Abstract
Expansion of a hexanucleotide repeat (HRE), GGGGCC, in the C9ORF72 gene is recognized as the most common cause of familial amyotrophic lateral sclerosis (FALS), frontotemporal dementia (FTD) and ALS-FTD, as well as 5–10% of sporadic ALS. Despite the location of the HRE in the non-coding region (with respect to the main C9ORF72 gene product), dipeptide repeat proteins (DPRs) that are thought to be toxic are translated from the HRE in all three reading frames from both the sense and antisense transcript. Here, we identified a CUG that has a good Kozak consensus sequence as the translation initiation codon. Mutation of this CTG significantly suppressed polyglycine-alanine (GA) translation. GA was translated when the G4C2 construct was placed as the second cistron in a bicistronic construct. CRISPR/Cas9-induced knockout of a non-canonical translation initiation factor, eIF2A, impaired GA translation. Transfection of G4C2 constructs induced an integrated stress response (ISR), while triggering the ISR led to a continuation of translation of GA with a decline in conventional cap-dependent translation. These in vitro observations were confirmed in chick embryo neural cells. The findings suggest that DPRs translated from an HRE in C9ORF72 aggregate and lead to an ISR that then leads to continuing DPR production and aggregation, thereby creating a continuing pathogenic cycle.
Keywords: C9ORF72, Dipeptide protein repeats (DPRs), Hexanucleotide repeat expansions (HREs), Repeat associated non-AUG (RAN) translation, Unconventional translation, Internal ribosome entry site (IRES), eIF2A, Integrated stress response (ISR)
1. Introduction
Expansion of a hexanucleotide repeat (HRE), GGGGCC, in the C9ORF72 gene is recognized as the most common cause of familial amyotrophic lateral sclerosis (FALS), frontotemporal dementia (FTD) and ALS-FTD (reviewed in (Taylor et al., 2016)). This mutation is also the cause of 5–10% of sporadic ALS, presumably because of incomplete penetrance of the mutation. A normal repeat number in healthy individuals expands in patients to hundreds or thousands of repeats that vary in number in different cells and different tissues. The HRE is present in the non-coding region (with respect to the main C9ORF72 gene product) in either the first intron or the promoter region of the mRNA depending on which transcription start site is used. Despite the presence of the repeat in the non-coding region, dipeptide repeat proteins (DPRs) are translated from the expanded repeat in all three reading frames from both the sense and antisense transcript (Gendron et al., 2013; Mori et al., 2013; Zu et al., 2013). DPR synthesis is a result of a poorly understood unconventional mechanism of translation that has been referred to as repeat associated non-AUG (RAN) translation. Polyglycine-proline (GP), polyglycine-alanine (GA), and polyglycine-arginine (GR) are produced by the sense strand, while polyproline-glycine (PG), polyproline-alanine (PA), and polyproline-arginine (PR) are produced from the anti-sense strand.
Results from experiments in model systems have suggested that the DPRs translated from the HRE of C9ORF72 are toxic and play a role in disease pathogenesis (reviewed in Taylor et al. (2016)). Although the DPRs are present in neurons and glia in various regions of the central nervous system (CNS) early in disease (Freibaum and Taylor, 2017), they surprisingly tend to be more frequent in CNS regions that have less pathology, such as the cerebellum, rather than the main targeted tissue – namely the motor system (MacKenzie et al., 2014). GA is the most common DPR found in autopsy tissue, and also the most aggregate-prone and amyloidogenic (reviewed in (Freibaum and Taylor, 2017). The mechanism for toxicity remains unclear, but might relate to its aggregate formation. PR and GR are thought to be the most toxic DPRs (reviewed in Freibaum and Taylor (2017)). GP can be detected in the cerebrospinal fluid of patients with C9ORF72-associated FALS as well as asymptomatic carriers of the HRE (Gendron et al., 2017); however, the levels of GP are relatively stable over time, showing no fluctuation related to disease progression. Of note, GR is recruited by GA into cytoplasmic inclusions (Yang et al., 2015). PA and PR are the least abundant, perhaps because they are derived from the antisense transcript or perhaps because the antibodies used for their detection have suboptimal sensitivity.
In the present study we investigated translation of DPRs following transfection of cells with a construct that encoded 75 repeats of C9ORF72 and flanking sequence. We found that translation initiation of GA occurred from a CUG codon upstream from the repeat. Interestingly, translation of GA involved internal ribosome entry and partly depended on eIF2A, a non-canonical initiation factor that is used in translation by certain viruses (Kim et al., 2011) and by cells experiencing various stress conditions (Sendoel et al., 2017; Starck et al., 2012). Furthermore, translation of GA was associated with induction of the integrated stress response (ISR), while the ISR led to continuing GA translation, suggesting that the interaction of GA translation and the ISR leads to a self-sustaining pathogenic cycle.
2. Materials and methods
2.1. Cell culture
NSC34 and HEK293 cells were cultured in DMEM supplemented with 10% FBS, 2 mM L-Glutamine, 100 U/ml Penicillin and 100 μg/ml Streptomycin. Eif2a-knockout (KO) and non-targeted control mouse keratinocyte cell lines (Sendoel et al., 2017) were cultured in E medium supplemented with 15% FBS and 50 μM CaCl2. DF-1 chicken embryonic fibroblast cell lines (ATCC; CRL-12003) were cultured in DMEM supplemented with 10% FBS, 100 U/ml Penicillin and 100 μg/ml Streptomycin. In experiments related to ER stress, cells were pretreated with 500 nM thapsigargin, 1 mM 1,4-dithiothreitol, and 100 μM sodium arsenite (Sigma).
2.2. Plasmids
2.2.1. HRE constructs with 75 repeats of G4C2 and 113 bp 5′, and 99 bp 3′ flanking sequences
All oligonucleotides were obtained from Integrated DNA Technologies. Oligonucleotide I (see Table S1 for list and sequence of oligonucleotides), which contained part of a HindIII site followed by 113 nucleotides upstream of the HRE and then by three G4C2 repeats, and oligonucleotide III, which contained 10 G4C2 repeats followed by part of a NotI site, were phosphorylated, annealed, and then ligated into restriction sites of HindIII and NotI of pAG (Gendron et al., 2013). In order to generate stop GA plasmids, oligonucleotide II rather than oligonucleotide I and oligonucleotide III was ligated into restriction sites of HindIII and NotI of pAG3. The plasmid was sequenced, then digested with HindIII and BamHI. The HindIII-BamHI fragment was digested with BanII, and the HindIII-BanII fragment was then ligated with oligonucleotide III into pAG3. This approach was serially repeated with similar digestions and ligations of oligonucleotide III in order to increase the number of G4C2 repeats to 73. Finally, the HindIII-BanII fragment was ligated with oligonucleotide IV (which contains 2 G4C2 repeats followed by a 99 bp flanking sequence and then part of the NotI site), into the pAG3 vector. The resultant insert and surrounding nucleotides were sequenced and the repeat digested out of the construct to confirm the number of repeats and the absence of any mutation (Figs. S1, S2).
2.2.2. GA, GP, and GR nLuc reporter constructs
The open reading frame of nLuc (Promega), without an AUG start codon, was PCR amplified using primer nLuc-R along with GA-nLuc-F, GP-nLuc-F or GR-nLuc-F (see Table S2 for list and sequence of primers). The amplified products were cloned into the Not I and BamHI sites of pAG3. The ΔC9 construct was prepared using the primer set of GAnLuc-F and nLuc-R.
2.2.3. Mutations of the CUG and Kozak consensus sequence
In order to generate mutations of the GA putative CUG initiation site, which is upstream from the HRE and in the GA reading frame, the GA-nLuc construct was digested with HindIII and NotI. The HindIII-NotI insert was then recovered and digested with BfaI, which cuts in the HRE 5′ flanking sequence. The HindIII-BfaI fragment was then replaced by oligonucleotides V, VI, VII, VIII, IX, X, XI, or XII which have various mutations of the CUG and Kozak consensus sequence, and the fragment was ligated into the HindIII and NotI sites of pAG3.
2.2.4. Bicistronic constructs
In order to prepare bicistronic constructs with fLuc (Promega) in the first cistron and either ΔC9-nLuc or C9-nLuc in the second cistron, fLuc was first PCR amplified using primers fLuc-F and fLuc-R. The amplified product was then cloned into the NheI and AflII sites of pcDNA6 (Thermo Fisher Scientific). The ΔC9-nLuc and C9-nLuc fragments were then digested with HindIII and BamHI, and the desired fragments were cloned into the HindIII and BamHI sites of pcDNA6 containing fLuc.
2.3. In vitro transcription
In vitro transcription was carried out with 10 μg of EcoRV-linearized plasmids using RiboMAX Large Scale RNA Production System (Promega) with 4:1 ratio of cap analog and anti-reverse cap analog [3′O-Me-m7G(5′)ppp(5′)G] (New England Biolabs) to GTP. Template DNA was digested with RNase-free DNaseI (Promega) for 15 min at 37 °C. Polyadenylation of mRNAs was performed with E. coli poly(A) polymerase (New England Biolabs) for 30 min at 37 °C. The mRNAs were isolated by RNA Clean and Concentrator-25 (Zymo Research). The integrity of the RNA was assessed by agarose gel electrophoresis.
2.4. Luciferase assay
NSC34 and HEK293 cells were planted in 24 well plates at 4 × 104 and 8 × 104 per well, respectively. In some experiments, cells were cotransfected using Lipofectamine LTX (Thermo Fisher Scientific) either with 500 ng pGL4.51 fLuc plasmid with 500 ng nLuc reporter plasmids or with 500 ng of bicistronic plasmids. In other experiments, cells were transfected with 500 ng of in vitro transcribed mRNA using TransIT-mRNA transfection kit (Mirus Bio). At various times after transfection, cells were lysed with 1 × passive lysis buffer (Promega). The levels of nLuc and fLuc were assessed using a Nano-Glo Dual-Luciferase Reporter assay system (Promega) and a Wallac 1420 VICTOR 3 V luminometer (Perkin Elmer) according to the manufacturer’s protocol.
2.5. Western blotting
NSC34 and HEK293 cells were planted in 6 well plates at 2 × 105 and 4 × 105 per well, respectively. Cells were transfected with 2.5 μg nLuc reporter plasmids using Lipofectamine LTX (Thermo Fisher Scientific). The cells were lysed 48 h later with TNES lysis buffer (50 mM Tris-HCl at pH 7.6, 1% Nonidet P-40, 150 mM NaCl, 2 mM EDTA) or a lysis buffer consisting of 4 M Urea and 0.5% SDS containing a protease inhibitor (cOmplete Mini EDTA-free, Roche) and phosphatase inhibitor cocktail (Sigma). Five to thirty micrograms of total protein were subjected to electrophoresis on 4–20% SDS polyacrylamide gels (Mini-PROTEAN TGX Gels, BIO-RAD) and transferred to Amersham Hybond P 0.45 μm PVDF membrane (GE Healthcare). The membrane was first blocked with 1% non-fat skim milk in Tris-buffered saline containing 0.05% Tween-20 for 1 h at room temperature. Following this treatment, it was then incubated with primary antibodies against poly-GA (1:1000, MABN889, EMD Millipore), β-actin (1:5000, AC-15, Sigma), phospho-eIF2α (1:1000, 119A11, Cell Signaling Technology), eIF2α (1:1000, D7D3, Cell Signaling Technology), or eIF2A (1:1000, EPR11042, Abcam) overnight at 4 °C. Finally the membrane was incubated with anti-mouse or anti-rabbit horseradish peroxidase–conjugated secondary antibodies (1:5000, both from GE Healthcare) for 1 h at room temperature. The signals were detected using SuperSignal West Dura Extended Duration Substrate (Thermo Fisher Scientific) and analyzed using ChemiDoc MP Imaging System (BIO-RAD).
2.6. Indirect immunofluorescence and confocal laser microscopy
HEK293 cells on coverslips were transfected with the GA-nluc or ΔC9-nluc plasmids. After 48 h, the cells were fixed with 4% paraformaldehyde for 5 min at room temperature, and then permeabilized with 100% ice-cold methanol or phosphate buffered saline with 0.1% Tween-20 for 30 min at 4 °C. The coverslips were incubated overnight at 4 °C with mouse monoclonal poly-GA antibody (1:150, MABN889, Millipore), mouse monoclonal HA antibody (1:100, 6E2, Cell Signaling Technology), rabbit polyclonal HA antibody (1:100, C29F4, Cell Signaling Technology), rabbit anti-phosphorylated eIF2α antibody (1:100, 119A11, Cell Signaling Technology), rabbit anti-G3BP1 antibody (1:200, ab181149, Abcam), rabbit anti-eIF3A antibody (1:200, ab128996, Abcam), rabbit anti-TIA-1 antibody (1:250, 12133–2-AP, Proteintech) or rabbit anti-ATF4 antibody (1:200, D4B8, Cell Signaling Technology). After rinsing, cells were incubated with Alexa 594-conjugated goat anti-mouse IgG (1:1000, Invitrogen) or Alexa 488-conjugated goat anti-rabbit IgG (1:1000, Invitrogen), and then counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Images were captured using a confocal laser microscope system (Leica TCS SP5, Leica Microsystems, Wetzlar, Germany). A sequential multiple fluorescence scanning mode was used to avoid nonspecific overlap of signals.
2.7. RT-PCR
Total RNA was extracted using TRIzol (Thermo Fisher Scientific) according to the manufacturer’s protocol. The samples were treated with TURBO DNase (Thermo Fisher Scientific) for 30 min at 37 °C, and RNA was isolated using RNeasy Mini kit (Qiagen). cDNA was generated using SuperScript III First-Strand Synthesis System (Thermo Fisher Scientific). Quantitative RT-PCR was performed using Power SYBR Green PCR Master Mix (Thermo Fisher Scientific) and primer sets of RT-fLuc-F and RT-fLuc-R or RT-nLuc-F and RT-nLuc-R (Table S2) or Taqman Gene Expression Master Mix and Taqman gene expression assays (Gg03815934_s1 for chicken β-actin and Gg03356565_m1 for chicken eIF2A) in a CFX96 Real-Time System (Bio-Rad).
2.8. Generation of EIF2A-knock out cell lines by CRISPR/Cas9 system
sgRNAs, selected from the GeCKO library, were as follows: non targeting (NT) sgRNA - 5′-GCGAGGTATTCGGCTCCGCG-3′; Eif2a sgRNA - 5′-AAGAGTTTCATCTTCTGACC-3′. The sgRNAs were cloned into a lenti-CRISPR v2 plasmid (Addgene plasmid repository). HEK293 cells were plated into 6 well plates at 4 × 105 cells per well, and transfected with 2.5 μg lenti-CRISPR v2 plasmids using Lipofectamine LTX. Transfected cells were selected using 2 μg/ml puromycin for 3 days. EIF2A-KO cell clones were obtained by limited dilution.
2.9. shRNA constructs
The sequence used for the eIF2A siRNA knockdown, 5′-CTGCACT ACATTGCAACAAAT, was obtained from the Genetic Perturbation Platform (GPP) Web portal of the Broad Institute GPP; this sequence is the same in mouse and chicken. Oligonucleotides including siRNA sequence were cloned into BamHI and Hind III site of psilencer 2.1-U6 neo vector (Thermo Fisher Scientific) according to the manufacturer’s protocol. The control shRNA vector was provided by this vector kit. The knockdown of chick eIF2A by shRNA directed against eIF2A was tested in the DF-1 chick fibroblast cell line.
2.10. Chicken embryo spinal cord electroporation
Three days before electroporation, pathogen-free fertilized White Leghorn chicken eggs (Sunnyside Hatchery, Beaver Dam, WI) were transferred to an egg incubator (39 °C, 60 to 70% humidity) for development of the embryos to stage 11–12 (Hamburger-Hamilton stage). The chick embryo spinal cords were electroporated as previously described (Guest et al., 2004). Embryos were dissected from the egg at 24–48 h post-electroporation, placed in 250 μl of PLB, and homogenized with a Kontes pestle (Fischer, Hampton, N.H.). Homogenates were centrifuged (Eppendorf centrifuge 5417C) for 1 min at 14,000 rpm. The supernatant was removed and assayed in a dual-luciferase assay.
2.11. Statistical analysis
Statistical analysis was performed by an unpaired t-test, one-way ANOVA with Tukey’s multiple comparisons test or two-way ANOVA with Tukey’s multiple comparisons test using GraphPad Prism version 7.0a. A P-value of < 0.05 was considered significant. The data are presented as mean ± standard error of the mean (SEM).
3. Results
3.1. Translation of GA dipeptides from the expanded repeats
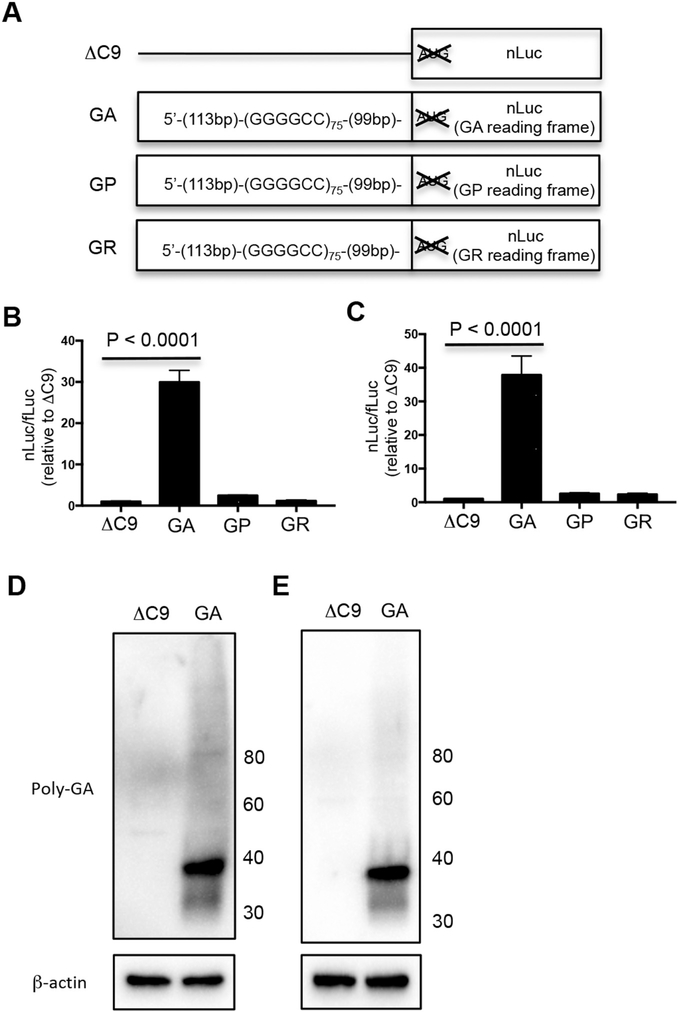
To begin to investigate translation of the DPRs, we prepared a construct that had 75 G4C2 repeats with 113 nucleotides upstream of the repeat and 99 nucleotides downstream of the repeat followed by a nanoluciferase (nLuc) reporter in the reading frame of GA, GP, or GR (Fig. 1A). Following transfection separately of each of these constructs into NSC34 (Fig. 1B) (a motor neuron-like cell line) and HEK293 cells (Fig. 1C), there was robust GA-nLuc expression compared to control ΔC9-nLuc (P < 0.0001), but relatively little GP-nLuc and GR-nLuc expression. The synthesis of GA was confirmed on western blots in which anti-GA antiserum immunostained lysates from NSC34 (Fig. 1D) and HEK293 cells (Fig. 1E) transfected with the GA-nLuc construct (Fig. 1D, E). These results confirm that the GA is translated from the C9ORF72 expanded repeat.
Fig. 1.
GA is the most abundantly translated DPR from G4C2 expanded repeat. (A) Schematic diagram showing ΔC9-nLuc, GA-nLuc, GP-nLuc, and GR-nLuc plasmids. NSC34 (B, D) and HEK293 (C, E) cells were cotransfected with fLuc plasmid along with either ΔC9-nLuc, GA-nLuc, GP-nLuc, or GR-nLuc plasmids. (B, C) Cells were harvested after 48 h and processed for dual luciferase assays. Unless noted in other figure legends, the nLuc/fLuc ratio was normalized to that of ΔC9-nLuc, which was set to 1. (D, E) Cell lysates were processed for western blots and immunostained with poly-GA and β-actin antibody. Molecular weight in kDa is shown on the side of the western blot.
3.2. GA translation is initiated from a CUG upstream of the expanded repeat
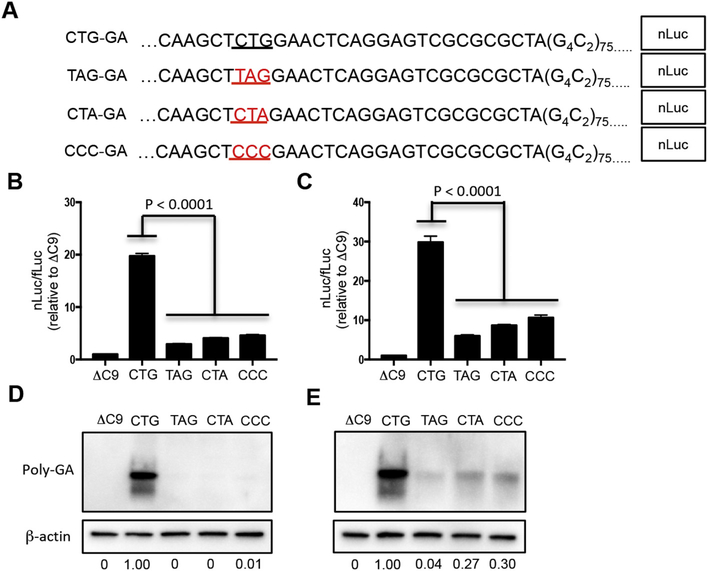
Of note, no AUG is present in the 113 nucleotides upstream of the expanded repeat (Figs. 2A, S1). Since near cognate initiation codons, especially CUG, are used in translation of cellular upstream open reading frames (uORF) (Starck et al., 2012), we examined the sequence upstream of the repeats for CTGs. Four CTGs are located upstream of the repeat; however, three of them (colored in Fig. S1) will encounter stop codons in their respective reading frame (shown in Fig. S1 with the same color for the reading frame as used for the putative CUG initiation codon). We therefore tested whether the fourth CTG, which is located 24 nucleotides upstream of the expanded G4C2 repeat (Figs. 2A, S1) and has a good Kozak consensus sequence, is the initiation codon.
Fig. 2.
Translation of GA is dependent on a CUG codon located upstream of the G4C2 expanded repeat. (A) Part of DNA sequence of CTG-GA-nLuc, TAG-GA-nLuc, CTA-GA-nLuc or CCC-GA-nLuc showing either the underlined putative CUG translation start codon upstream of the expanded repeat or with mutated CTG shown in red. NSC34 (B, D) and HEK293 (C, E) cells were cotransfected with fLuc plasmid along with either ΔC9-nLuc, CTG-GA-nLuc, TAG-GA-nLuc, CTA-GA-nLuc or CCC-GA-nLuc plasmids. Cells were harvested after 48 h and assessed by dual luciferase assays (B, C) or western blot (D, E) of cell lysates that were immunostained with poly-GA and β-actin antibody. Molecular weight in kDa is shown on the side of the western blot. The ratio of the GA band to the β-actin band is shown at the bottom of the western blots with the ratios normalized to the ratio in the CTG lane, which was set to 1.
In order to investigate this issue, we made several mutations, separately changing the CTG to TAG, CTA, and CCC (Fig. 2A). A firefly luciferase (fLuc) construct was cotransfected in NSC34 (Fig. 2B) and HEK293 cells (Fig. 2C) along with GA-nLuc that had either an upstream CTG or mutated CTG. The nLuc/fLuc ratio was much greater following transfection of GA-nLuc that had an upstream CTG compared to the ratio with constructs that had a mutated CTG (P < 0.0001) (Fig. 2B, C).
In order to confirm that GA was no longer translated following mutation of the upstream CTG, we western blotted homogenates of NSC34 (Fig. 2D) or HEK293 cells (Fig. 2E) that had been transfected with ΔC9 or a GA-nLuc construct that contained a wild type or mutated CTG. There was robust anti-GA antibody immunostaining of cells transfected with GA-nluc that had wild type CTG, but none in the case of ΔC9, virtually none with TAG-GA-nLuc, and minimal immunostaining with CTA-GA-nLuc and CCC-GA-nLuc. The finding that both of the two protein products seen on the western blots in Fig. 2 (and Fig. 1D, E) were significantly decreased by transfection of constructs with CTG mutations suggests that the lower band on the western blots is a degradative product of CUG translation-initiated GA.
To further confirm that translation of GA occurs by scanning from the upstream CUG rather than a translation initiation site for GA within the expanded repeat, a TAG stop was inserted in the GA reading frame immediately upstream of the repeat (Fig. S3). Cotransfection of this stop-GA-nLuc construct along with the fLuc construct into NSC34 (Fig. S3B) and HEK293 cells (Fig. S3C) led to a significant decrease in the nLuc/fLuc ratio compared to that seen with the GA-nLuc construct (P < 0.0001). This result confirms that there is little translation initiation in the GA reading frame from the actual G4C2 expanded repeat.
Of interest, transfection of GP-nLuc or GR-nLuc that had the same TAG insertion in the GA reading frame led to a significant increase in the nLuc/fLuc ratio for GP-nLuc (Fig. S3D, E) and for GR-nLuc (S3F, S3G). These results suggest that ribosomes that meet a stop codon shortly after translation initiation at the GA CUG initiation codon may reinitiate at translation start sites for GP and GR, presumably within the HRE. Additional evidence supporting the presence of the GP initiation codon in the expanded repeat is supported by the presence of a stop codon in the GP reading frame 2 nucleotides upstream from the repeat (Fig. S1).
We compared the efficiency of translation of GA-nLuc using CUG as the start codon with translation of GA-nLuc using the more conventional AUG translation initiation codon by replacing the CTG with an ATG (Fig. S4A). Cotransfection of fLuc with ATG-GA-nLuc into NSC34 (Fig. S4B) and HEK293cells (Fig. S4C) showed a very significant increase in the nLuc/fLuc ratio compared to that seen following transfection of GA-nLuc with a CTG. This is not surprising considering prior studies that have shown increased translation efficiency with AUG compared to CUG as the start codon (Kearse and Wilusz, 2017).
We tested whether GA translation could be initiated upstream of the CUG by inserting a TAG stop codon 4 nucleotides upstream of the CUG (Fig. S5A). As a result of the insertion, there was a small but statistically significant decrease in GA translation following transfection into NSC34 cells (P < 0.05) (Fig. S5B), but not into HEK293 cells (Fig. S5C). The latter results suggest that the majority of GA translation occurs following translation initiation at a CUG −24 nucleotides upstream from the repeat; however, a small amount of translation initiation may occur upstream of this CUG in certain cell types.
We next tested the importance of nucleotides that resemble a Kozak consensus sequence that normally flanks the AUG initiation codon, C(A/G)CCAUGG, immediately upstream and downstream of the putative CUG initiation codon. We compared GA translation following mutation (with respect to the CUG) of the G in the −3 position to T and also mutation of the G in the +4 position to A and to C (Fig. S6A). NSC34 (Fig. S6B) and HEK293 (Fig. S6C) cells were cotransfected with the fLuc construct along with either ΔC9, GA-nLuc or GA-nLuc that had mutations in the −3 and +4 position. The nLuc/fLuc ratio was significantly decreased (P < 0.0001) when mutations were made in this Kozak sequence, supporting the identification of CUG as the initiation codon.
One question that arose is whether the CUG that is −24 nucleotides upstream of the 75 repeats is also used for translation initiation for a construct with a lesser number of repeats. In order to test this we prepared a 5R-GA construct identical to GA-nLuc, but with 5 rather than 75 repeats. The CUG that is −24 nucleotides upstream of the 5 repeats was then mutated to TAG, CTA or CCC (Fig. S7A). There was a statistically significant decrease (P < 0.05) in the nLuc/fLuc ratio following cotransfection of NSC34 cells with fLuc along with any of the 5 repeat constructs that had a mutated CTG when compared to the ratio found after cotransfection of fLuc with the 5 repeat construct that had a CTG (Fig. S7B). Although there was also a decrease in the ratio following cotransfection in HEK293 cells, it was not statistically significant (Fig. S7C). Of note, the decrease in the ratio that was seen in both cell lines following cotransfection of fLuc along with the 5 repeat mutant constructs was much less than the decrease seen following cotransfection of fLuc with the 75 repeat construct (GA-nLuc) that had similar mutations of the CTG (compare Fig. S7B, C with Fig. 2B, C) – suggesting that other translation initiation sites are used in addition to CTG in the case of the 5 repeat construct.
We then compared the efficiency of translation of 5R-GA with GAnLuc following cotransfection of fLuc separately with each construct into NSC34 (Fig. S7D) and HEK293 cells (Fig. S7E). Cotransfection of 5R-GA had a significantly increased nLuc/fLuc ratio compared to 75RGA. We suspect that this may be because the 5R-GA has the multiple translation start sites or perhaps different rates of transcription or translation.
3.3. GA is translated from an internal ribosome entry site (IRES)
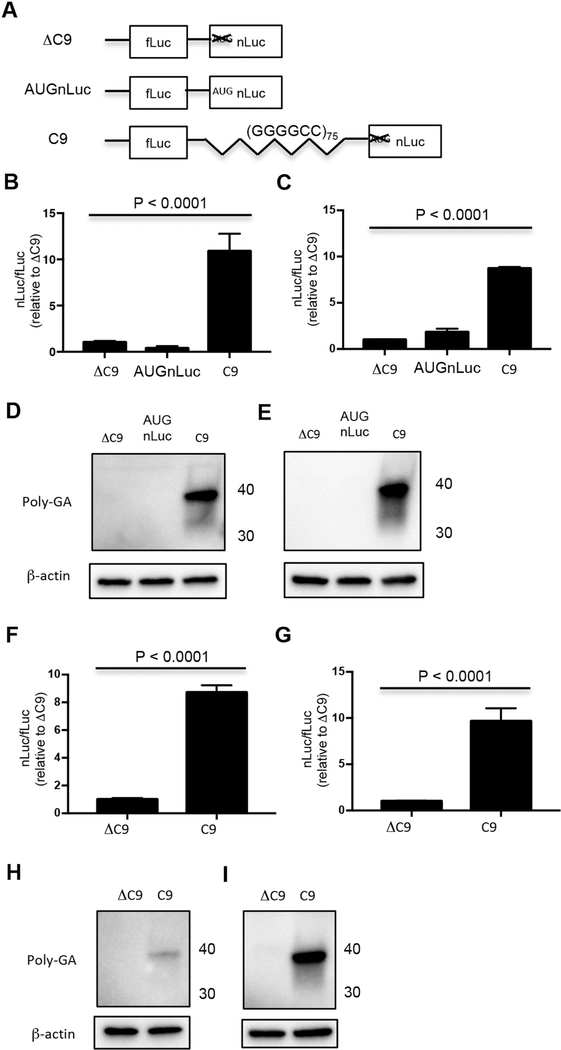
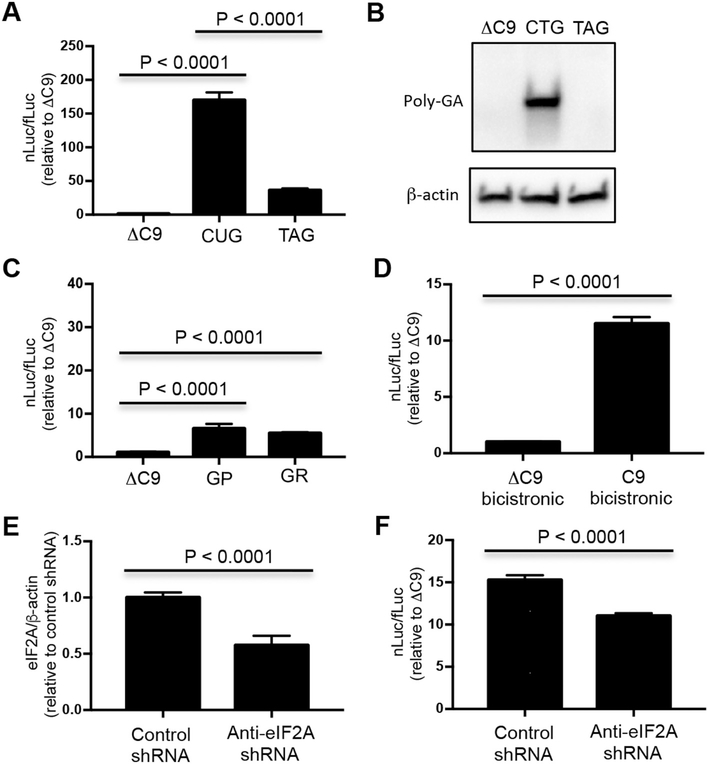
To test whether GA was translated from an IRES, a C9 bicistronic construct was prepared: fLuc cDNA was placed in the first cistron, while the second cistron contained GA (which contained 75 G4C2 repeats and the previously described nucleotides upstream and downstream of the repeats followed by a nLuc reporter in the reading frame of GA) (Fig. 3A). There were stop codons in the fLuc and two other reading frames between the first and second cistron. We compared results seen with the C9 bicistronic construct with those seen with control ΔC9 bicistronic constructs that had fLuc in the first cistron followed by nLuc with an AUG initiation codon (AUGnLuc) or without an AUG initiation codon (ΔC9) in the second cistron (Fig. 3A). The ratio of the nLuc/fLuc was significantly increased following transfection of NSC34 (Fig. 3B) and HEK293 (Fig. 3C) cells with the C9 bicistronic construct compared to that seen following transfection of the control bicistronic constructs (P < 0.0001).
Fig. 3.
GA is translated from an IRES. (A) Schematic diagram of ΔC9, AUGnLuc, and C9 bicistronic constructs. NSC34 (B, D, F, H) and HEK293 (C, E, G, I) cells were transfected with ΔC9, AUGnLuc, or C9 bicistronic plasmids (B–E) or in vitro derived transcripts from ΔC9 and C9 bicistronic plasmids (F–I). Cells were harvested 48 h later for dual luciferase assays (B, C, F, G) or western blots (D, E, H, I). Western blots were immunostained with poly-GA and β-actin antibody. Molecular weight in kDa is shown on the side of the western blot.
In order to test whether the −24 upstream CUG was the initiation codon for translation of GA following transfection of the bicistronic construct, the CTG was changed to TAG, CTA, and CCC (Fig. S8A). Transfection of these mutated bicistronic constructs into NSC34 (Fig. S8B) showed that the nLuc/fLuc ratio decreased from the level seen with C9 bicistronic constructs to baseline levels (P < 0.0001). Transfection of HEK293 (Fig. S8C) cells with the mutated bicistronic constructs also significantly decreased the nLuc/fLuc ratio compared to the level seen with C9 bicistronic constructs (P < 0.0001); however, the ratios were higher than baseline levels. The latter results suggest that alternative translation initiation codons may be used in HEK293 cells following mutation of the −24 CTG.
Additional experiments were carried out with in vitro derived transcripts from the bicistronic constructs. There was a similar increase in the nLuc/fLuc ratio following transfection of NSC34 (Fig. 3F) and HEK293 (Fig. 3G) cells with RNAs derived from the C9 bicistronic construct compared to transcripts derived from the control bicistronic constructs (P < 0.0001). The robust translation of GA-nLuc when placed in the second cistron suggests that translation of GA occurs by means of an IRES. A western blot showed that the translated product from the second cistron of C9 following transfection of both the C9 bicistronic construct as well as in vitro derived transcripts from the C9 bicistronic construct was similar in size to that seen with monocistronic GA-nLuc (Fig. 3D, E, H, I), suggesting that the monocistronic and bicistronic constructs are utilizing the same translation initiation site and stop codon.
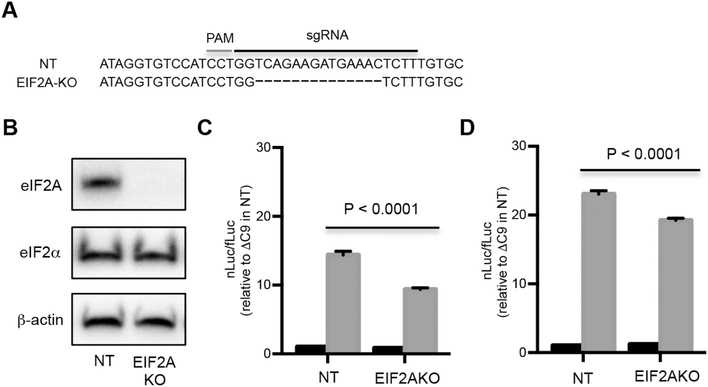
3.4. eIF2A is used for GA translation
eIF2A rather than eIF2α has been reported to be a key initiation factor for translation of uORFs that have CUG as the initiation codon (Starck et al., 2012). In order to assess the importance of eIF2A in GA IRES-mediated translation, we made use of CRISPR/Cas9 to prepare EIF2A-knockout (KO) HEK293 cells (Fig. 4A). By western blot analysis, expression of eIF2A appeared to be abrogated in EIF2A-KO cells compared to control non-targeted CRISPR/Cas9 HEK293 cells (Fig. 4B).
Fig. 4.
Translation of GA utilizes eIF2A. (A) Part of DNA sequence of non-targeted (NT) HEK293 cells and EIF2A-knockout (EIF2A-KO) cell lines showing protospacer adjacent motif (PAM) and sgRNA sequence. (B) Western blot of lysates of NT and EIF2A-KO cell lines immunostained with eIF2A, eIF2α, and β-actin antibody. NT and EIF2A-KO cells were transfected with either ΔC9 (black) or C9 (gray) bicistronic plasmids (C), or with in vitro derived transcripts from ΔC9 (black), or C9 (gray) bicistronic plasmids (D).
The EIF2A-KO and control non-targeted cells were transfected with C9 and ΔC9 bicistronic constructs to determine whether translation of the second cistron depended on eIF2A. RT-PCR showed that non-targeted and EIF2A-KO cells had comparable amounts of RNA following transfection of C9 and ΔC9 bicistronic constructs (Fig. S9). Importantly, there was a decrease in the nLuc/fLuc ratio following transfection of the C9 bicistronic construct in EIF2A-KO cells compared to transfection of the control non-targeted HEK293 (P < 0.0001) (Fig. 4C). This decrease in the ratio was a result of a more prominent decrease in nLuc than the change in fLuc. Similar results were also obtained using an independently-made Eif2a-null keratinocyte line from a mouse squamous cell carcinoma that has been previously described (Sendoel et al., 2017) (Fig. S10).
In order to test whether comparable results were seen following transfection of RNA, the C9 and control ΔC9 bicistronic constructs were in vitro transcribed, and the RNAs were then transfected into EIF2A-KO and control HEK293 cells. There was a statistically significant decrease in the nLuc/fLuc ratio following transfection of the in vitro derived transcripts in EIF2A-KO HEK293 cells compared to transfection of control cells (P < 0.0001) (Fig. 4D). This decrease was not as significant as that seen following transfection of the C9 bicistronic plasmid, perhaps related to degradation of the RNA transcripts or the timing of the cell harvest. These results suggest that translation of GA depends, at least partly, on eIF2A.
3.5. GA translation and the integrated stress response (ISR)
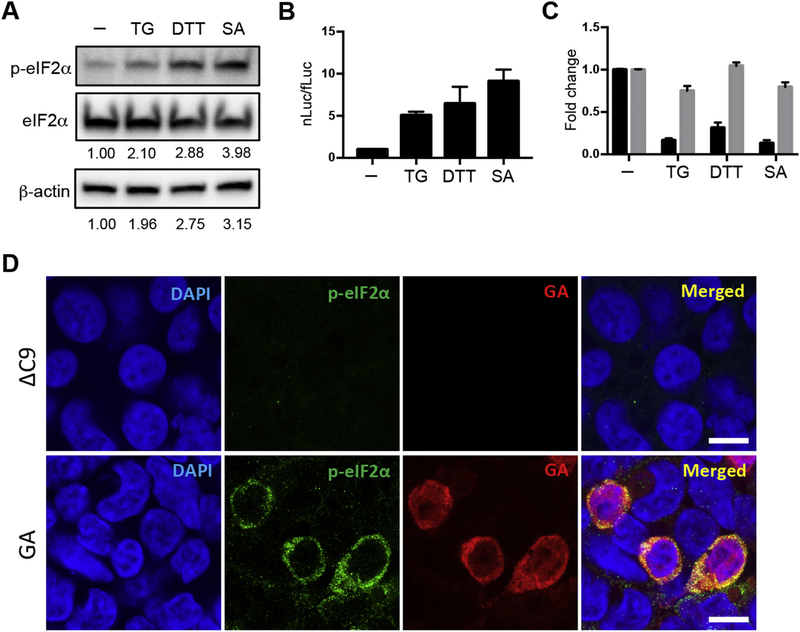
Misfolded proteins and oxidative stress, which are known to be present in ALS and FTD patient tissues (Palluzzi et al., 2017; Shahheydari et al., 2017) and have been implicated in the pathogenesis of these diseases, trigger an integrated stress response (ISR) in cells. The ISR leads to phosphorylation of eIF2α with a subsequent decrease in cap dependent translation. Since eIF2A is thought to be used for translation in the case of cellular stresses, we sought to determine the effect of ER stress on GA translation. To this end, we separately treated HEK293 cells with thapsigargin, dithiothreitol, and sodium arsenite, which are known to induce ER stress. A representative western blot of HEK293 cells following administration of each of these ER stressors showed that, as expected, these agents lead to increased phosphorylated eIF2α (Fig. 5A). These agents were then individually administered to HEK293 cells that had been transfected with C9 bicistronic constructs. All three agents led to a striking increase in the nLuc/fLuc ratio following transfection of the C9 bicistronic construct (Fig. 5B). The change in the ratio was a result of a prominent decrease in translation of fLuc with no statistically significant change in GA-nLuc translation (Fig. 5C). This result confirms that phosphorylation of eIF2α interferes with cap-dependent conventional translation, but spares IRES-dependent translation of GA.
Fig. 5.
Translation of GA and the ISR. (A) Western blot of HEK293 cells that were either untreated or pretreated with 500 nM thapsigargin (TG), 1 mM 1,4-dithiothreitol (DTT), or 100 μM sodium arsenite (SA) for 30 min. Western blots of lysates were immunostained with antibody against phosphorylated eIF2α (p-eIF2α) or eIF2α. The ratio of p-eIF2α/eIF2α is shown at the bottom of the eIF2α western blot, and the ratio of p-eIF2α/β-actin is shown at the bottom of the β-actin western blot, with the ratios normalized to the ratio of untreated HEK293 cells, which was set to 1. (B, C) Thirty minutes following treatment with TG, DTT or SA or no treatment, HEK293 cells were transfected with transcripts in vitro derived from the C9 bicistronic construct. After 2 h the cells were harvested for dual luciferase assays. In (B), the nLuc/fLuc ratio is shown normalized to those in untreated samples. In (C), the level of fLuc (black) and nLuc (gray) in the untreated cells is normalized to 1, and the level for each treated sample is normalized to the level in the untreated sample of nLuc and fLuc respectively. (D) Double immunofluorescent staining for p-eIF2α (green) and poly-GA (red) in HEK293 cells transfected with ΔC9-nluc (upper panel) or GA-nluc plasmid (lower panel). Cells transfected with GAnluc plasmid that had poly-GA staining also had p-eIF2α staining. The p-eIF2α staining was significantly above the background staining seen in cells that were not transfected or cells that were transfected with ΔC9-nluc plasmid. All images shown have the same magnification, laser intensity, gain and offset values, and pinhole setting. Scale bars, 10 μm.
We then tested whether transfection of the G4C2 expanded repeat induces an ISR. All cells that were transfected with GA-nluc plasmid that had evidence of GA expression also had evidence of phosphorylated eIF2α (Fig. 5D); the phosphorylated eIF2α immunostaining was significantly above the background staining in cells that were not transfected or cells that were transfected with ΔC9-nluc plasmid. High magnification of cells expressing GA identified small aggregates of GA in some transfected cells (Fig. S11A). We also examined other indicators of activation of the ISR. Cells transfected with GA-nLuc plasmid that express GA had evidence of stress granule formation, as demonstrated by the appearance of focal staining of three stress granule markers, TIA-1, G3BP1, and eIF3A (Fig. S11B); there was also upregulation of ATF4 (Fig. S11C). These studies suggest that the ISR is induced in cells following transfection of GA-nluc plasmid.
We questioned whether a 5 G4C2 repeat construct with identical flanking sequence to that in GA-nLuc induced the ISR, as is true with GA-nLuc, the 75 repeat construct. Since we were unable by immunofluorescence assay to detect GA following transfection of cells with a 5 repeat construct, we prepared a 5 and 75 repeat construct similar to GAnLuc, but with a hemagglutinin (HA) epitope tag replacing the nLuc. Immunofluorescence assay showed that HA aggregated following transfection of GA75-HA into HEK293 cells, while transfection of GA5-HA had a homogenous appearance of HA in the cytoplasm (Fig. S12). Importantly, cells that expressed HA following transfection of GA75-HA had abundant evidence of phosphorylated eIF2α, ATF4, and eIF3A, while cells that received GA5-HA had little (Fig. S13).
3.6. GA is translated from the upstream CUG in chick embryo spinal cord neural cells
To more reliably assess translation of DPRs from G4C2 expanded repeats in a neuron in an organism, we took advantage of a method involving electroporation of DNA into the spinal cords of developing chicken embryos. The bicistronic constructs were microinjected into the neural tube of a chicken embryo, followed by the placement of electrodes on each side of the neural tube and subsequent delivery of current. Our previous studies showed that electroporation efficiently delivers genes into neurons on one side of the caudal half of the spinal cord (Ghadge et al., 2006; Guest et al., 2004). The tissues were then processed for dual luciferase assays.
Coelectroporation of the GA-nLuc construct along with the fLuc construct demonstrated a significantly increased ratio of nLuc/fLuc compared to the ΔC9 construct or C9 that had a TAG stop rather than CTG at position −24 (Fig. 6A). Homogenates from chicks that had been electroporated with the GA-nLuc were immunostained with anti-GA antibody confirming the identity of the product as this dipeptide (Fig. 6B). In contrast to the expression of GA, there was only a slight increase in the nLuc/fLuc ratio following coelectroporation of fLuc with GP-nLuc and of coelectroporation of fLuc with GR-nLuc (Fig. 6C).
Fig. 6.
Translation of poly-GA in chick embryo spinal cord neural cells is dependent on a CUG codon and mediated by eIF2A. (A–C) The fLuc plasmid was coelectropolated into the central canal of chick embryos along with either: ΔC9-nLuc, CTG-GA-nLuc (C9), TAG-GA-nLuc (C9 with a TAG mutation replacing the upstream CTG), GP-nLuc, or GR-nLuc plasmids. The samples were harvested after 24 h for dual luciferase assays (A, C) or western blotted with immunostaining with anti-polyGA antibody (B). (D) The ΔC9 or C9 bicistronic plasmid was electroporated into the central canal of chick embryos. The samples were harvested after 48 h and assessed by dual luciferase assays. The nLuc/fLuc ratio was normalized to that of ΔC9, which was set to 1. (E) The control or anti-eIF2A shRNA was transfected into DF-1 chick fibroblast cell line. The levels of β-actin and eIF2A mRNAs were assessed by RT-PCR. (F) C9 bicistronic plasmid along with either control or anti-eIF2A shRNA plasmids were coelectroporated into the central canal of chick embryos. The samples were harvested after 48 h and assessed by dual luciferase assays. The nLuc/fLuc ratio was normalized to that of ΔC9, which was set to 1.
We then tested the translation of the C9 bicistronic construct in chick embryo spinal cord neural cells. The ratio of nLuc/fLuc was significantly increased following electroporation of the C9 bicistronic construct compared to that seen following transfection of the control ΔC9 bicistronic construct (P < 0.0001) (Fig. 6D). In order to determine whether the eIF2A plays a role in GA translation we prepared an shRNA directed against eIF2A that targets an identical sequence in mouse and chick eIF2A. In order to confirm the knockdown, we transfected the eIF2A shRNA into DF-1 chick fibroblast cell line. There was a significant knockdown of eIF2A mRNA with the eIF2A shRNA compared to a control shRNA (P < 0.0001) (Fig. 6E). We then coelectroporated shRNA directed against eIF2A along with the C9 bicistronic construct. There was a significant decrease in the nLuc/fLuc ratio as a result of the shRNA eIF2A knockdown (P < 0.0001) (Fig. 6F) that was similar to that seen in cultured cells.
4. Discussion
The identification of the C9ORF72 G4C2 HRE as the most common genetic cause of FALS, FTD, and ALS-FTD as well as the cause of 5–10% of sporadic ALS was a sentinel finding that has raised a number of questions, many of which remain unanswered. Several mechanisms by which this repeat expansion cause disease have been proposed including toxicity from DPRs translated from the HREs. Although evidence of toxicity from the DPRs comes from varied experimental models, it still remains controversial.
Although the C9ORF72 HRE is in a non-coding region with respect to the main gene product, DPRs are synthesized from the repeat from both the positive and negative strand of mRNA. DPRs are an authentic product of translation of C9ORF72 HREs because they are present in neural cells in autopsy tissue from patients with this mutation. This unconventional translation, known as RAN translation, can occur in a number of expanded repeat diseases from repeats in both the noncoding and coding region of the main gene product. For example, translation of DPRs from the CAG repeat in the coding region of huntingtin mRNA occurs in addition to translation of polyglutamines in huntingtin (Bañez-Coronel et al., 2015). Unconventional cap-independent translation has also been increasingly found in cellular genes, frequently from an uORF or IRES (Gebauer and Hentze, 2016; Hinnebusch et al., 2016; Weingarten-Gabbay et al., 2016).
Transfection of constructs containing 75 G4C2 repeats with 113 nucleotides 5′ to the repeats and 99 nucleotides 3′ to the repeats showed that GA can be translated in both HEK293 and NSC34 cells as well as chick embryo spinal cord neural cells. A CUG with a good Kozak consensus sequence upstream of the HRE provided a potential candidate for an initiation codon, especially since this codon has been recognized as a major alternative translation start site for uORFs in cells (Starck et al., 2012). Of note, the Kozak sequence may have a greater influence on non-AUG initiation codons than AUG initiation codons (Kearse and Wilusz, 2017). Other studies have recently shown that this upstream CUG is involved in GA translation in cultured cells (Green et al., 2017; Tabet et al., 2018). Our mutagenesis studies demonstrated that this upstream CUG was indeed the GA translation initiation site both in cultured cells as well as in vivo in chick embryo spinal cord neural cells. The HRE was translated in cultured cells as well as chick embryo spinal cord neural cells when placed as the second cistron in a bicistronic construct, suggesting that this translation occurred by means of an IRES and was independent of the mRNA 5′-end. The location of a translation initiation site upstream of a repeat expansion has also been described with the CGG expanded repeats in FMR1 (Sellier et al., 2017).
Despite robust translation of GA, only small amounts of GR and GP were translated in cultured cells and chick embryo spinal cord neural cells from the 75 copies of G4C2 in monocistronic constructs. Interestingly, GA is also the most common dipeptide found in autopsy tissue in FALS patients with C9ORF72 HRE; however, the difference in the frequency of GA deposits compared to the other DPRs in autopsy tissue is not as extreme as in our results. Since the length of the repeat in other diseases is important in the efficiency of DPR translation (Bañez-Coronel et al., 2015; Kearse et al., 2016), small amounts of GR and GP may have been seen in our study because the repeat number of 75 is far less that the 100 s and possibly 1000s in the CNS of patients with C9ORF72 HREs. Furthermore, the presence of only 99 nucleotides 3′ to the expanded repeats may be too few to allow efficient translation of GR and GP since RNA secondary structure downstream of a non-AUG initiation codon can influence the translation efficiency (Kearse and Wilusz, 2017). It may also be the cell cultures and chick embryo neural cells lack (human neural) cell-specific IRES-transacting factors (ITAFs) that are critical for efficient translation of GR and GP.
Of note, transfection of GP-nLuc and GR-nLuc constructs that had a stop codon just upstream of the HRE in the GA reading frame displayed enhanced nLuc expression. These results suggest that ribosomes that meet a stop codon after binding to the CUG initiation codon may reinitiate in the HRE. Therefore, factors that decrease CUG translation initiation or stall ribosomes, such as a paucity of a particular ITAF, can lead to inefficient translation of GA, with subsequent enhancement of downstream translation initiation of GP or GR within the HRE; it is known that translation of non-AUG uORFs can have varied effects on translation of more downstream ORFs (Young and Wek, 2016).
Importantly, our results show that GA translation partly depends on eIF2A since the ratio of the first (fLuc) cistron to the second cistron (GAnLuc) of the C9 bicistronic plasmid or of in vitro-derived transcripts from this bicistronic construct was significantly lowered in cells that had eIF2A knocked out. Knockdown of eIF2A in the chick embryo also decreased translation of GA-nLuc in chick embryo neural cells; however, the knockout of eIF2A did not completely abolish translation of GA, suggesting that GA translation initiation depends on other factors in addition to eIF2A.
eIF2A is known to be used under cellular stress conditions as demonstrated in the case of translation initiation of hepatitis C virus RNA (Kim et al., 2011) and of cellular uORFs (Kwon et al., 2017; Sendoel et al., 2017; Starck et al., 2012, 2016). Interestingly, eIF2A has recently been implicated in the translational regulation of genes associated with cancer (Sendoel et al., 2017), presumably because of stresses in cancer cells. Cells respond to stressors, such as ER stress from misfolded proteins, oxidative stress, amino acid deprivation, and viral infections, with an ISR. The ISR triggers various kinases to phosphorylate eIF2α, leading to a decrease in cap-dependent translation. When conventional cap-dependent translation is limited by phosphorylation of eIF2α, eIF2A is able to play an increased role in translation of certain mRNAs, especially ones that have an uORF. Of note, cellular uORFs that depend on eIF2A for translation can influence the efficiency of downstream translation of the gene’s main open reading frame, suggesting that the DPRs translated from the C9ORF72 HRE may also influence expression of the main C9ORF72 product (Kearse and Wilusz, 2017).
The identification in the present study that eIF2A is important in GA translation and that ER stressors and inducers of the ISR lead to attenuation of conventional cap-dependent translation with no change in RAN translation of the G4C2 expanded repeats raised questions as to whether the ISR plays a role in translation of GA as well as the other DPRs in C9ORF72-induced disease. In fact, a possible role for ER stress in the pathogenesis of ALS has been previously proposed (Dafinca et al., 2016; Lee et al., 2016; Shahheydari et al., 2017; Taylor et al., 2016; Wang et al., 2011, 2014; Zhang et al., 2014), and recent studies have suggested a relationship between ISR and GA translation (Cheng et al., 2018; Green et al., 2017).
We not only found that the ISR leads to attenuation of conventional cap-dependent translation with no change in RAN translation of the G4C2 expanded repeat, but also that GA translation from the expanded repeat (but not from 5 G4C2 repeats, perhaps because the protein product does not aggregate) is associated with induction of the ISR. These observations suggest that the following events could occur to enhance pathology in C9ORF72-induced ALS and FTD. Oxidative stress as well as misfolded/aggregated DPRs and TDP43 in neural cells of ALS and FTD patients could trigger the ISR to phosphorylate eIF2α and favor the use of eIF2A over eIF2α in translation initiation, a step that is critical in translation regulation. This preferential use of eIF2A in translation initiation as a result of phosphorylation of eIF2α could lead to a relative increase in DPR synthesis compared to the synthesis of other proteins dependent on conventional translation from a capped mRNA. The decrease in conventional cap-dependent translation could lead to a decrease in proteins that normally limit DPR aggregate formation. Furthermore, DPR synthesis may increase because various stresses can lead to increases in eIF2A protein (Kwon et al., 2017; Starck et al., 2012, 2016), and because DPRs are reported to interfere with nucleocytoplasmic transport, thereby potentially bringing more TDP-43 into the cytoplasm over time as well as more eIF2A from its normal predominantly nuclear localization to the cytoplasm (Kim et al., 2011). The increase in DPR aggregation over time, especially in non-replicating motor neurons, could then lead to further ER stress, induction of the ISR, as well as further DPR interference with nucleocytoplasmic transport (resulting in more TDP43 aggregation and more eIF2A in the cytoplasm) in a continuing pathogenic cycle. Eventually, when the ISR is overwhelmed by oxidative stress and misfolded proteins, apoptosis could ensue from overexpression of C/EBP homologous protein or HIPK2 (Lee et al., 2016). In addition, UPR/ISR activation of NF-κB could occur, which has been reported to be pathogenic in ALS (Picher-Martel et al., 2015; Swarup et al., 2011). The recognition that eIF2A plays a potentially important role in DPR translation and pathogenicity in ALS and FTD suggests that this initiation factor and the unconventional translation of the G4C2 expanded repeat could be targets for therapy.
Subsequent to the submission of the present study, three other investigations were published describing DPR translation from the C9ORF72 expanded repeat (Cheng et al., 2018; Green et al., 2017; Tabet et al., 2018). There were a number of similarities in all of these publications, but also some differences. Two publications as well as the present study identified the same translation initiation codon for GA, a CUG upstream of the repeats (Green et al., 2017; Tabet et al., 2018). Tabet et al. found that translation of GP and GR was affected by this CUG, suggesting that there was frameshifting. Two publications (Cheng et al., 2018; Green et al., 2017) as well as the present study identified a relationship between DPR translation and the ISR and suggested that there was a self-sustaining feedforward loop. Two of the publications found that translation of the DPRs was cap-dependent (Green et al., 2017; Tabet et al., 2018). Although the third study (Cheng et al., 2018) found that DPR translation was cap-independent, the authors did not rule out the possibility that there may be additional cap-dependent DPR translation.
Why were there differences in some of the results from four laboratories, and how accurate are these results in characterizing DPR translation in patients? It may be that the differences seen are related to different constructs with different numbers of repeats and different flanking sequences in different translation assays. For example, translation results found in the rabbit reticulocyte system may differ depending on how the lysates are made, and may differ from translation results in cultured cells and in cells in living animals. Furthermore, transfection of cells will lead to a transient overexpression that has a different effect than that seen with normal expression levels in the cell. Although our investigation was the only one that included studies of translation in the CNS of a living organism, our findings in the electroporated spinal cord of chick embryos may not faithfully mirror the situation in neural cells of patients. In patients, varying RNAs with varying numbers of repeats and possible variations in sequence may act as templates for translation for years. Despite these shortcomings, the investigations are important in clarifying the varied mechanisms of C9ORF72 unconventional translation of DPRs and in identifying possible novel targets for treatments.
Supplementary Material
Acknowledgments
RPR, YS, GG and KM are funded by a grant from the ALS Association and gifts from Steps 4 Doug, Families Unite Against ALS, John and Patricia McDonald, Gerry Kaufman, Arnold Sarnoff, Susan Warso, and Marc and Barbara Posner. This work was also partly supported by grants to EF from NYSTEM CO29559. AS is currently supported by the People Programme (Marie Curie Actions) of the European Union 7th Framework Programme FP7 under REA grant agreement no. 629861. EF is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Conflict of interests
The authors declare no conflict of interests.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nbd.2018.05.009.
References
- Bañez-Coronel M, Ayhan F, Tarabochia AD, Zu T, Perez BA, Tusi SK, Pletnikova O, Borchelt DR, Ross CA, Margolis RL, Yachnis AT, Troncoso JC, Ranum LP, 2015. RAN translation in Huntington disease. Neuron 88, 667–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W, Wang S, Mestre AA, Fu C, Makarem A, Xian F, Hayes LR, Lopez-Gonzalez R, Drenner K, Jiang J, Cleveland DW, Sun S, 2018. C9ORF72 GGGGCC repeat-associated non-AUG translation is upregulated by stress through eIF2α phosphorylation. Nat. Commun 9, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dafinca R, Scaber J, Ababneh N, Lalic T, Weir G, Christian H, Vowles J, Douglas AGL, Fletcher-Jones A, Browne C, Nakanishi M, Turner MR, Wade-Martins R, Cowley SA, Talbot K, 2016. C9orf72 hexanucleotide expansions are associated with altered endoplasmic reticulum calcium homeostasis and stress granule formation in induced pluripotent stem cell-derived neurons from patients with amyotrophic lateral sclerosis and frontotemporal dementia. Stem Cells 34, 2063–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, Taylor JP, 2017. The role of dipeptide repeats in C9ORF72-related ALS-FTD. Front. Mol. Neurosci 10, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Hentze MW, 2016. IRES unplugged. Science 351, 228. [DOI] [PubMed] [Google Scholar]
- Gendron TF, Bieniek KF, Zhang YJ, Jansen-West K, Ash PEA, Caulfield T, Daughrity L, Dunmore JH, Castanedes-Casey M, Chew J, Cosio DM, van Blitterswijk M, Lee WC, Rademakers R, Boylan KB, Dickson DW, Petrucelli L, 2013. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 126, 829–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron TF, Chew J, Stankowski JN, Hayes LR, Zhang YJ, Prudencio M, Carlomagno Y, Daughrity LM, Jansen-West K, Perkerson EA, O’Raw A, Cook C, Pregent L, Belzil V, van Blitterswijk M, Tabassian LJ, Lee CW, Yue M, Tong J, Song Y, Castanedes-Casey M, Rousseau L, Phillips V, Dickson DW, Rademakers R, Fryer JD, Rush BK, Pedraza O, Caputo AM, Desaro P, Palmucci C, Robertson A, Heckman MG, Diehl NN, Wiggs E, Tierney M, Braun L, Farren J, Lacomis D, Ladha S, Fournier CN, McCluskey LF, Elman LB, Toledo JB, McBride JD, Tiloca C, Morelli C, Poletti B, Solca F, Prelle A, Wuu J, Jockel-Balsarotti J, Rigo F, Ambrose C, Datta A, Yang W, Raitcheva D, Antognetti G, McCampbell A, Van Swieten JC, Miller BL, Boxer AL, Brown RH, Bowser R, Miller TM, Trojanowski JQ, Grossman M, Berry JD, Hu WT, Ratti A, Traynor BJ, Disney MD, Benatar M, Silani V, Glass JD, Floeter MK, Rothstein JD, Boylan KB, Petrucelli L, 2017. Poly(GP) proteins are a useful pharmacodynamic marker for C9ORF72-associated amyotrophic lateral sclerosis. Sci. Transl. Med 9, eaai7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghadge GD, Wang L, Sharma K, Monti AL, Bindokas V, Stevens FJ, Roos RP, 2006. Truncated wild-type SOD1 and FALS-linked mutant SOD1 cause neural cell death in the chick embryo spinal cord. Neurobiol. Dis 21, 194–205. [DOI] [PubMed] [Google Scholar]
- Green KM, Glineburg MR, Kearse MG, Flores BN, Linsalata AE, Fedak SJ, Goldstrohm AC, Barmada SJ, Todd PK, 2017. RAN translation at C9orf72-associated repeat expansions is selectively enhanced by the integrated stress response. Nat. Commun 8, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guest S, Pilipenko E, Sharma K, Chumakov K, Roos RP, 2004. Molecular mechanisms of attenuation of the Sabin strain of poliovirus type 3. J. Virol 78, 11097–11107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG, Ivanov IP, Sonenberg N, 2016. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science 352, 1413–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse MG, Wilusz JE, 2017. Non-AUG translation: a new start for protein synthesis in eukaryotes. Genes Dev. 31, 1717–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse MG, Green KM, Krans A, Rodriguez CM, Linsalata AE, Goldstrohm AC, Todd PK, 2016. CGG repeat-associated non-AUG translation utilizes a cap-dependent scanning mechanism of initiation to produce toxic proteins. Mol. Cell 62, 314–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Park SM, Park JH, Keum SJ, Jang SK, 2011. eIF2A mediates translation of hepatitis C viral mRNA under stress conditions. EMBO J 30, 2454–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon OS, An S, Kim E, Yu J, Hong KY, Lee JS, Jang SK, 2017. An mRNA-specific tRNAi carrier eIF2A plays a pivotal role in cell proliferation under stress conditions: stress-resistant translation of c-Src mRNA is mediated by eIF2A. Nucleic Acids Res 45, 296–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Shang Y, Redmond SA, Urisman A, Tang AA, Li KH, Burlingame AL, Pak RA, Jovicic A, Gitler AD, Wang J, Gray NS, Seeley WW, Siddique T, Bigio EH, Lee VMY, Trojanowski JQ, Chan JR, Huang EJ, 2016. Activation of HIPK2 promotes ER stress-mediated neurodegeneration in amyotrophic lateral sclerosis. Neuron 91, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie IR, Frick P, Neumann M, 2014. The neuropathology associated with repeat expansions in the C9ORF72 gene. Acta Neuropathol. 127, 347–357. [DOI] [PubMed] [Google Scholar]
- Mori K, Arzbeger T, Grässer FA, Gijselinck I, May S, Rentzsch K, Weng SM, Schludi MH, van der Zee J, Cruts M, Van Broeckhoven C, Kremmer E, Kretzschmar HA, Haass C, Edbauer D, 2013. Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol. 126, 881–893. [DOI] [PubMed] [Google Scholar]
- Palluzzi F, Ferrari R, Graziano F, Novelli V, Rossi G, Galimberti D, Rainero I, Benussi L, Nacmias B, Bruni AC, Cusi D, Salvi E, Borroni B, Grassi M, 2017. A novel network analysis approach reveals DNA damage, oxidative stress and calcium/cAMP homeostasis-associated biomarkers in frontotemporal dementia. PLoS One 12, e0185797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picher-Martel V, Dutta K, Phaneuf D, Sobue G, Julien JP, 2015. Ubiquilin-2 drives NF-kappaB activity and cytosolic TDP-43 aggregation in neuronal cells. Mol. Brain 8, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellier C, Buijsen RAM, He F, Natla S, Jung L, Tropel P, Gaucherot A, Jacobs H, Meziane H, Vincent A, Champy MF, Sorg T, Pavlovic G, Wattenhofer-Donze M, Birling MC, Oulad-Abdelghani M, Eberling P, Ruffenach F, Joint M, Anheim M, Martinez-Cerdeno V, Tassone F, Willemsen R, Hukema RK, Viville S, Martinat C, Todd PK, Charlet-Berguerand N, 2017. Translation of expanded CGG repeats into FMRpolyG is pathogenic and may contribute to fragile X tremor ataxia syndrome. Neuron 93, 331–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendoel A, Dunn JG, Rodriguez EH, Naik S, Gomez NC, Hurwitz B, Levorse J, Dill BD, Schramek D, Molina H, Weissman JS, Fuchs E, 2017. Translation from unconventional 5′ start sites drives tumour initiation. Nature 541, 494–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahheydari H, Shahheydari H, Ragagnin A, Walker AK, Toth RP, Vidal M, Jagaraj CJ, Perri ER, Konopka A, Sultana JM, Atkin JD, 2017. Protein quality control and the amyotrophic lateral sclerosis/frontotemporal dementia continuum. Front. Mol. Neurosci 10, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck SR, Jiang V, Pavon-Eternod M, Prasad S, McCarthy B, Pan T, Shastri N, 2012. Leucine-tRNA initiates at CUG start codons for protein synthesis and presentation by MHC class I. Science 336, 1719–1723. [DOI] [PubMed] [Google Scholar]
- Starck SR, Tsai JC, Chen K, Shodiya M, Wang L, Yahiro K, Martins-Green M, Shastri N, Walter P, 2016. Translation from the 5′ untranslated region shapes the integrated stress response. Science 351, aad3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup V, Phaneuf D, Dupré N, Petri S, Strong M, Kriz J, Julien JP, 2011. Deregulation of TDP-43 in amyotrophic lateral sclerosis triggers nuclear factor kappaB-mediated pathogenic pathways. J. Exp. Med 208, 2429–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabet R, Schaeffer L, Freyermuth F, Jambeau M, Workman M, Lee CZ, Lin CC, Jiang J, Jansen-West K, Abou-Hamdan H, Désaubry L, Gendron T, Petrucelli L, Martin F, Lagier-Tourenne C, 2018. CUG initiation and frameshifting enable production of dipeptide repeat proteins from ALS/FTD C9ORF72 transcripts. Nat. Commun 9, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JP, Brown RH Jr., Cleveland DW, 2016. Decoding ALS: from genes to mechanism. Nature 539, 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Popko B, Roos RP, 2011. The unfolded response in familial amyotrophic lateral sclerosis. Hum. Mol. Genet 20, 1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Popko B, Roos RP, 2014. An enhanced integrated stress response ameliorates mutant SOD1-induced ALS. Hum. Mol. Genet 23, 2629–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingarten-Gabbay S, Elias-Kirma S, Nir R, Gritsenko AA, Stern-Ginossar N, Yakhini Z, Weinberger A, Segal E, 2016. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science 351, aad4939. [DOI] [PubMed] [Google Scholar]
- Yang D, Abdallah A, Li Z, Lu Y, Almeida S, Gao FB, 2015. FTD/ALS-associated poly(GR) protein impairs the Notch pathway and is recruited by poly(GA) into cytoplasmic inclusions. Acta Neuropathol. 130, 525–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SK, Wek RC, 2016. Upstream open reading frames differentially regulate genespecific translation in the integrated stress response. J. Biol. Chem 291, 16927–16935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Jansen-West K, Xu YF, Gendron TF, Bieniek KF, Lin WL, Sasaguri H, Caulfield T, Hubbard J, Daughrity L, Chew J, Belzil VV, Prudencio M, Stankowski JN, Castanedes-Casey M, Whitelaw E, Ash PEA, DeTure M, Rademakers R, Boylan KB, Dickson DW, Petrucelli L, 2014. Aggregation-prone c9FTD/ALS poly(GA) RAN-translated proteins cause neurotoxicity by inducing ER stress. Acta Neuropathol. 128, 505–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Liu Y, Bañez-Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook AE, Subramony SH, Ostrow LW, Rothstein JD, Troncoso JC, Ranum LP, 2013. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc. Natl. Acad. Sci. U. S. A 110, E4968–E4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.