Significance
Acute myeloid leukemia (AML) is a heterogeneous disease commonly treated with azacitidine (AZA), but additional therapeutic strategy is needed. We found that AZA treatment in four AML cell lines had diverse effects at the individual gene and protein level, and these changes converged to common responses at the pathway level. The most prominent responses were the down-regulation of metabolism and up-regulation of immune defense. Given the heterogeneous responses in the four cell lines, we discuss serval potential therapeutic strategies for AML in combinations with AZA. In addition, the surface proteomics experiment has identified the greatest number of surface proteins for these cell lines to date and represents a valuable resource to others who use these cell lines as AML models.
Keywords: AML, azacitidine, target discovery, multiomics, surface proteomics
Abstract
Myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML) are diseases of abnormal hematopoietic differentiation with aberrant epigenetic alterations. Azacitidine (AZA) is a DNA methyltransferase inhibitor widely used to treat MDS and AML, yet the impact of AZA on the cell-surface proteome has not been defined. To identify potential therapeutic targets for use in combination with AZA in AML patients, we investigated the effects of AZA treatment on four AML cell lines representing different stages of differentiation. The effect of AZA treatment on these cell lines was characterized at three levels: the DNA methylome, the transcriptome, and the cell-surface proteome. Untreated AML cell lines showed substantial overlap at all three omics levels; however, while AZA treatment globally reduced DNA methylation in all cell lines, changes in the transcriptome and surface proteome were subtle and differed among the cell lines. Transcriptome analysis identified five commonly up-regulated coding genes upon AZA treatment in all four cell lines, TRPM4 being the only gene encoding a surface protein, and surface proteome analysis found no commonly regulated proteins. Gene set enrichment analysis of differentially regulated RNA and surface proteins showed a decrease in metabolic pathways and an increase in immune defense response pathways. As such, AZA treatment led to diverse effects at the individual gene and protein levels but converged to common responses at the pathway level. Given the heterogeneous responses in the four cell lines, we discuss potential therapeutic strategies for AML in combination with AZA.
Myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML) are hematopoietic malignancies that are genetically and epigenetically diverse in nature. As myeloid lineage cells differentiate from their hematopoietic stem/progenitor cells, aberrant epigenetic changes can occur at any differentiation stage, driving cells into cancerous phenotypes (1). As such, AML is routinely classified according to hematopoietic lineages by cell morphology or by cytometry using sparse surface markers (2, 3). Among many epigenetic changes that occur in MDS and AML, the best-characterized change is the DNA methylation of cytosine bases in CpG islands (4). In fact, a hallmark of epigenetic changes in AML is the redistribution of methylated CpG dinucleotides with loss of methylation across intergenic regions, primarily transposable elements and repeats, and gain of aberrant methylation near the promoters of a number of genes, including well-known tumor suppressors such as p16INK4a (5). As such, it is believed that these diseases are more sensitive to hypomethylating agents such as DNA methyltransferase inhibitors (DMNTi) (6, 7). One such DMNTi, azacitidine (AZA), has been efficaciously used for over a decade to treat MDS and AML (8, 9). At high doses, AZA induces rapid DNA damage and is cytotoxic; at lower doses, AZA induces DNA hypomethylation by covalent trapping and degradation of DNA methyltransferases, leading to loss of methylation in newly synthesized DNA (10, 11). It was recently shown that AZA treatment of cervical (12, 13) and colorectal (14) cancer cells can induce interferon responses through reactivation of endogenous retroviruses. This phenomenon, termed viral mimicry, is thought to induce antitumor effects by activating and engaging the immune system.
Although AZA treatment has demonstrated clinical benefit in AML patients, additional therapeutic options are needed (8, 9). Our group has recently generated antibodies toward potential targets in RAS-driven cancers, and there is significant interest in identifying surface protein targets for antibody-derived therapeutic strategies in combination with AZA for the treatment of AML (15). Currently, there are numerous antibody-based therapeutics in development for AML patients, targeting about a dozen cell-surface proteins, but it is not clear if AZA changes the expression levels of these proteins (16). Furthermore, over 15 ongoing clinical trials are investigating the combination of AZA and checkpoint inhibitors in various leukemias and solid tumors, since AZA induces checkpoint inhibitory molecules on both tumor and immune cells (7, 17). To identify cell-surface markers, cell-surface capture proteomics has recently emerged as a highly sensitive target discovery technology and has been used to define a large number of common and distinct markers in AML (18–20). Taken together, a broader understanding of how AZA treatment remodels the cell-surface proteome in AML cells could aid in identifying surface protein targets for antibody-based therapy, leading to unique immunotherapies for use in combination with AZA.
Using a multiomics approach, we characterized four AML cell lines, representing different stages of differentiation, and studied the changes in DNA methylation, RNA expression, and surface proteome induced by AZA treatment. Across the four cell lines, AZA reduced DNA methylation in nearly all of the hypermethylated CpG sites probed, but surprisingly the changes in gene expression and surface protein expression were few and diverse. Transcriptome analysis identified only one gene encoding a surface protein that is commonly up-regulated in all four cell lines, and surface proteomics analysis did not identify any commonly regulated proteins. Despite little overlap, functional analysis revealed some common responses among the four cell lines—down-regulation of genes and proteins in metabolism and up-regulation of genes in immune response. Collectively, our study detailed the distinct impact of AZA treatment in four AML cell line at the individual gene level and illustrated that functional networks are commonly regulated.
Results
Methylome in AML Cells and its Regulation by AZA.
Four well-characterized AML cell lines, KG1a, HL60, HNT34, and AML193, were chosen to reflect a gradient of differentiation stages along the myeloid lineage, according to the French–American–British (FAB) classification system (SI Appendix, Rationale for Cell Line). The four cell lines exhibited varied mutation profiles; PHF6 (PHD Finger Protein 6) was the only gene mutated among all four cell lines, while genes like TET1 (Tet Methylcytosine Dioxygenase 1), DNMT3B (DNA Methyltransferase 3B), and NRAS (NRAS Proto-Oncogene) showed distinct mutation patterns in the four cell lines (Dataset S1).
We determined the DNA methylome of each cell line using the Illumina Infinium EPIC array. The baseline DNA methylation profile for each cell line exhibited a bimodal distribution, representing hypermethylated and hypomethylated CpG sites (Fig. 1A). The number of hypermethylated sites (beta value >0.8) was highest for AML193 and lowest for HL60, following a general trend of increasing hypermethylation from the least to most differentiated cell line (Fig. 1B). KG1a does not follow this trend of increasing baseline hypermethylation, potentially due to its differentiation from the KG1 parental cell line and a deviation from the annotated early progenitor lineage. Comparing the methylation status across the four cell lines, 55% (342,320/628,240) of loci were commonly hypermethylated (beta value >0.8) and 39% (108,968/278,579) of loci were commonly hypomethylated (beta value <0.2) (Fig. 1B). This indicates a high degree of similarity in the methylomes among the four cell lines.
Fig. 1.
AZA treatment drives global DNA demethylation among all four AML cell lines. (A) Vehicle-treated cell lines have a bimodal distribution of genome-wide beta values (kernel density estimation). (B) Vehicle-treated cells share a high proportion of hypermethylated and hypomethylated sites. Overlapping hypermethylated sites (red, beta values >0.8) and hypomethylated sites (blue, beta values <0.2) are indicated by upward and downward bars, respectively, in the vertical bar graph. The specific overlapping groups are indicated by the black solid points below the bar graph. Total hypermethylated and hypomethylated sites found in each cell line are indicated in the horizontal bar graph. (C) AZA-treated cells have decreased hypermethylated beta values indicating DNA demethylation. (D) DNA demethylation in AZA-treated cells is shown by median change in beta value for KG1a (−0.097), HL60 (−0.28), HNT34 (−0.132), and AML193 (−0.099). (E) A high proportion of demethylated sites are common among the four cell lines, indicated by downward bars in the vertical bar graph (decrease in beta value >0.1, false discovery rate adjusted P < 0.05).
To study the effects of AZA treatment on the regulation of DNA methylation, each of the four cell lines was treated with AZA (0.5 M) for 3 d, followed by a 4-d drug holiday to maximally reduce DNA methylation (SI Appendix, Fig. S1A). With this treatment regimen, AZA inhibited cell growth to varying degrees in the AML cell lines, with ∼85% growth inhibition in AML193 cells (most sensitive) and ∼30% inhibition in HL60 cells (least sensitive) (SI Appendix, Table S1). Remarkably, AZA treatment reduced methylation in nearly all of the hypermethylated sites probed (Fig. 1 C and D and SI Appendix, Fig. S2). The median change in methylation across all CpG sites ranged from −0.097 for KG1a cells to −0.28 for HL60 cells (Fig. 1D). The greater reduction of DNA methylation seen in HL60 compared with the other three cell lines could be due to a lower basal expression of the de novo DNA methyltransferases (DNMT3A and DNMT3B) in the HL60 cells, as detected in the transcriptome data (SI Appendix, Fig. S3). Comparing the effects of AZA across all four cell lines, a large proportion of CpG sites with reduced DNA methylation were shared by all four cell lines (37% or 247,715 shared out of 657,868 total sites probed) (Fig. 1E). The loci with reduced methylation induced by AZA were uniformly distributed across the genome (SI Appendix, Fig. S4). Further gene set enrichment analysis (GSEA) of the AZA-regulated DNA methylation loci did not identify any functionally enriched gene sets.
Transcriptome in AML Cell Lines and Its Regulation by AZA.
As DNA methylation at promoter CpG islands can be associated with transcriptional silencing, we next assessed RNA expression profiles of the four cell lines. Using an approach similar to comparing baseline DNA methylation status, highly and lowly expressed genes were defined by the expression levels from the highest and lowest tertiles in each cell line. At baseline, 53% (10,971 out of 20,517 total) of all high- and low-expressing genes were common to all four cell lines (Fig. 2A). Despite a shared gene expression profile, functional analysis using GSVA with a hallmark set of key pathways indicated that each AML cell line had a distinct biological state. Specifically, cell-cycle-related genes were highly expressed in AML193 and KG1a cells, MYC pathway genes in KG1a and HL60 cells, and TGF-, WNT, and Notch signaling genes in HNT34 cells (Fig. 2B). Similarly, GSVA using an expanded gene set list of Gene Ontology (GO) term analysis also revealed differences in differentiation and cell death activation pathways (SI Appendix, Fig. S5).
Fig. 2.
AZA treatment induces unique and subtle transcriptome changes in the four AML cell lines. (A) Gene expression profiles are similar among the four cell lines at baseline. Overlapping highly (top tertile) and lowly (bottom tertile) expressed genes are indicated by upward (red) and downward (blue) bars, respectively, in the vertical bar graph (n = 20,517 unique genes mapped from >54,000 probesets). (B) Distinct biological states shown by gene set variation analysis (GSVA) of the four cell lines at baseline levels using the MSigDB hallmark gene sets (variation score >0.2 in at least one cell line). (C) Most differentially regulated genes induced by AZA are unique to each cell line, and only six genes are common among the four cell lines. Up-regulated genes and down-regulated genes are indicated by the upward (red) and downward (blue) bars, respectively (fold change >2 and adjusted P value <0.05). Total differentially regulated genes for each cell line are indicated in the horizontal bar graph. (D) Common biological processes are enriched upon AZA treatment. Gene set enrichment analysis (GSEA) was performed using Hallmark (H), Reactome (R), and KEGG (K) pathways (GSEA normalized effect size >1 or <−1).
Upon AZA treatment, only a small percentage (∼0.8 to 2.4% of all genes) of the transcriptome was differentially regulated in each AML cell line, despite the large changes observed in the DNA methylome. Hierarchical clustering of the top variable gene expression profiles showed clustering by cell line and not by AZA treatment, suggesting that AZA did not have a dominant effect on the transcriptome (SI Appendix, Fig. S6). The number of differentially regulated genes varied from ∼160 genes for KG1a cells to ∼500 genes for HL60 and HNT34 cells (Fig. 2C). The changes in gene expression in each cell line (∼0.8 to 2.4%) were much lower compared with those observed for DNA methylation (45 to 70% of all CpG sites were significantly demethylated). The effects on gene expression were also much more stochastic and appeared to be divergent among the cell lines, with only five coding and one noncoding gene uniformly up-regulated in all four cell lines (Fig. 2C). The commonly up-regulated coding genes were TRPM4 (Transient Receptor Potential Cation Channel subfamily M member 4), PPBP (Pro-Platelet Basic Protein), MAGEB2 (MAGE family member B2), CREB5 (cAMP Responsive Element Binding Protein 5), and GCNA (Germ Cell Nuclear Acidic Peptidase). The noncoding gene LINC01088 found to be up-regulated in all four cell lines was recently implicated as a tumor suppressor found in reduced levels in ovarian tumors and acts by targeting miR-24-1-5p–mediated regulation of PAK4 expression (21). Despite the unique gene regulation in each cell line, functional-level analysis with GSEA showed several common pathways regulated by AZA in all four cell lines (Fig. 2D and SI Appendix, Fig. S7 A and B). The enriched gene sets included increased expression of epithelial mesenchymal transition, apoptosis, coagulation, complement, interferon gamma response, hemostasis, platelet activation signaling and aggregation, and decreased expression of tRNA aminoacylation and amino acid synthesis. In general, these gene sets represent activation of immune response and repression of metabolism.
Unique Cell-Surface Proteome Regulation by AZA.
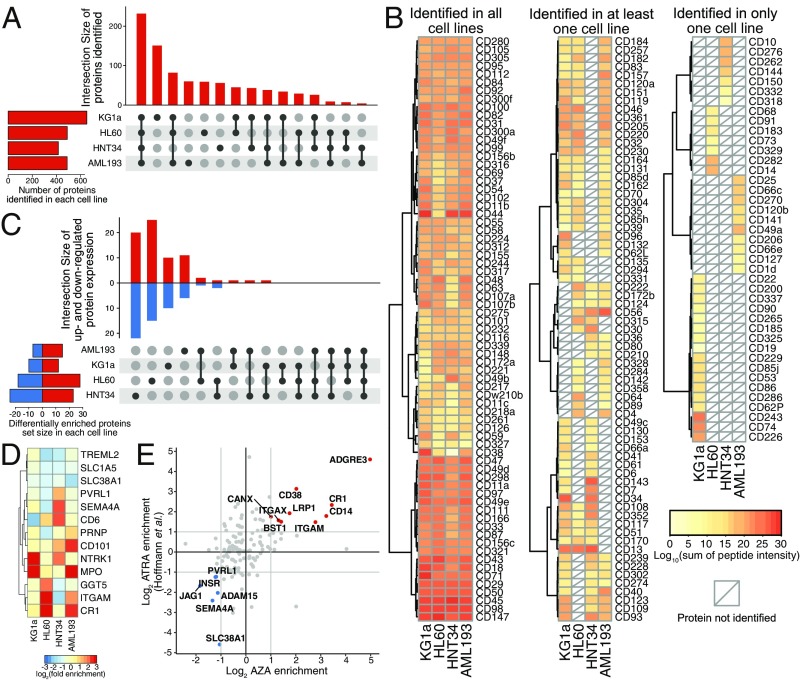
In an effort to identify novel therapeutic targets induced by AZA, we probed the surface proteomes of the four cell lines using a modified cell-surface capture of N-linked glycosylated proteins protocol and quantified the changes induced by AZA treatment using stable isotope labeling by amino acids in cell culture (SILAC) (22, 23) (SI Appendix, Fig. S1B). Each experiment was performed in both the forward and reverse SILAC mode, and pairwise comparisons of the enrichment ratios for the biological replicates showed good reproducibility (SI Appendix, Fig. S8 A and B). To compare the baseline surface protein profiles among the four cell lines, we extrapolated a subset of proteomics data from heavy-labeled vehicle-treated cells (Dataset S2). At baseline, a total of 875 unique surface membrane proteins were identified among the four cell lines, and a common set of 232 proteins were detected in all four cell lines (Fig. 3A). Historically, classification of cell lineage is dependent on staining of CD markers on cells. Here, we detected an extensive number of CD markers expressed on the four cell lines and reported an estimate of protein abundance for these markers (Fig. 3B). Among the common CD markers identified were several therapeutic targets of AML, such as CD33 (SIGLEC-3) and CD47 (integrin associated protein) (16). Comparison between our proteomics and transcriptome data to known immunophenotyping CD markers of the four cell lines also showed remarkable agreement (SI Appendix, Table S2).
Fig. 3.
Surface proteome changes induced by AZA treatment in the four AML cell lines. (A) Surface proteins identified in the four vehicle-treated AML cell lines. Overlapping proteins identified are indicated in the vertical bar graph and the specific overlapping groups are indicated by the black solid points below the bar graph. Total surface proteins identified in each cell line are indicated in the horizontal bar graph. (B) CD markers identified by surface proteomics in vehicle-treated sample. The heat map is shaded from yellow to red to reflect estimated abundance (logarithmic sum of peptide intensities for each protein). (C) AZA induced unique changes on the cell-surface proteome. Overlapping up-regulated and down-regulated proteins are indicated by upward and downward bars, respectively, in the vertical bar graph [median stable isotope labeling by amino acids in cell culture (SILAC) ratio >2 or <2, P value <0.05]. The specific overlapping groups are indicated by the black solid points below the bar graph. Total differentially regulated surface proteins for each cell line are indicated in the horizontal bar graph showing variable surface proteome regulation by AZA. No commonly regulated protein was identified among the AZA-treated cell lines. (D) Proteins with significant changes in at least two cell lines are shown to illustrate distinct regulation of surface proteins by AZA (n = 13). (E) Comparison of surface proteomics data between AZA treatment and all-trans retinoic acid (ATRA) treatment in HL60 cells. Pearson correlation between the two datasets is 0.44. Data for ATRA treatment in HL60 was obtained from Hofmann et al. (18).
We next explored how AZA treatment affected the surface expression of membrane proteins using SILAC quantification (SI Appendix, Fig. S1A). The number of differentially regulated proteins ranged from 22 to 47, or 5 to 10% across the cell lines, and no protein was commonly regulated upon AZA treatment (Fig. 3C). In fact, the majority of the changes were unique to each cell line, and only 13 proteins were significantly differentially regulated in at least two cell lines (Fig. 3D). Some proteins, such as CR1 (Complement C3b/C4b Receptor 1), ITGAM (Integrin Subunit Alpha M), and MPO (Myeloperoxidase), appeared to be generally up-regulated by AZA treatment, but this was statistical significance in only two or three cell lines. Western blot of the regulation of ITGAM (CD11b), a common marker of neutrophil/monocyte differentiation, was consistent with SILAC quantification (SI Appendix, Fig. S9). Specifically, expression of ITGAM was up-regulated in HL60 and AML193 cells, did not change in KG1a, and was down-regulated in HNT34. Together, the surface proteomics analysis, similar to gene expression analysis, suggests that the effects of AZA are largely dependent on the inherent differences among the AML cell lines. Hierarchical clustering of significantly enriched protein expression showed a distinctive protein regulation profile in each cell line (SI Appendix, Fig. S8C). Further functional analysis using GSEA with GO terms indicated an increase in immune response and a decrease of various transmembrane transporters (SI Appendix, Fig. S10). The pathway analysis of proteomics was consistent with the pathway analysis of transcriptomics, showing activation of immune response and repression of metabolism.
Previously, Hofmann et al. (18) used three cell-surface capture techniques to identify a total of ∼500 surface proteins between two AML cell lines (HL60 and NB4) that represent the M2 and M3 stages of AML. Indeed, comparison of our HL60 datasets showed an overlap of 230 identified proteins (SI Appendix, Fig. S11). To understand cellular differentiation, Hofmann et al. (18) further characterized the surface proteome in response to all-trans retinoic acid (ATRA). Despite different mechanism of action, both ATRA and AZA treatment of HL60 cells are known to induce granulocytic and monocytic differentiation. To this end, we compared our HL60 dataset to the existing dataset and observed a considerable overlap of changes in the surface proteome (Pearson correlation of 0.44; Fig. 3F). Among the up-regulated proteins identified in both datasets are several known monocytic differentiation markers such as ITGAM (CD11b), CD14, and CD38, as well as some previously undefined markers such as ADGRE3 and CR1. It is remarkable that even though the two molecules target different cellular functions a number of common targets emerged. As such, these proteins are potential therapeutic targets for subtypes of AML that undergo differentiation upon AZA or ATRA treatment.
Comparisons of Methylome, Transcriptome, and Surface Proteome Profiles.
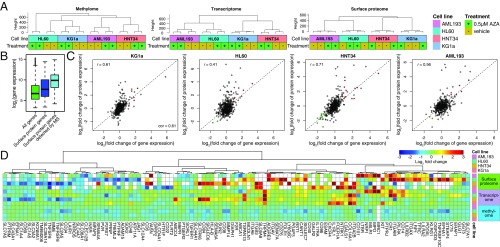
Having all three omics datasets allowed for comparisons of cellular states of the four cell lines with and without AZA at the DNA, RNA, and surface protein levels. Hierarchical clustering of each omics dataset was dominated by variation among the individual cell lines rather than variation due to AZA treatment (Fig. 4A). At the DNA methylation level, KG1a (M1) clustered with HL60 (M2), while AML193 (M5) clustered with HNT34 (M4), consistent with their FAB classifications. At the transcriptome and surface proteome levels, however, KG1a (M1) and HNT34 (M4) clustered together, while AML193 (M5) and HL60 (M2) clustered together. KG1a and HNT34 are cell lines known to be nonresponsive to differentiation agents such as GM-CSF (24, 25), while AML193 and HL60 have been shown to differentiate in response to various reagents (26, 27). Therefore, the four cell lines defined by cellular morphology may correlate with the methylome, but the functional state of these cells is more closely correlated to transcriptome and cell-surface proteome.
Fig. 4.
Omics comparison between methylome, transcriptome, and surface proteome in AML cells treated by AZA. (A) Hierarchal clustering of methylome, transcriptome, and surface proteome data were dominated by differences between cell lines rather than differences between vehicle and AZA treatment. (B) Representative RNA expression profiles of all genes (green), of all genes annotated to be surface proteins (blue), and of genes identified by mass spectrometry experiment (cyan) illustrate that surface proteins have higher gene expression levels. (C) Correlation of changes between gene and protein expression range from 0.44 for HL60 cells to 0.71 for HNT34 cells (r, Pearson correlation). Significantly up- and down-regulated genes and proteins are highlighted in red and green, respectively (P value <0.05 for both gene and protein expression profile). For KG1a and HL60, correlation was calculated after removing two and one outlier points with (fold change of protein expression) >0, respectively. Dashed lines (y = x) are drawn for reference. (D) Comparison of omics datasets for genes with significant protein changes in at least one cell line. fold changes are plotted for protein and gene expression, and scaled average changes in beta values for CpG sites within 1,500 bp of the transcriptional start site are plotted for methylation changes.
Comparison between gene and surface protein expression showed that surface proteins identified in the proteomic datasets tend to have a higher transcriptional signal overall (Fig. 4B and SI Appendix, Fig. S12A). This likely reflects the nature of mass spectrometry in detecting proteins with the highest abundance. Quantitative comparison between the changes in RNA and protein expression after AZA treatment showed Pearson correlation coefficients (r) ranging from 0.41 to 0.71 (Fig. 4C). Such correspondence has been previously reported in other whole-cell proteomics experiments using cell lines (28–30). The differences between mRNA and protein levels are likely due to protein regulation and steady-state turnover.
Next, we compared the magnitude of changes in DNA methylation, gene expression, and surface protein expression in a subset of cell-surface proteins that were significantly regulated in at least one of the cell lines (Fig. 4D and SI Appendix, Fig. S12B). Comparing the three omics datasets for these genes/proteins of interest, we found that while protein and transcript levels tracked reasonably well, changes in methylation status were not correlated with either transcript or protein level. Studies have shown that methylation effects on transcription can be both positive and negative (31, 32). This may explain why only a small subset of RNA changes can be directly accounted for by DNA methylation changes at either the corresponding gene promoter regions or gene body.
In an effort to identify AZA-induced functional modules that might be missing from analyzing one type of data, we adopted a recently developed algorithm, SMITE (Significance-based Modules Integrating the Transcriptome and Epigenome) to further probe for the functional consequence that might be exerted jointly at methylation and transcription levels (33). Through this analysis, we found that although methylation and transcription regulation by AZA might not be synchronized at the gene level across cell lines, functional networks appeared to be commonly regulated (SI Appendix, SMITE Analysis and Dataset S3).
Discussion
As omics technology becomes more widely accessible, integrating data analysis from orthogonal omics sources will be crucial to understanding any biological question. In this study, we asked how AZA affects four different AML cell lines at three omics levels: the DNA methylome, RNA transcriptome, and surface proteome. This allowed us to compare the AML cell lines at the epigenetic level and how that manifests into gene expression and surface protein expression. Our multiomics study of the four AML cell lines showed that ∼80% CpG sites, ∼53% transcripts, and ∼50% surface proteins overlap in methylation or expression pattern in vehicle-treated cells. AZA treatment led to global reduction in DNA methylation, ranging from 45 to 70% of all probed CpG sites, while changes in mRNA and surface protein expression were much more subdued, ranging from 5 to 10%. Although we focused on the surfaceomics of the surviving cells, it could also be of interest to study the apoptotic cell population in the future. One gene encoding a surface protein, TRPM4, was found to be commonly up-regulated by AZA treatment in all four cell lines and may represent a potential novel therapeutic target for AML in combination with AZA. Comparing to previously published data of ATRA treatment in HL60, we identified several previously undefined markers such as ADGRE3 and CR1 that are potential therapeutic targets for subtypes of AML that undergo differentiation with AZA or ATRA treatment.
Despite relatively few changes observed at the transcriptome and surface proteome levels after AZA treatment, functional analysis of RNA and protein regulation showed a general repression of metabolism and activation of immune response across the four cell lines. The repression of metabolism was consistent with a general inhibition of cell growth, albeit to different degrees, suggesting a common response of the cells toward AZA treatment (SI Appendix, Table S1). We also observed activation of immune-responsive genes, which is consistent with previous studies showing that AZA treatment in cells of epithelial origin led to the transcription of endogenous retrovirus, and an induction of a number of immune response genes (AIM genes) related to antiviral response (13). Even though most of the defined AZA-induced immune genes (AIM genes) were activated in the current study, the magnitude of induction was variable among all four cell lines (SI Appendix, Fig. S13). Recently, a number of clinical trials using combination therapy with AZA and checkpoint inhibitors have shown some clinical efficacy, and it has been postulated that this antiviral response induced by AZA can sensitize various cancers (7, 17). Given the common functional impact among the different subtypes of AML cell lines as well as cervical (12, 13) and colorectal (14) cancer, combination therapy with AZA and checkpoint inhibitor is a promising strategy for cancer types of hematopoietic origin.
AZA and other DNMTi have been approved for treatment of MDS and AML for the past decade; however, unmet medical need remains for these patients. Here, we used a multiomics approach to detail the impact of AZA on AML at the individual-gene level as well as the functional-pathway level. The heterogeneous response of AZA treatment reflects the heterogeneity of cell types, implicating that a subtype-specific therapeutic strategy would be more suitable than a general antibody-based therapy against all AML. Three therapeutic candidate targets, TRPM4, ADGRE3, and CR1, were identified for treatment of AML in combination with AZA, and we hope to validate these targets in matched patient samples before and after AZA treatment using more sensitive targeted proteomics methods such as parallel reaction monitoring. Beyond specific validation of these targets, further experiments using a similar integrated omics approach to analyze clinical specimens will advance our understanding of how DNMTi affect AML. Given the heterogenous response we observed, developing techniques toward single-cell parallel analysis of epigenome, transcriptome, and proteome will be paramount to deciphering the interaction between cell types and different cellular states.
Materials and Methods
AML193, HL60, KG1a, and HNT34 cells were cultured in RPMI SILAC media and treated with vehicle (DMSO) or 0.5 M AZA daily for 3 d. Cells were cultured for another 4 d in drug-free RPMI SILAC media before they were harvested for DNA methylation, gene expression, and surface proteomics analyses. Detailed materials and methods are included in SI Appendix.
Supplementary Material
Acknowledgments
This study was funded by the Celgene Corporation. K.K.L. was funded by a Canadian Institutes of Health Research Postdoctoral Fellowship Award. J.A.W. was funded by NIH Grants R35GM122451 and P41CA196276 and the Chan Zuckerberg Biohub Investigator award program.
Footnotes
Conflict of interest statement: This study was funded by the Celgene Corporation. A.N., T.S., L.T., X.N., L.E., K.J.M., and J.D. are employees of Celgene Corporation. K.K.L. and J.A.W. received research funding from Celgene Corporation but no personal financial gain or equity.
Data deposition: Proteomics data have been deposited to ProteomeXchange Consortium (proteomecentral.proteomexchange.org) via the MassIVE partner repository (dataset identifier PXD011298). Methylome and transcriptome datasets have been deposited to the National Center for Biotechnology Information Gene Expression Omnibus database, https://www.ncbi.nlm.nih.gov/geo (SuperSeries GSE123211, accession nos. GSE123140 and GSE123207).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1813666116/-/DCSupplemental.
References
- 1.Issa J-PJ. The myelodysplastic syndrome as a prototypical epigenetic disease. Blood. 2013;121:3811–3817. doi: 10.1182/blood-2013-02-451757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Döhner H, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Döhner H, et al. Diagnosis and management of acute myeloid leukemia in adults: Recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115:453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 4.David Allis C, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17:487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- 5.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 6.Mund C, Brueckner B, Frank L. Reactivation of epigenetically silenced genes by DNA methyltransferase inhibitors: Basic concepts and clinical applications. Epigenetics. 2006;1:7–13. doi: 10.4161/epi.1.1.2375. [DOI] [PubMed] [Google Scholar]
- 7.Wolff F, Leisch M, Greil R, Risch A, Pleyer L. The double-edged sword of (re)expression of genes by hypomethylating agents: From viral mimicry to exploitation as priming agents for targeted immune checkpoint modulation. Cell Commun Signaling CCS, 2017;15:13. doi: 10.1186/s12964-017-0168-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dombret H, et al. International phase 3 study of azacitidine vs conventional care regimens in older patients with newly diagnosed AML with 30 blasts. Blood. 2015;126:291–299. doi: 10.1182/blood-2015-01-621664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silverman LR, et al. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: A study of the cancer and leukemia group B. J Clin Oncol Official J Am Soc Clin Oncol, 2002;20:2429–2440. doi: 10.1200/JCO.2002.04.117. [DOI] [PubMed] [Google Scholar]
- 10.Stresemann C, Frank L. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int J Cancer. 2008;123:8–13. doi: 10.1002/ijc.23607. [DOI] [PubMed] [Google Scholar]
- 11.Tsai H-C, et al. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell. 2012;21:430–446. doi: 10.1016/j.ccr.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, et al. Immune regulation by low doses of the DNA methyltransferase inhibitor 5-azacitidine in common human epithelial cancers. Oncotarget. 2014;5:587–598. doi: 10.18632/oncotarget.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiappinelli KB, et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. 2015;162:974–986. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roulois D, et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell. 2015;162:961–973. doi: 10.1016/j.cell.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinko AJ, et al. Targeting RAS-driven human cancer cells with antibodies to upregulated and essential cell-surface proteins. eLife. 2018;7:e31098. doi: 10.7554/eLife.31098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoseini SS, Cheung NK. Acute myeloid leukemia targets for bispecific antibodies. Blood Cancer J. 2017;7:e522. doi: 10.1038/bcj.2017.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daver N, et al. Hypomethylating agents in combination with immune checkpoint inhibitors in acute myeloid leukemia and myelodysplastic syndromes. Leukemia. 2018;32:1094–1105. doi: 10.1038/s41375-018-0070-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofmann A, et al. Proteomic cell surface phenotyping of differentiating acute myeloid leukemia cells. Blood. 2010;116:e26–34. doi: 10.1182/blood-2010-02-271270. [DOI] [PubMed] [Google Scholar]
- 19.Perna F, et al. Integrating proteomics and transcriptomics for systematic combinatorial chimeric antigen receptor therapy of AML. Cancer Cell. 2017;32:506–519.e5. doi: 10.1016/j.ccell.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haverland NA, et al. 2017. Cell surface proteomics of N-linked glycoproteins for typing of human lymphocytes. Proteomics 17, 10.1002/pmic.201700156.
- 21.Zhang W, et al. LINC01088 inhibits tumorigenesis of ovarian epithelial cells by targeting miR-24-1-5p. Sci Rep. 2018;8:2876. doi: 10.1038/s41598-018-21164-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ong S-E, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics MCP. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 23.Wollscheid B, et al. Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins. Nat Biotechnol. 2009;27:378–386. doi: 10.1038/nbt.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furley AJ, et al. Divergent molecular phenotypes of KG1 and KG1a myeloid cell lines. Blood. 1986;68:1101–1107. [PubMed] [Google Scholar]
- 25.Yamamoto K, et al. Establishment of a novel human acute myeloblastic leukemia cell line (YNH-1) with t(16;21), t(1;16) and 12q13 translocations. Leukemia. 1997;11:599–608. doi: 10.1038/sj.leu.2400594. [DOI] [PubMed] [Google Scholar]
- 26.Gallagher R, et al. Characterization of the continuous, differentiating myeloid cell line (HL-60) from a patient with acute promyelocytic leukemia. Blood. 1979;54:713–733. [PubMed] [Google Scholar]
- 27.Valtieri M, Boccoli G, Testa U, Barletta C, Peschle C. Two-step differentiation of AML-193 leukemic line: Terminal maturation is induced by positive interaction of retinoic acid with granulocyte colony-stimulating factor (CSF) and vitamin D3 with monocyte CSF. Blood. 1991;77:1804–1812. [PubMed] [Google Scholar]
- 28.Lundberg E, et al. Defining the transcriptome and proteome in three functionally different human cell lines. Mol Syst Biol. 2010;6:450. doi: 10.1038/msb.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 30.Wiita AP, et al. Global cellular response to chemotherapy-induced apoptosis. eLife. 2013;2:e01236. doi: 10.7554/eLife.01236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klco JM, et al. Genomic impact of transient low-dose decitabine treatment on primary AML cells. Blood. 2013;121:1633–1643. doi: 10.1182/blood-2012-09-459313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin Y, et al. Impact of cytosine methylation on dna binding specificities of human transcription factors. Science. 2017;356:eaaj2239. doi: 10.1126/science.aaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wijetunga NA, et al. SMITE: An R/Bioconductor package that identifies network modules by integrating genomic and epigenomic information. BMC Bioinformatics. 2017;18:41. doi: 10.1186/s12859-017-1477-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.