Abstract
There is a strong connection between inflammation, altered microRNA (miRNA) expression and colon cancer. Longstanding inflammatory bowel diseases-related colitis leads to increased risk for the development of colorectal cancer (CRC), while sporadic CRC is in part driven by the inflammatory microenvironment This supports a causative role for inflammation in colon carcinogenesis. miRNAs are a class of small noncoding RNAs that have recently emerged as key players in both inflammation and cancer. Some miRNAs act as inflammatory mediators, others can act as either oncogenes or tumor suppressors depending on the cellular environment in which they are expressed. In particular, miR-21 is an oncogenic miRNA that has been implicated as an inflammatory mediator and may promote inflammation-associated colon carcinogenesis. miRNAs have potential as biomarkers and therapeutic targets in CRC. They are currently being evaluated as early detection biomarkers and prognostic classifiers. Polymorphisms in miRNAs and miRNA-binding sites may alter one’s risk of CRC. This review will focus on the role of inflammation and miRNAs in colon carcinogenesis and discuss the potential for miRNAs and Inflammatory genes to be used as biomarkers and therapeutic targets of CRC.
Keywords: MicroRNA, Colorectal cancer, Inflammation, Biomarker, miR-21
Introduction
Colorectal cancer (CRC) is the third most common cancer and the fourth leading cause of cancer-related death worldwide. More than 1.2 million new cases and more than 600,000 deaths are estimated to occur annually [1]. Discovering risk factors, biomarkers and therapeutic targets for CRC may help reduce the burden of this cancer.
The connection between inflammation and cancer has been well established [2, 3]. Several chronic inflammatory diseases increase the risk of cancer, while certain anti-inflammatory therapies can decrease the risk of cancer. For example, chronic inflammation caused by inflammatory bowel disease (IBD), i.e. ulcerative colitis (UC) and Crohn’s disease, is associated with increased risks of CRC [4, 5]. Furthermore, nonsteroidal anti-inflammatory drugs reduce CRC risk, and are considered to be chemopreventive agents [4, 5], Infiltrating inflammatory cells and inflammatory mediators in the tumor microenvironment play crucial roles in colon carcinogenesis [3]. These inflammatory mediators include cytokines, chemokines, transcription factors, reactive oxygen and nitrogen species, prostaglandins, and microRNAs (miRNAs).
MiRNAs are small (19-25 nucleotides) noncoding RNAs, which regulate the translation of specific genes through sequence-specific binding to the 3’ untranslated region of target mRNAs. Expression of miRNAs has been shown to be altered in every type of human cancer that has been examined, including CRC. Specific miRNAs have been shown to have oncogenic or tumor suppressive properties, which implicates these miRNAs as key players in carcinogenesis [6]. miRNAs also have important roles in inflammatory pathways. Inflammatory stimuli lead to altered miRNA expression and certain miRNAs act as mediators of inflammation [2], miRNAs are more stable than mRNAs; therefore, they are easily detectable in formalin-fixed paraffin-embedded tissues as well as in plasma/serum by using reliable methods, including quantitative reverse-transcription PCR and microarray analysis. In view of that, miRNAs are highlighted as potentially useful biomarkers that may be sensitive and specific for early detection, prognostic classification and therapeutic decision of CRC.
This review discusses the interactive role of inflammation and altered miRNA expression in CRC, and provides a potential usefulness of miRNAs as biomarkers on the management of patients with CRC. We emphasize that oncogenic miR-21 may have a key role in colon carcinogenesis and is a potential prognostic classifier for CRC.
Inflammation and miRNAs, Linking to Colon Carcinogenesis
Colon Carcinogenesis in Sporadic and Colitis-Associated Cancer
CRCs develop through the sequential accumulation of genetic and epigenetic alterations, driving tumor initiation and progression from adenoma to adenocarcinoma. Loss of adenomatous polyposis coli (APC) tumor suppressor gene function (and subsequent β-catenin activation involved in Wnt signaling) occurs as an initiating event of adenoma, followed by activating mutations of KRAS oncogene for growth of the adenoma. Subsequently, additional pathways are dysregulated for adenoma to carcinoma progression, which include inactivation of p53 tumor suppressor as well as increased expression of an inducible inflammatory mediator, cyclooxygenase 2 (COX2). Genomic instability, including chromosomal instability and microsatellite instability (MSI), is recognized as an essential feature of cancer cells that accompanies the acquisition of these gene alterations. Colitis-associated cancer (CAC) is colon cancer that arises in the IBD patients. The risk of developing colon cancer increases with longer duration and larger extent of colitis in IBD patients. Unlike sporadic CRCs, CACs develop primarily through a cohtis-dysplasia-carcinoma sequence without the formation of adenoma. Inactivation of p53 is an important early event in CAC development [5], COX2 expression also occurs early in CAC, followed by KRAS activation and APC inactivation [5,7]. Therefore, there is considerable overlap in genetic mechanisms of the pathogenesis between CRC and CAC, despite the difference in the timing and the frequency. In addition, DNA hyper- methylation and genetic instability (chromosomal instability and MSI) are observed in both types [5,7]. Inducible nitric oxide synthese (NOS2, iNOS) is also involved in the pathogenesis of CAC and CRC. NOS2 activity is positively correlated with the TP53 mutations of G:C to A:C transition at 5-methylcytosme sites in sporadic CRC and with increased TP53 mutations in inflamed colonic mucosa of UC patients [8, 9]. In addition to producing nitric oxide, NOS2 can bind to COX2 and S-nitrosylate, enhancing its activity [10]. Recent studies have highlighted the role of cellular senescence as a physiological barrier against progression from premalignant to malignant tumor, and two main pathways, p53 and Rb, are critically involved in this process (fig. 1) [11,12]. The specific expression of p53 isoforms may signal an escape from the senescence barrier during the progression from colon adenoma to carcinoma [13], In UC, senescence may also act as an antitumorigenic to prevent the transition from low-grade to high-grade dysplasia [14].
Fig. 1.
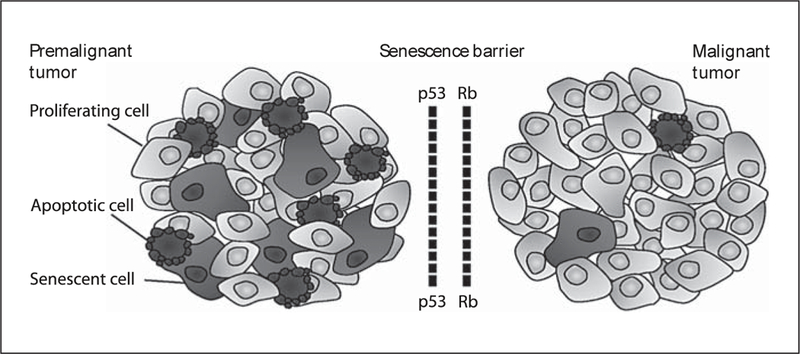
p53 and Rb tumor suppressor networks are senescence barriers to progression from premalignant to malignant tumor. Premalignant tumors can be characterized by the cells undergoing apoptosis and/or senescence. Senescence barriers may be bypassed by inactivation of p53 and Rb tumor suppressor pathways.
Inflammation has a clear role in both sporadic CRC and CAC. Both CRC and CAC tumors contain infiltrating inflammatory cells within the tumor microenvironment and these cells produce a variety of inflammatory mediators that can influence tumor progression. Both sporadic CRC and CAC tumors display increased expression of proinflammatory cytokines and exhibit constitutive activation of multiple inflammatory pathways, including nuclear factor-kappa B (NF-kB) and signal transducer and activator of transcription 3 (STAT3), which are activated by tumor necrosis factor-a (TNF-α), interleukin-1β (IL-1β) and IL-6 [5,15]. Transcription factors NF- kB and STAT3 are key players in both CRC and CAC, which can induce positive signaling loops that increase cytokines, chemokines and recruitment of inflammatory cells in the tumor microenvironment. These components of cancer-associated inflammation can promote colon carcinogenesis by regulating angiogenesis, cell proliferation and apoptosis, leading to tumor progression and metastasis [3,5,15]. Consistent with the fact that inflammation contributes to colon carcinogenesis, we have reported that the expression signature of inflammatory genes can predict CRC prognosis [16].
miRNAs Contribute to Inflammation and Colon Carcinogenesis
miRNA expression patterns are altered in inflammatory diseases, including IBD. Wu et al. [17] reported that active inflammation in UC was associated with the differential expression of 11 miRNAs, particularly, increased miR-21 and decreased miR-192 expression. Increased miR-21 was further confirmed in inflamed UC tissues as well as intestinal inflammation in Crohn’s disease [18]. Notably, elevated miR-21 expression is observed not only in the site of inflammation, including colitis, but also in many types of malignancies, including CRC, suggesting that elevated miR-21 may have a causative role in inflammation-associated carcinogenesis [2,6,19]. In fact, many in vitro studies indicate that miR-21 is an oncogenic miRNA (oncomiR), which can promote cell proliferation, inhibit apoptosis, and enhance invasion and metastasis by targeting putative tumor suppressive genes, including phosphatase and tensin homolog (PTEN), programmed cell death 4 (PDCD4), sprouty 2 (SPRY2) and others [20, 21]. Recently, Hatley et al. [22] revealed an in vivo miR-21 oncogenic pathway by using gain-of-function and loss- of-function mice of miR-21 in combination with a KRAS- induced lung cancer model. Medina et al. [23] demonstrated that inducible miR-21 overexpression was sufficient to induce pre-B cell lymphoma and was required to maintain malignancy, suggesting that tumors can be ‘addicted’ to high miR-21 expression. Consistent with its oncomiR functions, miR-21 expression can be enhanced by oncogenic signaling, including epidermal growth factor receptor (EGFR) and KRAS pathways [22, 24]. It is noteworthy that oncogenic KRAS induces not only miR-21, but also proinflammatory cytokines IL-6 and IL-8 [2]. Furthermore, various inflammatory stimuli can also induce miR-21 expression. Proinflammatory cytokines IL-6 and interferon can induce miR-21 expression in a STAT3-dependent manner [25, 26], in agreement with our report demonstrating a positive correlation between miR- 21 and IL-6 expression in colon cancer tissues [16]. Ilio-poulos et al. [27] suggested a positive-feedback loop involving inflammation and colon cancer, in which IL-6- induced STAT3 activates miR-21 and miR-181b-l, leading to NF-kB activation required to maintain the transformed state by inhibiting PTEN and cylindromatosis (CYLD) tumor suppressors, respectively. In view of those findings, evidence is accumulating that miR-21 plays an interactive role in both inflammation and carcinogenesis.
Several studies implicate altered expression of miRNAs as a causal factor in colon carcinogenesis. miR- 143 and miR-145 are downregulated in CRC and can act as tumor suppressors in colon cancer cells in vitro. These miRNAs can affect cell growth by directly targeting genes involved in multiple oncogenic pathways, such as KRAS and MYC [20, 28, 29]. Also, miR-143 can inhibit DNA methyltransferase 3A [30], and miR-145 can repress pluripotency genes in embryonic stem cells [31]. The let-7 miRNA family is another regulator of KRAS by binding to the 3´ untranslated region of KRAS, thereby blocking subsequent RAS pathway activation and inhibiting cell growth in colon cancer cell lines [6, 20]. Increased expression of COX2, a frequent event in CRC, can be negatively regulated by miR-101, suggesting that an impairment of miR-101 expression might contribute to colon carcinogenesis [32]. The oncogenic miR-17-92 cluster is transactivated by MYC and inhibits E2F1 to contribute to CRC development [33]. MiR-135a/b can directly suppress the expression of APC and induce Wnt signaling activity, suggesting their role in CRC pathogenesis [34], p53 induces miR-34a, leading to apoptosis, cell cycle arrest and senescence, while its expression is repressed by promoter hypermethylation, allelic loss or TP53 mutations [6], Overexpression of miR-155 can downregulate mismatch repair enzymes, including MSH2, MSH6 and MLH1, resulting in a mutator phenotype and MSI [35]. In addition, MSH2 expression is negatively regulated by miR-21 [36]. Collectively, there is growing evidence that miRNAs can affect crucial pathways responsible for initiation and progression of CRC.
miRNAs as Biomarkers for CRC
Circulating miRNAs and Early Detection Biomarkers
Early diagnosis can provide the increased opportunity for successful curative resection. Colonoscopy is the most reliable tool to detect CRC and has been shown to be effective at reducing deaths caused by CRC. However, the invasiveness and high cost of colonoscopies reduce screening rates. Fecal occult blood test have lower cost and are less invasiveness, but fecal occult blood test has limitations of low sensitivity which make it a less than ideal screening biomarker. Noninvasive biomarkers with high sensitivity and specificity are required to increase CRC screening rates.
Given that miRNAs are stable and detectable in serum and plasma, it raises the possibility of circulating miRNAs as noninvasive biomarkers for CRC detection [2]. Ng et al. [37] discovered that miR-I7-3p and miR-92a, both members of the oncogenic miR-17-92 cluster, were elevated in plasma in patients with CRC. These miRNAs could discriminate CRC from control subjects with 89% sensitivity and 70% specificity, demonstrating some potential of circulating miRNAs as a noninvasive test to detect CRC. Plasma levels of these two miRNAs were reduced after surgical resection, suggesting that circulating miRNAs may be potential biomarkers to detect relapse following surgery. The diagnostic value of miR-92a, along with a second candidate miR-29a, in plasma was further validated by Huang et al. [38]. Circulating miR-221 in plasma was also reported as a potential CRC biomarker [39]. Recently, a circulating miRNA study which employed two independent CRC cohorts suggested that the expression of miR-141 in plasma could be a prognostic biomarker for advanced CRC patients [40]. Future studies will have to develop standardized, sensitive and specific detection methods with relatively low cost for CRC screening.
Prognostic Utility
Although surgical resection is the only curative treatment for patients with localized CRC, many of these patients will recur and die from this disease. Adjuvant chemotherapy after surgery improves survival for TNM stage III patients, while its clinical benefit for stage II patients remains controversial [41], Therefore, developing prognostic biomarkers that can identify CRC patients at high risk for disease recurrence may identify patients that will benefit from additional chemotherapy.
We have previously published a large-scale study of miRNA expression analysis on paired tumor and nontumor tissues from two independent CRC cohorts: an American cohort using microarray to identify miRNAs that were associated with survival and a Chinese cohort using quantitative reverse-transcription PCR to validate those findings [42]. We identified five highly expressed miRNAs in tumor (miR-21, miR-20a, miR-lSlb, miR-203 and miR-106a) that were each associated with poor outcome in the American cohort miR-21 expression was validated in the Chinese cohort as it was significantly associated with worse cancer specific mortality in each cohort, independent of other clinical parameters. High miR-21 expression was also associated with worse prognosis in TNM stage II patients which demonstrates that miR-21 expression may be a promising prognostic biomarker for early-stage CRC patients. Our findings were further verified by other studies [20]. Also, miR-21 is one of the most dysregulated miRNAs and is associated with prognosis in many human cancers, thus, miR-21 might act as an oncomiR in broad malignancies, which can contribute to tumor aggressiveness and influence clinical outcome [21].
There have been a number of miRNAs identified as potential prognostic biomarkers. Decreased expression of miR-320 and miR 498 were each associated with poor survival in stage II CRC patients [43]. Other studies indicated that higher levels of miR-2Q0c, miR-17, miR-125b, miR-185, miR-215 and lower levels of miR-106a, miR- 133b were each associated with poor prognosis [20, 44-47]. However, these studies still remain to be validated. Although the association between miRNAs and CRC outcome has been largely explored, prospective studies based on standardized methods for mi RNA analysis are required to determine their usefulness in clinic.
Combining multiple validated biomarkers may provide a more accurate prognostic risk stratification of cancer patients than using single biomarkers. We have reported that the combination of an inflammatory gene signature with miR-21 expression improved predictions of cancer-specific mortality in CRC patients, including stage II patients [16]. This indicates a potential utility of protein coding and noncoding gene biomarkers, alone or in combination, to identify high-risk patients with early- stage CRC (fig. 2).
Fig. 2.
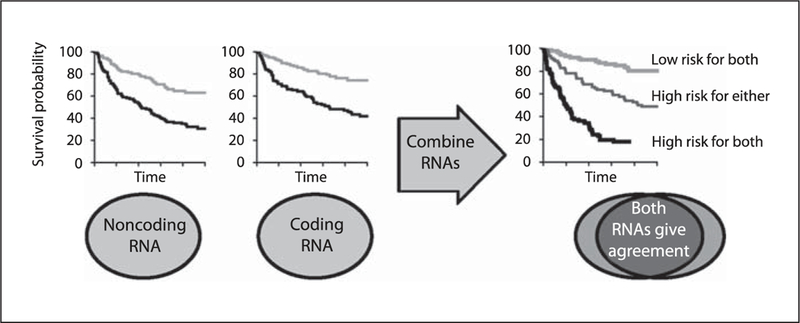
The combination of multiple, validated biomarkers may improve predictions of clinical outcomes. In this example, protein noncoding RNA and protein coding RNA expression are statistically and mechanistically independent and can be combined to improve associations with prognosis. Each biomarker misclassifies a different subset of patients and combining them provides a more robust prediction.
Therapeutic Response
Each patient may have a different susceptibility to chemotherapeutic drugs, depending on their genetic background and acquired drug resistance during tumor progression. Chemotherapy in general is costly and has significant side effects, thus highlighting the need for predictive biomarkers that can individualize treatment for each patient. For instance, KRAS mutation testing has become routine in the clinic to determine eligibility for EGFR targeting therapy, as KRAS-mutant tumors do not respond to the anti-EGFR treatment [48].
We previously reported that high miR-21 expression was associated with poor therapeutic outcome in CRC patients who received 5-fluorouracil (5-FU)-based adjuvant chemotherapy [42]. CRC with MSI phenotype, caused by the impairment of mismatch repair genes, including MSH2, are resistant to 5-FU [49]. Thus, it may be possible that miR-21 can repress MSH2, leading to che-moresistance to 5-FU [36]. The expression of let-7g and miR-181b are associated with chemoresponse to S-l, which contains a 5-FU prodrug [50].
Single-nucleotide polymorphisms in miRNAs or miRNA-binding sites may also influence prognosis and response to therapy [2, 20]. A KRAS 3´ untranslated region polymorphism in a let-7 miRNA complementary site was found to be positively associated with anti-EGFR agent cetuximab responsiveness in metastatic CRC patients with KRAS-wild-type tumor [51]. Single-nucleotide polymorphisms in pre-miR-423 and pre-miR-608 were associated with prognosis, especially in patients receiving 5-FU and platinum-based chemotherapy [52]. Single-nucleotide polymorphisms in pri-miR-26a-l and pri-miR-100 are associated with treatment outcome in metastatic CRC patients treated with 5-FU and irinote-can [53], These studies should be validated in independent populations and the underlying mechanisms responsible for the response to specific chemotherapies need to be examined before these discoveries can be translated into the clinic.
Conclusions and Future Perspectives
Although there is little doubt that miRNAs can link inflammation and colon cancer, further mechanistic studies are required to elucidate the mechanisms by which miRNAs contribute to colon carcinogenesis along with modifying the inflammatory tumor microenvironment. Also, more in vivo evidence for a causative role of miRNAs in CRC is needed.
Although recent studies have highlighted the potential usefulness of miRNAs in CRC management, further validation in prospective independent clinical panels would be the next desirable step. Future efforts will also be required to develop clinical tools based on the expression and/or genetic polymorphisms of miRNAs that can serve as sensitive, specific and noninvasive biomarkers for CRC detection as well as personalized therapeutic strategies. Among a large number of candidate miRNAs, miR-21 in particular not only represents one of the most important oncomiRs involved in CRC development and progression, but also has the potential utility as a CRC biomarker that predicts prognosis and therapeutic outcome. miR-21 is also a potential target supported by preclinical studies.
Footnotes
Disclosure Statement
The authors have no conflict of interest to declare.
References
- 1.Ferlay J, Shic HR, Bray F, et al. : Estimates of worldwide burden of cancer in 2008: Globo- can 2808. Int J Cancer 2010;127:2893–2917. [DOI] [PubMed] [Google Scholar]
- 2.Schetter AJ, Heegaard NH, Harris CC: Inflammation and cancer: interweaving mi- croRNA, free radical, cytokine and p53 path-ways. Carcinogenesis 2010;31:37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mantovaai A, Alkvena P, Sica A, et al. : Cancer-related inflammation. Nature 2008;454: 436–444. [DOI] [PubMed] [Google Scholar]
- 4.Farraye FA, Qdze RD, Eaden J, et al. : AGA technical review on the diagnosis and management of colorectal neoplasia in inflam-matory bowel disease. Gastroenterology 2010;08:746–774. 774.el-4, quiz e712-743. [DOI] [PubMed] [Google Scholar]
- 5.Uliman TA, Itakowitü SH: Intestinal inflammation and cancer. Gastroenterology 2011; 140:1807–1816. [DOI] [PubMed] [Google Scholar]
- 6.Kasinski AL, Slack FJ: MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer 2011;11:843–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goel GA, Kandiel A, Achkar JP, et al. ; Molecular pathways underlying IBD-associated colorectal neoplasia: therapeutic implications. Am J Gastroenterol 2011;106:719–730. [DOI] [PubMed] [Google Scholar]
- 8.AmbsS Bennett WP, Mercians WG, et al. : Relationship between p53 mutations and inducible nitric oxide synthase expression in human colorectal cancer. J Natl Cancer Inst 1999;91:86–88. [DOI] [PubMed] [Google Scholar]
- 9.Hussain SP, Amatad P, Raja K, et al. : Increased p53 mutation load in noncancerous colon tissue from ulcerative colitis: a cancer- prone chronic inflammatory disease. Cancer Res 2000;60:3333–3337. [PubMed] [Google Scholar]
- 10.Kim SF, Huri DA, Snyder SH: Inducible nitric oxide synthase binds, S-nitrosylates, and activates cydoaxygenase-2. Science 2005; 310:1966–1970. [DOI] [PubMed] [Google Scholar]
- 11.Collado M, Serrano M: The power and the promise of oncogene-induced senescence markers. Nat Rev Cancer 2006;6:472–476. [DOI] [PubMed] [Google Scholar]
- 12.Sohn JJ, Schetter AJ, Yfantis HG, et al. : Macrophages, nitric oxide and microRNAs are associated with DNA damage response pathway and senescence in inflammatory bowel disease. PLoS ONE 2012;7:e44156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujita K, Mondai AM, Horikawa I, et al. : p53 isoforms Dehal33p53 and p53beta are endogenous regulators of replicative cellular senescence. Nat Cell Biol 2009;11:1135–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Risqaes RA, Lai LA, Himmetoglu C, et al. : Ulcerative colitis-associated colorectal cancer arises in a field of short telomeres, senescence, and inflammation. Cancer Res 2011; 71:1669–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teradc J, Grivennikov S, Karin E, et al. : Inflammation and colon cancer. Gastroenterology 2010; 138:2101–2114.e5. [DOI] [PubMed] [Google Scholar]
- 16.Schetter AJ, Nguyen GH, Bowman ED, et al. : Association of infkmmation-rdated and microRNA gene expression with caacer-spe- cific mortality of colon adenocarcinoma. Clin Cancer Res 2009;15:5878–5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu F, Zikusoka M, Trindade A, et al. : MicroRNAs are differentially expressed in ulcerative colitis and alter expression of tnac- mphage inflammatory peptide-2 alpha. Gastroenterology 2008; 135:1624–1635.e24. [DOI] [PubMed] [Google Scholar]
- 18.Pekow JR, Kwon JH: MicroRNAs in inflammatory bowel disease. Inflamm Bowel Dis 2012;18:187–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Volinia S, Calin GA, Liu CG, et al. : A micro- H.NA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 2006;103:2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schetter AJ, Harris CC: Alterations of microRNAs contribute to colon carcinogenesis. Semin Oncol 2011;38:734–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krichevsky AM, Gabriely G: miR-21: a small multi-faceted RNA. J Cdl Mol Mad 2009;13: 39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatley ME, Patrick DM, Garcia MR, et al. : Modulation of K-Ras-dependent lung tu- morigenesis by micioRNA-21. Cancer Cell 2010;18:282–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Medina PP, Nolds M, Slack FJ: OncomiR addiction in an in vivo model of microRNA-21- induced pre-B-cell lymphoma. Nature 2010; 467:86–90. [DOI] [PubMed] [Google Scholar]
- 24.Seike M, Goto A, Okano T, et al. : miR-21 is an EGFR-regukted anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sri USA 2009; 106:12085–12090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang CH, Yue J, Fan M, et al. : IFN induces miR-21 through a signal transducer and activator of transcription 3-dependent pathway as a suppressive negative feedback on IFN-induced apoptosis. Cancer Res 2010;70: 8108–8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Löffler D, Brocke-Heidrich K, Pfeifer G, et al. : Interleukin-6 dependent survival of multiple myeloma cdls involves the STAT3-mediated induction ofmicroRNA-21 through a highly conserved enhancer. Blood 2007;110:1330–1333. [DOI] [PubMed] [Google Scholar]
- 27.IliopouLos D, Jaeger SA, Hirsch HA, et al. : STAT3 activation of miR-21 and miR-181b-l via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell 2010;39:493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sadideva M, Zhu S, Wu F, et al. : p53 represses c-Mye through induction of the tumor suppressor miR-145. ProcNstl Acad Sci USA 2009;106:3207–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X, Guo X. Zhang H, et al. : Role of miR- 143 targeting KRAS in colorectal tumori- genoris. Oncogene 2009;28:1385–1392. [DOI] [PubMed] [Google Scholar]
- 30.Ng EK, Tsang WP, Ng SS, et al. : Micro- RNA-143 targets DNA methyltransferases 3A in colorectal cancer. Br J Cancer 2009; 101:699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu N, Papagiannakopoulos T, Pan G, et al. : MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. cell 2009:137:647–658. [DOI] [PubMed] [Google Scholar]
- 32.Strillacci A, Griffoni C, Sansone P, et al. : miR- 101 downregulatioa is involved in cydooxy- genase-2 overexpression in human colon cancer cells. Exp Cdl Res 2009;315:1439–1447. [DOI] [PubMed] [Google Scholar]
- 33.Diosdado B, van de Wiel MA, Terhaar Stve Droste JS, et al. : miR-17-92 duster is associated with 13q gain and c-myc expression during colorectal adenomata adenocarcinoma progression. Br J Cancer 2009:101:707–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagel R, le Sage C, Diosdado B, et al. : Regulation of the adenomatous polyposis coli gene by the mLR-125 family in colorectal cancer. Cancer Res 2008;68:5795–5802. [DOI] [PubMed] [Google Scholar]
- 35.Valeri N, Gasparini P, Fabbri M, et al. : Modulation of mismatch repair and genomic stability by miR-155. Proc Natl Acad Sci USA 2010;107:6982–6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu Y, Wang Y, Ren X, et al. : Cantext-depen- dent bidirectional regulation of the MutS ho¬molog 2 by transforming growth factor β contributes to chemoresiftance in breast cancer cells. Mol Cancer Res 2010;8:1633–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.NgEK Chong WW, Jin H,et al. : Differential expression of mkroRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut 2009;58:1375–1381. [DOI] [PubMed] [Google Scholar]
- 38.Huang Z, Huang D, Ni S, et al. : Plasma mi- croRNAs are promising novel biomarkers for early detection of colorectal, cancer. Int J Cancer 2010;127:118–126. [DOI] [PubMed] [Google Scholar]
- 39.Pu XX, Huang GL, Guo HQ, et al. : Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J Gastroenterol Hepatol 2010;25:1674–1680. [DOI] [PubMed] [Google Scholar]
- 40.Cheng H, Zhang L, Cogdell DE, et al. : Circu lating plasma miR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS One 2011;6:e17745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dotan E, Cohen SJ: Challenges in the min- agetnent of stage II colon cancer. Semin Oncol 2011;38:511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schetter AJ, Leung SY, Sohn JJ, et al. : Micro- RNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma, JAMA 2008;299:425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sdiepder T, Reinert JT, Ostenfeid MS, et al. : Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res 2008;68: 6416–6424, [DOI] [PubMed] [Google Scholar]
- 44.Karaayvaz M, Pal T, Song B, et al. : Prognostic significance of miR-215 in colon cancer. Clin Colorectal Cancer 2011;10:340–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nishida N, Yokobori T, Mimori K, et al. : Mi- croRNA miR-125b is a prognostic marker in human colorectal cancer. Int J Oncol 2011; 38:1437–1443, [DOI] [PubMed] [Google Scholar]
- 46.Diaz R, Silva J, Garcia JM, et al. ; Deregulated oppression of miR-106a predicts survival in human colon cancer patients. Genes Chromosomes Cancer 2008;47:794–802. [DOI] [PubMed] [Google Scholar]
- 47.Akcakaya P, Ekehmd S, Kolosenko I, et al. : miR-185 and miR-133b deregulation is associated with overall survival arid metastasis in colorectal cancer. Ini J Oncol 2011:39:311–318. [DOI] [PubMed] [Google Scholar]
- 48.Engstrom PF, Arnoletti JP, Benson AB 3rd, et al. : NCCN dinted practice guidelines in oncology: colon cancer. J Natl Compr Cane Nelw 2009;7:778–831. [DOI] [PubMed] [Google Scholar]
- 49.Vilar E, Gruber SB: Microsatellite instability in colorectal cancer-the stable evidence. Nat Rev Clin Oncol 2010:7:153–162, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakajima G, Hsyashi K, Xi Y, et al. : Non-cod- ingimcroRNAs hsa-let-7g and haa-miR-lSlb are associated with chemoresponse to S-l in colon cancer. Cancer Genomics Proteomics 2006;3:317–324. [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang W, Winder T, Ning Y, et al. : A let-7 microRNA-binding site polymorphism in 3’-untranslated region of KRAS gene predicts response in wild-type KRAS patients with metastatic colorectal cancer treated with cetuximab monotherapy. Ann Oncol 2011;22:104–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xing J, Wan S, Zhou F, et al. : Genetic poly morphisms in pre-microRNA genes as prognostic markers of colorectal cancer. Cancer EpidemiolBiomarkersPrev2012;21:217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boni V, Zarate R, Villa JC, et al. : Role of primary miRNA polymorphic variants in metastatic colon cancer patients treated with 5-fhiorauradl and irinotecan. Pharmacoge- nomics J 2010;11:429–436. [DOI] [PubMed] [Google Scholar]


