Abstract
Obesity and type 2 diabetes are increasing in prevalence around the world, and there is a clear need for new and effective strategies to promote metabolic health. A low protein (LP) diet improves metabolic health in both rodents and humans, but the mechanisms that underlie this effect remain unknown. The gut microbiome has recently emerged as a potent regulator of host metabolism and the response to diet. Here, we demonstrate that a LP diet significantly alters the taxonomic composition of the gut microbiome at the phylum level, altering the relative abundance of Actinobacteria, Bacteroidetes, and Firmicutes. Transcriptional profiling suggested that any impact of the microbiome on liver metabolism was likely independent of the microbiome-farnesoid X receptor (FXR) axis. We therefore tested the ability of a LP diet to improve metabolic health following antibiotic ablation of the gut microbiota. We found that a LP diet promotes leanness, increases energy expenditure, and improves glycemic control equally well in mice treated with antibiotics as in untreated control animals. Our results demonstrate that the beneficial effects of a LP diet on glucose homeostasis, energy balance, and body composition are unlikely to be mediated by diet-induced changes in the taxonomic composition of the gut microbiome.
Introduction
Around the world, approximately 425 million people have diabetes, and that number is expected to grow by 50% over the next three decades1. Beyond the direct effects of diabetes on mortality, its impact is amplified by its association with other causes of morbidity and mortality, such as cardiovascular disease2, cancer3, and Alzheimer’s disease4. Type 2 diabetes, which is associated with diet and obesity, accounts for the vast majority of diabetes cases, and the epidemic rise in obesity has fueled the development of this health crisis.
Dietary interventions to control or prevent type 2 diabetes could be highly effective and affordable, but long-term reduced calorie diets have not proven to be sustainable for most people. Diets that alter the level of specific macronutrients without a decrease in caloric consumption may be more sustainable5; one variety of such diets are high protein, low carbohydrate diets such as the Atkins diet, which promise rapid weight loss without restricting calories6. Some clinical trials have observed that high protein diets can promote weight loss7–9, at least in highly compliant subjects10. However, long-term prospective cohort studies have observed that high protein consumption is associated with increased insulin resistance, diabetes, cancer, and cardiovascular disease, and an overall increase in mortality11–13.
In agreement with these findings, recent long-term studies in Drosophila and mice, as well as a short-term randomized control trial conducted in humans, find that low protein (LP) diets are associated with improvements in health, survival, and insulin sensitivity13–18. Reducing dietary protein largely blocks the effect of a high-fat diet on glucose tolerance18, and we recently showed that in a mouse model of pre-existing diet-induced obesity, reducing dietary protein rapidly restored metabolic health, dramatically reduced adiposity, and improved glucose tolerance and insulin sensitivity19. While some of these phenotypes are mediated in part by the insulin-sensitizing and energy expenditure promoting hormone fibroblast growth factor 21 (FGF21), it is likely that other mechanisms are also involved18–24.
Over the last decade, numerous studies have found that the composition of the gut microbiome plays an important role in regulating the metabolic health of both rodents and humans25,26 by mediating the response to drugs, diet, and aging27–33. One major pathway by which the gut microbiota regulates glycemic control is by altering bile acid metabolism and activating the farnesoid X receptor (FXR) – FGF15 signaling axis34,35. Recent work suggests that at least in rodents, the major dietary factors that regulate the taxonomic composition of the gut microbiome are protein and carbohydrate intake36. However, the source of dietary protein – e.g. red meat, white meat, dairy, or plant protein – also has an important effect on the taxonomic composition of the gut microbiome37. It remains unknown if the effect of a LP diet on the composition and function of the gut microbiome plays a role in its beneficial metabolic effects.
In this study, we determined that an amino acid defined LP diet, which has similar metabolic benefits to a LP diet containing natural protein22, alters the taxonomic and functional composition of the gut microbiome. We found that a LP diet significantly alters the hepatic transcriptome, possibly reducing FXR-FGF15 signaling. Finally, we found that ablation of the gut microbiome with antibiotics does not significantly alter the metabolic response to a LP diet. Our data suggests that while dietary protein plays an important role in shaping the taxonomic and functional composition of the gut microbiome, these diet-induced changes do not mediate the beneficial metabolic effects of a LP diet in young, lean mice.
Materials and Methods
Animals and Treatments
For all experiments, male C57BL/6J mice were purchased from The Jackson Laboratory and group housed in static microisolator cages in a specific-pathogen free animal facility. For experiments investigating the composition of the gut microbiome and transcriptional profiling of the liver, mice were purchased at 8 weeks of age, and diet changes occurred at 9 weeks of age. Approximately 4 months later, cecal contents and livers were collected from mice sacrificed in the morning following an overnight, approximately ~16 hr fast. For experiments in which the gut microbiome was ablated with antibiotics, mice were purchased at 5 weeks of age; starting at 6 weeks of age, mice were randomized at the cage level to receive water or water containing antibiotics as described below. Diet changes occurred at 9 weeks of age, and antibiotic treatment was continued for the duration of the experiment. All procedures involving animals were approved by the Institutional Animal Care and Use Committee of the William S. Middleton Memorial Veterans Hospital (Madison, WI), and all experiments were performed in accordance with relevant guidelines and regulations.
Diets
Prior to 9 weeks of age, animals were fed the standard facility chow (Purina 5001; Purina Mills, Richmond, IN, USA). Amino acid defined animal diets (non-irradiated) were obtained from Envigo (formerly Harlan Laboratories). At 9 weeks of age, animals were switched to either a Control amino acid defined diet (TD.140711; 22.0% of calories derived from amino acids; 59.4% from carbohydrate; 18.6% from fat) or a Low Protein amino acid defined diet (TD.140712; 7.1% of calories derived from amino acids; 74.4% from carbohydrates; 18.5% from fat). The complete composition of these diets has been previously described22.
Antibiotic Treatment
The gut microbiome was ablated using an antibiotic treatment protocol previously described to efficiently ablate the gut microbiome of mice38. Briefly, mice were provided with free access to autoclaved water containing 1 g/L ampicillin, 500 mg/L vancomycin, and 1 g/L neomycin; however, in contrast to the protocol followed in38, aspartame was omitted due to its negative effects on glucose homeostasis and body composition in mice39. The mice and the water bottles were weighed and changed biweekly to monitor water intake. Control mice were provided with autoclaved water not containing antibiotics. To verify the efficacy of the antibiotic treatment, fecal pellets were collected and total DNA was extracted using a modification of a previously described protocol40. Briefly, fecal pellets (~30–50 mg) were resuspended in a solution containing 500 µL of extraction buffer [200 mM Tris (pH 8.0), 200 mM NaCl, 20 mM EDTA], 210 µL of 20% SDS, 500 µL phenol:chloroform:isoamyl alcohol (pH 7.9, 25:24:1) and 500 µL of Fisher Scientific 1.4 mm diameter ceramic beads (Cat# 15340159). Following mechanical disruption using a FastPrep 24 (M.P. Biomedicals), the solution was centrifuged at 8,000 rpm at 4 °C for three minutes. The aqueous layer was then sequentially precipitated using sodium acetate/isopropanol and sodium acetate/ethanol. DNA samples were then quantified using a Nanodrop 2000c.
Mouse metabolic phenotyping
Glucose and alanine tolerance tests (GTT and ATT) were performed by fasting the mice overnight for 16 hr and then injecting either glucose (1 g/kg) or alanine (2 g/kg) intraperitoneally as previously described41,42. Glucose measurements were taken using a Bayer Contour blood glucose meter and test strips. Mouse body composition was determined using an EchoMRI 3-in-1 Body Composition Analyzer. For assay of multiple metabolic parameters (O2, CO2, food consumption) and activity tracking, mice were acclimated to housing in a Columbus Instruments Oxymax/CLAMS metabolic chamber system for ~24 hr, and data from a continuous 24-hr period was then recorded and analyzed.
Gut microbial community DNA preparation
Approximately 20–50 mg of cecal matter was added to an autoclaved Sarstedt 2 m micro screw-cap tube (Ref# 72.693.005) containing approximately 500 μL of BioSpec Zirconia/Silica beads (Cat# 11079101z) and one large Bio Spec bead (Cat# 11079132ss). To this, 500 μL of 200 mM Tris-HCl, pH 8.0/200 mM NaCl/20 mM EDTA was added, as well as 210 μL 20% SDS. 500 μL of Phenol/Chloroform/isoamyl alcohol, pH 7.9, 25:24:1, was added before bead beating using a FastPrep 24 (M.P. Biomedicals) until sample was fully homogenized in solution. Tubes were centrifuged at 8,000 rpm at 4 °C for three minutes. The aqueous layer, approximately 500 µL, was transferred to a new microcentrifuge tube (Axygen). 60 μL of 3 M NaAcetate was added, then 600 μL of isopropanol, then inverted to mix. The samples were placed on ice for one hour before centrifuging at 13,000 rpm at 4 °C for 20 minutes. Samples were decanted, and pellet was rinsed with 200 μL of 100% EtOH, then decanted and briefly dried. The pellet was dissolved in 100–200 μL of TE buffer. 100 μL of DNA was cleaned using the Macherey-Nagel PCR Clean-up kit, using 2 NT3 washes and eluting with 50–100 μL of elution buffer.
Construction and Sequencing of v3-v4 16S Metagenomic libraries
Purified genomic DNA was submitted to the University of Wisconsin-Madison Biotechnology Center. DNA concentration was verified fluorometrically using either the Qubit® dsDNA HS Assay Kit or Quant-iT™ PicoGreen® dsDNA Assay Kit (ThermoFisher Scientific, Waltham, MA, USA). Samples were prepared in a similar process to the one described in Illumina’s 16 S Metagenomic Sequencing Library Preparation Protocol, Part # 15044223 Rev. B (Illumina Inc., San Diego, California, USA) with the following modifications: The 16 S rRNA gene V3/V4 variable region was amplified with fusion primers (forward primer 341 f: 5′-ACACTCTTTCCCTACACGACGCTCTTCCGATCT(N)3/6CCTACGGGNGGCWGCAG-3′, reverse primer 805r: 5′-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT(N)3/6GACTACHVGGGTATCTAATCC-3′). Region specific primers were previously described (43; underlined sequences above), and were modified to add 3–6 random nucleotides ((N)3/6) and Illumina adapter overhang nucleotide sequences 5′ of the gene‐specific sequences. Following initial amplification, reactions were cleaned using a 0.7x volume of AxyPrep Mag PCR clean-up beads (Axygen Biosciences, Union City, CA). In a subsequent PCR, Illumina dual indexes and Sequencing adapters were added using the following primers (Forward primer: 5′-AATGATACGGCGACCACCGAGATCTACAC[55555555]ACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′, Reverse Primer: 5′-CAAGCAGAAGACGGCATACGAGAT[77777777]GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT-3′, where bracketed sequences are equivalent to the Illumina Dual Index adapters D501-D508 and D701-D712,N716,N718-N724,N726-N729). Following PCR, reactions were cleaned using a 0.7x volume of AxyPrep Mag PCR clean-up beads (Axygen Biosciences). Quality and quantity of the finished libraries were assessed using an Agilent DNA 1000 kit (Agilent Technologies, Santa Clara, CA) and Qubit® dsDNA HS Assay Kit (ThermoFisher Scientific), respectively. Libraries were pooled in an equimolar fashion and appropriately diluted prior to sequencing. Paired end, 300 bp sequencing was performed using the Illumina MiSeq Sequencer and a MiSeq. 600 bp (v3) sequencing cartridge. Images were analyzed using the standard Illumina Pipeline, version 1.8.2. OTU assignments and diversity plots were created using QIIME analysis pipeline44.
Microbiota analysis using QIIME
Microbiota analysis to obtain OTU assignments and diversity plots were performed using Quantitative Insights Into Microbial Ecology (QIIME)44 version 1.9.1. Illumina sequencing reads were adapter and quality trimmed using the Skewer45 trimming program to remove low quality (<Q25) bases and sequencing adapters. Reads shorter than 100 nucleotides after trimming were discarded. Flash46 was used to merge paired end reads into amplicon sequences using a minimum overlap of 10 nucleotides. Amplicons were then PCR primer trimmed and quality filtered. Sequences were then clustered in OTUs using an open-reference OTU picking protocol based on 97% identity using UCLUST46 against the Greengenes reference database47. Representative sequences (most abundant sequence in OTUs) were picked, aligned to GreenGenes47 Core reference alignment using PyNAST48. Taxonomic assignments were associated with OTUs based on the taxonomy associated with the Greengenes reference sequence defining each OTU. UniFrac distances between samples were calculated using the Greengenes reference tree (ftp://greengenes.microbio.me/greengenes_release/gg_13_5/gg_13_8_otus.tar.gz). The resulting biom-formatted OTU table was filtered to remove singletons and OTUs that could not be aligned using PyNAST. Alpha rarefaction curves were calculated for all samples with a rarefaction upper limit of (median depth/sample count). Samples were removed from further characterization if they did not contain sufficient reads at a depth where the Good’s coverage value for most samples was greater than 0.9. Beta diversity was calculated using weighted and unweighted unifrac on OTU data leveled according to the lowest sample depth. An alternative normalization by CSS49 is also provided for additional downstream analysis.
Liver mRNA preparation and transcriptional profiling
Total liver RNA was extracted with Trireagent (Sigma) as previously described19. Concentration and purity of RNA was initially determined using a Nanodrop 2000c, and then submitted to the University of Wisconsin- Madison Biotechnology Center Gene Expression Center & DNA Sequencing Facility. RNA quality was then assayed using an Agilent RNA NanoChip, and stranded mRNA libraries with polyA enrichment were prepared as described in the Illumina TruSeq Stranded mRNA Sample Preparation Guide Rev. E. DNA sequencing was performed using an Illumina HiSeq. 2500 1 × 100 (TruSeq v3) full flowcell. Pathway enrichment was performed using the “edgeR”50 and “org.Mm.eg.db”51 packages in R version 3.4.452.
Gene expression (qPCR)
Total RNA was isolated from liver with Tri-Reagent, and cDNA was generated as previously described24. Oligo dT primers and primers for real-time PCR were obtained from Integrated DNA Technologies (Coralville, IA,USA). Primer sequences used for qPCR were as follows: Cyp8b1: F: GTTTCTGGGTCCTCTTATTCCTG, R: TGGGAGTGAAAGTGAACGAC; Zfp36l1: F: CACACCAGATCCTAGTCCTTG, R: CTGGGAGTGCTGTAGTTGAG; Cyp7a1: F: AACGATACACTCTCCACCTTT, R: CTGCTTTCATTGCTTCAGGG; Ppp2cb: F: ATGGAAGGATATAACTGGTGCC, R: AGGTGCTGGGTCAAACTG; Map2k1: F: CGTACATCGTGGGCTTCTAC, R: CAGAACTTGATCCAAGGACCC; Shp: F: CTACCCTCAAGAACATTCCAGG, R: CACCAGACTCCATTCCACG; Abcb11: F: CCTCATACGGAAACCCAAGATC, R: CTGACTGTTGATAGGCGATGG; Actb: F: ACCTTCTACAATGAGCTGCG, R: CTGGATGGCTACGTACATGG. Reactions were run on an Applied Biosystems StepOnePlus Real-Time PCR System (Thermo Fisher Scientific) with Invitrogen SYBR Green PCR Master Mix (ThermoFisher Scientific). Actin (Actb) was used to normalize the results from gene-specific reactions.
Statistics
Statistics were carried out in Prism 7 (Graphpad Prism). For each measured parameter, we conducted a two‐factor ANOVA which included an effect of drug treatment (vehicle or antibiotics), an effect of diet (Control or Low AA), and an interaction between diet and treatment. A Sidak’s post-test was performed to determine the significance of factors identified as significant in the two-factor ANOVA. PCA analysis was performed using Clustvis53.
Results
A Low Amino Acid Diet Alters the Taxonomic Composition of the Gut Microbiome
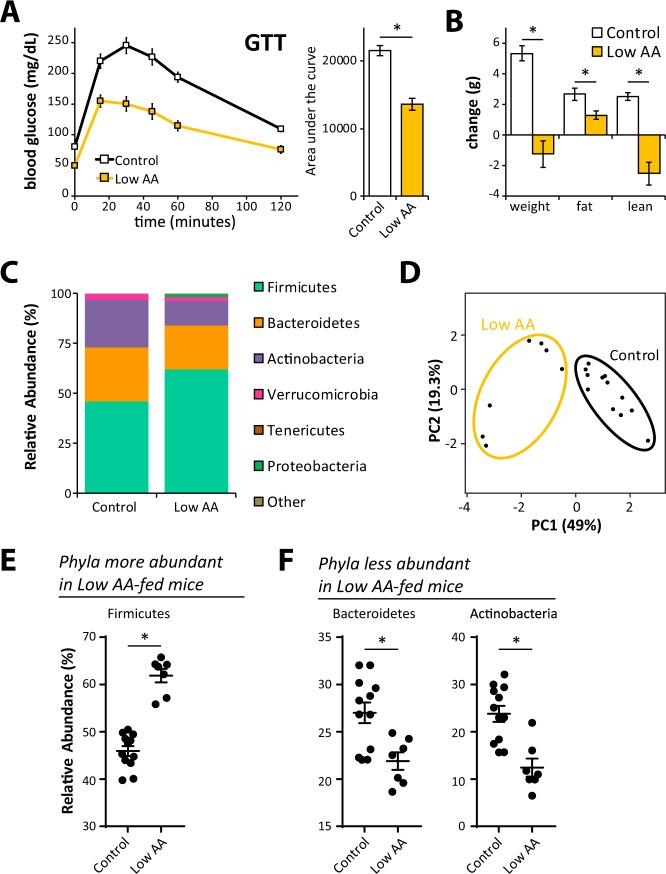
To determine if the taxonomic composition of the microbiome is altered by consuming reduced dietary protein, we fed mice amino acid (AA) defined diets modeled on the AA profiles of naturally sourced control and low protein diets. The Control diet is modeled on a naturally sourced 21% protein diet, while the Low AA diet is based on a naturally sourced 7% protein diet, and we have previously shown that low protein and Low AA diets are comparable in their effect on glycemic control and body composition22. As shown in Fig. 1, mice fed a Low AA diet for four months have improved glucose tolerance (Fig. 1A) and reduced weight and fat mass gain relative to mice fed the Control diet (Fig. 1B).
Figure 1.
A low protein diet promotes metabolic health and alters the taxonomic composition of the cecal microbiome. (A) Glucose tolerance test on male C57BL/6J mice fed a Control (22% of calories from amino acids) or Low AA (7% of calories from amino acids) diet for 4 months (n = 8–10/group; *p < 0.05, t-test). (B) Weight and body composition were measured immediately prior to diet start and after 10 weeks on the indicated diets (n = 8–10/group; *p < 0.05, = t-test). (C) Bar plot of average relative abundance at the phylum taxonomic level. Top 6 phyla are shown. (D) Principle component analysis of demonstrating the effect of diet on taxonomic composition. (E,F) Bacterial phyla differentially represented in cecal contents from mice fed the specified diets for 4 months (n = 7–12/group; Sidak’s test following ANOVA, *p < 0.05). Error bars represent SEM.
We sacrificed mice fed the Control and Low AA diets after four months, collecting cecal contents and the liver. We prepared DNA from the contents of the cecum, and performed 16 S ribosomal RNA (rRNA) sequencing in order to identify alterations in the microbial composition of the gut microbiome. We utilized QIIME to determine relative taxonomic composition of each sample at the phylum and family levels (Figs 1C and S1A). While the alpha diversity did not significantly differ between Control and Low AA diet-fed mice (Fig. S1B), we observed a major shift in the taxonomic composition of the gut microbiome. Utilizing principal component analysis, we determined that the first two principle components explained a majority of the variability in the taxonomic composition of the gut microbiome, and individuals clustered by diet (Fig. 1D). We observed major differences at the phylum level, with an increase in the relative abundance of Firmicutes (Fig. 1E), and a decrease in the relative abundance of Bacteroidetes and Actinobacteria (Fig. 1F). At the family level, we found that the increased relative abundance of Firmicutes was primarily driven by an increase in the order Clostridiales (Fig. S1c); we also observed a decrease in the abundance of the Bacteroidetes families S24–7 and Odoribacteraceae, and the Actinobacteria families Bifidobacteriaceae and Coriobacteriaceae (Fig. S1D).
A Low Amino Acid Diet Alters the Hepatic Transcriptome, but does not activate FXR-FGF15 signaling
A low protein diet improves glucose tolerance in part by improving hepatic insulin sensitivity22,54. While induction of the insulin-sensitizing hormone FGF21 has been shown to play a role in this response20,21, possibly via inhibition of hepatic mTORC1 (mechanistic Target Of Rapamycin Complex 1)55, we recently observed that dietary methionine restriction, which mimics many of the effects of a low protein diet, can improve glucose tolerance independently of changes in FGF21 and hepatic mTORC124.
Over the past decade, it has become clear that the composition and function of the gut microbiome can regulate host metabolism, including glycemic control56,57. One recently characterized pathway by which the microbiome can regulate host glucose metabolism is through bile acids; alterations in the amount and type of secondary bile acids can regulate glycemic control by activating or repressing the FXR – FGF15 signaling axis34,35.
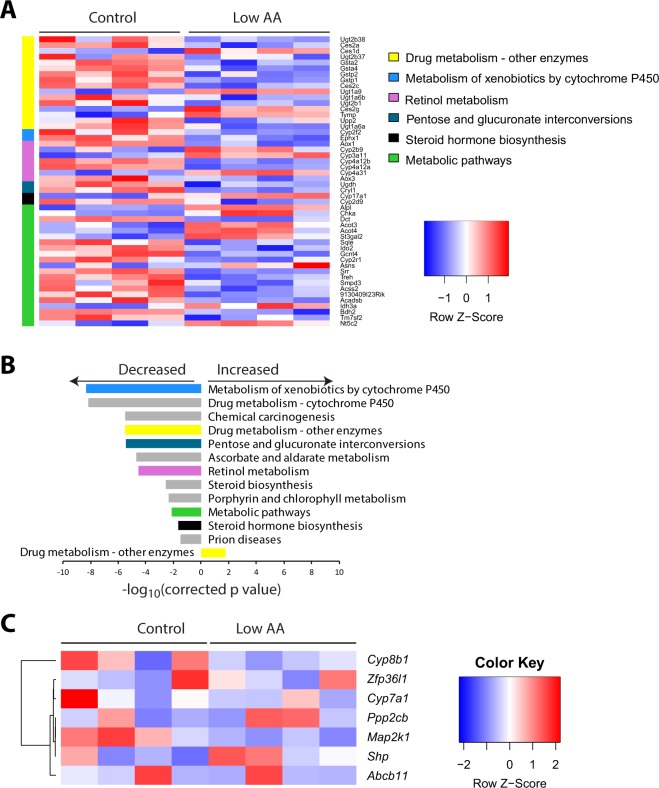
We used RNA-Seq to identify gene expression changes induced by a low protein diet. We identified several differentially expressed metabolic pathways when mice were fed a Low AA diet (Fig. 2A,B). In particular, we observed altered expression of many genes involved in xenobiotic or drug metabolism as well as steroid hormone biosynthesis. However, our analysis did not identify bile acid signaling as significantly altered, and we did not observe significantly altered expression of Shp, Cyp7a1, or several other genes that have been shown to be regulated by the FXR-FGF15 signaling axis (Fig. 2C)58–60. Targeted qPCR analysis of mRNA from a larger, additional cohort of mice confirmed that a Low AA diet did not significantly alter the expression of any of one of these genes (Fig. S2). However, we observed an overall effect of diet consistent with reduced FXR-FGF15 signaling (Fig. S2).
Figure 2.
A low protein diet alters the hepatic transcriptome and shows distinct changes in biological pathways. (A–C) RNA-Seq was performed on the livers for mice fed a Control diet or a Low AA for four months. (A) A heatmap indicating the relative expression of genes involved in the most significantly enriched biological KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways based on genes differentially expressed in the livers of Control and Low AA fed mice (q < 0.05, FDR). Genes in more than one significantly enriched KEGG pathway are listed only once, and assigned to the most significantly affected pathway. (B) Pathway enrichment analysis was performed using g:Profiler (g:GOSt)73, and the p-values of KEGG pathways significantly up- and downregulated by Low AA diet feeding were determined. Colors are matched to that of pathways in (A). (C) Heatmap representing the relative expression of liver genes known to be altered by FXR-FGF15 bile acid signaling.
The metabolic effects of protein restriction are not mediated by the gut microbiota
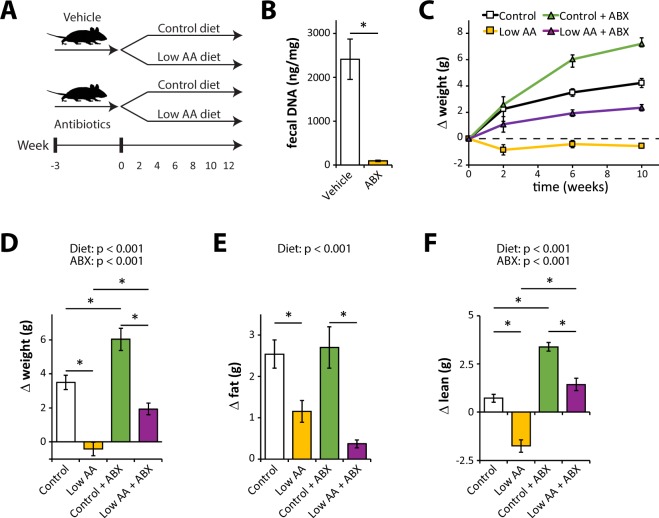
In order to directly assess if the altered taxonomic composition of the gut microbiome contributes to the metabolic effects of a low protein diets, we pretreated mice with either vehicle or antibiotics (ABX) for three weeks; mice were then randomized to either Control or Low AA diets. Antibiotic-treated mice continued to receive antibiotics throughout the course of the experiment (Fig. 3A). As expected, mice treated with antibiotics had significantly reduced fecal DNA content (Fig. 3B). Over the course of the experiment, we tracked weight and body composition of mice in each group (Fig. 3C–F). As expected based on our previous studies, mice fed the Low AA diet gained less weight, less fat mass, and less lean mass than mice fed a Control diet. While antibiotic administration increased weight gain and lean mass gain compared to vehicle treated mice, the Low AA diet had similar effects on weight and body composition in the presence and absence of antibiotics.
Figure 3.
A low protein diet alters body composition similarly in vehicle and antibiotic-treated mice. (A) Schematic representation of the experimental plan; mice were pretreated with antibiotics or vehicle for three weeks, and then randomized to either a Control or Low AA diet. (B) Fecal DNA content was determined following 3 weeks of antibiotic treatment (n = 8/group; *p < 0.05, t-test). (C) Weight of the mice in each group was tracked following randomization to each diet. (D–F) Weight and body composition were determined immediately prior to diet start and after 6 weeks on the indicated diets, and the change in (D) weight, (E) fat mass, and (F) lean mass was determined (n = 12/group; statistics for the overall effects of diet, antibiotic (ABX) treatment, and the interaction represent the p-value from a two-way ANOVA; *p < 0.05 from a Sidak’s post-test examining the effect of parameters identified as significant in the two-way ANOVA). Error bars represent SEM.
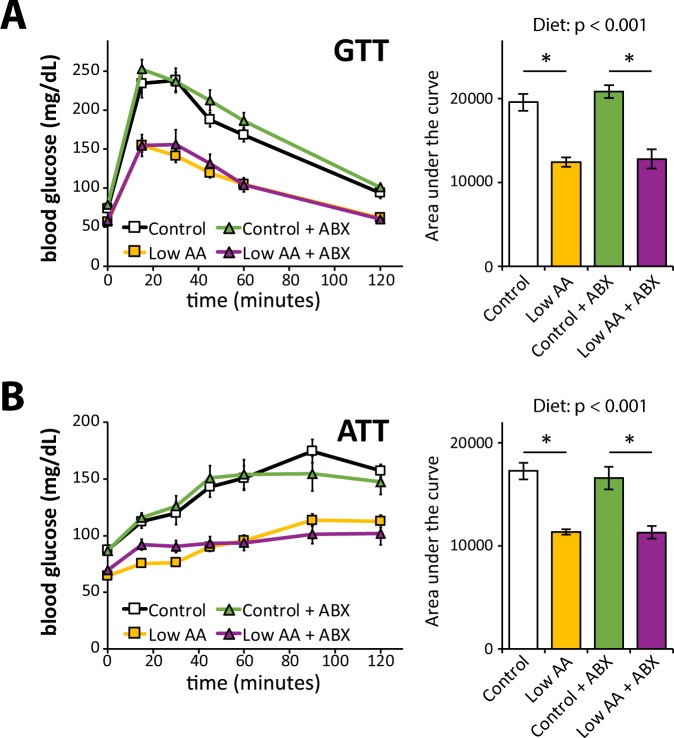
As we determined previously, mice fed a Low AA diet had improved glucose tolerance compared to mice fed a Control diet (Fig. 4A). We also specifically assessed gluconeogenesis in the liver by performing an alanine tolerance test; as in our previous studies utilizing pyruvate, we observed a decrease in the area under the curve (AUC) in mice fed a Low AA diet, indicating improved suppression of gluconeogenesis (Fig. 4B). We saw equivalent reductions in AUC in response to a Low AA diet in both vehicle and antibiotic fed mice; and we did not observe any differences in AUC resulting from antibiotic treatment.
Figure 4.
A low protein diet improves glucose homeostasis similarly in vehicle and antibiotic-treated mice. (A) Glucose and (B) alanine tolerance tests were conducted in mice fed the indicated diets for 8 weeks and 4 weeks, respectively (n = 12/group; statistics for the overall effects of diet, antibiotic (ABX) treatment, and the interaction represent the p-value from a two-way ANOVA; *p < 0.05 from a Sidak’s post-test examining the effect of parameters identified as significant in the two-way ANOVA). Error bars represent SEM.
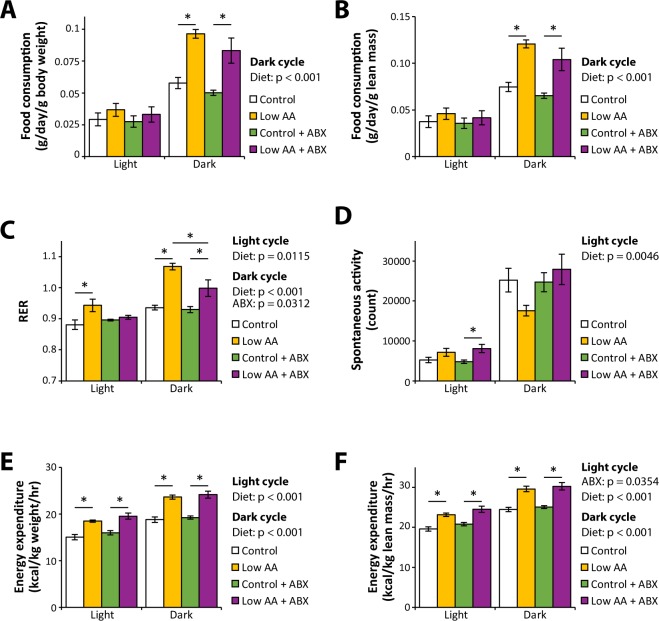
Rodents fed a low protein diet have increased food consumption and increased energy expenditure15,18,20–22,61. We examined the effect of antibiotics on these phenotypes by placing mice in metabolic chambers and assessing food consumption, spontaneous activity, respiratory exchange ratio (RER), and energy expenditure (Fig. 5A–F). In agreement with our previous results, we observed increased food consumption (Fig. 5A,B) and energy expenditure (Fig. 5E,F) in mice fed a Low AA diet, with equivalent effects in both vehicle and antibiotic treated mice. Surprisingly, while a Low AA diet increased both the daytime and nighttime RER of vehicle-treated mice, mice treated with antibiotics did not have increased daytime RER when fed a Low AA diet, and had a reduced increase in nighttime RER (Fig. 5C). The effects of diet on spontaneous activity were relatively small, with increased activity in Low AA fed mice during the day (Fig. 5D).
Figure 5.
A low protein diet increases food consumption and energy expenditure similarly in vehicle and antibiotic-treated mice. (A–F) Metabolic chambers were used to assess (A,B) food consumption, (C) spontaneous activity, (D) respiratory exchange ratio (RER), and (E,F) energy expenditure in mice fed the indicated diets for approximately two months. (n = 5–7/group; statistics for the overall effects of diet, antibiotic (ABX) treatment, and the interaction represent the p-value from a two-way ANOVA; *p < 0.05 from a Sidak’s post-test examining the effect of parameters identified as significant in the two-way ANOVA). Error bars represent SEM.
Discussion
Understanding the mechanisms by which dietary choices impact metabolic health is an area of significant research interest due to the increasing prevalence of diabetes and obesity in the population. Recently, we and others have shown that reducing dietary protein can promote metabolic health in both humans and rodents, but the molecular mechanisms that mediate these effects are unclear.
Here, we examine the effect of a low protein diet on the gut microbiome. In general agreement with the results of Holmes and colleagues36, we find that reducing dietary amino acid levels (and increasing dietary carbohydrates) results in an increased Firmicutes-to-Bacteroidetes ratio. These findings demonstrate the robustness of this intervention on the gut microbiome, as the same effect can be observed in two different laboratories on different continents, using either naturally sourced or defined diets. These results are somewhat surprising, as some previous studies have suggested a link between obesity and an increased prevalence of Firmicutes62. However, they are consistent with the model of Holmes and colleagues that increased caloric intake promotes Firmicutes abundance36.
Since the gut microbiome has proven to be a potent regulator of metabolic health, obesity, and glycemic control, we tested the hypothesis that diet-induced changes in the gut microbiome mediate the beneficial effects of a low protein diet on metabolism. We first used a targeted approach, examining the response of the liver, which is responsive to microbiome-mediated alterations in the amount and type of secondary bile acids via the FXR-FGF15 signaling axis34,35, at the transcriptional level. While we observed altered expression of many genes in response to reduced dietary protein, we found no evidence of altered bile acid signaling and no evidence of increased signaling by the FXR-FGF15 signaling axis. However, a follow-up qPCR analysis of a targeted panel of genes regulated by FXR-FGF15 signaling in a second cohort of mice suggested that a low protein diet might reduce hepatic FXR-FGF15 signaling.
To directly test the role of gut microbiome, we then took an unbiased approach, testing the requirement for an intact gut microbiome in the metabolic response to reduced dietary protein. Following three weeks of high-dose antibiotic treatment, a regimen previously shown to ablate the gut microbiome and dramatically reduce fecal DNA content, we placed mice on either Control or Low AA diets. As anticipated based on previous studies in mice and many other mammals63–66, antibiotics had an overall positive effect on growth and lean mass. However, we observed that protein restriction had very similar effects on weight, body composition, glucose tolerance, and energy expenditure in both the presence and absence of antibiotics. The one major difference we observed was that antibiotic treated, Low AA-fed mice had a lower RER relative to their vehicle treated counterparts, suggesting increased utilization of lipids and decreased utilization of carbohydrates as a fuel source.
A significant caveat of our studies is that they were conducted in young, lean C57BL/6 J mice, which have relatively low intestinal permeability; this may limit the ability of the gut microbiome composition to affect host metabolism. The microbiome could play a larger role in the metabolic response of obese or older animals, which have increased gut permeability67,68; therefore, investigating the role of the gut microbiome in response to reduced dietary protein in aged or obese animals might be an important area for future study. In addition, we did not examine the taxonomic composition of the gut microbiome of antibiotic treated mice, which could help clarify if there are any antibiotic-resistant microbes which might mediate the metabolic effects of a low protein diet. Finally, we did not examine the metabolic effects of protein restriction in germ-free mice; examination in these animals, which completely lack all bacteria, could conceivably reveal subtle effects of the gut microbiome on the response to dietary protein that were not detectable using our antibiotic-ablation model.
Our study also examined only a limited number of metabolic phenotypes associated with a low protein diet, over a relatively short period of time; other phenotypes associated with reduced protein consumption, including increased longevity and healthspan69, may be more directly linked to composition of the gut microbiota. A more detailed investigation of the transcriptional response to protein restriction in antibiotic-treated or germ-free mice may provide valuable clues to identify specific phenotypes that are dependent upon changes in the gut microbiome. There is also growing understanding that the specific amino acid composition of the diet mediates metabolic health70–72, and there may be a role for the microbiome in the metabolic response to other diets with unusual amino acid profiles or from particular dietary sources.
Our findings highlight that dietary macronutrient composition plays an important role in determining the taxonomic composition of the gut microbiome. Yet, while the effects of a low protein diet on the gut microbiome are dramatic, at least in the short term, an intact gut microbiome is not required to realize the metabolic benefits of a low protein diet on glucose homeostasis, body composition, or energy balance. Identifying the physiological and molecular mechanisms by which reducing dietary protein can promote metabolic health remains critical to developing drugs which can take advantage of these pathways to combat obesity and diabetes.
Supplementary information
Acknowledgements
We thank Dr. Federico Rey and Julia Kreznar (Rey lab) for assistance with experimental design and protocols, and Dr. Rey for critical reading of the manuscript. The authors thank the University of Wisconsin-Madison Biotechnology Center Gene Expression Center & DNA Sequencing Facility for providing library preparation and next generation sequencing services. This research was supported, in part, by the NIH through grants to DWL (AG041765, AG050135, AG051974, AG056771), and a pilot grant to DWL from the Diabetes Research Center at Washington University, Grant No. 2 P30 DK020579. This work was also supported by a New Investigator Program Award from the Wisconsin Partnership Program, an Innovator Award from the Progeria Research Foundation, and a Glenn Foundation Award for Research in the Biological Mechanisms of Aging to DWL; an Endocrinology and Reproductive Physiology (ERP) Research Supplies Award from the ERP Graduate Training Program to NEC; and startup funds from the UW-Madison School of Medicine and Public Health and Department of Medicine. This research was conducted while DWL was an American Federation for Aging Research (AFAR) grant recipient. NEC is supported by a training grant from the UW Institute on Aging (NIA T32 AG000213). DY is supported in part by a fellowship from the American Heart Association (17PRE33410983). IO was supported by the University of Wisconsin Carbone Cancer Center Support Grant P30 CA014520 and the Clinical and Translational Science Award (CTSA) program, through the NIH National Center for Advancing Translational Sciences (NCATS), grant UL1TR002373. The Lamming laboratory is supported in part by the U.S. Department of Veterans Affairs (I01-BX004031), and this work was supported using facilities and resources from the William S. Middleton Memorial Veterans Hospital. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This work does not represent the views of the Department of Veterans Affairs or the United States Government.
Author Contributions
H.P., N.E.C. and D.W.L. conceived the experiments and secured funding. H.P., N.E.C., J.A.B., D.Y., J.L.T., S.E.Y., C.B. and E.N.K. performed the experiments. H.P., N.E.C., C.L.G., I.M.O. and D.W.L. analyzed the data. H.P., N.E.C., C.L.G., I.M.O. and D.W.L. wrote the manuscript.
Data Availability
Liver transcriptional profiling data has been deposited at GEO, accession number GSE115683. All other datasets generated during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-37177-3.
References
- 1.International Diabetes Federation. IDF Diabetes Atlas, 8th edn. Brussels, Belgium: International Diabetes Federation, http://www.diabetesatlas.org (2017).
- 2.Diabetes mellitus: a major risk factor for cardiovascular disease A joint editorial statement by the American Diabetes Association; The National Heart, Lung, and Blood Institute; The Juvenile Diabetes Foundation International; The National Institute of Diabetes and Digestive and Kidney Diseases; and The American Heart Association. Circulation. 1999;100:1132–1133. doi: 10.1161/01.CIR.100.10.1132. [DOI] [PubMed] [Google Scholar]
- 3.Giovannucci E, et al. Diabetes and cancer: a consensus report. Diabetes Care. 2010;33:1674–1685. doi: 10.2337/dc10-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbagallo M, Dominguez LJ. Type 2 diabetes mellitus and Alzheimer’s disease. World J Diabetes. 2014;5:889–893. doi: 10.4239/wjd.v5.i6.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weickert MO. Nutritional modulation of insulin resistance. Scientifica. 2012;2012:424780. doi: 10.6064/2012/424780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malik VS, Hu FB. Popular weight-loss diets: from evidence to practice. Nat Clin Pract Cardiovasc Med. 2007;4:34–41. doi: 10.1038/ncpcardio0726. [DOI] [PubMed] [Google Scholar]
- 7.Due A, Toubro S, Skov AR, Astrup A. Effect of normal-fat diets, either medium or high in protein, on body weight in overweight subjects: a randomised 1-year trial. Int J Obes Relat Metab Disord. 2004;28:1283–1290. doi: 10.1038/sj.ijo.0802767. [DOI] [PubMed] [Google Scholar]
- 8.Skov AR, Toubro S, Ronn B, Holm L, Astrup A. Randomized trial on protein vs carbohydrate in ad libitum fat reduced diet for the treatment of obesity. Int J Obes Relat Metab Disord. 1999;23:528–536. doi: 10.1038/sj.ijo.0800867. [DOI] [PubMed] [Google Scholar]
- 9.Weigle DS, et al. A high-protein diet induces sustained reductions in appetite, ad libitum caloric intake, and body weight despite compensatory changes in diurnal plasma leptin and ghrelin concentrations. Am J Clin Nutr. 2005;82:41–48. doi: 10.1093/ajcn/82.1.41. [DOI] [PubMed] [Google Scholar]
- 10.Campos-Nonato I, Hernandez L, Barquera S. Effect of a High-Protein Diet versus Standard-Protein Diet on Weight Loss and Biomarkers of Metabolic Syndrome: A Randomized Clinical Trial. Obes Facts. 2017;10:238–251. doi: 10.1159/000471485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sluijs I, et al. Dietary intake of total, animal, and vegetable protein and risk of type 2 diabetes in the European Prospective Investigation into Cancer and Nutrition (EPIC)-NL study. Diabetes Care. 2010;33:43–48. doi: 10.2337/dc09-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lagiou P, et al. Low carbohydrate-high protein diet and mortality in a cohort of Swedish women. J Intern Med. 2007;261:366–374. doi: 10.1111/j.1365-2796.2007.01774.x. [DOI] [PubMed] [Google Scholar]
- 13.Levine ME, et al. Low protein intake is associated with a major reduction in IGF-1, cancer, and overall mortality in the 65 and younger but not older population. Cell Metab. 2014;19:407–417. doi: 10.1016/j.cmet.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Solon-Biet SM, et al. The ratio of macronutrients, not caloric intake, dictates cardiometabolic health, aging, and longevity in ad libitum-fed mice. Cell Metab. 2014;19:418–430. doi: 10.1016/j.cmet.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Solon-Biet SM, et al. Dietary Protein to Carbohydrate Ratio and Caloric Restriction: Comparing Metabolic Outcomes in Mice. Cell reports. 2015;11:1529–1534. doi: 10.1016/j.celrep.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mair W, Piper MD, Partridge L. Calories do not explain extension of life span by dietary restriction in Drosophila. PLoS Biol. 2005;3:e223. doi: 10.1371/journal.pbio.0030223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee KP, et al. Lifespan and reproduction in Drosophila: New insights from nutritional geometry. Proc Natl Acad Sci USA. 2008;105:2498–2503. doi: 10.1073/pnas.0710787105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maida A, et al. A liver stress-endocrine nexus promotes metabolic integrity during dietary protein dilution. J Clin Invest. 2016;126:3263–3278. doi: 10.1172/JCI85946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cummings NE, et al. Restoration of metabolic health by decreased consumption of branched-chain amino acids. The Journal of physiology. 2018;596:623–645. doi: 10.1113/JP275075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laeger T, et al. FGF21 is an endocrine signal of protein restriction. J Clin Invest. 2014;124:3913–3922. doi: 10.1172/JCI74915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laeger T, et al. Metabolic Responses to Dietary Protein Restriction Require an Increase in FGF21 that Is Delayed by the Absence of GCN2. Cell reports. 2016;16:707–716. doi: 10.1016/j.celrep.2016.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fontana L, et al. Decreased Consumption of Branched-Chain Amino Acids Improves Metabolic Health. Cell reports. 2016;16:520–530. doi: 10.1016/j.celrep.2016.05.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keipert S, et al. Long-Term Cold Adaptation Does Not Require FGF21 or UCP1. Cell Metab. 2017;26:437–446 e435. doi: 10.1016/j.cmet.2017.07.016. [DOI] [PubMed] [Google Scholar]
- 24.Yu, D. et al. Short-term methionine deprivation improves metabolic health via sexually dimorphic, mTORC1-independent mechanisms. FASEB J, fj201701211R, 10.1096/fj.201701211R (2018). [DOI] [PMC free article] [PubMed]
- 25.Kreznar JH, et al. Host Genotype and Gut Microbiome Modulate Insulin Secretion and Diet-Induced Metabolic Phenotypes. Cell reports. 2017;18:1739–1750. doi: 10.1016/j.celrep.2017.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ridaura VK, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu H, et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med. 2017;23:850–858. doi: 10.1038/nm.4345. [DOI] [PubMed] [Google Scholar]
- 28.Sung MM, et al. Improved Glucose Homeostasis in Obese Mice Treated With Resveratrol Is Associated With Alterations in the Gut Microbiome. Diabetes. 2017;66:418–425. doi: 10.2337/db16-0680. [DOI] [PubMed] [Google Scholar]
- 29.Rabot S, et al. High fat diet drives obesity regardless the composition of gut microbiota in mice. Scientific reports. 2016;6:32484. doi: 10.1038/srep32484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greiner TU, Hyotylainen T, Knip M, Backhed F, Oresic M. The gut microbiota modulates glycaemic control and serum metabolite profiles in non-obese diabetic mice. PLoS One. 2014;9:e110359. doi: 10.1371/journal.pone.0110359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vogt NM, et al. Gut microbiome alterations in Alzheimer’s disease. Scientific reports. 2017;7:13537. doi: 10.1038/s41598-017-13601-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turnbaugh PJ, et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Filippo C, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie C, et al. An Intestinal Farnesoid X Receptor-Ceramide Signaling Axis Modulates Hepatic Gluconeogenesis in Mice. Diabetes. 2017;66:613–626. doi: 10.2337/db16-0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sayin SI, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 36.Holmes AJ, et al. Diet-Microbiome Interactions in Health Are Controlled by Intestinal Nitrogen Source Constraints. Cell Metab. 2017;25:140–151. doi: 10.1016/j.cmet.2016.10.021. [DOI] [PubMed] [Google Scholar]
- 37.Zhu Y, et al. Meat, dairy and plant proteins alter bacterial composition of rat gut bacteria. Scientific reports. 2015;5:15220. doi: 10.1038/srep15220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pluznick JL, et al. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc Natl Acad Sci USA. 2013;110:4410–4415. doi: 10.1073/pnas.1215927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gul SS, et al. Inhibition of the gut enzyme intestinal alkaline phosphatase may explain how aspartame promotes glucose intolerance and obesity in mice. Applied physiology, nutrition, and metabolism = Physiologie appliquee, nutrition et metabolisme. 2017;42:77–83. doi: 10.1139/apnm-2016-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krautkramer KA, et al. Diet-Microbiota Interactions Mediate Global Epigenetic Programming in Multiple Host Tissues. Mol Cell. 2016;64:982–992. doi: 10.1016/j.molcel.2016.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mutel E, et al. Control of blood glucose in the absence of hepatic glucose production during prolonged fasting in mice: induction of renal and intestinal gluconeogenesis by glucagon. Diabetes. 2011;60:3121–3131. doi: 10.2337/db11-0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lamming DW, et al. Depletion of Rictor, an essential protein component of mTORC2, decreases male lifespan. Aging Cell. 2014;13:911–917. doi: 10.1111/acel.12256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klindworth A, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang H, Lei R, Ding SW, Zhu S. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics. 2014;15:182. doi: 10.1186/1471-2105-15-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caporaso JG, et al. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paulson JN, Stine OC, Bravo HC, Pop M. Differential abundance analysis for microbial marker-gene surveys. Nature methods. 2013;10:1200–1202. doi: 10.1038/nmeth.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.org.Mm.eg.db: Genome wide annotation for Mouse v. R package version 3.6.0 (2018).
- 52.R: A language and environment for statistical computing (R Foundation for Statistical Computing, Vienna, Austria, 2018).
- 53.Metsalu T, Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015;43:W566–570. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harputlugil E, et al. The TSC complex is required for the benefits of dietary protein restriction on stress resistance in vivo. Cell reports. 2014;8:1160–1170. doi: 10.1016/j.celrep.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gong Q, et al. Fibroblast growth factor 21 improves hepatic insulin sensitivity by inhibiting mammalian target of rapamycin complex 1 in mice. Hepatology. 2016;64:425–438. doi: 10.1002/hep.28523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Utzschneider KM, Kratz M, Damman CJ, Hullar M. Mechanisms Linking the Gut Microbiome and Glucose Metabolism. J Clin Endocrinol Metab. 2016;101:1445–1454. doi: 10.1210/jc.2015-4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuipers F, Bloks VW, Groen AK. Beyond intestinal soap–bile acids in metabolic control. Nature reviews. Endocrinology. 2014;10:488–498. doi: 10.1038/nrendo.2014.60. [DOI] [PubMed] [Google Scholar]
- 58.Fu T, et al. FXR Primes the Liver for Intestinal FGF15 Signaling by Transient Induction of beta-Klotho. Mol Endocrinol. 2016;30:92–103. doi: 10.1210/me.2015-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jung D, et al. FXR agonists and FGF15 reduce fecal bile acid excretion in a mouse model of bile acid malabsorption. J Lipid Res. 2007;48:2693–2700. doi: 10.1194/jlr.M700351-JLR200. [DOI] [PubMed] [Google Scholar]
- 60.Tarling EJ, et al. RNA-binding protein ZFP36L1 maintains posttranscriptional regulation of bile acid metabolism. J Clin Invest. 2017;127:3741–3754. doi: 10.1172/JCI94029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang X, et al. Effects of dietary protein to carbohydrate balance on energy intake, fat storage, and heat production in mice. Obesity (Silver Spring) 2013;21:85–92. doi: 10.1002/oby.20007. [DOI] [PubMed] [Google Scholar]
- 62.Chakraborti CK. New-found link between microbiota and obesity. World J Gastrointest Pathophysiol. 2015;6:110–119. doi: 10.4291/wjgp.v6.i4.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McEwen SA, Fedorka-Cray PJ. Antimicrobial use and resistance in animals. Clin Infect Dis. 2002;34(Suppl 3):S93–S106. doi: 10.1086/340246. [DOI] [PubMed] [Google Scholar]
- 64.Ozawa E. Studies on growth promotion by antibiotics. I. Effects of chlortetracycline on growth. J Antibiot (Tokyo) 1955;8:205–211. [PubMed] [Google Scholar]
- 65.Cho I, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621–626. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Butaye P, Devriese LA, Haesebrouck F. Antimicrobial growth promoters used in animal feed: effects of less well known antibiotics on gram-positive bacteria. Clin Microbiol Rev. 2003;16:175–188. doi: 10.1128/CMR.16.2.175-188.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thevaranjan N, et al. Age-Associated Microbial Dysbiosis Promotes Intestinal Permeability, Systemic Inflammation, and Macrophage Dysfunction. Cell Host Microbe. 2017;21:455–466 e454. doi: 10.1016/j.chom.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Spengler E, Loomba R. The Gut Microbiota, Intestinal Permeability, Bacterial Translocation, and Nonalcoholic Fatty Liver Disease: What Comes First? Cell Mol Gastroenterol Hepatol. 2015;1:129–130. doi: 10.1016/j.jcmgh.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fontana L, Partridge L. Promoting health and longevity through diet: from model organisms to humans. Cell. 2015;161:106–118. doi: 10.1016/j.cell.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cummings NE, Lamming DW. Regulation of metabolic health and aging by nutrient-sensitive signaling pathways. Molecular and cellular endocrinology. 2017;455:13–22. doi: 10.1016/j.mce.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brown-Borg HM, Buffenstein R. Cutting back on the essentials: Can manipulating intake of specific amino acids modulate health and lifespan? Ageing Res Rev. 2017;39:87–95. doi: 10.1016/j.arr.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lees EK, et al. Direct comparison of methionine restriction with leucine restriction on the metabolic health of C57BL/6J mice. Scientific reports. 2017;7:9977. doi: 10.1038/s41598-017-10381-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reimand J, et al. g:Profiler-a web server for functional interpretation of gene lists (2016 update) Nucleic Acids Res. 2016;44:W83–89. doi: 10.1093/nar/gkw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Liver transcriptional profiling data has been deposited at GEO, accession number GSE115683. All other datasets generated during the current study are available from the corresponding author on reasonable request.