Abstract
Physical interaction of regulatory elements in three-dimensional space poses a challenge for studies of disease because noncoding risk variants may be substantial distances from the genes they regulate. Experimental methods to capture these interactions, such as chromosome conformation capture (3C), usually cannot assign causal direction of effect between regulatory elements, an important component of disease fine-mapping. We developed a Bayesian hierarchical approach that uses two-stage least squares and applied it to a novel set of ATAC-seq from 100 individuals, to identify over 15,000 high confidence causal interactions. Most (60%) interactions occurred over <20Kb, where 3C-based methods perform poorly. For a fraction of loci, we identified a single variant that alters accessibility across multiple regions, and experimentally validated the BLK locus associated with multiple autoimmune diseases using CRISPR genome editing. Our study highlights how association genetics of chromatin state is a powerful approach for identifying interactions between regulatory elements.
Introduction
Three-dimensional (3D) interactions between regulatory elements are a fundamental process in gene regulation1. Understanding the guiding principles that control these interactions is a major research interest in genomics2,3. Long-range regulation poses a challenge for studies of human disease because risk variants may be located many kilobases (Kb) from the genes they regulate, making causal variant identification difficult4,5. Chromosome conformation capture (3C)-based techniques have enabled the generation of genome-scale maps of 3D contacts in human cells6–8. These maps have provided valuable insights into large-scale structure and organisation of chromosomes9,10, and often also provide useful information linking distal disease risk alleles with putatively regulated genes11,12. However, it can be hard to distinguish functional interactions, such as enhancer-promoter looping, detected using 3C-based methods from a background of random collisions13, which are particularly pronounced over distances of less than 20Kb11.
A complementary approach to mapping genome-wide 3D interactions is to utilise germline genetic variation. Quantitative trait locus (QTL) mapping of chromatin traits can identify genetic variants that regulate chromatin both locally and distally, sometimes over distances of hundreds of kilobases14–17. These distal QTLs are known to be enriched in topologically associating domains14,15,17 (TADs), suggesting regulatory regions mapped by chromatin QTLs do indeed physically interact with one other. For fine-mapping of putative causal variants identified in human disease studies, this approach has some attractive features. First, unlike 3C-based techniques, our ability to detect interactions between regulatory elements is not correlated with the distance between them. Second, QTLs identified in these studies can be naturally aligned with those from disease studies using colocalisation18. Third, causal interactions between different regulatory elements can be potentially deduced by Mendelian Randomisation19–21 (MR), where germline genetic variants are used as instrument variables to resolve relationships between different active regions. Here we develop a pairwise hierarchical model (PHM) that incorporates a technique from MR in a Bayesian framework to map causal regulatory interactions using ATAC-seq data set from 100 unrelated individuals of British ancestry.
Results
The model
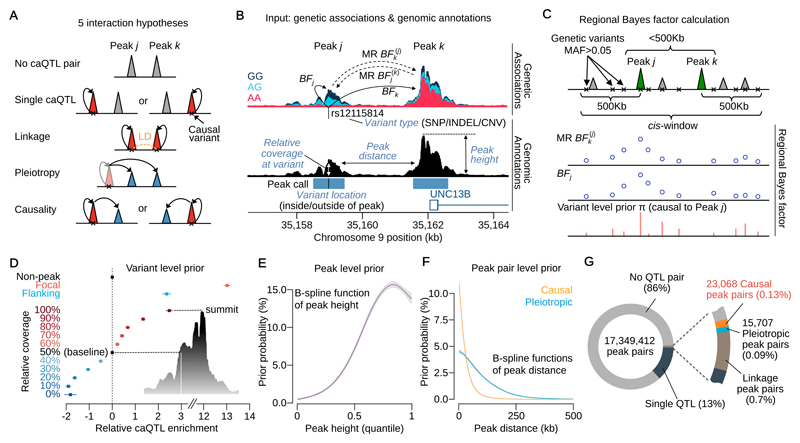
Associations between genotype at the same genetic variant and chromatin accessibility often appear spread across multiple independent “peaks” of open chromatin16 and can arise for multiple reasons. Two or more variants in linkage disequilibrium can drive independent associations at different peaks (hereafter, “linkage”). Alternatively, a single variant might independently drive association signals at multiple peaks (“pleiotropy”). Finally, individual variants may alter accessibility at one regulatory element that in turn alters accessibility elsewhere in the genome, an indication that these elements functionally interact in 3D space (“causality”). Our PHM classifies peak pairs within 500Kb of one another into hypotheses of linkage, pleiotropy, causality, a single QTL at either of the modelled peaks or a null hypothesis of no QTLs in either peak (Fig. 1A). To compute the pairwise likelihood (Online Methods) for a given peak pair j and k, we calculate Bayes Factors (BFj and BFk) for the association between genotype at a putative causal genetic variant and chromatin accessibility at each member of the pair (Fig. 1B). For the hypothesis of causality we compute Mendelian Randomisation Bayes Factors (MR BFj(k) and MR BFk(j)) for the regression of chromatin accessibility in peak j on peak k (or vice versa) using two stage least squares22 (2SLS), with genotype at the given genetic variant as the instrumental variable (Fig. 1B). We compute BFs for all variants in a cis window extending 500Kb 5’ and 3’, marginalising by the appropriate prior probabilities to derive a “regional” BF (RBF) (Fig. 1C). We use a “variant-level” prior probability of being a causal regulatory variant within the cis window (Fig. 1D) assuming a single causal variant23. We also model a “peak-level” prior probability on the probability of observing a caQTL, which is a function of peak height (Fig. 1E), and a “peak-pair-level” prior probability that adjusts the support for pleiotropy or causality between two peaks, as a function of the distance between them (Fig. 1F). Both the peak-level and peak-pair-level priors are conceptually similar to independent hypothesis weighting24. The model outputs a posterior probability that a peak pair belongs to one of the interaction categories, including the posterior probability of a causal interaction (PPC). Hereafter PPCjk denotes the posterior probability that peak j regulates, or is “upstream” of peak k, while PPCkj denotes the converse (j is “downstream” of k). PPC without a subscript refer to the sum of PPCjk and PPCkj.
Figure 1. Overview of the pairwise hierarchical model and summary statistics.
(A) The five main hypotheses of interaction between peaks. (B) Genetic associations with chromatin accessibility and related genomic annotations as input data. The Bayes factor (BF) of genetic associations with peaks (solid lines) are computed from the simple linear regression. The BFs of association between peaks (dashed lines) are computed using two stage least square method in the Mendelian Randomisation (MR). (C) For the j-k peak pair, BFs obtained in Fig. 1B are calculated for all variants in a cis-window and averaged as the regional Bayes factor (RBF). The schematic shows the two types of BFs across all variants were averaged by the variant level prior probability that the peak j is upstream of k (genetic variant is causal to peak j) to map causal interaction from j to k. (D) The estimated relative caQTL enrichments for genomic annotations used to compute the variant level prior probability in Fig. 1C. (E) The estimated prior probability of a peak being a caQTL as a function of the peak height quantile among 277,128 peaks. The B-spline function was applied to capture non-linear relationship. (F) The estimated prior probability that a peak pair is pleiotropic or causal as a function of peak distance. Two different B-spline functions were applied. (G) The breakdown of mapped interactions according to Fig. 1A. The numbers are based on the sum of posterior probabilities.
Mapped causal interactions
Our consensus set contained 277,128 peaks of open chromatin, corresponding to 17 million peak pairs. We found that 14% of peak pairs showed some evidence of genetic control (Fig. 1G). Summing over the posterior probabilities, we estimated that 23,036 peak pairs (0.13%) causally interact (for example, Fig. S1A); 15,487 we refer to as “high confidence” (PPC>0.5) with a Bayesian false discovery probability25 (BFDP) of 18.4% on average.
Following the initial round of interaction detection, we performed a post-hoc summarisation to identify directed acyclic graphs (DAGs) of causally interacting peaks in our high confidence call set. Because an exhaustive search of all possible DAGs in a cis window was computationally intractable, we used an ad-hoc algorithm (Online Methods). We identified 3,557 independent DAGs (Fig. S1B), of which 1,366 DAGs involve between 2 peaks and 60 peaks, with the maximum at the MB21D2 locus (Fig. S1C) that we previously reported16.
Our empirical prior suggested that the probability of any two peaks within 500Kb of each another interacting was 1.4% (Fig. 1F) suggesting that 1.23% or over 220,000 causal interactions remain to be discovered. However, analysis of down sampled data suggested that the number of interactions was far from saturated, with many real interactions below our detection limit (Fig. S1D).
Model performance assessment by simulation
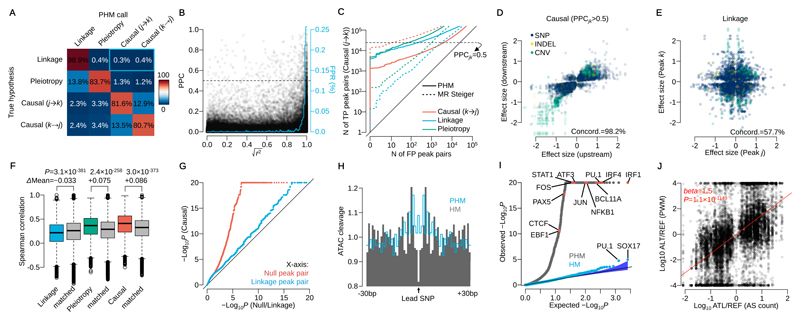
To test model performance, we simulated data with one causal variant per focal peak, under one of the 5 hypotheses in Fig. 1A (Online Methods). The false positive rate (FPR) of causality when linkage or pleiotropy were simulated was 0.7% or 1.5%, respectively (Fig. 2A). The model found it more challenging to correctly assign the direction of causal effects (Fig. 2A; 18.9% incorrect directionality on average). Under simulated linkage, the FPR of causality increased with increasing linkage disequilibrium (LD) between the two variants (Fig. 2B), but overall was low even for variants in high LD (0.0025 for variants in |r|>0.99).
Figure 2. Model performance assessment by simulation and real data.
(A) Confusion matrix of mapped interactions under the 4 hypotheses. Percentages are calculated from peak pairs with posterior probability was greater than 0.5. The blue rectangle highlights the false positive rate (0.7%) for mislabelling linkage as causality. (B) Posterior probability of causality (PPC) versus r2 between two caQTL variants simulated under linkage. The blue line shows the average false positive rate (mislabelling linkage as causality) in 1% r2 bins (area under this curve is 0.7%, equivalent to the blue rectangle in Fig. 2A). (C) Sensitivity and specificity of causal interactions for PHM and MR Steiger in simulated data. The y-axis shows the number of true positive (TP; simulated causal (j→k) model) peak pairs against the number of false positive on the x-axis (FP; simulated under the causal (k→j), linkage or pleiotropy model) peak pairs. The horizontal dashed line illustrates PPCjk=0.5 for PHM. (D) Effect sizes of the lead variant at upstream and downstream peaks in confident causal peak pairs. (E) Effect sizes of two independent caQTLs at peaks in linkage (posterior probability greater than 0.5). Linkage peaks with lead variants with LD index r2>0.25 were used. (F) Distribution of Spearman’s rank correlation coefficient of DNaseI-seq read count across 53 cell types from the Roadmap Epigenomics Project stratified by the mapped interaction categories (Online Methods). Tow-sided t-test was performed with the distance matched control for linkage, pleiotropy and causality, respectively (n=98,963, 12,233 and 15,487 peak pairs). (G) QQ-plot of –log10 P-values for allele-specific accessibility of downstream peak for the high confidence set of 15,487 causal peak pairs (y-axis), and for 15,487 randomly chosen, distance-matched controls where the posterior probability of either null or linkage hypothesis was greater than 0.5 (x-axis). (H) Aggregated ATAC-seq cleavage across 1,577 regions around the lead SNPs detected by pairwise hierarchical model (PHM; grey) and simple hierarchical model (HM; blue line). (I) QQ-plot of Binomial test P-values for 2,570 motifs in CISBP (Online Methods). Blue points correspond to the HM and grey points correspond to the PHM. (J) The ratio of putative TF binding affinities between reference and alternative allele at each lead SNP versus the ratio of ATAC-seq allele-specific (AS) counts (n=14,642 SNPs). AS counts were generated by aggregating only heterozygous individuals at each lead variant. The red line shows the linear regression line.
We extended our simulations to include two causal variants in the focal peak for each scenario (Online Methods). Multiple causal variants did not substantially increase the false positive rate for any scenario (Fig. S2A). Finally, we simulated hybrid hypotheses of linkage, pleiotropy and causality. Here, our power to detect causality reduced to 62.9% (hybrid pleiotropy, causality (j→k)) or 37.5% (hybrid linkage, causality (j→k)) and the false positive rate also became 5.3% on average across all hypotheses (Fig. S2A).
We also compared our model’s performance on simulated data with MR Steiger, an alternative approach to identifying causal interactions26. We note that MR Steiger assumes that the causal variant is known, while our model attempts to infer the causal variant from the data. Despite this, the PHM produced a lower false positive rate for causality when data were simulated under the linkage or pleiotropy models (Fig. 2C), but MR Steiger was better at identifying the causal direction of effect. For example, at a PPC>0.5, the PHM correctly called 24,332 causal (j→k) peak pairs and incorrectly called 216, 73 and 3,978 linkage, pleiotropy, or causality (k→j) under the true hypothesis of causality (j→k), respectively. For the same number of true positives (same power to detect the true causal (j→k) interactions), the equivalent numbers were 2,998, 1,014 and 9 for MR Steiger. We observed that the misclassification rate of causal direction for the PHM decreased significantly when the causal variant was more strongly associated with the focal peak (less than 1% for BFj greater than 100) (Fig. S2B).
Model performance assessment using real data
Next, we investigated model performance on real data. Effect directions for inferred causal peak pairs were substantially more likely to be in the same direction than peaks in linkage (Fig. 2D-E), with 98.2% of peak pairs concordant in the confident causal set compared with 57.5% in confident linkage hypothesis (posterior probability of linkage > 0.5). Using RoadMap Epigenomics data on chromatin state from 53 different cell types (Online Methods) we also observed that the activity of causally interacting or pleiotropic peak pairs was more highly correlated across tissues than distance matched controls, but significantly lower than matched control for linkage peak pairs (Fig. 2F). An example of cross-tissue correlation is shown in Fig. S2C.
Because allele-specific signals were not used in the original model, they can provide independent confirmation of a genetic effect. We used RASQUAL16 to map caQTLs using allele-specific counts only at feature SNPs in the downstream peak region. A QQ-plot of P-values for the allele-specific signals were significantly skewed toward 0 compared with the distance matched controls of null peak pairs or linkage peak pairs (posterior probability of null or linkage hypothesis > 0.5) (Fig. 2G, Fig. S2D-E).
Finally, we examined the overlap between transcription factor (TF) binding site footprints and the lead variants detected by our PHM, and compared this with results from a hierarchical model that did not consider interactions between peaks (HM, as used in the first stage of optimisation). Compared with the HM, lead variants detected by the PHM were highly enriched in TF footprints (Fig. 2H), particularly B-cell specific TF motifs (Fig. 2I), such as IRF1 and PU.1. The ratio of putative binding affinity between reference and alternative alleles at the PHM lead variant was also more highly correlated with allelic imbalance of ATAC-seq reads at PHM lead SNPs (Fig. 2J) than HM lead SNPs (Fig. S2F).
Comparison with 3C-based assays
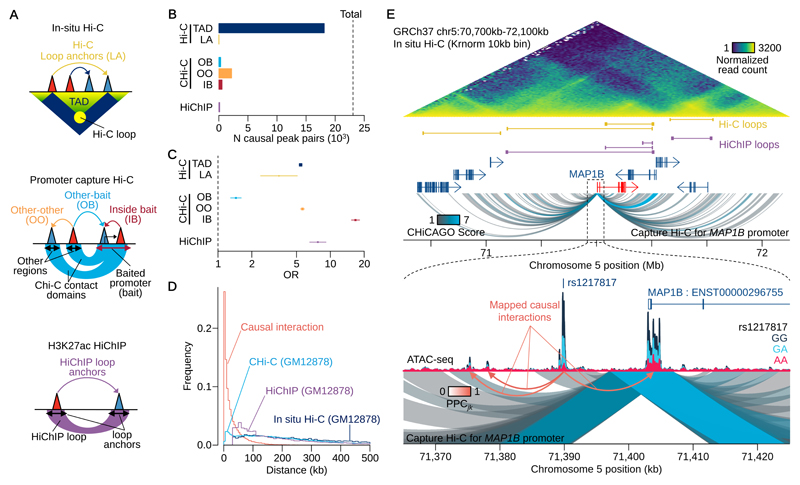
We compared causal interactions inferred from our model with chromatin loops inferred from Hi-C, promoter Capture Hi-C (Chi-C) and H3K27ac HiChIP applied to GM1287810–12 (Fig. 3A). 74% of causal interactions were between peaks located within the same TAD called from Hi-C, a 5-fold enrichment over genomic background (Fig. 3B-C). The remaining causal interactions (26%) were primarily in non-TAD regions (Fig. S3A-B), with peaks spanning TAD boundaries being significantly (15-fold) depleted (Fig. S3C) Although rare, the TAD boundary-spanning interactions we did detect were as strongly supported by allele-specific accessibility analysis as those found within TADs (“Across TADs” panel, Fig. S3D). Effect directions of lead variants were less concordant when peak pairs spanned one or more insulators or TAD boundaries (Fig. S3E), with average concordance of 89.0% and 86.3% respectively (P=4.5x10-7 and 2.9x10-13) compared with background sets (91.8% and 91.6%). An example of a causal interaction spanning a TAD boundary is shown in Fig. S2D. Causal interactions were also enriched for loops inferred from H3K27ac HiChIP and CHi-C data (7.7 and 1.4-fold, respectively), although the absolute numbers of overlaps were small (152 and 324, Fig. 3B-C). Our model also highlighted interactions that could be missed by promoter capture-based techniques. Of the 49,579 peak regions linked to baited promoters, we estimated that there were 2,208 causal interactions between the non-promoter elements and a further 561 between two peaks located within the same CHi-C bait region (Fig. 3B-C).
Figure 3. Comparison with 3C-based assays.
(A) Schematics of Hi-C, CHi-C and HiChIP annotations. (B) The numbers of causal peak pairs overlapping with the annotations in Fig. 3A. Topologically associating domains (TAD), loop anchors (LA), other-other (OO), other-bait (OB), inside bait (IB). (C) Enrichment Odds Ratio (OR) with 95% confidence interval of causal interactions with annotations (Online Methods). For Hi-C and HiChIP annotations, n=15,884,515 peak pairs (peak distance > 35Kb) were used to compute the odds ratio and all peak pairs (n=17,349,412) were used for CHi-C annotations. (D) Distribution of interaction length. For our interactions, we computed the distance between all 17 million peak pairs considered, and weighted these by the posterior probability of causality (PPC). (E) An example of causal interactions found in the promoter flanking region of MAP1B gene. There is a caQTL peak (with the QTL SNP rs1217817:G>A) 10Kb upstream of MAP1B promoter affects multiple open chromatin peaks including the promoter peak.
Most causal interactions occur over sub 20Kb distances
We found causal interactions inferred from the PHM occurred over much shorter distances than those captured by 3C-based techniques (Fig. 3D): 63% were less than 20Kb distant from one another, compared with 7% of CHi-C interactions (Fig. 3D). We confirmed that the distance distribution did not reflect interactions between pseudo “sub-peaks” that were part of the same broad peak (Fig. S3F-G). Our results suggest that many functional three-dimensional interactions may be below the resolution of conventional 3C-based techniques. One example is shown at the promoter region of the MAP1B gene (Fig. 3E). Here, a high confidence (PPCjk>0.99) causal interaction occurs between a promoter and an enhancer that is less than 13Kb distal, but the contact domain inferred from CHi-C, has weak statistical support (CHICAGO score 1.87).
Enhancer-enhancer and promoter-enhancer interactions are common
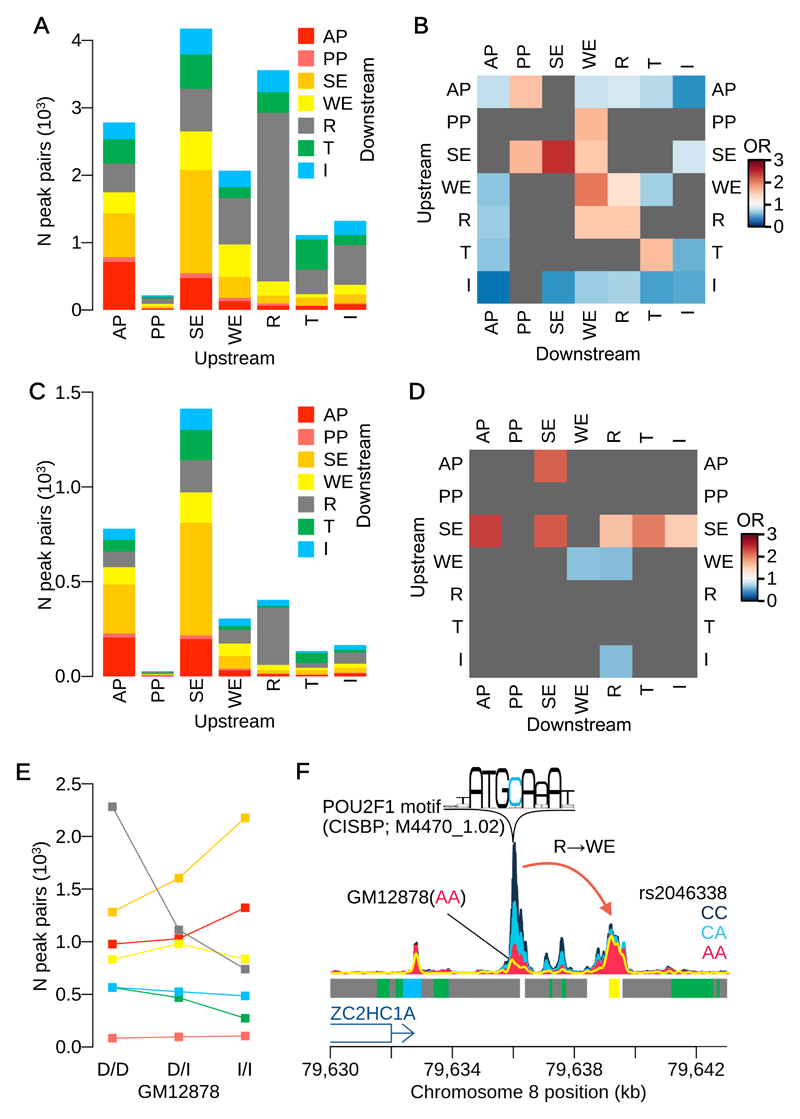
We next examined the functional classes to which the members of causally interacting regulatory elements belonged, using the ENCODE genome segmentation annotations for LCLs27,28 (Online Methods). The most frequent class of interactions (5,061 peak pairs, 22% of all interactions) were strong enhancers (SEs) that appeared to regulate other element types, including other SEs (1,531 peak pairs, 6.6%), a 2.5-fold enrichment (Fig. 4A, B). The effect directions of the lead variant between SE-SE interactions were significantly more concordant compared with the background (Fig. S4A), a 95.0% concordance (P=0.0043) compared with the complement set (82.2%), suggesting those regions may work in a coordinated manner.
Figure 4. Comparison with genome segmentation.
(A) The numbers of causal peak pairs overlapping ENCODE genome segmentation. Numbers of interactions were computed weighting by PPCjk. The ATAC-seq peaks are classified by 7 different regulatory categories: active promoter (AP); poised promoter (PP); strong enhancer (SE); weak enhancer (WE); repressor (R); transcribed region (T); and insulator (I). Each bar indicates upstream peak category and the colour code indicates downstream peak category. (B) Enrichment of ENCODE segmentation category pairs for our causal interaction. Heatmap shows the odds ratios (see Online Methods for computation of enrichment using PPCjk) for all combinations of segmentation categories at upstream and downstream peaks (among n=17,349,412 total peak pairs). The segmentation category pairs that were above FDR 10% or supported by less than 10 causal peak pairs are masked by grey. (C) The numbers of causal peak pairs that are jointly colocalised with one or more eQTLs overlapped with the ENCODE segmentation. (D) Enrichment of ENCODE segmentation category pairs for our causal interactions that are jointly colocalised with one or more eQTLs (among n=23,068 causal peak pairs) (see Online Methods for computation of enrichment using PPCjk). (E) The number of peak pairs whose upstream peak overlaps with one of the seven segmentation categories, stratified by the genotypes of GM12878 at lead QTL variant (Online Methods). Each genotype is labelled as a combination of decreasing “D” and increasing “I” alleles according to the sign of QTL signal at the lead variant. Colour code is same as in Fig. 4A. (F) An example of causal interaction from a repressed region to a weak enhancer. The normalised ATAC-seq coverage is stratified by three genotype groups at rs2046338:C>A. The yellow line shows ATAC-seq coverage of GM12878 whose genotype is AA (decreasing homozygote) at rs2046338.
When we focussed only on variants that also altered gene expression, using 4,670 interacting peak pairs that jointly colocalised with an eQTL from the GEUVADIS data set (Online Methods), we found these were enriched (2.4-fold, P=6.4x10-19) for SE to active promoter (AP) interactions (Fig. 4C, D). However, expression-associated variants were also enriched for interactions from APs to SEs (2.2-fold) or between pairs of SEs (2.2-fold enrichment) (Fig. 4D). One hypothesis is that many of these interactions are mediated by transcriptionally induced changes in chromatin accessibility over the gene body, creating apparent interactions between a single upstream functional element and chromatin peaks throughout the transcribed region. Consistent with this idea, peaks downstream of an AP were significantly enriched in the gene body (2.3-fold enrichment, P=8.1x10-24; Fig. S4B) compared with peaks to the 5’ of the promoter. This hypothesis is also consistent with the observation that chromatin accessibility over the gene body is highly correlated with gene expression level (Fig. S4C). A striking example of this potential phenomenon is found at the MB21D2 locus (Fig. S1C).
Genetically-driven changes in the reference epigenome
We found a surprisingly large number of interactions (4,134 peak pairs) originating from within repressed regions (Fig. 4A). Preliminary analysis suggested that these might arise due to genotype effects on the reference epigenome annotation derived from a single individual (GM12878). To test this, we stratified all upstream peaks in causally interacting pairs based on whether their lead caQTL genotype in GM12878 was an increasing homozygote, decreasing homozygote or heterozygote (Online Methods). Upstream repressed regions were highly enriched (3.1-fold) for decreasing homozygotes compared with increasing homozygotes (Fig. 4E), suggesting that in these cases a strong caQTL almost completely removes a region of open chromatin in GM12878, an example of which is shown in Fig. 4F. We found that 1.4% of repressed regions overlapped a caQTL where GM12878 was a decreasing homozygote. This estimate is also likely to be a lower bound due to incomplete power to detect caQTLs.
Causal interactions improve fine-mapping
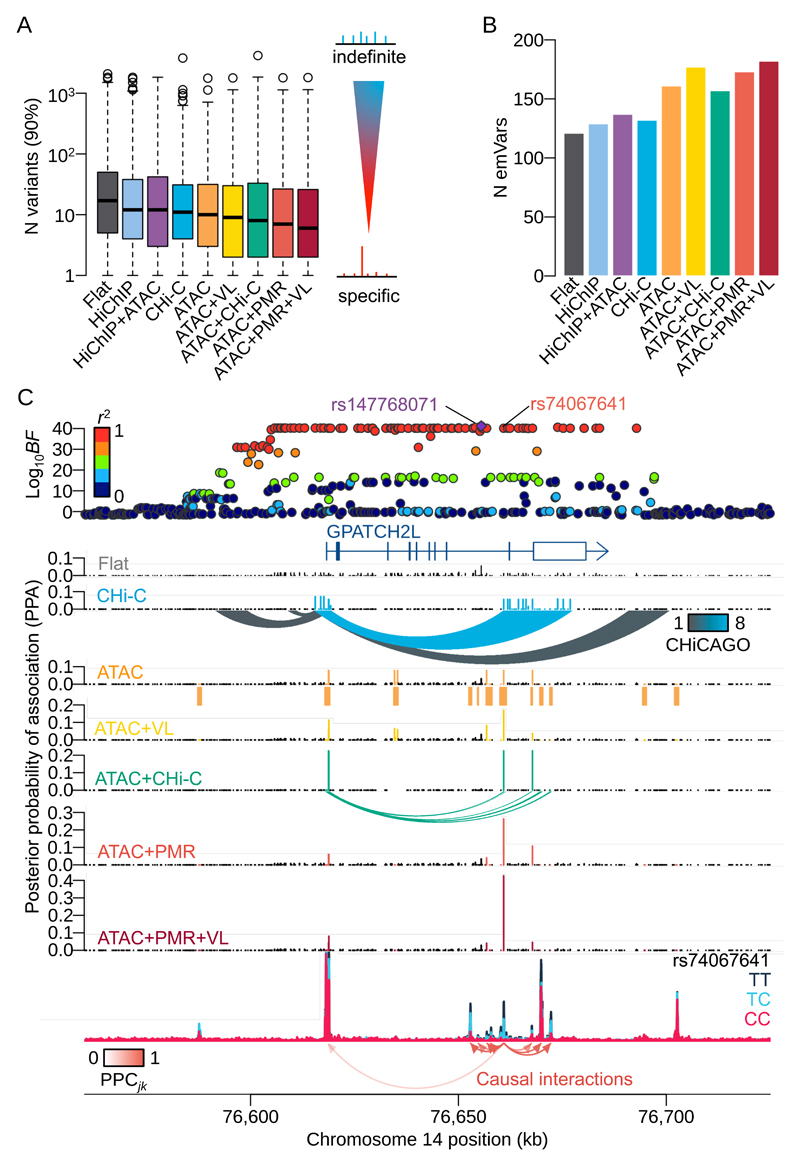
Next we examined whether the information on causal direction of variant effects could be used to improve fine-mapping accuracy, using gene expression as a model quantitative trait. For each peak within a 1Mb cis-window around a gene TSS, we first computed the probability of master regulator (PMR) for each peak (Online Methods). We then used a hierarchical model23 to compute the posterior probabilities of association (PPA) for eQTL variants with PMR and the following four other annotations: (1) inside or outside an ATAC peak, (2) eQTL variant location (VL), relative to an ATAC peak coverage, (3) promoter CHi-C contacts, (4) HiChIP loops from promoter regions (Online Methods for details). Genome-wide, the best performing annotation was the combination of PMR with ATAC peak status and VL, which reduced the 90% credible set of eQTL variants by 65%, from 17 to 6 variants on average, compared with 11 variants for CHi-C, 10 for ATAC peaks and 8 for Chi-C combined with ATAC peaks (Fig. 5A). The effect of adding information on the causal direction, by prioritising the most upstream variant via the PMR, significantly reduced the credible set size compared to the ATAC peak annotation alone (P<10-49, paired t-test). We then compared our results with data from massively parallel reporter assay (MPRA) performed in LCLs29 (Online Methods). We found the highest overlap (21.6% or 182 emVars) for the combined PMR, ATAC peak and VL annotations (Fig. 5B). We applied this approach to a challenging locus, where a strong eQTL for the GPATCH2L gene is associated with more than 100 candidate regulatory variants in almost perfect LD (Fig. 5C). With no annotation information, the 90% credible set size at this locus is large, at 65 variants. Although different annotations produce varying effects, our model proposes a SNP (rs74067641:T>C) as the likely causal variant with the highest PPA=0.42. This variant is located within a predicted master regulatory peak located furthest upstream in the regulatory cascade (Fig. S5A-C). We note that reduction in credible set size is an imperfect measure of fine-mapping accuracy in cases where multiple causal variants are segregating.
Figure 5. Fine-mapping eQTLs using mapped causal interactions as an annotation.
(A) Distribution of the number of variants in the 90% credible set, across all protein coding genes with more than one colocalised ATAC peaks (N genes=1,207) over nine different annotation combinations. Non-informative prior (FLAT); inside/outside an ATAC peak (ATAC); HiChIP anchor regions (HiChIP); CHi-C contact domains (CHi-C); variant location (VL); probability of master regulator (PMR). In the boxplots, the box represents the interquartile range (IQR), the black line is the median, the whiskers are 1.5 times the IQR above or below the first and third quartiles, with data points outside the whiskers shown by open circles. (B) The number of expression-modulating variants (emVars) overlapping lead eQTL variants detected by the eQTL hierarchical model with various annotations. (C) An example of fine-mapped region with more than hundred of significant variants in almost perfect LD. The top panel shows negative Log10 Bayes factors of eQTL for GPATCH2L gene using gEUVADIS RNA-seq data. Each point is coloured by the degree of LD index (r2 value) with the index variant (rs147768071:AGTTTT>A). The SNP (rs74067641:T>C) in the master regulatory peak shows the highest PPA with ATAC+PMR+VL annotation.
Causally interacting caQTLs are enriched in autoimmune GWAS hits and eQTLs
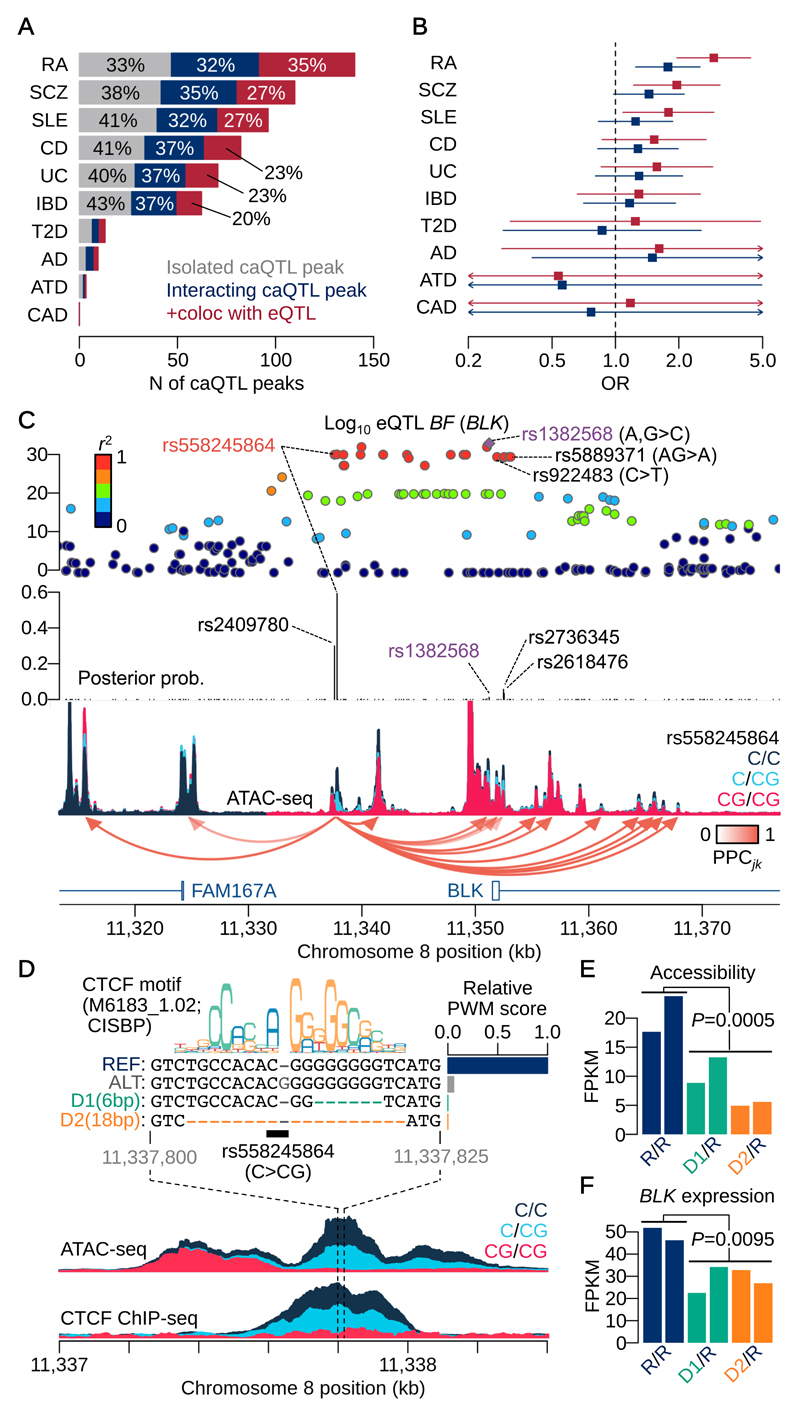
We performed an enrichment analysis of causally interacting caQTL peaks for disease GWAS hits. We colocalised our caQTLs with 10 genome-wide association studies (GWAS) whose genome-wide summary statistics were available30–37. caQTLs detected in LCLs strongly colocalised with autoimmune disease, including Rheumatoid Arthritis (RA; 140 colocalised caQTL-GWAS loci) or systemic lupus erythematosus (SLE; 96 loci) (Fig. 6A). Using RA as an example trait, we found that causally-interacting loci were significantly more likely to colocalise (1.8-fold, P=1.4x10-3) with risk loci than non-interacting caQTLs (Fig. 6B). Interacting peaks that also colocalised with an eQTL were further enriched (2.9-fold, P=1.7x10-7). This suggests causal interactions were more often involved in a gene regulatory cascade leading to downstream consequences.
Figure 6. Enrichment analysis and fine-mapping of GWAS associations and a CRISPR validation.
(A) Number of caQTLs colocalised with GWAS hits. The numbers are based on the sum of posterior probabilities of colocalisation between a caQTL peak and a GWAS trait. Rheumatoid arthritis (RA); schizophrenia (SCZ); systemic lupus erythematosus (SLE); Crohn’s disease (CD); ulcerative colitis (UC); inflammatory bowel diseases (IBD); type 2 diabetes (T2D); Alzheimer’s disease (AD); atopic dermatitis (ATD); coronary artery disease (CAD). (B) Enrichment odds ratio (OR) with 95% confidence interval of causally interacting caQTL peaks (blue) and those that are colocalised with one or more eQTLs (red) relative to isolated caQTLs colocalised with GWAS hits (among n=277,128 peaks). (C) A chromatin accessibility altering variant at the BLK/FAM167A locus. The top panel shows negative log10 Bayes factors of eQTL mapping for BLK gene using gEUVADIS RNA-seq data. Each point is coloured by the degree of LD index (r2 value) with the index variant (rs1382568:A>C,G). The middle panel shows the posterior probability of association (PPA) obtained from the full annotation model (Online Methods) in which the insertion variant rs558245864:C>CG shows the highest PPA. The bottom panel shows ATAC-seq coverage depth stratified by the insertion variant (rs558245864) with the causal interactions from the peak in which the insertion variant exists. (D) CRISPR engineered locus around the insertion variant. The insertion variant disrupts the CTCF binding site and attenuates the binding affinity (bar plot) calculated from the canonical CTCF binding motif (CISBP: M6183_1.02). Independent analysis of CTCF ChIP-seq binding QTL supports the result. CRISPR engineering was performed to generate two different deletions (D1 and D2) from the parental line (HG00142) whose genotype is reference homozygote at the insertion variant. The maximum CTCF binding affinity around the region after extracting the deleted sequences is lower than that of the alternative allele. (E) FPKMs at the focal peak for the two heterozygous deletion lines (D1: green and D2: orange) compared with the parental line with reference homozygote (R/R: navy). All lines were replicated twice as different cell cultures (n=6 replicates in total; see Online Methods for P-value calculation). (F) FPKMs of BLK gene expression for the same lines in Fig. 6E (n=6 replicates; see Online Methods for P-value calculation).
CRISPR validation of a putative causal variant at the BLK locus
Finally, we applied our method in an attempt to fine-map a challenging GWAS locus with contradictory evidence for multiple causal variants in previous studies. The BLK/FAM167A locus on 8p21 has a strong eQTL (gEUVADIS P<10-26 and 10-46 for BLK and FAM167A genes, respectively) in LCLs (Fig. 6C) that colocalises well with genome wide significant associations for SLE and RA (Fig. S6A-B). Previous attempts to fine-map this locus have been hampered by multiple genetic variants in tight LD (Fig. S6C-D). Two SNP variants, rs1382568:A>C,G and rs922483:C>T, located near the promoter of the BLK gene, have previously been reported as putative causal variants of SLE that alter BLK expression in various B and T cell lines38. However, MPRA studies have pinpointed a different deletion variant (rs5889371:AG>A) that might also potentially alter BLK expression in LCLs29. Two of the previously reported variants (rs5889371 and rs1382568) are located in regions of low chromatin accessibility (Fig. S6E-G) and less likely to causally influence BLK expression in LCLs.
We detected a single base pair insertion variant (rs558245864:C>CG) located in a strong caQTL peak 14Kb upstream of the BLK promoter that interacted with 15 flanking peaks including several promoter peaks (Fig. 6C). The insertion variant showed the highest posterior probability (PPA=0.59) of any putative causal eQTL variant for BLK gene (Fig. 6C). This variant is located at the middle of a canonical CTCF binding motif, with an extra “G” nucleotide decreasing the predicted CTCF binding affinity to almost to background (Fig. 6D). The direction of binding affinity change was consistent with the caQTL signal. This variant was also a CTCF ChIP-seq QTL (Fig. 6D), with a 99.7% the probability of colocalisation between the CTCF QTL and caQTL for this peak (Online Methods). We used CRISPR/Cas9 genome engineering to generate two different heterozygous deletion lines from a parental line that was homozygous for the high CTCF binding allele (Online Methods). These deletions overlapped the CTCF binding site: the 6bp deletion disrupts the right hand side of the binding site and the 18bp deletion that removes almost the entire motif (Fig. 6D). ATAC-seq and RNA-seq in the deletion lines revealed a significant down-regulation of chromatin accessibility at the focal peak compared with the parental line (P=0.0005) (Fig. 6E), and a concomitant down-regulation of BLK expression (P=0.0095) (Fig. 6F). We observed decreases in accessibility at some neighbouring peaks around BLK promoter region (Fig. S6H-I). We also observed an increase in accessibility around FAM167A promoter region (Fig. S6H-I) and in FAM167A expression (Fig. S6J), although this was not significant (P=0.18).
Discussion
We have presented a novel approach to detect interactions between regulatory elements, which uses principles of Mendelian Randomisation embedded within a Bayesian hierarchical model. We show that the majority of causal interactions within 500Kb occur over short distances (<20Kb), typically a region of low sensitivity for 3C-based techniques. Many of the interactions we detect are between enhancers, which we assemble into hierarchies of interacting regulatory elements. We demonstrate that our model can be used to identify hierarchies of regulatory elements within a region and prioritise putative causal variants, validating a single locus using CRISPR/Cas9 editing.
The low frequency of long-range interactions we observed agrees with previous estimates from eQTL studies23,39,40. One question is, given that most regulatory interactions detected using 3C-based methods occur over distances of 100Kb and above (Fig. 3D), why have large numbers of genetic variants operating at these distances not also been detected? Although QTL studies typically test variants in a restricted cis window of 1Mb39–41, this does not completely explain the lack of signal: the number of eQTL associations detected decreases dramatically by approximately 20Kb distant from the gene TSS23,40. A possible explanation is that there may be an underlying relationship between interaction distance and cellular frequency, such that long-range interactions occur in a relatively small number of cells in the population39. This is consistent with the negative correlation between read coverage and distance in Chi-C data (Fig. S7A). It seems plausible that 3C-based methods could be more sensitive to rare, long-range regulatory interactions while variants residing in these elements have relatively weak effects40, requiring large sample sizes to detect when averaged across the entire cell population. An alternative hypothesis is that short-range interactions may not be driven by chromatin looping, but instead reflect transcriptional activity and the movement of polymerase across the sequence (Fig. S4B-C).
Our study also revealed the genomic architecture of causal interactions between regulatory elements. In particular, we detected frequent interactions between annotated enhancer elements, many of which we hypothesise are mediated by an intermediate eQTL that alters chromatin accessibility globally across the gene body. Nonetheless, the enrichment of these interactions in gene bodies was modest, and we also found many examples of interactions that were not colocalised with eQTLs, and were located far from annotated genes (an example is shown in Fig. S7B). In a small number of cases (18 DAGs) we also found strong evidence (PPC > 0.5 for each enhancer pairs) that these occurred between multiple enhancers upstream of a promoter (i.e., SE→SE→AP). It is possible that some of these represent enhancer “seeding” events, where individual enhancers drive progressive activation of additional nearby elements42.
One of the limitations of our method is that regulatory elements lacking a common genetic variant that perturbs their function will be missed. Additionally, interactions between genotype and regulatory elements further downstream appear to become harder to detect, perhaps due to additional biological noise. One example of this is the systematically lower genetic effect sizes (14% decreasing) we found at downstream promoters (Fig. S7C-D).
Our approach allows for a natural prioritisation of variants in disease-associated loci. Although overlapping of those variants with open chromatin can reduce credible sets, this frequently leaves many loci with tens of variants to characterise by direct experimental follow up. Assignment of the direction of effect between different peaks allowed us to identify smaller sets of plausible candidate variants by identifying “master regulatory” regions. Although we have focused on ATAC-seq data, we believe our model can be readily extended to other types of chromatin-based assay, in particular ChIP-seq for histone modifications14,15,17. Some limitations of this approach might include a greater difficulty in assigning causal variants based on their location within a ChIP-seq peak, which will typically be in a nucleosome depleted region and therefore low read coverage14 (for an example, see Fig. S7C). However, we anticipate that, applied to existing data sets from primary cells, such as that generated by the BLUEPRINT initiative43, that our approach will be a valuable tool in dissecting the molecular architecture of specific GWAS loci.
Online methods
ATAC-seq in LCLs
We collected 100 lymphoblastoid cell line (LCL) samples of British ancestry (1000 Genomes Project, GBR cohort) from Coriell. ATAC-seq library preparation was performed for each line (except for the 24 lines we previously performed16) as previously described16. We performed 75bp paired end sequencing in 4.4 billion sequence fragments on a HiSeq 2500 (Illumina). Although data from the 24 lines has been previously sequenced16, we performed additional sequencing to increase the coverage. We called 277,128 chromatin accessibility peaks on autosomes from the aggregated data (Supplementary Note, Section 1), from which we map chromatin accessibility QTLs (caQTLs). We also performed an additional ATAC-seq experiment in GM12878 that was not used for QTL mapping, but was used to assess genotypic effects on the reference epigenome.
Sequencing data preprocessing
All sequence data sets were aligned to human genome assembly GRCh37. We performed adapter trimming for our ATAC-seq data using skewer44 (version 0.1.127; see URLs) before alignment. FASTQ files of GEUVADIS RNA-seq data41 (N=372) were downloaded from ArrayExpress (Accession E-GEUV-3) and ChIP-seq data for CTCF binding45 (N=50) were downloaded from the European Nucleotide Archive (Accession ERP002168). Our ATAC-seq data and the CTCF ChIP-seq data were aligned using bwa 0.7.446. RNA-seq data were aligned using Bowtie247 (version 2.2.4; see URLs) and reads mapped to splice junctions using TopHat248 (version 2.0.13; see URLs), using ENSEMBL human gene assembly 69 as the reference transcriptome. Following alignment, we performed peak calling in the CTCF ChIP-seq and ATAC-seq data by pooling all samples. Fragment counts of ATAC-seq, CTCF ChIP-seq and RNA-seq for each feature (a called peak or an union of exons for each gene) were normalised into FPKMs using length referred to the peak length in kilobases. Batch effects were adjusted by GC contents and principal components. See Section 2.1-2.5 of Supplementary Note for more detail.
SNP genotype data
We downloaded VCF files from the 1000 Genomes Phase III integrated variant set from the project website. For the ATAC-seq, RNA-seq and CTCF ChIP-seq samples that did not overlap with the 1000 Genomes Phase III samples, we extracted genotype data from the 1000 Genomes Phase I data or 1000 Genomes high density SNP chip data (performed on the Illumina Omni platform). We then performed whole genome imputation for the extracted genotype data by using the Beagle software49 (version 4.0; see URLs). See Section 2.6 in Supplementary Note for details.
Genomic annotations
To compute ATAC-seq peak height, we pooled ATAC-seq data for the 100 samples. The peak height was defined as the highest value of the coverage depth within each peak region. Peak height was quantile normalised across all peaks. The relative coverage at each variant location (VL) was calculated by the absolute coverage depth divided by the peak height inside the peak. This value was used as the VL prior probability for both caQTL mapping and eQTL mapping. Peak distance was calculated based on the midpoint of a peak region.
We also used various external genomic annotations for comparison. The Hi-C contact map and Hi-C loops for GM12878 were obtained from Rao et al. (2004)10. TAD boundaries were defined as the anchor regions of a Hi-C loop. Capture Hi-C data for GM12878 was obtained from Cairns et al. (2016)13 and CHiCAGO13 (version 1.1.8; see URLs) was used to extract CHi-C interactions with CHiCAGO score > 1. The H3K27ac HiChIP data for GM12878 was obtained from Mumback et al. (2017)12. The JuiceBox output was processed by HiCCUPS50 implemented in the Juicer Tools (version 0.7.5; see URLs) with default parameter setting to obtain the HiChIP loops. The integrated genomic segmentation annotation28 combining Segway51 and ChromHMM52 results was downloaded from ENCODE Project27 website (URLs). Each ATAC peak was labelled by one of the 7 different segmentation categories at the peak midpoint. See Section 2.7 in Supplementary Note for details.
Roadmap Epigenomics Project data analysis
We downloaded all DNaseI-seq data from 53 cell types from the project web page (URLs). We counted the number of reads that mapped to the 277,128 annotated peaks from our ATAC-seq data. This count matrix was normalized in the same way as our ATAC-seq data (Section 2.3-2.4, Supplementary Note). We computed Spearman’s rank correlation between all peak pairs within 500Kb distance of one another.
Pairwise hierarchical model
There are three key features of the model. First, support for the hypothesis of a causal relationship between two peaks is computed using two stage least squares22 (2SLS). Second, we use a hierarchical model23 in which prior probabilities depend on genomic annotations at multiple model levels. Third, the model is empirical, such that the prior probabilities are learned as the penalised likelihood is maximised across all peak pairs simultaneously.
The pairwise hierarchical model is a product of finite mixture probabilities over all j-k peak pairs in 500Kb (1 ≤ j < k ≤ J; J = 277,128). The finite mixture model comprises the regional Bayes factor to observe chromatin accessibility yj and yk at peak j and k across 100 samples under the different interaction hypotheses h (Fig. 1A). The pairwise likelihood is given by
where Φ(0) denotes the mixture probability that j-k peak pair is no caQTLs, Φ(h) denotes the mixture probability for the alternative hypothesis h and H1 is the set of alternative hypotheses, so that Φ(0) + ∑h∈H1 Φ(h) = 1. RBF is obtained from the joint regression model p(yj, yk|h) which comprises two independent regression models that also depend on the hypothesis h. For the causality hypotheses (H4.1 for the causal interaction from peak j to k and H4.2 for peak k to j), we used 2SLS to estimate the causal effect between peaks with each genetic variant in the cis window as the instrumental variable (Fig. 1C).
We note that our model is not strictly Bayesian because we do not perform any Bayesian inference on the model parameters. Instead, to reduce the computational complexity, we employed a two-stage optimisation of the likelihood using the EM algorithm. In the first stage we estimated hyperparameters for the variant-level and peak-level prior probabilities. We used the standard hierarchical model23 to learn these prior probabilities by temporarily assuming peaks are independent. In the second stage, we estimated hyperparameters in the peak-pair-level prior regarding Φ(h). We used the Expectation-Maximisation algorithm to iteratively estimate hyperparameters while updating the following posterior probabilities
in the E-step. Because all model distributions belong to exponential family, we can utilise the penalised iteratively reweighted least square (P-IRLS) method53 in the M-step, which does not require calculation of the gradient and Hessian of the log likelihood. All subsequent analyses were performed based on the posterior probabilities without any threshold. Note that the posterior probability of causality (PPC) is denoted by PPCjk and PPCkj (corresponding to and respectively) in the main text. Mathematical rationale and implementation of the pairwise hierarchical model are fully described in Section 3.1-3.5 of Supplementary Note. A software package (“PHM”) that computes the BFs, RBFs and maximises the pairwise likelihood is available from GitHub (URLs).
Mapping multi-way interactions
Multi-way interactions were also constructed from PPCjk and PPCkj by finding a DAG among more than 2 peaks. We first used only confident causal interactions with PPCjk>0.5, then found the most likely parent for each peak, and finally solve the cyclic graphs by discarding an interaction with the lowest PPCjk. See Section 3.6 of Supplementary Note for details.
Detection of lead caQTL variant
Within each cis-regulatory window (500Kb on either side of a peak), we calculated a posterior probability of each variant being the causal caQTL and obtained the maximum a posteriori variant as the lead variant. We used the pairwise likelihood to solve the problem that multiple caQTL variants are associated with chromatin accessibility due to strong linkage disequilibrium. The central assumption here is that variants predicted by our model to be upstream in the regulatory cascade are more likely to be causal. See Section 3.7 in Supplementary Note for details.
Effect size calculation
For j-k peak pairs, we identified single lead variants under the hypothesis of causality or pleiotropy and two causal variants for the linkage hypothesis based on the variant-level posterior probability (see Section 3.8, Supplementary Note for more detail). We computed effect sizes of the lead variant(s) against the two peaks (j and k) using simple linear regression. Under the linkage hypothesis, if the genotypes of the two causal variants were negatively correlated (LD index r<0), the effect size of peak k was multiplied by -1 to align the effect direction.
Probability of Master regulator
We defined the master regulatory peak as a peak with more than one interacting downstream peak and no interacting upstream peaks. We computed the product of the following two posterior probabilities: the probability that the peak regulates at least one other peak in the cis-window, and the probability the peak is not regulated by any other peak within the cis-window, which we referred to as the probability of being master regulator (PMR). See Section 3.9 of Supplementary Note for details.
Hierarchical model for eQTL fine-mapping
The standard hierarchical model23 was applied to the gEUVADIS RNA-seq data (372 European samples) with various combinations of the following five annotations: (1) inside or outside an ATAC peak (referred to as ATAC); (2) eQTL variant location, relative to an ATAC peak (referred to as VL); (3) promoter capture Hi-C contacts (CHi-C); (4) HiChIP loops from baited promoter regions (HiChIP); and (5) PMR value at each ATAC peak. The variant-level prior was learned and the posterior probability of association (PPA) was calculated for each variant in 1Mb cis-window centred at TSS. For the eQTL fine-mapping of BLK/FAM167A locus, we incorporated all the genomic annotations used in the caQTL mapping in conjunction with the colocalisation probability of caQTL and eQTL as the weight of the prior probability. See Section 3.10 of Supplementary Note for details.
Colocalisation with expression QTLs
The pairwise hierarchical model can be utilised to colocalise caQTLs with other cellular QTLs, such as expression QTLs (eQTLs). The reduced model without causality hypothesis (H4.1 and H4.2) was applied to colocalise caQTL-eQTL as well as CTCF binding QTL-caQTL. We assumed a non-informative prior probability for the three different levels of hierarchy and estimated the posterior probability of pleiotropy between caQTL and eQTL/CTCF binding QTLs as the colocalisation signal. Joint colocalisation probability between eQTL and a peak pair is also calculated from the result. See Section 3.11 of Supplementary Note for details.
Colocalisation with GWAS summary data
We downloaded the following 10 GWAS summary statistics (see Section 2.9 of Supplementary Note for details): Rheumatoid arthritis (RA), schizophrenia (SCZ), systemic lupus erythematosus (SLE), Crohn’s disease (CD), ulcerative colitis (UC), inflammatory bowel diseases (IBD), type 2 diabetes (T2D), Alzheimer’s disease (AD), atopic dermatitis (ATD) and coronary artery disease (CAD). The asymptotic Bayes factors were calculated and colocalised with caQTLs using the same model as was used in colocalisation with eQTLs. Posterior probability of each caQTL peak colocalised with a GWAS trait was calculated and used for the subsequent enrichment analysis. See Section 3.12 in the Supplementary Note for details.
Allele-specific accessible chromatin
We used the lead caQTL variant for each peak identified by PHM as the putative causal variant. We confirmed allelic imbalance (AI) at feature SNPs inside the downstream peak in the confident set of 15,487 causal interactions. If there was a true causal interaction, allelic imbalance is observed for individuals who are heterozygous for the lead variant. To assess statistical significance of AI we used RASQUAL (URLs) with the “—as-only” option to map caQTLs using allele specific counts at feature SNPs.
Overlap of lead SNPs with TFBS
In the high confidence set of 15,487 mapped causal interactions, we detected the lead variant for each downstream peak using the HM and PHM (Supplementary Note Section 3.8) and selected 1,577 downstream peaks where lead SNP differed between the two models, excluding any peak where the lead variant was an INDEL or CNV). Then we generated the ATAC-seq cleavage (Transposase cut site) around the lead SNPs (30bp on either side).
To investigate motif disruption of the lead variants, we downloaded the 3,059 motifs from CISBP54 (version 1.02; see URLs). Within each chromatin accessibility peak, we generated all possible personal genome sequences using phased haplotypes of SNPs and INDELs for our 100 samples. We computed the position weight matrix (PWM) score for each motif and the posterior probability of transcription factor (TF) binding as follows:
where PWM1 denotes the PWM score for the motif given a part of sequence within the peak and PWM0 denotes the PWM score with background probability (0.25 for each nucleotide). We set the prior probability of TF binding as 0.001 for any TF, and defined a TF as bound if p(TF binding|sequence) was greater than 0.5.
Enrichment analysis with posterior probability of causal interaction
Any enrichment analysis was carried out based on PPC for all j-k peak pairs. We compute a 2 × 2 table of a binary annotation Xjk (e.g., if j-k peak pair within TAD then Xjk = 1 otherwise 0) and the existence of causality between j-k peak, such that
where (PPC for peak j and k). We compute the odds ratio from the table T to perform hypothesis testing. See Section 3.13 of Supplementary Note for details.
Simulation strategy
We simulated 17,349,412 peak pairs under each of the 4 hypotheses: causality (j→k), causality (k→j), linkage and pleiotropy. To simulate realistic linkage disequilibrium, we used real genotype data of 100 samples. To simulate a caQTL at peak j, a causal variant was chosen at random, weighted by the estimated variant-level prior from the real data. The effect size and standard deviation of the error of the simulated causal variant were the same as the estimated effect size and standard deviation for that variant from the simple linear regression and 2SLS of the real data. Chromatin accessibility for each sample at peak j was then simulated as a draw from a Normal distribution, with mean set to the effect size times the genotype dose, and variance equal to the squared standard deviation. For the linkage hypothesis, we repeated this procedure for peak k. For the pleiotropy hypothesis, we generated chromatin accessibility at peak k with the same causal variant at peak j. For causality from peak j to k, we used the 2SLS estimator of effect size and standard deviation to generate chromatin accessibility at peak k.
We also assessed two other scenarios where our model assumptions are potentially violated. First, generated chromatin accessibility with two causal variants for the focal peak under causality or pleiotropy, or four causal variants (two in each peak) under linkage. In addition, we simulated hybrid hypotheses where combinations of linkage, pleiotropy and causality were considered. See Section 3.14 of Supplementary Note for more detail.
To compare PHM to MR Steiger, we fit both models (PHM and MR Steiger) to the simulated data under the 4 different hypotheses (causality (j→k), causality (k→j), linkage and pleiotropy), and tested their ability to distinguish the causality (j→k) peak pairs from each of the 3 other scenarios in turn. A positive call set was defined if the PPCjk > T1 for PHM (where T1 was a variable threshold), or for PMR and PSteiger both < T2 and Steiger Z statistics > 0 for MR Steiger (where T2 was also a variable threshold). Importantly, in these tests, the MR Steiger model was given the correct causal variant, while PHM attempted to infer the putative causal variant from the data.
Comparison with massively parallel reporter assay (MPRA)
We downloaded the table of combined LCL analysis for all 39,478 variants with MPRA29. We extracted 842 variants that showed significant allelic imbalance according to the criteria applied in the paper (referred to as expression-modulating variant; emVar). We then selected the lead eQTL variant for each gene based on the eQTL PPA and asked how many overlapped the validated emVar. When there were ties in PPA for the lead eQTL variants, we randomly selected one variant.
Knock-out of BLK-FAM167A locus (rs558245864:C>CG) using CRISPR/Cas9
The lymphoblastoid cell line HG00146, which is homozygous for the reference rs558245864 allele, was nucleofected with an enhanced Cas9-2a-GFP plasmid and a guide RNA expression plasmid targeting the rs558245864 locus. Deletion clones were selected, expanded and then subjected to ATAC-seq and RNA-seq. Methods for engineering of the rs558245864 locus are described in full in the Section 4.1-4.6 of Supplementary Note.
Differential chromatin accessibility and expression analyses
We used DESeq55 (see URLs) to perform differential chromatin accessibility and differential expression analyses. We compared the two replicates of the parental line against the four replicates of the deletion lines (two replicates for D1 and D2 heterozygous lines, respectively). See Section 4.7 of Supplementary Note for more detail.
Supplementary Material
Acknowledgements
We thank Christopher Brown and two anonymous reviewers for their helpful comments. N.K., A.J.K. and D.J.G. were funded by Wellcome Trust grant WT206194.
Accession Codes
ATAC-seq data in 100 LCLs (European Nucleotide Archive: ERP110508).
GEUVADIS RNA-seq data (ArrayExpress: E-GEUV-3)
CTCF ChIP-seq data (European Nucleotide Archive: ERP002168).
Data Availability
ATAC-seq data for the 100 LCLs used in this study are available from European Nucleotide Archive (Accession ID: ERP110508). All summary statistics are available from Zenodo (DOI: 10.5281/zenodo.1405945).
Code Availability
The Pairwise Hierarchical Model is available from GitHub (https://github.com/natsuhiko/PHM).
Author contributions
D.J.G. and N.K. conceived and designed the experiments. A.J.K. performed the experiments. N.K. performed statistical analysis and analysed the data. N.K. and A.J.K. contributed reagents, materials and analysis tools. D.J.G., N.K. and A.J.K. wrote the manuscript.
Competing Interests Statement
The authors declare no competing financial interests.
URLs
Pairwise Hierarchical Model (https://github.com/natsuhiko/PHM)
RASQUAL (https://github.com/natsuhiko/rasqual)
1000 Genomes Phase III integrated variant set (http://www.internationalgenome.org/data)
Combined Segway and ChromHMM results from ENCODE Project website (http://hgdownload.soe.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeAwgSegmentation/)
Roadmap Epigenomics Project (http://www.roadmapepigenomics.org/)
CHiCAGO 1.1.8 (http://regulatorygenomicsgroup.org/chicago)
Juicer Tools 0.7.5 (https://github.com/theaidenlab/juicer/wiki/Juicer-Tools-Quick-Start)
Beagle 4.0 (https://faculty.washington.edu/browning/beagle/b4_0.html)
DESeq (https://bioconductor.org/packages/release/bioc/html/DESeq.html)
bwa 0.7.4 (https://sourceforge.net/projects/bio-bwa/files/)
Bowtie2 2.2.4 (http://bowtie-bio.sourceforge.net/bowtie2/manual.shtml)
TopHat2 2.0.13 (http://ccb.jhu.edu/software/tophat/index.shtml)
skewer 0.1.127 (https://github.com/relipmoc/skewer)
References
- 1.Pombo A, Dillon N. Three-dimensional genome architecture: players and mechanisms. Nat Rev Mol Cell Biol. 2015;16:245–57. doi: 10.1038/nrm3965. [DOI] [PubMed] [Google Scholar]
- 2.Haarhuis JHI, et al. The Cohesin Release Factor WAPL Restricts Chromatin Loop Extension. Cell. 2017;169:693–707 e14. doi: 10.1016/j.cell.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fudenberg G, et al. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep. 2016;15:2038–49. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claussnitzer M, et al. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N Engl J Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smemo S, et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507:371–5. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denker A, de Laat W. The second decade of 3C technologies: detailed insights into nuclear organization. Genes Dev. 2016;30:1357–82. doi: 10.1101/gad.281964.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Wit E, de Laat W. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 2012;26:11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonev B, Cavalli G. Organization and function of the 3D genome. Nat Rev Genet. 2016;17:661–678. doi: 10.1038/nrg.2016.112. [DOI] [PubMed] [Google Scholar]
- 9.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rao SS, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–80. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mifsud B, et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet. 2015;47:598–606. doi: 10.1038/ng.3286. [DOI] [PubMed] [Google Scholar]
- 12.Mumbach MR, et al. Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nat Genet. 2017 doi: 10.1038/ng.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cairns J, et al. CHiCAGO: robust detection of DNA looping interactions in Capture Hi-C data. Genome Biol. 2016;17:127. doi: 10.1186/s13059-016-0992-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grubert F, et al. Genetic Control of Chromatin States in Humans Involves Local and Distal Chromosomal Interactions. Cell. 2015;162:1051–65. doi: 10.1016/j.cell.2015.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waszak SM, et al. Population Variation and Genetic Control of Modular Chromatin Architecture in Humans. Cell. 2015;162:1039–50. doi: 10.1016/j.cell.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 16.Kumasaka N, Knights AJ, Gaffney DJ. Fine-mapping cellular QTLs with RASQUAL and ATAC-seq. Nat Genet. 2016;48:206–13. doi: 10.1038/ng.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delaneau O, et al. Intra- and inter-chromosomal chromatin interactions mediate genetic effects on regulatory networks. bioRxiv. 2017 [Google Scholar]
- 18.Giambartolomei C, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voight BF, et al. Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. Lancet. 2012;380:572–80. doi: 10.1016/S0140-6736(12)60312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Do R, et al. Common variants associated with plasma triglycerides and risk for coronary artery disease. Nat Genet. 2013;45:1345–52. doi: 10.1038/ng.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Day FR, et al. Genomic analyses identify hundreds of variants associated with age at menarche and support a role for puberty timing in cancer risk. Nat Genet. 2017;49:834–841. doi: 10.1038/ng.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burgess S, Thompson SG. Mendelian randomization : methods for using genetic variants in causal estimation. Chapman & Hall/CRC interdisciplinary statics series 1. CRC Press, Taylor & Francis Group; Boca Raton, FL: 2015. online resource. [Google Scholar]
- 23.Veyrieras JB, et al. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4:e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ignatiadis N, Klaus B, Zaugg JB, Huber W. Data-driven hypothesis weighting increases detection power in genome-scale multiple testing. Nat Methods. 2016;13:577–80. doi: 10.1038/nmeth.3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wakefield J. A Bayesian measure of the probability of false discovery in genetic epidemiology studies. Am J Hum Genet. 2007;81:208–27. doi: 10.1086/519024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hemani G, Tilling K, Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017;13:e1007081. doi: 10.1371/journal.pgen.1007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Encode Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoffman MM, et al. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 2013;41:827–41. doi: 10.1093/nar/gks1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tewhey R, et al. Direct Identification of Hundreds of Expression-Modulating Variants using a Multiplexed Reporter Assay. Cell. 2016;165:1519–1529. doi: 10.1016/j.cell.2016.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okada Y, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–81. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bentham J, et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat Genet. 2015;47:1457–1464. doi: 10.1038/ng.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu JZ, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lambert JC, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet. 2013;45:1452–8. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paternoster L, et al. Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat Genet. 2015;47:1449–1456. doi: 10.1038/ng.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scott RA, et al. An Expanded Genome-Wide Association Study of Type 2 Diabetes in Europeans. Diabetes. 2017;66:2888–2902. doi: 10.2337/db16-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nikpay M, et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guthridge JM, et al. Two functional lupus-associated BLK promoter variants control cell-type- and developmental-stage-specific transcription. Am J Hum Genet. 2014;94:586–98. doi: 10.1016/j.ajhg.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.GTEx Consortium et al. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Battle A, et al. Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals. Genome Res. 2014;24:14–24. doi: 10.1101/gr.155192.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lappalainen T, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–11. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shin HY, et al. Hierarchy within the mammary STAT5-driven Wap super-enhancer. Nat Genet. 2016;48:904–911. doi: 10.1038/ng.3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen L, et al. Genetic Drivers of Epigenetic and Transcriptional Variation in Human Immune Cells. Cell. 2016;167:1398–1414 e24. doi: 10.1016/j.cell.2016.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang H, Lei R, Ding SW, Zhu S. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics. 2014;15:182. doi: 10.1186/1471-2105-15-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ding Z, et al. Quantitative genetics of CTCF binding reveal local sequence effects and different modes of X-chromosome association. PLoS Genet. 2014;10:e1004798. doi: 10.1371/journal.pgen.1004798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Browning BL, Browning SR. Genotype Imputation with Millions of Reference Samples. Am J Hum Genet. 2016;98:116–26. doi: 10.1016/j.ajhg.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Durand NC, et al. Juicer Provides a One-Click System for Analyzing Loop-Resolution Hi-C Experiments. Cell Syst. 2016;3:95–8. doi: 10.1016/j.cels.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoffman MM, et al. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat Methods. 2012;9:473–6. doi: 10.1038/nmeth.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–6. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wood SN. Generalized additive models : an introduction with R. Chapman & Hall/CRC; Boca Raton, Fla.; London: 2006. p. 391. xvii. [Google Scholar]
- 54.Weirauch MT, et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014;158:1431–1443. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.