chromatin to alter histone-DNA interactions and re-position nucleosomes towards controlling gene expression.
DNA regulatory elements – Include promoters, enhancers, silencers and insulators which interact with transcription factors and other proteins to regulate levels of gene expression.
Transcription factors – DNA-binding proteins that regulate gene expression, either by working alone or in a complex with other proteins to activate or repress recruitment of RNA polymerase that will transcribe DNA to RNA.
Promoters – 100–1000 bp regions of DNA that assist in gene transcription and located upstream of transcriptional start sites. Can work in concert with other regulatory elements to control levels of transcription of a gene.
Enhancers – 50–1500 bp region of DNA that can be found up to 1 Mbp upstream or downstream of transcription start sites. Enhancers bind to transcriptional activators that helps increase levels of transcription for a particular gene.
Silencers – Region of DNA that binds to transcriptional repressors and inhibits gene transcription. Can be located upstream of transcriptional start sites, or downstream within an exon or intron of the gene or in the 3’-untranslated region.
Insulators – Also known as boundary elements, mediate intra- and interchromosomal interactions. Regions of DNA that partition the genome into distinct regions of gene expression by blocking enhancer-promoter interactions and restricting heterochromatin spread, thereby confining activities of regulatory elements to defined regions.
Topologically Associated Domains (TADs) – Self-interacting chromosomal domains. Promoter-enhancer interactions are restricted by TAD boundaries, thus affecting gene expression.
DNA loop – DNA loops are created when transcriptional machinery at promoters interacts with activating proteins at enhancers, bringing the 2 regulatory elements into close physical proximity.
Polycomb proteins – Group of proteins that epigenetically silence genes via repressive histone modifications, affecting cellular processes such as development, proliferation, and differentiation.
Histone readers – Proteins that contain specialized domains that can recognize modifications on histones (for example, Bromodomain-containing proteins that can bind to acetylated lysines on histones). Downstream regulation of gene expression depends on the functions carried out by the specific reader proteins.
Single Nucleotide Polymorphism (SNP) – Variation in a single nucleotide present at a specific position within the genome. Single nucleotide polymorphisms can be present in the coding and noncoding regions of genes, as well as in the intergenic region between genes, and may or may not affect gene expression. Single nucleotide polymorphisms can be used as biomarkers to identify particular traits or diseases.
Genome Wide Association Studies (GWAS) – Study of genetic variation between individuals with the goal of associating variants/SNPs with defined traits or diseases.
Chromosome conformation capture (3C) – Technique used to study the 3-dimensional organization of chromatin within the nucleus. Explores interactions between 2 specific chromosomal fragments, such as a promoter-enhancer interaction.
Chromosome conformation capture-on-chip (4C) – Captures interactions between one defined genomic locus and all other genomic regions associated with it.
Hi-C – High-throughput sequencing to detect all pairwise interactions between all genomic fragments tested.
Promoter capture Hi-C – Enrichment of promoter-containing fragments from Hi-C libraries to explore long range chromosomal interactions with enhancers, distal regulatory elements or promoters in nuclear 3D space.
Supported by: The Longworth lab is funded by a grant from the Department of Defense (PR150084 to MSL).
REFERENCES
- 1. Aleksandrova K, Romero-Mosquera B, Hernandez V. Diet, gut microbiome and epigenetics: emerging links with inflammatory bowel diseases and prospects for management and prevention. Nutrients. 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gajendran M, Loganathan P, Catinella AP, et al. . A comprehensive review and update on Crohn’s disease. Dis Mon. 2018;64:20–57. [DOI] [PubMed] [Google Scholar]
- 3. Cho JH, Weaver CT. The genetics of inflammatory bowel disease. Gastroenterology. 2007;133:1327–1339. [DOI] [PubMed] [Google Scholar]
- 4. Ananthakrishnan AN, Bernstein CN, Iliopoulos D, et al. . Environmental triggers in IBD: a review of progress and evidence. Nat Rev Gastroenterol Hepatol. 2018;15:39–49. [DOI] [PubMed] [Google Scholar]
- 5. Loftus EV., Jr Update on the incidence and prevalence of inflammatory bowel disease in the United States. Gastroenterol Hepatol (N Y). 2016;12:704–707. [PMC free article] [PubMed] [Google Scholar]
- 6. Stenke E, Bourke B, Knaus U. Crohn’s strictures-moving away from the knife. Front Pediatr. 2017;5:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Paeschke A, Erben U, Kredel LI, et al. . Role of visceral fat in colonic inflammation: from Crohn’s disease to diverticulitis. Curr Opin Gastroenterol. 2017;33:53–58. [DOI] [PubMed] [Google Scholar]
- 8. Hindryckx P, Jairath V, D’Haens G. Acute severe ulcerative colitis: from pathophysiology to clinical management. Nat Rev Gastroenterol Hepatol. 2016;13:654–664. [DOI] [PubMed] [Google Scholar]
- 9. Jostins L, Ripke S, Weersma RK, et al. ; International IBD Genetics Consortium (IIBDGC) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu JZ, van Sommeren S, Huang H, et al. ; International Multiple Sclerosis Genetics Consortium; International IBD Genetics Consortium Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang H, Fang M, Jostins L, et al. ; International Inflammatory Bowel Disease Genetics Consortium Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature. 2017;547:173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mokry M, Middendorp S, Wiegerinck CL, et al. . Many inflammatory bowel disease risk loci include regions that regulate gene expression in immune cells and the intestinal epithelium. Gastroenterology. 2014;146:1040–1047. [DOI] [PubMed] [Google Scholar]
- 13. Vignali M, Hassan AH, Neely KE, et al. . ATP-dependent chromatin-remodeling complexes. Mol Cell Biol. 2000;20:1899–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kalitsis P, Zhang T, Marshall KM, et al. . Condensin, master organizer of the genome. Chromosome Res. 2017;25:61–76. [DOI] [PubMed] [Google Scholar]
- 15. Rankin S, Dawson DS. Recent advances in cohesin biology. F1000Res. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Phillips-Cremins JE, Sauria ME, Sanyal A, et al. . Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yi JM, Kim TO. Epigenetic alterations in inflammatory bowel disease and cancer. Intest Res. 2015;13:112–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Low D, Mizoguchi A, Mizoguchi E. DNA methylation in inflammatory bowel disease and beyond. World J Gastroenterol. 2013;19:5238–5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Karatzas PS, Gazouli M, Safioleas M, et al. . DNA methylation changes in inflammatory bowel disease. Ann Gastroenterol. 2014;27:125–132. [PMC free article] [PubMed] [Google Scholar]
- 20. MacAlpine DM, Almouzni G. Chromatin and DNA replication. Cold Spring Harb Perspect Biol. 2013;5:a010207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Scarpa M, Stylianou E. Epigenetics: concepts and relevance to IBD pathogenesis. Inflamm Bowel Dis. 2012;18:1982–1996. [DOI] [PubMed] [Google Scholar]
- 22. Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kimura K, Hirano T. ATP-dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell. 1997;90:625–634. [DOI] [PubMed] [Google Scholar]
- 24. Kimura K, Rybenkov VV, Crisona NJ, et al. . 13S condensin actively reconfigures DNA by introducing global positive writhe: implications for chromosome condensation. Cell. 1999;98:239–248. [DOI] [PubMed] [Google Scholar]
- 25. Ladurner R, Bhaskara V, Huis in ‘t Veld PJ, et al. . Cohesin’s atpase activity couples cohesin loading onto DNA with smc3 acetylation. Curr Biol. 2014;24:2228–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Elbatsh AMO, Haarhuis JHI, Petela N, et al. . Cohesin releases DNA through asymmetric atpase-driven ring opening. Mol Cell. 2016;61:575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Parelho V, Hadjur S, Spivakov M, et al. . Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. [DOI] [PubMed] [Google Scholar]
- 28. Rubio ED, Reiss DJ, Welcsh PL, et al. . CTCF physically links cohesin to chromatin. Proc Natl Acad Sci U S A. 2008;105:8309–8314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wutz G, Várnai C, Nagasaka K, et al. . Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. Embo J. 2017;36:3573–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Losada A, Hirano M, Hirano T. Identification of xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 1998;12:1986–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ono T, Yamashita D, Hirano T. Condensin II initiates sister chromatid resolution during S phase. J Cell Biol. 2013;200:429–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Longworth MS, Herr A, Ji JY, et al. . RBF1 promotes chromatin condensation through a conserved interaction with the Condensin II protein dcap-D3. Genes Dev. 2008;22:1011–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Longworth MS, Walker JA, Anderssen E, et al. . A shared role for RBF1 and dCAP-D3 in the regulation of transcription with consequences for innate immunity. PLoS Genet. 2012;8:e1002618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang F, Graham WV, Wang Y, et al. . Interferon-gamma and tumor necrosis factor-alpha synergize to induce intestinal epithelial barrier dysfunction by up-regulating myosin light chain kinase expression. Am J Pathol. 2005;166:409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Poritz LS, Garver KI, Tilberg AF, et al. . Tumor necrosis factor alpha disrupts tight junction assembly. J Surg Res. 2004;116:14–18. [DOI] [PubMed] [Google Scholar]
- 36. Bruewer M, Luegering A, Kucharzik T, et al. . Proinflammatory cytokines disrupt epithelial barrier function by apoptosis-independent mechanisms. J Immunol. 2003;171:6164–6172. [DOI] [PubMed] [Google Scholar]
- 37. Schmitz H, Barmeyer C, Fromm M, et al. . Altered tight junction structure contributes to the impaired epithelial barrier function in ulcerative colitis. Gastroenterology. 1999;116:301–309. [DOI] [PubMed] [Google Scholar]
- 38. Westendorf AM, Fleissner D, Hansen W, et al. . T cells, dendritic cells and epithelial cells in intestinal homeostasis. Int J Med Microbiol. 2010;300:11–18. [DOI] [PubMed] [Google Scholar]
- 39. Barnes MJ, Powrie F. Regulatory T cells reinforce intestinal homeostasis. Immunity. 2009;31:401–411. [DOI] [PubMed] [Google Scholar]
- 40. Beumer J, Clevers H. Regulation and plasticity of intestinal stem cells during homeostasis and regeneration. Development. 2016;143:3639–3649. [DOI] [PubMed] [Google Scholar]
- 41. Chiacchiera F, Pasini D. Control of adult intestinal identity by the polycomb repressive machinery. Cell Cycle. 2017;16:243–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Benoit YD, Lepage MB, Khalfaoui T, et al. . Polycomb repressive complex 2 impedes intestinal cell terminal differentiation. J Cell Sci. 2012;125:3454–3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chiacchiera F, Rossi A, Jammula S, et al. . PRC2 preserves intestinal progenitors and restricts secretory lineage commitment. Embo J. 2016;35:2301–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu Y, Peng J, Sun T, et al. . Epithelial EZH2 serves as an epigenetic determinant in experimental colitis by inhibiting TNFα-mediated inflammation and apoptosis. Proc Natl Acad Sci U S A. 2017;114:E3796–E3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Roostaee A, Benoit YD, Boudjadi S, et al. . Epigenetics in intestinal epithelial cell renewal. J Cell Physiol. 2016;231:2361–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zimberlin CD, Lancini C, Sno R, et al. . HDAC1 and HDAC2 collectively regulate intestinal stem cell homeostasis. Faseb J. 2015;29:2070–2080. [DOI] [PubMed] [Google Scholar]
- 47. Turgeon N, Gagné JM, Blais M, et al. . The acetylome regulators Hdac1 and Hdac2 differently modulate intestinal epithelial cell dependent homeostatic responses in experimental colitis. Am J Physiol Gastrointest Liver Physiol. 2014;306:G594–G605. [DOI] [PubMed] [Google Scholar]
- 48. Gonneaud A, Turgeon N, Boudreau F, et al. . Distinct roles for intestinal epithelial cell-specific Hdac1 and Hdac2 in the regulation of murine intestinal homeostasis. J Cell Physiol. 2016;231:436–448. [DOI] [PubMed] [Google Scholar]
- 49. Sakatani T, Kaneda A, Iacobuzio-Donahue CA, et al. . Loss of imprinting of igf2 alters intestinal maturation and tumorigenesis in mice. Science. 2005;307:1976–1978. [DOI] [PubMed] [Google Scholar]
- 50. Schuster AT, Homer CR, Kemp JR, et al. . Chromosome-associated protein D3 promotes bacterial clearance in human intestinal epithelial cells by repressing expression of amino acid transporters. Gastroenterology. 2015;148: 1405–1416.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Small CL, Reid-Yu SA, McPhee JB, et al. . Persistent infection with crohn’s disease-associated adherent-invasive Escherichia coli leads to chronic inflammation and intestinal fibrosis. Nat Commun. 2013;4:1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gradel KO, Nielsen HL, Schønheyder HC, et al. . Increased short- and long-term risk of inflammatory bowel disease after salmonella or campylobacter gastroenteritis. Gastroenterology. 2009;137:495–501. [DOI] [PubMed] [Google Scholar]
- 53. García Rodríguez LA, Ruigómez A, Panés J. Acute gastroenteritis is followed by an increased risk of inflammatory bowel disease. Gastroenterology. 2006;130:1588–1594. [DOI] [PubMed] [Google Scholar]
- 54. Ternhag A, Törner A, Svensson A, et al. . Short- and long-term effects of bacterial gastrointestinal infections. Emerg Infect Dis. 2008;14:143–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Larmonier CB, Shehab KW, Ghishan FK, et al. . T lymphocyte dynamics in inflammatory bowel diseases: role of the microbiome. Biomed Res Int. 2015;2015:504638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chen S, Ma J, Wu F, et al. . The histone H3 lys 27 demethylase JMJD3 regulates gene expression by impacting transcriptional elongation. Genes Dev. 2012;26:1364–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Agger K, Cloos PA, Christensen J, et al. . UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. [DOI] [PubMed] [Google Scholar]
- 58. Burgold T, Spreafico F, De Santa F, et al. . The histone H3 lysine 27-specific demethylase Jmjd3 is required for neural commitment. Plos One. 2008;3:e3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Li Q, Zou J, Wang M, et al. . Critical role of histone demethylase Jmjd3 in the regulation of CD4+ T-cell differentiation. Nat Commun. 2014;5:5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liu Z, Cao W, Xu L, et al. . The histone H3 lysine-27 demethylase Jmjd3 plays a critical role in specific regulation of Th17 cell differentiation. J Mol Cell Biol. 2015;7:505–516. [DOI] [PubMed] [Google Scholar]
- 61. Luckheeram RV, Zhou R, Verma AD, et al. . CD4⁺T cells: differentiation and functions. Clin Dev Immunol. 2012;2012:925135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Liu HP, Cao AT, Feng T, et al. . TGF-β converts Th1 cells into Th17 cells through stimulation of Runx1 expression. Eur J Immunol. 2015;45:1010–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Antignano F, Burrows K, Hughes MR, et al. . Methyltransferase G9A regulates T cell differentiation during murine intestinal inflammation. J Clin Invest. 2014;124:1945–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Baine I, Basu S, Ames R, et al. . Helios induces epigenetic silencing of IL2 gene expression in regulatory T cells. J Immunol. 2013;190:1008–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ribeiro de Almeida C, Heath H, Krpic S, et al. . Critical role for the transcription regulator CCCTC-binding factor in the control of Th2 cytokine expression. J Immunol. 2009;182:999–1010. [DOI] [PubMed] [Google Scholar]
- 66. Guan Q, Zhang J. Recent advances: the imbalance of cytokines in the pathogenesis of inflammatory bowel disease. Mediators Inflamm. 2017;2017:4810258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sanchez-Munoz F, Dominguez-Lopez A, Yamamoto-Furusho JK. Role of cytokines in inflammatory bowel disease. World J Gastroenterol. 2008;14:4280–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhang Q, Zhao K, Shen Q, et al. . Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature. 2015;525:389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ishiguro K, Watanabe O, Nakamura M, et al. . Inhibition of KDM4A activity as a strategy to suppress interleukin-6 production and attenuate colitis induction. Clin Immunol. 2017;180:120–127. [DOI] [PubMed] [Google Scholar]
- 70. Keubler LM, Buettner M, Häger C, et al. . A multihit model: colitis lessons from the interleukin-10-deficient mouse. Inflamm Bowel Dis. 2015;21:1967–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bai AH, Wu WK, Xu L, et al. . Dysregulated lysine acetyltransferase 2B promotes inflammatory bowel disease pathogenesis through transcriptional repression of interleukin-10. J Crohns Colitis. 2016;10:726–734. [DOI] [PubMed] [Google Scholar]
- 72. Gong D, Shi W, Yi SJ, et al. . Tgfβ signaling plays a critical role in promoting alternative macrophage activation. BMC Immunol. 2012;13:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Papadakis KA, Krempski J, Svingen P, et al. . Krüppel-like factor KLF10 deficiency predisposes to colitis through colonic macrophage dysregulation. Am J Physiol Gastrointest Liver Physiol. 2015;309:G900–G909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Guarente L. Sirtuins in aging and disease. Cold Spring Harb Symp Quant Biol. 2007;72:483–488. [DOI] [PubMed] [Google Scholar]
- 75. Lo Sasso G, Menzies KJ, Mottis A, et al. . SIRT2 deficiency modulates macrophage polarization and susceptibility to experimental colitis. Plos One. 2014;9:e103573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Agyekum S, Church A, Sohail M, et al. . Expression of lymphotoxin-beta (LT-beta) in chronic inflammatory conditions. J Pathol. 2003;199:115–121. [DOI] [PubMed] [Google Scholar]
- 77. Cohen JC, Horton JD, Hobbs HH. Human fatty liver disease: old questions and new insights. Science. 2011;332:1519–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gustafson B, Hammarstedt A, Andersson CX, et al. . Inflamed adipose tissue: a culprit underlying the metabolic syndrome and atherosclerosis. Arterioscler Thromb Vasc Biol. 2007;27:2276–2283. [DOI] [PubMed] [Google Scholar]
- 80. Haybaeck J, Zeller N, Wolf MJ, et al. . A lymphotoxin-driven pathway to hepatocellular carcinoma. Cancer Cell. 2009;16:295–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Watanabe T, Ishihara K, Hirosue A, et al. . Higher-order chromatin regulation and differential gene expression in the human tumor necrosis factor/lymphotoxin locus in hepatocellular carcinoma cells. Mol Cell Biol. 2012;32:1529–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hadjur S, Williams LM, Ryan NK, et al. . Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Scheinman EJ, Avni O. Transcriptional regulation of GATA3 in T helper cells by the integrated activities of transcription factors downstream of the interleukin-4 receptor and T cell receptor. J Biol Chem. 2009;284:3037–3048. [DOI] [PubMed] [Google Scholar]
- 84. Taniguchi Y. The bromodomain and extra-terminal domain (BET) family: functional anatomy of BET paralogous proteins. Int J Mol Sci. 2016;17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wang CY, Filippakopoulos P. Beating the odds: bets in disease. Trends Biochem Sci. 2015;40:468–479. [DOI] [PubMed] [Google Scholar]
- 86. Floyd SR, Pacold ME, Huang Q, et al. . The bromodomain protein brd4 insulates chromatin from DNA damage signalling. Nature. 2013;498:246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hsu SC, Gilgenast TG, Bartman CR, et al. . The BET protein BRD2 cooperates with CTCF to enforce transcriptional and architectural boundaries. Mol Cell. 2017;66:102–116.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Doroshow DB, Eder JP, LoRusso PM. BET inhibitors: a novel epigenetic approach. Ann Oncol. 2017;28:1776–1787. [DOI] [PubMed] [Google Scholar]
- 89. Zhang W, Prakash C, Sum C, et al. . Bromodomain-containing protein 4 (BRD4) regulates RNA polymerase II serine 2 phosphorylation in human CD4+ T cells. J Biol Chem. 2012;287:43137–43155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Cheung K, Lu G, Sharma R, et al. . BET N-terminal bromodomain inhibition selectively blocks Th17 cell differentiation and ameliorates colitis in mice. Proc Natl Acad Sci U S A. 2017;114:2952–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Schilderink R, Bell M, Reginato E, et al. . BET bromodomain inhibition reduces maturation and enhances tolerogenic properties of human and mouse dendritic cells. Mol Immunol. 2016;79:66–76. [DOI] [PubMed] [Google Scholar]
- 92. Franke A, McGovern DP, Barrett JC, et al. . Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Mehta S, Cronkite DA, Basavappa M, et al. . Maintenance of macrophage transcriptional programs and intestinal homeostasis by epigenetic reader SP140. Sci Immunol. 2017;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Tehranchi AK, Myrthil M, Martin T, et al. . Pooled chip-seq links variation in transcription factor binding to complex disease risk. Cell. 2016;165:730–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Spielmann M, Mundlos S. Looking beyond the genes: the role of non-coding variants in human disease. Hum Mol Genet. 2016;25:R157–R165. [DOI] [PubMed] [Google Scholar]
- 96. Javierre BM, Burren OS, Wilder SP, et al. ; BLUEPRINT Consortium Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell. 2016;167:1369–1384.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Meddens CA, Harakalova M, van den Dungen NA, et al. . Systematic analysis of chromatin interactions at disease associated loci links novel candidate genes to inflammatory bowel disease. Genome Biol. 2016;17:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Gheldof N, Leleu M, Noordermeer D, et al. . Detecting long-range chromatin interactions using the chromosome conformation capture sequencing (4C-seq) method. Methods Mol Biol. 2012;786:211–225. [DOI] [PubMed] [Google Scholar]
- 99. Schofield EC, Carver T, Achuthan P, et al. . CHiCP: a web-based tool for the integrative and interactive visualization of promoter capture hi-C datasets. Bioinformatics. 2016;32:2511–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Piechaczyk M, Blanchard JM. C-fos proto-oncogene regulation and function. Crit Rev Oncol Hematol. 1994;17:93–131. [DOI] [PubMed] [Google Scholar]
- 101. McKay S, Bromhaar MM, de Jongste JC, et al. . Pro-inflammatory cytokines induce c-fos expression followed by IL-6 release in human airway smooth muscle cells. Mediators Inflamm. 2001;10:135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Simpson-Abelson MR, Hernandez-Mir G, Childs EE, et al. . CCAAT/enhancer-binding protein β promotes pathogenesis of EAE. Cytokine. 2017;92:24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Schroder K, Hertzog PJ, Ravasi T, et al. . Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75:163–189. [DOI] [PubMed] [Google Scholar]
- 104. Jiang Y, Zhu Y, Qiu W, et al. . Structural and functional analyses of human DDX41 DEAD domain. Protein Cell. 2017;8:72–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Zhang Z, Yuan B, Bao M, et al. . The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12:959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Moliterno AR, Resar LM. AKNA: another AT-hook transcription factor “hooking-up” with inflammation. Cell Res. 2011;21:1528–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Weiser M, Simon JM, Kochar B, et al. . Molecular classification of crohn’s disease reveals two clinically relevant subtypes. Gut. 2018;67:36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Buenrostro JD, Wu B, Chang HY, et al. . ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol. 2015;109:21.29.1–21.29.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Nebbioso A, Carafa V, Benedetti R, et al. . Trials with ‘epigenetic’ drugs: an update. Mol Oncol. 2012;6:657–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Andrieu G, Belkina AC, Denis GV. Clinical trials for BET inhibitors run ahead of the science. Drug Discov Today Technol. 2016;19:45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Lee C, Kim BG, Kim JH, et al. . Sodium butyrate inhibits the NF-kappa B signaling pathway and histone deacetylation, and attenuates experimental colitis in an IL-10 independent manner. Int Immunopharmacol. 2017;51:47–56. [DOI] [PubMed] [Google Scholar]
- 112. Liu Y, Dey M. Dietary phenethyl isothiocyanate protects mice from colitis associated colon cancer. Int J Mol Sci. 2017;18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Boyd M, Thodberg M, Vitezic M, et al. . Characterization of the enhancer and promoter landscape of inflammatory bowel disease from human colon biopsies. Nat Commun. 2018;9:1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Takahashi H, Kato S, Murata M, et al. . CAGE (cap analysis of gene expression): a protocol for the detection of promoter and transcriptional networks. Methods Mol Biol. 2012;786:181–200. [DOI] [PMC free article] [PubMed] [Google Scholar]