Among the possible environmental reservoirs in a patient care environment, sink drains are increasingly recognized as a potential reservoir to hospitalized patients of multidrug-resistant health care-associated pathogens. With increasing antimicrobial resistance limiting therapeutic options for patients, a better understanding of how pathogens disseminate from sink drains is urgently needed. Once this knowledge gap has decreased, interventions can be engineered to decrease or eliminate transmission from hospital sink drains to patients. The current study further defines the mechanisms of transmission for bacteria that colonize sink drains.
KEYWORDS: antimicrobial resistance, dispersion, GFP-E. coli, sink lab, carbapenem-resistant Enterobacteriaceae, hand washing, infection control, nosocomial transmission, premise plumbing, sinks
ABSTRACT
An alarming rise in hospital outbreaks implicating hand-washing sinks has led to widespread acknowledgment that sinks are a major reservoir of antibiotic-resistant pathogens in patient care areas. An earlier study using green fluorescent protein (GFP)-expressing Escherichia coli (GFP-E. coli) as a model organism demonstrated dispersal from drain biofilms in contaminated sinks. The present study further characterizes the dispersal of microorganisms from contaminated sinks. Replicate hand-washing sinks were inoculated with GFP-E. coli, and dispersion was measured using qualitative (settle plates) and quantitative (air sampling) methods. Dispersal caused by faucet water was captured with settle plates and air sampling methods when bacteria were present on the drain. In contrast, no dispersal was captured without or in between faucet events, amending an earlier theory that bacteria aerosolize from the P-trap and disperse. Numbers of dispersed GFP-E. coli cells diminished substantially within 30 minutes after faucet usage, suggesting that the organisms were associated with larger droplet-sized particles that are not suspended in the air for long periods.
IMPORTANCE Among the possible environmental reservoirs in a patient care environment, sink drains are increasingly recognized as a potential reservoir to hospitalized patients of multidrug-resistant health care-associated pathogens. With increasing antimicrobial resistance limiting therapeutic options for patients, a better understanding of how pathogens disseminate from sink drains is urgently needed. Once this knowledge gap has decreased, interventions can be engineered to decrease or eliminate transmission from hospital sink drains to patients. The current study further defines the mechanisms of transmission for bacteria that colonize sink drains.
INTRODUCTION
Recent reports have implicated hand-washing sinks as a primary reservoir of antibiotic-resistant pathogens within patient care environments (1–27). Many of these reports have been published since 2016, highlighting the global recognition and vital role that biofilms located in and on sinks can have in disseminating clinically important, drug-resistant, Gram-negative bacteria (14–27). Retrospective and prospective surveillance investigations affirm that hospital sinks provide habitats for several opportunistic pathogens, raising serious concerns (8, 24, 26, 28). It is not the mere presence of these drug-resistant pathogens in the hospital wastewater that is of concern, but the ability of these organisms to colonize biofilms on the luminal surfaces of wastewater plumbing and thereby withstand routine cleaning practices. While several Gammaproteobacteria species detected from the sinks in hospitals have been linked to health care-associated infections, opportunistic pathogens like Pseudomonas aeruginosa, Acinetobacter baumannii, and Stenotrophomonas maltophilia are typically known to be found in water environments (29–31). In contrast, emerging pathogens, such as carbapenemase-producing Enterobacteriaceae (CPE), many of which have fecal origins, may survive within the biofilm formed on sink surfaces and wastewater premise plumbing (32, 33). CPE infections are especially threatening because they often acquire mobile resistance elements through horizontal gene transfer and are more frequent causes of highly antibiotic-resistant infections with reduced treatment options.
In outbreak investigations, species and strain matches between patient and sink isolates are often attributed to sink source contamination; however, the direction (sink to patient versus patient to sink) and precise mode of transmission remain inconsistent and elusive (29, 30). Even with increased recognition of transmission, a knowledge gap exists with regard to the precise mechanism of transmission from sink reservoirs to the patient. Using a model of sink colonization with green fluorescent protein (GFP)-expressing Escherichia coli (GFP-E. coli), we recently demonstrated the source and the degree of dispersion from sink wastewater to the surrounding environment (34). Factors effecting the rate and extent of droplet-mediated dispersion were investigated, but particle size involved in dispersion was not measured in this study. Studies that claim aerosols as the primary dispersion mechanism of bacteria from sinks are based on rudimentary findings (2, 23, 35, 36) or on assumptions drawn based on these unsubstantiated findings (3, 6, 10, 13, 21, 37). Airborne particles originating from sinks can vary in size and composition. The World Health Organization and Healthcare Infection Control Practices Advisory Committee (HICPAC) guidelines use a particle diameter of 5 μm to delineate between bioaerosol (≤5 μm) and droplet (>5 μm) transmission (38, 39).
Aerosol-mediated transmission and droplet-mediated transmission in the health care environment require conceptually different infection control strategies. Clarity regarding aerosol- versus droplet-mediated dispersion in the context of sinks is critical. In the present study, we aim to further define the following outstanding knowledge gaps: (i) the dispersion mechanism of bacteria (aerosol-sized particles or droplets from biofilms in handwashing sinks), (ii) factors triggering dispersion from a colonized sink drain, and (iii) the role of biologically active aerosols spontaneously dispersing from the drain or P-trap without a triggering event. GFP-E. coli as the surrogate organism for Enterobacteriaceae was used in this model study to investigate these questions.
RESULTS
Free and total chlorine concentrations in the faucet water were consistent across the experiments (Table 1), as were the water and air temperatures (Table 2). In comparison, the relative humidity recorded across the experiments varied, with the highest recorded in the case of P-trap inoculation experiments (Table 2).
TABLE 1.
Chlorine concentrations and faucet water temperaturea
| Parameter | Initial (first catch) | Final (after 2 min) |
|---|---|---|
| Total chlorine (mg/liter) | 0.77 ± 0.26 | 0.63 ± 0.07 |
| Free chlorine (mg/liter) | 0.62 ± 0.25 | 0.54 ± 0.13 |
| Water temperature (°C) | 22.53 ± 1.87 | 36.69 ± 2.17 |
Means and standard deviations of total and free chlorine residual concentrations and water temperature, measured at different time points and from water collected from faucets supplying different sinks over the course of this study.
TABLE 2.
Air temperature and relative humidity recorded across the experimentsa
| Type of experiment | Air temperature (°C) | Relative humidity (%) |
|---|---|---|
| Drain inoculation | 19.8 ± 0.2 | 31.4 ± 4.9 |
| P-trap inoculation | 19.5 ± 0.4 | 54.4 ± 11.5 |
| Drain + P-trap colonization | 19.9 ± 0.2 | 44.7 ± 20.3 |
| Control | 19.7 ± 0.2 | 46.5 ± 16.3 |
Shown are means and standard deviations.
Dispersion immediately following P-trap inoculation.
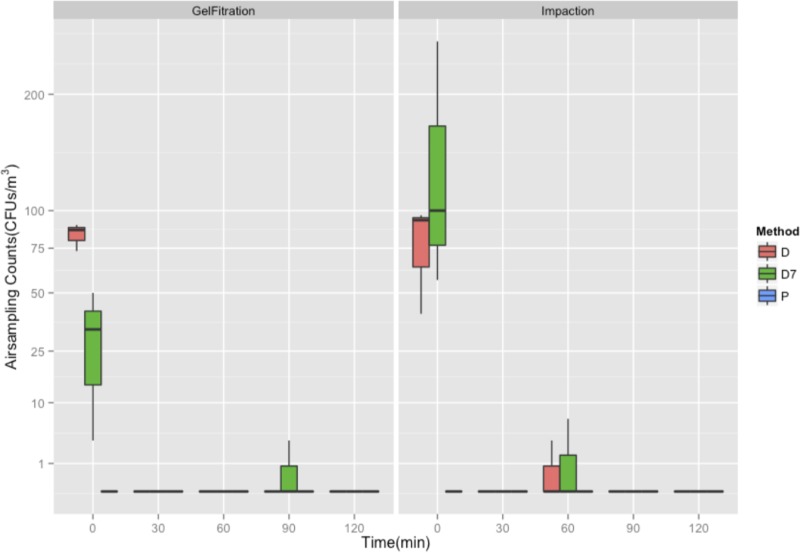
No GFP-E. coli dispersion was detected on settle plates immediately following inoculation of P-traps with 1010 CFU GFP-E. coli (Fig. 1a). No dispersion was detected using impaction, impingement, or filtration air sampling methods (Fig. 2). Furthermore, no GFP-E. coli cells were recorded in the P-trap water, P-trap, tailpiece, or sink bowl surface samples collected at the end of the dispersion experiment.
FIG 1.
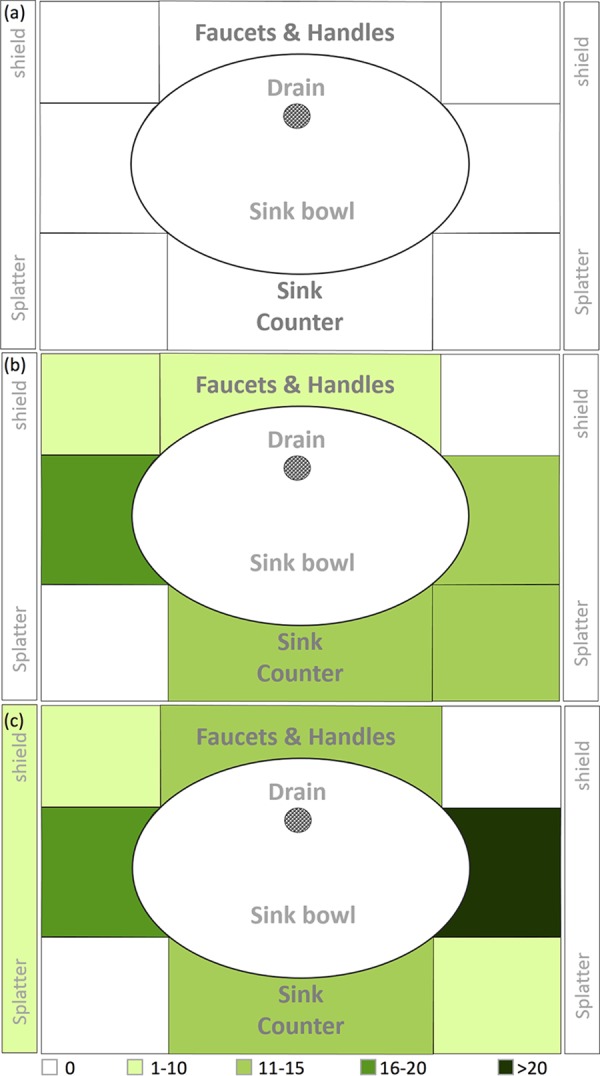
Heat map representation of GFP-E. coli dispersion captured on tryptic soy agar (TSA) settle plates following (a) P-trap, (b) sink drain inoculation, and (c) drain line colonization.
FIG 2.
GFP-E. coli as measured by impaction (SAS90) and gel filtration (MD8) across drain inoculation (D), drain line colonization (D7), and P-trap inoculation (P) methods.
Dispersion immediately following drain inoculation.
When sink drains were inoculated with 1010 CFU GFP-E. coli, dispersion was detected on settle plates and by impaction and filtration. Dispersion detected by settle plates across the three sinks ranged from 35 to 107 CFU/plate. GFP-E. coli levels were higher on the counter space surrounding the sink bowl than near the faucets (Fig. 1b). Dispersion was not detected on side-splatter shields. With the exception of the detection of 1 CFU/m3 at 60 min, using the impaction method, dispersion of GFP-E. coli was detected only at the first faucet event (t = 0 min), using both impaction and filtration methods (Fig. 2). Average dispersions captured at the first faucet event (t = 0 min) were 77 and 83 CFU/m3 using impaction and filtration methods, respectively. GFP-E. coli was not detected at any time point using liquid impingement.
Dispersion following growth for 7 days in an amended P-trap biofilm.
We allowed colonization of GFP-E. coli in drain lines between the strainer and P-trap with nutrient exposure over time, and dispersion was detected on settle plates (Fig. 1c), with counts ranging from 49 to 107 CFU/plate. The counter space surrounding the sink bowl received the largest amount of droplet dispersion, followed by faucet, faucet handle surfaces, and splatter shields. GFP-E. coli levels were highest at the first faucet event (t = 0 min) and not detectable afterwards, with the exception of a 2 CFU/m3 count at 60 min using air impaction and a 1 CFU/m3 count at 90 min using filtration, similar to what was observed for the drain inoculation experiment (Fig. 2). Dispersion captured at the first faucet event (t = 0 min) was 138 and 29 CFU/m3 using impaction and filtration methods, respectively. GFP-E. coli was not detected using liquid impingement. GFP-E. coli was detected on the sink bowl, drain grate, tailpiece, and P-trap and in P-trap water at the completion of this experiment (data not shown).
Dispersion without faucet events (control experiments).
Without a faucet event, GFP-E. coli was not detected on settle plates or by impaction, impingement, or filtration air sampling methods in any experiment. Without a faucet event, fungal and nonfluorescent CFU were occasionally recorded on settle plates, subsequently identified as Staphylococcus spp., nonhemolytic streptococci, Paenibacillus spp., yeast, and small Gram-positive rods.
Total viable heterotrophic organisms in laboratory air.
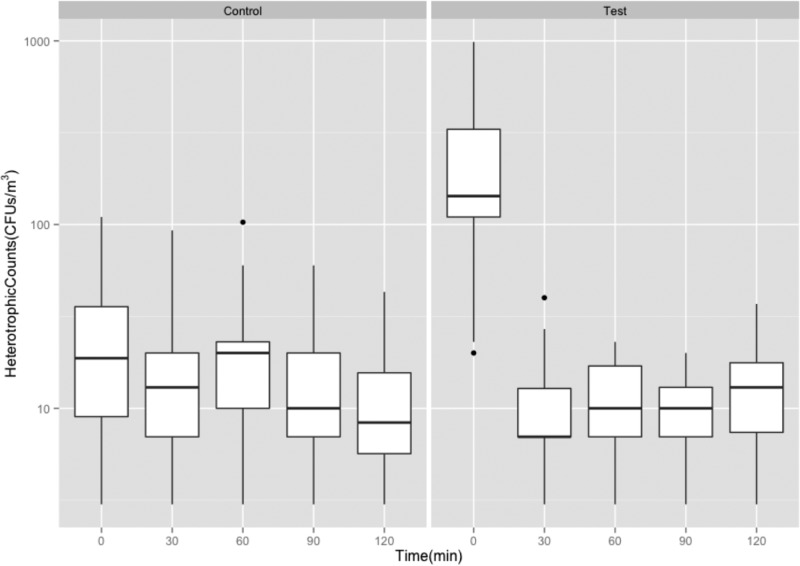
GFP-E. coli was not detected in the faucet water at any time during the experiments. Air samples were collected by impaction on Reasoner’s 2A (R2A) medium, with and without faucet events, in order to quantify total heterotrophic organisms in the air space in proximity to the sink during each experiment. Across the experiments, the heterotrophic organisms in the air ranged from 4 to 578 and 2 to 69 CFU/m3 with (test) and without (control) faucet events, respectively (Fig. 3). Dispersion captured at the first faucet event (t = 0 min) when the faucets were turned on (test) was 1 log10 higher than the same recorded in the case of the control experiment (without faucet event). Heterotrophic organisms captured from the air steadily declined between the time points 0, 30, 60, 90, and 120 min, with and without faucet events. Particle concentrations in the air, with and without faucet events, were found to be consistent during the day of the experiment. However, compared across the experiments, no correlation could be established (Fig. S1).
FIG 3.
Heterotrophic plate counts as measured by impaction air sampling without faucet event (Control) and with faucet events (Test).
Dispersion in the presence of mannequin hands.
When mannequin hands, which created a barrier for water flow onto the drain, were positioned under the faucet and the sink drain was inoculated with ∼1010 CFU GFP-E. coli, 10-fold-lower dispersion was captured on settle plates. No dispersion in air sampling using the impaction or filtration methods was observed. Dispersion pattern and load on settle plates varied considerably across the three sinks and experiments with the mannequin hands in place.
DISCUSSION
The objective of the present study was to characterize the mechanism of bacterial dispersion from handwashing sinks, using a GFP plasmid-containing E. coli strain as a surrogate for multidrug-resistant Enterobacteriaceae. Very few studies have investigated the dispersion from sinks using methods to sample aerosol-associated microorganisms (23, 36, 40, 41); however, several studies have drawn subjective interpretations about aerosol-mediated transmission from contaminated sinks (3, 6, 10, 13, 21, 37). Doring et al. (41) used an impaction method to examine P. aeruginosa dispersion from the sink bowl surface during faucet use. P. aeruginosa was detected 15 cm from the sink drain when counts in the sink drains exceeded 105 CFU/ml (41). Kramer et al. detected 439 CFU/m3 P. aeruginosa in the air sampled 10 cm above the sink drain, with 105 CFU/ml bacteria in the “sink fluid” (P-trap water) (40). In these studies, dispersion was not measured without faucet usage (control samples). De Geyter et al. used an MAS-100 air sampler to measure carbapenem-resistant Enterobacteriaceae (CRE) dispersal from contaminated sinks, with and without faucet usage. Several species in the family Enterobacteriaceae were detected during faucet usage, but results for control samples (without faucet usage) were not provided (23). Fusch et al., in contrast, sampled air around sinks with and without faucet events (control) and did not detect P. aeruginosa in air samples collected without running water (36). We had previously provided a quantitative assessment of dispersal as a function of faucet usage and reported that GFP-E. coli could be dispersed up to 30 in. beyond the sink drain during faucet usage (34). However, dispersion was assessed using a gravity method, and bioaerosol production was not evaluated.
The air sampling methods chosen and tested in the present study were three of the most widely used methods previously reported (23, 35, 36, 40, 42, 43) and were selected to assess bioaerosol production during sink usage. In the present study, GFP-E. coli dispersion was detected during a faucet event but was not detected in the absence of faucet events using either settle plates or impaction and filtration air sampling methods. This finding corroborates those of previous studies (2, 21, 34, 36). It also implies that the shear forces of the faucet water flowing directly onto the sink drain and/or bowl surfaces results in dispersion of bacteria. Detection of dispersed GFP-E. coli during a faucet event and nondetection at subsequent time points (after 30 min) suggested that dispersed cells were associated with larger, heavier droplets that would quickly settle onto surfaces due to gravity, rather than to aerosol-sized particles that remain in the air (44). Dispersion of GFP-E. coli from sinks does not appear to be associated with the production of bioaerosols, that is, of particles smaller than 5 μm (35, 40, 45, 46). Studies that measured air sampling lacked resolution between aerosols and droplets (35, 40, 41). Air was sampled significantly closer (∼4 in.) to the sink drain or impact point of faucet water on the sink and, therefore, samples might have picked up droplets rather than aerosols.
A consistent result from this work that is worth reemphasizing is the finding that for dispersion to occur, the presence of bacteria on the drain and/or bowl surface is necessary (34). When GFP-E. coli was inoculated into a new P-trap, dispersion was not detected using settle plates or air sampling methods. This underscores the fact that as long as the sink drain and bowl remain free of the target organisms (e.g., CPE or other antibiotic-resistant Gammaproteobacteria), dispersion can be controlled. However, under favorable conditions, bacteria can grow or mobilize from the P-trap into the drain piping (tailpiece) and colonize the sink drain surfaces, with the potential for a dispersion event to occur. This further underlines the importance of sanitary hygiene practices and strategic surveillance paired with hand-washing-only use of hand-washing sinks in the patient care environment to reduce the risk of hand-washing sink contamination by multidrug-resistant microorganisms (8). This also emphasizes the necessity to implement stricter measures to prohibit disposal into the sinks of nutrients, body fluids, and anything that could be a nutrient source for maintenance of microorganism biofilms in drains (23). Dispersion from a contaminated sink reservoir can result in transmission to patients, either directly or indirectly mediated through numerous contact surfaces. Herruzo and colleagues demonstrated the potential for microbial transfer from contaminated hands, which continued to disperse microorganisms after more than 10 successive contacts with surfaces (25).
The droplet dispersion load observed on settle plates was similar to and consistent with our previous work (34). Total dispersion measured in corresponding experiments in the previous study was higher, which may be attributed to one or more of the following factors: (i) fewer settle plates were used in the present study (22 versus 90), (ii) a higher water flow rate was used in the present study (8 versus 1.8 to 3.0 liters/min), and (iii) air sampling methods performed in conjunction with the settle plate method may have captured a portion of the dispersed droplets. Settle plates were found to be a reliable method to assess the large-droplet dispersion from sinks. In this study, 22 settle plates (11.24 m2) were used, which accounted for a defined surface area and locations on the sink counter. Dispersion could have been higher in locations on the sink counter other than those chosen in the present study, and the dispersion load recorded in this study may not be the absolute value. Of the three methods investigated for air sampling, impaction and filtration were found to be reliable and consistent. In the same amount of air sampled using impaction and filtration, comparable counts were recorded; however, air sampled using the impinger method was unable to capture the dispersion of GFP-E. coli under similar testing conditions.
Mannequin hands functioned as obstructions to the faucet water stream directly impacting the sink drain, and therefore no dispersion was detected. This rationale behind testing mannequin hands was to simulate hand washing, but in reality, the water would be flowing before, after, and during a hand washing event. In other words, an actual handwashing event is more dynamic than that of static mannequin hands and is likely to have a direct impact on water on the sink drain at least for brief periods when the water is running. There is also the scenario where the sinks and faucets may be used outside of hand washing (e.g., dumping liquid wastes) (5, 8, 23). We think this finding further defines and supports another important dynamic that may minimize dispersion in health care settings (i.e., avoid faucet water flow directly onto drains to minimize dispersion). All of these findings must be taken in the context of an experimental water stream that directly hits the drain, which is outside of the Facility Guidelines Institute (FGI) guidance but is thought to be frequently found in health care sink design. This finding further supports the importance of enforcement of avoidance of this design in a health care sink.
This study has several limitations. First, the dispersion experiments were not performed in a controlled environment. Each dispersion experiment lasted at least 12 h, and it was therefore not possible to maintain precisely the same conditions with regard to airflow velocity, air temperature, relative humidity, and bacterial and/or fungal burden in the laboratory space harboring the sinks. These parameters may have a direct or indirect influence on the dispersion pattern and load recorded across experiments (47). To address this issue, we monitored the heterotrophic plate counts, relative humidity, and particle concentration in the air. Particle counts recorded in the absence of a faucet event (control) were higher than or equal to that in the presence of a faucet event (test). This observation implies that particle concentrations in the air were driven by the relative humidity and/or temperature of the air. This trend was observed in all the experimental methods (drain, P-trap inoculation, and drain colonization) (Fig. S1). In other words, particle counts were largely consistent across the day for a given experiment (control preceding test). Furthermore, the particle counter used in the study could not resolve or measure particles of >5 μm, which defined droplet particles. Another limitation was that air samples were collected at only one location relative to the sink bowl, so it is not possible for this data set to define a “splash zone” pattern without additional measurements collected from various positions and distance from the source of dispersed organisms.
We have provided data to support the position that microorganisms will disperse from contaminated sink bowl and drain surfaces primarily as large droplets that are generated during faucet usage. These droplet-associated organisms remain viable, with the potential to contaminate surfaces surrounding the sink bowl. However, with this simulated sink design and study, it does not appear that dispersion results in the production of bioaerosols with sustained dispersion characteristics that disperse into the patient’s room space.
Based on the findings of this study and other contextual studies, we have some further resolution around the dynamics of bacterial dispersion from sinks. A few recommendations with regard to hospital sink design and usage practices should be strictly implemented and practiced to minimize sink-related transmissions in hospitals, namely, (i) faucet spouts should not flow directly into the drain; (ii) new sink designs should limit counter space surrounding the sink bowl, and usage of counter space around existing sinks to place or store patient care items should be prohibited; (iii) placement of sinks in patient care area should be strategically determined, keeping in mind a 1-m droplet dispersion zone around the sinks; and (iv) patient sink usage should be limited to hand washing, and disposal of nutrients or contaminated wastes into the sinks should be prohibited.
MATERIALS AND METHODS
Sink gallery operation and automation.
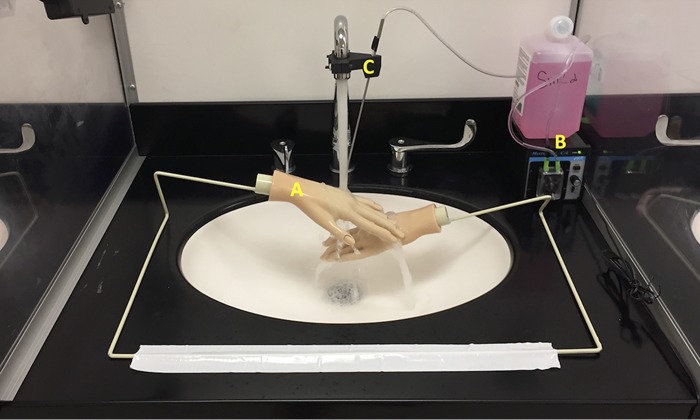
A dedicated sink gallery at the University of Virginia that was described in an earlier study (34) was used in the present study. Sinks were operated in automated mode using a microprocessor. The microprocessor activated the faucets for 30 s each hour via an inline solenoid valve in the hot water supply line (faucet event) and also turned on a peristaltic pump (Masterflex pump HV-77120-42; Cole-Parmer, Vernon Hills, IL) to dispense 1 ml of soap (Kleenex foam skin cleaner; Kimberly-Clark Worldwide Inc., Roswell, GA). A steel tube discharging the soap was held in a clamp attached to the faucet and positioned directly below the discharge of the faucet water. Water flow rate from the faucet was 8 liters/min and the stream of water hit directly onto the drain. In one experiment, mannequin hands (Dianne Practice Hand, D902; Fromm International, Mt. Prospect, IL) attached to a metal rack and positioned between the faucet head and sink drain were used (Fig. 4), preventing the stream of water from directly hitting the drain. To facilitate access to the luminal surface of the drain line, sampling ports were drilled along the length of the tailpiece (between the P-trap and the drain) and the trap arm (between the P-trap and the common line). These holes were fitted with size 00 silicone stoppers (Cole-Parmer, Vernon Hills, IL). Temperature and total and free chlorine residual concentrations of the faucet water at (i) first catch and (ii) after 2 min flushing of faucets were measured at regular intervals using the N,N-diethyl-p-phenylenediamine (DPD) method (Hach Model; Hach, Loveland, CO).
FIG 4.
Test sink, showing mannequin hands positioned directly below the faucet (A), and a peristaltic pump on the right to deliver hand soap (B) via a steel tube attached to the faucet (C).
Inoculation, growth, and establishment of GFP-E. coli colonies in sink P-traps.
A single isolated colony of GFP-E. coli (ATCC 25922GFP) cells grown from −80°C stock was inoculated in 5 ml tryptic soy broth (TSB) containing 100 μg/ml ampicillin (ATCC medium 2855). The method of inoculation varied for each experiment. For P-traps, a 10-ml mid-log-phase culture of GFP-E. coli (109 CFU/ml) was added into the P-trap water (∼150 ml) through the lowermost sampling port on the tailpiece, using a 60-ml syringe attached to silicone tubing (Cole-Parmer, Vernon Hills, IL). The inoculum was mixed with the P-trap water by repeated withdrawal and injection of the inoculum, with precautions taken to avoid unintentional inoculation of the drain (strainer) or sink bowl (bowl). For the drain inoculation, a 10-ml mid-log-phase culture of GFP-E. coli (109 CFU/ml) was evenly applied on the surface of the drain using a sterile pipette. For establishment of GFP-E. coli P-trap biofilm, a 10-ml mid-log-phase culture of GFP-E. coli (109 CFU/ml) was added into an unused P-trap and, following inoculation, 25 ml TSB and 25 ml (×2) 0.85% saline was added on a daily basis through the drain for 7 days to facilitate biofilm growth on the luminal surface of the drain line without additional water every hour. Seven days later, P-trap water and swab samples from the inner surfaces of the drain, tailpiece, and the P-trap were plated on tryptic soy agar (TSA) containing 100 μg/ml ampicillin. TSA plates were incubated overnight at 37°C, and the CFU fluorescing under UV light were enumerated. All preparatory culturing of GFP-E. coli cells took place in a separate room from the sink gallery to avoid unintentional contamination.
Sampling and enumeration of GFP-E. coli.
To monitor the growth of GFP-E. coli within the plumbing, sterile cotton swabs (Covidien, Mansfield, MA) presoaked in 0.85% sterile saline were inserted through sampling ports, and biofilm samples were collected by turning the swab in a circular motion on the inner surface (∼20 cm2). Sample swabs were pulse-vortexed in 3 ml saline, and serial dilutions were plated on TSA. The entire surface area of the drain, the faucet aerator, and sink bowl surfaces were sampled using environmental sponge wipes (sponge-stick with neutralizing buffer, 3M, St. Paul, MN), using overlapping and multidirectional motions. The sponge wipes were expressed in 90 ml of phosphate-buffered saline containing Tween 80 (0.02%) (PBST) using a stomacher (400 Circulator; Seward Ltd., UK). The eluate was concentrated by centrifugation, and plate counts were performed on TSA and R2A agar (Becton, Dickinson and Company, Franklin Lakes, NJ). TSA plates were incubated at 35°C for 48 h, and fluorescent CFU were enumerated; R2A plates were incubated at 25°C for 7 days, and CFU were counted.
Experimental approach for dispersion studies.
Each dispersion experiment comprised a 30-s faucet event repeated three times at 60-min intervals (Fig. 5). Each experiment also comprised a control, (without faucet events) followed by a test (with faucet events). During the control experiments, automation was turned off to withhold the faucet events. Three sinks were tested concurrently but staggered by a few minutes to account for the variability in dispersion driven by faucet water flow rate, airflow dynamics in the room, contact angle, and wastewater drainage/water backup rate.
FIG 5.
Experimental set-up used for different air sampling methods. (a) Impaction, (b) impinger, and (c) gel filtration. Air samples were collected at the initial faucet event (0 min) and every 30 min thereafter. Faucet events (faucet activation) occurred at 0, 60, and 120 min under test conditions. Faucets were not activated in control experiments.
Sampling droplet dispersion.
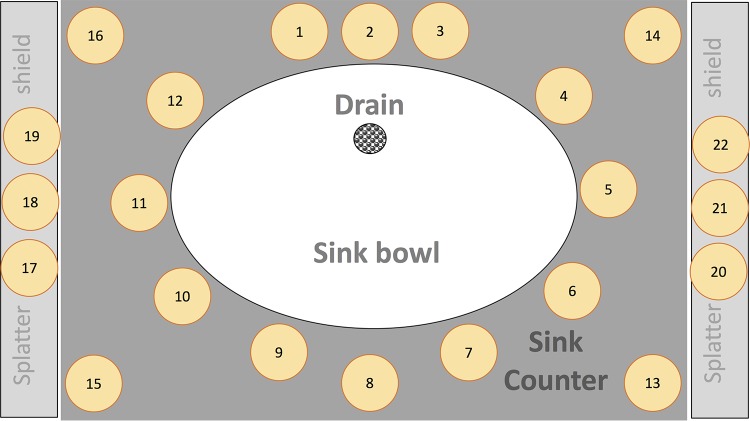
TSA settle plates were used to capture the droplet dispersion. Numbered TSA plates were laid out radially around the sink bowl. A fixed layout and number of settle plates around the sinks was used for each dispersal experiment (Fig. 6). The counter space of each sink was thoroughly disinfected with Caviwipes-1 (Metrex Research, LLC, Orange, CA) prior to each experiment. TSA plates were then positioned on the sink counter surrounding the sink bowl. Additional plates were attached to the faucets, plexiglass partitions, and faucet handles using adhesive tape. Plates were not placed in the sink bowl. TSA plates were also placed >3 m away from the sink as negative controls. Lids of the TSA plates were removed only for the duration of the dispersal experiment. Dispersion per defined area (CFU/cm2) for settle plates was determined by dividing the CFU counts from the TSA plate by the surface area of the plate.
FIG 6.
Graphical representation depicting the layout of the settle plates positioned around the sink and used to capture droplet dispersion.
Air sampling and particle counts.
Each dispersion experiment comprised the collection of air samples at five separate time points, as follows: time t = 0 (first faucet event), t = 30 min after the first faucet event, t = 60 min (second faucet event), t = 90 min, and t = 120 min (third faucet event) (Fig. 5). Individual sink sampling was staggered by a few minutes to provide time for air sampler installation and sampling of each sink. A control experiment period, in which faucets were not activated for the entire 120 min, was also performed for each sink. Three air sampling methods were tested: impaction, impingement, and filtration. For the impaction method, two SAS90 air samplers (Bioscience International, Rockville, MD) containing one TSA plate and one R2A plate each were positioned 12 in. from the sink bowl and set for a 300-liter sample (at 90 liters/min for 200 s) (Fig. 5a). TSA and R2A plates from each air sampling event were incubated as described earlier. A gel filtration device (MD8 portable air sampler; Sartorius AG, Goettingen, Germany) fitted with disposable gelatin filters (Sartorius AG, Goettingen, Germany) was positioned 12 in. from the sink bowl and set for a 300-liter sample (at 100 liters/min for 180 s) (Fig. 5c). Gelatin filters were carefully overlaid on TSA plates, which were as already described. Liquid impingers (Ace Glass Inc. Vineland, NJ) were autoclaved and filled with 20 ml sterile phosphate-buffered saline (PBS) prior to each experiment. Each was connected via a flow meter (Cole-Parmer, Vernon Hills, IL) and vacuum pump (Cole-Parmer, Vernon Hills, IL). The impinger was positioned 12 in. from the sink bowl (set at 6 liters/min for 50 min) to collect a 300-liter air sample (Fig. 5b). In a biological safety cabinet, the liquid from the impinger was transferred to a sterile tube, vortexed, and filtered through 0.22-μm membrane filters (Pall Laboratories, Port Washington, NY), and 5 ml duplicate samples were plated on TSA and R2A plates. Fluorescent CFU were enumerated after TSA plates were incubated at 35°C for 48 h and counted. R2A plates were incubated for 7 days at 25°C and counted. Plates from air impaction samples and samples collected from liquid impingement were shipped via overnight courier to the Centers for Disease Control and Prevention (CDC) laboratories for processing and counting. Gel filtration plates were processed and counted at the University of Virginia. Paired with air sampling, particles of 0.3, 0.5, 0.7, 1.0, 2.0, and 5.0 μm were measured using a particle counter (GT-526; Met One Instruments, Inc., Grants Pass, OR) placed 12 in. from the sink bowl (Fig. 5). With a runtime of 660 s each, 3 successive runs of the particle counter were performed, with the first run coinciding with t = 0. The particle counter also recorded relative humidity and air temperature.
Verification of GFP-E. coli.
Fluorescent colonies on TSA-ampicillin plates were counted under a long-wavelength UV light source. To verify that fluorescent colonies were GFP-E. coli, two fluorescent colonies from each sample were randomly selected and first screened on MacConkey II agar (BD, Franklin Lakes, NJ). The MacConkey II plates were incubated at 35°C for 24 h, and lactose fermenters were isolated on tryptic soy agar with 5% sheep blood (TSA II) (BD, Franklin Lakes, NJ) and incubated under the same conditions. Once colonies were isolated, they were identified using matrix assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS; Bruker, Billerica, MA), or using the Vitek 2 system (bioMérieux, Durham, NC).
GFP-E. coli detection in sink plumbing and faucet water.
Prior to each experiment, 500 ml of first-catch faucet water and a 500-ml sample collected after 2 min of flushing were collected in sterile bottles containing sodium thiosulfate (0.18 g/liter) for dechlorination. Aliquots of faucet water samples were filtered through a 0.45-μm membrane filter, placed on TSA, and incubated at 35°C for 48 h. This was done to test for GFP-E. coli in the faucet water supplied to each sink and to ensure that cross contamination of GFP-E. coli had not occurred. First-catch and 2-min flush samples were diluted, plated on R2A, incubated at 25°C for 7 days, and counted. After the experiment was completed, samples were collected and processed to detect GFP-E. coli in the sink plumbing. A 50-ml aliquot of the P-trap water was collected using a 60-ml syringe connected to sterile silicone tubing, samples were thoroughly vortex mixed, and 1-ml and 5-ml aliquots were filtered through 0.22-μm membrane filters and placed on TSA and R2A media, respectively. The sink P-trap and tailpiece were removed from the sink lab units, filled with faucet water, plugged, and shipped by overnight courier to the CDC for analysis. Swab samples from the tailpipe and P-trap and sponge wipes were processed as already described to recover and quantify biofilm organisms. Samples were placed on TSA and R2A media and counted, as described above.
Supplementary Material
ACKNOWLEDGMENTS
This research was performed under a Research Collaboration Agreement between University of Virginia and the CDC. A.J.M. was supported in part by an IPA agreement between the CDC and the University of Virginia School of Medicine (15IPA1508992).
The findings and conclusions in this presentation are those of the author(s) and do not necessarily represent the views of the Centers for Disease Control and Prevention.
We thank Alexander Kallen for helpful discussions; James Matheson, Maria Burgos-Garay, Amanda Lyons, and Terri Davy for assistance with sample processing; and Will Guilford for design of the sink laboratory and automation.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01997-18.
REFERENCES
- 1.Brooke J. 2008. Pathogenic bacteria in sink exit drains. J Hosp Infect 70:198–199. doi: 10.1016/j.jhin.2008.06.017. [DOI] [PubMed] [Google Scholar]
- 2.Hota S, Hirji Z, Stockton K, Lemieux C, Dedier H, Wolfaardt G, Gardam M. 2009. Outbreak of multidrug-resistant Pseudomonas aeruginosa colonization and infection secondary to imperfect intensive care unit room design. Infect Control Hosp Epidemiol 30:25–33. doi: 10.1086/592700. [DOI] [PubMed] [Google Scholar]
- 3.La Forgia C, Franke J, Hacek D, Thomson R, Robicsek A, Peterson L. 2010. Management of a multidrug-resistant Acinetobacter baumannii outbreak in an intensive care unit using novel environmental disinfection: A 38-month report. American J Infection Control 38:259–263. doi: 10.1016/j.ajic.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 4.Breathnach AS, Cubbon MD, Karunaharan RN, Pope CF, Planche TD. 2012. Multidrug-resistant Pseudomonas aeruginosa outbreaks in two hospitals: association with contaminated hospital waste-water systems. J Hosp Infect 82:19–24. doi: 10.1016/j.jhin.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 5.Lowe C, Willey B, O’Shaughnessy A, Lee W, Lum M, Pike K, Larocque C, Dedier H, Dales L, Moore C, McGeer A. 2012. Outbreak of extended-spectrum beta-lactamase-producing Klebsiella oxytoca infections associated with contaminated handwashing sinks. Emerg Infect Dis 18:1242–1247. doi: 10.3201/eid1808.111268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Starlander G, Melhus A. 2012. Minor outbreak of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae in an intensive care unit due to a contaminated sink. J Hosp Infect 82:122–124. doi: 10.1016/j.jhin.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 7.Kotsanas D, Wijesooriya WR, Korman TM, Gillespie EE, Wright L, Snook K, Williams N, Bell JM, Li HY, Stuart RL. 2013. “Down the drain”: carbapenem-resistant bacteria in intensive care unit patients and handwashing sinks. Med J Aust 198:267–269. doi: 10.5694/mja12.11757. [DOI] [PubMed] [Google Scholar]
- 8.Roux D, Aubier B, Cochard H, Quentin R, van der Mee-Marquet N. 2013. Contaminated sinks in intensive care units: an underestimated source of extended-spectrum beta-lactamase-producing Enterobacteriaceae in the patient environment. J Hosp Infect 85:106–111. doi: 10.1016/j.jhin.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 9.Tofteland S, Naseer U, Lislevand J, Sundsfjord A, Samuelsen O. 2013. A long-term low-frequency hospital outbreak of KPC-producing Klebsiella pneumoniae involving intergenus plasmid diffusion and a persisting environmental reservoir. PLoS One 8:e59015. doi: 10.1371/journal.pone.0059015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vergara-López S, Domínguez MC, Conejo MC, Pascual Á, Rodríguez-Baño J. 2013. Wastewater drainage system as an occult reservoir in a protracted clonal outbreak due to metallo-beta-lactamase-producing Klebsiella oxytoca. Clin Microbiol Infect 19:E490–E498. doi: 10.1111/1469-0691.12288. [DOI] [PubMed] [Google Scholar]
- 11.Knoester M, de Boer M, Maarleveld J, Claas E, Bernards A, Jonge E, van Dissel J, Veldkamp K. 2014. An integrated approach to control a prolonged outbreak of multidrug-resistant Pseudomonas aeruginosa in an intensive care unit. Clin Microbiol Infect 20:O207–O215. doi: 10.1111/1469-0691.12372. [DOI] [PubMed] [Google Scholar]
- 12.Wolf I, Bergervoet P, Sebens F, van den Oever H, Savelkoul P, van der Zwet W. 2014. The sink as a correctable source of extended-spectrum beta-lactamase contamination for patients in the intensive care unit. J Hosp Infect 87:126–130. doi: 10.1016/j.jhin.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Leitner E, Zarfel G, Luxner J, Herzog K, Pekard-Amenitsch S, Hoenigl M, Valentin T, Feierl G, Grisold AJ, Hogenauer C, Sill H, Krause R, Zollner-Schwetz I. 2015. Contaminated handwashing sinks as the source of a clonal outbreak of KPC-2-producing Klebsiella oxytoca on a hematology ward. Antimicrob Agents Chemother 59:714–716. doi: 10.1128/AAC.04306-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ambrogi V, Cavalie L, Mantion B, Ghiglia MJ, Cointault O, Dubois D, Prere MF, Levitzki N, Kamar N, Malavaud S. 2016. Transmission of metallo-beta-lactamase-producing Pseudomonas aeruginosa in a nephrology-transplant intensive care unit with potential link to the environment. J Hosp Infect 92:27–29. doi: 10.1016/j.jhin.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 15.Chapuis A, Amoureux L, Bador J, Gavalas A, Siebor E, Chretien ML, Caillot D, Janin M, de Curraize C, Neuwirth C. 2016. Outbreak of extended-spectrum beta-lactamase producing Enterobacter cloacae with high MICs of quaternary ammonium compounds in a hematology ward associated with contaminated sinks. Front Microbiol 7:1070. doi: 10.3389/fmicb.2016.01070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clarivet B, Grau D, Jumas-Bilak E, Jean-Pierre H, Pantel A, Parer S, Lotthe A. 2016. Persisting transmission of carbapenemase-producing Klebsiella pneumoniae due to an environmental reservoir in a university hospital, France, 2012 to 2014. Euro Surveill 21:pii=30213. doi: 10.2807/1560-7917.ES.2016.21.17.30213. [DOI] [PubMed] [Google Scholar]
- 17.Stjärne Aspelund A, Sjöström K, Olsson Liljequist B, Mörgelin M, Melander E, Påhlman LI. 2016. Acetic acid as a decontamination method for sink drains in a nosocomial outbreak of metallo-beta-lactamase-producing Pseudomonas aeruginosa. J Hosp Infect 94:13–20. doi: 10.1016/j.jhin.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 18.Swan JS, Deasy EC, Boyle MA, Russell RJ, O'Donnell MJ, Coleman DC. 2016. Elimination of biofilm and microbial contamination reservoirs in hospital washbasin U-bends by automated cleaning and disinfection with electrochemically activated solutions. J Hosp Infect 94:169–174. doi: 10.1016/j.jhin.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Zhou Z, Hu B, Gao X, Bao R, Chen M, Li H. 2016. Sources of sporadic Pseudomonas aeruginosa colonizations/infections in surgical ICUs: association with contaminated sink trap. J Infect Chemother 22:450–455. doi: 10.1016/j.jiac.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 20.Amoureux L, Riedweg K, Chapuis A, Bador J, Siebor E, Pechinot A, Chretien M, de Curraize C, Neuwirth C. 2017. Nosocomial infections with IMP-19-producing Pseudomonas aeruginosa linked to contaminated sinks, France. Emerg Infect Dis 23:304–307. doi: 10.3201/eid2302.160649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baranovsky S, Jumas-Bilak E, Lotthe A, Marchandin H, Parer S, Hicheri Y, Romano-Bertrand S. 2017. Tracking the spread routes of opportunistic premise plumbing pathogens in a haematology unit with water points-of-use protected by antimicrobial filters. J Hosp Infect 98:52–59. doi: 10.1016/j.jhin.2017.07.028. [DOI] [PubMed] [Google Scholar]
- 22.Bousquet A, van der Mee-Marquet N, Dubost C, Bigaillon C, Larréché S, Bugier S, Surcouf C, Mérat S, Blanchard H, Mérens A. 2017. Outbreak of CTX-M-15–producing Enterobacter cloacae associated with therapeutic beds and syphons in an intensive care unit. Am J Infect Control 45:1160–1164. doi: 10.1016/j.ajic.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 23.De Geyter D, Blommaert L, Verbraeken N, Sevenois M, Huyghens L, Martini H, Covens L, Pierard D, Wybo I. 2017. The sink as a potential source of transmission of carbapenemase-producing Enterobacteriaceae in the intensive care unit. Antimicrob Resist Infect Control 6:24. doi: 10.1186/s13756-017-0182-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lalancette C, Charron D, Laferriere C, Dolce P, Deziel E, Prevost M, Bedard E. 2017. Hospital drains as reservoirs of Pseudomonas aeruginosa: multiple-locus variable-number of tandem repeats analysis genotypes recovered from faucets, sink surfaces and patients. Pathogens 6:E36. doi: 10.3390/pathogens6030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herruzo R, Ruiz G, Vizcaino MJ, Rivas L, Perez-Blanco V, Sanchez M. 2017. Microbial competition in environmental nosocomial reservoirs and diffusion capacity of OXA48-Klebsiella pneumoniae: potential impact on patients and possible control methods. J Prev Med Hyg 58:E34–E41. [PMC free article] [PubMed] [Google Scholar]
- 26.Varin A, Valot B, Cholley P, Morel C, Thouverez M, Hocquet D, Bertrand X. 2017. High prevalence and moderate diversity of Pseudomonas aeruginosa in the U-bends of high-risk units in hospital. Int J Hyg Environ Health 220:880–885. doi: 10.1016/j.ijheh.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 27.Deasy EC, Moloney EM, Boyle MA, Swan JS, Geoghegan DA, Brennan GI, Fleming TE, O’Donnell MJ, Coleman DC. 2018. Minimizing microbial contamination risk simultaneously from multiple hospital washbasins by automated cleaning and disinfection of U-bends with electrochemically activated solutions. J Hosp Infect 100:e98–e104. doi: 10.1016/j.jhin.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 28.Dewi R, Tarini A, Sukrama D. 2013. Phenotypic detection of carbapenemase-producing gram-negative bacteria in hospital environment. Abstr ASID Gram Negative “Superbugs” Meeting, Queensland, Australia. [Google Scholar]
- 29.Loveday H, Wilson J, Kerr K, Pitchers R, Walker J, Browne J. 2014. Association between healthcare water systems and Pseudomonas aeruginosa infections: a rapid systematic review. J Hosp Infect 86:7–15. doi: 10.1016/j.jhin.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 30.Bloomfield S, Exner M, Flemming HC, Goroncy-Bermes P, Hartemann P, Heeg P, Ilschner C, Krämer I, Merkens W, Oltmanns P, Rotter M, Rutala WA, Sonntag HG, Trautmann M. 2015. Lesser-known or hidden reservoirs of infection and implications for adequate prevention strategies: Where to look and what to look for. GMS Hyg Infect Control 10:Doc04. doi: 10.3205/dgkh000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guyot A, Turton JF, Garner D. 2013. Outbreak of Stenotrophomonas maltophilia on an intensive care unit. J Hosp Infect 85:303–307. doi: 10.1016/j.jhin.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 32.Parkes LO, Hota SS. 2018. Sink-related outbreaks and mitigation strategies in healthcare facilities. Curr Infect Dis Rep 20:42. doi: 10.1007/s11908-018-0648-3. [DOI] [PubMed] [Google Scholar]
- 33.Gordon A, Mathers A, Cheong E, Gottlieb T, Kotay S, Walker A, Peto T, Crook D, Stoesser N. 2017. The hospital water environment as a reservoir for carbapenem-resistant organisms causing hospital-acquired infections—a systematic review of the literature. Clin Infect Dis 64:1435–1444. doi: 10.1093/cid/cix132. [DOI] [PubMed] [Google Scholar]
- 34.Kotay S, Chai W, Guilford W, Barry K, Mathers AJ. 2017. Spread from the sink to the patient: in situ study using green fluorescent protein (GFP) expressing-Escherichia coli to model bacterial dispersion from hand washing sink trap reservoirs. Appl Environ Microbiol 83:e03327-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneider H, Geginat G, Hogardt M, Kramer A, Durken M, Schroten H, Tenenbaum T. 2012. Pseudomonas aeruginosa outbreak in a pediatric oncology care unit caused by an errant water jet into contaminated siphons. Pediatr Infect Dis J 31:648–650. doi: 10.1097/INF.0b013e31824d1a11. [DOI] [PubMed] [Google Scholar]
- 36.Fusch C, Pogorzelski D, Main C, Meyer C, el Helou S, Mertz D. 2015. Self-disinfecting sink drains reduce the Pseudomonas aeruginosa bioburden in a neonatal intensive care unit. Acta Paediatr 104:e344–e349. doi: 10.1111/apa.13005. [DOI] [PubMed] [Google Scholar]
- 37.Pitten FA, Panzig B, Schröder G, Tietze K, Kramer A. 2001. Transmission of a multiresistant Pseudomonas aeruginosa strain at a German university hospital. J Hospital Infection 47:125–130. doi: 10.1053/jhin.2000.0880. [DOI] [PubMed] [Google Scholar]
- 38.Siegel JD, Rhinehart E, Jackson M, Chiarello L. 2007. 2007 guideline for isolation precautions: preventing transmission of infectious agents in health care settings. Am J Infect Control 35:S65–164. doi: 10.1016/j.ajic.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.WHO. 2014. Infection prevention and control of epidemic- and pandemic-prone acute respiratory infections in health care: WHO guidelines. World Health Organization, Geneva, Switzerland: http://www.who.int/csr/bioriskreduction/infection_control/publication/en/. [PubMed] [Google Scholar]
- 40.Kramer A, Daeschlein G, Niesytto C, Sissoko B, Sütterlin R, Blaschke M, Fusch C. 2005. Contamination of sinks and emission of nosocomial gramnegative pathogens in a NICU—outing of a reservoir as risk factor for nosocomial colonization and infection. Umweltmed Forsch Prax 10. [Google Scholar]
- 41.Doring G, Horz M, Ortelt J, Grupp H, Wolz C. 1993. Molecular epidemiology of Pseudomonas aeruginosa in an intensive care unit. Epidemiol Infect 110:427–436. doi: 10.1017/S0950268800050858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zemouri C, de Soet H, Crielaard W, Laheij A. 2017. A scoping review on bio-aerosols in healthcare and the dental environment. PLoS One 12:e0178007. doi: 10.1371/journal.pone.0178007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haig C, Mackay W, Walker J, Williams C. 2016. Bioaerosol sampling: sampling mechanisms, bioefficiency and field studies. J Hosp Infect 93:242–255. doi: 10.1016/j.jhin.2016.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morawska L. 2006. Droplet fate in indoor environments, or can we prevent the spread of infection? Indoor Air 16:335–347. doi: 10.1111/j.1600-0668.2006.00432.x. [DOI] [PubMed] [Google Scholar]
- 45.Eames I, Shoaib D, Klettner C, Taban V. 2009. Movement of airborne contaminants in a hospital isolation room. J R Soc Interface 6:S757–S766. doi: 10.1098/rsif.2009.0319.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stetzenbach L. 2002. Introduction to aerobiology In Hurst C, Crawford R, Knudsen G, McInerney M, Stetzenbach L (ed), Manual of environmental microbiology, 2nd ed ASM Press, Washington, DC. [Google Scholar]
- 47.Mohr A. 2002. Fate and transport of microorganisms in the air, p 827–838. In Hurst C, Crawford R, Knudsen G, McInerney M, Stetzenbach L (ed), Manual of environmental microbiology, 2nd ed ASM Press, Washington, DC. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.