Significance
Mesenchymal progenitor cells play important roles in the formation of skeletal tissues; however, how cell fates of mesenchymal progenitor cells are regulated remains largely unclear. We made use of the dental follicle surrounding the developing tooth bud, which critically regulates tooth eruption and tooth root formation. Dental follicle mesenchymal progenitor cells express parathyroid hormone-related peptide (PTHrP), a locally acting autocrine/paracrine ligand, and become essential skeletal cell types establishing the root–bone interface. These PTHrP+ mesenchymal progenitors maintained their physiological cell fates through the PTH/PTHrP receptor, a deficiency of which resulted in failure of tooth eruption phenotypes closely resembling human genetic conditions. We conclude that proper cell fates of mesenchymal progenitor cells are maintained by autocrine signaling to achieve functional formation of skeletal tissues.
Keywords: mesenchymal progenitor cells, parathyroid hormone-related peptide, dental follicle, tooth eruption, in vivo lineage-tracing experiment
Abstract
Formation of functional skeletal tissues requires highly organized steps of mesenchymal progenitor cell differentiation. The dental follicle (DF) surrounding the developing tooth harbors mesenchymal progenitor cells for various differentiated cells constituting the tooth root–bone interface and coordinates tooth eruption in a manner dependent on signaling by parathyroid hormone-related peptide (PTHrP) and the PTH/PTHrP receptor (PPR). However, the identity of mesenchymal progenitor cells in the DF and how they are regulated by PTHrP-PPR signaling remain unknown. Here, we show that the PTHrP-PPR autocrine signal maintains physiological cell fates of DF mesenchymal progenitor cells to establish the functional periodontal attachment apparatus and orchestrates tooth eruption. A single-cell RNA-seq analysis revealed cellular heterogeneity of PTHrP+ cells, wherein PTHrP+ DF subpopulations abundantly express PPR. Cell lineage analysis using tamoxifen-inducible PTHrP-creER mice revealed that PTHrP+ DF cells differentiate into cementoblasts on the acellular cementum, periodontal ligament cells, and alveolar cryptal bone osteoblasts during tooth root formation. PPR deficiency induced a cell fate shift of PTHrP+ DF mesenchymal progenitor cells to nonphysiological cementoblast-like cells precociously forming the cellular cementum on the root surface associated with up-regulation of Mef2c and matrix proteins, resulting in loss of the proper periodontal attachment apparatus and primary failure of tooth eruption, closely resembling human genetic conditions caused by PPR mutations. These findings reveal a unique mechanism whereby proper cell fates of mesenchymal progenitor cells are tightly maintained by an autocrine system mediated by PTHrP-PPR signaling to achieve functional formation of skeletal tissues.
Stem and progenitor cells of the skeletal cell lineage, particularly skeletal stem cells (SSCs) and mesenchymal progenitor cells, are considered to play important roles in the formation, maintenance, and repair of skeletal tissues (1). These mesenchymal progenitor cell populations reside in a variety of tissues, including bone marrow (2), growth plates (3), and craniofacial sutures (4). In postnatal growth plates, the resting zone harbors skeletal stem cells expressing parathyroid hormone-related peptide (PTHrP) and maintains the integrity of growth plates (3). Cells in the dental follicle (DF), a sac-like membranous tissue surrounding the developing tooth bud, also express PTHrP abundantly and coordinate tooth eruption and root formation by facilitating the formation of osteoclasts that resorb alveolar bones to create the eruption pathway and providing a source of cementoblasts, periodontal ligament (PDL) cells, and alveolar bone osteoblasts to establish the root–bone interface anchoring the tooth to the bone.
PTHrP is a locally acting autocrine/paracrine ligand, and signaling by its receptor, PTH/PTHrP receptor (PPR), critically regulates the processes of tooth eruption and root formation. PTHrP is absolutely required for tooth eruption (5), whereas PPR is essential for tooth root formation (6). In humans, primary failure of tooth eruption (PFE; OMIM 125350), a rare autosomal dominant disorder that exclusively affects tooth eruption (7), is characterized by a cessation of tooth eruption before emergence despite an unobstructed eruption path. PFE is caused by loss-of-function mutations in PPR (8–11). Despite these lines of evidence, the identity of mesenchymal progenitor cells in the DF and how they are regulated by PTHrP-PPR signaling remain unknown, however.
In this study, we set out to reveal cell fates of PTHrP+ DF mesenchymal progenitor cells during tooth root formation by in vivo lineage-tracing experiments based on a DF-specific PTHrP-creER line, and also to define the roles of PPR in this process by specifically deleting the receptor in PTHrP+ DF cells. Our findings reveal that PTHrP-PPR autocrine regulation is essential for maintaining the proper cell fates of DF mesenchymal progenitor cells and critically supports tooth eruption.
Results
Characterization of PTHrP+ DF Cells.
We first made use of a PTHrP-mCherry knock-in allele to delineate the formation of PTHrP+ cells in the DF. PTHrP-mCherry+ cells were found predominantly in the DF surrounding the tooth bud during the fetal stage and the early postnatal stage before initiation of tooth root formation (SI Appendix, Fig. S1 A and B), and subsequently on the root surface on tooth eruption, with a majority of these cells also positive for osteocalcin (Oc)-GFP (SI Appendix, Fig. S1 C and D, arrows in Inset), confirming our previous findings based on a PTHrP-LacZ allele (6, 12). The PTHrP-mCherry reporter accurately reflects endogenous PTHrP mRNA expression patterns (SI Appendix, Fig. S1 E and F).
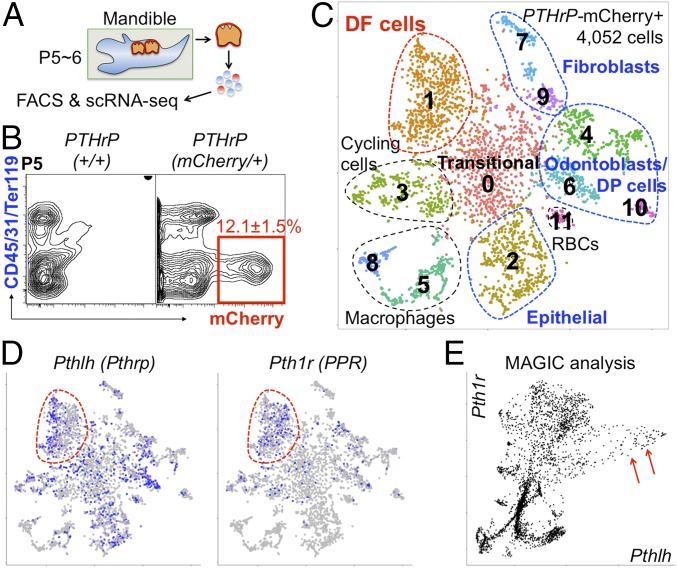
We then characterized PTHrP-expressing cells in developing molars based on a single-cell RNA-seq analysis. Flow cytometry analysis using a cell dissociation protocol to harvest cells containing DF (Fig. 1A and SI Appendix, Fig. S2) revealed that PTHrP-mCherry+ cells were found exclusively in a CD45−Ter119−CD31− fraction and accounted for 12.1 ± 1.5% of total harvested cells (Fig. 1B). We subsequently profiled fluorescence-activated cell sorting (FACS)-isolated 4,052 PTHrP-mCherry+ single cells at P6 using the 10X Genomics Chromium Single-Cell Gene Expression Solution platform. An initial t-distributed stochastic neighbor embedding (t-SNE) analysis revealed 12 clusters, including three impurified clusters (468 cells in clusters 5, 8, and 11) and one cluster abundant in cell cycle genes (396 cells in cluster 3) (Fig. 1C and SI Appendix, Fig. S3). Of the eight clusters of interest, we discovered three clusters of odontoblasts/dental papilla cells abundant in Tubb3, Dmp1, and Sp7 (672 cells in clusters 4, 6, and 10) and two clusters of fibroblasts abundant in Fn1 and Mmp13 (267 cells in clusters 7 and 9). Among the remaining three major clusters, we found that cells in cluster 2 (595 cells) expressed epithelial markers Krt5, Krt14, and Cdh1, whereas cells in cluster 1 (738 cells) expressed DF markers Spon1 (13), Mkx (14), and Acta2 (Fig. 1C). Interestingly, cells in cluster 0 (916 cells) exhibited transitional states between epithelial and DF cells. Furthermore, both Pthlh (Pthrp) and Pth1r (Pth/Pthrp receptor; PPR) were broadly expressed in cluster 1 (Fig. 1D, red contour). To reveal the relationship between Pthlh and Pth1r in DF, we performed a MAGIC imputation analysis (15). Cells expressing Pth1r abundantly (>0.2) coexpressed Pthlh at a high level (>0.5), wherein a population of Pthlh-abundant cells expressed Pth1r at a unanimously high level (Fig. 1E, red arrows). Therefore, these analyses reveal fundamental cell heterogeneity and characteristics of PTHrP-expressing cells in DF and its surrounding tissues.
Fig. 1.
Characterization of PTHrP-mCherry+ DF cells. (A and B) PTHrP-mCherry expression in PTHrP+/+ and PTHrPmCherry/+ DF cells at P5. (C and D) t-SNE–based visualization of major classes of FACS-sorted PTHrP-mCherry+ single cells at P6 (clusters 0–11). Cluster 0, transitional cells; cluster 1, DF cells; cluster 2, epithelial cells; cluster 3, cycling cells; clusters 4, 6, and 10, odontoblasts and dental papilla (DP) cells; clusters 5, 8, and 11, impurified cells; clusters 7 and 9, fibroblasts. Dots indicate individual cells; color indicates cell type. (D) Pthlh and Pth1r expression. Blue, high expression; gray, no expression; red contour, DF cells. (E) MAGIC imputation analysis showing the Pthlh-Pth1r relationship (DF cells). Red arrows indicate PthlhhighPth1r+ cells.
PTHrP+ DF Cells Become Coronal Cementoblasts, Ligament Fibroblasts, and Cryptal Bone Osteoblasts.
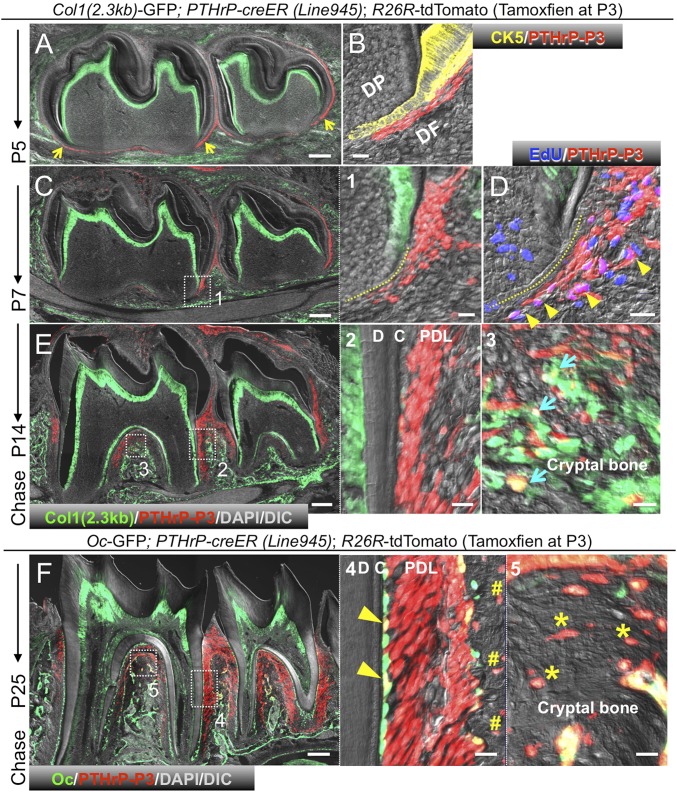
To reveal physiological cell fates of PTHrP+ DF cells during tooth root formation, we performed in vivo lineage-tracing experiments based on a tamoxifen-inducible PTHrP-creER bacterial artificial chromosome (BAC) transgenic line (L945) (5). Analysis of PTHrPmCherry/+; PTHrP-creER; R26R-ZsGreen mice at P5 after a tamoxifen pulse at P3 revealed that 96.0 ± 0.8% of ZsGreen+ cells were mCherry+, and that 8.3 ± 3.7% of mCherry+ cells expressed ZsGreen (SI Appendix, Fig. S4A), indicating that PTHrP-creER can accurately mark a subset of PTHrP-mCherry+ cells on tamoxifen injection. We also analyzed Col1(2.3kb)-GFP; PTHrP-creER; R26R-tdTomato mice at P5 shortly after tamoxifen injection at P3. We consistently found tdTomato+ cells exclusively in DF but not in other portions of the tooth bud; in particular, DF cells of the middle and periapical portion were tdTomato+ (Fig. 2A, yellow arrows). Immunostaining for cytokeratin 5 (an epithelial cell marker) revealed that tdTomato+ cells were located outside the Hertwig epithelial root sheath (HERS) (Fig. 2B). No tdTomato+ cells were observed in alveolar bones. Importantly, no tdTomato+ cells were observed without tamoxifen at any time points observed (SI Appendix, Fig. S4 B–E). Therefore, our PTHrP-creER line (L945) can specifically mark a subset of DF mesenchymal cells when tamoxifen is administered at the onset of tooth root formation.
Fig. 2.
PTHrP-creER marks DF mesenchymal progenitor cells in vivo. (A–F) Lineage tracing of P3 PTHrP-creER+ DF cells. Yellow arrowheads in A indicate tdTomato+ DF cells in periapical areas. (B) Cytokeratin 5 (CK5) staining. (C) Yellow dotted line in (1) denotes HERS. (D) EdU was administered shortly before analysis. Arrowheads in D denote EdU+tdTomato+ DF cells. (E) Blue arrows in (3) indicate Col1-GFP+tdTomato+ osteoblasts in cryptal bones. (F) Yellow arrowheads in (4) indicate GFP+tdTomato+ cementoblasts on root surface. Pound signs in (4) denote tdTomato+ osteocytes in interseptic cryptal bones. Asterisks in (5) indicate tdTomato+ osteocytes in interradicular cryptal bones. C, cementum; D, dentin. PDL, periodontal ligament. [Scale bars: 200 µm (Left), 20 µm (Right).]
Subsequently, we traced the fate of these PTHrP+ DF cells (PTHrPCE-P3 cells) in vivo. After 4 d of chase at P7, PTHrPCE-P3 cells increased their number as the root elongated (Fig. 2C, area 1); tdTomato+ cells proliferated in close proximity to the HERS as they incorporated 5-ethynyl-2′-deoxyuridine (EdU) administered shortly before analysis (Fig. 2D, arrowheads; 69.0 ± 18.0 EdU+tdTomato+ cells among 212.3 ± 22.0 tdTomato+ cells per 200-µm thickness; EdU+: 32.1 ± 5.5% of tdTomato+ cells per 200-µm thickness; n = 3). After 11 d of chase at P14 when the root was half-formed, PTHrPCE-P3 cells actively participated in the formation of periodontal tissues (Fig. 2E) by differentiating predominantly to nascent PDL cells and cementoblasts (area 2), as well as alveolar cryptal bone osteoblasts (area 3, blue arrows). After 22 d of chase at P25, when tooth root formation was complete, PTHrPCE-P3 cells continued to contribute substantially to PDL cells and Oc-GFP+ cementoblasts on the acellular cementum (70.4 ± 1.3% and 17.3 ± 5.0%, respectively; n = 3) (Fig. 2F, area 4, pound signs) as well as cryptal bone osteoblasts/cytes in the interradicular (area 5, asterisks) and interseptal region (area 4, arrowheads). In contrast, PTHrPCE-P3 cells did not become Oc-GFP+ cementoblasts on the cellular cementum near the apices or in the furcation. Therefore, PTHrP-creER–marked DF cells include mesenchymal progenitors for PDL cells, cementoblasts on the acellular cementum, and alveolar cryptal osteoblasts in vivo.
PPR Deletion Causes a Cell Fate Shift of PTHrP+ DF Cells.
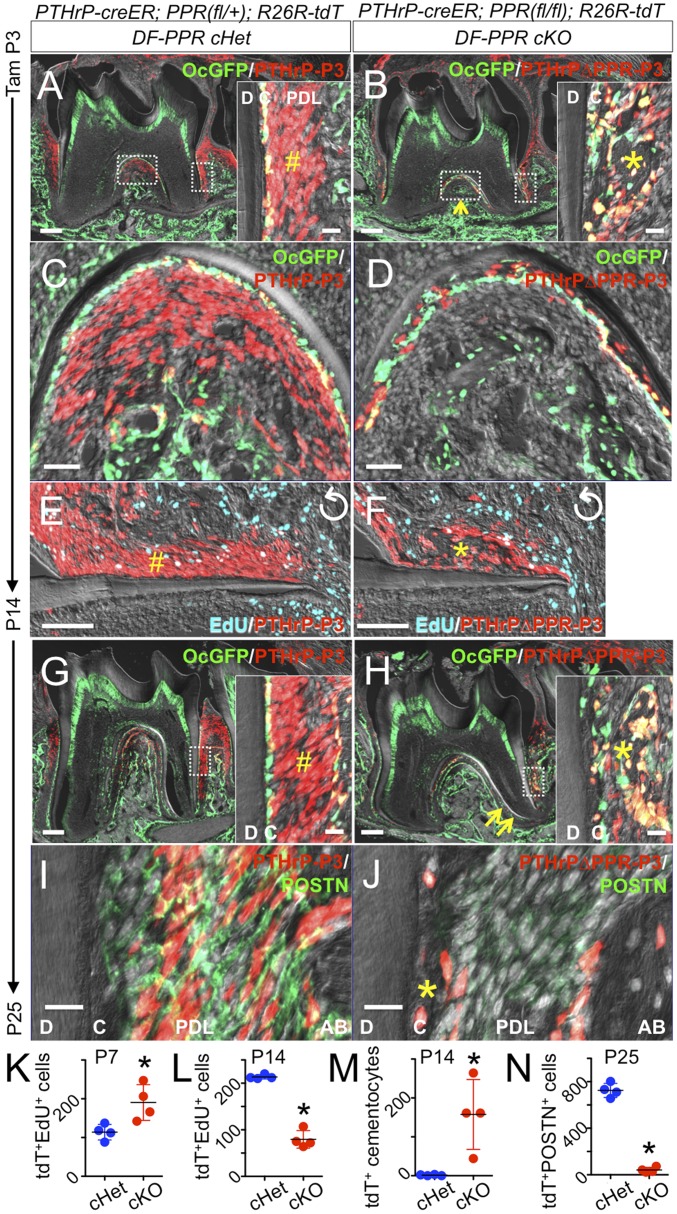
PTHrP+ DF cells express PPR (Fig. 1D) and PPR (SI Appendix, Fig. S5A). To test the hypothesis that PTHrP-PPR autocrine signaling regulates DF mesenchymal cells, we conditionally inactivated PPR using PTHrP-creER and PPR-floxed alleles at the onset of tooth root formation and simultaneously traced the fate of PPR-deficient DF mesenchymal progenitor cells using an R26R-tdTomato reporter allele. Littermate triple transgenic mice with two corresponding genotypes—DF-PPR cHet (PTHrP-creER; PPRfl/+; R26RtdTomato/+/PTHrPCEPPRHet-P3 cells) and DF-PPR cKO (PTHrP-creER; PPRfl/fl; R26RtdTomato/+/PTHrPCE∆PPR-P3 cells)—were pulsed with a single dose of tamoxifen (0.25 mg) at P3. Some of these mice also carried Oc-GFP as a marker of cementoblasts.
At P14, PTHrPCE∆PPR-P3 cells did not form the acellular cementum without becoming PDL cells, but rather differentiated prematurely into Oc-GFP+ cementoblast-like cells in the cellular cementum ectopically formed on the external root surface (Fig. 3B, asterisk and SI Appendix, Fig. S6). This was in sharp contrast to PTHrPCEPPRHet-P3 cells, which robustly contributed to cells in the periodontal space and on the acellular cementum (Fig. 3A, pound sign; quantification of tdTomato+-embedded cementocytes shown in Fig. 3M). PTHrPCE∆PPR-P3 cells also failed to differentiate into cryptal bone osteoblasts (Fig. 3B, arrow, C, and D). Proliferation of PTHrPCE∆PPR-P3 cells was transiently increased compared with that of PTHrPCEPPRHet-P3 cells after 4 d of chase at P7 (SI Appendix, Fig. S5 B–H) but was significantly decreased after 11 d of chase at P14 (Fig. 3 E and F; quantification of EdU+tdTomato+ cells shown in Fig. 3 K and L), indicating premature exhaustion of a progenitor cell pool.
Fig. 3.
PTHrP-PPR autocrine signaling orchestrates PDL differentiation of DF mesenchymal progenitor cells. Conditional deletion of PPR in PTHrP-creER+ DF cells. (A–F) P14. (Insets) Distal PDL space. The pound sign in A indicates tdTomato+ PDL cells. In B, the asterisk denotes the precocious cellular cementum-like structure, and the arrow denotes underdeveloped cryptal bones. In E and F, EdU was administered shortly before analysis. Perpendicularly rotated images. (G–J) P25. The pound sign in G indicates tdTomato+ PDL cells. In H, the asterisk denotes the cellular cementum-like structure, and the arrows indicate root dilacerations. (I and J) POSTN staining. The asterisk in J indicates the cellular cementum-like structure. C, cementum; D, dentin. (Scale bars: 200 µm, except 20 µm in Insets and I and J, 50 µm in C and D, and 100 µm in E and F.) (K–N) Quantification of tdTomato+EdU+ cells and tdTomato+ cells per 200-µm thickness. *P < 0.05, Mann–Whitney U test. All data are mean ± SD.
After 22 d of chase at P25, DF-PPR cKO molars were truncated and dilacerated, associated with hypercementosis on the external root surface (Fig. 3 G and H and SI Appendix, Fig. S6B). A majority of PTHrPCE∆PPR-P3 cells were associated with the precocious cellular cementum and expressed Oc-GFP, but these cells were scarce in the periodontal space or in the alveolar cryptal bone (∼11% of control cells and ∼7% of tdTomato+ cells) (Fig. 3H, asterisk in Inset). Interestingly, such hypercementosis was not evident in the furcation, indicating that PPR deficiency does not affect the cellular cementum. To investigate the ligament differentiation potential of PPR-deficient DF progenitor cells, we performed immunohistostaining for periostin (POSTN), a marker for PDL fibroblasts (16). PDL cells of DF-PPR cHet molars, including those descended from DF progenitor cells (PTHrPCEPPRHet-P3 cells), abundantly expressed POSTN (Fig. 3I). In contrast, only a small number of cells in the periodontal space of DF-PPR cKO molars expressed POSTN (Fig. 3J; quantification of POSTN+ tdTomato+ cells shown in Fig. 3N), indicating that PPR deficiency impaired PDL differentiation of DF progenitor cells. Therefore, PPR-deficiency in PTHrP+ DF mesenchymal progenitor cells causes a cell fate shift away from cementoblasts on the acellular cementum, PDL cells, and alveolar osteoblasts to cementoblast-like cells precociously forming the cellular cementum on the root surface, resulting in loss of the proper periodontal attachment apparatus.
We also performed tartrate-resistant acid phosphatase (TRAP) staining to ascertain whether alterations in osteoclasts accounted for the lack of eruption. No apparent difference was observed in terms of the location and the number of TRAP+ cells around molars before and after root formation and eruption (SI Appendix, Fig. S7 A–C), consistent with the previous study (5).
PPR Deletion in PTHrP+ DF Cells in Mice Recapitulates Human PFE Conditions.
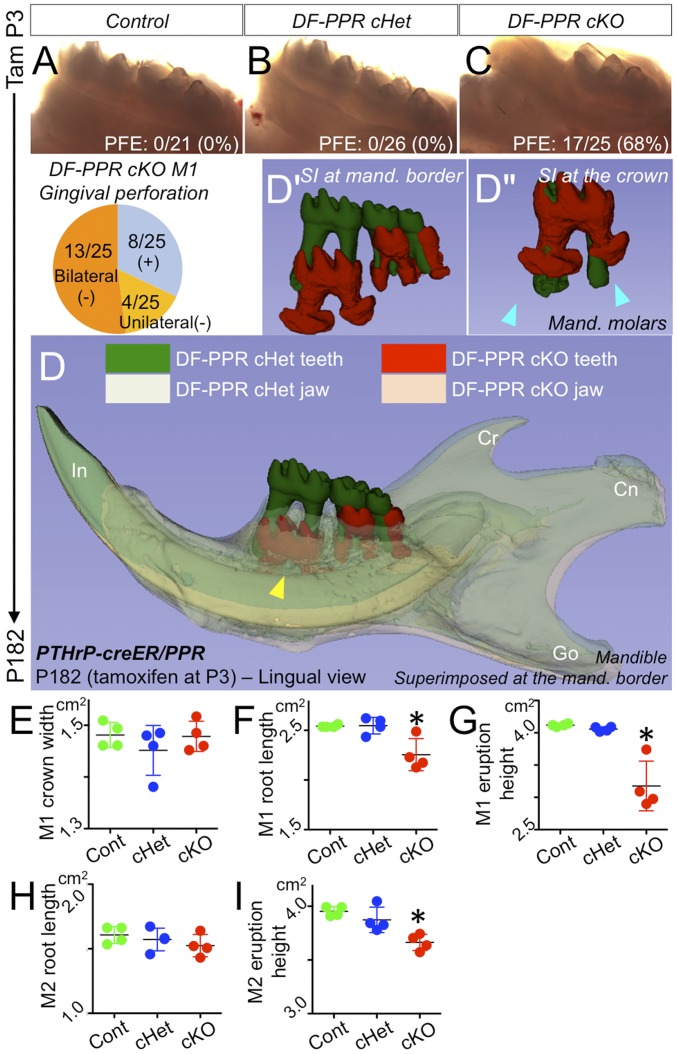
To investigate how the cell fate shift caused by PPR deficiency in PTHrP-creER+ DF progenitor cells leads to a phenotype in tooth eruption, we further chased P3-pulsed control (PPRfl/fl; R26RtdTomato/+), DF-PPR cHet (PTHrP-creER; PPRfl/+; R26RtdTomato/+), and DF-PPR cKO (PTHrP-creER; PPRfl/fl; R26RtdTomato/+) mice for an extended period of 6 mo. While control and DF-PPR cHet molars emerged and reached the occlusal plane normally, as expected (Fig. 4 A and B), a substantial fraction of DF-PPR cKO first molars were submerged below the gingiva (17/25; 68%), exhibiting a characteristic phenotype of primary failure of eruption (Fig. 4C). A majority of DF-PPR cKO first molars demonstrated bilateral involvement (13/25; 52%), whereas a smaller fraction of these molars showed unilateral involvement (4/25; 16%). These phenotypes in tooth eruption were essentially identical to those caused by PPR deficiency in Osx-creER+ dental mesenchymal cells (SI Appendix, Fig. S7 D–F). A substantial fraction of Osx-PPR cKO first molars exhibited characteristic phenotypes in primary of failure eruption (6/9, 67%) (SI Appendix, Fig. S7F). Therefore, PTHrP-creER+ DF cells are a functional subset of Osx-creER+ mesenchymal cells, in which PPR deficiency is sufficient to induce failure of molar eruption.
Fig. 4.
PPR deletion in PTHrP-creER+ DF cells recapitulates human primary failure of eruption conditions. (A–C) PFE phenotypes at 6 mo. (Pie chart) Incidence of failure of eruption (lack of gingival perforation). (D) 3D micro CT surface model overlays: superimposition of registered DF-PPR cHet and DF-PPR cKO mandibles. The yellow arrowhead indicates the first molar associated with pronounced PFE phenotypes and tooth root anomalies. Cn, condyle; Cr, coronoid process; Go, gonial angle; In, incisor. (D′ and D″) Composite 3D surface model overlay of the mandibular first molars. Blue arrowheads indicate truncation (short roots) associated with dilacerations (curved roots). (F–I) Quantitative 3D micro CT analysis. *P < 0.05, one-way ANOVA followed by the Mann–Whitney U test. All data are mean ± SD.
In addition, we closely analyzed four littermate mice with three corresponding DF-PPR genotypes with 3D mandibular registration on 3D micro computed tomography (CT) images. First, we superimposed the DF-PPR cHet and DF-PPR cKO mandibles using the mandibular border as a reference structure. Most strikingly, mandibular molars of the DF-PPR cKO, especially the first molar, were significantly undererupted compared with DF-PPR cHet molars (Fig. 4 D and D′), whereas other components of the mandible were almost identical between the two genotypes (Fig. 4D). We superimposed the DF-PPR cHet and DF-PPR cKO molars using the crown as a reference structure. DF-PPR cKO first molars exhibited pronounced tooth root anomalies compared with DF-PPR cHet counterparts, including dilacerations and truncation (Fig. 4D″, arrowheads). Quantitative 3D micro CT analyses revealed that although the crown width was not altered, the root length of DF-PPR cKO first molars (M1) was significantly shorter than that of the other two genotypes (Fig. 4 E and F). In addition, the eruption height of DF-PPR cKO first molars was significantly reduced compared with that of the other two genotypes (Fig. 4G). Interestingly, the root length of DF-PPR cKO second molars (M2) was not altered (Fig. 4H); however, their eruption height was significantly reduced compared with that of the other two genotypes (Fig. 4I), indicating that the failure of eruption is not attributed solely to the root truncation. No ankylosis was found in DF-PPR cKO mice, excluding the possibility that the root-bone fusion accounts for failure of tooth eruption (SI Appendix, Fig. S7G). Taken together, these findings demonstrate that PPR deletion in PTHrP+ DF mesenchymal progenitor cells results in tooth eruption and root phenotypes that resemble human PFE conditions.
Accelerated Differentiation and Precocious Matrix Production in PPR-Deficient PTHrP+ DF Cells.
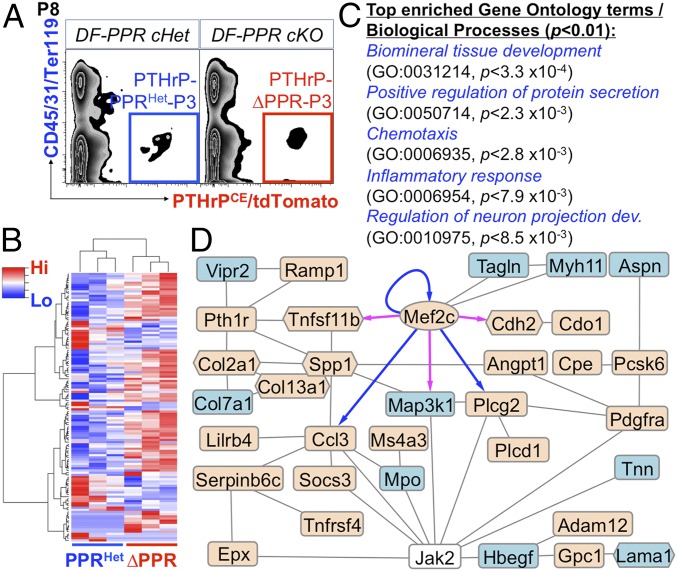
We further performed an RNA-seq analysis to identify how defective PTHrP-PPR autocrine signaling alters the gene expression profiles of DF mesenchymal progenitor cells, We isolated tdTomato+ DF cells at P8 from DF-PPR cHet (PTHrP-creER; PPRfl/+; R26RtdTomato/+/PTHrPCEPPRHet-P3 cells) and DF-PPR cKO (PTHrP-creER; PPRfl/fl; R26RtdTomato/+/PTHrPCE∆PPR-P3 cells) molars (tamoxifen pulse at P3) using FACS (Fig. 5A). Harvested RNAs were further amplified and subjected to deep sequencing (SI Appendix, Fig. S8 A and B). An unsupervised clustering analysis revealed that PTHrPCEPPRHet-P3 and PTHrPCE∆PPR-P3 triplicate samples each clustered together, suggesting that PPR deficiency induced a consistent change in the transcriptomes of DF mesenchymal progenitor cells (Fig. 5B). A close examination on the Pth1r exons revealed that a significant reduction of reads was observed only in the second exon of PTHrPCE∆PPR-P3 samples (∼80% reduction), which is flanked by loxP sites in the PPR floxed allele (SI Appendix, Fig. S8C), confirming the significant loss of functional PPR genes in these samples.
Fig. 5.
RNA-seq analysis of PPR-deficient PTHrP-creER+ DF mesenchymal progenitor cells. (A) FACS-sorting of PTHrP-creER+ DF cells. Blue box, PTHrP-PPRHet-P3 cells (cHet); red box, PTHrP-∆PPR-P3 cells (cKO). (B–D) RNA-seq analysis of DF cells. (B) Heatmap of 122 DEGs with hierarchical clustering. Red indicates higher expression; blue, lower expression. (C) GO terms/biological processes overrepresented in DEGs (P[elim] < 0.01; minimum number of DEGs, 6). (D) STRING, Cytoscape, and Genomatix network and transcription factor analysis of DEGs. Oval denotes transcription factor; hexagons, matrix genes. Beige indicates up-regulated genes; blue, down-regulated genes. Blue arrows indicate validated targets; magenta arrows, predicted targets.
Analyses of differentially expressed genes (DEGs) revealed that 122 genes were differentially expressed between the two groups (fold change ≥ ±1.5), of which 84 genes were up-regulated in PTHrPCE∆PPR-P3 cells (Fig. 5B and SI Appendix, Fig. S8D). Gene Ontology (GO) enrichment analysis of DEGs revealed significant enrichment of several GO terms, including biomineral tissue development (GO:0031214; adjusted P < 3.3 × 10−4) and positive regulation of protein secretion (GO:0050714; P < 2.3 × 10−3) (Fig. 5C and SI Appendix, Fig. S8E). Several genes encoding regulators of mineralization were up-regulated in PTHrPCE∆PPR-P3 cells, including dentin matrix acidic phosphoprotein 1 (Dmp1), secreted phosphoprotein 1 (Spp1), and IFN-induced transmembrane protein 5 (Ifitm5), suggesting that PPR deficiency accelerates osteoblast/cementoblast differentiation of DF mesenchymal progenitor cells. In addition, myocyte enhancer factor 2c (Mef2c), a transcription factor putatively located downstream of the PPR-Gsα-cAMP-protein kinase A pathway (17), was up-regulated in PTHrPCE∆PPR-P3 cells. A network analysis of DEGs based on the STRING and Cytoscape software connected 34 genes with an integration of one non-DEG (Jak2), spanning a wide variety of genes involved in extracellular matrix organization and cytokine regulation (Fig. 5D). Further transcription factor analysis based on the Genomatix software identified that MEF2C has validated or predicted binding sites with several DEGs, including Mef2c, Ccl3, Plcg2, Tnfrsf11b, Map3k1, and Cdh2 (Fig. 5D), supporting the idea that MEF2C may function as the linchpin for the phenotypes caused by PPR deficiency in DF progenitor cells. Therefore, these RNA-seq analyses revealed that loss of PTHrP-PPR autocrine signaling induced accelerated differentiation and precocious matrix production, presumably through defective Mef2c autoregulation.
We then set out to confirm these RNA-seq analysis results using an independent approach. RNAscope analysis revealed that Mef2c was significantly up-regulated in DF cells of DF-PPR cKO molars (SI Appendix, Fig. S9C). Cd200 encodes OX-2 membrane glycoprotein CD200, a cell surface protein expressed by transplantable skeletal stem cells (18). Flow cytometry analysis revealed that CD200 was significantly up-regulated in PTHrPCE∆PPR-P3 cells compared with PTHrPCEPPRHet-P3 cells, as measured by fluorescence intensity (SI Appendix, Fig. S9 A and B). Therefore, the transcriptome change identified in our RNA-seq analysis is translated into the change in mRNA and protein expression levels. Interestingly, flow cytometry analysis of DF cells dissociated from Col1(2.3kb)-GFP; PTHrP-creER; R26R-tdTomato molars at P5 (tamoxifen pulse at P3) with CD200 staining revealed approximately 20-fold greater CD200 expression in Col1(2.3kb)-GFP+ cells compared with tdTomato+ cells (SI Appendix, Fig. S10A). Histologically, CD200 was abundantly expressed by matrix-producing osteoblasts on the bone surface and mesenchymal cells among the trabeculae, but not by odontoblasts (SI Appendix, Fig. S10B).
To further define how CD200 expression develops during differentiation of DF cells, we analyzed cells dissociated from P3-pulsed PTHrP-creER; R26R-tdTomato molars carrying Col1(2.3kb)-GFP or Oc-GFP reporters sequentially after the pulse. The tdTomato+ cells maintained an intermediate level of CD200 expression relatively homogenously at P5 and P8, when no root developed (SI Appendix, Fig. S10C), but demonstrated a polarized CD200 expression profile at P18, when the root developed, bifurcating into CD200high and CD200low populations (SI Appendix, Fig. S10 C–E). Approximately one-half of the CD200high cells expressed Col1(2.3kb)-GFP or Oc-GFP (47.5 ± 8.9% or 46.9 ± 11.2% of CD200high cells, respectively). In contrast, while 35.2 ± 0.1% of CD200low cells expressed Col1(2.3kb)-GFP, only 5.9 ± 3.2% of these cells expressed Oc-GFP (SI Appendix, Fig. S10C and SI Appendix, Fig. S10 D and E). Therefore, PTHrP-creER/tdTomato+ DF cells differentiate into CD200high cementoblasts and osteoblasts and CD200lowPDL fibroblasts during tooth root formation and eruption.
Taken together, these findings indicate that the PTHrP-PPR autocrine signaling suppresses premature up-regulation of CD200 and other bone/cementum matrix proteins in DF progenitor cells by suppressing Mef2c expression, thereby preventing their precocious differentiation into cementoblasts forming the cellular cementum, and also maintains ligament cell fates and acellular cementum formation (SI Appendix, Fig. S11).
Discussion
Collectively, our data indicate that PTHrP-PPR autocrine regulation of DF mesenchymal progenitor cells is essential to maintaining their physiological cell fates and proper formation of the acellular cementum, the PDL, and the cryptal bone, which constitute the functional tooth–bone interface. A defect in this autoregulatory loop causes a cell fate shift, wherein DF progenitor cells choose a nonphysiological cell fate by becoming cementoblast-like cells forming the cellular cementum while failing to become ligament cells and cryptal bone osteoblasts that establish the periodontal attachment apparatus. This pathological differentiation is associated with up-regulation of bone/cementum matrix protein and a key transcription factor, Mef2c. In addition, loss of responsiveness to PTHrP released from the root surface obliterates a ligament cell pool, preventing proper insertion of periodontal fibers into the cementum, as well as a supply for cryptal bone osteoblasts, resulting in significant loss of functionality in the periodontal attachment apparatus. Indirect interactions between PTHrP+ cells and cryptal bone osteocytes may also contribute to this phenotype, suggesting that PTHrP-based regulation might not be entirely autocrine.
How cementoblasts on the acellular cementum differ fundamentally from those on the cellular cementum remains unknown. PTHrP+ DF cells appear to play specific roles in formation of “acellular” cementoblasts and absolutely require PTHrP-PPR signaling in their proper differentiation. In contrast, “cellular” cementoblasts do not require PTHrP-PPR signaling and may be composed of a heterogeneous group of cells with diverse origins, including dental epithelial cells from the root sheath (19), PTHrP− DF cells, and dental papilla mesenchymal cells. Our findings identify the DF derivative trio of the acellular cementum, PDL, and cryptal bone as essential to establishing the functional periodontal attachment apparatus and proper tooth eruption.
Our mouse genetic study highlights several possible mechanisms of failure of tooth eruption. Deficient PTHrP-PPR signaling may render the DF “nonfunctional,” supporting the classical Marks and Cahill theory (20, 21). Root dilacerations may also contribute indirectly by mechanically preventing proper tooth eruption. Our data collectively indicate that failure of molar eruption observed in human genetic conditions due to PPR mutations may occur through a combination of these mechanisms.
Methods
Mice.
PTHrP-mCherry/null knockin, PTHrP-creER BAC (3), PPR-floxed, Col1a1(2.3kb)-GFP (JAX013134), Oc-GFP, and Osx-creER mice have been described elsewhere. Rosa26-CAG-loxP-stop-loxP-tdTomato (Ai14: R26R-tdTomato; JAX007914) mice were acquired from The Jackson Laboratory. All procedures were conducted in compliance with the Guidelines for the Care and Use of Laboratory Animals approved by the University of Michigan’s Institutional Animal Care and Use Committee, protocol 7000 (Ono). All mice were housed in a specific pathogen-free conditions and analyzed in a mixed background. Tamoxifen (T5648; Sigma-Aldrich) was dissolved in sunflower seed oil (S5007; Sigma-Aldrich) and injected i.p. into P3 mice. Mice were euthanized by overdosage of CO2 or decapitation under inhalation anesthesia in a drop jar (isoflurane USP; Fluriso; VetOne).
Three-Dimensional Micro Computed Tomography Analysis of Mouse Samples.
Mandibles including molars and incisors were placed in a 19-mm diameter specimen holder and scanned using a micro CT system (µCT100; Scanco Medical). Scan settings were as follows: voxel size, 12 µm; 70 kVp; 114 µA; 0.5-mm AL filter; and integration time, 500 ms.
Histology, Flow Cytometry, and FACS Analyses.
Frozen sections at 16 µm were analyzed using a fluorescence microscope (Zeiss Axio Observer Z1 with the ApoTome.2 system). Enzymatically digested DF cells were analyzed using a five-laser LSR Fortessa cell analyzer (BD Biosciences) or sorted using a five-laser FACSAriaIII (BD Biosciences) or a six-laser Synergy SY3200 (Sony) high-speed cell sorter with a 100-µm nozzle. More details are provided in SI Appendix, Methods.
Single-Cell and Bulk RNA-Seq Analysis.
For single-cell RNA-seq analysis, FACS-sorted cells were loaded onto the Chromium Single-Cell Gene Expression Solution platform (10X Genomics). For bulk RNA-seq analysis, complementary DNAs were prepared using the SMART-Seq v4 Ultra-Low-Input RNA Kit for Sequencing (Clontech) using 150–800 pg of total RNA. cDNA libraries were sequenced using an Illumina HiSeq 4000 system. More details are provided in SI Appendix, Methods. The data presented herein have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus under accession nos. GSE117936 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE117936) and GSE120108 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE120108).
Statistical Analysis.
Results are presented as mean ± SD. Statistical analysis were done using one-way ANOVA followed by a post hoc test or Mann–Whitney U test. P < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank R. Tagett (University of Michigan Bioinformatics Core) for assistance with bioinformatics analysis. This study was supported by National Institutes of Health Grants R03 DE027421 (to W.O.) and R01 DE026666 (to N.O.) and an American Association of Orthodontists Foundation postdoctoral research award (to W.O.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Microarray data are available at Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession nos. GSE117936 and GSE120108).
See Commentary on page 353.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810200115/-/DCSupplemental.
References
- 1.Ono N, Kronenberg HM. Bone repair and stem cells. Curr Opin Genet Dev. 2016;40:103–107. doi: 10.1016/j.gde.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sacchetti B, et al. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131:324–336. doi: 10.1016/j.cell.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 3.Mizuhashi K, et al. Resting zone of the growth plate houses a unique class of skeletal stem cells. Nature. 2018;563:254–258. doi: 10.1038/s41586-018-0662-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao H, et al. The suture provides a niche for mesenchymal stem cells of craniofacial bones. Nat Cell Biol. 2015;17:386–396. doi: 10.1038/ncb3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Philbrick WM, Dreyer BE, Nakchbandi IA, Karaplis AC. Parathyroid hormone-related protein is required for tooth eruption. Proc Natl Acad Sci USA. 1998;95:11846–11851. doi: 10.1073/pnas.95.20.11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ono W, Sakagami N, Nishimori S, Ono N, Kronenberg HM. Parathyroid hormone receptor signalling in osterix-expressing mesenchymal progenitors is essential for tooth root formation. Nat Commun. 2016;7:11277. doi: 10.1038/ncomms11277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frazier-Bowers SA, Simmons D, Wright JT, Proffit WR, Ackerman JL. Primary failure of eruption and PTH1R: The importance of a genetic diagnosis for orthodontic treatment planning. Am J Orthod Dentofacial Orthop. 2010;137:160.e1–160.e7; discussion 160–161. doi: 10.1016/j.ajodo.2009.10.019. [DOI] [PubMed] [Google Scholar]
- 8.Decker E, et al. PTHR1 loss-of-function mutations in familial, nonsyndromic primary failure of tooth eruption. Am J Hum Genet. 2008;83:781–786. doi: 10.1016/j.ajhg.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamaguchi T, et al. Exome resequencing combined with linkage analysis identifies novel PTH1R variants in primary failure of tooth eruption in Japanese. J Bone Miner Res. 2011;26:1655–1661. doi: 10.1002/jbmr.385. [DOI] [PubMed] [Google Scholar]
- 10.Risom L, et al. Identification of six novel PTH1R mutations in families with a history of primary failure of tooth eruption. PLoS One. 2013;8:e74601. doi: 10.1371/journal.pone.0074601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jelani M, et al. A novel homozygous PTH1R variant identified through whole-exome sequencing further expands the clinical spectrum of primary failure of tooth eruption in a consanguineous Saudi family. Arch Oral Biol. 2016;67:28–33. doi: 10.1016/j.archoralbio.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 12.Chen X, et al. Initial characterization of PTH-related protein gene-driven lacZ expression in the mouse. J Bone Miner Res. 2006;21:113–123. doi: 10.1359/JBMR.051005. [DOI] [PubMed] [Google Scholar]
- 13.Nishida E, et al. Transcriptome database KK-periome for periodontal ligament development: Expression profiles of the extracellular matrix genes. Gene. 2007;404:70–79. doi: 10.1016/j.gene.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 14.Koda N, et al. The transcription factor mohawk homeobox regulates homeostasis of the periodontal ligament. Development. 2017;144:313–320. doi: 10.1242/dev.135798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Dijk D, et al. Recovering gene interactions from single-cell data using data diffusion. Cell. 2018;174:716–729.e27. doi: 10.1016/j.cell.2018.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kruzynska-Frejtag A, et al. Periostin is expressed within the developing teeth at the sites of epithelial-mesenchymal interaction. Dev Dyn. 2004;229:857–868. doi: 10.1002/dvdy.10453. [DOI] [PubMed] [Google Scholar]
- 17.Kozhemyakina E, Cohen T, Yao TP, Lassar AB. Parathyroid hormone-related peptide represses chondrocyte hypertrophy through a protein phosphatase 2A/histone deacetylase 4/MEF2 pathway. Mol Cell Biol. 2009;29:5751–5762. doi: 10.1128/MCB.00415-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan CK, et al. Identification and specification of the mouse skeletal stem cell. Cell. 2015;160:285–298. doi: 10.1016/j.cell.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang X, Bringas P, Jr, Slavkin HC, Chai Y. Fate of HERS during tooth root development. Dev Biol. 2009;334:22–30. doi: 10.1016/j.ydbio.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cahill DR, Marks SC., Jr Tooth eruption: Evidence for the central role of the dental follicle. J Oral Pathol. 1980;9:189–200. doi: 10.1111/j.1600-0714.1980.tb00377.x. [DOI] [PubMed] [Google Scholar]
- 21.Wang XP. Tooth eruption without roots. J Dent Res. 2013;92:212–214. doi: 10.1177/0022034512474469. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.