Abstract
The International Genomics of Alzheimer’s Project (IGAP) is a consortium for characterizing the genetic landscape of Alzheimer’s disease (AD). The identified and/or confirmed 19 single nucleotide polymorphisms (SNPs) are located on non-coding DNA regions, and their functional impacts on AD are as yet poorly understood. We evaluated the roles of the IGAP SNPs by integrating data from many resources, based on whether the IGAP SNP was (A) a proxy for a coding SNP or (B) associated with altered mRNA transcript levels. For (A), we confirmed that 12 AD-associated coding common SNPs and five nonsynonymous rare variants are in linkage disequilibrium with the IGAP SNPs. For (B), the IGAP SNPs in CELF1 and MA4A6A were associated with expression of their neighboring genes, MYBPC3 and MA4A6A respectively, in blood. The IGAP SNP in DSG2 was an expression quantitative trait loci (eQTL) for DLGAP1 and NETO1 in human frontal cortex. The IGAP SNPs in ABCA7, CD2AP, and CD33 each acted as eQTL for AD-associated genes in brain. Our approach for identifying proxies and examining eQTL highlighted potentially impactful, novel gene regulatory phenomena pertinent to the AD phenotype.
Keywords: WES, ADSP, ADGC, GWAS, neuroinflammation
1. Introduction
Dementia is a clinical state characterized by a loss of function in memory and behavior, and the clinical state is associated with underlying degeneration of central nervous system synapses and cells. Alzheimer’s disease (AD) is the most common form of dementia, accounting for over 50% of dementia cases (Querfurth and LaFerla, 2010). Although it has been more than 100 years since Alois Alzheimer published “About a Peculiar Disease of the Cerebral Cortex” (Alzheimer, 1907), the exact cause of AD has not yet been defined. Amyloid β (Aβ) protein and hyperphosphorylated tau aggregates in brain are considered the key pathological hallmarks (Reitz, et al., 2011,Selkoe, et al., 2004). A widely held mechanistic hypothesis for AD pathogenesis is the “amyloid cascade hypothesis” wherein a key early pathogenetic role is played by parenchymal Aβ peptide accumulation, which causes or exacerbates downstream neuronal injury, enhanced neuroinflammation, tau hyperphosphorylation, and eventually the clinical symptoms of AD (Hardy and Selkoe, 2002).
Familial AD, which often occurs early in life, is linked to mutations in three genes: the amyloid precursor protein (APP) gene and the presenilin protein (PSEN1 and PSEN2) genes (van Es and van den Berg, 2009). These genes are associated with altered processing of the APP protein, including a shift in Aβ peptide production from Aβ40 to more neurotoxic Aβ42 (e.g., Volga German mutation in PSEN2 and Iberian mutation in APP) (Jayadev, et al., 2010,Levy-Lahad, et al., 1995,Lichtenthaler, et al., 1999,Walker, et al., 2005), increased total Aβ levels (Swedish mutation in APP) (Mullan, et al., 1992), and increased Aβ protofibril formation (Arctic mutation in APP) (Nilsberth, et al., 2001). In contrast, late-onset AD, which accounts for > 95% of all AD cases (Mancuso, et al., 2008), has a more complex genetic architecture. The ε4 allele of apolipoprotein E (APOE) gene is the most well-established susceptibility risk factor for late-onset AD.
A series of genome-wide association studies (GWAS) have identified AD-associated single nucleotide polymorphisms (SNPs) in addition to the APOE alleles (Harold, et al., 2009,Hollingworth, et al., 2011,Lambert, et al., 2009,Lambert, et al., 2013,Naj, et al., 2011,Seshadri, et al., 2010). The study with the largest number of AD and non-AD individuals was the International Genomics of Alzheimer’s Project (IGAP), which capitalized on a large, multicenter study design to include 74,046 individuals (Lambert, et al., 2013). This study extended associations between the AD phenotype and genetics, finding 21 SNPs as significant by meta-analyzing genetic and phenotype data from four component consortia (Lambert, et al., 2013). These SNPs are in or close to CR1, BIN1, INPP5D, MEF2C, CD2AP, NME8, EPHA1, PTK2B, PICALM, SORL1, FERMT2, SLC24A4-RIN3, DSG2, CASS4, HLA-DRB5-DBR1, CLU, MS4A6A, ABCA7, CD33, ZCWPW1, and CELF1 (Supplementary Table 1). Although GWAS have succeeded in revealing numerous susceptibility variants for AD, determining the functional impact of those gene variants and understanding how they contribute to AD pathogenesis represents a barrier to progress in the field.
Genetic variants located in coding regions constitute only ~1% of gene polymorphisms seen in humans (Rabbani, et al., 2014). However, there are many ways that genetic variants in non-coding regions can affect protein expression and structure, and thereby exert a protective or disease-inducing impact. Functional variants may be located in a coding region, an alternative splicing region, or a regulatory region such as promoter, operator, insulator, enhancer or silencer.
Nonsynonymous variants, by definition, alter the primary amino acid sequence of a protein and may have effects on the protein structure and function. Synonymous mutations occur in the coding region, but do not change the amino acid sequence. These variants were referred to as “silent mutations” until recently (Sauna and Kimchi-Sarfaty, 2011). Several synonymous mutations have been reported to affect mRNA splicing and stability, gene expression, and protein folding and function (Sauna and Kimchi-Sarfaty, 2011). Most of the non-APOE AD-associated genetic variants described to date are located in intronic or intergenic regions (i.e., non-coding regions), which may contain regulatory elements. Intronic and intergenic SNPs may act by regulating expression of disease-associated genes and/or modulating translation efficiency and stability (Mockenhaupt and Makeyev, 2015).
In the present study, we analyzed data from multiple sources to gain insights into the roles of non-coding SNPs identified in the IGAP study (including CD33 and DSG2, although their associated SNPs, rs3865444 and rs8093731, respectively, did not reach statistical significance in the combined stages of that study (Lambert, et al., 2013)), hereafter referred to as “IGAP SNPs”. We hypothesized that each IGAP SNP is potentially: (1) a proxy for an exonic (coding) variant (Supplementary Figure 1A and 1B) that has not yet been identified; or (2) associated with altered transcript/mRNA levels (Supplementary Figure 1C). One approach to test the first hypothesis is to identify coding variants in strong linkage disequilibrium (LD) with the variant identified by GWAS, which indicates that the two gene loci are commonly co-inherited. For the second hypothesis, expression quantitative trait loci (eQTL) analyses can be used to assess the association between the gene variant and mRNA levels of various transcripts. Thus, eQTL are genetic loci that contribute to variation in gene expression. By mapping eQTL, we investigated how the variants regulate gene expression. Using these combined methods, and multiple data sources, we discovered new evidence of complex gene expression regulation mechanisms in association with previously identified IGAP SNPs.
2. Material and methods
2.1. Genetic datasets
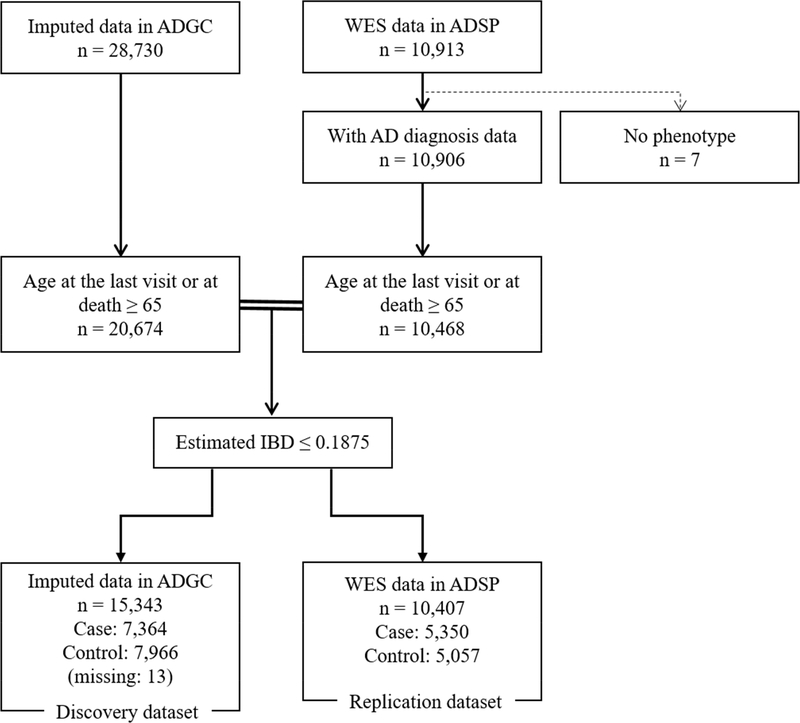
Genetic data were obtained from multiple sources. Whole exome sequence (WES) data came from the Alzheimer’s Disease Sequencing Project (ADSP), composed of 18 cohorts from the Alzheimer’s Disease Genetic Consortium (ADGC) and six cohorts from the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium (Beecham, et al., 2017). We also used imputed SNP data (Supplementary Method) from the ADGC comprising 23 different cohorts (Supplementary Table 2). From these sources, there were 28,730 unrelated subjects with imputed GWAS SNP data in ADGC and 10,913 unrelated subjects with WES data in ADSP. We estimated identity-by-descent (IBD) to identify any relatedness and duplicate individuals in the two datasets. Individuals were excluded with estimated IBD ≥ 0.1875 from ADGC datasets, and two independent datasets were created: an imputed ADGC dataset that excluded related individuals and those that potentially overlapped with those in ADSP (hereafter referred to as “ADGC”), and WES data in ADSP (hereafter referred to as “ADSP”) (Figure 1). We limited the included subjects to those who had AD diagnosis information and who were 65 years or older at the last visit or at death, yielding a total of 15,343 ADGC subjects with imputed SNP data in the discovery analysis and a total of 10,407 ADSP subjects with WES data in the replication analysis.
Figure 1.
Flow diagram of the subjects included in the analyses.
Key: ADGC, Alzheimer’s Disease Genetics Consortium; ADSP, Alzheimer’s Disease Sequencing Project; IBD, identity-by-descent
2.2. Gene expression datasets
Data were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). The ADNI was launched in 2003 as a public-private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer’s disease (AD). Data were included from 763 subjects aged 65 years or older who had both gene expression data from blood (Affymetrix Human Genome U219 Array platform) and whole genome sequencing (WGS) data available. Clinical status was determined based on the clinical evaluation at the last examination.
Human brain gene expression and genotype data were obtained from the North American Brain Expression Consortium (NABEC) (Hernandez, et al., 2012) and United Kingdom Brain Expression Consortium (UKBEC) (Trabzuni, et al., 2011). Details were as described in our previous report (Katsumata, et al., 2017). Briefly, the NABEC expression data for FCTX were available at Gene Expression Omnibus (GEO: https://www.ncbi.nlm.nih.gov/geo/) public repository and the genotype data were obtained from the database of Genotypes and Phenotypes (dbGaP: http://www.ncbi.nlm.nih.gov/gap). Standard quality control (QC) procedures were performed on the NABEC genotype data using PLINK v1.90a (Purcell, et al., 2007). Markers were excluded based on the following criteria: (1) minor allele frequency (MAF) < 1%; (2) call rate per variant (SNPs and indels) < 95%, (3) Hardy-Weinberg equilibrium test in controls < 10−5. Samples were excluded based on the following criteria: (1) call rate per individual < 95%, (2) a high degree of relatedness per an estimated proportion of IBD > 0.1875, (3) excess of ± 3.0 standard deviations of heterozygosity rate. After performing QC, we imputed using the Michigan Imputation Server (https://imputationserver.sph.umich.edu/start.html) (Das, et al., 2016,Loh, et al., 2016) with the following parameters: 1000 Genome Phase 3 v5 reference panel, Eagle v2.3 phasing (Loh, et al., 2016), and EUR population. Of the 455 neurologically normal donors, 85 subjects who died at age 65 years or older and passed QC were included in the analysis (all were US Caucasians). The UKBEC gene expression for three brain regions (frontal cortex, FCTX; hippocampus, HIPP; and temporal cortex, TCTX) and genotype data were obtained from the BRAINEAC website (http://www.braineac.org/). Dosage genotype data were converted into PLINK file format using Genome-wide Complex Trait Analysis (GCTA) software version 1.24.4 (Yang, et al., 2011). Among the 134 neuropathologically normal individuals, 49 subjects who died at age 65 years or older were included in the present analyses.
Since the NABEC and UKBEC datasets do not have AD diagnosis information, we retrieved microarray datasets generated from Affymetrix Human Genome U133 Plus 2.0 Array platform (GPL570) regarding AD status from GEO to examine whether the levels of gene expression were different in association with AD status versus controls. We focused on gene expression in four brain regions affected by AD (entorhinal cortex (EC), FCTX, HIPP, and TCTX). We obtained two datasets for EC (GSE48350 (Berchtold, et al., 2008) and GSE5281 (Liang, et al., 2007)), four datasets for FCTX (GSE48350 (Berchtold, et al., 2008), GSE5281 (Liang, et al., 2007), GSE66333 (Simpson, et al., 2016), and GSE53890 (Lu, et al., 2014)), three datasets for HIPP (GSE48350 (Berchtold, et al., 2008), GSE5281 (Liang, et al., 2007), GSE28146 (Blalock, et al., 2011)), and two datasets for TCTX (GSE5281 (Liang, et al., 2007) and GSE29652 (Simpson, et al., 2011)). Included were 25 AD cases and 29 controls for EC, 52 cases and 56 controls for FCTX, 50 cases and 44 controls for HIPP, and 34 cases and 11 controls for TCTX, who were 65 years or older at death (Supplementary Table 3). The raw expression data downloaded from GEO (Affymetrix CEL files) were background-corrected and normalized by the RmaBackgroundCorrection and QuantileNormalization functions in “aroma.affymetrix” Bioconductor R package (Bengtsson, et al., 2008), and then log2-transformed. The normalized and log2-transformed expression data in each brain region were merged by the Combat function in “sva” Bioconductor R package (Leek, et al., 2012). Using principal component analysis (PCA), we confirmed that Combat successfully eliminated batch effects in each brain region. We removed one outlier identified in the PCA from GSE5281 in FCTX (Supplementary Figure 2).
Probes were excluded that targeted transcripts from different genes (i.e., probes with “_x” suffix) if a more reliable probe was available. We also excluded mono-allelically expressed genes including genes on chromosomes X and Y, and HLA- genes (i.e., HLA-A, HLA-B, HLA-C, HLA-DMA, HLA-DMB, HLA-DOA, HLA-DOB, HLA-DPA1, HLA-DPB1, HLA-DQA1, HLA-DQA2, HLA-DQB1, HLA-DQB2, HLA-DRA, HLA-DRB1, HLA-DRB3, HLA-DRB4, HLA-DRB5, HLAE, HLA-F, HLA-G, HLA-J, HLA-P, and HLA-T). The number of probes in each study is shown in Supplementary Table 4.
2.3. Statistical analysis
2.3.1. Hypothesis 1: identified IGAP SNPs are proxies for exonic/coding variants
We applied two separate methods to identify potentially co-inherited SNPs with the IGAP SNPs: one was for common SNPs and one for rare variants. We first identified common SNPs in the nearby coding regions showing strong (r2 ≥ 0.8) or moderate (0.4 ≤ r2 < 0.8) LD with each of the IGAP SNPs by using 1000 Genomes Project Phase 3 in individuals of European ancestry (1000 Genomes EUR) (1000 Genomes Project Consortium, 2010). In the discovery analysis for common SNPs in ADGC, we performed association tests under an additive mode of inheritance (MOI) using logistic regression adjusted for age at last visit or death, sex, and the top five principal components (PCs) computed in PLINK v1.90a (Chang, et al., 2015,Purcell, et al., 2007). The coding SNPs were evaluated for replication in independent individuals (within the ADSP dataset) to limit the possibility of imputation errors. We then identified potentially co-inherited rare variants with each of the IGAP SNPs using the Lewontin’s D’ estimates (Lewontin, 1964) in 1000 Genomes EUR (1000 Genomes Project Consortium, 2010). Due to the properties of LD metrics, a given common SNP can exhibit disparate patterns of LD (large D’ but low r2) between it and many rare variants. We thus focused on nonsynonymous rare variants which are more likely functional, and then applied the following criteria: MAF < 0.05, minor allele count ≥ 5, D’ ≥ 0.9, the same direction of effect on AD, and within 1 Mb from the IGAP SNP. Fisher’s exact test was used to examine the association between rare variants and AD in ADSP.
Variant Effect Predictor (VEP) (McLaren, et al., 2010) was used to annotate functional consequences of the common coding SNPs and rare variants identified in the association tests. The pathogenetic nature of nonsynonymous common SNPs/rare variants associated with AD was predicted by SIFT (http://sift.jcvi.org/) (Ng and Henikoff, 2003), PolyPhen-2 with HumDiv classifier (http://genetics.bwh.harvard.edu/pph2/) (Adzhubei, et al., 2010), and PROVEAN (http://provean.jcvi.org/index.php) (Choi, et al., 2012) to evaluate the effect of amino acid substitution on a protein function. We also used Genomic Evolutionary Rate Profiling (GERP)++ (http://mendel.stanford.edu/SidowLab/downloads/gerp/) (Davydov, et al., 2010) to examine evolutionary conservation for each of the associated nonsynonymous SNPs/rare variants. Higher score of rejected substitutions (RS) score indicates that a site is inferred to have a greater level of evolutionary constraint. We implemented these in silico algorithm tools except for PROVEAN for canonical transcripts that are defined as either the longest coding sequence or the longest cDNA in the UCSC Genome Browser (https://genome.ucsc.edu/) (Kent, et al., 2002).
2.3.2. Hypothesis 2: identified IGAP SNPs are eQTLs
The goal of these analyses was to evaluate whether the IGAP SNPs were eQTL. We first tested association between the IGAP SNPs and gene expression on the same chromosome as each of the SNPs, assuming an additive MOI as implemented in PLINK v1.90a (Chang, et al., 2015,Purcell, et al., 2007). We then examined whether the levels of gene expression associated with the IGAP SNP status were different from the association with AD phenotype. An analysis of covariance (ANCOVA) with age at the death and sex as covariates was applied to test for statistical significance.
For all analyses, we defined associations with false discovery rate (FDR) adjusted p-value < 0.05 as statistically significant using the Benjamini-Hochberg procedure (Benjamini and Hochberg, 1995).
3. Results
For the current study, individuals with either prevalent or incident AD were considered as AD cases in ADSP. Descriptive characteristics of individuals in the two genetic datasets are shown in Supplementary Table 5. In ADGC and ADSP, 7,364 (48.0%) and 5,374 (51.4%) were AD cases, respectively.
3.1. Hypothesis 1: identified IGAP SNPs are proxies of coding variants
In the common SNP analyses, 10 exonic SNPs were in strong LD (r2 ≥ 0.8) and 16 exonic SNPs in moderate LD (0.4 ≤ r2 < 0.8) with IGAP SNPs based on 1000 Genomes EUR (Supplementary Table 6). We first analyzed the imputed genotype data from ADGC to replicate IGAP SNP association with AD and extended the analyses to exonic common SNPs. We confirmed that several proxy SNPs located in coding regions demonstrated statistically significant associations with AD phenotype. Of these coding SNPs, we replicated 12 loci (rs2296160 in CR1, rs1049086 in HLA-DQB1, rs2722372 and rs2598044 in NME8, rs2405442 and rs1859788 in PILRA, rs7982 in CLU, rs12453 and rs7232 in MS4A6A, and rs3752246, rs4147930, and rs4147934 in ABCA7) that surpassed the statistical significance level with FDR adjustment in the separate ADSP dataset (Table 1 and Supplementary Table 7). Of these 12 coding SNPs, six SNPs (rs2296160 in CR1, rs2722372 in NME8, rs1859788 in PILRA, rs7232 in MS4A6A, and rs3752246 and rs4147934 in ABCA7) are missense mutations on at least one of their transcripts (Supplementary Table 8).
Table 1.
Association of IGAP SNPs and the coding SNPs strongly correlated with the IGAP SNPs with Alzheimer’s disease in two datasets: ADGC a and ADSP b
| IGAP SNP |
Exonic SNP |
|||||||
|---|---|---|---|---|---|---|---|---|
| SNP | Closest gene | ADGC a |
SNP ID | ADGC a |
ADSP b |
|||
| OR | P-value | OR | P-value | OR | P-value | |||
| Strong LD (r2 ≥ 0.8) | ||||||||
| rs6656401 | CR1 | 1.17 | 8.49 ×10−7 | rs4844600 | 1.17 | 4.22×10−7 | - | - |
| rs2296160 | 1.18 | 4.24×10−8 | 1.11 | 8.56×10−3 | ||||
| rs9271192 | HLA-DRB5 | 1.11 | 2.66×10−4 | rs9270303 | - | - | 1.15 | 2.50×10−4 |
| rs1476679 | ZCWPW1 | 0.92 | 1.41×10−3 | rs2405442 | 0.92 | 2.12×10−3 | 0.89 | 1.09×10−3 |
| rs1859788 | 0.93 | 5.62×10−3 | 0.89 | 1.12×10−3 | ||||
| rs9331896 | CLU | 0.92 | 1.02×10−3 | rs7982 | 0.91 | 3.64×10−4 | 0.90 | 1.62×10−3 |
| rs10838725 | CELF1 | 1.05 | 0.054 | rs2293576 | 1.03 | 0.24 | 1.07 | 0.040 |
| rs983392 | MS4A6A | 0.86 | 7.22×10−9 | rs12453 | 0.86 | 1.34×10−9 | 0.89 | 3.60×10−4 |
| rs4147929 | ABCA7 | 1.12 | 1.39×10−3 | rs3752246 | 1.11 | 2.70×10−3 | 1.18 | 9.89×10−5 |
| rs3865444 | CD33 | 0.91 | 4.49 ×10−4 | rs12459419 | 0.91 | 4.02×10−4 | 0.94 | 0.090 |
Imputed genotype data from ADGC
Whole exome sequencing data from ADSP
Bold p-value represents the statistical significance after false discovery rate adjustment.
Key: IGAP, International Genomics of Alzheimer’s Project; SNP, single nucleotide polymorphism; ADSP, Alzheimer’s Disease Sequencing Project; ADGC, Alzheimer’s Disease Genetic Consortium; OR, odds ratio; LD, linkage disequilibrium
In rare variant analyses using ADSP, five rare variants were identified that (i) had D’ = 1 and the same direction of effect as an IGAP SNP (rs11575848 in LY6G6C, rs2070600 in AGER, rs62483572 in EPO, rs74547795 in SYTL2, and rs111986709 in DSG3) (Supplementary Table 9), and (ii) were missense mutations on at least one of their transcripts (Supplementary Table 10).
The nonsynonymous SNPs and rare variants in the canonical transcript were analyzed in silico with SIFT, PolyPhen-2, PROVEAN, and GERP++. Supplementary Table 11 shows the pathogenetic nature prediction only for the canonical transcripts. None of the common nonsynonymous SNPs except for rs7232 in MS4A6A were predicted to have functional impact; the minor allele of rs7232 was predicted to be possibly damaging to the MS4A6A protein according to PolyPhen-2. In contrast, all of the rare variants were predicted to have deleterious effects on protein function.
3.2. Hypothesis 2: identified IGAP SNPs are eQTLs
Table 2 shows transcript levels (gene expression) in blood that were significantly associated with the IGAP SNPs, each reaching FDR adjusted significance level. The risk allele of rs10838725 in CELF1 was associated with increased MYBPC3 expression, and the protective allele of rs983392 in MA4A6A was associated with decreased expression of MS4A6A itself. MYBPC3 expression (probe ID: 11725151_at) and MS4A6A expression (probe ID: 11716846_a_at) were also significantly associated with AD status.
Table 2.
Significant association of the IGAP SNPs with gene expression in blood in ADNI
| IGAP SNP | Closest gene | Probe set ID a | Gene | eQTL association |
AD association |
|
|---|---|---|---|---|---|---|
| P-valueb | P-value c | |||||
| rs6733839 | BIN1 | 11719631_s_at | BIN1 | 0.071 | 1.51 × 10−7 | 0.28 |
| 11746895_a_at | BIN1 | 0.084 | 2.05×10−6 | 0.56 | ||
| rs111418223 | HLA-DRB5 | 11730933_a_at | AGPAT1 | 0.120 | 1.83×10−11 | 0.91 |
| 11750187_a_at | AGPAT1 | 0.114 | 6.96×10−9 | 0.68 | ||
| 11751668_a_at | AGPAT1 | 0.119 | 1.71×10−8 | 0.94 | ||
| rs1476679 | ZCWPW1 | 11722909_a_at | GATS | 0.177 | 1.53×10−17 | 0.53 |
| 11736388_a_at | TRIM4 | −0.129 | 3.53×10−9 | 0.95 | ||
| 11743311_a_at | PILRB | −0.115 | 4.15×10−9 | 0.85 | ||
| 11730023 s at | PILRB | −0.107 | 1.58×10−8 | 0.78 | ||
| 11730022_a_at | PILRB | −0.128 | 3.77×10−8 | 0.89 | ||
| 11760665_at | ZKSCAN1 | 0.147 | 2.30×10−7 | 0.15 | ||
| 11730247_a_at | PVRIG | 0.086 | 6.07×10−5 | 0.82 | ||
| rs11771145 | EPHA1 | 11755327 s at | LOC154761 | 0.109 | 3.68×10−6 | 0.40 |
| rs28834970 | PTK2B | 11720981_a_at | PTK2B | 0.114 | 6.86×10−18 | 0.40 |
| 11720982 s at | PTK2B | 0.086 | 1.18×10−17 | 0.91 | ||
| 11720980_a_at | PTK2B | 0.094 | 7.33×10−12 | 0.47 | ||
| 11723344_at | TRIM35 | −0.070 | 2.31×10−6 | 0.45 | ||
| rs10838725 | CELF1 | 11725151_at | MYBPC3 | 0.140 | 1.07×10−7 | 8.13×10−5 |
| rs983392 | MS4A6A | 11716846_a_at | MS4A6A | −0.082 | 2.34×10−12 | 4.97×10−3 |
| 11751570_a_at | MS4A4A | −0.150 | 1.15×10−6 | 0.85 | ||
| 11732865_a_at | MS4A4A | −0.179 | 1.59×10−6 | 0.68 | ||
Probe set IDs on Affymetrix Human Genome U219 Array
P-values less than significance level after false discovery rate adjustment were displayed
P-values calculated by analysis of covariance with the outcome of gene expression and the predictor of Alzheimer’s disease status (normal/mild cognitive impairment/AD)
Key: IGAP, International Genomics of Alzheimer’s Project; SNP, single nucleotide polymorphism; ADNI, Alzheimer’s Disease Neuroimaging Initiative; eQTL, expression quantitative trait locus
The significant associations between the IGAP SNPs and brain gene expression in NABEC and UKBEC are shown in Table 3. In FCTX data of NABEC, rs8093731 in DSG2 acted as an eQTL for two genes, DLGAP1 and NETO1, which were highly correlated (r2 = 0.69). In UKBEC, the risk allele of rs4147929 in ABCA7, the risk allele of rs10948363 in CD2AP, and the protective allele of rs3865444 in CD33 were associated with increased EID2B expression in FCTX, increased AK9 expression in FCTX, and decreased IER2 expression in TCTX, respectively.
Table 3.
Significant association of the IGAP SNPs with brain gene expression in NABEC and UKBEC
| IGAP SNP | Closest gene |
Probe set ID a | Gene expression |
Brain region |
P-value b | |
|---|---|---|---|---|---|---|
| NABEC | ||||||
| rs8093731 | DSG2 | ILMN_23 80779 | DLGAP1 | FCTX | 0.770 | 1.36×10−8 |
| ILMN_1783168 | NETO1 | FCTX | 0.706 | 1.11×10−5 | ||
| UKBEC | ||||||
| rs4147929 | ABCA7 | t3862068 | EID2B | FCTX | 0.259 | 3.94×10−6 |
| rs10948363 | CD2AP | t2969159 | AK9 | FCTX | 0.343 | 1.64×10−5 |
| rs3865444 | CD33 | t3822216 | IER2 | TCTX | -0.255 | 1.96×10−5 |
Probe set IDs on HumanHT-12_v3 Expression BeadChips in NABEC (platform = GPL6947) and on Affymetrix Exon 1.0 ST Arrays in UKBEC (platform = GPL5175)
P-values less than significance level after false discovery rate adjustment are displayed.
Key: IGAP, International Genomics of Alzheimer’s Project; SNP, single nucleotide polymorphism; NABEC, North American Brain Expression Consortium; UKBEC, United Kingdom Brain Expression Consortium; FCTX, frontal cortex; TCTX, temporal cortex
In comparison with non-AD samples using the merged datasets, AD cases had significantly lower expressions of DLGAP1 and NETO1 (potentially regulated by the DSG2 SNP) in the four brain regions and of EID2B (potentially regulated by the ABCA7 SNP) in HIPP. Significantly higher expression of AK9 (potentially regulated by the CD2AP SNP) and IER2 (potentially regulated by the CD33 SNP) in AD cases was seen in EC/HIPP and EC/TCTX, respectively (Table 4).
Table 4.
Associations between gene expressions identified in NABEC and UKBEC and Alzheimer’s disease status in the merged dataset
| Probe set ID a | EC |
FCTX |
HIPP |
TCTX |
||||
|---|---|---|---|---|---|---|---|---|
| P-value | P-value | P-value | P-value | |||||
| Identified in NABEC | ||||||||
| DLGAP1 | ||||||||
| 206489_s_at | −0.495 | 5.68× 10-3 | −0.294 | 0.013 | −0.635 | 2.77×10−6 | −0.318 | 1.58 × 10−4 |
| 206490_at | −0.225 | 0.071 | −0.202 | 0.039 | −0.280 | 3.18×10−4 | −0.212 | 0.052 |
| 210750 s at | −0.343 | 0.013 | −0.135 | 0.13 | −0.084 | 0.27 | 0.214 | 0.071 |
| NETO1 | ||||||||
| 1552736_a_at | −0.433 | 5.36×10−3 | −0.210 | 0.14 | −0.597 | 1.53×10−5 | −0.088 | 0.58 |
| 1552904 at | −0.411 | 2.70×10−4 | −0.115 | 0.085 | −0.359 | 5.23×10−6 | −0.119 | 0.085 |
| 1562713_a_at | −0.548 | 2.95×10−3 | −0.255 | 8.99×10−3 | −0.615 | 1.97×10−5 | −0.276 | 4.73×10−4 |
| Identified in UKBEC | ||||||||
| EID2B | ||||||||
| 242470_at | −0.383 | 0.051 | −0.319 | 0.015 | −0.424 | 6.70×10−4 | −0.166 | 0.33 |
| AK9 | ||||||||
| 1552299_at | 0.039 | 0.54 | 0.014 | 0.82 | 0.001 | 0.99 | −0.179 | 0.048 |
| 1564002_a_at | 0.197 | 5.23×10−3 | 0.092 | 0.084 | 0.287 | 6.38×10−3 | 0.083 | 0.41 |
| IER2 | ||||||||
| 202081 at | 0.292 | 7.11×10−4 | 0.141 | 0.060 | 0.061 | 0.52 | 0.949 | 4.48×10−5 |
Probe set IDs on Affymetrix U133 Plus 2.0 array (platform = GPL570)
Bold p-value represents the statistical significance after FDR adjustment.
Key: NABEC, North American Brain Expression Consortium; UKBEC, United Kingdom Brain Expression Consortium; EC, entorhinal cortex; HIPP, hippocampus; FCTX, frontal cortex; TCTX, temporal cortex
4. Discussion
Although large GWAS have identified novel loci that are associated with altered AD risk, we have a relatively poor understanding of the functional impact of these loci. In this study, we examined possible functional effects of the IGAP SNPs on AD under two hypotheses: “the IGAP SNP is a proxy of a coding variant” and “the IGAP SNP is an eQTL”. For the first hypothesis, rs6656401 in CR1, rs9271192 in HLA-DRB5-DRB1, rs2718058 in NME8, rs1476679 in ZCWPW1, rs9331896 in CLU, rs983392 in MS4A6A, and rs4147929 in ABCA7 are proxies of common coding SNPs. Additionally, the IGAP SNPs rs9271192 in HLA-DRB5-DRB1, rs1476679 in ZCWPW1, rs10792832 in PICALM, and rs8093731 in DSG2 may reflect the net effect of nonsynonymous rare variants. For the second hypothesis, rs8093731 in DSG2, rs4147929 in ABCA7, rs10948363 in CD2AP, and rs3865444 in CD33 are associated with gene expression, although whether these SNPs are proxies for the functional regulatory SNP or functional themselves requires further studies.
4.1. CR1 SNPs
There were two common coding SNPs, rs4844600 (synonymous) and rs2296160 (nonsynonymous), in strong LD with the IGAP SNP, although the association between rs4844600 and AD could not be assessed for replication because of the lack of WES data in ADSP. CR1, located on chromosome 1q32.2 within a cluster of complement-related genes, encodes complement receptor 1 which typically acts to bind complement-labeled proteins or complexes for their clearance by the immune system (Khera and Das, 2009). Regarding AD, the CR1 protein acts a receptor for complement fragments bound to Aβ, and thus the change in CR1 protein structure and expression levels may be related to Aβ clearance (Rogers, et al., 2006). The synonymous SNP rs4844600 (E60E) is located on exon 2, the IGAP SNP rs6656401 is located between exon 4 and 5, and the nonsynonymous SNP rs2296160, which causes an alanine-tothreonine amino acid substitution at codon position 2419 (A2419T), is located on exon 44. The SNP rs1408077 in CR1, which is in strong LD with the IGAP SNP rs6656401, was reported to be associated with loss of EC thickness (Biffi, et al., 2010), and carriers of the IGAP SNP rs6656401_A had smaller local gray matter volume in the EC in young health adults, which may lead to or reflect increased risk of late-onset AD (Bralten, et al., 2011). These results may indicate a causal relationship between CR1 SNPs and AD development.
4.2. HLA-DRB5-DRB1 SNPs
The IGAP SNP rs9271192 is located in an intergenic region (chromosome 6p21.32), near HLA class II genes (HLA-DR, -DQ and -DP). There were four common coding SNPs which were in strong or moderate LD with the IGAP SNP; one nonsynonymous SNP rs9270303 in HLA-DRB1, three synonymous SNPs rs2308759 in HLA-DRB1, rs1049092 and rs1049086 in HLA-DQB1. Because of missing data in ADGC, the association tests of rs9270303, rs2308759, and rs1049092 with AD could not be performed in the discovery analysis (Table 1 and Supplementary Table 7). We also identified two nonsynonymous rare variants, rs11575848 and rs2070600, that are potentially co-inherited with the IGAP SNP (D’ = 1) and are located in the major histocompatibility complex (MHC) class III region. LY6G6C (rs11575848) encodes a leukocyte antigen-6 superfamily member, and AGER (rs2070600) encodes a receptor for advanced glycosylation end product (RAGE). The RAGE protein is a member of the immunoglobulin superfamily and may regulate Aβ transport across the blood brain barrier (Tarasoff-Conway, et al., 2015).
At least three classes of genes expressed from a single allele (mono-allelic) are recognized to exist (Chess, 2012,Gimelbrant, et al., 2007). One class is the autosomal imprinted genes regulated in a parent-of-origin specific manner. The second class is X-inactivated. The third class of these genes are located randomly in autosomes and include several immune system genes (Chess, 2012,Gimelbrant, et al., 2007). Because we excluded from the analysis HLA-genes that are randomly mono-allelically expressed, we did not examine the associations between the IGAP SNP rs9271192 and expression of HLA-genes including HLA-DRB5 and HLA-DRB1. Given epigenetic association between DNA methylation in HLA-DRB5 and AD pathology (Yu, et al., 2015), allele specific expression at these loci may have a strong impact on immunobiological function related to AD.
4.3. CD2AP SNPs
Increased expression of AK9 in FCTX was associated with the risk allele of the CD2AP IGAP SNP, and AK9 was significantly over-expressed in EC and HIPP brain regions of AD cases. AK9, located in chromosome 6q21 and more than 60Mb away from CD2AP locus, encodes a member of adenylate kinase family of enzymes. Adenylate kinase reversibly catalyzes the interconversion of adenine nucleotides (ATP + AMP ↔ 2 ADP) (Amiri, et al., 2013).
4.4. NME8 SNPs
Two common coding SNPs were in moderate LD with the IGAP NME8 SNP; one nonsynonymous SNP rs2722372 and one synonymous SNP rs2598044. The nonsynonymous SNP rs2722372 causes arginine-to-lysine amino acid substitution at codon position 43 (R43K). These coding SNPs were significantly associated with AD and the associations were replicated. NME8, located on chromosome 7p14.1, encodes a protein with an N-terminal thioredoxin domain and C-terminal nucleoside diphosphate kinase domains. The NME8 protein is a member of NME/NM23 family. Although the function of this gene is poorly characterized, Liu et al. showed that the IGAP SNP rs2718058 had a neuroprotective effect against cognitive decline, elevated tau levels in cerebrospinal fluid (CSF), and hippocampal atrophy (Liu, et al., 2014).
4.5. ZCWPW1 SNPs
There were three common coding SNPs which were in strong or moderate LD with the IGAP ZCWPW1 SNP; one nonsynonymous SNP rs1859788 (in PILRA) and two synonymous SNP rs2405442 (in PILRA) and rs909152 (in LRCH4). The nonsynonymous SNP rs1859788 causes glycine-to-arginine amino acid substitution at codon position 78 (G78R). The coding SNPs which are in strong LD with the IGAP SNP were significantly associated with AD and we confirmed associations in the replication analysis. There was also one nonsynonymous rare variant rs62483572 (in EPO) with D’ = 1, that causes an amino acid substitution of aspartic acid to asparagine at codon position 70 (D70N). This variant had a more protective effect than the nonsynonymous common SNP rs18597955 (OR = 0.53 for rs62483572 vs. OR = 0.89 for rs1859788 in ADSP). Erythropoietin (EPO) exhibits a neuroprotective effect under various conditions of neuronal damage such as hypoxia-ischemia, and thus the nonsynonymous rare variants may be involved in promoting maintenance of homeostasis (Siren, et al., 2001).
The IGAP SNP was associated with the expression of several genes including GATS, TRIM4, PILRB, ZKSCAN1, and PVRIG in blood. However, we did not find significant associations between expression of these genes and AD. ZCWPW1, located on chromosome 7q22.1, encodes zinc finger CW (zf-CW)-type and PWWP domain containing 1. Although the function(s) of this gene are unknown, zf-CW may be involved in epigenetics as it is regarded as a member of histone modification reader modules (He, et al., 2010).
The IGAP SNP was associated with the expression of several genes including GATS, TRIM4, PILRB, ZKSCAN1, and PVRIG in blood. However, we did not find significant associations between expression of these genes and AD. ZCWPW1, located on chromosome 7q22.1, encodes zinc finger CW (zf-CW)-type and PWWP domain containing 1. Although the function(s) of this gene are unknown, zf-CW may be involved in epigenetics as it is regarded as a member of histone modification reader modules (He, et al., 2010).
4.6. CLU SNPs
We confirmed that the synonymous CLU SNP rs7982 is in strong LD with the IGAP SNP rs9331896 and was protectively associated with AD. However, we found no evidence of gene expression regulation that was associated with either the CLU IGAP SNP or the proxy, synonymous SNP. CLU is located in chromosome 8p21.1, and encodes clusterin, also known as apolipoprotein J. Clusterin directly influences Aβ, regulating the conversion of Aβ into insoluble forms (Desikan, et al., 2014,Yu and Tan, 2012). CLU has two main isoforms, nuclear CLU (nCLU, isoform 1) and secretory CLU (sCLU, isoform 2) with different functions. The sCLU form is pro-survival, while nCLU is pro-apoptotic (Shannan, et al., 2006). Since the coding SNP rs7982 is synonymous, it may affect alternative splicing as Ling et al. showed that the protective SNP rs11136000 (which is in almost perfect LD with rs7982 in 1000 genomes EUR) was associated with increased nCLU expression level (Ling, et al., 2012).
4.7. CELF1 SNPs
We did not find evidence that the CELF1 SNP rs10838725 and the proxy coding SNPs in LD with the IGAP SNP were associated with AD. However, rs10838725 acted as eQTL for MYBPC3 expression in blood which was associated with AD. MYBPC3 is located on chromosome 11p11.2, and encodes cardiac myosin binding protein C expressed in heart muscle (Gautel, et al., 1995). Huang et al. reported that the protective allele of rs1057233 in CELF1 (r2 = 0.17 and D’ = 0.97 with the IGAP SNP rs10838725 as shown in Supplementary Table 1) was associated with decreased MYBPC3 and SPI1 expressions and with the higher CSF Aβ42 levels (Huang, et al., 2017). Our finding that the risk allele of IGAP SNP rs10838725 in CELF1 was associated with the increased MYBPC3 expression and also correlated with AD status is concordant with the previous study.
4.8. MS4A6A SNPs
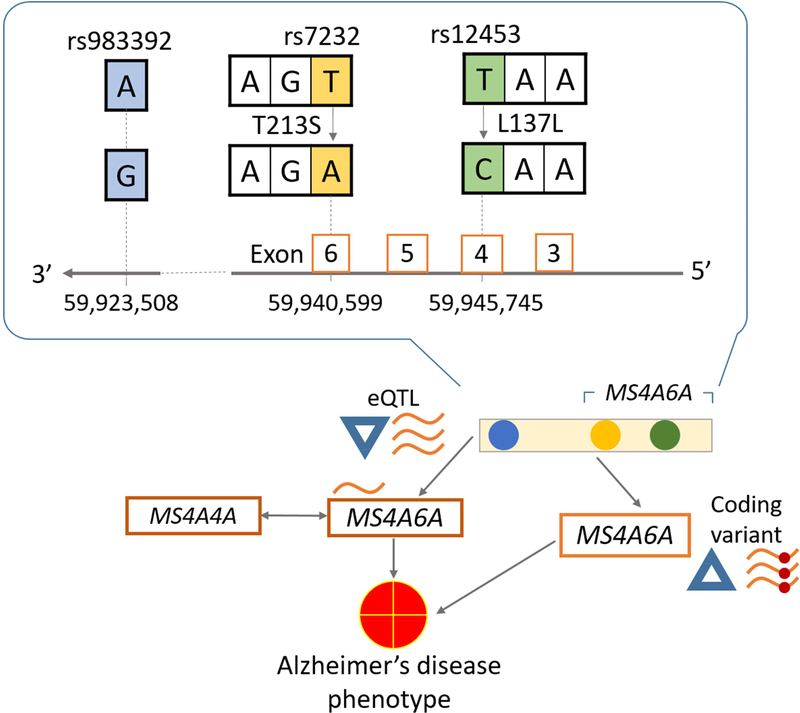
The IGAP SNP rs983392 located downstream of MS4A6A was associated with several striking gene regulatory features. We found two coding SNPs, rs12453 and rs7232 in LD with the protective IGAP SNP rs983392. The coding SNP rs7232 is nonsynonymous, causing threonineto-serine amino acid substitution at codon position 213 (T213S), while the SNP rs12453 is synonymous (L137L). The IGAP SNP was associated with expression in blood of MS4A6A as well as other members of the MS4A gene family, Moreover, decreased MS4A6A expression was associated with AD risk (Figure 2). These results help illustrate that the two basic hypotheses we were testing (SNP is a proxy for a coding variant; and, SNP is an eQTL) are not mutually exclusive.
Figure 2.
Potential pathway for the relationship between MS4A6A SNPs, blood gene expression and Alzheimer’s disease phenotype. The relationship between MS4A6A IGAP SNP rs983392, blood gene expression, and Alzheimer’s disease phenotype are complex and possibly multifactorial. The rs983392 SNP constitutes an eQTL for MS4A6A, and also a proxy for a nonsynonymous exonic MS4A6A SNP rs7232. It is possible that the nonsynonymous SNP rs7232 is not, or is only partially responsible, for the eQTL phenomena, which may indicate parallel gene regulatory mechanism(s).
Key: SNP, single nucleotide polymorphism, IGAP, International Genomics of Alzheimer’s Project, eQTL, expression quantitative trait locus
MS4A6A, located on chromosome 11q12.2, encodes a member of membrane-spanning 4A gene family (membrane-spanning 4A domains, subfamily A, member 6A). MS4A genes are highly expressed in hematopoietic cells, and involved in the regulation of calcium signaling (Ma, et al., 2015). Although functions of the MS4A6A protein are still incompletely understood, it is possible that the MS4A6A SNPs are linked to AD via deregulation of calcium signaling implicated in neurodegenerative diseases (LaFerla, 2002,Marambaud, et al., 2009).
4.9. PICALM SNPs
There was one nonsynonymous rare variant rs74547795 in SYTL2 with D’ = 1 for the IGAP SNP, that causes amino acid substitution of alanine to aspartic acid at codon position 825 (A825D). PICALM is located on chromosome 11q14.2, and encodes a phosphatidylinositol cinding clathrin assembly protein that may be involved in Aβ clearance (Zhao, et al., 2015) and synaptic neurotransmission release (Sleegers, et al., 2010). On the other hand, there is no evidence that SYTL2 is associated with AD.
4.10. DSG2 SNPs
There was one nonsynonymous rare variant rs111986709 located in DSG3 with D’ = 1 for the IGAP SNP. The variant causes serine-to-phenylalanine amino acid substitution at codon position 771 (S771F), and the mutation was predicted to have an impact on the DSG3 protein. DSG2 and DSG3 encode members of the desmoglein family. The role of these genes in AD is unknown.
The expression of both DLGAP1 and NETO1 were strongly associated with the DSG2 IGAP SNP and were highly correlated with each other in FCTX. Interestingly, these genes were significantly under-expressed in four brain regions of AD cases. Located in chromosome 18p11.31 more than 25Mb away from DSG2, DLGAP1 encodes disks large-associated protein 1 (also known as guanylate kinase- associated protein (GKAP)). NETO1 is located on chromosome 18q22.3, more than 40Mb away from DSG2, and encodes neuropilin and tolloid like 1. Both DLGAP1 and NETO1 are mainly expressed in neurons of human brains (http://web.stanford.edu/group/barres_lab/brainseqMariko/brainseq2.html) (Bennett, et al., 2016), and may be involved in N-methyl-D-aspartate receptor-dependent synaptic plasticity (Ng, et al., 2009,Shin, et al., 2012).
4.11. ABCA7 SNPs
Of six coding SNPs in strong or moderate LD with the IGAP SNP, we confirmed that two nonsynonymous SNPs (rs3752246 causing alanine-to-glycine amino acid substitution at codon position 1527 (A1527G) and rs4147934 causing serine-to-alanine amino acid substitution at codon position 2045 (S2045A)) and one synonymous SNP (rs4147930 (L1995L)) were associated with AD. The IGAP SNP acted as an eQTL for EID2B expression (the risk allele was associated with increased EID2B expression in FCTX). However, decreased expression of EID2B in HIPP was associated with AD risk (i.e., the association directions are in conflict). ABCA7, located in chromosome 19p13.3, encodes a member of the super family of ATP-binding cassette transporters. ABCA7 is expressed in hippocampal CA1 neurons and in microglia (Kim, et al., 2006). ABCA7 is involved in lipid efflux from cells to lipoproteins and has been associated with Aβ accumulation (Kim, et al., 2013).
4.12. CD33 SNPs
There were two nonsynonymous SNPs in strong or moderate LD with the IGAP CD33 SNP, rs12459419 causing alanine-to-valine amino acid substitution at codon position 14 (A14V) and rs35112940 causing glycine-to-arginine amino acid substitution at codon position 304 (G304R). Although we did not find sufficient evidence that the proxy coding SNPs in LD with the IGAP SNP rs3865444 were associated with AD, the protective allele of rs3865444 was associated with decreased IER2 expression in TCTX and, further, decreased IER2 expression in EC and TCTX had a protective effect on AD. IER2 is located on chromosome 19p13.2 more than 35Mb away from CD33, and encodes immediate early response 2. IER2 may function as a transcription factor (Takaya, et al., 2009).
Malik et al. reported that the IGAP SNP rs3865444 modulated exon 2 splicing by showing that the proportion of CD33 expressed as a CD33 isoform lacking exon 2 was increased in the protective allele of rs3865444; the proxy nonsynonymous SNP rs12459419 was shown to modulate exon 2 splicing efficiency (Malik, et al., 2013). Additional studies are warranted to examine the association between CD33 isoform and IER2 expression.
There are limitations to this study. We aggregated data from many rich resources that aid in establishing a confluence of related information; however, these datasets are heterogeneous and can exhibit biases from their respective parent study designs, analytic protocols, and participant pools. A major limitation of our study is that we have limited our assessment to subjects with European-type genomic characteristics, which is connected to the fact that many research centers and clinics that contribute to the study share this underlying bias. Also, according to the commonly used, but inexact convention, we focused on genes closest to the identified IGAP SNP.
Supplementary Material
The IGAP SNPs are located on non-coding regions.
The functional impacts of the IGAP SNPs are poorly understood.
Some of the IGAP SNPs are proxies of coding SNPs.
Some of the IGAP SNPs acted as eQTL for AD-related genes’ expression.
Acknowledgments
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12–2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
The National Institutes of Health, National Institute on Aging (NIH-NIA) supported this work through the following grants: ADGC, U01 AG032984, RC2 AG036528; Samples from the National Cell Repository for Alzheimer’s Disease (NCRAD), which receives government support under a cooperative agreement grant (U24 AG21886) awarded by the National Institute on Aging (NIA), were used in this study. We thank contributors who collected samples used in this study, as well as patients and their families, whose help and participation made this work possible; Data for this study were prepared, archived, and distributed by the National Institute on Aging Alzheimer’s Disease Data Storage Site (NIAGADS) at the University of Pennsylvania (U24-AG041689–01); NACC, U01 AG016976; NIA LOAD, U24 AG026395, R01AG041797; Banner Sun Health Research Institute P30 AG019610; Boston University, P30 AG013846, U01 AG10483, R01 CA129769, R01 MH080295, R01 AG017173, R01 AG025259, R01AG33193; Columbia University, P50 AG008702, R37 AG015473; Duke University, P30 AG028377, AG05128; Emory University, AG025688; Group Health Research Institute, UO1 AG006781, UO1 HG004610, UO1 HG006375; Indiana University, P30 AG10133; Johns Hopkins University, P50 AG005146, R01 AG020688; Massachusetts General Hospital, P50 AG005134; Mayo Clinic, P50 AG016574; Mount Sinai School of Medicine, P50 AG005138, P01 AG002219; New York University, P30 AG08051, UL1 RR029893, 5R01AG012101, 5R01AG022374, 5R01AG013616, 1RC2AG036502, 1R01AG035137; Northwestern University, P30 AG013854; Oregon Health & Science University, P30 AG008017, R01 AG026916; Rush University, P30 AG010161, R01 AG019085, R01 AG15819, R01 AG17917, R01 AG30146; TGen, R01 NS059873; University of Alabama at Birmingham, P50 AG016582; University of Arizona, R01 AG031581; University of California, Davis, P30 AG010129; University of California, Irvine, P50 AG016573; University of California, Los Angeles, P50 AG016570; University of California, San Diego, P50 AG005131; University of California, San Francisco, P50 AG023501, P01 AG019724; University of Kentucky, P30 AG028383, AG05144; University of Michigan, P50 AG008671; University of Pennsylvania, P30 AG010124; University of Pittsburgh, P50 AG005133, AG030653, AG041718, AG07562, AG02365; University of Southern California, P50 AG005142; University of Texas Southwestern, P30 AG012300; University of Miami, R01 AG027944, AG010491, AG027944, AG021547, AG019757; University of Washington, P50 AG005136; University of Wisconsin, P50 AG033514; Vanderbilt University, R01 AG019085; and Washington University, P50 AG005681, P01 AG03991. The Kathleen Price Bryan Brain Bank at Duke University Medical Center is funded by NINDS grant # NS39764, NIMH MH60451 and by Glaxo Smith Kline. Genotyping of the TGEN2 cohort was supported by Kronos Science. The TGen series was also funded by NIA grant AG041232 to AJM and MJH, The Banner Alzheimer’s Foundation, The Johnnie B. Byrd Sr. Alzheimer’s Institute, the Medical Research Council, and the state of Arizona and also includes samples from the following sites: Newcastle Brain Tissue Resource (funding via the Medical Research Council, local NHS trusts and Newcastle University), MRC London Brain Bank for Neurodegenerative Diseases (funding via the Medical Research Council),South West Dementia Brain Bank (funding via numerous sources including the Higher Education Funding Council for England (HEFCE), Alzheimer’s Research Trust (ART), BRACE as well as North Bristol NHS Trust Research and Innovation Department and DeNDRoN), The Netherlands Brain Bank (funding via numerous sources including Stichting MS Research, Brain Net Europe, Hersenstichting Nederland Breinbrekend Werk, International Parkinson Fonds, Internationale Stiching Alzheimer Onderzoek), Institut de Neuropatologia, Servei Anatomia Patologica, Universitat de Barcelona. ADNI data collection and sharing was funded by the National Institutes of Health Grant U01 AG024904 and Department of Defense award number W81XWH-12–2-0012. ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. We thank Drs. D. Stephen Snyder and Marilyn Miller from NIA who are ex-officio ADGC members. Support was also from the Alzheimer’s Association (LAF, IIRG-08–89720; MP-V, IIRG-05–14147) and the US Department of Veterans Affairs Administration, Office of Research and Development, Biomedical Laboratory Research Program. P.S.G.-H. is supported by Wellcome Trust, Howard Hughes Medical Institute, and the Canadian Institute of Health Research.
The Alzheimer’s Disease Sequencing Project (ADSP) is comprised of two Alzheimer’s Disease (AD) genetics consortia and three National Human Genome Research Institute (NHGRI) funded Large Scale Sequencing and Analysis Centers (LSAC). The two AD genetics consortia are the Alzheimer’s Disease Genetics Consortium (ADGC) funded by NIA (U01 AG032984), and the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) funded by NIA (R01 AG033193), the National Heart, Lung, and Blood Institute (NHLBI), other National Institute of Health (NIH) institutes and other foreign governmental and non-governmental organizations. The Discovery Phase analysis of sequence data is supported through UF1AG047133 (to Drs. Schellenberg, Farrer, Pericak-Vance, Mayeux, and Haines); U01AG049505 to Dr. Seshadri; U01AG049506 to Dr. Boerwinkle; U01AG049507 to Dr. Wijsman; and U01AG049508 to Dr. Goate and the Discovery Extension Phase analysis is supported through U01AG052411 to Dr. Goate, U01AG052410 to Dr. Pericak-Vance and U01 AG052409 to Drs. Seshadri and Fornage. Data generation and harmonization in the Follow-up Phases is supported by U54AG052427 (to Drs. Schellenberg and Wang).
The ADGC cohorts include: Adult Changes in Thought (ACT), the Alzheimer’s Disease Centers (ADC), the Chicago Health and Aging Project (CHAP), the Memory and Aging Project (MAP), Mayo Clinic (MAYO), Mayo Parkinson’s Disease controls, University of Miami, the Multi-Institutional Research in Alzheimer’s Genetic Epidemiology Study (MIRAGE), the National Cell Repository for Alzheimer’s Disease (NCRAD), the National Institute on Aging Late Onset Alzheimer’s Disease Family Study (NIA-LOAD), the Religious Orders Study (ROS), the Texas Alzheimer’s Research and Care Consortium (TARC), Vanderbilt University/Case Western Reserve University (VAN/CWRU), the Washington Heights-Inwood Columbia Aging Project (WHICAP) and the Washington University Sequencing Project (WUSP), the Columbia University Hispanic- Estudio Familiar de Influencia Genetica de Alzheimer (EFIGA), the University of Toronto (UT), and Genetic Differences (GD).
The CHARGE cohorts are supported in part by National Heart, Lung, and Blood Institute (NHLBI) infrastructure grant HL105756 (Psaty), RC2HL102419 (Boerwinkle) and the neurology working group is supported by the National Institute on Aging (NIA) R01 grant AG033193. The CHARGE cohorts participating in the ADSP include the following: Austrian Stroke Prevention Study (ASPS), ASPS-Family study, and the Prospective Dementia Registry-Austria (ASPS/PRODEM-Aus), the Atherosclerosis Risk in Communities (ARIC) Study, the Cardiovascular Health Study (CHS), the Erasmus Rucphen Family Study (ERF), the Framingham Heart Study (FHS), and the Rotterdam Study (RS). ASPS is funded by the Austrian Science Fond (FWF) grant number P20545-P05 and P13180 and the Medical University of Graz. The ASPS-Fam is funded by the Austrian Science Fund (FWF) project I904),the EU Joint Programme - Neurodegenerative Disease Research (JPND) in frame of the BRIDGET project (Austria, Ministry of Science) and the Medical University of Graz and the Steiermärkische Krankenanstalten Gesellschaft. PRODEM-Austria is supported by the Austrian Research Promotion agency (FFG) (Project No. 827462) and by the Austrian National Bank (Anniversary Fund, project 15435. ARIC research is carried out as a collaborative study supported by NHLBI contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C). Neurocognitive data in ARIC is collected by U01 2U01HL096812, 2U01HL096814, 2U01HL096899, 2U01HL096902, 2U01HL096917 from the NIH (NHLBI, NINDS, NIA and NIDCD), and with previous brain MRI examinations funded by R01-HL70825 from the NHLBI. CHS research was supported by contracts HHSN268201200036C, HHSN268200800007C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086, and grants U01HL080295 and U01HL130114 from the NHLBI with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided by R01AG023629, R01AG15928, and R01AG20098 from the NIA. FHS research is supported by NHLBI contracts N01-HC-25195 and HHSN268201500001I. This study was also supported by additional grants from the NIA (R01s AG054076, AG049607 and AG033040 and NINDS (R01 NS017950). The ERF study as a part of EUROSPAN (European Special Populations Research Network) was supported by European Commission FP6 STRP grant number 018947 (LSHG-CT-2006–01947) and also received funding from the European Community’s Seventh Framework Programme (FP7/2007–2013)/grant agreement HEALTH-F4–2007-201413 by the European Commission under the programme “Quality of Life and Management of the Living Resources” of 5th Framework Programme (no. QLG2-CT-2002–01254). High-throughput analysis of the ERF data was supported by a joint grant from the Netherlands Organization for Scientific Research and the Russian Foundation for Basic Research (NWO-RFBR 047.017.043). The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, the Netherlands Organization for Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the municipality of Rotterdam. Genetic data sets are also supported by the Netherlands Organization of Scientific Research NWO Investments (175.010.2005.011, 911–03-012), the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, the Research Institute for Diseases in the Elderly (014–93-015; RIDE2), and the Netherlands Genomics Initiative (NGI)/Netherlands Organization for Scientific Research (NWO) Netherlands Consortium for Healthy Aging (NCHA), project 050–060-810. All studies are grateful to their participants, faculty and staff. The content of these manuscripts is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the U.S. Department of Health and Human Services.
The three LSACs are: the Human Genome Sequencing Center at the Baylor College of Medicine (U54 HG003273), the Bro Alzheimer’s disease Institute Genome Center (U54HG003067), and the Washington University Genome Institute (U54HG003079).
Biological samples and associated phenotypic data used in primary data analyses were stored at Study Investigators institutions, and at the National Cell Repository for Alzheimer’s Disease (NCRAD, U24AG021886) at Indiana University funded by NIA. Associated Phenotypic Data used in primary and secondary data analyses were provided by Study Investigators, the NIA funded Alzheimer’s Disease Centers (ADCs), and the National Alzheimer’s Coordinating Center (NACC, U01AG016976) and the National Institute on Aging Genetics of Alzheimer’s Disease Data Storage Site (NIAGADS, U24AG041689) at the University of Pennsylvania, funded by NIA, and at the Database for Genotypes and Phenotypes (dbGaP) funded by NIH. This research was supported in part by the Intramural Research Program of the National Institutes of health, National Library of Medicine. Contributors to the Genetic Analysis Data included Study Investigators on projects that were individually funded by NIA, and other NIH institutes, and by private U.S. organizations, or foreign governmental or nongovernmental organizations.
The work done by the North American Brain Expression Consortium (NABEC) was supported in part by the Intramural Research Program of the National Institute on Aging, National Institutes of Health, part of the US Department of Health and Human Services (project number ZIA AG000932–04), and by a Research Grant from the Department of Defense (W81XWH-092–0128).
Funding
This work was supported by the National Cell Repository for Alzheimer’s Disease (U24 AG21886), and National Institute on Aging (K25 AG043546, UL1TR000117, the UK-ADC P30 AG028383, and R01 AG057187).
Abbreviations
- ADGC
Alzheimer’s Disease Genetic Consortium
- ADNI
Alzheimer’s Disease Neuroimaging Initiative
- ADSP
Alzheimer’s Disease Sequencing Project
- GWAS
Genome-wide association studies
- IGAP
International Genomics of Alzheimer’s Project
- LD
Linkage disequilibrium
- NABEC
North American Brain Expression Consortium
- UKBEC
United Kingdom Brain Expression Consortium
Footnotes
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
Disclosure statement
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1000 Genomes Project Consortium. 2010. A map of human genome variation from population-scale sequencing. Nature 467(7319), 1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR 2010. A method and server for predicting damaging missense mutations. Nat Methods 7(4), 248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzheimer A 1907. Über eine eigenartige Erkrankung der Hirnrinde. Allgemeine Z Psychiatrie Psychisch-Gerichtliche Med 64, 146–8. [Google Scholar]
- Amiri M, Conserva F, Panayiotou C, Karlsson A, Solaroli N 2013. The human adenylate kinase 9 is a nucleoside mono- and diphosphate kinase. Int J Biochem Cell Biol 45(5), 925–31. doi: 10.1016/j.biocel.2013.02.004. [DOI] [PubMed] [Google Scholar]
- Beecham GW, Bis JC, Martin ER, Choi SH, DeStefano AL, van Duijn CM, Fornage M, Gabriel SB, Koboldt DC, Larson DE, Naj AC, Psaty BM, Salerno W, Bush WS, Foroud TM, Wijsman E, Farrer LA, Goate A, Haines JL, Pericak-Vance MA, Boerwinkle E, Mayeux R, Seshadri S, Schellenberg G 2017. The Alzheimer’s Disease Sequencing Project: Study design and sample selection. Neurol Genet 3(5), e194. doi: 10.1212/NXG.0000000000000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson H, Simpson K, Bullard J, Hansen K 2008. aroma.affymetrix: A generic framework in R for analyzing small to very large Affymetrix data sets in bounded memory. Tech Report #745, Department of Statistics, University of California, Berkeley. [Google Scholar]
- Benjamini Y, Hochberg Y 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser A Stat Soc 57, 289–300. [Google Scholar]
- Bennett ML, Bennett FC, Liddelow SA, Ajami B, Zamanian JL, Fernhoff NB, Mulinyawe SB, Bohlen CJ, Adil A, Tucker A, Weissman IL, Chang EF, Li G, Grant GA, Hayden Gephart MG, Barres BA 2016. New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci U S A 113(12), E1738–46. doi: 10.1073/pnas.1525528113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchtold NC, Cribbs DH, Coleman PD, Rogers J, Head E, Kim R, Beach T, Miller C, Troncoso J, Trojanowski JQ, Zielke HR, Cotman CW 2008. Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc Natl Acad Sci U S A 105(40), 15605–10. doi: 10.1073/pnas.0806883105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi A, Anderson CD, Desikan RS, Sabuncu M, Cortellini L, Schmansky N, Salat D, Rosand J, Alzheimer’s Disease Neuroimaging, I. 2010. Genetic variation and neuroimaging measures in Alzheimer disease. Arch Neurol 67(6), 677–85. doi: 10.1001/archneurol.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blalock EM, Buechel HM, Popovic J, Geddes JW, Landfield PW 2011. Microarray analyses of laser-captured hippocampus reveal distinct gray and white matter signatures associated with incipient Alzheimer’s disease. J Chem Neuroanat 42(2), 118–26. doi: 10.1016/j.jchemneu.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bralten J, Franke B, Arias-Vasquez A, Heister A, Brunner HG, Fernandez G, Rijpkema M 2011. CR1 genotype is associated with entorhinal cortex volume in young healthy adults. Neurobiol Aging 32(11), 2106 e7–11. doi: 10.1016/j.neurobiolaging.2011.05.017. [DOI] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chess A 2012. Mechanisms and consequences of widespread random monoallelic expression. Nat Rev Genet 13(6), 421–8. doi: 10.1038/nrg3239. [DOI] [PubMed] [Google Scholar]
- Choi Y, Sims GE, Murphy S, Miller JR, Chan AP 2012. Predicting the functional effect of amino acid substitutions and indels. PLoS One 7(10), e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Forer L, Schonherr S, Sidore C, Locke AE, Kwong A, Vrieze SI, Chew EY, Levy S, McGue M, Schlessinger D, Stambolian D, Loh PR, Iacono WG, Swaroop A, Scott LJ, Cucca F, Kronenberg F, Boehnke M, Abecasis GR, Fuchsberger C 2016. Next-generation genotype imputation service and methods. Nat Genet 48(10), 1284–7. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S 2010. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput Biol 6(12), e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, Thompson WK, Holland D, Hess CP, Brewer JB, Zetterberg H, Blennow K, Andreassen OA, McEvoy LK, Hyman BT, Dale AM, Alzheimer’s Disease Neuroimaging Initiative, G. 2014. The role of clusterin in amyloid-beta-associated neurodegeneration. JAMA Neurol 71(2), 180–7. doi: 10.1001/jamaneurol.2013.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautel M, Zuffardi O, Freiburg A, Labeit S 1995. Phosphorylation switches specific for the cardiac isoform of myosin binding protein-C: a modulator of cardiac contraction? EMBO J 14(9), 1952–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimelbrant A, Hutchinson JN, Thompson BR, Chess A 2007. Widespread monoallelic expression on human autosomes. Science 318(5853), 1136–40. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ 2002. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science 297(5580), 353–6. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, Heun R, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O’Donovan M, Owen MJ, Williams J 2009. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet 41(10), 1088–93. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Umehara T, Saito K, Harada T, Watanabe S, Yabuki T, Kigawa T, Takahashi M, Kuwasako K, Tsuda K, Matsuda T, Aoki M, Seki E, Kobayashi N, Guntert P, Yokoyama S, Muto Y 2010. Structural insight into the zinc finger CW domain as a histone modification reader. Structure 18(9), 1127–39. doi: 10.1016/j.str.2010.06.012. [DOI] [PubMed] [Google Scholar]
- Hernandez DG, Nalls MA, Moore M, Chong S, Dillman A, Trabzuni D, Gibbs JR, Ryten M, Arepalli S, Weale ME, Zonderman AB, Troncoso J, O’Brien R, Walker R, Smith C, Bandinelli S, Traynor BJ, Hardy J, Singleton AB, Cookson MR 2012. Integration of GWAS SNPs and tissue specific expression profiling reveal discrete eQTLs for human traits in blood and brain. Neurobiol Dis 47(1), 20–8. doi: 10.1016/j.nbd.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Jones N, Stretton A, Thomas C, Richards A, Ivanov D, Widdowson C, Chapman J, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Beaumont H, Warden D, Wilcock G, Love S, Kehoe PG, Hooper NM, Vardy ER, Hardy J, Mead S, Fox NC, Rossor M, Collinge J, Maier W, Jessen F, Ruther E, Schurmann B, Heun R, Kolsch H, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Gallacher J, Hull M, Rujescu D, Giegling I, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Pankratz VS, Sando SB, Aasly JO, Barcikowska M, Wszolek ZK, Dickson DW, Graff-Radford NR, Petersen RC, Alzheimer’s Disease Neuroimaging, I., van Duijn CM, Breteler MM, Ikram MA, DeStefano AL, Fitzpatrick AL, Lopez O, Launer LJ, Seshadri S, consortium, C., Berr C, Campion D, Epelbaum J, Dartigues JF, Tzourio C, Alperovitch A, Lathrop M, consortium E, Feulner TM, Friedrich P, Riehle C, Krawczak M, Schreiber S, Mayhaus M, Nicolhaus S, Wagenpfeil S, Steinberg S, Stefansson H, Stefansson K, Snaedal J, Bjornsson S, Jonsson PV, Chouraki V, Genier-Boley B, Hiltunen M, Soininen H, Combarros O, Zelenika D, Delepine M, Bullido MJ, Pasquier F, Mateo I, Frank-Garcia A, Porcellini E, Hanon O, Coto E, Alvarez V, Bosco P, Siciliano G, Mancuso M, Panza F, Solfrizzi V, Nacmias B, Sorbi S, Bossu P, Piccardi P, Arosio B, Annoni G, Seripa D, Pilotto A, Scarpini E, Galimberti D, Brice A, Hannequin D, Licastro F, Jones L, Holmans PA, Jonsson T, Riemenschneider M, Morgan K, Younkin SG, Owen MJ, O’Donovan M, Amouyel P, Williams J 2011. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet 43(5), 429–35. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KL, Marcora E, Pimenova AA, Di Narzo AF, Kapoor M, Jin SC, Harari O, Bertelsen S, Fairfax BP, Czajkowski J, Chouraki V, Grenier-Boley B, Bellenguez C, Deming Y, McKenzie A, Raj T, Renton AE, Budde J, Smith A, Fitzpatrick A, Bis JC, DeStefano A, Adams HHH, Ikram MA, van der Lee S, Del-Aguila JL, Fernandez MV, Ibanez L, International Genomics of Alzheimer’s, P., Alzheimer’s Disease Neuroimaging, I., Sims R, Escott-Price V, Mayeux R, Haines JL, Farrer LA, Pericak-Vance MA, Lambert JC, van Duijn C, Launer L, Seshadri S, Williams J, Amouyel P, Schellenberg GD, Zhang B, Borecki I, Kauwe JSK, Cruchaga C, Hao K, Goate AM 2017. A common haplotype lowers PU.1 expression in myeloid cells and delays onset of Alzheimer’s disease. Nat Neurosci 20(8), 1052–61. doi: 10.1038/nn.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayadev S, Leverenz JB, Steinbart E, Stahl J, Klunk W, Yu CE, Bird TD 2010. Alzheimer’s disease phenotypes and genotypes associated with mutations in presenilin 2. Brain 133(Pt 4), 1143–54. doi: 10.1093/brain/awq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumata Y, Nelson PT, Ellingson SR, Fardo DW 2017. Gene-based association study of genes linked to hippocampal sclerosis of aging neuropathology: GRN, TMEM106B, ABCC9, and KCNMB2. Neurobiol Aging 53, 193 e17- e25. doi: 10.1016/j.neurobiolaging.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D 2002. The human genome browser at UCSC. Genome Res 12(6), 996–1006. doi: 10.1101/gr.229102. Article published online before print in May 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khera R, Das N 2009. Complement Receptor 1: disease associations and therapeutic implications. Mol Immunol 46(5), 761–72. doi: 10.1016/j.molimm.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WS, Guillemin GJ, Glaros EN, Lim CK, Garner B 2006. Quantitation of ATP-binding cassette subfamily-A transporter gene expression in primary human brain cells. Neuroreport 17(9), 891–6. doi: 10.1097/01.wnr.0000221833.41340.cd. [DOI] [PubMed] [Google Scholar]
- Kim WS, Li H, Ruberu K, Chan S, Elliott DA, Low JK, Cheng D, Karl T, Garner B 2013. Deletion of Abca7 increases cerebral amyloid-beta accumulation in the J20 mouse model of Alzheimer’s disease. J Neurosci 33(10), 4387–94. doi: 10.1523/JNEUROSCI.4165-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFerla FM 2002. Calcium dyshomeostasis and intracellular signalling in Alzheimer’s disease. Nat Rev Neurosci 3(11), 862–72. doi: 10.1038/nrn960. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fievet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, European Alzheimer’s Disease Initiative, I., de Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossu P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanche H, Dartigues JF, Tzourio C, Gut I, Van Broeckhoven C, Alperovitch A, Lathrop M, Amouyel P 2009. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet 41(10), 1094–9. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thorton-Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y, Lin CF, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau MT, Choi SH, Reitz C, Pasquier F, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston JA, Evans D, Lovestone S, Letenneur L, Moron FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate AM, Fievet N, Huentelman MW, Gill M, Brown K, Kamboh MI, Keller L, BarbergerGateau P, McGuiness B, Larson EB, Green R, Myers AJ, Dufouil C, Todd S, Wallon D, Love S, Rogaeva E, Gallacher J, St George-Hyslop P, Clarimon J, Lleo A, Bayer A, Tsuang DW, Yu L, Tsolaki M, Bossu P, Spalletta G, Proitsi P, Collinge J, Sorbi S, Sanchez-Garcia F, Fox NC, Hardy J, Deniz Naranjo MC, Bosco P, Clarke R, Brayne C, Galimberti D, Mancuso M, Matthews F, European Alzheimer’s Disease, I., Genetic, Environmental Risk in Alzheimer’s, D., Alzheimer’s Disease Genetic, C., Cohorts for, H., Aging Research in Genomic, E., Moebus S, Mecocci P, Del Zompo M, Maier W, Hampel H, Pilotto A, Bullido M, Panza F, Caffarra P, Nacmias B, Gilbert JR, Mayhaus M, Lannefelt L, Hakonarson H, Pichler S, Carrasquillo MM, Ingelsson M, Beekly D, Alvarez V, Zou F, Valladares O, Younkin SG, Coto E, Hamilton-Nelson KL, Gu W, Razquin C, Pastor P, Mateo I, Owen MJ, Faber KM, Jonsson PV, Combarros O, O’Donovan MC, Cantwell LB, Soininen H, Blacker D, Mead S, Mosley TH Jr., Bennett DA, Harris TB, Fratiglioni L, Holmes C, de Bruijn RF, Passmore P, Montine TJ, Bettens K, Rotter JI, Brice A, Morgan K, Foroud TM, Kukull WA, Hannequin D, Powell JF, Nalls MA, Ritchie K, Lunetta KL, Kauwe JS, Boerwinkle E, Riemenschneider M, Boada M, Hiltuenen M, Martin ER, Schmidt R, Rujescu D, Wang LS, Dartigues JF, Mayeux R, Tzourio C, Hofman A, Nothen MM, Graff C, Psaty BM, Jones L, Haines JL, Holmans PA, Lathrop M, Pericak-Vance MA, Launer LJ, Farrer LA, van Duijn CM, Van Broeckhoven C, Moskvina V, Seshadri S, Williams J, Schellenberg GD, Amouyel P 2013. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 45(12), 1452–8. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD 2012. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28(6), 882–3. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Lahad E, Wasco W, Poorkaj P, Romano DM, Oshima J, Pettingell WH, Yu CE, Jondro PD, Schmidt SD, Wang K, et al. 1995. Candidate gene for the chromosome 1 familial Alzheimer’s disease locus. Science 269(5226), 973–7. [DOI] [PubMed] [Google Scholar]
- Lewontin RC 1964. The Interaction of Selection and Linkage. I. General Considerations; Heterotic Models. Genetics 49(1), 49–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang WS, Dunckley T, Beach TG, Grover A, Mastroeni D, Walker DG, Caselli RJ, Kukull WA, McKeel D, Morris JC, Hulette C, Schmechel D, Alexander GE, Reiman EM, Rogers J, Stephan DA 2007. Gene expression profiles in anatomically and functionally distinct regions of the normal aged human brain. Physiol Genomics 28(3), 311–22. doi: 10.1152/physiolgenomics.00208.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler SF, Wang R, Grimm H, Uljon SN, Masters CL, Beyreuther K 1999. Mechanism of the cleavage specificity of Alzheimer’s disease gamma-secretase identified by phenylalanine-scanning mutagenesis of the transmembrane domain of the amyloid precursor protein. Proc Natl Acad Sci U S A 96(6), 3053–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling IF, Bhongsatiern J, Simpson JF, Fardo DW, Estus S 2012. Genetics of clusterin isoform expression and Alzheimer’s disease risk. PLoS One 7(4), e33923. doi: 10.1371/journal.pone.0033923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Yu JT, Wang HF, Hao XK, Yang YF, Jiang T, Zhu XC, Cao L, Zhang DQ, Tan L 2014. Association between NME8 locus polymorphism and cognitive decline, cerebrospinal fluid and neuroimaging biomarkers in Alzheimer’s disease. PLoS One 9(12), e114777. doi: 10.1371/journal.pone.0114777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh PR, Danecek P, Palamara PF, Fuchsberger C, Y AR, H KF, Schoenherr S, Forer L, McCarthy S, Abecasis GR, Durbin R, A LP 2016. Reference-based phasing using the Haplotype Reference Consortium panel. Nat Genet 48(11), 1443–8. doi: 10.1038/ng.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Aron L, Zullo J, Pan Y, Kim H, Chen Y, Yang TH, Kim HM, Drake D, Liu XS, Bennett DA, Colaiacovo MP, Yankner BA 2014. REST and stress resistance in ageing and Alzheimer’s disease. Nature 507(7493), 448–54. doi: 10.1038/nature13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Yu JT, Tan L 2015. MS4A Cluster in Alzheimer’s Disease. Mol Neurobiol 51(3), 1240–8. doi: 10.1007/s12035-014-8800-z. [DOI] [PubMed] [Google Scholar]
- Malik M, Simpson JF, Parikh I, Wilfred BR, Fardo DW, Nelson PT, Estus S 2013. CD33 Alzheimer’s risk-altering polymorphism, CD33 expression, and exon 2 splicing. J Neurosci 33(33), 13320–5. doi: 10.1523/JNEUROSCI.1224-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso M, Orsucci D, Siciliano G, Murri L 2008. Mitochondria, mitochondrial DNA and Alzheimer’s disease. What comes first? Curr Alzheimer Res 5(5), 457–68. [DOI] [PubMed] [Google Scholar]
- Marambaud P, Dreses-Werringloer U, Vingtdeux V 2009. Calcium signaling in neurodegeneration. Mol Neurodegener 4, 20. doi: 10.1186/1750-1326-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F 2010. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics 26(16), 2069–70. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockenhaupt S, Makeyev EV 2015. Non-coding functions of alternative pre-mRNA splicing in development. Semin Cell Dev Biol 47–48, 32–9. doi: 10.1016/j.semcdb.2015.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullan M, Crawford F, Axelman K, Houlden H, Lilius L, Winblad B, Lannfelt L 1992. A pathogenic mutation for probable Alzheimer’s disease in the APP gene at the Nterminus of beta-amyloid. Nat Genet 1(5), 345–7. doi: 10.1038/ng0892-345. [DOI] [PubMed] [Google Scholar]
- Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, Boeve BF, Graff-Radford NR, De Jager PL, Evans D, Schneider JA, Carrasquillo MM, Ertekin-Taner N, Younkin SG, Cruchaga C, Kauwe JS, Nowotny P, Kramer P, Hardy J, Huentelman MJ, Myers AJ, Barmada MM, Demirci FY, Baldwin CT, Green RC, Rogaeva E, St George-Hyslop P, Arnold SE, Barber R, Beach T, Bigio EH, Bowen JD, Boxer A, Burke JR, Cairns NJ, Carlson CS, Carney RM, Carroll SL, Chui HC, Clark DG, Corneveaux J, Cotman CW, Cummings JL, DeCarli C, DeKosky ST, Diaz-Arrastia R, Dick M, Dickson DW, Ellis WG, Faber KM, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Gilman S, Giordani B, Glass JD, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha GA, Jin LW, Johnson N, Karlawish J, Karydas A, Kaye JA, Kim R, Koo EH, Kowall NW, Lah JJ, Levey AI, Lieberman AP, Lopez OL, Mack WJ, Marson DC, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Parisi JE, Perl DP, Peskind E, Petersen RC, Poon WW, Quinn JF, Rajbhandary RA, Raskind M, Reisberg B, Ringman JM, Roberson ED, Rosenberg RN, Sano M, Schneider LS, Seeley W, Shelanski ML, Slifer MA, Smith CD, Sonnen JA, Spina S, Stern RA, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Williamson J, Woltjer RL, Cantwell LB, Dombroski BA, Beekly D, Lunetta KL, Martin ER, Kamboh MI, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Montine TJ, Goate AM, Blacker D, Tsuang DW, Hakonarson H, Kukull WA, Foroud TM, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD 2011. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet 43(5), 436–41. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng D, Pitcher GM, Szilard RK, Sertie A, Kanisek M, Clapcote SJ, Lipina T, Kalia LV, Joo D, McKerlie C, Cortez M, Roder JC, Salter MW, McInnes RR 2009. Neto1 is a novel CUB-domain NMDA receptor-interacting protein required for synaptic plasticity and learning. PLoS Biol 7(2), e41. doi: 10.1371/journal.pbio.1000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S 2003. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res 31(13), 3812–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsberth C, Westlind-Danielsson A, Eckman CB, Condron MM, Axelman K, Forsell C, Stenh C, Luthman J, Teplow DB, Younkin SG, Naslund J, Lannfelt L 2001. The ‘Arctic’ APP mutation (E693G) causes Alzheimer’s disease by enhanced Abeta protofibril formation. Nat Neurosci 4(9), 887–93. doi: 10.1038/nn0901-887. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81(3), 559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querfurth HW, LaFerla FM 2010. Alzheimer’s disease. N Engl J Med 362(4), 329–44. doi: 10.1056/NEJMra0909142. [DOI] [PubMed] [Google Scholar]
- Rabbani B, Tekin M, Mahdieh N 2014. The promise of whole-exome sequencing in medical genetics. J Hum Genet 59(1), 5–15. doi: 10.1038/jhg.2013.114. [DOI] [PubMed] [Google Scholar]
- Reitz C, Brayne C, Mayeux R 2011. Epidemiology of Alzheimer disease. Nat Rev Neurol 7(3), 137–52. doi:nrneurol.2011.2[pii] 10.1038/nrneurol.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J, Li R, Mastroeni D, Grover A, Leonard B, Ahern G, Cao P, Kolody H, Vedders L, Kolb WP, Sabbagh M 2006. Peripheral clearance of amyloid beta peptide by complement C3-dependent adherence to erythrocytes. Neurobiol Aging 27(12), 1733–9. doi: 10.1016/j.neurobiolaging.2005.09.043. [DOI] [PubMed] [Google Scholar]
- Sauna ZE, Kimchi-Sarfaty C 2011. Understanding the contribution of synonymous mutations to human disease. Nat Rev Genet 12(10), 683–91. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ, American College of, P., American Physiological, S. 2004. Alzheimer disease: mechanistic understanding predicts novel therapies. Ann Intern Med 140(8), 627–38. [DOI] [PubMed] [Google Scholar]
- Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, Bis JC, Smith AV, Carassquillo MM, Lambert JC, Harold D, Schrijvers EM, Ramirez-Lorca R, Debette S, Longstreth WT Jr., Janssens AC, Pankratz VS, Dartigues JF, Hollingworth P, Aspelund T, Hernandez I, Beiser A, Kuller LH, Koudstaal PJ, Dickson DW, Tzourio C, Abraham R, Antunez C, Du Y, Rotter JI, Aulchenko YS, Harris TB, Petersen RC, Berr C, Owen MJ, Lopez-Arrieta J, Varadarajan BN, Becker JT, Rivadeneira F, Nalls MA, Graff-Radford NR, Campion D, Auerbach S, Rice K, Hofman A, Jonsson PV, Schmidt H, Lathrop M, Mosley TH, Au R, Psaty BM, Uitterlinden AG, Farrer LA, Lumley T, Ruiz A, Williams J, Amouyel P, Younkin SG, Wolf PA, Launer LJ, Lopez OL, van Duijn CM, Breteler MM, Consortium, C., Consortium, G., Consortium, E. 2010. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA 303(18), 1832–40. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannan B, Seifert M, Boothman DA, Tilgen W, Reichrath J 2006. Clusterin and DNA repair: a new function in cancer for a key player in apoptosis and cell cycle control. J Mol Histol 37(5–7), 183–8. doi: 10.1007/s10735-006-9052-7. [DOI] [PubMed] [Google Scholar]
- Shin SM, Zhang N, Hansen J, Gerges NZ, Pak DT, Sheng M, Lee SH 2012. GKAP orchestrates activity-dependent postsynaptic protein remodeling and homeostatic scaling. Nat Neurosci 15(12), 1655–66. doi: 10.1038/nn.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JE, Ince PG, Minett T, Matthews FE, Heath PR, Shaw PJ, Goodall E, Garwood CJ, Ratcliffe LE, Brayne C, Rattray M, Wharton SB, Function MRCC, Ageing Neuropathology Study, G. 2016. Neuronal DNA damage response-associated dysregulation of signalling pathways and cholesterol metabolism at the earliest stages of Alzheimer-type pathology. Neuropathol Appl Neurobiol 42(2), 167–79. doi: 10.1111/nan.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JE, Ince PG, Shaw PJ, Heath PR, Raman R, Garwood CJ, Gelsthorpe C, Baxter L, Forster G, Matthews FE, Brayne C, Wharton SB, Function, M.R.C.C., Ageing Neuropathology Study, G. 2011. Microarray analysis of the astrocyte transcriptome in the aging brain: relationship to Alzheimer’s pathology and APOE genotype. Neurobiol Aging 32(10), 1795–807. doi: 10.1016/j.neurobiolaging.2011.04.013. [DOI] [PubMed] [Google Scholar]
- Siren AL, Knerlich F, Poser W, Gleiter CH, Bruck W, Ehrenreich H 2001. Erythropoietin and erythropoietin receptor in human ischemic/hypoxic brain. Acta Neuropathol 101(3), 271–6. [DOI] [PubMed] [Google Scholar]
- Sleegers K, Lambert JC, Bertram L, Cruts M, Amouyel P, Van Broeckhoven C 2010. The pursuit of susceptibility genes for Alzheimer’s disease: progress and prospects. Trends Genet 26(2), 84–93. doi: 10.1016/j.tig.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Takaya T, Kasatani K, Noguchi S, Nikawa J 2009. Functional analyses of immediate early gene ETR101 expressed in yeast. Biosci Biotechnol Biochem 73(7), 1653–60. doi: 10.1271/bbb.90162. [DOI] [PubMed] [Google Scholar]
- Tarasoff-Conway JM, Carare RO, Osorio RS, Glodzik L, Butler T, Fieremans E, Axel L, Rusinek H, Nicholson C, Zlokovic BV, Frangione B, Blennow K, Menard J, Zetterberg H, Wisniewski T, de Leon MJ 2015. Clearance systems in the brain-implications for Alzheimer disease. Nat Rev Neurol 11(8), 457–70. doi: 10.1038/nrneurol.2015.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabzuni D, Ryten M, Walker R, Smith C, Imran S, Ramasamy A, Weale ME, Hardy J 2011. Quality control parameters on a large dataset of regionally dissected human control brains for whole genome expression studies. J Neurochem 119(2), 275–82. doi: 10.1111/j.1471-4159.2011.07432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Es MA, van den Berg LH 2009. Alzheimer’s disease beyond APOE. Nat Genet 41(10), 1047–8. doi:ng1009-1047[pii] 10.1038/ng1009-1047. [DOI] [PubMed] [Google Scholar]
- Walker ES, Martinez M, Brunkan AL, Goate A 2005. Presenilin 2 familial Alzheimer’s disease mutations result in partial loss of function and dramatic changes in Abeta 42/40 ratios. J Neurochem 92(2), 294–301. doi: 10.1111/j.1471-4159.2004.02858.x. [DOI] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM 2011. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 88(1), 76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JT, Tan L 2012. The role of clusterin in Alzheimer’s disease: pathways, pathogenesis, and therapy. Mol Neurobiol 45(2), 314–26. doi: 10.1007/s12035-012-8237-1. [DOI] [PubMed] [Google Scholar]
- Yu L, Chibnik LB, Srivastava GP, Pochet N, Yang J, Xu J, Kozubek J, Obholzer N, Leurgans SE, Schneider JA, Meissner A, De Jager PL, Bennett DA 2015. Association of Brain DNA methylation in SORL1, ABCA7, HLA-DRB5, SLC24A4, and BIN1 with pathological diagnosis of Alzheimer disease. JAMA Neurol 72(1), 15–24. doi: 10.1001/jamaneurol.2014.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Sagare AP, Ma Q, Halliday MR, Kong P, Kisler K, Winkler EA, Ramanathan A, Kanekiyo T, Bu G, Owens NC, Rege SV, Si G, Ahuja A, Zhu D, Miller CA, Schneider JA, Maeda M, Maeda T, Sugawara T, Ichida JK, Zlokovic BV 2015. Central role for PICALM in amyloid-beta blood-brain barrier transcytosis and clearance. Nat Neurosci 18(7), 978–87. doi: 10.1038/nn.4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.