Abstract
Pro-protein convertase subtilisin/kexin type 9 (PCSK9), a secreted serine protease, regulates serum low-density lipoprotein (LDL) cholesterol levels by targeting the degradation of LDL receptor (LDLR) in the liver. Although previous reports describe elevated levels of PCSK9 in patients with periodontitis, the mechanisms that trigger this increase in serum PCSK9 levels and induce the related inflammatory response remain unclear. In an unc93b1-deficient mouse of Porphyromonas gingivalis infection, nucleic acid antigen recognition via Toll-like receptors was found to promote PCSK9 production, suggesting an indirect role for tumor necrosis factor-α as an inducer of PCSK9 in contrast to that reported in previous studies. Furthermore, PCSK9 production was independent of the TIR domain-containing adapter-inducing interferon-β-dependent signaling pathway. These results indicate that changes in LDLR expression precede an increase in the serum PCSK9 level in the context of an infectious disease such as periodontitis.
Keywords: Immunology
1. Introduction
Infection and inflammation affect lipid metabolism. In this process, inflammation and lipid synthesis are linked and modulated by a shared liver X receptor (LXR)-mediated signaling cascade that provides cholesterol to functional immune cells, as demonstrated by the ability of infections to alter serum lipid profiles [1, 2]. The low-density lipoprotein (LDL) receptor (LDLR), which mediates the uptake of LDL cholesterol (LDL-C) by hepatocytes, is an essential regulator of plasma cholesterol levels. Intercellular cholesterol controls the expression of LDLR at the transcriptional level via the sterol regulatory element-binding protein (SREBP) [3] and at the post-transcriptional level by inducible degrader of LDLR (IDOL)-dependent ubiquitination of the LDLR, which is mediated via LXRs [4].
Pro-protein convertase subtilisin/kexin type 9 (PCSK9) is a secreted serine protease that degrades LDLR in the liver and thus regulates serum LDL-C levels [5, 6, 7, 8]. Therefore, PCSK9 acts as a post-transcriptional regulator of LDLR and has been identified as a major determinant of plasma LDL-C levels [5, 9]. Human genetic studies of PCSK9 have validated this critical function by identifying gain-of-function mutations associated with elevated serum LDL-C levels and premature coronary heart disease (CHD), as well as loss-of-function mutations associated with low plasma LDL-C levels and a reduced CHD incidence over a 15-year follow-up [6, 7].
Previous case-control and community studies reported significantly elevated serum PCSK9 concentrations in patients with periodontitis, compared with healthy subjects [10, 11]. These findings suggest that the chronic bacterial infections or chronic inflammation associated with periodontitis can influence the circulating levels of PCSK9. In a mouse model study of peritoneal infection with the representative periodontal bacterium Porphyromonas gingivalis, PCSK9 production simultaneously increased with the upregulated expression of Srebf2 in the liver, leading to an increase in the serum concentration of LDL-C [12]. Despite these findings, the mechanism by which serum PCSK9 levels increase in periodontitis patients, as well as the precise means by which PCSK9 expression is induced in response to inflammation in vitro, remains unclear. In the literature, inflammatory stimuli such as lipopolysaccharide and zymosan, which are respective ligands for Toll-like receptor (TLR) 4 and TLR2, reduced the hepatic expression of LDLR protein while concomitantly increasing hepatic PCSK9 mRNA expression [13]. Additionally, one study reported that recombinant tumor necrosis factor (TNF)-α protein could induce PCSK9 production in HepG2 cells in a suppressor of cytokine signaling 3-dependent manner [14]. In other words, a TLR-mediated innate immune response can induce PCSK9 expression in the liver in the context of a bacterial infection. However, the effects of other TLR-mediated inflammatory stimuli on the induction of PCSK9 expression have been partially elucidated. Herein, we report a newly discovered effect of nucleic acid antigen recognition, namely, the promotion of PCSK9 production. Additionally, we report findings that contradict a previous report regarding the role of TNF-α in PCSK9 production, as clarified in a P. gingivalis infection model based on Unc93b1-deficient mice.
2. Materials and methods
2.1. Reagents
Poly (I:C) and CpG-DNA were purchased from InvivoGen (San Diego, CA, USA), and resiquimod (R848) was obtained from Sigma-Aldrich (St. Louis, MO, USA). Recombinant human TNF-α (Sigma-Aldrich) was used for in vitro experiments.
2.2. Mice
We obtained 6- to 8-week-old male C57BL/6 mice from Japan SLC, Inc. (Shizuoka, Japan). Unc93b1-mutant mice (3d) deficient in the TLR3, TLR7, and TLR9 pathways preventing recognition of nucleic acid antigens such as Poly (I:C), R848, and CpG-DNA were generated as previously described [15, 16]. All experiments were performed using 8-week-old mice in accordance with the Regulations and Guidelines on the Scientific and Ethical Care and Use of Laboratory Animals of the Science Council of Japan as enforced on June 1st, 2006, and were approved by the Institutional Animal Care and Use Committee at Niigata University (permit number 27-282-1, SA00325, SA00198). The mice were kept in an animal room under specific-pathogen-free conditions with 55 ± 10% relative humidity and a 12/12 h light/dark cycle at 22 ± 1 °C. They were housed in groups of 2–3 in a sterilized cage and were fed regular chow and sterile water. Experiments were initiated at 8-week-old.
2.3. Bacterial cultures and infection
P. gingivalis strain W83 was cultured in modified Gifu anaerobic medium broth (Nissui, Tokyo, Japan) in an anaerobic jar (Becton Dickinson Microbiology System, Cockeysville, MD, USA) and in the presence of an AnaeroPackTM (Mitsubishi Gas Chemical Co. Inc., Tokyo, Japan) at 37 °C for 48 h. Bacterial suspensions were prepared in phosphate-buffered saline (PBS) without Mg2+/Ca2+ using established growth curves and spectrophotometric analysis. The number of colony-forming units (CFU) was standardized by measuring the optical density at 600 nm. For the in vivo experiment, each mouse received a single intraperitoneal inoculum of P. gingivalis (109 CFU), Poly (I:C) (100 μg; InvivoGen), R848 (100 μg; Sigma-Aldrich), or CpG-DNA (ODN1668; 100 μg; InvivoGen). All treatments were administered in 0.5 mL sterile PBS. Control mice were inoculated with PBS alone. The mice were euthanized 16 h after infection and analyzed.
2.4. Cell preparation and culture
The hepatic cell line, HepG2 was obtained from the Riken BioResource Center (Tsukuba, Japan) and maintained in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin (Invitrogen). Twenty-four hours prior to stimulation, the medium was switched to DMEM supplemented with 10% lipoprotein deficient serum (#S5394-50ML, Sigma-Aldrich) to avoid unexpected influences of lipoprotein in serum. The monocytic cell line THP-1 was maintained in 25 mM HEPES-buffered RPMI 1640 (Sigma-Aldrich) supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin. Forty-eight hours prior to stimulation, THP-1 was incubated with 200 nM phorbol myristate acetate (PMA) to induce differentiation into macrophage-like cells. All cell cultures were incubated at 37 °C in an atmosphere of 5% CO2 in air. For the monoculture experiments, HepG2 cells were cultured in a 24-well culture plate (TPP, Trasadingen, Switzerland) at a concentration of 2 × 105 cells/mL in medium supplemented with lipoprotein free serum for 24 h, then the cells were treated with the indicated stimulants for 6 h. For the co-culture experiments, THP-1 cells and HepG2 cells were cultured individually in separated plates before beginning co-cultures. THP-1 monocytes were seeded into upper compartment (Transwell® inserts; membrane pore size: 0.4 μm; #3450; Corning, Corning, NY, USA) and differentiated to macrophage-like cells by treatment with PMA. HepG2 cells were seeded and cultured in the lower compartment at a density of 2 × 105 cells/mL. The medium was switched to DMEM supplemented with lipoprotein free serum 24 h before beginning co-cultures. The co-culture was started by transferring the THP-1-seeded upper compartment into HepG2-seeded lower compartment. THP-1 cells were stimulated by adding the P. gingivalis (MOI: 100) in co-culture system. Twenty-four hours after co-culturing, the mRNA expression profiles of HepG2 cells were analyzed.
2.5. Real-time PCR
Total RNA was isolated from cells and liver tissues using TRI Reagent® (Molecular Research Center, Inc., Cincinnati, OH, USA). Subsequently, cDNA was synthesized using a Transcriptor Universal cDNA Master (Roche Molecular Systems, Inc., Pleasanton, CA, USA). The following TaqMan Probes® for real-time PCR were purchased from Applied Biosystems (Foster City, CA, USA): Hs00545399_m1 (human PCSK9), Hs00181192_m1 (human LDLR), Mm00443258_m1 (mouse Tnfα), Mm00656927_g1 (mouse Saa-1), Mm04207507_gH (mouse Ifnα), and Mm00439552_s1 (mouse Ifnβ). Quantitative PCR was performed using a FastStart Essential DNA Green Master and LightCycler® 480 (Roche Molecular Systems, Inc.). The relative expression levels of each mRNA were normalized to that of GAPDH mRNA using the delta-delta Ct method.
2.6. Transfection with siRNA
The HepG2 cells were transfected in suspension using the SF Cell Line 4D-Nucleofector™ X kit (Lonza, Basel, Switzerland) and Amaxa™ 4D-Nucleofector™ X Unit (Lonza) following the manufacturer's instructions. Briefly, 2 × 105 cells/mL cells were resuspended in 100 μl Nucleofector™ SF solution containing 100 nM TNFRI siRNA (Applied biosystems; #s14266) and transferred to a Nucleocuvette to be transfected using the EH-100 program. After 24 h transfection, the medium was replaced with fresh medium for the experiment. Specific silencing of TNFRI was confirmed by real-time PCR.
2.7. Enzyme-linked immunosorbent assay
The levels of serum PCSK9 were measured using commercially available enzyme-linked immunosorbent assay kits (R & D Systems, Minneapolis, MN, USA). Serum samples were collected from mice prior to euthanasia. Serum cholesterol profiles were analyzed by Skylight Biotech Inc. (Akita, Japan).
2.8. Statistical analysis
All experiments were independently repeated at least twice on separate days. All data are expressed as means ± standard errors of the means (SEM). The Mann–Whitney U-test and analysis of variance were used to determine the statistical significance of differences between experimental animal groups and in multiple-group comparisons, respectively. The unpaired t-test was used to compare two unpaired in vitro experimental groups. GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA) was used to conduct the statistical analyses. A p-value of <0.05 was considered statistically significant; these differences are indicated in the text by asterisks (*, P < 0.05, **, P < 0.01).
3. Results
3.1. Induction of serum PCSK9 by P. gingivalis infection and selective use of nucleic acid sensing pathway, through TLR7 and TLR9 but not TLR3
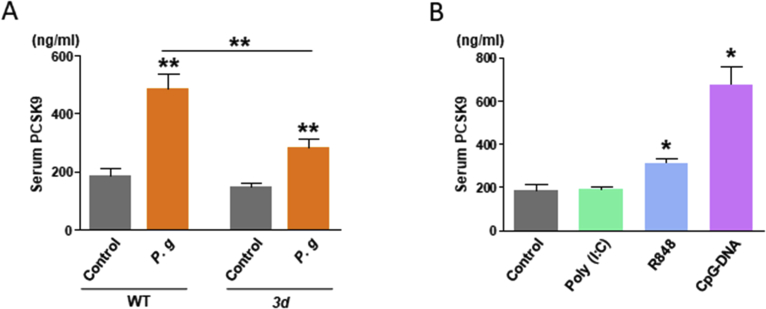
First, this study evaluated the antigenicity of nucleic acid antigens of P. gingivalis and the abilities of these antigens to induce PCSK9 expression. Accordingly, serum PCSK9 concentrations were evaluated in wild-type (WT) and 3d mice after peritoneal infection with live P. gingivalis. Although P. gingivalis infection triggered increased serum levels of PCSK9 in both WT and 3d mice (P < 0.01, respectively), this increase was significantly abrogated in the 3d mice compared with the WT mice (P < 0.01); in other words, nucleic acid antigen recognition enhances the production of PCSK9 in the context of bacterial infection (Fig. 1A).
Fig. 1.
(A) Serum PCSK9 concentrations in wild-type (WT) and 3d mice after Porphyromonas gingivalis infection. N = 5 mice per group. Results are expressed as means ± SEM. **, P < 0.01. (B) Serum PCSK9 concentrations in WT mice after the administration of nucleic acid antigens (Poly (I:C), R848, and CpG-DNA). N = 5 mice per group. Results are expressed as means ± SEM. *, P < 0.05.
Next, the representative antigens Poly (I:C), R848, and CpG-DNA, which are respectively recognized by TLR3, TLR7, and TLR9, were peritoneally administrated to the WT mice to identify the TLR pathway responsible for enhancing the production of PCSK9. R848 and CpG-DNA, which mimic bacterial DNA antigens, enhanced the serum levels of PCSK9 (P < 0.05 and P < 0.01, respectively). However, Poly (I:C) had no effect on PCSK9 levels. In summary, PCSK9 production is induced via TLR7 and TLR9 signaling pathways in a myeloid differentiation primary response 88 (MyD88)-dependent manner, but not by TLR3 (Fig. 1B).
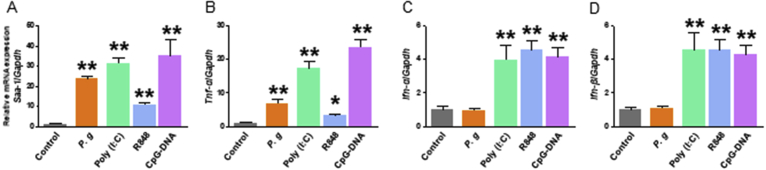
3.2. Elevated expression of genes associated with the acute phase inflammatory response and LDL-C
Serum PCSK9 levels are known to increase during the acute phases of inflammatory responses [12]. Quantitative PCR was, therefore, used to evaluate the hepatic expression of the genes encoding SAA, and TNF-α was analyzed in the context of elevated serum PCSK9 levels. Herein, SAA-1 and TNF-α mRNA expression were induced in the WT mice by P. gingivalis and the TLR ligands Poly (I:C), R848, and CpG-DNA (P < 0.05 and P < 0.01, respectively; Fig. 2A and B). Remarkably, despite the inability of Poly (I:C) to induce PCSK9 expression (Fig. 1B), treatment with this TLR ligand led to increased SAA-1 and TNF-α mRNA expression, suggesting that TNF-α is not a direct inducer of PCSK9 in contrast to a previous report [14].
Fig. 2.
Expression levels of genes encoding inflammatory mediators (relative to Gapdh) in the livers of wild-type mice: (A) Saa1, (B) Tnfα, (C) Ifnα, and (D) Ifnβ. N = 5 mice per group. Results are expressed as means ± SEM. *, P < 0.05; **, P < 0.01.
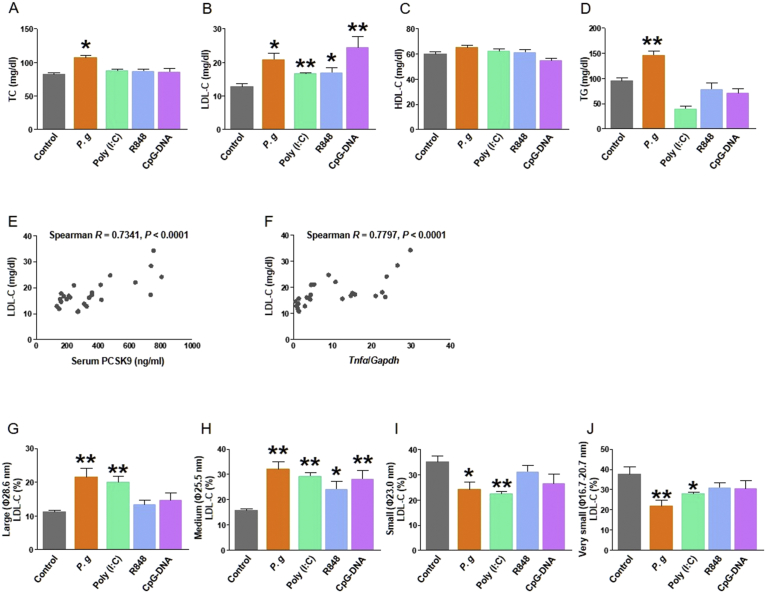
In the liver, the expression of IFN-α and IFN-β mRNA increased in response to Poly (I:C), R848, and CpG-DNA administration (P < 0.01, respectively; Fig. 2C and D). By contrast, P. gingivalis was not sufficiently potent to induce INF-α and INF-β mRNA expression in the liver, suggesting that the induction of PCSK9 in this model does not depend on Type I interferons. During the same experimental time course, significantly higher serum levels of total cholesterol (TC) and LDL-C were observed in the P. gingivalis-infected mice, compared with the sham-infected mice, concordant with our previous study findings (Fig. 3A and B). Notably, treatment with Poly (I:C) enhanced the level of serum LDL-C, despite having no effect on the serum PCSK9 level as shown in Fig. 1B. This also suggests that inflammatory cytokines are not direct inducers of PCSK9. No effect of treatment with antigens was observed on high-density lipoprotein cholesterol (HDL-C), and only P. gingivalis enhanced the level of triglycerides (Fig. 3C and D).
Fig. 3.
Changes in serum lipid levels following the administration of P. gingivalis and nucleic acid antigens: (A) total cholesterol (TC), (B) low-density lipoprotein cholesterol (LDL-C), (C) high-density lipoprotein cholesterol (HDL-C), and (D) triglycerides (TG). N = 5 mice per group. Results are expressed as means ± SEM. *, P < 0.05; **, P < 0.01. Analysis of serum cholesterol levels.: Correlations between the serum lipids and serum PCSK9 level or hepatic TNF-α gene expression in wild-type mice. (E) LDL-C versus PCSK9, (F) HDL-C levels versus PCSK9, (G) TC versus PCSK9, (H) LDL-C versus Tnfα, (I) HDL-C versus Tnfα, and (J) TC versus Tnfα. The ratio of each LDL-C fraction level to the total: (K) large (φ28.6 nm), (L) medium (φ25.5 nm), (M) small (φ23.0 nm), or (N) very small (φ16.7–20.7 nm). N = 5 mice per group. Results are expressed as means ± SEM. *, P < 0.05; **, P < 0.01.
3.3. Correlations between serum lipid and PCSK9 levels
As shown in Fig. 3E and H, the serum LDL cholesterol concentration correlated with the serum PCSK9 concentration and hepatic TNF-α mRNA expression, even in the absence of a direct induction of PCSK9 production (e.g., peritoneal administration of Poly (I:C), suggesting that LDL-C levels are indirectly regulated by PCSK9 under inflammatory conditions. By contrast, the serum levels of TC and HDL-C were not correlated with the serum levels of PCSK9 or TNF-α (Fig. 3F, G, I and J). In a fractional analysis of LDL-C, the production of large and medium LDL-C increased in response to the administration of P. gingivalis or nucleic acid antigens. Simultaneously, decrease of small and very small LDL-C, more atherogenic subclasses, was observed in P. gingivalis and Poly (I:C) treatment. In other words, the inflammatory response increased LDL-C levels but did not specifically exacerbate the profile of LDL-C subclass (Fig. 3K, L, M and N).
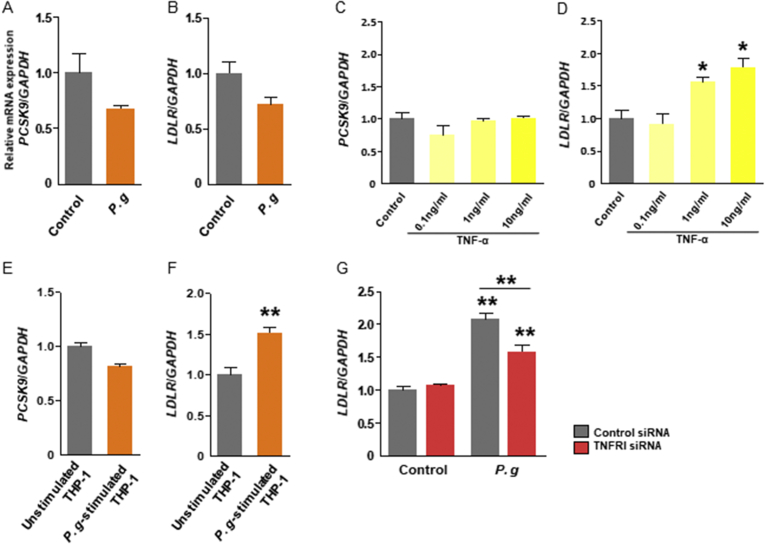
3.4. Indirect induction of PCSK9 by P. gingivalis infection or TNF-α
As the liver is a major source of circulating PCSK9, the ability of P. gingivalis to promote PCSK9 and LDLR gene expression in vitro was analyzed in the hepatic cell line HepG2. Unexpectedly, gene expressions of PCSK9 and LDLR were not directly promoted by P. gingivalis (Fig. 4A and B). Under the condition of incubation with recombinant TNF-α, the expression of PCSK9 was not significantly promoted but that of LDLR was significantly promoted in dose-dependent manner (Fig. 4C and D). Additionally, the co-culture of HepG2 with P. gingivalis-stimulated THP-1 in a Transwell® system enhanced the expression of LDLR but not of PCSK9 (Fig. 4E and F). In the same experiment, knockdown of TNFRI by specific siRNA transfection suppressed the gene expression of LDLR (Fig. 4G). These observations suggest that neither P. gingivalis infection nor the subsequent production of TNF-α directly induces PCSK9 expression. However, TNF-α, and potentially other soluble components produced by THP-1, modulate LDLR expression, which may further affect lipid homeostasis and thus regulate PCSK9 expression.
Fig. 4.
Expression of PCSK9 and LDLR relative to GAPDH in HepG2 cells. The cells were directly stimulated with Porphyromonas gingivalis (A and B) or recombinant TNF-α at the indicated concentration (C and D). The cells were co-cultured with macrophage-like differentiated THP-1 cells and then stimulated with P. gingivalis (MOI: 100) (E and F). The cells are transfected with TNFRI specific siRNA and then co-cultured with THP-1 cells (G). N = 3 wells per group. Results are expressed as means ± SEM. *, P < 0.05; **, P < 0.01.
4. Discussion
In this study, we identified a unique role of inflammation in the induction of PCSK9 expression in an Unc93b1 mutant mouse model, 3d, which exhibits deficiencies in the ability to sense nucleic acid antigens via the TLR3, TLR7, and TLR9 pathways [15, 16]. To the best of our knowledge, this is the first report to demonstrate that nucleic acid antigen recognition can enhance serum PCSK9 expression in the context of bacterial infection.
Infection and inflammation have been shown to affect multiple aspects of lipid and lipoprotein metabolism [1]. Our previous study showed that peritoneal P. gingivalis infection led to increased serum PCSK9 levels and concomitantly decreased expression of Ldlr and Srebp2 in the liver [12]. Therefore, one might expect that the defective nucleic acid antigen sensing characteristic of the 3d mouse model would lead to reduced serum levels of PCSK9, as a loss of TLR9 signaling in response to bacterial DNA should reduce the downstream transcription of inflammatory cytokines such as TNF-α [17, 18]. Indeed, peritoneal injection of CpG-DNA promoted the elevation of serum PCSK9 in WT mice. On the contrary, one critical observation of this study was that the administration of Poly (I:C) failed to increase serum PCSK9 levels, despite the concurrent induction of Saa1 and Tnfα expression.
In contrast to previous studies [14, 19], our findings revealed that neither TNF-α nor the TRIF-dependent signaling pathway was a direct inducer of PCSK9. P. gingivalis administration was not sufficiently potent to induce the expression of Ifnα and Ifnβ in the liver concurrently with the enhanced expression of PCSK9, indicating that Type I interferons do not induce PCSK9.
Furthermore, we observed significant increases in serum LDL-C levels following the administration of nucleic acid antigens or P. gingivalis. The finding that Poly (I:C) enhanced LDL-C but not PSCK9 levels indicated that the former might initially be regulated in a PCSK9-independent manner in an inflammatory response. We additionally analyzed the LDL-C particle diameters to determine whether P. gingivalis affected this parameter. Etiologically, P. gingivalis infection may be linked to the development of atherosclerosis, as suggested by a previous finding that an increased small LDL-C concentration may enhance the risk of Cardiovascular disease (CVD) in humans [20]. However, we observed that the administration of P. gingivalis or nucleic acid antigens similarly had no effect on small and very small LDL-C levels, suggesting that the bacterial infection did not particularly promote atherogenesis by altering the LDL-C profile.
In our in vitro analysis of HepG2 cells, TNF-α promoted LDLR but not PCSK9 mRNA expression under normal and lipid-depleted conditions, in contrast to that observed in a previous report [14]. Our analysis, therefore, suggests that inflammatory cytokines are also unlikely to be direct regulators of PCSK9 production in vivo. In other words, the transcription of LDLR is independent of the change in serum PCSK9 levels in response to infection. As the transcription of both PCSK9 and LDLR is regulated by SREBPs [21, 22], a feedback-regulated cholesterol synthesis mechanism may enhance the production of PCSK9 via an increase in LDLR expression [22].
We note that our finding that PCSK9 expression did not increase in HepG2 cells in response to inflammatory stimuli differed from previous findings [14, 19]; this discrepancy might be due to differences in characteristics between laboratories or unknown technical issues affecting the experimental conditions. This topic should be explored in future studies. We also note that several investigations reported an association between periodontitis and atherosclerotic vascular disease [23, 24, 25, 26, 27, 28], and studies have attributed this relationship to changes in the lipid profiles of patients with periodontitis, which manifest as reduced HDL-C and increased LDL-C levels [29, 30, 31]. However, the mechanisms underlying the elevation of LDL-C levels in patients with periodontitis have not yet been clearly elucidated. As PCSK9 regulates serum LDL-C levels and is thus critical to the development of atherogenesis and the incidence of CVD [5, 6, 7, 8], the elevated serum PCSK9 levels detected in patients with periodontitis may play a key role in the relationship between periodontitis and CVD. Burdens such as P. gingivalis infection may promote elevated levels of PCSK9 in patients with periodontitis, as shown in this study and the previous literature [12]. However, the present study was limited by the discrepancy between periodontitis in human patients and an animal model comprising mice infected via intraperitoneal inoculation. Another limitation of the study is based on the observation in vitro models using cultured macrophage derived from THP-1 cell line for the source of inflammatory molecules in innate response which mimic Kupffer cells in liver. Therefore, future studies should investigate how gingival infection contributes to the elevation of serum PCSK9 levels in humans.
Declarations
Author contribution statement
Mai Yokoji-Takeuchi, Koichi Tabeta: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Naoki Takahashi: Contributed reagents, materials, analysis tools or data; Wrote the paper.
Kei Arimatsu, Haruna Miyazawa, Yumi Matsuda-Matsukawa, Keisuke Sato: Contributed reagents, materials, analysis tools or data.
Miki Yamada: Performed the experiments; Contributed reagents, materials, analysis tools or data.
Kazuhisa Yamazaki: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Funding statement
This work was supported by JSPS (Japan Society for the Promotion of Science) grants KAKENHI JP15H0502 and JP15KK0338 to KT and JP15H02578 to KY.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Contributor Information
Koichi Tabeta, Email: koichi@dent.niigata-u.ac.jp.
Kazuhisa Yamazaki, Email: kaz@dent.niigata-u.ac.jp.
References
- 1.Khovidhunkit W., Kim M.S., Memon R.A., Shigenaga J.K., Moser A.H., Feingold K.R. Effects of infection and inflammation on lipid and lipoprotein metabolism: mechanisms and consequences to the host. J. Lipid Res. 2004;45(7):1169–1196. doi: 10.1194/jlr.R300019-JLR200. PubMed PMID: 15102878. [DOI] [PubMed] [Google Scholar]
- 2.Maekawa T., Takahashi N., Tabeta K., Aoki Y., Miyashita H., Miyauchi S. Chronic oral infection with Porphyromonas gingivalis accelerates atheroma formation by shifting the lipid profile. PLoS One. 2011;6(5):e20240. doi: 10.1371/journal.pone.0020240. PubMed PMID: 21625524; PubMed Central PMCID: PMC3098290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yokoyama C., Wang X., Briggs M.R., Admon A., Wu J., Hua X. SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell. 1993;75(1):187–197. PubMed PMID: 8402897. [PubMed] [Google Scholar]
- 4.Zelcer N., Hong C., Boyadjian R., Tontonoz P. LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor. Science. 2009;325(5936):100–104. doi: 10.1126/science.1168974. PubMed PMID: 19520913; PubMed Central PMCID: PMCPMC2777523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lambert G., Charlton F., Rye K.A., Piper D.E. Molecular basis of PCSK9 function. Atherosclerosis. 2009;203(1):1–7. doi: 10.1016/j.atherosclerosis.2008.06.010. PubMed PMID: 18649882. [DOI] [PubMed] [Google Scholar]
- 6.Abifadel M., Varret M., Rabes J.P., Allard D., Ouguerram K., Devillers M. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 2003;34(2):154–156. doi: 10.1038/ng1161. PubMed PMID: 12730697. [DOI] [PubMed] [Google Scholar]
- 7.Cohen J.C., Boerwinkle E., Mosley T.H., Jr., Hobbs H.H. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N. Engl. J. Med. 2006;354(12):1264–1272. doi: 10.1056/NEJMoa054013. PubMed PMID: 16554528. [DOI] [PubMed] [Google Scholar]
- 8.Zhang D.W., Lagace T.A., Garuti R., Zhao Z., McDonald M., Horton J.D. Binding of proprotein convertase subtilisin/kexin type 9 to epidermal growth factor-like repeat A of low density lipoprotein receptor decreases receptor recycling and increases degradation. J. Biol. Chem. 2007;282(25):18602–18612. doi: 10.1074/jbc.M702027200. PubMed PMID: 17452316. [DOI] [PubMed] [Google Scholar]
- 9.Lambert G., Ancellin N., Charlton F., Comas D., Pilot J., Keech A. Plasma PCSK9 concentrations correlate with LDL and total cholesterol in diabetic patients and are decreased by fenofibrate treatment. Clin. Chem. 2008;54(6):1038–1045. doi: 10.1373/clinchem.2007.099747. PubMed PMID: 18436719. [DOI] [PubMed] [Google Scholar]
- 10.Tabeta K., Hosojima M., Nakajima M., Miyauchi S., Miyazawa H., Takahashi N. Increased serum PCSK9, a potential biomarker to screen for periodontitis, and decreased total bilirubin associated with probing depth in a Japanese community survey. J. Periodontal. Res. 2018;53(3):446–456. doi: 10.1111/jre.12533. PubMed PMID: 29516504. [DOI] [PubMed] [Google Scholar]
- 11.Miyazawa H., Honda T., Miyauchi S., Domon H., Okui T., Nakajima T. Increased serum PCSK9 concentrations are associated with periodontal infection but do not correlate with LDL cholesterol concentration. Clin. Chim. Acta. Int. J. Clin. Chem. 2012;413(1-2):154–159. doi: 10.1016/j.cca.2011.09.023. Epub 2011/10/18. PubMed PMID: 22001517. [DOI] [PubMed] [Google Scholar]
- 12.Miyazawa H., Tabeta K., Miyauchi S., Aoki-Nonaka Y., Domon H., Honda T. Effect of Porphyromonas gingivalis infection on post-transcriptional regulation of the low-density lipoprotein receptor in mice. Lipids Health Dis. 2012;11:121. doi: 10.1186/1476-511X-11-121. PubMed PMID: 22992388; PubMed Central PMCID: PMCPMC3503659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feingold K.R., Moser A.H., Shigenaga J.K., Patzek S.M., Grunfeld C. Inflammation stimulates the expression of PCSK9. Biochem. Biophys. Res. Commun. 2008;374(2):341–344. doi: 10.1016/j.bbrc.2008.07.023. PubMed PMID: 18638454; PubMed Central PMCID: PMCPMC2571081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruscica M., Ricci C., Macchi C., Magni P., Cristofani R., Liu J. Suppressor of cytokine signaling-3 (SOCS-3) induces proprotein convertase subtilisin kexin type 9 (PCSK9) expression in hepatic HepG2 cell line. J. Biol. Chem. 2016;291(7):3508–3519. doi: 10.1074/jbc.M115.664706. PubMed PMID: 26668321; PubMed Central PMCID: PMCPMC4751391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tabeta K., Hoebe K., Janssen E.M., Du X., Georgel P., Crozat K. The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat. Immunol. 2006;7(2):156–164. doi: 10.1038/ni1297. PubMed PMID: 16415873. [DOI] [PubMed] [Google Scholar]
- 16.Janssen E., Tabeta K., Barnes M.J., Rutschmann S., McBride S., Bahjat K.S. Efficient T cell activation via a Toll-Interleukin 1 Receptor-independent pathway. Immunity. 2006;24(6):787–799. doi: 10.1016/j.immuni.2006.03.024. PubMed PMID: 16782034. [DOI] [PubMed] [Google Scholar]
- 17.Tabeta K., Georgel P., Janssen E., Du X., Hoebe K., Crozat K. Toll-like receptors 9 and 3 as essential components of innate immune defense against mouse cytomegalovirus infection. Proc. Natl. Acad. Sci. U. S. A. 2004;101(10):3516–3521. doi: 10.1073/pnas.0400525101. PubMed PMID: 14993594; PubMed Central PMCID: PMCPMC373494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hemmi H., Takeuchi O., Kawai T., Kaisho T., Sato S., Sanjo H. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408(6813):740–745. doi: 10.1038/35047123. PubMed PMID: 11130078. [DOI] [PubMed] [Google Scholar]
- 19.Ding Z., Liu S., Wang X., Deng X., Fan Y., Shahanawaz J. Cross-talk between LOX-1 and PCSK9 in vascular tissues. Cardiovasc. Res. 2015;107(4):556–567. doi: 10.1093/cvr/cvv178. PubMed PMID: 26092101. [DOI] [PubMed] [Google Scholar]
- 20.Rizzo M., Berneis K. Lipid triad or atherogenic lipoprotein phenotype: a role in cardiovascular prevention? J. Atherosclerosis Thromb. 2005;12(5):237–239. doi: 10.5551/jat.12.237. PubMed PMID: 16205019. [DOI] [PubMed] [Google Scholar]
- 21.Espenshade P.J. SREBPs: sterol-regulated transcription factors. J. Cell Sci. 2006;119(Pt 6):973–976. doi: 10.1242/jcs.02866. PubMed PMID: 16525117. [DOI] [PubMed] [Google Scholar]
- 22.Brown M.S., Goldstein J.L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89(3):331–340. doi: 10.1016/s0092-8674(00)80213-5. PubMed PMID: 9150132. [DOI] [PubMed] [Google Scholar]
- 23.Aimetti M., Romano F., Nessi F. Microbiologic analysis of periodontal pockets and carotid atheromatous plaques in advanced chronic periodontitis patients. J. Periodontol. 2007;78(9):1718–1723. doi: 10.1902/jop.2007.060473. PubMed PMID: 17760541. [DOI] [PubMed] [Google Scholar]
- 24.Beck J.D., Couper D.J., Falkner K.L., Graham S.P., Grossi S.G., Gunsolley J.C. The Periodontitis and Vascular Events (PAVE) pilot study: adverse events. J. Periodontol. 2008;79(1):90–96. doi: 10.1902/jop.2008.070223. PubMed PMID: 18166097. [DOI] [PubMed] [Google Scholar]
- 25.Buduneli N., Kinane D.F. Host-derived diagnostic markers related to soft tissue destruction and bone degradation in periodontitis. J. Clin. Periodontol. 2011;38(Suppl 11):85–105. doi: 10.1111/j.1600-051X.2010.01670.x. PubMed PMID: 21323706. [DOI] [PubMed] [Google Scholar]
- 26.Buhlin K., Hultin M., Norderyd O., Persson L., Pockley A.G., Pussinen P.J. Periodontal treatment influences risk markers for atherosclerosis in patients with severe periodontitis. Atherosclerosis. 2009;206(2):518–522. doi: 10.1016/j.atherosclerosis.2009.03.035. PubMed PMID: 19411077. [DOI] [PubMed] [Google Scholar]
- 27.Tabeta K., Yoshie H., Yamazaki K. Current evidence and biological plausibility linking periodontitis to atherosclerotic cardiovascular disease. Jpn Dent Sci Rev. 2014;50(3):55–62. [Google Scholar]
- 28.Tonetti M.S. Periodontitis and risk for atherosclerosis: an update on intervention trials. J. Clin. Periodontol. 2009;36(Suppl 10):15–19. doi: 10.1111/j.1600-051X.2009.01417.x. PubMed PMID: 19432627. [DOI] [PubMed] [Google Scholar]
- 29.Katz J., Flugelman M.Y., Goldberg A., Heft M. Association between periodontal pockets and elevated cholesterol and low density lipoprotein cholesterol levels. J. Periodontol. 2002;73(5):494–500. doi: 10.1902/jop.2002.73.5.494. PubMed PMID: 12027250. [DOI] [PubMed] [Google Scholar]
- 30.Losche W., Karapetow F., Pohl A., Pohl C., Kocher T. Plasma lipid and blood glucose levels in patients with destructive periodontal disease. J. Clin. Periodontol. 2000;27(8):537–541. doi: 10.1034/j.1600-051x.2000.027008537.x. PubMed PMID: 10959778. [DOI] [PubMed] [Google Scholar]
- 31.D'Aiuto F., Nibali L., Parkar M., Suvan J., Tonetti M.S. Short-term effects of intensive periodontal therapy on serum inflammatory markers and cholesterol. J. Dent. Res. 2005;84(3):269–273. doi: 10.1177/154405910508400312. PubMed PMID: 15723869. [DOI] [PubMed] [Google Scholar]