Understanding factors that control microbial communities is essential for designing and supporting microbiome-based agriculture. In this study, we used a grafted tomato system to study the effect of rootstock genotypes and grafting on bacterial communities colonizing the endosphere and rhizosphere. To compare the bacterial communities in control treatments (nongrafted and self-grafted plants) with the hybrid rootstocks used by farmers, we evaluated the effect of rootstocks on overall bacterial diversity and composition. These findings indicate the potential for using plant genotype to indirectly select bacterial taxa. In addition, we identify taxa responsive to each rootstock treatment, which may represent candidate taxa useful for biocontrol and in biofertilizers.
KEYWORDS: BHN589, Maxifort, Solanum lycopersicum, endosphere, grafting, hybrid, microbiome, rhizosphere, rootstocks
ABSTRACT
Root-associated microbes are critical to plant health and performance, although understanding of the factors that structure these microbial communities and the theory to predict microbial assemblages are still limited. Here, we use a grafted tomato system to study the effects of rootstock genotypes and grafting in endosphere and rhizosphere microbiomes that were evaluated by sequencing 16S rRNA. We compared the microbiomes of nongrafted tomato cultivar BHN589, self-grafted BHN589, and BHN589 grafted to Maxifort or RST-04-106 hybrid rootstocks. Operational taxonomic unit (OTU)-based bacterial diversity was greater in Maxifort compared to the nongrafted control, whereas bacterial diversity in the controls (self-grafted and nongrafted) and the other rootstock (RST-04-106) was similar. Grafting itself did not affect bacterial diversity; diversity in the self-graft was similar to that of the nongraft. Bacterial diversity was higher in the rhizosphere than in the endosphere for all treatments. However, despite the lower overall diversity, there was a greater number of differentially abundant OTUs (DAOTUs) in the endosphere, with the greatest number of DAOTUs associated with Maxifort. In a permutational multivariate analysis of variance (PERMANOVA), there was evidence for an effect of rootstock genotype on bacterial communities. The endosphere-rhizosphere compartment and study site explained a high percentage of the differences among bacterial communities. Further analyses identified OTUs responsive to rootstock genotypes in both the endosphere and rhizosphere. Our findings highlight the effects of rootstocks on bacterial diversity and composition. The influence of rootstock and plant compartment on microbial communities indicates opportunities for the development of designer communities and microbiome-based breeding to improve future crop production.
IMPORTANCE Understanding factors that control microbial communities is essential for designing and supporting microbiome-based agriculture. In this study, we used a grafted tomato system to study the effect of rootstock genotypes and grafting on bacterial communities colonizing the endosphere and rhizosphere. To compare the bacterial communities in control treatments (nongrafted and self-grafted plants) with the hybrid rootstocks used by farmers, we evaluated the effect of rootstocks on overall bacterial diversity and composition. These findings indicate the potential for using plant genotype to indirectly select bacterial taxa. In addition, we identify taxa responsive to each rootstock treatment, which may represent candidate taxa useful for biocontrol and in biofertilizers.
INTRODUCTION
The root-associated microbiome, or “rhizobiome,” is essential for plant health and performance (1, 2). Some microbes in the rhizobiome are especially important for plants during harsh and unfavorable growing conditions (3). Microbe-mediated nutrient uptake, disease resistance, and stress tolerance (1, 4, 5) are some examples of microbial functions important to agriculture and, more generally, to host biology. While the importance of microbes in provisioning ecosystem services is clear, greater understanding of factors that control the microbial community and microbial processes is needed to achieve the potential of microbial management in agricultural systems. Both biotic and abiotic factors and their interactions may control microbial community composition (6–10), but our ability to predict the key factors and their magnitude of influence on microbial community structure and function is limited. Understanding the factors that control plant-associated microbiomes could offer a novel opportunity to engineer microbiomes to support microbiome-based agriculture.
Plant species and genotypes influence root microbiomes, regardless of soil type or geographic location (11–17). However, the effect of plant species or genotype on the microbiome is generally smaller than the effect of environmental and edaphic factors (7, 12, 18, 19). Despite the relatively small magnitude of effect, plant genotypic effects on microbiome composition are particularly important because they indicate the potential for harvesting benefits from microbes indirectly, through the choice of crop genotypes. Plant genotypes with desired phenotypes can be used as an engineering tool to select candidate taxa (20, 21). For example, microbial assemblages that are directly associated with high-yield genotypes may represent candidate taxa for designing microbial consortia with a potential to serve as biofertilizers or biocontrol (22), while exploring the host genes associated with microbial selection may provide insight to support microbiome-focused crop breeding.
Compared to aerial plant surfaces, roots are particularly important for microbe-microbe and host-microbe interactions (12). The endosphere and rhizosphere are active in the exchange of nutrients and microbes between soil and plant (11, 15, 17, 19, 23, 24). Microbial diversity and community composition often differ between the rhizosphere and endosphere (11, 15, 17, 19, 25). Models of microbial acquisition in plant roots posit a gradual enrichment of microbial communities in the rhizosphere and rhizoplane, followed by pronounced exclusions of microbial communities in the endosphere (25, 26). It seems that the endosphere is more directly under host control and serves as a stronger filter than the rhizosphere, where control is less direct and perhaps driven by chemical cues or abiotic filters, along with trophic and nontrophic microbial interactions (25).
Evaluation of microbial diversity in designed experiments provides an avenue to generate hypotheses about the mechanisms of treatment effects on host phenotype and performance (27). Under the insurance hypothesis, systems with higher species diversity may be more likely to maintain community functions during perturbation, for example (28). Additionally, greater microbial diversity may increase host performance and system robustness. For instance, a host plant lacking resistance to a pathogen under sterile experimental conditions gained the disease-resistant phenotype through the introduction of a phyllosphere microbiome (29), highlighting the potential importance of microbial diversity for host phenotypes/traits (21, 30). In some cases, microbiome composition varied with the level of pathogen infection (30, 31), and particular microbial taxa may be “driver microbes” or “passenger microbes,” as reflected by the magnitude of their effect on host performance (32). In agricultural systems, an increase in overall diversity of microbial populations has been reported in disease-suppressive soils, even though few key microbes have been deemed crucial in regulating suppression (33, 34). Greater diversity, along with key functional species, might drive some ecological functions, and failure to select for microbial associates that are antagonistic to pathogens could result in lower host fitness (35, 36). Together, these findings indicate that microbiomes can modulate host traits and disease phenotypes, and they suggest the potential to design agricultural practices to manipulate microbiomes in microbiome-focused crop improvement programs (20, 21).
Our project addresses central questions about how grafting and the choice of tomato genotype may affect the tomato rhizobiome. Although tomato grafting is a fairly new practice among farmers in the United States, it is an ancient propagation practice in agriculture that is commonly used in vegetable production. In the case of tomato, interspecific rootstocks, where rootstock and scion belong to different species of Solanum, generally have a rootstock resistant to soilborne diseases (e.g., Fusarium wilt, Verticillium wilt, bacterial wilt, and root-knot nematodes) grafted with a scion that produces higher-quality fruit (37–39). Soilborne pathogens are difficult to manage and can result in up to 90% yield loss (40). In addition to the effectiveness of grafted plants in managing soilborne diseases, grafted plants are often more vigorous and more efficient in nutrient uptake and resource utilization, as well as resistant to abiotic stresses (41–43). Thus, plants grafted with effective and vigorous rootstocks often provide higher fruit yield and plant biomass (44, 45). These phenotypic traits of grafted plants appear to be influenced in part by modification of the scion through migration of molecules such as proteins, mRNA, and small RNA from rootstocks to scion (46, 47) or by epigenetic modifications (48, 49). Restriction of pathogen migration in resistant rootstocks due to pit membrane architecture (50) and the mobility of nucleic acids and proteins (46, 47) are among the many physiological and molecular responses of grafted plants during infection.
In our study, we address how hybrid rootstocks affect the tomato rhizobiome, and we directly assess the role of rootstock genotypes and grafting (or artificial selection/breeding) on the rhizobiome. Given the great potential of microbes in plant health and production and the economic value of grafted tomato, understanding how grafting and rootstock treatments modulate root-associated microbial diversity and community composition will lay the groundwork for future microbiome-based systems with rootstocks supporting higher plant biomass and fruit yield. Furthermore, identification of microbes associated with desirable host traits is a first step to select candidates for synthetic microbiomes. We evaluated the effects of rootstocks on tomato rhizobiomes under Midwestern (Kansas, USA) growing conditions. We characterized and compared the composition and diversity of bacterial communities associated with tomato rootstocks by sampling the rhizobiome—microbes within roots (in the endosphere) and surrounding the root (in the rhizosphere). Based on the previous studies of microbial communities in related systems that we review above, our expectation was that (i) effective rootstocks, defined in terms of fruit yield and plant biomass, will be associated with higher microbial diversity, (ii) differences in microbiome composition will be greater in the endosphere than in the rhizosphere, (iii) the number of taxa responsive to rootstock genotypes (operational taxonomic units [OTUs] whose proportion is different than in the nongrafted and self-grafted controls) will correlate with rootstock performance, and (iv) responsive taxa will be more frequent in the endosphere than in the rhizosphere. In total, we address the effects of (i) an agricultural practice (grafting), (ii) rootstock genotypes, (iii) farm sites, and (iv) root compartment (rhizosphere or endosphere) on microbial community composition and diversity.
(This article was submitted to an online preprint archive [51].)
RESULTS
General bacterial community data description.
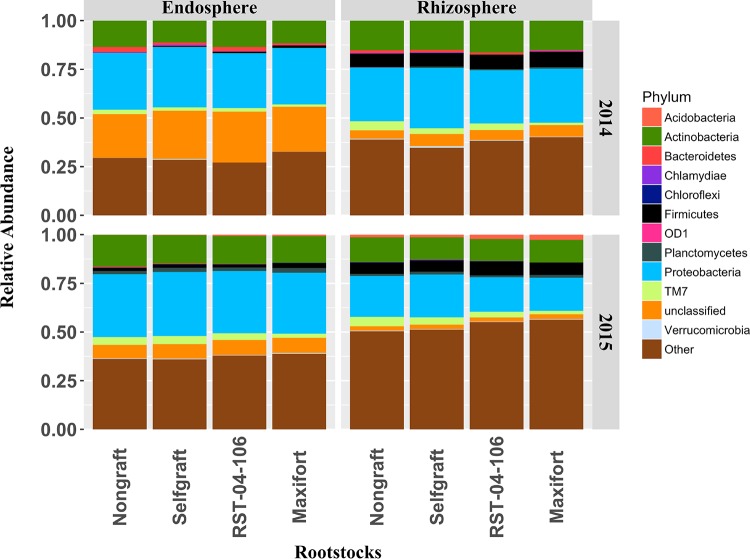
The final curated data set consisted of 1,282,843 sequences from the endosphere- and rhizosphere-associated bacterial communities in tomato. The bacterial communities were dominated by Proteobacteria (37.9%), which was the most abundant phylum across all the rootstock treatments and in both the rhizosphere and endosphere (Fig. 1 and Fig. S1 and Data Set S4 in the supplemental material). Approximately 14.5% of the sequences in the data set were unclassified at the phylum level. Among the rootstocks, the self-graft had the highest percentage of Proteobacteria (39.0%). Actinobacteria (16.9%) and Firmicutes (8.9%) were the other dominant phyla observed in the overall community. Firmicutes and Planctomycetes were enriched in the hybrid rootstocks compared to in the nongraft and self-graft, whereas the Bacteroidetes were depleted in the hybrid rootstocks. At the class level, the communities were largely dominated by Alphaproteobacteria (18.5%). Bacteria of class Sphingobacteria were found at a lower percentage in the hybrid rootstocks than in the self-grafts and nongrafts. Bacteria of class Gammaproteobacteria (6.9%) were less dominant in the Maxifort rootstock than in other rootstocks. Rhizobiales (13.8%) was the most dominant bacterial order in the overall community, including all rootstock treatments and both compartments. Bacillales species were more frequent in Maxifort (7.2%) than in RST-04-106 (6.8%), self-graft (6.4%), and nongraft (6.2%). At the family level, Planctomycetaceae species were more frequent in Maxifort and RST-04-106 (7.4%) than in nongrafted (6.9%) and self-grafted (7.0%) treatments. Taxa in the order Myxococcales were more frequent in Maxifort (2.6%) than in the other rootstocks. Analysis at the genus level revealed Pasturia spp. as the most dominant taxa in the overall community, as well as in Maxifort. Comparison of community profiles of bacteria between the endosphere and rhizosphere showed that Proteobacteria, Actinobacteria, and Bacteroidetes species were more abundant in the endosphere than in the rhizosphere, whereas Planctomycetes, Firmicutes, and TM7 bacteria were more abundant in the rhizosphere.
FIG 1.
Profile of bacterial communities from tomato at the phylum level. The relative abundances of bacterial phyla recovered from the endosphere and rhizosphere from two hybrid rootstocks (RST-04-106 and Maxifort) and nongrafted and self-grafted controls (BHN589) for the years 2014 and 2015. The colored area of each bar represents the relative abundance of the corresponding phylum. The horizontal facet in the graph represents root compartments, and the vertical facet divides the plot by year. OTUs with relative sequence abundance <1% are summed as “other.”
Effects of grafting and rootstock on α diversity.
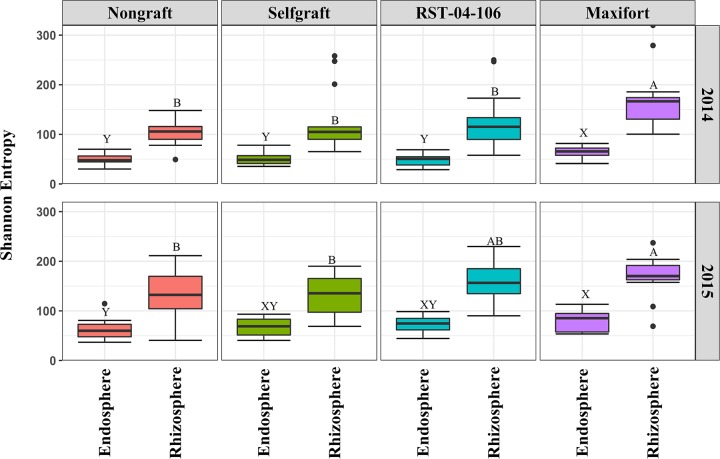
There was strong evidence for a rootstock treatment effect on the diversity of bacteria in the endosphere and rhizosphere compartments of tomato plants (P < 0.01). Among the four treatments in 2014, plants grafted with the Maxifort rootstock had the highest diversity in both the endosphere and the rhizosphere compared to self-grafted and nongrafted plants (P < 0.05; Fig. 2). In 2015, we observed similar results, with the highest diversity in the Maxifort-grafted plants for both the compartments; however, there was less evidence (P = 0.1) for a difference in the endosphere between the self-grafted and Maxifort-grafted treatments (Fig. 2). The effect of RST-04-106 on bacterial diversity was similar to that of the nongrafted and self-grafted treatments (P > 0.05). Additionally, there was no evidence for a difference between the self-grafted and the nongrafted plants in the endosphere (P = 0.9) or in the rhizosphere (P = 0.2). When the effect of rootstock treatments (a fixed effect) on bacterial diversity was evaluated for each compartment, study site, and year separately in a factorial design, the effects of rootstocks varied (see Table S2 in the supplemental material), although there was never strong evidence for an interaction between rootstock treatments and study sites (P > 0.05).
FIG 2.
Diversity of bacterial communities associated with tomato. Bacterial diversity was evaluated for the endosphere and rhizosphere from two hybrid rootstocks (RST-04-106 and Maxifort) and nongrafted and self-grafted controls (BHN589) in a mixed-model analysis of variance (ANOVA). Rootstock treatments were compared with the study site as a random factor within each year. The plot is horizontally faceted by rootstock treatments, and the vertical facets represent sample years. Shannon entropy (a measure of α-diversity) was higher for Maxifort than that of the nongrafted controls (P < 0.05). There was strong evidence that Shannon entropy differed for treatment combinations with different letters (P < 0.05).
Effects of grafting and rootstock on bacterial composition.
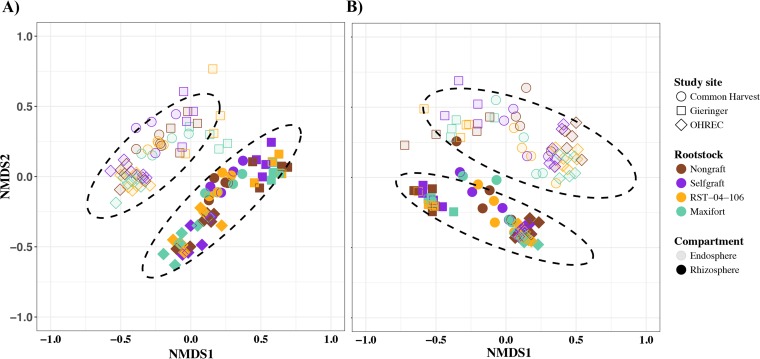
Permutational analysis of variance (PERMANOVA) based on the Bray-Curtis dissimilarity matrix identified rootstock treatment, study site, and endosphere-rhizosphere compartments as factors explaining the variation in the bacterial community. The percentage of variation explained by the rootstock treatment was small (3%) but consistent across the 2 years (PERMANOVA in year 2014: df = 3, Fmodel = 2.0, R2 = 0.03, P = 0.02; PERMANOVA in year 2015: df = 3, Fmodel = 1.9, R2 = 0.03, P = 0.01). Visualization of the distance matrix for samples using nonmetric multidimensional scaling (NMDS) ordination plots (Fig. 3) indicated overlapping confidence regions for the centroids for the rootstock genotypes, whereas compartment and study site were the primary factors partitioning communities in both years. Endosphere-rhizosphere compartments (PERMANOVA in year 2014: df = 1, Fmodel = 58, R2 = 0.28, P = 0.001; PERMANOVA in year 2015: df = 1, Fmodel = 53, R2 = 0.25, P = 0.001) and study site (PERMANOVA in year 2014: df = 2, Fmodel = 18, R2 = 0.17, P = 0.001; PERMANOVA in year 2015: df = 2, Fmodel = 21, R2 = 0.20, P = 0.001) explained about 45% of the variation in the bacterial community.
FIG 3.
Nonmetric multidimensional scaling (NMDS) ordination of samples from tomato rootstock treatments. NMDS ordination of samples was based on the Bray-Curtis dissimilarity matrix of OTUs from bacterial communities inhabiting the endosphere and rhizosphere compartments in the years 2014 (A) and 2015 (B). Color indicates rootstock treatment (two hybrid rootstocks (RST-04-106 and Maxifort) and nongrafted and self-grafted controls (BHN589)), shape represents study site, and solid and lighter fill colors represent the rhizosphere and endosphere compartments, respectively. Ellipses indicate 95% confidence regions around the centroids of the endosphere and rhizosphere samples.
Comparison of DAOTUs.
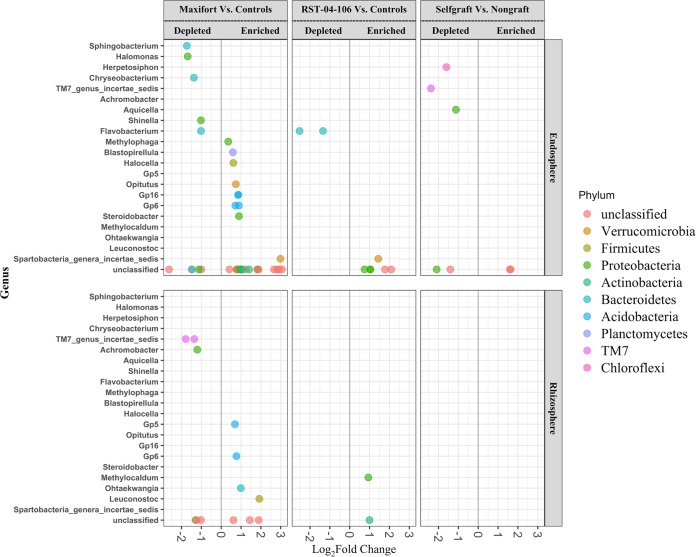
Bacterial diversity and community composition in the tomato endosphere and rhizosphere differed among rootstock treatments. To identify taxa that responded to rootstocks, we used a differential abundance test. Although we consistently observed higher α diversity in the rhizosphere than in the endosphere, the total number of differentially abundant OTUs (DAOTUs), either enriched or depleted, was greater in the endosphere (n = 56) than in the rhizosphere (n = 15; Fig. 4). The analysis of contrasts designed to compare OTU proportion to controls (nongrafts and self-grafts) found a higher number of responsive taxa in Maxifort in both the endosphere (n = 41) and rhizosphere (n = 13). Enriched OTUs in the Maxifort rhizosphere included OTUs assigned to the following taxa: Gp5 and Gp6 within the phylum Actinobacteria, Ohtaekwangnia spp. in Bacteroidetes, Leuconostoc spp. in Firmicutes, and three unclassified OTUs (Fig. 4). In contrast, depleted OTUs in the rhizosphere included OTUs assigned to the following taxa: two in the TM7 group, two in Proteobacteria, and two that remained unclassified. OTUs such as Methylophaga spp. (Proteobacteria), Blastopirellula spp. (Planctomycetes), Halocella spp. (Firmicutes), Opitutus spp. (Verrucomicrobia), Gp16 (Acidobacteria), Gp6 (Acidobacteria), and Steroidobacter spp. (Proteobacteria) were enriched in the Maxifort endosphere. In contrast, OTUs with taxonomic affinities to Sphingobacterium spp. (Bacteroidetes), Halomonas spp. (Proteobacteria), Chryseobacterium spp. (Bacteroidetes), Shigella spp. (Proteobacteria), and Flavobacterium spp. (Bacteroidetes) were depleted in the Maxifort endosphere. Abundances of eight OTUs changed significantly in the endosphere community of RST-04-106, where six OTUs were enriched and two were depleted, compared to the controls. OTUs enriched in the RST-04-106 endosphere included Spartobacteria spp. (Verrucomicrobia), and five OTUs unclassified at the genus level, whereas the proportion of Flavobacterium spp. (Bacteroidetes) was significantly reduced. The RST-04-106 rhizosphere community was resilient to the grafting treatment; only two OTUs were enriched compared to the controls. Enriched OTUs in the rhizosphere belonged to Methylocaldum spp. (Proteobacteria) and an unclassified OTU in phylum Actinobacteria. The bacterial communities in the self-grafted and nongrafted controls were surprisingly similar in the rhizosphere—none of the OTU abundances changed significantly. Although no effect was observed in the rhizosphere, an effect of grafting was observed in the endosphere community, where the proportions of six OTUs changed compared to those in the nongrafted control, out of which one (unclassified) was enriched and five were depleted (Fig. 4).
FIG 4.
Comparisons of differentially abundant OTUs. Differentially abundant OTUs (DAOTUs) were evaluated across tomato rootstocks for the endosphere and rhizosphere, with OTU counts from self-grafts and nongrafts (BHN589) as controls. All of the tests were adjusted to control for the false-discovery rate (FDR) (P value < 0.05) using the Benjamini-Hochberg method. Each point represents an OTU labeled at the genus level and colored based on the associated phylum. The position along the x axis represents the frequency fold change contrasted with that of the two controls (self-graft and nongraft [BHN589]) for the two hybrid rootstocks (Maxifort and RST-04-106) and a self-graft versus nongraft (BHN589) contrast.
DISCUSSION
Rootstocks affected both microbial diversity and community composition. We observed higher bacterial diversity in the endosphere and rhizosphere of the high-yielding Maxifort rootstock, compared to nongrafted and self-grafted controls of BHN589. Previous studies have reported host genotypic effects on microbiome composition, but the host effect has generally been smaller than the effect of environmental and edaphic factors (7, 52). Consistent with these published results, tomato rootstocks explained roughly 3% of the compositional variation in the bacterial community, whereas there was strong evidence that the compartment (endosphere versus rhizosphere) and the study site explained a major portion of the variation (25% to 28% and 17% to 20%, respectively). A study evaluating six different rice genotypes found that only 1.5% to 2.5% of community composition variation was attributable to rice genotype (19). However, it is important to keep in mind that variation in taxonomic composition may not be strongly reflective of variation in function; plant genotypes may have substantial effects specifically on microbial taxa that are particularly important to plant health.
Differential abundance tests of OTUs across treatments found a greater number of responsive taxa in Maxifort than in the other rootstocks. Interestingly, the endosphere had a greater number of responsive OTUs than the rhizosphere, while α diversity was higher in the rhizosphere, consistent with the results from grafting studies with grapevine rootstocks (52). We expected that hybrid rootstocks would have higher diversity as well as more DAOTUs than the control treatments, and this was the case for Maxifort compared to the nongrafted control (BHN589) but not for the other hybrid rootstock, RST-04-106. Note that the RST-04-106 rootstock also had lower performance in terms of yield and biomass in our field trial (53), consistent with the idea that bacterial diversity and host performance may be associated with graft performance.
Earlier agricultural microbiome studies have primarily focused on how a particular management strategy—such as organic versus conventional farming (54, 55) or tillage practices (56)—influences soil microbial communities and rarely incorporated host genotypic effects and their interaction with agricultural practices. Using tomato as a grafting model, our study provides a new perspective on the effects of host genotype on microbial communities. Our results for microbial diversity and composition suggest that grafting with specific rootstocks influences microbiome assembly, as well as yield and biomass. Our studies included only two hybrid rootstocks and one scion (BHN589), so research that includes more rootstocks and scions along with hybrid rootstock specific self-grafted and nongrafted controls would help to generalize these results. Given the economic importance of grafted tomatoes, and the need to develop sustainable production systems, our findings indicate new opportunities for improving microbiome-based practices in agriculture.
The rootstocks included in our study represent different genetic backgrounds and are specifically bred to provide resistance to multiple soilborne diseases (Table S1 in the supplemental material). Variation in a plant’s genetic background influences the acquisition of root-associated microbial communities (57), as shown in rice (19), maize (16), and Arabidopsis (11, 15, 58). Physiologically, hosts vary in their root exudates and rhizodeposits (59–61), thereby creating host-specific cues to select microbial associates from the surrounding soil (25, 62, 63). Root exudates and rhizodeposits not only contain carbon and other nutrients that support the belowground food web, but also contain chemicals, such as cytokinins (64), phytotoxins, antibiotics, and hormones (65), that are key to supporting some microbial assemblages while deterring others (66, 67). These findings indicate an active and host-specific microbiome filter, although the extent of such selection may differ across plant types and studies (15, 17–19, 21, 23). Usually, when plant types are closely related, their microbial communities are more similar (25), and smaller differences are observed in selection of microbial communities by plant type (68). Besides the genotypic effects, we observed some effects of grafting itself in the microbial communities, which might be consequences of wounding stress. However, field studies suggest that the use of self-grafted plants would not be practical for disease management (39). Our current studies focused on options that were practical for farmers, but future studies that include nongrafted and self-grafted treatments for a range of rootstocks would be helpful to better understand how rootstock and scion interact to structure the rhizobiome.
The effect of tomato rootstocks on rhizobiome diversity and composition depends on which root compartment is being considered. The tomato rhizosphere supported a more diverse microbial community than the endosphere. These results were consistent with other studies of plant-based selection of microbial communities, and with a proposed model of microbial acquisition (25), where the endosphere microbial community is under more direct host control (69, 70) and is more strongly filtered than the rhizosphere, where control is less direct and may be driven by chemical cues or intermicrobe interactions (71). These results confirm the effect of agricultural practices on the acquisition of endosphere and rhizosphere microbiomes and are particularly interesting from a practical standpoint. Microbes in the rhizosphere microbiome could represent candidate taxa for biofertilizers or plant supplements, and understanding the mechanisms underlying the host-based selection of endosphere microbiome members could guide microbiome-based breeding programs. While research to support such breeding programs is still limited, plant loci identified for host genotype-dependent structuring of microbial community via a genome-wide association study (GWAS) (72) are a promising first step.
Root architecture and anatomy change to mediate plant responses to biotic and abiotic stresses (73, 74), and these changes can vary within and between plant species and genotypes (71, 75). If we learn the extent to which host genotypes affect root architecture and physiology, such as exudation, then we may also understand the potential mechanisms by which host genotypes control microbiomes via metabolites and exudates (62, 63). Genotypic variation in root traits and functions selects for different microbial communities by modulating the quality as well as the quantity of root exudates and by modifying the physical and chemical properties of the surrounding soil environment (76). For instance, a greater root mass with more abundant fine roots was reported for Maxifort, along with greater arbuscular mycorrhizal colonization than that of the self-graft (77). We did not evaluate root architecture in our experiments, but future studies of microbially diverse populations for root types (e.g., primary roots, secondary roots, and root hairs) and the effects of root exudates will help to clarify the process of microbial acquisition by rootstock genotypes.
Previous studies in our experimental system (53) and other grafted tomato systems (45, 78, 79) have indicated that tomatoes with effective rootstocks gain greater above-ground biomass and have higher overall photosynthetic activity, a result often attributed to the root system. A particular pairing of scion and rootstock may define the root nutrient and metabolite profile (42) through absorption and shoot feedbacks. This aboveground gain increases the total leaf area, likely resulting in a greater supply of photosynthates belowground and modifying the nutrient profile in root systems. Translocation of fixed carbon from shoots to roots may allow plant roots to actively recruit and sustain diverse microbiomes (80). It is generally accepted that 30% to 60% of plant photosynthate is transported belowground, much of which (40% to 90%) is excreted into the rhizosphere, supporting diverse root-associated microorganisms (81, 82). Thus, the higher microbial diversity observed in the rhizosphere of plants grafted with the Maxifort rootstock may be a result of its vigorous root system (77), resulting in greater shoot biomass as well. Grafting may provide a boost in the continuous feedback between the aboveground and belowground compartments and consequently greater investment of the photosynthetic capital in recruiting diverse microbial communities.
Using differential abundance tests, we identified OTUs that were sensitive to the rootstock treatments. These OTUs were either enriched or depleted in contrast to those in the controls, and potentially represent taxa that were under direct selection by rootstocks and their chemical cues. We expected that there would be a higher number/percentage of responsive OTUs (or DAOTUs) in the endosphere than in the rhizosphere, and our results support this, corroborating results from other studies (19, 52). More DAOTUs in the endosphere relative to those in the rhizosphere might be a consequence of direct host control in the endosphere microbiome (83, 84). In addition to the compartment-specific effect on the number of sensitive taxa, it was interesting to observe the higher number of sensitive OTUs in Maxifort compared to that in the other treatments. The results are consistent with the expectation that enhanced host performance is associated with an increase in responsive taxa. Responsive taxa could be important for host performance, as differentially abundant taxa are often functionally associated with host physiology and immune system (58, 85). However, many of the responsive taxa in this study are unclassified and may be unculturable. Future experiments are needed to evaluate the influence of DAOTUs on host phenotypes.
Taxa that were enriched in Maxifort included representatives of Firmicutes, Verrucomicrobia, Planctomycetes, Proteobacteria, and Acidobacteria. We observed significant depletion of TM7, also known as “Candidatus Saccharibacteria,” in the rhizosphere of Maxifort. TM7 bacteria, previously reported as antagonists to the antibiotic-producing Actinobacteria (86), can also be involved in suppressing host immune systems and causing disease (87, 88). The observed proportion of Actinomycetes was 2-fold higher in both the Maxifort endosphere and rhizosphere compared to that in TM7 (P < 0.001 in both cases; Mann-Whitney-Wilcoxon test). It is possible that the productivity of Maxifort is due, in part, to selection for antibiotic-producing bacteria that are detrimental to pathogenic bacteria. Further studies with taxa identified in this study would be necessary to evaluate this hypothesis, especially in the presence of the relevant pathogens. Note that we did not observe any obvious disease symptoms in our experiments. Under disease pressure, greater effects of rootstock treatments on microbiomes would be likely, and the microbes identified in such scenarios could be useful for disease suppression (89).
The other OTUs enriched for Maxifort in DAOTU analysis included members of the Planctomycetes. Members of this phylum are efficient crossfeeders of exopolysaccharides (EPS), commonly found with nitrogen-fixing bacteria (90) and in environments rich in organic matter and nitrate (91). In addition, some members of the Planctomycetes are highly tolerant of environmental stressors, such as seawater, acidic peat bogs, hot springs, and low temperatures (92–94). Among other enriched phyla were the Proteobacteria, which include many important plant growth-promoting (PGP) organisms, but note that some of the OTUs from Proteobacteria were depleted as well, especially Halomonas spp. and Shinella spp. This analysis also identified some oligotrophic groups in Verrucomicrobia that have been reported in association with pre-agricultural tallgrass prairie soil (95). Past studies have found a decline in Verrucomicrobia with nutrient amendments (96), while they were dominant and functionally active in undisturbed soil with recalcitrant carbon compounds (95, 97). Many of the rhizobiome taxa in our study currently lack information about biological function, pointing out the need for further development of culturing methods, along with experiments to test biological functions, to support the design of synthetic communities in microbiome-based crop production.
Conclusions.
Multiple factors define the structure and function of plant-associated microbiomes. Understanding these factors and their control of plant microbiome assembly will support future strategies to augment specific microbes for crop production and disease management. Our studies of a grafted tomato system found evidence for a small contribution of rootstocks in determining the microbial community. The effect attributable to plant compartment (endosphere versus rhizosphere) was 9- to 10-fold greater, whereas the effect of study site was 6- to 7-fold greater than the rootstock effect. We also identified microbes specific to rootstock treatments. Further study of select microbes could help to identify candidate taxa for synthetic communities and support exploration of other layers of microbial information in association-based single and multilayer networks (98). In the long term, identifying specific plant alleles/genes that correlate with target microbial taxa can inform plant breeding to meet goals beyond fruit traits and yield, perhaps including means to facilitate low-input sustainable agriculture through plant-mediated selection of desirable microbiome components. These observations of how rootstock treatments, environment, and plant compartment (endosphere versus rhizosphere) can structure microbial communities help to lay the groundwork for the development of designer communities and microbiome-based breeding to improve crop production.
MATERIALS AND METHODS
Rootstocks and experimental design.
Plants sampled for this analysis were part of a larger study of grafted tomato plant yield. The main objectives of the larger study were to identify rootstocks that are more productive in Midwestern (Kansas, USA) growing conditions and to evaluate the effects of rootstocks on the rhizobiome (53). Plants sampled for rhizobiome analyses included three rootstock genotypes (BHN589, RST-04-106, and Maxifort) representing four different treatments, as follows: (i) nongrafted BHN589 plants, (ii) self-grafted BHN589 plants (plants grafted to their own rootstock), (iii) BHN589 grafted on RST-04-106, and (iv) BHN589 grafted on Maxifort. The choice of BHN589 as scion was primarily based on the popularity of BHN589 for its high fruit yield and quality and long shelf life. We selected Maxifort because it is a common and popular rootstock and RST-04-106 as a new rootstock variety based on breeders’ recommendations. In the initial field trials that evaluated rootstock performance, plants grafted with Maxifort had higher yield and biomass, whereas the performance of plants grafted with RST-04-106 was similar to that of the nongrafted and self-grafted controls (53). More information about these rootstocks and their potential disease resistance profiles is available in a USDA resource database (http://www.vegetablegrafting.org/resources/rootstock-tables/tomato-rootstock-table/) and also listed in Table S1 in the supplemental material.
We established field trials at the Olathe Horticulture Research and Extension Center (OHREC) and in collaboration with local farmers at Common Harvest Farm and Gieringer’s Orchard (Table 1). At each of three study sites, the four graft treatments were assigned to four plots per block in a randomized complete block design. Each plot consisted of 5 to 8 plants, and one of the middle plants from each plot was sampled for the rhizobiome characterization. The number of blocks varied from one study site to another, depending on the area available for growing tomato. There were six blocks at the OHREC and four each at Gieringer's Farm and Common Harvest Farm, such that for each year, all treatments were replicated 14 times. The experiments were conducted in 2014 and 2015 with similar experimental designs and management. However, at each study site, the blocks were randomly assigned separately for each of the two years.
TABLE 1.
Sites included in the study, with geographic coordinates and soil type
| Study site | Kansas county | Latitude | Longitude | Soil type |
|---|---|---|---|---|
| Olathe Horticulture Research and Extension Center (OHREC) | Johnson | 38.88 N | −94.99 W | Chase silt loam |
| Common Harvest | Douglas | 38.96 N | −95.20 W | Eudora-Kimo complex |
| Gieringer's Orchard | Johnson | 38.79 N | −95.04 W | Sibleyville loam |
Tomato grafting and high-tunnel production.
Tomato plants were grafted in-house using a tube grafting technique. Details about the grafting and postgrafting management were provided by Meyer (53). Briefly, young seedlings (9 to 12 days old) of the scion and the rootstocks were grafted and kept in a healing chamber (dark with high relative humidity, 85% to 95%) for an additional seven days to facilitate the healing process. Darkness minimizes photosynthesis, and high humidity prevents the scion from wilting by maintaining sufficient turgor pressure. Healed plants were transferred to full sunlight in a greenhouse for a week. To maintain similar initial plant sizes across treatments, the nongrafted plants were kept at 12.2°C while the grafted plants healed. All plants were then transplanted to high tunnels along with the original potting mix.
All of our experiments were conducted in high tunnels, a popular system for tomato production. A high tunnel is an unheated greenhouse covered with plastic or acrylic glass (plexiglass) with partial ventilation on the sides, and it is a relatively new production system among small-scale farmers in the Midwest. High-tunnel production systems protect crops against biotic and abiotic stresses, extend the growing season, and improve fruit quality and yield (99). Details about the high-tunnel design and management used in our experiments are provided by Meyer (53).
Endosphere and rhizosphere sample preparation.
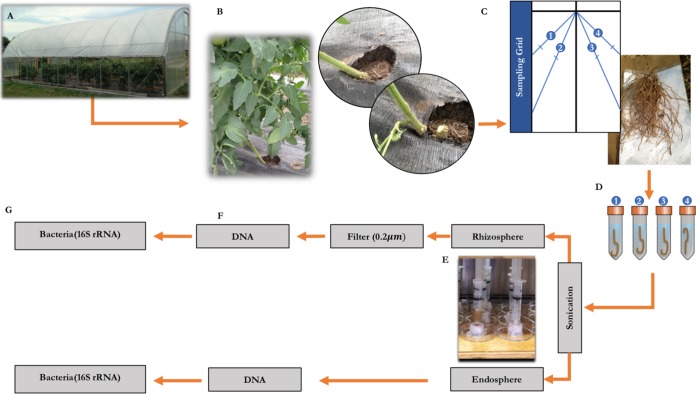
To evaluate microbial communities at the most productive phase of tomato plant development, we sampled during the peak harvest. From each plot, one of the middle plants was carefully dug up such that the majority of roots were intact. The intact root mass was shaken ten times to dislodge bulk soil and placed on top of a marked sampling grid to randomly select four roots, each about 10 to 12 cm in length (Fig. 5). In most plants, we sampled mainly secondary roots, with few primary roots. In other words, we sampled only roots ∼1 to 2 mm in diameter, as these higher-order lateral roots are the active organs of nutrient absorption, exudation, and bacterial colonization (71, 100, 101). Each root piece was transferred to an individual sterile 15-ml Falcon tube (i.e., one piece per tube and four tubes per plant) containing 10 ml 0.1% Triton X solution, stored on ice in a cooler, and transported back to the lab within 6 h, then stored overnight at 4°C. On the following day, roots were sonicated for 10 min at high intensity in an ultrasonic bath cleaner (Fisher Scientific, Waltham, MA). High-intensity sonication effectively separates rhizoplane communities from endosphere communities (102); in our case, the separated rhizoplane communities were included in the rhizosphere samples. The sonicated roots were pressed between sheets of Kimtech Kimwipes (Kimberly-Clark, Roswell, GA), dried, and stored in a 1.5-ml Rino screw-cap tube (Next Advance, Averill Park, NY) at −20°C for DNA extraction. The buffer solution containing the dislodged rhizosphere material was collected into a sterile 20-ml BD Luer lock disposable syringe (Becton, Dickinson and Company, NJ), and passed through a 0.2-μm Whatman Nucleopore polycarbonate track-etched membrane filter with a 25 mm diameter (Fisher Scientific, Waltham, MA) to collect bacteria and suspended particles. The membrane filters containing rhizosphere contents were stored in a 1.5-ml Rino screw-cap tube at −20°C until DNA extraction. The four root pieces per plant were processed individually until the secondary PCR amplification, and pooled in a single unit prior to sequencing. The Luer lock filter holders were cleaned with 10% bleach, rinsed with deionized water, and autoclaved after each run to prevent cross contamination between samples.
FIG 5.
Flow chart of methods for sampling the rhizobiome. Tomato plants grown in high tunnels (A) were trimmed at the graft line (B), and four pieces of roots (1 to 2 mm in diameter) were sampled systematically using a marked sampling grid (C). Each root piece was transferred to a sterile 15-ml Falcon tube containing 10 ml 0.1% Triton X solution (D), sonicated, and filtered through a 0.2-μm filter using a Luer filter holder and disposable syringe (E). DNA was extracted from the endosphere and rhizosphere samples (F) using an extraction kit, and amplicon libraries representing the bacterial community were prepared using primer sets for the V4 region of 16S rRNA (G).
DNA extraction and amplicon generation.
Total genomic DNA was extracted from both the rhizosphere and root tissue samples using a MoBio Ultra clean soil DNA extraction kit (MoBio, Carlsbad, CA) as per the manufacturer’s protocol, with slight modification during the homogenization step. Due to the toughness of the tomato roots, they were cut into smaller pieces using a sterile razor blade and then homogenized using stainless steel beads in a Bullet blender (Next Advance, Averill Park, NY) at 4°C. First, we ran a dry homogenization for 10 min, and then a wet homogenization for another 10 min in inhibitor removal solution (IRS) and bead solutions from the extraction kit. We ran the extraction process for empty tubes and used the obtained product as a negative control to assess any contamination of reagents and PCR amplification using gel electrophoresis. After extraction, DNA was quantified using a ND1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) and normalized to 2 ng/μl. Amplicons for the variable region V4 within the bacterial rRNA gene were generated using the primers 515F-GTGCCAGCMGCCGCGGTAA and 806R-GGACTACHVGGGTWTCTAAT (103). Prior to sequencing, amplicons from different plant samples were multiplexed by incorporating unique molecular identifier tags (MIDs) at the 5′ end of the reverse primer.
PCR amplicons were generated in 50-μl reaction mixtures under the following conditions: 1 μM forward and reverse primers, 10 ng template DNA, 25 μl 5× Phusion high-fidelity (HF) buffer (Finnzymes, Vantaa, Finland) containing 200 μM each deoxynucleotide and 1.5 mM MgCl2 in a master mix, 15 μl molecular biology grade water, and 1 unit (0.5 μl) Phusion high-fidelity DNA polymerase (Finnzymes, Vantaa, Finland). PCR cycle conditions consisted of a 94°C initial denaturing step for 3 min, followed by 30 cycles at 94°C for 45 s, 50°C annealing for 1 min, and a 72°C extension for 1.5 min, followed by a final extension at 72°C for 10 min. To incorporate MIDs into the PCR amplicons, secondary PCR was run using similar reagents and PCR cycling conditions as in the primary PCR, with the amplification cycles reduced to 10. All DNA samples were amplified in triplicate to minimize PCR stochasticity, pooled, and cleaned using a Diffinity RapidTip (Diffinity Genomics, West Chester, PA). Similarly, the secondary PCR was run in triplicate, pooled by experimental unit, and cleaned with an Agencourt AmPure cleanup kit using a SPRIPlate 96-ring magnet (Beckman Coulter, Beverly, MA, USA) as per the manufacturer’s protocol. Prior to cleaning the secondary amplicons with the Agencourt AmPure kit, the amplicons of four pieces of root samples from a particular plant (i.e., an experimental unit) were pooled to a single unit. Then, 500 ng of cleaned, barcoded PCR amplicons were combined per experimental unit, and the final pool was cleaned again, using an Agencourt AmPure cleanup kit as above. Illumina MiSeq adaptors were ligated to the library and paired-end sequenced on a MiSeq personal sequencing system (Illumina, San Diego, CA) using a MiSeq reagent kit v2 with 500 cycles. The endosphere and the rhizosphere amplicon libraries were sequenced separately in two runs. Adaptor ligation and sequencing were performed at the Integrated Genomics Facility at Kansas State University.
Bioinformatics and OTU designation.
The sequence data were curated using mothur v1.33.3 (104), following steps outlined in the MiSeq Standard Operating Protocol (SOP; www.mothur.org/wiki/MiSeq_SOP). Briefly, the forward and the reverse sequence reads were assembled into contigs using default criteria, as specified in the SOP. Any sequence shorter than 250 bp or containing ambiguous bases, more than eight homopolymers, more than one mismatch in primer or MIDs, or missing MIDs was removed (Data Sets S1 to S3 in the supplemental material). Barcoded sequences were assigned to experimental units, and fasta and groups files for the endosphere and the rhizosphere libraries were merged and processed together for the remaining steps in mothur. The cleaned sequences were aligned against the curated 16S rRNA gene SILVA alignment v123, chimeric sequences identified using UCHIME (105) were removed, and the remaining sequences were assigned to taxonomic groups using a naive Bayesian classifier (106) at 60% bootstrap confidence score against the 16S rRNA gene training set (v9) of the Ribosomal Database Project (107). Sequences without known affinities or assigned to mitochondria or chloroplasts were removed. Pairwise distances (less than 0.10) between aligned DNA sequences were used to cluster the sequences into OTUs at 97% similarity, using the nearest neighbor joining algorithm. However, because of the large distance matrix, sequences were split into bins by taxonomy prior to clustering into OTUs. Finally, the clustered OTUs were assigned to consensus taxonomy and used in community analyses. Due to variation in the sequence yield per sample over 2 years, we analyzed the libraries for each year separately. To minimize the bias resulting from unequal sequence counts per sample, samples within each year were rarified at the sequence frequency of the sample yielding the lowest count (2,568 per sample in 2014, and 8,885 per sample in 2015).
Statistical analyses.
To evaluate diversity, we calculated Shannon entropy in R (108), using the vegan package (109) implemented as a part of the phyloseq package (110). The observed Shannon entropy was compared among rootstock treatments using a mixed analysis of variance (ANOVA) model in the lme4 package (111) in R. Rootstock treatments were compared with study sites as a random factor. Differences in the bacterial communities across rootstocks, compartments, and study sites were visualized in nonmetric multidimensional scaling (NMDS) plots, based on the Bray-Curtis dissimilarity matrix. The observed variation in the bacterial community was partitioned using a permutational multivariate analysis of variance (PERMANOVA, at 1,000 permutations), using the adonis function in the vegan package. To identify the OTUs that were depleted or enriched as a function of rootstock treatments, differentially abundant OTUs (DAOTUs) were evaluated by fitting a generalized linear model (GLM) with a negative binomial distribution. Likelihood ratio tests and contrast analyses were performed on the fitted GLM to identify DAOTUs. We used OTU counts from self-grafts and nongrafts as controls, and compared them with other rootstocks in a contrast analysis. All of the tests were adjusted to control for the false-discovery rate (FDR) (P value < 0.05) using the Benjamini-Hochberg method. Similarly, a differential abundance test was performed comparing the controls (self-graft versus nongraft) to identify the OTUs responsive to grafting. General community profiles were constructed using OTUs labeled at the phylum level, split by rootstocks and compartment (endosphere or rhizosphere), and visualized in a bar graph.
Accession number(s).
All sequence data generated in this study were deposited in the NCBI Sequence Read Archive depository under the BioProject identifier PRJNA496268.
Supplementary Material
ACKNOWLEDGMENTS
We thank Alison Cioffi, Bryan S. Cordova, and Jeanelle L. Brisbane for their assistance during the sample processing and DNA extraction; Lani Meyer and Kimberly Oxley at OHREC for coordinating the field visits and sampling; and Tom Buller and Jill Elmers of Common Harvest Farms and Alicia Ellingsworth and Katherine Kelly of Gibbs Road Farm for their collaboration. We appreciate the helpful and insightful comments from AEM reviewers.
We are extremely appreciative of support from the Ceres Trust, a USDA NCR SARE Research and Education Grant (grant LNC13-355), the Kansas Agricultural Experiment Station, the National Institute for Mathematical and Biological Synthesis (NIMBioS), and the University of Florida. A.J. was supported in part by USDA-NIFA capacity project KS-495.
K.A.G., A.J., M.M.K., C.L.R., and R.P. designed the study. R.P. and L.G.M. collected and processed the samples. R.P. analyzed and interpreted the data. R.P., K.A.G., and A.J. wrote the manuscript. R.P., K.A.G., A.J., M.M.K., C.L.R., and L.G.M. revised the manuscript. All authors read and approved the final manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01765-18.
REFERENCES
- 1.Berendsen RL, Pieterse CM, Bakker PA. 2012. The rhizosphere microbiome and plant health. Trends Plant Sci 17:478–486. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Marasco R, Rolli E, Ettoumi B, Vigani G, Mapelli F, Borin S, Abou-Hadid AF, El-Behairy UA, Sorlini C, Cherif A, Zocchi G, Daffonchio D. 2012. A drought resistance-promoting microbiome is selected by root system under desert farming. PLoS One 7:e48479. doi: 10.1371/journal.pone.0048479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meena KK, Sorty AM, Bitla UM, Choudhary K, Gupta P, Pareek A, Singh DP, Prabha R, Sahu PK, Gupta VK, Singh HB, Krishanani KK, Minhas PS. 2017. Abiotic stress responses and microbe-mediated mitigation in plants: the omics strategies. Front Plant Sci 8:172. doi: 10.3389/fpls.2017.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berendsen RL, Vismans G, Yu K, Song Y, de Jonge R, Burgman WP, Burmolle M, Herschend J, Bakker P, Pieterse CMJ. 2018. Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J 12:1496–1507. doi: 10.1038/s41396-018-0093-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evelin H, Kapoor R, Giri B. 2009. Arbuscular mycorrhizal fungi in alleviation of salt stress: a review. Ann Bot 104:1263–1280. doi: 10.1093/aob/mcp251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bokati D, Herrera J, Poudel R. 2016. Soil influences colonization of root-associated fungal endophyte communities of maize, wheat, and their progenitors. J Mycol 2016:1–9. doi: 10.1155/2016/8062073. [DOI] [Google Scholar]
- 7.Fierer N. 2017. Embracing the unknown: disentangling the complexities of the soil microbiome. Nat Rev Microbiol 15:579–590. doi: 10.1038/nrmicro.2017.87. [DOI] [PubMed] [Google Scholar]
- 8.Herrera J, Poudel R, Nebel KA, Collins SL. 2011. Precipitation increases the abundance of some groups of root-associated fungal endophytes in a semiarid grassland. Ecosphere 2:art50. doi: 10.1890/ES11-00001.1. [DOI] [Google Scholar]
- 9.Herrera J, Poudel R, Khidir HH. 2011. Molecular characterization of coprophilous fungal communities reveals sequences related to root-associated fungal endophytes. Microb Ecol 61:239–244. doi: 10.1007/s00248-010-9744-0. [DOI] [PubMed] [Google Scholar]
- 10.Niu B, Paulson JN, Zheng XQ, Kolter R. 2017. Simplified and representative bacterial community of maize roots. Proc Natl Acad Sci U S A 114:E2450–E2459. doi: 10.1073/pnas.1616148114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bulgarelli D, Rott M, Schlaeppi K, Ver Loren van Themaat E, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E, Peplies J, Gloeckner FO, Amann R, Eickhorst T, Schulze-Lefert P. 2012. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488:91–95. doi: 10.1038/nature11336. [DOI] [PubMed] [Google Scholar]
- 12.Coleman-Derr D, Desgarennes D, Fonseca-Garcia C, Gross S, Clingenpeel S, Woyke T, North G, Visel A, Partida-Martinez LP, Tringe SG. 2016. Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytol 209:798–811. doi: 10.1111/nph.13697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrera J, Poudel R, Bokati D. 2013. Assessment of root-associated fungal communities colonizing two species of tropical grasses reveals incongruence to fungal communities of North American native grasses. Fungal Ecol 6:65–69. doi: 10.1016/j.funeco.2012.08.002. [DOI] [Google Scholar]
- 14.Leff JW, Lynch RC, Kane NC, Fierer N. 2017. Plant domestication and the assembly of bacterial and fungal communities associated with strains of the common sunflower, Helianthus annuus. New Phytol 214:412–423. doi: 10.1111/nph.14323. [DOI] [PubMed] [Google Scholar]
- 15.Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Tremblay J, Engelbrektson A, Kunin V, Del Rio TG, Edgar RC, Eickhorst T, Ley RE, Hugenholtz P, Tringe SG, Dangl JL. 2012. Defining the core Arabidopsis thaliana root microbiome. Nature 488:86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL, Buckler ES, Ley RE. 2013. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci U S A 110:6548–6553. doi: 10.1073/pnas.1302837110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang P, Marsh EL, Ainsworth EA, Leakey ADB, Sheflin AM, Schachtman DP. 2017. Shifts in microbial communities in soil, rhizosphere and roots of two major crop systems under elevated CO2 and O3. Sci Rep 7:15019. doi: 10.1038/s41598-017-14936-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cregger MA, Veach AM, Yang ZK, Crouch MJ, Vilgalys R, Tuskan GA, Schadt CW. 2018. The Populus holobiont: dissecting the effects of plant niches and genotype on the microbiome. Microbiome 6:31. doi: 10.1186/s40168-018-0413-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edwards J, Johnson C, Santos-Medellin C, Lurie E, Podishetty NK, Bhatnagar S, Eisen JA, Sundaresan V. 2015. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci U S A 112:E911–E920. doi: 10.1073/pnas.1414592112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mueller UG, Sachs JL. 2015. Engineering microbiomes to improve plant and animal health. Trends Microbiol 23:606–617. doi: 10.1016/j.tim.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 21.Panke-Buisse K, Poole AC, Goodrich JK, Ley RE, Kao-Kniffin J. 2015. Selection on soil microbiomes reveals reproducible impacts on plant function. ISME J 9:980–989. doi: 10.1038/ismej.2014.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poudel R, Jumpponen A, Schlatter DC, Paulitz TC, Gardener BBM, Kinkel LL, Garrett KA. 2016. Microbiome networks: a systems framework for identifying candidate microbial assemblages for disease management. Phytopathology 106:1083–1096. doi: 10.1094/PHYTO-02-16-0058-FI. [DOI] [PubMed] [Google Scholar]
- 23.Fonseca-García C, Coleman-Derr D, Garrido E, Visel A, Tringe SG, Partida-Martínez LP. 2016. The cacti microbiome: interplay between habitat-filtering and host-specificity. Front Microbiol 7:150. doi: 10.3389/fmicb.2016.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mendes LW, Kuramae EE, Navarrete AA, van Veen JA, Tsai SM. 2014. Taxonomical and functional microbial community selection in soybean rhizosphere. ISME J 8:1577–1587. doi: 10.1038/ismej.2014.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Heijden MG, Schlaeppi K. 2015. Root surface as a frontier for plant microbiome research. Proc Natl Acad Sci U S A 112:2299–2300. doi: 10.1073/pnas.1500709112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hacquard S, Garrido-Oter R, Gonzalez A, Spaepen S, Ackermann G, Lebeis S, McHardy AC, Dangl JL, Knight R, Ley R, Schulze-Lefert P. 2015. Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe 17:603–616. doi: 10.1016/j.chom.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 27.Shade A. 2017. Diversity is the question, not the answer. ISME J 11:1. doi: 10.1038/ismej.2016.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McNaughton SJ. 1977. Diversity and stability of ecological communities: a comment on the role of empiricism in ecology. Am Nat 111:515–525. doi: 10.1086/283181. [DOI] [Google Scholar]
- 29.Ritpitakphong U, Falquet L, Vimoltust A, Berger A, Metraux JP, L'Haridon F. 2016. The microbiome of the leaf surface of Arabidopsis protects against a fungal pathogen. New Phytol 210:1033–1043. doi: 10.1111/nph.13808. [DOI] [PubMed] [Google Scholar]
- 30.Zhao J, Liu J, Liang H, Huang J, Chen Z, Nie Y, Wang C, Wang Y. 2018. Manipulation of the rhizosphere microbial community through application of a new bio-organic fertilizer improves watermelon quality and health. PLoS One 13:e0192967. doi: 10.1371/journal.pone.0192967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ardanov P, Sessitsch A, Haggman H, Kozyrovska N, Pirttila AM. 2012. Methylobacterium-induced endophyte community changes correspond with protection of plants against pathogen attack. PLoS One 7:e46802. doi: 10.1371/journal.pone.0046802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogel C, Bodenhausen N, Gruissem W, Vorholt JA. 2016. The Arabidopsis leaf transcriptome reveals distinct but also overlapping responses to colonization by phyllosphere commensals and pathogen infection with impact on plant health. New Phytol 212:192–207. doi: 10.1111/nph.14036. [DOI] [PubMed] [Google Scholar]
- 33.Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JHM, Piceno YM, DeSantis TZ, Andersen GL, Bakker PAHM, Raaijmakers JM. 2011. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332:1097–1100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- 34.Yin C, Hulbert SH, Schroeder KL, Mavrodi O, Mavrodi D, Dhingra A, Schillinger WF, Paulitz TC. 2013. Role of bacterial communities in the natural suppression of Rhizoctonia solani bare patch disease of wheat (Triticum aestivum L.). Appl Environ Microbiol 79:7428–7438. doi: 10.1128/AEM.01610-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Theis KR, Dheilly NM, Klassen JL, Brucker RM, Baines JF, Bosch TCG, Cryan JF, Gilbert SF, Goodnight CJ, Lloyd EA, Sapp J, Vandenkoornhuyse P, Zilber-Rosenberg I, Rosenberg E, Bordenstein SR. 2016. Getting the hologenome concept right: an eco-evolutionary framework for hosts and their microbiomes. mSystems 1:e00028-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vorholt JA, Vogel C, Carlstrom CI, Muller DB. 2017. Establishing causality: opportunities of synthetic communities for plant microbiome research. Cell Host Microbe 22:142–155. doi: 10.1016/j.chom.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 37.Kubota C, McClure MA, Kokalis-Burelle N, Bausher MG, Rosskopf EN. 2008. Vegetable grafting: history, use, and current technology status in North America. HortScience 43:1664–1669. [Google Scholar]
- 38.Louws FJ, Rivard CL, Kubota C. 2010. Grafting fruiting vegetables to manage soilborne pathogens, foliar pathogens, arthropods and weeds. Sci Hortic 127:127–146. doi: 10.1016/j.scienta.2010.09.023. [DOI] [Google Scholar]
- 39.Rivard CL, O'Connell S, Peet MM, Louws FJ. 2010. Grafting tomato with interspecific rootstock to manage diseases caused by Sclerotium rolfsii and southern root-knot nematode. Plant Dis 94:1015–1021. doi: 10.1094/PDIS-94-8-1015. [DOI] [PubMed] [Google Scholar]
- 40.Yuliar Nion YA, Toyota K. 2015. Recent trends in control methods for bacterial wilt diseases caused by Ralstonia solanacearum. Microbes Environ 30:1–11. doi: 10.1264/jsme2.ME14144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He Y, Zhu ZJ, Yang J, Ni XL, Zhu B. 2009. Grafting increases the salt tolerance of tomato by improvement of photosynthesis and enhancement of antioxidant enzymes activity. Environ Exp Bot 66:270–278. doi: 10.1016/j.envexpbot.2009.02.007. [DOI] [Google Scholar]
- 42.Savvas D, Papastavrou D, Ntatsi G, Ropokis A, Olympios C, Hartmann H, Schwarz D. 2009. Interactive effects of grafting and manganese supply on growth, yield, and nutrient uptake by tomato. HortScience 44:1978–1982. [Google Scholar]
- 43.Melnyk CW. 2017. Plant grafting: insights into tissue regeneration. Regeneration 4:3–14. doi: 10.1002/reg2.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nawaz MA, Imtiaz M, Kong QS, Cheng F, Ahmed W, Huang Y, Bie ZL. 2016. Grafting: a technique to modify ion accumulation in horticultural crops. Front Plant Sci 7:1457. doi: 10.3389/fpls.2016.01457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rivard CL, Louws FJ. 2008. Grafting to manage soilborne diseases in heirloom tomato production. HortScience 43:2104–2111. [Google Scholar]
- 46.Goldschmidt EE. 2014. Plant grafting: new mechanisms, evolutionary implications. Front Plant Sci 5:727. doi: 10.3389/fpls.2014.00727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haroldsen VM, Szczerba MW, Aktas H, Lopez-Baltazar J, Odias MJ, Chi-Ham CL, Labavitch JM, Bennett AB, Powell AL. 2012. Mobility of transgenic nucleic acids and proteins within grafted rootstocks for agricultural improvement. Front Plant Sci 3:39. doi: 10.3389/fpls.2012.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu R, Wang X, Lin Y, Ma Y, Liu G, Yu X, Zhong S, Liu B. 2013. Inter-species grafting caused extensive and heritable alterations of DNA methylation in Solanaceae plants. PLoS One 8:e61995. doi: 10.1371/journal.pone.0061995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Melnyk CW, Molnar A, Bassett A, Baulcombe DC. 2011. Mobile 24 nt small RNAs direct transcriptional gene silencing in the root meristems of Arabidopsis thaliana. Curr Biol 21:1678–1683. doi: 10.1016/j.cub.2011.08.065. [DOI] [PubMed] [Google Scholar]
- 50.Nakaho K, Hibino H, Miyagawa H. 2000. Possible mechanisms limiting movement of Ralstonia solanacearum in resistant tomato tissues. J Phytopathol 148:181–190. doi: 10.1046/j.1439-0434.2000.00476.x. [DOI] [Google Scholar]
- 51.Poudel R, Jumpponen A, Kennelly M, Rivard C, Gomez-Montano L, Garrett K. 2018. Rootstocks shape the rhizobiome: rhizosphere and endosphere bacterial communities in the grafted tomato system. bioRxiv 10.1101/375444. [DOI] [PMC free article] [PubMed]
- 52.Marasco R, Rolli E, Fusi M, Michoud G, Daffonchio D. 2018. Grapevine rootstocks shape underground bacterial microbiome and networking but not potential functionality. Microbiome 6:3. doi: 10.1186/s40168-017-0391-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meyer L. 2016. Grafting to increase high tunnel tomato productivity in the central United States. Master’s thesis. Kansas State University, Manhattan, KS: https://krex.k-state.edu/dspace/handle/2097/32736. [Google Scholar]
- 54.Hartmann M, Frey B, Mayer J, Mader P, Widmer F. 2015. Distinct soil microbial diversity under long-term organic and conventional farming. ISME J 9:1177–1194. doi: 10.1038/ismej.2014.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van der Heijden MGA, Hartmann M. 2016. Networking in the plant microbiome. PLoS Biol 14:e1002378. doi: 10.1371/journal.pbio.1002378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gomez-Montano L, Jumpponen A, Gonzales MA, Cusicanqui J, Valdivia C, Motavalli PP, Herman M, Garrett KA. 2013. Do bacterial and fungal communities in soils of the Bolivian Altiplano change under shorter fallow periods? Soil Biol Biochem 65:50–59. doi: 10.1016/j.soilbio.2013.04.005. [DOI] [Google Scholar]
- 57.Mendes LW, Raaijmakers JM, de Hollander M, Mendes R, Tsai SM. 2018. Influence of resistance breeding in common bean on rhizosphere microbiome composition and function. ISME J 12:212–224. doi: 10.1038/ismej.2017.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lebeis SL, Paredes SH, Lundberg DS, Breakfield N, Gehring J, McDonald M, Malfatti S, del Rio TG, Jones CD, Tringe SG, Dangl JL. 2015. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349:860–864. doi: 10.1126/science.aaa8764. [DOI] [PubMed] [Google Scholar]
- 59.Näsholm T, Huss-Danell K, Hogberg P. 2000. Uptake of organic nitrogen in the field by four agriculturally important plant species. Ecology 81:1155–1161. doi: 10.2307/177188. [DOI] [Google Scholar]
- 60.Reeve JR, Smith JL, Carpenter-Boggs L, Reganold JP. 2008. Soil-based cycling and differential uptake of amino acids by three species of strawberry (Fragaria spp.) plants. Soil Biol Biochem 40:2547–2552. doi: 10.1016/j.soilbio.2008.06.015. [DOI] [Google Scholar]
- 61.U’Ren N. 2007. Types, amounts, and possible functions of compounds released into the rhizosphere by soil-grown plants, p 1–21. In Pinton R, Varanini Z, Nannipieri P, The rhizosphere. CRC Press, Boca Raton, FL. doi: 10.1201/9781420005585. [DOI] [Google Scholar]
- 62.Beattie GA. 2018. Metabolic coupling on roots. Nat Microbiol 3:396–397. doi: 10.1038/s41564-018-0139-1. [DOI] [PubMed] [Google Scholar]
- 63.Zhalnina K, Louie KB, Hao Z, Mansoori N, da Rocha UN, Shi SJ, Cho HJ, Karaoz U, Loque D, Bowen BP, Firestone MK, Northen TR, Brodie EL. 2018. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat Microbiol 3:470–480. doi: 10.1038/s41564-018-0129-3. [DOI] [PubMed] [Google Scholar]
- 64.Guan WJ, Zhao X, Hassell R, Thies J. 2012. Defense mechanisms involved in disease resistance of grafted vegetables. HortScience 47:164–170. [Google Scholar]
- 65.Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. 2006. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol 57:233–266. doi: 10.1146/annurev.arplant.57.032905.105159. [DOI] [PubMed] [Google Scholar]
- 66.Hassan S, Mathesius U. 2012. The role of flavonoids in root-rhizosphere signalling: opportunities and challenges for improving plant-microbe interactions. J Exp Bot 63:3429–3444. doi: 10.1093/jxb/err430. [DOI] [PubMed] [Google Scholar]
- 67.Shaw LJ, Morris P, Hooker JE. 2006. Perception and modification of plant flavonoid signals by rhizosphere microorganisms. Environ Microbiol 8:1867–1880. doi: 10.1111/j.1462-2920.2006.01141.x. [DOI] [PubMed] [Google Scholar]
- 68.Redford AJ, Bowers RM, Knight R, Linhart Y, Fierer N. 2010. The ecology of the phyllosphere: geographic and phylogenetic variability in the distribution of bacteria on tree leaves. Environ Microbiol 12:2885–2893. doi: 10.1111/j.1462-2920.2010.02258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boller T, Felix G. 2009. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 70.Jones JD, Dangl JL. 2006. The plant immune system. Nature 444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 71.Badri DV, Vivanco JM. 2009. Regulation and function of root exudates. Plant Cell Environ 32:666–681. doi: 10.1111/j.1365-3040.2008.01926.x. [DOI] [PubMed] [Google Scholar]
- 72.Horton MW, Bodenhausen N, Beilsmith K, Meng D, Muegge BD, Subramanian S, Vetter MM, Vilhjalmsson BJ, Nordborg M, Gordon JI, Bergelson J. 2014. Genome-wide association study of Arabidopsis thaliana leaf microbial community. Nat Commun 5:5320. doi: 10.1038/ncomms6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Koevoets IT, Venema JH, Elzenga JT, Testerink C. 2016. Roots withstanding their environment: exploiting root system architecture responses to abiotic stress to improve crop tolerance. Front Plant Sci 7:1335. doi: 10.3389/fpls.2016.01335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lynch JP, Chimungu JG, Brown KM. 2014. Root anatomical phenes associated with water acquisition from drying soil: targets for crop improvement. J Exp Bot 65:6155–6166. doi: 10.1093/jxb/eru162. [DOI] [PubMed] [Google Scholar]
- 75.Warschefsky EJ, Klein LL, Frank MH, Chitwood DH, Londo JP, von Wettberg EJB, Miller AJ. 2016. Rootstocks: diversity, domestication, and impacts on shoot phenotypes. Trends Plant Sci 21:418–437. doi: 10.1016/j.tplants.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 76.Sasse J, Martinoia E, Northen T. 2018. Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci 23:25–41. doi: 10.1016/j.tplants.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 77.Kumar P, Lucini L, Rouphael Y, Cardarelli M, Kalunke RM, Colla G. 2015. Insight into the role of grafting and arbuscular mycorrhiza on cadmium stress tolerance in tomato. Front Plant Sci 6:477. doi: 10.3389/fpls.2015.00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Albacete A, Martinez-Andujar C, Martinez-Perez A, Thompson AJ, Dodd IC, Perez-Alfocea F. 2015. Unravelling rootstock×scion interactions to improve food security. J Exp Bot 66:2211–2226. doi: 10.1093/jxb/erv027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Barrett CE, Zhao X, McSorley R. 2012. Grafting for root-knot nematode control and yield improvement in organic heirloom tomato production. HortScience 47:614–620. [Google Scholar]
- 80.Pieterse CMJ, de Jonge R, Berendsen RL. 2016. The soil-borne supremacy. Trends Plant Sci 21:171–173. doi: 10.1016/j.tplants.2016.01.018. [DOI] [PubMed] [Google Scholar]
- 81.Hernandez M, Dumont MG, Yuan Q, Conrad R. 2015. Different bacterial populations associated with the roots and rhizosphere of rice incorporate plant-derived carbon. Appl Environ Microbiol 81:2244–2253. doi: 10.1128/AEM.03209-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lynch JM, Whipps JM. 1990. Substrate flow in the rhizosphere. Plant Soil 129:1–10. doi: 10.1007/BF00011685. [DOI] [Google Scholar]
- 83.Fitzpatrick CR, Copeland J, Wang PW, Guttman DS, Kotanen PM, Johnson MTJ. 2018. Assembly and ecological function of the root microbiome across angiosperm plant species. Proc Natl Acad Sci U S A 115:E1157–E1165. doi: 10.1073/pnas.1717617115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Naylor D, DeGraaf S, Purdom E, Coleman-Derr D. 2017. Drought and host selection influence bacterial community dynamics in the grass root microbiome. ISME J 11:2691–2704. doi: 10.1038/ismej.2017.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cekanaviciute E, Yoo BB, Runia TF, Debelius JW, Singh S, Nelson CA, Kanner R, Bencosme Y, Lee YK, Hauser SL, Crabtree-Hartman E, Sand IK, Gacias M, Zhu Y, Casaccia P, Cree BAC, Knight R, Mazmanian SK, Baranzini SE. 2017. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc Natl Acad Sci U S A 114:E8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.He X, McLean JS, Edlund A, Yooseph S, Hall AP, Liu SY, Dorrestein PC, Esquenazi E, Hunter RC, Cheng G, Nelson KE, Lux R, Shi W. 2015. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc Natl Acad Sci U S A 112:244–249. doi: 10.1073/pnas.1419038112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brinig MM, Lepp PW, Ouverney CC, Armitage GC, Relman DA. 2003. Prevalence of bacteria of division TM7 in human subgingival plaque and their association with disease. Appl Environ Microbiol 69:1687–1694. doi: 10.1128/AEM.69.3.1687-1694.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kuehbacher T, Rehman A, Lepage P, Hellmig S, Folsch UR, Schreiber S, Ott SJ. 2008. Intestinal TM7 bacterial phylogenies in active inflammatory bowel disease. J Med Microbiol 57:1569–1576. doi: 10.1099/jmm.0.47719-0. [DOI] [PubMed] [Google Scholar]
- 89.Kwak MJ, Kong HG, Choi K, Kwon SK, Song JY, Lee J, Lee PA, Choi SY, Seo M, Lee HJ, Jung EJ, Park H, Roy N, Kim H, Lee MM, Rubin EM, Lee SW, Kim JF. 2018. Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat Biotechnol 36:1100–1109. doi: 10.1038/nbt.4232. [DOI] [PubMed] [Google Scholar]
- 90.Wang X, Sharp CE, Jones GM, Grasby SE, Brady AL, Dunfield PF. 2015. Stable-isotope probing identifies uncultured Planctomycetes as primary degraders of a complex heteropolysaccharide in soil. Appl Environ Microbiol 81:4607–4615. doi: 10.1128/AEM.00055-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Buckley DH, Huangyutitham V, Nelson TA, Rumberger A, Thies JE. 2006. Diversity of Planctomycetes in soil in relation to soil history and environmental heterogeneity. Appl Environ Microbiol 72:6429–6429. doi: 10.1128/AEM.01601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fuerst JA, Sagulenko E. 2011. Beyond the bacterium: Planctomycetes challenge our concepts of microbial structure and function. Nat Rev Microbiol 9:403–413. doi: 10.1038/nrmicro2578. [DOI] [PubMed] [Google Scholar]
- 93.Kulichevskaya IS, Ivanova AO, Baulina OI, Bodelier PL, Damste JS, Dedysh SN. 2008. Singulisphaera acidiphila gen. nov., sp. nov., a non-filamentous, Isosphaera-like planctomycete from acidic northern wetlands. Int J Syst Evol Microbiol 58:1186–1193. doi: 10.1099/ijs.0.65593-0. [DOI] [PubMed] [Google Scholar]
- 94.Kulichevskaya IS, Ivanova AO, Belova SE, Baulina OI, Bodelier PL, Rijpstra WI, Sinninghe Damste JS, Zavarzin GA, Dedysh SN. 2007. Schlesneria paludicola gen. nov., sp. nov., the first acidophilic member of the order Planctomycetales, from Sphagnum-dominated boreal wetlands. Int J Syst Evol Microbiol 57:2680–2687. doi: 10.1099/ijs.0.65157-0. [DOI] [PubMed] [Google Scholar]
- 95.Fierer N, Ladau J, Clemente JC, Leff JW, Owens SM, Pollard KS, Knight R, Gilbert JA, McCulley RL. 2013. Reconstructing the microbial diversity and function of pre-agricultural tallgrass prairie soils in the United States. Science 342:621–624. doi: 10.1126/science.1243768. [DOI] [PubMed] [Google Scholar]
- 96.Ramirez KS, Craine JM, Fierer N. 2012. Consistent effects of nitrogen amendments on soil microbial communities and processes across biomes. Glob Chang Biol 18:1918–1927. doi: 10.1111/j.1365-2486.2012.02639.x. [DOI] [Google Scholar]
- 97.Herlemann DP, Lundin D, Labrenz M, Jurgens K, Zheng Z, Aspeborg H, Andersson AF. 2013. Metagenomic de novo assembly of an aquatic representative of the verrucomicrobial class Spartobacteria. mBio 4:e00569-12. doi: 10.1128/mBio.00569-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Garrett KA, Alcalá-Briseño RI, Andersen KF, Buddenhagen CE, Choudhury RA, Fulton JC, Hernandez Nopsa JF, Poudel R, Xing Y. 2018. Network analysis: a systems framework to address grand challenges in plant pathology. Annu Rev Phytopathol 56:559–580. doi: 10.1146/annurev-phyto-080516-035326. [DOI] [PubMed] [Google Scholar]
- 99.Janke RR, Altamimi ME, Khan M. 2017. The use of high tunnels to produce fruit and vegetable crops in North America. Agric Sci 08:692–715. doi: 10.4236/as.2017.87052. [DOI] [Google Scholar]
- 100.Hodge A, Berta G, Doussan C, Merchan F, Crespi M. 2009. Plant root growth, architecture and function. Plant Soil 321:153–187. doi: 10.1007/s11104-009-9929-9. [DOI] [Google Scholar]
- 101.van Egeraat AWSM. 1975. The growth of Rhizobium leguminosarum on the root surface and in the rhizosphere of pea seedlings in relation to root exudates. Plant Soil 42:367–379. doi: 10.1007/BF00010012. [DOI] [Google Scholar]
- 102.Richter-Heitmann T, Eickhorst T, Knauth S, Friedrich MW, Schmidt H. 2016. Evaluation of strategies to separate root-associated microbial communities: a crucial choice in rhizobiome research. Front Microbiol 7:773. doi: 10.3389/fmicb.2016.00773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.R Core Team. 2018. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org. [Google Scholar]
- 109.Oksanen J, Blanchet F, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin P, O'Hara R, Simpson G, Solymos P, Stevens M, Szoecs E, Wagner H. 2018. vegan: community ecology package. R package version 2.4-6.
- 110.McMurdie PJ, Holmes S. 2013. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bates D, Machler M, Bolker BM, Walker SC. 2015. Fitting linear mixed-effects models using lme4. J Stat Softw 67:1–48. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.