Abstract
Acute myeloid leukemia (AML) is a heterogeneous disease caused by a variety of mutations in transcription factors, epigenetic regulators and signaling molecules. To determine how different mutant regulators establish AML subtype-specific transcriptional networks we performed a comprehensive global analysis of cis-regulatory element activity and interaction, transcription factor occupancy and gene expression patterns in purified leukemic blast cells. Here, we focussed on specific sub-groups of patients carrying mutations in genes encoding transcription factors (RUNX1, CEBPA) and signaling molecules (FTL3-ITD, RAS, NPM1). Integrated analyses of these data demonstrates that each mutant regulator establishes a specific transcriptional and signaling network unrelated to that seen in normal cells, sustaining the expression of unique sets of genes required for AML growth and maintenance.
Acute myeloid leukemia (AML) is characterized by blocked myeloid lineage differentiation and accumulation of leukemic blast cells. AML is a highly heterogeneous disease caused by different types of mutations affecting signaling pathways as well as transcriptional and epigenetic regulators1–3. Recurrent mutations include loss of function mutations in transcription factors (TFs) controlling hematopoietic development, such as RUNX1, GATA2 or C/EBPα4, and gain of function mutations in signaling molecules such as FLT3, KIT, JAK2 and NRAS regulating inducible TFs such as NF-κB, STAT or AP-1 family members5,6,7. The most common FLT3 mutations are internal tandem duplications (FLT3-ITD), which give rise to a constitutively active growth factor receptor8,9 and often occur together with nucleophosmin1 mutations (NPM1). Another major group of mutations alters genes encoding epigenetic and chromatin regulators10,11 which play widespread roles in development and differentiation by controlling establishment, maintenance and extinction of lineage-specific gene expression programs. These include regulators of histone and DNA methylation such as MLL, EZH2, TET2, DNMT3A and IDH1/211–17. In normal cells, all common mutation targets cooperate to control the finely balanced gene expression changes essential for cell differentiation and lineage commitment.
TFs interact with defined target gene sequences and recruit epigenetic regulators to program specific chromatin states and mediate the coordinated activation or de-activation of cis-regulatory elements driving gene expression18,19. Distal cis-regulatory elements interact directly with promoter elements, an arrangement that is both dynamic and robust20,21. From global studies examining a few selected types of AML we know that gene expression patterns and the epigenetic landscape differ from normal cells22–27. However, how the disruption of specific TF activity leads to a specific pattern of aberrant chromatin programming and changes in gene expression in AML remained unclear. It was not established at the global level which cis-regulatory elements are affected in their activity in different types of AML, how their activity is altered in patients carrying specific TF and signaling mutations or which factors maintain their transcriptional networks.
Here we addressed these questions by collecting transcriptome, digital footprinting and chromatin conformation capture data from purified leukemic blasts from AML patients with defined transcription factor and signaling molecule mutations and defined the components of AML subtype-specific regulatory circuitries. Our study comprises a comprehensive resource of the transcriptional networks of different AML subtypes, highlighting pathways required for tumour maintenance
Results
AML subtypes adopt unique chromatin landscapes
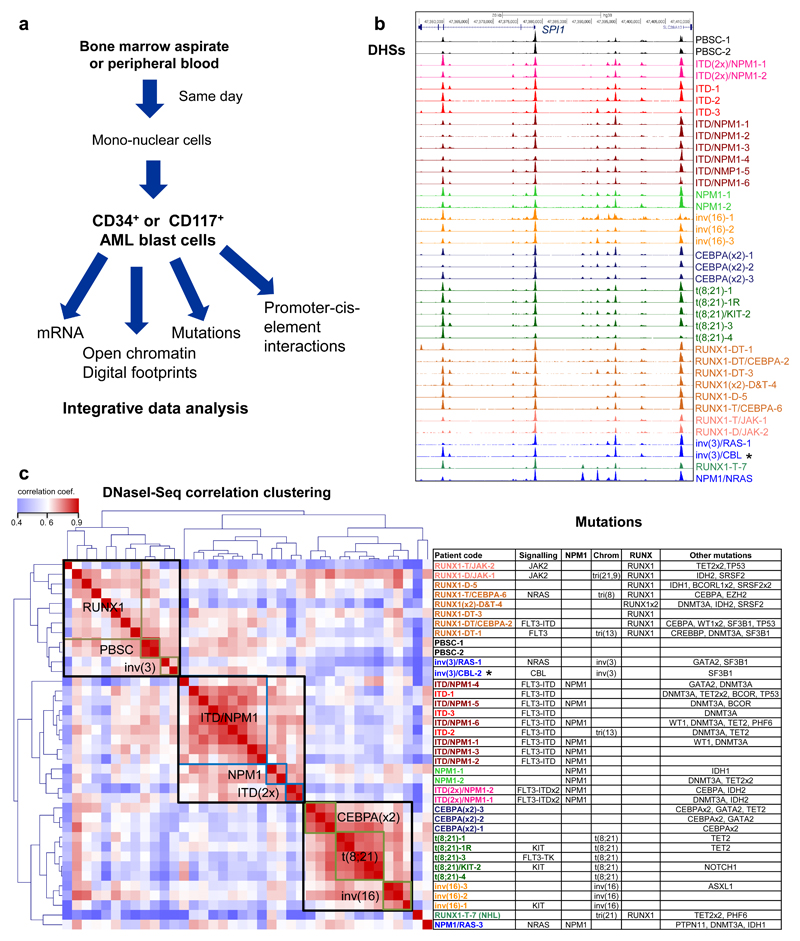
In order to examine how specific TF and signaling mutations alter the epigenome of AML, we purified leukemic blast cells from bone marrow or peripheral blood samples from AML patients (Fig. 1a). After determining the mutation status (Table S1), we selected a cohort of patients with defined mutations, which included: RUNX1 mutations affecting DNA-binding (D-type) or lacking the trans-activating domain (T-Type), t(8;21) translocations fusing the DNA binding domain of RUNX1 to the co-repressor ETO, inv(16) which fuses CBFβ to smooth muscle myosin heavy chain 11 protein, mutations of both alleles of the CEBPA gene whereby one mutation leads to loss of DNA-binding activity28 and FLT3-ITD with or without NPM1 mutations. We performed RNA-Seq (Supplementary Fig. 1a) and high read depth DNaseI-Seq (Fig. 1b) to map DNaseI hypersensitive sites (DHSs) on 29 samples comprising seven major groups, and at least one analysis on 12 additional samples, including mutations such as NRAS, CBL, JAK2, SRFS2, or inv(3) (Supplementary Table 1). One patient with Non-Hodgkin Lymphoma (NHL) carried a RUNX1 mutation (RUNX1-T-7). Samples were compared to CD34+ mobilized peripheral blood stem cells (PBSCs) from two healthy individuals and to cord blood (CB) CD34+ cells. To provide the community with a data resource, we established an online database containing multiple data-sets including a genome browser (see Data Availability).
Figure 1. Different types of AML adopt unique transcriptome and chromatin landscapes.
(a) Experimental strategy. (b) UCSC Genome browser tracks of DNaseI-seq mapping in purified AML cells. (c) Hierarchical clustering of Pearson correlation coefficients of DNaseI accessible sequences from all patient samples with normalized read counts of DNaseI-Seq data for the different classes of mutations (left panel), right panel: list of mutations in cells from each patient
Unsupervised clustering revealed that distal DHSs formed different groups according to their mutations class (Fig. 1c). Samples with FLT3-ITD and/or NPM1 mutations represented one major group with sub-clusters for patients with NPM1 mutations or carrying two FLT3-ITD alleles, but excluding a FLT3-ITD patient carrying a RUNX1 mutation. DHSs from the t(8;21), inv(16) and CEBPA double mutant patients clustered as discrete groups within a larger group, indicating that these mutations affect similar pathways. Examples of these patterns can be seen in Supplementary Fig. S1b. DHSs from patients with RUNX1 mutations were more heterogeneous and clustered with the PBSCs and the inv(3) patients. The NHL (RUNX1-T-7) and the NPM1/RAS-3 patterns were unrelated to any of the others. We further validated our findings by analysing an independently derived published ATAC-Seq data-set25, confirming that mutations in FLT3 underpin one major component of the clustering (Supplementary Fig. 2a). In contrast, the presence or absence of epigenetic mutations such as DNMT3A did not influence chromatin accessibility levels (Supplementary Fig. 2b) or gene expression (data not shown). Our mutation analyses showed no indications for the presence of confounding major sub-clones in purified undifferentiated AML cell populations, as mutations were present at close to either a 50% or a 100% allele frequency (Supplementary Dataset 1).
Unsupervised clustering analysis of RNA-Seq data from the same patients as well as direct comparisons between individual samples (Supplementary Figs. 1c, 2c) revealed strong correlations between mutation-specific chromatin landscapes and mutation-specific differential gene expression as exemplified by POU4F1 and FOXC1 where mRNA patterns correlated well with chromatin profiles (Supplementary Fig. 1d). We identified distinct patterns of expression for specific TF-encoding genes in different AML types (Supplementary Fig. 2d). For example, various homeo-domain gene family members (HOX, NKX, IRX and PBX families) were up-regulated in the FLT3-ITD and NPM1-mutated patients. Our comparative analyses show that aberrant TFs and chronic signaling impose distinct mutation group-specific patterns of chromatin accessibility and gene expression, irrespective of the presence of other classes of mutations.
AML-specific clustering of distal cis-regulatory elements
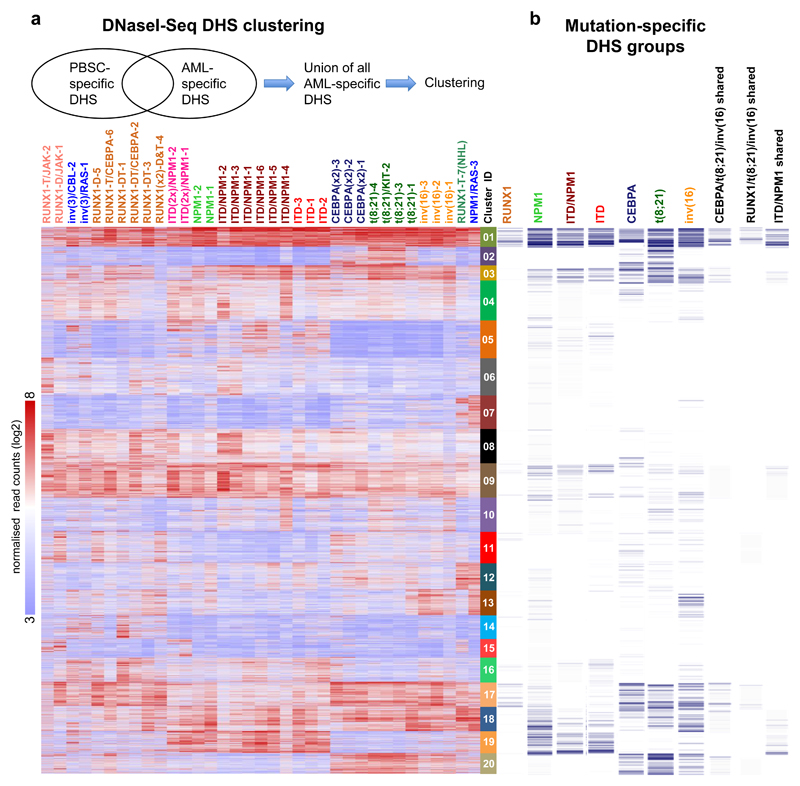
We next examined the active cis-regulatory elements specific for each AML subtype by defining the union of all AML-specific DHSs as compared to CD34+ PBSCs and performing k-mean clustering to identify unique and common DHSs shared between patients, which identified 20 distinct DHS clusters (Fig. 2a). Less than half of these DHSs were found in any of the Corces et al. progenitor data ATAC-Seq sets25 and the percentage overlap varied substantially between clusters (range 2 – 40%; b). We verified mutation-specific clustering behaviour of our samples by comparing them with a recently published AML histone H3K27 acetylation data set26 showing similar patterns for FLT3-ITD, RUNX1 and CEBPA double mutations (Supplementary Fig. 3d). We also defined mutation specific groups of deregulated DHSs that were shared between the specific members of each of the seven major mutation groups defined in Table S1 (Supplementary Figs. 4a and 4b) which were distributed between both the mutation-specific clusters and the shared clusters (Fig. 2b) and were associated with differentially expressed genes (Dataset S2). Again, the t(8;21), inv(16), and CEBPA groups showed similar patterns whereby 914 upregulated DHSs were shared between the three groups (Supplementary Fig. 4a). The FLT3-ITD, FLT3- ITD/NPM1 and NPM1 mutation groups showed substantial overlap with 942 shared DHSs, and with only 19% of these DHSs included in the 914 ITD/NPM1-specific group. These AML-specific patterns showed little similarity to normal myeloid differentiation as the majority of these specific sites were not up-regulated in GMPs relative to PBSCs (Fig. SN2b).
Figure 2. AML-specifically active cis-regulatory elements cluster into common and unique chromatin landscapes.
(a) Heatmap depicting unsupervised K-mean clustering of the DNaseI-Seq log2 signals seen in each AML specific distal DHS peak in each AML sample compared to PBSCs. Clustering was done only on rows (DHS peaks) while samples were ranked based on the clustering in Fig, 1c. The diagram on top of the heatmap shows the DHS peak population used for clustering. (b) Binary heatmap showing the overlap between the clusters from A and the DHSs of the 7 mutation classes which are deregulated compared to CD34+ PBSCs as described in Supplementary Fig 4a.
The presence of specific DHSs strongly correlated with the up-regulation of their associated genes (Supplementary Dataset 3, Supplementary Fig.4c), as exemplified by two DHS at POU4F1 (Supplementary Fig 4d). Supplementary Fig 4e shows examples of AML type-specific up-regulated TF genes, and growth factor or receptor genes, which were associated with AML type-specific DHSs (Supplementary Data 4). The gene expression patterns of such genes were validated using publicly available data-sets (Supplementary Fig 5).
AML-specific transcription factor binding patterns
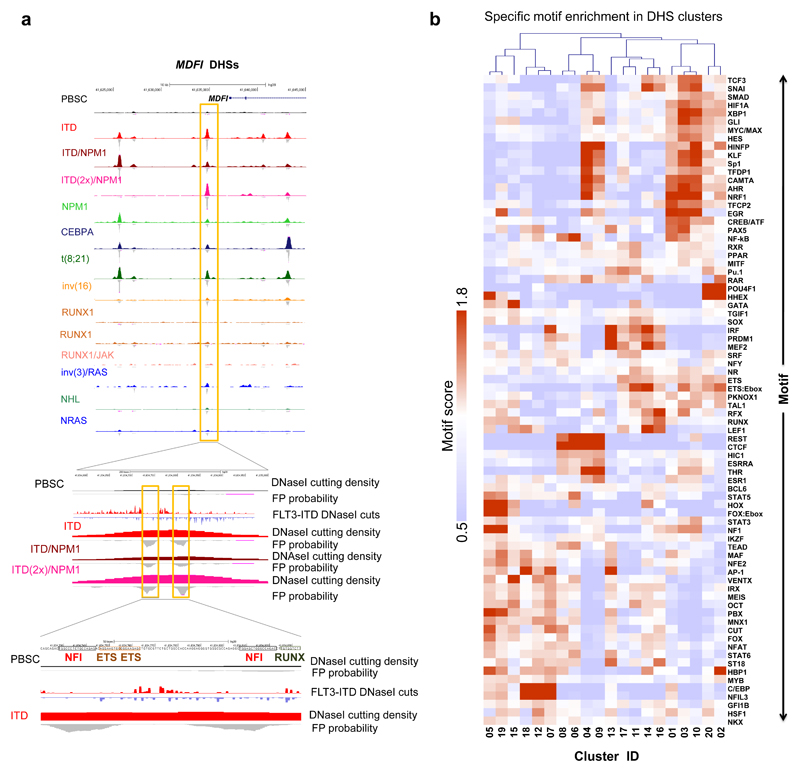
To identify TFs associated with the different chromatin patterns we analysed high-read depth DNaseI-Seq data using our Wellington digital footprinting algorithm29. Since closely related factors recognise identical sequences, compiling a non-redundant database of motifs (Supplementary table 2) and selected representative motifs encompassing each transcription factor family as defined in more detail on our web server (see URL list). Examples of footprints are depicted for NFI and ETS motifs at the MDFI locus in FLT3-ITD/NPM1-mutated AML (Fig. 3a) and for RUNX, NFAT and C/EBP motifs at the C3AE1 locus in t(8;21) and CEBPA-mutated AML (Supplementary Fig. 6a). The majority of AML type-specific DHSs within the 20 AML-specific DHS clusters contained footprints (Supplementary Fig. 6b). For validation, we compared RUNX motif footprints with publicly available RUNX1 ChIP data from our studies (FLT3-ITD/NPM124, t(8;21)30) and others (inv(16)31) (Supplementary Fig. 6c). Between 60% and 85% of footprinted RUNX motifs occurred in regions shown to bind RUNX1.
Figure 3. AML-specifically active cis-regulatory elements display AML type-specific transcription factor occupancy patterns.
(a) UCSC browser screen shot of the MDFI locus zooming in on an AML type-specific DHS (box) with occupied NFI, ETS and RUNX sites. (b) Heatmap depicting the degree of motif enrichment after hierarchical clustering of motif occupancy in each of the 20 AML DHS clusters. Enrichment score was calculated by the level of motif enrichment in all the footprints of all high read-depth samples for each cluster, as compared to union of footprints in all experiments.
We next evaluated occupied motif enrichment in the 20 AML subtype-specific DHS clusters (Fig. 3b) which showed that motif occupancy patterns are highly AML type-specific. For example, the FLT3-ITD/NPM1-specific clusters 5 and 19 are enriched for occupied HOX, FOX/E-box and NFI motifs and correlate with up-regulation of FOXC1, NFIX and multiple homeo-domain genes (Supplementary Fig. 2d). Occupied AP-1 motifs are enriched in multiple clusters (01, 05, 07, 12, 13, 18, 19), many of which in AML with signaling mutations. Because AP-1 factors mediate MAP kinase (MAPK) signaling this indicates wide-spread activation of this signaling pathway in FLT3-ITD AML24 and other AML types. Finally, we observed significant POU4F1 motif occupancy in clusters 02 and 20 containing samples from t(8;21) and CEBPA double mutant patients but nowhere else (Fig. 3b). POU4F1 is aberrantly expressed in t(8;21) cells32, but has so far not been linked to CEBPA double mutations. A similar differential occupancy pattern was seen when footprints were clustered according to mutation-specific groups of DHSs (Supplementary Fig. 6d). Note that motif occupancy of C/EBP motifs in AMLs with CEBPA double mutations showed no reduction in overall motif occupancy, suggesting compensation by other C/EBP family members.
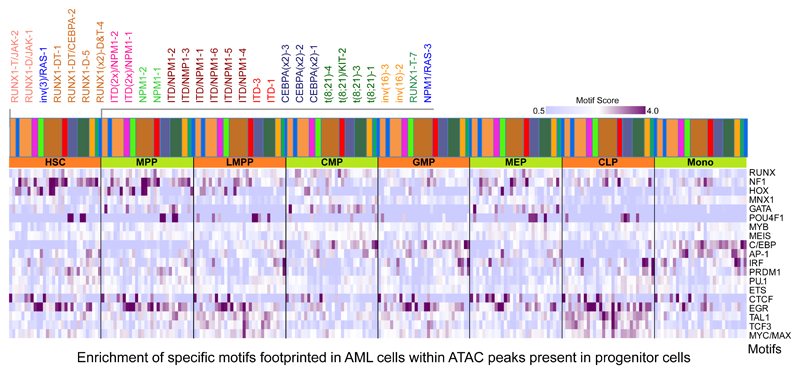
To examine the position of transcription factor occupancy patterns within the hematopoietic hierarchy, we correlated the presence of footprints specific for AML-subtypes with accessible chromatin regions present in precursor cells (Fig 4)25. This analysis revealed unique factor occupancy patterns in AML cells compared to normal progenitors. For example, HOX motifs within open chromatin regions observed in HSCs, MPPs and MEPs are occupied in the FLT3-ITD/NPM1 and RUNX1 groups, but not in the t(8;21) group, indicating an early block in differentiation (see also Fig SN1b). Many of the samples, including NPM1, FLT3-ITD/NPM1 and t(8;21) cells, display high AP-1 motif occupancy which is normally only seen in monocytes. POU4F1 is expressed in HSCs, MPPs, MEPs and in CLPs25 and its binding motifs are occupied in t(8;21) and CEBPA double mutant cells, yet these AML cells also show strong occupancy of C/EBP motifs, which is normally a hallmark of GMPs and monocytes. In summary, our digital footprinting analysis shows (i) that each AML subtype employs a different combination of factors binding to elements shared with different types of precursor cells and that (ii) lineage unrelated expressed TFs such as FOXC1, NFIX and POU4F1 participate in such cooperation.
Figure 4. AML cells show occupied motif patterns unrelated to normal progenitor cells.
Enrichment analysis of footprinted motifs in AML subgroups which overlap with ATAC-Seq peaks present in precursor cells25. The bars with the colour code above the heatmap reflect the type of mutation and the order of the different patients is depicted in the enlarged panel above the colour bars. Additional explanations are provided in Supplementary methods.
AML subtype-specific cis-element interactions
The construction of gene regulatory networks relies on linking cis-regulatory elements to their respective promoters33. We therefore examined (i) whether the differential activity of cis-regulatory elements in AML sub-types resulted in the formation of alternate cis-element interactions, and (ii) which TF families were involved in such interactions, by employing promoter-capture chromosomal structure analysis (CHi-C)34. We analysed cells from relapse patient sample t(8;21)-1R (Supplementary table 1), which maintained a gene regulation network similar to the presentation sample t(8;21)-1 (Figs. 1c, Supplementary Figs. 1c and 7a), and a patient carrying a FLT3-ITD/NPM1 mutation (ITD/NPM1-2, Supplementary Table 1). We compared these data to a dataset derived from human CD34+ cells34. Intra-chromosomal interactions did not differ at the global level (Supplementary Fig. 7b) and the organization into topologically associated domains (TADs) each containing the DHSs was unaffected by the type of AML (Supplementary Fig. 7c).
The proportion of DHSs involved in AML subtype-specific interactions varied between DHS-clusters (Supplementary Fig. 7d) and ~40% of all promoters showing differential interactions were associated with expressed genes (Supplementary Fig. 7e,f). A direct comparison between the CHi-C data from the two patients (Supplementary Fig. 7g) demonstrated that differential interactions (i) correlated with differential DHS patterns and that (ii) the expression of a differential set of genes with different GO terms (Supplementary Dataset 5) as exemplified by the KLF2 gene which is differentially expressed between FLT3-ITD and t(8;21) and CD34+ cells (Supplementary Fig. 7h).
On average 80% of all DHSs mapped in the t(8;21) AML, the FLT3-ITD AML and the CD34+ cells participated in interactions (Supplementary Fig. 8a). An average of 17% of interactions were AML type-specific and not present in CD34+ PBSCs (Supplementary Fig. 8b). To identify the TF families involved in regulating differential interactions we determined the proportions of enriched occupied motifs in the DHSs underlying interactions (Supplementary Fig. 8c). These analyses suggest that hematopoietic TFs such as RUNX, ETS and C/EBP family members and AML subtype-specifically expressed TFs participate in differential interactions in both AML types, together with the AP-1 factor family. In the FLT3-ITD AMLs this included HOX proteins and factors occupying FOX/E-box motifs. In the t(8;21) AMLs this included proteins binding to FOX and POU4F1 motifs.
Differential interactions drive AML-specific gene expression
The majority of DHSs underlying interactions between the three data-sets and those of individual patients were shared with an average level of 80% overlap (Supplementary Fig. 8d), confirming that the global transcriptional network of related cells is highly related35,36. Sub-type-specific DHSs participating in interactions were enriched within related groups, but not within unrelated groups (Supplementary Fig. 8e), confirming that the two patients were representative for those groups. For both the FLT3-ITD/NPM1 and the t(8;21) sample the nearest promoter accounted for 65-74% of AML type-specific interactions associated with genes that are up-regulated compared to CD34+ cells (Fig. 5a). Similar results were seen for each of the 20 DHS clusters (Supplementary Fig. 8f).
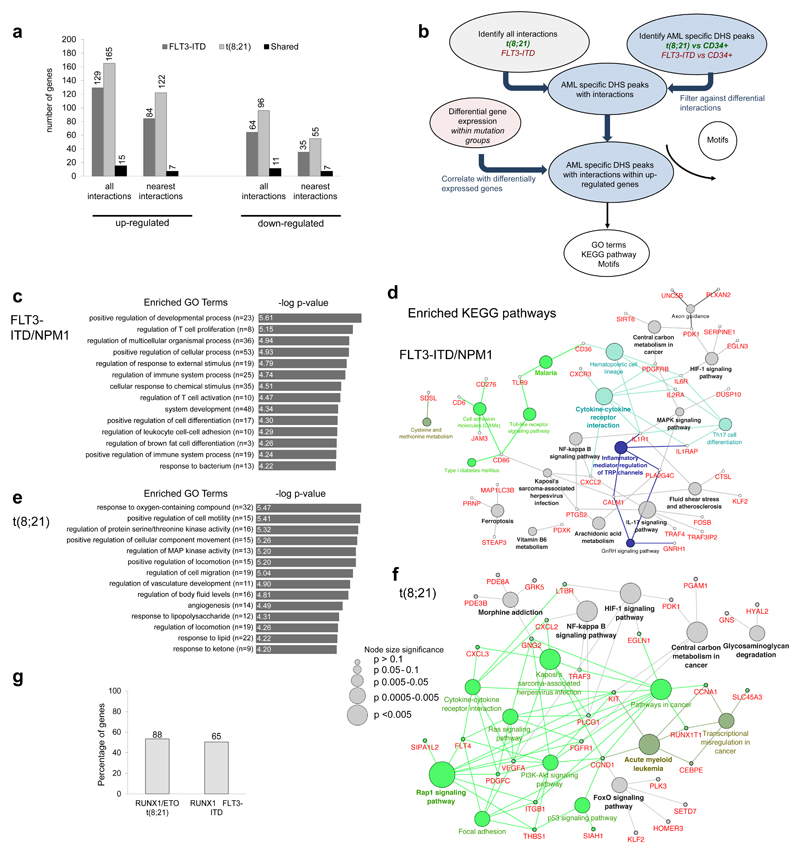
Figure 5. Capture HiC shows differences in locus-specific cis-regulatory interactions between different types of AML and normal cells.
(a) Percentage of up- and down-regulated genes with differential interactions from the FLT3-ITD and the t(8;21) compared to CD34+ PBSCs. The bar figure shows also the percentage of the common genes for the FLT3-ITD and the t(8;21), the number of differentially expressed genes (DEG) is shown on top of each bar. (b) Flow diagram showing the steps for identification of differential interactions and the downstream analysis. (c) Top enriched GO terms for the up-regulated genes of the FLT3-ITD compared to CD34+ PBSCs as outlined in (a). (d) Network diagram of top KEGG pathways for the up-regulated genes of the FLT3-ITD compared to CD34+ PBSCs as outlined in (a). (e) Top enriched GO terms for the up-regulated genes of the t(8;21) compared to to CD34+ PBSCs shown as outlined in (a) Network diagram of top KEGG pathways for the up-regulated genes of the t(8;21) compared to CD34+ PBSCs as outlined in (b). (h): Percentage of RUNX1-ETO and RUNX1 targets amongst up-regulated genes with differential interactions, the exact number of genes is shown above the bars.
To integrate differential interaction data, digital footprinting data and gene expression data, we assigned the respective DHSs to their interacting promoter as described in Fig 5b. GO-term and KEGG-pathway analyses of expressed genes in the two types of AML (Fig 5c-f) revealed an AML subtype-specific core signature of genes being driven by specific cis-regulatory elements (for an extended gene list see Supplementary Dataset 5). For both AML samples these included genes involved in regulating pro-inflammatory pathways. FLT3-ITD cells also displayed an activated MAPK signaling signature whereas the t(8;21) signature also included RAP, RAS, PI3K and FOXO signaling genes. FOXO1 is already known to be part of the t(8;21) pre-leukemic maintenance program37. More than 50% of all genes within these pathways were targets of RUNX1-ETO (Fig. 5g)30 thereby linking them to the actual driver mutation. A similar percentage of the genes within the FLT3-ITD/NPM1 core pathway are bound by RUNX1 (Fig. 5g) which is up-regulated in FLT3-ITD24 (Supplementary Dataset 5). Since ~83% of the DHSs involved in interactions in each of the 3 samples (Supplementary Fig. 8g) were shared, we merged all three CHi-C data-sets to use this data to assign the DHS from the 20 clusters (Fig 2a) to their respective promoters. The remaining 17% of DHS were assigned to the nearest promoter. GO-term and KEGG-pathway analyses of such genes (listed in Supplementary Dataset 6) again showed activation of genes connected with signaling processes, such as an inflammatory response, regulation of MAPK activity and cytokine regulation in all types of AML.
AML subtypes display unique transcription factor networks
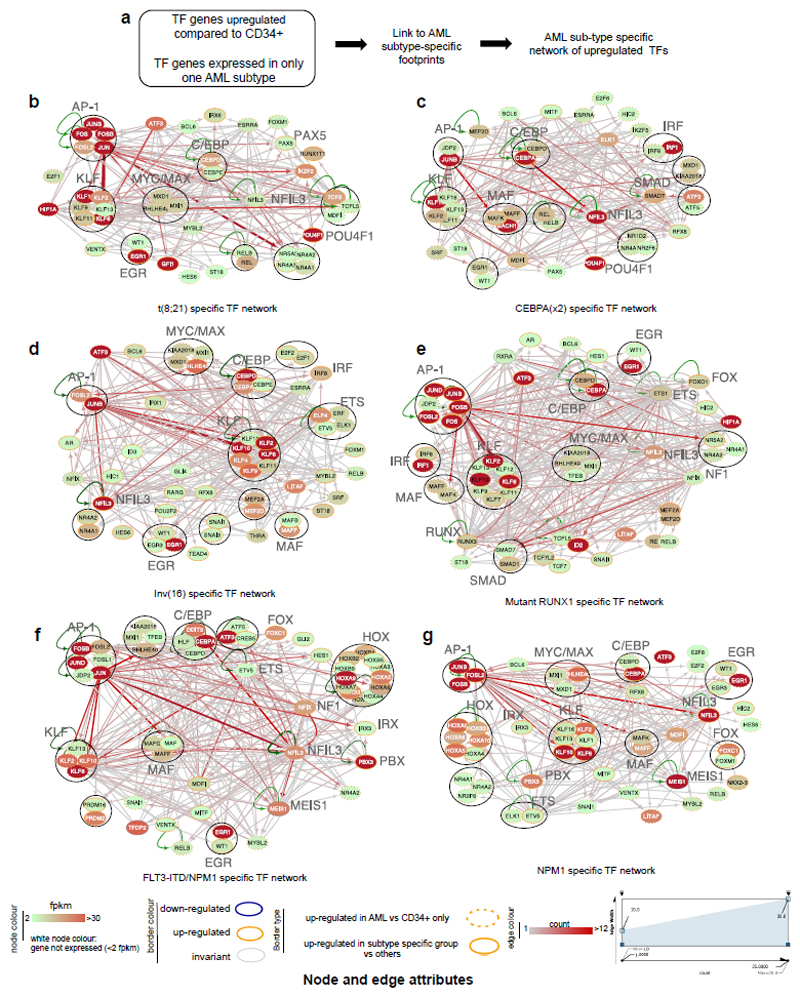
Constitutive and inducible TFs form regulatory networks by interacting with their own and/or other regulatory genes35. Cancer cells maintain a stable regulatory network, implying that the expression of each network member is tightly controlled and remains in balance. Consequently, perturbation of the network components maintaining this balance may destabilize leukemic cells thus offering novel therapeutic options. We therefore combined footprinting, TF gene expression and where possible, CHi-C data to construct TF networks in normal CD34+ cells and the different AML subtypes by linking occupied binding motifs within TF genes to the specific TF families recognising these motifs. The TF families involved and the full network structure for each cell type without filtering can be studied in detail via the weblink (see URL). Comparison between the different AML subtypes and normal CD34+ cells identified interactions between TF sub-sets that were either shared between AMLs and CD34+ cells (Supplementary Fig. 9) or were subtype-specific (Fig. 6), These results suggest that the AP-1 family network is of regulatory relevance for all AML subtypes (Fig. 6b-g). POU4F1 and HLH family factors recognising MYC/MAX type E-boxes formed prominent nodes in t(8;21) AML only, while HOX proteins, FOXC1, NFIX and the MAF family were exclusively highlighted in FLT3-ITD and NPM1 AML. Specific nodes and edges were also part of the normal precursor program (Supplementary Fig 9). For example the link between the C/EBP family and NFIL3 was shared between the FLT3-ITD/NPM1 cells (Fig. 6f) and CD34+ PBSCs (Supplemental Fig. S9f). A detailed discussion of the different network structures and the role of different TF families with more examples can be found in Supplemental Notes.
Figure 6. Identification of transcription factor networks driving the expression of AML type-specific up-regulated TF genes.
(a) Outline of analysis strategy. (b) t(8;21)-specific TF network, (c) CEBPA(x2)-specific TF, (d) Inv(16) specific TF network, (e) Mutant RUNX1-specific TF network, (f) FLT3-ITD/NPM1 specific TF network, (g) NPM1-specific TF network
Factor families binding to the same motif as shown in Supplementary table 2 form a node contained within a circle. Arrows going outwards from the entire node highlight footprinted motifs in individual genes generated by any member of this factor family whereby the footprint was annotated to the gene using the CHiC data where possible, otherwise to the nearest gene. For selected nodes, the name of the underlying motif is highlighted in large letters. The expression level (FKPM) for the individual genes is depicted in white (low)/red (high) colour. An orange smooth ring around the circle indicates that this gene is specifically up-regulated in this type of AML compared to CD34+ PBSCs and/or other AML types, a dotted circle indicates a gene that is up-regulated as compared to CD34+ cells. Genes without outgoing arrows due to a lack of know binding motifs are highlighted by their octagon shapes.
Transcription factors contributing to AML-specific growth
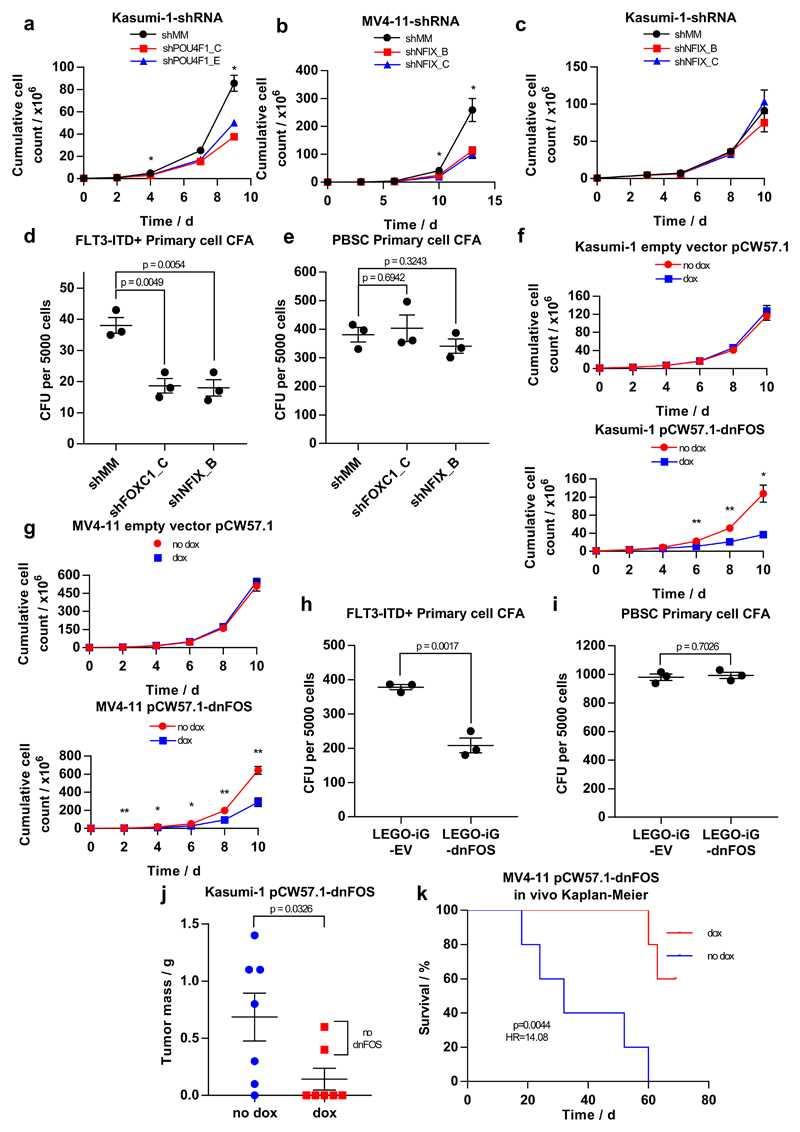
We next validated the role of TFs forming network nodes by transducing three different AML cell lines and primary FLT3-ITD AML cells with lentiviral vectors coding shRNAs targeting POU4F1 (specific for t(8;21)) or NFIX and FOXC1 (specific for FLT3-ITD). NFIX is known to play a role in myeloid lineage specification38 but has not been linked to specific mutation types. FOXC1 is an oncogene in its own right39 and overexpression is observed in AMLs with FLT3-ITD mutations24. However, NFIX and FOXC1 have not been linked to the maintenance of the FLT3-ITD AML. We transduced two distinct shRNA constructs per gene into FLT3-ITD and t(8;21) cell lines, significantly reducing the corresponding TF transcript and protein levels (Supplementary Fig 10a-f). Knockdown of POU4F1 (Supplementary Figs. 10a and d) inhibited the proliferation of t(8;21)-positive Kasumi-1 cells (Fig. 7a, Supplementary Fig. 10g), confirming our previous findings32. Expression of NFIX shRNAs (Supplementary Figs. 10b and e) impaired the proliferation of FLT3-ITD-positive MV4-11, but not FLT3-ITD-negative Kasumi-1 cells (Figs. 7b,c, Supplementary Fig. 10h,i). We next tested the effect of shRNA knock-down of these genes on the colony forming ability of CD34+ FLT3-ITD/NPM1 mutant patient cells and on CD34+ PBSCs. Both NFIX and FOXC1 shRNA constructs reduced the colony forming ability of AML cells, but not that of normal CD34+ HSP cells (Figs. 7d, e).
Figure 7. Identification of AML type-specific TFs required for maintaining leukemic growth and colony forming ability.
(a - c) Scatter plot showing the growth curves of (a) Kasumi-1 cells after transduction with shPOU4F1 or (c) shNFIX and (b) of MV4-11 cells after transduction with shNFIX. (d, e) Dot plot showing the number of colonies formed by a FLT3-ITD+ primary AML cell samples (d) or PBSCs (e) after transduction with shRNA targeting FOXC1, NFIX or a mismatch control. (f) Scatter plot showing the growth curve of Kasumi-1 cells transduced with either a doxycycline-inducible dnFOS or an empty vector control with and without 1.5 mcg/ml doxycycline. (g) Dot plots showing the growth curve of MV4-11 cells transduced with either a doxycycline-inducible dnFOS or an empty vector control (right panel) with and without 1.5 μg/ml doxycycline. (h,i) The expression of a dnFOS causes a reduction in the colony forming ability of CD34+ FLT3-ITD+ primary AML cells (H) but not CD34+ PBSCs (i (j) Granulosarcoma formation in RG mice by Kasumi-1 expressing a doxycycline-inducible dnFOS. dnFOS was induced by intraperitoneal injection of doxycycline. (k) Survival curve for RG mice transplanted with MV4-11 cells expressing doxycycline-inducible dnFOS. The induction of dnFOS significantly increased the survival time of mice.
Significance in all experiments was tested using a two-tailed Student’s t-test (n=3) with * p<0.05, **p<0.01 in both samples compared to the mismatch control where indicated. Error bars show standard error of the mean. Further detail on statistical analysis can be found in Online Methods.
In addition to subtype-specific TFs such as POU4F1 or NFIX, our network analysis suggests that the AP-1 family is of general significance for all AML subtypes examined. AP-1 is a heterodimer formed by members of the FOS, JUN, ATF, CREB and JDP families of transcription factors making it challenging to target by defined RNAi approaches. To this end, we transduced AML cells with an inducible version of a dominant negative FOS (dnFOS) protein40,41. Doxycyclin-mediated induction of dnFOS significantly inhibited proliferation of both t(8;21)-positive Kasumi-1 cells and FLT3-ITD expressing MV4 11 cell lines (Figs. 7f, g, Supplementary Fig. 10j, k) and the colony-forming ability of primary CD34+ FLT3-ITD cells but not of CD34+ HPSCs (Figs. 7h,i, Supplementary Fig. 10l).
Finally, we examined the significance of AP-1 for leukemia propagation in vivo by transplanting either Kasumi-1 or MV4-11 cells expressing a doxycycline-inducible dnFOS into immunodeficient RG mice, followed by randomization into a doxycycline and untreated arm. In the case of Kasumi-1 transplantation, 6 out of 7 animals of the control group, but only 2 animals of doxycyclin-treated group developed granulosarcomas (Fig. 7j). Neither of the latter two tumours expressed dnFOS after DOX treatment (data not shown). Doxycycline treatment of mice transplanted with FLT3-ITD MV4-11 cells harboring the dnFOS transgene inhibited the development of leukemia while all untreated mice rapidly developed tumours and had to be sacrificed (Fig. 7k). Taken together, these findings demonstrate the significance of AP-1 for several AML subtypes and emphasize the potential of transcriptional network analyses to predict TFs crucial for malignant propagation.
Discussion
In this study we defined how aberrantly expressed TFs and signaling molecules shape the epigenetic landscape of different sub-types of human AML. We show (i) that it is possible to use high-quality DNaseI footprinting analyses of purified AML blast cells to identify AML subtype-specific TF networks, (ii) that such TF networks point towards a dependency on specific factors for leukemic growth and (iii) that the global activation of signaling pathways parallels a growth dependency on AP-1 activity in multiple types of AML. Our comprehensive integration of multi-omics data reveals a strong connection between leukemic classifier mutations, networks of TFs and signaling components in primary AML. Moreover, mapping of cis-element promoter interactions by CHiC enabled assigning the majority of genes of all analysed subtypes to their correct promoter. Different types of AML have previously been classified by their gene expression and DNA-methylation patterns42,43 revealing the existence of specific gene regulatory networks. Our work now defines these networks in detail, and convincingly proves that leukemic drivers determine the regulatory phenotype by establishing and maintaining specific gene regulatory and signaling networks distinct from normal cells. Induced and aberrantly expressed TFs are not bystanders, but are important for network maintenance and leukemic growth, thus highlighting novel therapeutic opportunities for targeted treatment.
The full set of target genes of RUNX1-ETO in t(8;21) is known and the t(8;21) specific epigenome and TF binding pattern has been extensively characterized44. Multiple target genes relevant for the maintenance of the leukemogenic state have been identified, including FOXO1, UBASH3B, POU4F1, and LAT2 together with the members of the RUNX1-ETO complex 22,32,37,45–47 and our current comparative study has identified multiple new network components. However, for the other types of AML, in particular for the FLT3-ITD such knowledge was not available. Here, we identified a number of signaling and transcriptional components distinguishing FLT3-ITD from normal blasts and from other types of AML, comprising a rich resource for combinatorial therapy approaches. We examined the contribution to leukemic growth for two genes with AML subtype-specific activity (NFIX and FOXC1) and showed that their elimination resulted in reduced growth and colony forming ability of AML but not normal cells, confirming that AML maintains an aberrant transcriptional network required for survival.
The AP-1 factor family plays an important role in many types of tumours48 and our study shows that it is of major importance for AML. FLT3-ITD MV4-11 cells have abundant levels of nuclear AP-1, and FLT3-ITD target genes such as CCNA1 are suppressed by MAPK inhibitors in these cells24. We have recently shown that JUN scores highly in a siRNA dropout screen examining the requirements for tumour development in t(8;21) AML (Martinez-Soria et al., in press). FOS plays an important role in the resistance against BCR-ABL inhibition in CML by activating compensatory signaling pathways49. Since several growth factor and stress signal cascades feed into AP-1, a targeted inhibition of all AP-1 binding may avoid resistance via rewiring of signalling pathways.
In the classical mechanism of two-step leukemogenesis a mutation altering a differentiation trajectory cooperates with signaling mutations directing leukemic growth10,11. Mutations in TFs and epigenetic regulators setting up the epigenetic landscape upon which TFs act, fall into the first category while the FLT3-ITD mutation falls into the second. However, these distinctions are becoming blurred as from the viewpoint of gene regulation, growth factor receptors strongly influence transcriptional activity via inducible TFs, as exemplified by AP-1 family members. These factors play a dominant role in driving the differentiation trajectory as they display an AML sub-type specific occupancy signature that is uninfluenced by the presence or absence of epigenetic regulator mutations (in this case DNMT3A)24. This is not to say that mutations in epigenetic regulators do not influence the developmental trajectory of AML and clinical outcomes, as shown in CBF AML50 since AML cells with such mutations acquire an altered DNA methylation landscape that is likely to influence TF binding51. However, our data show that leukemic phenotype and self-renewal of different types of AML are defined by differentially activating a multitude of different and often lineage-unrelated signaling pathways and by expressing lineage-unrelated TFs. From the viewpoint of finding therapeutic targets, identifying such mutation-specific pathways will offer an opportunity to eliminate their specific maintenance program by targeting multiple pathways simultaneously. Our study provides a first step towards this goal.
Accession code
URLs
Data Server: http://bioinformatics-bham.co.uk/tfinaml/
fastQC: (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/)
Bowtie: (http://bowtie-bio.sourceforge.net/bowtie2/manual.shtml)
MACS 2: (https://github.com/taoliu/MACS)
DEseq2: (https://bioconductor.org/packages/release/bioc/html/DESeq2.html)
Wellington algorithm: (http://pythonhosted.org/pyDNase/)
STAR: (https://github.com/alexdobin/STAR)
Cufflinks: (http://cole-trapnell-lab.github.io/cufflinks/)
LIMMA: (http://bioconductor.org/packages/release/bioc/html/limma.html)
JASPAR: http://jaspar.genereg.net/
HOMER: (http://homer.ucsd.edu/homer/index.html)
HiCUP: (https://www.bioinformatics.babraham.ac.uk/projects/hicup/)
GOTHiC: (http://bioconductor.org/packages/release/bioc/html/GOTHiC.html)
R scripting language: (https://www.r-project.org/)
GSEA: (http://software.broadinstitute.org/gsea/index.jsp)
Picard: (http://broadinstitute.github.io/picard/).
DFilter: (http://collaborations.gis.a-star.edu.sg/~cmb6/kumarv1/dfilter/)
Bedtools: (https://bedtools.readthedocs.io/en/latest/)
Cytoscape: (http://www.cytoscape.org/)
Online Methods
Patient samples and PBSC cell processing
Human tissue was obtained with the required ethical approval from the NHS National Research Ethics Committee. AML and PBSC samples used in this study were either surplus diagnostic samples, or were fresh samples obtained with specific consent from the patients. AML samples were obtained from either (i) the Haematological Malignancy Diagnostic Service (St James’s Hospital, Leeds, UK, (ii) the Centre for Clinical Haematology, Queen Elizabeth Hospital Birmingham, Birmingham, UK, (iii) the West Midlands Regional Genetics Laboratory, Birmingham Women’s NHS Foundation Trust, Birmingham, UK, or from iv) Erasmus University Medical Center, Rotterdam, The Netherlands. Mononuclear cells were purified on the same day that they were received, and in most cases also directly further purified using either CD34 or CD117 (KIT) magnetic antibodies, as previously described 24. For some samples with greater than 92% blast cells the column purification was not performed. Mobilized PBSCs were provided by NHS BT, Leeds, and NHS BT, Birmingham.
Mutation detection
Mutated genes identified in each patient are summarized in Supplementary Table 1, together with the age, gender and white blood cell count for each patient. Mutations were identified by one of two different methods. The first batch of patients were assayed by targeted exon sequencing of 55 cancer-associated genes using 1212 pairs of previously defined PCR primers 24 for amplification using a RainDance Technologies platform. The mutation sequence data from this screen was analyzed using algorithms to detect either (i) nucleotide variants using the Genome Analysis Toolkit (GATK) 52 or insertions and deletions using Pindel 52. Mutations were also screened against the COSMIC database of previously observed mutations (link see URL section). Subsequent samples were assayed using the Illumina Trusight myeloid panel of primers and processed by approaches similar to those used for the first batch. All identified mutations are listed in Table S1. Some of these patients were also included in a previous publication from our laboratory, using different patients identification codes 24 to those used in the current study.
Cell lines
Cell lines were cultured in an incubator at 37°C in GIBCO™ 1640 RPMI + Glutamax™ medium supplemented with 10% heat inactivated fetal calf serum (GIBCO), 100 U/ml Penicillin, 100 mg/ml Streptomycin.
Growth curve measurements
250000 MV4-11 or Kasumi-1 cells were cultured in RPMI supplemented with 10 % fetal calf serum, 2mM L-Glutamine, 100 U/ml penicillin and 100 mg/ml streptomycin. Cells were counted with Trypan Blue exclusion and split every 3 days to maintain them in the log phase of growth. For the inducible dnFOS, cells were counted and split every 2 days and 1.5 μg/ml of doxycycline was added.
Co-culture of Primary Cells with MS-5 feeders
Primary cells were maintained in co-culture with MS-5 cells 53 Briefly, cells were cultured in LTC medium (α-minimum essential medium (Lonza) supplemented with heat-inactivated 12.5% fetal calf serum (Gibco), heat-inactivated 12.5% horse serum (Gibco), penicillin and streptomycin, 200 mM glutamine, 57.2 μM β-mercaptoethanol (Sigma) and 1 μM hydrocortisone; (Sigma) supplemented with 20 ng/ml IL-3, granulocyte colony-stimulating factor (G-CSF) and thrombopoietin (TPO) in flasks pre-coated with MS-5 cells.
Lentiviral transduction and shRNA treatment
LEGO-iG-shRNA were generated by cloning shRNAs (Table S3) into the LEGO-iG vector54. LEGO-iG-dnFOS was generated by cloning the dnFOS insert, originally generated by Charles Vinson (National Cancer Institute, Bethesda, USA 40 into the LEGO-iG backbone. Inducible dnFOS was cloned into a pENTR backbone and then using Gateway Cloning to insert that into the Tet-on plasmid pCW57.1 (David Root, Addgene plasmid #41393). Backbone vectors LEGO-iG and Inducible dnFOS then used to generate lentiviral particles using packaging and envelope genes on four separate plasmids: TAT, REV, GAG/POL and VSV-G55.
For virus production, 293T Human Embryonic Kidney cells were cultured in Dulbecco’s Modified Eagle Medium supplemented with 10 % fetal calf serum, 2mM L-Glutamine, 100 U/ml penicillin, 100 mg/ml streptomycin and 0.11 mg/ml Sodium pyruvate; and were seeded to achieve 70-80% confluency at time of transfection. HEK293T cells were transfected using the calcium phosphate co-precipitation of the five-plasmids (LEGO-iG with TAT, REV, GAG/POL and VSV-G) at a mass ratio of 24 μg : 1.2 μg : 1.2 μg : 1.2 μg : 2.4 μg per 150 mm diameter plate of cells. Viral supernatant was harvested after 24 h and subsequently every 12 h for 36 h prior to concentration with Centricon Plus 70 100 kDa filter (Millipore, USA), using the manufacturer’s instructions. Concentrated viral particles were stored at 4 °C prior to lentiviral transduction. Cell lines were transduced with concentrated virus in the presence of 8 μg/ml polybrene by spinoculation at 1500 xG for 50 min. After 12 – 16 h incubation at 37 °C viral media was exchanged for fresh media. Cell sorting by FACS was performed to isolate GFP+ cells 3 days after transduction.
Primary cell samples were defrosted 24 h prior to transduction and co-cultured with MS-5 feeder cells in LTC medium. 6 well non-tissue culture treated plates were coated with 24 μg/ml retronectin (Takara Clontech) for 2 h prior to blocking with 2% BSA PBS for 30 min. The blocking buffer was washed off with HBSS (Gibco) containing 2.5% HEPES. 1 ml viral concentrate was applied to the retronectin coated plate by centrifugation at 2000 xG for 45 minutes, after which the concentrated viral supernatant was refreshed and the centrifugation repeated. Primary cells suspended to a concentration of 1 x 106 cells/ml in the remaining viral supernatant; supplemented with 20 ng/ul G-CSF, IL-3, TPO and 8 μg/ml polybrene, were then added to the plate and transduced by spinoculation at 1500 xG for 50 min. After 12 – 16 h incubation at 37 °C viral media was exchanged for fresh media. Cell sorting by FACS was performed to isolate GFP+ cells 3 days after transduction.
Colony Formation Assays of Primary Cells
Colony formation assays were performed on sorted cells by seeding at 2500 cells/ml in Methocult Express (Stem Cell Technologies). After 14 days colonies were counted.
Animal experiments
Immunodeficient Rag2-/-Il2rγ-/-129×Balb/c (RG) mice were housed in the Comparative Biology Centre (Newcastle University) under specific pathogen free conditions. All animal work was conducted in accordance with Home Office Project License PPL60/4552 by researchers who had completed approved Home Office training and held current Personal Licenses under the Animals (Scientific Procedures) Act 1986. Kasumi-1 pCW57.1-dnFOS cells were intrahepatically injected into 14 newborn (2 days old) RG mice at a cell dose of 2.5x105 cells/mouse as described previously (Martinez Soria et al., 2009). Twelve days later, mice were randomized into two treatment groups, one given doxycycline 50 mg/kg three times per week intraperitoneally in an unblended fashion till the experimental endpoint. MV4-11 pCW57.1-dnFOS cells were intrafemorally injected into RG mice at a cell dose of 5x105 cells/mouse followed by randomization into two groups. For the dox group doxycycline was added at a concentration of 2 mg/ml for the initial 3 days and at 0.2 mg/ml subsequently to drinking water containing 2% sucrose. Controls were given water containing 2% sucrose. Animals were humanely killed upon clinical signs of illness or at defined experimental endpoints.
RT- qPCR
RNA was extracted using the Machery-Nagel Nucleospin kit. 1μg RNA was used to make cDNA with 0.5μg OligoDT primer, Murine Moloney Reverse Transcriptase and RNase Inhibitor (Promega, USA) according to manufacturer’s protocol. RT-PCR was performed using Sybr Green mix (Applied Biosystems, UK), at 2x dilution. Primers were used at 100nM concentration. A 7900HT system (Applied Biosystems, UK) was used to perform qPCR. Analyses were performed in technical duplicates using a standard curve derived from the untreated cell line.
Western Blotting
Protein lysates from cell lines were analyzed by Western blot. Relevant primary antibodies against FOXC1 (Cell Signaling Technology - #8758), NFIX (Invitrogen - #PA5-31234), POU4F1 (Santa Cruz Biotechnology – sc-8429) were used to detect target genes and GAPDH (mouse αGAPDH – Abcam – ab8245; rabbit αGAPDH – Cell Signaling Technology – 2118L) was used as a housekeeping gene. Secondary antibodies mouse anti-rabbit HRP (Santa Cruz Biotechnology – sc-2054) and goat anti-mouse HRP (Jackson ImmunoResearch – 115-035-062) enabled detection and quantifications by densitometry using Imagelab software and a GelDoc imager.
Statistical analysis of validation experiments
Validation experiments were performed in triplicate (n=3) where each replicate represents a separately transduced cell line or primary cell sample cultured and analyzed independently. The exception are the in vitro inducible dnFOS cell line experiments where each replicate represents a separate culture of a clone stably expressing the dnFOS peptide in order to avoid variations in the degree of expression. The significance of differences in colony counts, doubling time, mRNA expression and cell concentrations at individual time points on growth curves were calculated using Student’s t-test where n=3 using 4 degrees of freedom, t values between significantly different groups (p<0.05) were all greater than t=3.2.
Standard error bars on graphs show standard error of the mean:
The tumor mass of mice transplanted with Kasumi-1 pCW57.1-dnFOS with and without dox was analyzed using a Mann-Whitney test as the distribution of data was not normal. This experiment was performed n of 7 engrafted mice in each group (p=0.0326, Mann-Whitney U = 8.5. The survival curve of mice engrafted with MV4-11 pCW57-dnFOS with and without dox was analyzed using a Kaplan-Meier plot (n=5 engrafted mice) using a Mantel-Cox Chi-square test (Chi square = 8.097, df=1, p=0.0044) and yielding a Mantel-Haenszel hazard ratio of 14.08.
RNA-Seq library preparation
RNA was extracted and analyzed from purified AML cells as previously described 23
DNaseI-Seq
DNaseI digestions of permeabilized cells were performed as previously described56. Briefly, live cells were added directly to a solution of DNaseI (DPFF, Worthington) in dilute Nonidet P40, digested for 3 min at 22°C, and the reactions then terminated by addition of SDS to 0.5%. DNaseI was typically used in the range of 2-6 μg/ml using a final 1.5 x 107 cells/ml. DNaseI-Seq libraries were then prepared and validated essentially as previously described 30. Libraries were run on Illumina sequencers.
Promoter capture HiC (CHi-C) from patient AML blasts
AML cells from patient peripheral blood were first purified by density gradient centrifugation (Lymphoprep™) and then using CD34 antibody coupled beads. 5× 107 t(8;21) blasts (patient t(8;21)-1R) and FLT3-ITD/NPM1 blasts (patient ITD/NPM1-2) were fixed in 37 ml of RPMI-1640 supplemented with 15% FBS and 2% formaldehyde for 10 minutes at room temperature. 6 ml of 1M glycine (0.125 M final concentration) was added to quench the reaction and cells were incubated at room temperature for 5 min, followed by 15 minutes on ice before pelleting the cells at 4 °C and washing them in ice cold PBS. Each sample was flash frozen in liquid nitrogen, and stored at −80 °C. Cells were lysed in a tight dounce homogeniser (ten cycles) with 3ml of cold lysis buffer (10 mM Tris-HCl pH 8, 10 mM NaCl, 0.2% Igepal CA-630, one tablet protease inhibitor cocktail (Roche complete, EDTA-free, 11873580001)). Cells were left on ice for five minutes then homogenised another ten times. The lysed cells, in 3 ml lysis buffer, were added to 47ml of lysis buffer and incubated on ice for 30 minutes with occasional mixing. Chromatin was pelleted and resuspended in 1ml of 1.25x NEBuffer 2 and split into four. Each sample was then pelleted at 1000 rpm and resuspended in 358 µl of 1.25x NEBuffer 2. 11 µl 10% SDS was added and each tube was incubated at 37°C for 60 minutes, rotating at 950 rpm. Samples were mixed by pipetting up and down every 15 minutes. SDS was quenched with 75µl 10% Triton X-100 and incubated at 37°C for 60 minutes. HindIII digestion, biotinylation, ligation, crosslink reversal, promoter capture and library preparation was performed exactly as described in34.
DNaseI-Seq data analysis
Alignment
DNaseI-seq sequences from all experiments were mapped onto the reference human genome version hg38, with Bowtie version 2.3.157,24 using default parameters. Low quality reads were trimmed prior to the alignment and the quality control (QC) statistics for the samples were obtained using FastQC tools. Reads that were aligned to unique chromosomal positions were retained.
Peak calling
DNaseI Hypersensitive Sites (DHSs) were called with MACS2 using callpeak function (nomodel, call-summits and q= 0.005 parameters)58. DHSs were allocated to genes and to the gene promoter if it was within 2kb of the gene transcription start site (TSS), and as distal otherwise. Overlaps between DHSs peaks were defined by requiring the summits of two peaks to lie within +/-200 bp.
DNaseI-Seq peak set definition
To define a common set of coordinates covering all of the significant distal DHSs investigated in this study, we merged all of the individual DNaseI-Seq reads for all of the AML samples assayed by DNaseI-Seq. This data set was then used to define the peak summits of 128,864 distal peaks, excluding promoters, which were detected in the merged data. This approach was designed to maximize the precision and sensitivity of the peak detection, allowing us to generate a single set of peak coordinates that (i) included all the regions where peaks might be found, thereby reducing the level of false negatives, and (ii) greatly diminished the number of false positives. The DNA read counts were then determined for 400 bp windows centered on each peak for each AML sample and for the PBSC samples. To account for the different number of reads in each of the samples; the read counts were initially normalized for total read depth using DEseq2 59. Because most of our individual DNaseI-Seq data sets encompassed in the range of 25,000 to 40,000 significant distal DHSs, we further normalized the values obtained on the basis of the midpoint (12.5 percentile) of the top 25% of peaks (32,216 peaks).
Mutation-specific DNaseI-Seq peak set definition
We determined the average log2 values for 7 distinct groups of AMLs that carried the same specific mutations in key regulators, and which shared similar patterns of DHSs based on the DHS clustering analysis. The samples included in each group are color-coded and listed in order in Table S1 for AML samples with the following mutations: (i) 3 samples with FLT3-ITD but not NPM1 (#1 to 3), (ii) 6 samples with FLT3-ITD and NPM1 (# 1 to 6), (iii) 2 samples with NPM1 but not FLT3-ITD (# 1 and 2), (iv) 4 samples with t(8;21) (1 to 4), (v) 3 samples with inv(16) (# 1 to 3); (vi) 6 samples with RUNX1 or RUNX1 and CEBPA (1 to 6), and (vii) 3 samples with 2 CEBPA mutations (#1 to 3). To define mutation-specific subsets of specific DHSs, we identified peaks where the average log2 value both was at least 64 and at least 3-fold higher than in PBSCs. Downregulated DHSs are defined as being at least 3-fold less than in PBSCs. Samples were not included in these 7 specific groups in cases where, for example, 2 copies of the FLT3-ITD mutation were present, the NPM1 mutation was paired with a NRAS instead of the FLT3-ITD, RUNX1 mutations were paired with a JAK2 mutation, or where only a single CEBPA allele was mutated.
Clustering of DNaseI-Seq data
Clustering of DNaseI-seq samples was carried out using the merged distal DHSs. The number of reads that mapped to these DHSs was counted in a 400bp window centered on the DHS summit, and subsequently normalized to total sample size using DEseq2 59. Pearson correlation coefficients were then calculated for each pair of samples using the log2 of the normalized read counts, and then hierarchically clustered using Euclidean distance and complete linkage clustering of the correlation matrix in R.
For further analysis methods see Supplementary Material.
RNA-seq data analysis
RNA-Seq reads were aligned to the human genome hg38 build with STAR60 using ENCODE recommend parameters. Separate density profiles for the positive and negative strand were generated using bedtools. Cufflinks61 was used to calculate the expression values as Fragments Per Kilobase per Million aligned reads (FPKM) from the aligned RNA-seq data. Mutation-specific group’s gene-wise expression values were obtained using the cuffdiff function of cufflinks. The correlation between any two AML samples was obtained as the Pearson correlation coefficient of expression values over all genes. A correlation matrix was thus generated for all the samples and hierarchically clustered to study the relationship among samples as given in Figure S1C. Smooth scatter plots were generated in R. For further analysis methods see Supplementary Material.
Promoter Capture HiC data analysis
The CHi-C paired-end sequencing reads from ITD/NMP1-2 and t(8;21)-1R patients and a publically available CD34+ dataset (accession numbers ERR436032 and ERR436025) were put through HiCUP pipeline62. The raw sequencing reads were separated and mapped against the human genome (hg38). The reads were then filtered for experimental artefacts and duplicate reads, and then re-paired. Statistically significant interactions were called using GOTHiC package63 and HOMER software. This uses a cumulative binomial test to detect interactions between distal genomic loci that have significantly more reads than expected by chance, by using a background model of random interactions. This analysis assigns each interaction with a p-value, which represents its significance. Differential interactions were determined with HOMER64 for t(8;21) using FLT3-ITD or CD34+ as background and FLT3-ITD using t(8;21) or CD34+ as a background. A difference with a p-value of less than 0.1 was deemed to be significant
Transcription Factor Gene Regulatory Network Construction
We identified a subset of 310 transcription factor (TF) genes that are expressed in one or more of our AML samples. The gene names for transcription factors in human were obtained from AnimalTFDB65. The 310 TFs were considered as nodes and the nodes coloured according to their expression values at each AML subtype (Fig 6, Supplementary Fig 9 and Fig SN5). Node border colour signifies whether the gene is up-regulated, down-regulated or invariant base on a 2-fold-change compared to CD34+ cells. The node border type indicates whether a gene is differentially expressed in one AML subtype as compared to other subtypes. A directed edge from TFa to TFb indicates motif binding of a TFa to the locus of the TFb and the edge is prominently displayed if TFa binds to the locus at that stage. The edge is classified and colour coded according to the significant of motif count enrichment.
Motif count enrichment for TFs network
Initially footprints for each AML subtype were identified by using the Wellington algorithm29 and were annotated to their related promoter using CHi-C data where possible. Motif search within footprint coordinates where performed using HOMER64. The number of motifs per TF gene were counted and the significance of motif enrichment was identified using bootstrapping on random sampling, a random set of mapped motif were extracted from all union footprinted motif of all AML subtypes and the CD34+ cells. After 1000 iterations the mean, standard deviation and the z-scores are computed. Motif (TFa) is linked to gene (TFb) with positive Z-score values only.
Motif count enrichment for up-regulated TFs
The correlations (re) between all TF genes based on FPKM values from the RNA-seq analysis were identified and the correlations (rm) between all TF genes based on motif count binding were identified. The correlation coefficients were z-transformed using Fisher Z-transformation with “FisherZ” function in R. The average of the transformed z-scores of both gene expression and motif were transformed to correlation (r). All TF genes with a correlation coefficient equal or greater than a cut-off of 0.3 were considered. Then for each AML cell type, the differentially expressed TF genes among these correlated genes were identified. First those of each AML subtype as compared to CD34+ cells and second those of all AML subtypes compared to the average expression of other subtypes including CD34+ cells. Up-regulated genes with a 2-fold-change in expression compared to CD34+ cells or compared to other AMLs were considered to construct the network. Motif enrichment for the correlated and up-regulated TF genes and edges were identified as described above.
List of used position weight matrices
A description of how the motifs were curated can be found in the legend of Table S2.
Data availability statement
Raw data have been deposited at GEO under the accession number GSE108316. Processed data are available from our Data server (see URL section)
Supplementary Material
Editorial Summary.
Integrated analysis of transcriptome, open chromatin region and chromatin conformation capture data from Acute Myeloid Leukemia patients with defined transcription factor and signaling molecule mutations provides insights into subtype-specific regulatory network in AML.
Acknowledgements
This research was funded by a programme grant from Bloodwise (15001) to C.B. and P.N.C, as well as studentship awards from Cancer Research UK and Bloodwise to A.Pickin and N.G, respectively, a Kay Kendall Clinical Training Fellowship for J.L. and a MRC/Leuka Clinical Training Fellowship for S.P.
Footnotes
Author contributions:
M.R.I., D.C., S.P., A.P, H.B., A. Pickin, N.G., J.L., P.S.C, S.R.J. performed experiments and generated data, R.D., M.R., S.R., M.G. P.J., A.U. provided patient samples, S.C., A,B, and P.N.C. conducted mutation analysis, S.A.A. and P.C. analysed data, O.H. supervised transplantation experiments and helped editing the manuscript, C.B. and P.N.C. conceived and directed the study and CB wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Cancer Genome Atlas Research N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–74. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papaemmanuil E, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. New England Journal of Medicine. 2016;374:2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonifer C, Cockerill PN. Chromatin Structure Profiling Identifies Crucial Regulators of Tumor Maintenance. Trends Cancer. 2015;1:157–160. doi: 10.1016/j.trecan.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Rosenbauer F, Tenen DG. Transcription factors in myeloid development: balancing differentiation with transformation. Nature reviews. Immunology. 2007;7:105–17. doi: 10.1038/nri2024. [DOI] [PubMed] [Google Scholar]
- 5.Cockerill PN. Receptor Signaling Directs Global Recruitment of Pre-existing Transcription Factors to Inducible Elements. Yale J Biol Med. 2016;89:591–596. [PMC free article] [PubMed] [Google Scholar]
- 6.Ward AF, Braun BS, Shannon KM. Targeting oncogenic Ras signaling in hematologic malignancies. Blood. 2012;120:3397–406. doi: 10.1182/blood-2012-05-378596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parikh C, Subrahmanyam R, Ren R. Oncogenic NRAS rapidly and efficiently induces CMML- and AML-like diseases in mice. Blood. 2006;108:2349–57. doi: 10.1182/blood-2004-08-009498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Masson K, Ronnstrand L. Oncogenic signaling from the hematopoietic growth factor receptors c-Kit and Flt3. Cell Signal. 2009;21:1717–26. doi: 10.1016/j.cellsig.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 9.Gerloff D, et al. NF-kappaB/STAT5/miR-155 network targets PU.1 in FLT3-ITD-driven acute myeloid leukemia. Leukemia. 2015;29:535–47. doi: 10.1038/leu.2014.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corces-Zimmerman MR, Hong WJ, Weissman IL, Medeiros BC, Majeti R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc Natl Acad Sci U S A. 2014;111:2548–53. doi: 10.1073/pnas.1324297111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shlush LI, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–33. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nature structural & molecular biology. 2013;20:1147–55. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- 13.Broske AM, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nature genetics. 2009;41:1207–15. doi: 10.1038/ng.463. [DOI] [PubMed] [Google Scholar]
- 14.Bonifer C, Bowen DT. Epigenetic mechanisms regulating normal and malignant haematopoiesis: new therapeutic targets for clinical medicine. Expert reviews in molecular medicine. 2010;12:e6. doi: 10.1017/S1462399410001377. [DOI] [PubMed] [Google Scholar]
- 15.Challen GA, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nature genetics. 2012;44:23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ko M, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–43. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nature reviews. Cancer. 2012;12:599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- 18.Goode DK, et al. Dynamic Gene Regulatory Networks Drive Hematopoietic Specification and Differentiation. Dev Cell. 2016;36:572–87. doi: 10.1016/j.devcel.2016.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Obier N, Bonifer C. Chromatin programming by developmentally regulated transcription factors: lessons from the study of haematopoietic stem cell specification and differentiation. FEBS Lett. 2016;590:4105–4115. doi: 10.1002/1873-3468.12343. [DOI] [PubMed] [Google Scholar]
- 20.Wilson NK, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–44. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 21.Rubin AJ, et al. Lineage-specific dynamic and pre-established enhancer-promoter contacts cooperate in terminal differentiation. Nat Genet. 2017;49:1522–1528. doi: 10.1038/ng.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ptasinska A, et al. Identification of a Dynamic Core Transcriptional Network in t(8;21) AML that Regulates Differentiation Block and Self-Renewal. Cell Reports. 2014;8:1974–1988. doi: 10.1016/j.celrep.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loke J, et al. RUNX1-ETO and RUNX1-EVI1 Differentially Reprogram the Chromatin Landscape in t(8;21) and t(3;21) AML. Cell Rep. 2017;19:1654–1668. doi: 10.1016/j.celrep.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cauchy P, et al. Chronic FLT3-ITD Signaling in Acute Myeloid Leukemia Is Connected to a Specific Chromatin Signature. Cell Rep. 2015;12:821–36. doi: 10.1016/j.celrep.2015.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corces MR, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet. 2016;48:1193–203. doi: 10.1038/ng.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKeown MR, et al. Superenhancer Analysis Defines Novel Epigenomic Subtypes of Non-APL AML, Including an RARalpha Dependency Targetable by SY-1425, a Potent and Selective RARalpha Agonist. Cancer Discov. 2017;7:1136–1153. doi: 10.1158/2159-8290.CD-17-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martens JH, et al. ERG and FLI1 binding sites demarcate targets for aberrant epigenetic regulation by AML1-ETO in acute myeloid leukemia. Blood. 2012;120:4038–48. doi: 10.1182/blood-2012-05-429050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pulikkan JA, Tenen DG, Behre G. C/EBPalpha deregulation as a paradigm for leukemogenesis. Leukemia. 2017;31:2279–2285. doi: 10.1038/leu.2017.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piper J, et al. Wellington: a novel method for the accurate identification of digital genomic footprints from DNase-seq data. Nucleic Acids Res. 2013;41:e201. doi: 10.1093/nar/gkt850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ptasinska A, et al. Depletion of RUNX1/ETO in t(8;21) AML cells leads to genome-wide changes in chromatin structure and transcription factor binding. Leukemia. 2012;26:1829–1841. doi: 10.1038/leu.2012.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mandoli A, et al. CBFB-MYH11/RUNX1 together with a compendium of hematopoietic regulators, chromatin modifiers and basal transcription factors occupies self-renewal genes in inv(16) acute myeloid leukemia. Leukemia. 2014;28:770–8. doi: 10.1038/leu.2013.257. [DOI] [PubMed] [Google Scholar]
- 32.Dunne J, et al. AML1/ETO proteins control POU4F1/BRN3A expression and function in t(8;21) acute myeloid leukemia. Cancer Res. 2010;70:3985–95. doi: 10.1158/0008-5472.CAN-09-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–11. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 34.Mifsud B, et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet. 2015;47:598–606. doi: 10.1038/ng.3286. [DOI] [PubMed] [Google Scholar]
- 35.Chasman D, Roy S. Inference of cell type specific regulatory networks on mammalian lineages. Curr Opin Syst Biol. 2017;2:130–139. doi: 10.1016/j.coisb.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neph S, et al. Circuitry and dynamics of human transcription factor regulatory networks. Cell. 2012;150:1274–86. doi: 10.1016/j.cell.2012.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin S, et al. A FOXO1-induced oncogenic network defines the AML1-ETO preleukemic program. Blood. 2017;130:1213–1222. doi: 10.1182/blood-2016-11-750976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Connor C, et al. Nfix expression critically modulates early B lymphopoiesis and myelopoiesis. PLoS One. 2015;10:e0120102. doi: 10.1371/journal.pone.0120102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Somerville TD, et al. Frequent Derepression of the Mesenchymal Transcription Factor Gene FOXC1 in Acute Myeloid Leukemia. Cancer Cell. 2015;28:329–42. doi: 10.1016/j.ccell.2015.07.017. [DOI] [PubMed] [Google Scholar]
- 40.Olive M, et al. A dominant negative to activation protein-1 (AP1) that abolishes DNA binding and inhibits oncogenesis. J Biol Chem. 1997;272:18586–94. doi: 10.1074/jbc.272.30.18586. [DOI] [PubMed] [Google Scholar]
- 41.Obier N, et al. Cooperative binding of AP-1 and TEAD4 modulates the balance between vascular smooth muscle and hemogenic cell fate. Development. 2016;143:4324–4340. doi: 10.1242/dev.139857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verhaak RGW, et al. Prediction of molecular subtypes in acute myeloid leukemia based on gene expression profiling. Haematologica. 2009;94:131–134. doi: 10.3324/haematol.13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Figueroa ME, et al. DNA Methylation Signatures Identify Biologically Distinct Subtypes in Acute Myeloid Leukemia. Cancer cell. 2010;17:13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin S, Mulloy JC, Goyama S. RUNX1-ETO Leukemia. Adv Exp Med Biol. 2017;962:151–173. doi: 10.1007/978-981-10-3233-2_11. [DOI] [PubMed] [Google Scholar]
- 45.Goyama S, et al. UBASH3B/Sts-1-CBL axis regulates myeloid proliferation in human preleukemia induced by AML1-ETO. Leukemia. 2016;30:728–39. doi: 10.1038/leu.2015.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun XJ, et al. A stable transcription factor complex nucleated by oligomeric AML1-ETO controls leukaemogenesis. Nature. 2013;500:93–7. doi: 10.1038/nature12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Essig A, Duque-Afonso J, Schwemmers S, Pahl HL, Lubbert M. The AML1/ETO target gene LAT2 interferes with differentiation of normal hematopoietic precursor cells. Leuk Res. 2014;38:340–5. doi: 10.1016/j.leukres.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trop-Steinberg S, Azar Y. AP-1 Expression and its Clinical Relevance in Immune Disorders and Cancer. Am J Med Sci. 2017;353:474–483. doi: 10.1016/j.amjms.2017.01.019. [DOI] [PubMed] [Google Scholar]
- 49.Kesarwani M, et al. Targeting c-FOS and DUSP1 abrogates intrinsic resistance to tyrosine-kinase inhibitor therapy in BCR-ABL-induced leukemia. Nat Med. 2017;23:472–482. doi: 10.1038/nm.4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Faber ZJ, et al. The genomic landscape of core-binding factor acute myeloid leukemias. Nat Genet. 2016;48:1551–1556. doi: 10.1038/ng.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Levis M, et al. Results from a randomized trial of salvage chemotherapy followed by lestaurtinib for patients with FLT3 mutant AML in first relapse. Blood. 2011;117:3294–301. doi: 10.1182/blood-2010-08-301796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Gosliga D, et al. Establishing long-term cultures with self-renewing acute myeloid leukemia stem/progenitor cells. Exp Hematol. 2007;35:1538–49. doi: 10.1016/j.exphem.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 54.Weber K, Bartsch U, Stocking C, Fehse B. A multicolor panel of novel lentiviral "gene ontology" (LeGO) vectors for functional gene analysis. Mol Ther. 2008;16:698–706. doi: 10.1038/mt.2008.6. [DOI] [PubMed] [Google Scholar]
- 55.Mostoslavsky G, et al. Efficiency of transduction of highly purified murine hematopoietic stem cells by lentiviral and oncoretroviral vectors under conditions of minimal in vitro manipulation. Mol Ther. 2005;11:932–40. doi: 10.1016/j.ymthe.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 56.Bert AG, Johnson BV, Baxter EW, Cockerill PN. A modular enhancer is differentially regulated by GATA and NFAT elements that direct different tissue-specific patterns of nucleosome positioning and inducible chromatin remodeling. Mol Cell Biol. 2007;27:2870–85. doi: 10.1128/MCB.02323-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wingett S, et al. HiCUP: pipeline for mapping and processing Hi-C data. F1000Res. 2015;4:1310. doi: 10.12688/f1000research.7334.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mifsud B, et al. GOTHiC, a probabilistic model to resolve complex biases and to identify real interactions in Hi-C data. PLoS One. 2017;12:e0174744. doi: 10.1371/journal.pone.0174744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heinz S, et al. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Molecular Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang HM, et al. AnimalTFDB: a comprehensive animal transcription factor database. Nucleic Acids Res. 2012;40:D144–9. doi: 10.1093/nar/gkr965. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data have been deposited at GEO under the accession number GSE108316. Processed data are available from our Data server (see URL section)