Abstract
The arylalkylamine N-acyltransferases (AANATs) are enzymes that catalyze the acyl-CoA-dependent formation of N-acylarylalkylamides: acyl-CoA + arylalkylamine → N-acylarylalkylamides + CoA-SH. Herein, we describe our study of a previously uncharacterized AANAT from Bombyx mori: Bm-iAANAT3. Bm-iAANAT3 catalyzes the direct formation of N-acylarylalkylamides and accepts a broad range of short-chain acyl-CoA thioesters and amines as substrates. Acyl-CoA thioesters possessing an acyl chain length >10 carbon atoms are not substrates for Bm-iAANAT3. We report that Bm-iAANAT3 is a “versatile generalist”, most likely, functioning in amine acetylation – a reaction in amine inactivation/excretion, cuticle sclerotization, and melanism. We propose a kinetic and chemical mechanism for Bm-iAANAT3 that is consistent with our steady-state kinetic analysis, dead-end inhibition studies, determination of the pH-rate profiles, and site-directed mutagenesis of a catalytically important amino acid in Bm-iAANAT3. These mechanistic studies of Bm-iAANAT3 will foster the development of novel compounds targeted against this enzyme and other insect AANATs for the control of insect pests.
Keywords: AANAT, Bombyx mori, N-acylarylakylamine, kinetic mechanism, chemical mechanism
1. Introduction
N-Acetylamides, CH3-CO-NH-R, have been isolated and characterized from a host of vertebrates and invertebrates and are usually produced in vivo by a reaction between acetyl-CoA, CoA-S-CO-CH3, and the corresponding amine, R-NH2 [1–5]. Amine N-acetylation is catalyzed by the arylalkylamine N-acetyltransferases (AANATs, also known as arylalkylamine N-acyltransferases) [6,7] which are members of the GCN5-related N-acetyltransferase enzyme superfamily [8–10]. In insects, the iAANATs (iAANATs = insect AANATs) produce N-acetylserotonin, an intermediate in melatonin biosynthesis, and are thought to function in controlling melanism [11,12], the inactivation of biogenic amines [13,14], the sclerotization of the insect cuticle [14,15], and photoperiodism [7]. Our interest in the AANATs is derived from our work on the biosynthesis of the fatty acid amides [16,17], a family of cell signaling lipids [18]. We and others have found that AANAT-like (AANATL) enzymes will catalyze the formation of fatty acid amides from fatty acyl-CoA thioesters and the corresponding amines [17,19–21]. Database searches of insect genomes indicate that a number of iAANATs and iAANATLs could be expressed in a variety of insects [7,14,22]. Thus, insects should serve as model organisms to better understand the possible role played by the iAANATs and iAANATLs in the biosynthesis of N-acetyl- and N-acylarylalkylamides and to further define the biological function of these molecules. Much remains unknown about the cellular function of the long-chain N-acylarylalkylamides and other fatty acid amides [23].
Herein, we report on the over-expression in E. coli, purification, and characterization of recombinant Bm-iAANAT3, a third iAANAT from B. mori. B. mori express two other iAANATs, Bm-iAANAT [24] and Bm-iAANAT2 [25]. Our work on Bm-iAANAT3 contributes to an understanding of N-acyltransferase chemistry and will foster the development of inhibitors targeted against insect N-acyltransferases, due to their emergence as intriguing targets for insecticide design. This is due, in part, to their low interspecies sequence identity between iAANATs (often ≤30%), very low sequence identity when aligned against AANATs from mammals and vertebrates (≤15%), their broad range of substrates from isoform to isoform, and their relative importance in the survival of the insect. [26–28]. In addition, we plan to extend our mechanistic work on the AANATs to focus on structural elucidation by NMR spectroscopy and X-ray crystallography. The other iAANATs we have over-expressed [20,21,29–32] have thwarted such experiments because these enzymes precipitate at high protein concentrations. Bm-iAANAT3 does not precipitate at high protein concentration enabling structural studies of this enzyme by NMR [33,34].
2. Material and Methods
2.1. Materials and general methods
Unless otherwise noted, all reagents were of the highest quality available from commercial sources. Codon-optimized Bm-iAANAT3 was purchased from Genscript. Oligonucleotides were purchased from Eurofins MWG Operon. BL21 (DE3) E. coli competent cells, XL-10 E. coli competent cells, and the pET-28a(+) vector were purchased from Novagen. PfuUltra High-Fidelity DNA polymerase was purchased from Agilent. XhoI, NdeI, Antarctic phosphatase, and T4 DNA ligase were purchased from New England Biolabs. Kanamycin monosulfate and isopropyl-β-D-thiogalactopyranoside (IPTG) were purchased from Gold Biotechnology. ProBond nickel-chelating resin was purchased from Invitrogen. N-acetyltryptamine was purchased from Cayman Chemical. Acyl-CoA and amine substrates were purchased from Sigma-Aldrich. Spectrophotometric analyses were performed on a Cary 300 Bio UV-Visible spectrophotometer.
2.2. Cloning of Bm-iAANAT3
Bm-iAANAT3 (Accession no. NM_001190842.1) was codon optimized for expression in E. coli, with the addition of terminal 5’-NdeI and 3’-XhoI restriction sites, and synthesized into a pUC57 vector. The full-length gene was excised from the pUC57 vector and cloned into the NdeI and XhoI restriction sites of the pET-28a vector. The Bm-iAANAT3-pET28a vector was then transformed into E. coli XL-10 competent cells, plated on a Luria Broth (LB) agar plate supplemented with 50 μg/mL kanamycin, and grown overnight at 37°C. A single colony from each vector transformation was cultured overnight in LB media supplemented with 50 μg/mL kanamycin overnight at 37°C. The Bm-iAANAT3-pET28a plasmid was purified from the overnight cultures using the Promega Wizard Plus SV Minipreps DNA purification kit, sequenced by Eurofins MWG Operon to confirm correct gene insertion, and, finally, transformed into E. coli BL-21 (DE3) cells for the expression of Bm-iAANAT3.
2.3. Expression and purification of wild-type Bm-iAANAT3
The E. coli BL-21 (DE3) competent cells containing the Bm-iAANAT3-pET28a vector were cultured in LB media supplemented with 50 μg/mL kanamycin at 37°C. Once the culture reached an OD600 of 0.6, the cells were induced with the addition of 1.0 mM IPTG for 4 hours at 37°C. The final culture was harvested by centrifugation at 6,000 × g for 10 min. at 4°C, and the pellet frozen at −80°C for later analysis.
Cell pellets were thawed and resuspended in binding buffer: 20 mM Tris pH 7.9, 500 mM NaCl, and 5 mM imidazole. Cells were lysed by sonication and the cellular debris was pelleted by centrifugation at 16,000 × g for 20 min. at 4°C. The supernatant was collected and loaded onto a 5 mL column of ProBond nickel-chelating resin. The column was washed with 5 column volumes (CVs) of binding buffer, followed by 10 CVs of wash buffer: 20 mM Tris pH 7.9, 500 mM NaCl, and 60 mM imidazole. Finally, purified enzyme was eluted from the column in 1 mL fractions with 2–3 CVs of elution buffer: 20 mM Tris pH 7.9, 500 mM NaCl, and 500 mM imidazole. Fractions were evaluated for purity using 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The protein concentration was determined using the Bradford binding assay [35]. Fractions containing the desired, purified Bm-iAANAT3 enzyme were pooled, dialyzed overnight into 20 mM Tris pH 7.4, 200 mM NaCl, and stored at −80°C.
2.4. Substrate identification for Bm-iAANAT3
An activity-based screening protocol was used to identify substrates for Bm-iAANAT3. First, the enzyme was evaluated using acetyl-CoA (representative of a short-chain CoA) or oleoyl-CoA (representative of a long-chain CoA) separately, with different groups of pooled amines. The groups of amines are listed in Table 1. Each assay consisted of 300 mM Tris pH 8.0, 150 μM DTNB (Ellman’s reagent, 5,5’-dithiobis(2-nitro-benzoic acid), 500 μM acyl-CoA, and individual amines at 20 mM or 60 mM (Table 1). Initial velocities were determined by measuring the release of CoA at 412 nm using Ellman’s reagent (ε412 = 13,600 M−1 cm−1) and were calculated after the background rate of CoA thioester hydrolysis was subtracted from the observed velocity. Any amine pool that displayed a background corrected specific activity of ≥0.1 μmoles/min/mg (≥0.04 M−1 s−1) in combination with acetyl-CoA or oleoyl-CoA was further evaluated to identify the specific Bm-iAANAT3 substrate(s) contained within that pool. Each amine within that group was individually interrogated at a relatively high concentration in the presence of acetyl-CoA or oleoyl-CoA to determine if the amine was truly a substrate for Bm-iAANAT3. The assay conditions were identical to that described for the screening protocol using the amine pools. Individual amines that displayed a background corrected specific activity of ≥0.1 μmoles/min/mg were included in our measurements for additional kinetic analysis.
Table 1.
Pooled Amine Substrates for the Initial Screening Assay
| Pool | Concentration of Each Amine (mM) | Amino Donors |
|---|---|---|
| 1 | 60 | Lysine, Threonine, Glycine, Histamine, and Dopamine |
| 2 | 60 | Alanine, Arginine, Histidine, Tyramine, and Norepinephrine |
| 3 | 60 | Serine, Proline, Methionine, Serotonin, and Tryptamine |
| 4 | 20 | Glutamate, Asparagine, Valine, Ethanolamine, and Octopamine |
| 5 | 20 | Aspartate, Isoleucine, Tryptophan, and Glutamine |
| 6 | 60 | Leucine, Phenylalanine, γ-Aminobutyric acid, and Taurine |
| 7 | 20 | Spermidine, Agmatine, Cadaverine, Putrescine, and β-Alanine |
2.5. Determination of the steady-state kinetic constants
Steady-state kinetic characterization of Bm-iAANAT3 was carried out using Ellman’s reagent to measure the release of CoA under the following conditions: 300 mM Tris pH 8.0, 150 μM DTNB, and varying concentrations of acyl-CoA and amine substrates at 22°C.
To determine the kinetic constants for the acyl-CoA substrates, the initial tryptamine concentration was 8.0 mM, while varying the concentration of the acyl-CoA substrates. To determine the kinetic constants for the amine substrates, the initial acetyl-CoA concentration was 500 μM, while varying the amine.
The kinetic constants for the acyl-CoA and amine substrates for Bm-iAANAT3 were determined by fitting the resulting data to Equation 1 using SigmaPlot 12.0: vo represents initial velocity, Vmax is the maximal velocity, Km is the Michaelis constant, and [S] is the concentration of the varied substrate. Note that the concentration of the other substrate was fixed at relatively high (saturating) concentration. Assays were performed in triplicate and the uncertainty for the kcat and (kcat/Km) values were calculated using Equation 2, with σ as the standard error [36].
| Equation 1 |
| Equation 2 |
2.6. Bm-iAANAT3 product characterization
The product of the reaction catalyzed by Bm-iAANAT3 was generated by incubating 100 μg of enzyme with 1 mM acetyl-CoA and 1 mM tryptamine ( ~ 10 × Km for both) and 300 mM Tris, pH 8.0, to a final volume of 2 mL, for 10 minutes at 30°C. The enzyme was then removed via centrifugation using a Millipore 10 kDa spin column. The remaining solution was diluted 500-fold and 20 μL injected into a Phenomenex Kinetex 2.6 μm C18 100 Å (50 mm × 2.1 mm) reverse phase column coupled to an Agilent 6540 liquid chromatography/quadrupole time-of-flight mass spectrometry (LC/QTOF-MS) in positive ion mode. The elution gradient for liquid chromatography was as described by Dempsey et al. [20]. The resulting retention time and high-resolution mass-to-charge ratio for the sample was compared with that of a commercial standard of N-acetyltryptamine to confirm the catalytic activity of the enzyme.
2.7. Kinetic mechanism for Bm-iAANAT3
2.7.1. Double reciprocal analysis
To differentiate between sequential and classic ping-pong kinetic mechanisms, double-reciprocal plots of the initial velocity data were generated for acetyl-CoA and tryptamine in SigmaPlot 12.0. Non-linear regression analysis of initial rates and model discrimination analyses were performed in WaveMetrics IGOR Pro 6.34A. Initial velocities were determined by varying the concentration of one substrate, while holding the other substrate at a fixed concentration. The two plots were generated by holding the tryptamine concentration constant (40 μM, 60 μM, 120 μM, 250 μM, and 500 μM) and varying the concentration of acetyl-CoA. Accordingly, the second plot held the concentration of acetyl-CoA constant (15 μM, 25 μM, 40 μM, 90 μM, and 200 μM) and varied the tryptamine concentration. The resulting initial velocity data was fit to Equation 3 for an ordered bi-bi mechanism and to Equation 4 for ping-pong mechanism, where vo is the initial velocity, Vmax is the maximal velocity, [A] is the concentration of substrate A, [B] is the concentration of substrate B, Kia is the dissociation constant for substrate A, Kb is the Michaelis constant for substrate B, and Ka is the Michaelis constant for substrate A. Kia, the dissociation constant for acetyl-CoA, was fixed at 0.7 μM, based on the direct measurement of acetyl-CoA binding to Bm-iAANAT3 by isothermal calorimetry (ITC) [34].
| Equation 3 |
| Equation 4 |
2.7.2. Dead-end inhibition with oleoyl-CoA, tyrosol, and N-acetyltryptamine
To further define the kinetic mechanism of Bm-iAANAT3, we performed dead-end and product inhibition analysis using oleoyl-CoA, tyrosol, and N-acetyltryptamine. The initial velocities were generated by holding one substrate (either acetyl-CoA or tryptamine) at a fixed concentration, varying the concentration of the other substrate, at different fixed concentrations of each inhibitor. All assays were performed in triplicate, and the resulting data was fit to Equations 5 – 7 in SigmaPlot 12.0 for competitive, noncompetitive, and uncompetitive inhibition, respectively. For Equations 5 – 7, vo is the initial velocity, Vmax is the maximal velocity, Km is the Michaelis constant, [S] is the substrate concentration, [I] is the inhibitor concentration, and Ki,s is the dissociation constant for the dissociation of I from the E•I complex, and Ki,i is the dissociation constant for the dissociation of I from the E•S•I complex.
| Equation 5 |
| Equation 6 |
| Equation 7 |
2.7.3. pH dependence of the steady-state kinetic constants
The pH dependence of the kinetic constants for acetyl-CoA (in the presence of saturating 8.0 mM tryptamine) were determined at intervals of 0.5 pH units ranging from pH 6.0 to 9.5. The buffers used were the following: MES (pH 6.0 – 6.5), Tris (pH 7.0 – 9.0) and AmeP (pH 9.0 – 9.5). The resulting kinetic data was fit to Equation 8 in WaveMetrics IGOR Pro 6.34A to delineate the pKa values of measured ionizable groups. In this equation, y represents either kcat/Km or Km, c is the pH-independent plateau, H is the hydrogen ion concentration, and Ka represents the two unresolvable hydrogen ion dissociation constants.
| Equation 8 |
2.8. Cloning, expression, and purification of site-directed mutants
The E27A mutant was produced to investigate the role of Glu-27 in substrate binding and catalysis. The generation of the E27A mutant was carried out using the overlap extension method [37]. Mutants were amplified with the PfuUltra High-Fidelity DNA polymerase with the following PCR conditions: an initial denaturation step of 95°C for 2 minutes, followed by 30 cycles of PCR amplification (95°C for 30 s, 58°C for 30 s, 72°C for 1 min), and a final extension step of 72°C for 10 minutes. Primers were designed on the Agilent QuickChange Primer Design tool and purchased from Eurofins MWG Operon (Table 2). PCR products were digested with NdeI and XhoI restriction enzymes and ligated into the pET28a vector with DNA ligase acquired from New England Biolabs. The E27A mutant was expressed and purified in the same manner as wild type Bm-iAANAT3.
Table 2.
Primers for the Creation of Site-directed Mutants of Bm-iAANAT3
| Clone | Primer | Sequence |
|---|---|---|
| Wildtype | Forward | GTAGCATATGGCTGATTTTGTTGTTGT |
| Reverse | GTAGCTCGAGTTACGGCAGTTTCAG | |
| E27A | Antisense | TCCTTCTTTGCCGATGCACCGCTGAACAAAGCC |
| Sense | GGCTTTGTTCAGCGGTGCATCGGCAAAGAAGGA | |
3. Results and discussion
3.1. Cloning, expression, and purification of Bm-iAANAT3
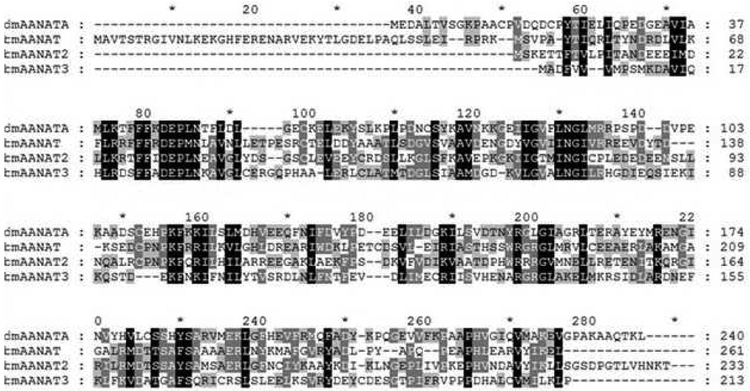
Bm-iAANAT3 from Bombyx mori was successfully cloned, expressed, and purified from E. coli using a codon-optimized gene, yielding 10.9 mg of Bm-iAANAT3 purified protein per liter of culture. Purity was ≥95% as assessed by SDS-PAGE (Figure S1, Supplementary Materials) and the molecular weight of the protein was in good agreement with its mass, 23.9 kDa for Bm-iAANAT3. Bm-iAANAT3 is a previously uncharacterized enzyme, and shares 32%, 24%, and 30% sequence identity with Bm-iAANAT, Bm-iAANAT2, and Dm-AANATA, respectively (Figure 1). It shares slightly less identity with the mammalian AANATs from human and sheep, at 21% and 17% identity, respectively. These data provide additional evidence that AANATs typically share low sequence identity when compared against each other, even between enzymes from organisms of the same phylum and class [9].
Figure 1.
Homology overlays of iAANAT enzymes from Bombyx mori and Drosophila melanogaster. The sequence data used in creating this figure are: D. melanogaster AANATA (Dm-AANATA, accession number is NP_523839), B. mori iAANAT _Bm-iAANAT, accession number is NP_001073122.1), B. mori iAANAT2 (Bm-iAANAT2, accession number is XP_004928436.2), and B. mori iAANAT3 (Bm-iAANAT3, accession number is NP_001177771.1).
3.2. Identification of amine and acyl-CoA substrates for Bm-iAANAT3
We have developed a screening strategy employing different pools of various amine substrates (arylalkylamines, polyamines, aminoglycosides, ethanolamine, histamine, and amino acids; see Table 1) that enables the rapid identification of substrates for any putative N-acyltransferase. We have successfully utilized this strategy to identify substrates for N-acyltransferases from Drosophila melanogaster [29–32].
In applying this strategy to identify amine substrates for Bm-iAANAT3, a relatively high concentration of each amine (20 mM or 60 mM, Table 1) was included in the pool because the Km value for glycine is reported at ≥6 mM for some of the glycine N-acyltransferases [38–40]. Pools of acyl-CoA substrates were not constructed because of the reports of long-chain acyl-CoAs acting as AANAT inhibitors [29,40,41]; therefore, we individually cross-screened amine pools against archetypal short and long-chain acyl-CoA thioesters, acetyl-CoA and oleoyl-CoA, because both have been reported as substrates for other GNAT family acyltransferases [20,21]. For Bm-iAANAT3, we found no initial velocity increase with oleoyl-CoA above the low enzyme-independent, background rate of oleoyl-CoA hydrolysis, suggesting that this enzyme does not catalyze the formation of long-chain N-acylamides. Employing acetyl-CoA as substrate, a broad range of amines were substrates exhibiting specific activities far above our background threshold specific activity of 0.1 μmoles/min/mg. Six arylalkylamines (tryptamine, tyramine, dopamine, octopamine, serotonin, and norepinephrine) and histamine were identified as substrates from the four amine pools that yielded Ellman-positive results. Ranking the amine substrates from the highest to the lowest (kcat/Km) values is as follows: tryptamine, tyramine, dopamine, octopamine, serotonin, norepinephrine, and histamine (Table 3). The (kcat/Km) value for the best amine substrate, tryptamine = (6.2 ± 0.6) × 105 M−1 s−1, is ~30-fold higher than the value for the worst amine substrate, histamine = (1.9 ± 0.1) × 104 M−1 s−1. Octopamine displayed the lowest Km value of 18 ± 1 μM, while histamine displayed the highest Km of 1.7 ± 0.1 mM. None of the polyamines, aminoglycosides, or amino acids we tested served as substrates for BmiAANAT3.
Table 3.
Kinetic Constants for the Amine Substrates for Bm-iAANAT3a
| Amine | Km (μM) |
kcat (s−1) |
(kcat/km) (M−1 s−1) |
Relative (kcat/km) |
|---|---|---|---|---|
| Tryptamine | 97 ± 8.8 | 60 ± 1.2 | (6.2 ± 0.6) × 105 | 33 |
| Tyramine | 63 ± 6.7 | 27 ± 0.6 | (4.3 ± 0.5) × 105 | 23 |
| Dopamine | 330 ± 23 | 55 ± 1.1 | (1.6 ± 0.1) × 105 | 8.4 |
| Octopamine | 18 ± 0.8 | 2.2 ± 0.02 | (1.2 ± 0.05) × 105 | 6.3 |
| Serotonin | 1100±63 | 59 ± 1.0 | (5.2 ± 0.3) × 104 | 2.7 |
| Norepinephrine | 140 ± 8.6 | 5.3 ± 0.07 | (3.7 ± 0.2) × 104 | 1.9 |
| Histamine | 1700±98 | 33 ± 0.68 | (1.9 ± 0.1) × 104 | 1.0 |
Kinetic constants for the amine substrates measured at one, fixed initial concentration of acetyl-CoA. Reactions were run at 22°C in 300 mM Tris pH 8.0, 150 μM DTNB, 500 μM acetyl-CoA, and the amine concentration was varied in the range 0.3 to 3.0 × Km.
Interestingly, acetyl-CoA displayed the highest Km value of the acyl-CoA substrates, 90 ± 4 μM. The other four acyl-CoA substrates, buytryl-CoA, hexanoyl-CoA, octanoyl-CoA, and decanoyl-CoA, exhibited nearly identical Km values of 10 to 14 μM (Table 5). As we have observed with other insect AANATs [20,29–32], the (kcat/Km)acyl-CoA values decreased as the length of the acyl chain increased for the acyl-CoA substrates: the (kcat/Km)acetyl-CoA is 170-fold higher than the (kcat/Km)decanocyl-CoA (Table 5). Decanoyl-CoA was the acyl-CoA substrate with the longest acyl-chain that showed rates of CoA-SH formation that were significantly above the background rate of activity of enzyme-independent acyl-CoA hydrolysis. In addition, we found that malonyl-CoA is a substrate for Bm-iAANAT3 with a (kcat/Km)malonyl-CoA that is ~6-fold lower than the (kcat/Km)decanocyl-CoA (Table 5). To the best of our knowledge, only one other GNAT enzyme, aminoglycoside 2’-N-acetyltransferase (AAC(2’)-Ic), will accept malonyl-CoA as a substrate [42]. N-Succinoylation, however, has been demonstrated in the detoxification of mammalian xenobiotics [43,44], N-succinoyl conjugates of octopomine, serotonin, dopamine, and tyramine have been identified in C. elegans [45], N-succinoylserotonin (bufobutanoic acid) is found in the venom of various toad species (the traditional Chinese medicine, Ch’an Su) [46], and crystallographic results identified succinoyl-CoA bound in two GNAT enzymes [47,48]. Our data showing that malonyl-CoA is a Bm-iAANAT3 substrate in combination with these other works hints that a panel of N-malonylated and N-succinoylated amines may exist in both vertebrates and invertebrates.
Table 5.
Kinetic Constants for the CoA Thioester Substrates (R-CO-S-CoA) for Bm-iAANAT3a
| Acyl-CoA | R | Km (μM) |
kcat (s−1) |
(kcat/km) (M−1 s−1) |
Relative (kcat/km) |
|---|---|---|---|---|---|
| Acetyl | CH3- | 90 ± 3.6 | 64 ± 0.7 | (7.1 ± 0.3) × 105 | 170 |
| Malonyl | HOOC-CH2- | 1900 ± 300 | 13 ± 1.5 | (7.0 ± 1.3) × 103 | 0.17 |
| Butyryl | CH3(CH2)2- | 14 ± 0.8 | 4.0 ± 0.04 | (2.9 ± 0.2) × 105 | 70 |
| Hexanoyl | CH3(CH2)4- | 13 ± 0.5 | 1.3 ± 0.01 | (9.5 ± 0.4) × 104 | 23 |
| Octanoyl | CH3(CH2)6- | 10 ± 1.0 | 0.36 ± 0.005 | (3.4 ± 0.3) × 104 | 8.1 |
| Decanoyl | CH3(CH2)8- | 12 ± 5.5 | 0.05 ± 0.004 | (4.2 ± 1.9) × 104 | 1.0 |
Kinetic constants for the amine substrates measured at one, fixed initial concentration of tryptamine. Reactions were run at 22°C in 300 mM Tris pH 8.0, 150 μM DTNB, 8.0 mM tryptamine, and the acyl-CoA concentration was varied in the range 0.3 to 3.0 × Km.
Our work on Bm-iAANAT3 follows earlier studies of two other iAANATs from B. mori, Bm-iAANAT [24] and Bm-iAANAT2 [25]. One of these, Bm-iAANAT, exhibited a wide expression pattern and accepted a diverse set of amines as substrates for N-acetylation [24]. The combination of its broad tissue distribution and amine substrate promiscuity led to the suggestion that Bm-iAANAT is the major enzyme of monoamine catabolism in B. mori [24]. The N-acetylation of dopamine is of particular significance in monoamine catabolism. The production of N-acetyldopamine is a reaction critical to cuticular sclerotization in insects [15]. Dopamine is required for the formation of the brown pigment, dopamine melanin, while N-acetyldopamine is the precursor for a colorless pigment [49]. In fact, B. mori defective in dopamine biosynthesis have a colorless and improperly formed cuticle [50]. The N-acetylation of serotonin is one of the reactions in the conversion of tryptophan to melatonin [2,6]. Since Bm-iAANAT catalyzes the acetylation of both serotonin and dopamine, the enzyme is likely involved in melatonin and dopamine melanin production in B. mori. Bm-iAANAT mutants exhibit higher than normal levels of dopamine and widespread black pigmentation in the larvae and adult moths [11,50]. Later, a second B. mori iAANAT was discovered and characterized, Bm-iAANAT2 [25]. Transgenic flies suppressing Bm-iAANAT2 expression exhibited elevated dopamine levels and melanin deposition in the head and integument. These results suggest that Bm-iAANATL2 contributes to the regulation of melanism in B. mori.
Our substrate specificity studies of Bm-iAANAT3 show that this enzyme is a “versatile generalist” capable of acylating a broad range of amines, with a preference for acetyl-CoA as a co-substrate. All amines included in Table 3, apart from tryptamine, have been identified in B. mori [51–55]; thus, it is possible that Bm-iAANAT3 does catalyze their acetylation in vivo. Information regarding the cellular concentration of the amines we have identified as Bm-iAANAT3 substrates is limited. The hemolymph concentrations of dopamine, tyramine, and serotonin from the fourth instar, fifth instar, and pupa stages are low, <10 μM [51], but it is not clear how these concentrations relate to their respective intracellular concentrations. There are two reports of the brain concentrations of biogenic amines in B. mori, all reported as pg/brain [51,52]. If the volume of a B. mori brain is ~1 μL, the highest concentration of dopamine and serotonin would be ~0.8 mM. These estimated intracellular concentrations for dopamine and serotonin are comparable to the Km values we determined for these amines, Km for dopamine = 0.33 and Km for serotonin = 1.1 mM (Table 3). Thus, Bm-iAANAT3 could have a role in the acetylation of dopamine and serotonin in vivo; therefore, functioning in sclerotization, pigmentation, and melatonin production.
3.3. Product Characterization
The product formed from by Bm-iAANAT3-catalyzed acetylation of tryptamine was confirmed via LC/QToF-MS in positive ion mode and compared to that of a commercial standard of N-acetyltryptamine. The enzymatically generated N-acetyltryptamine was successfully identified in the sample with an equivalent retention time and m/z compared to the commercial standard (Table 4).
Table 4.
Bm-iAANAT3 product characterization
| Sample | Retention Time (min) | Mass/Charge [M + H] (m/z) |
|---|---|---|
| N-acetyltryptamine Standard | 3.487 | 203.1188 |
| Bm-iAANAT3 Assaya | 3.438 | 203.1187 |
Assay conditions – 300 mM Tris, pH 8.0, 1 mM acetyl-CoA, 1 mM tryptamine, 100 μg Bm-iAANAT3
3.4. Kinetic mechanism
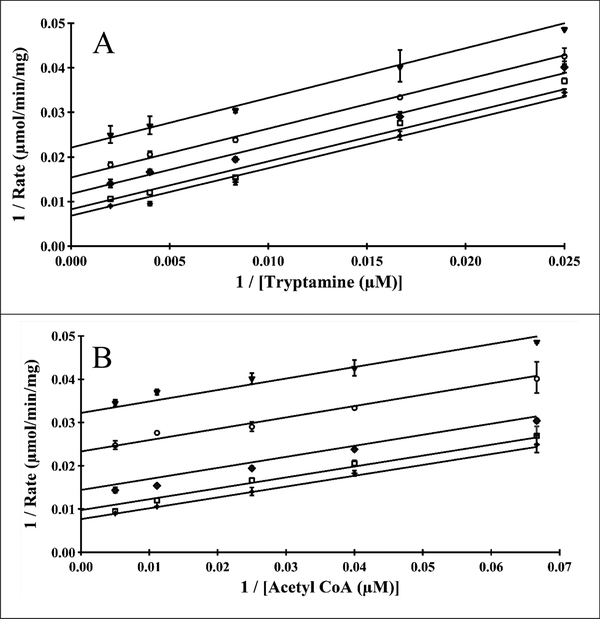
Two different experimental approaches were used to deduce the kinetic mechanism of Bm-iAANAT3. One set of experiments was to vary one substrate, acetyl-CoA or tryptamine, while holding the other substrate at a fixed initial concentration. Our initial velocity data were subjected to global non-linear regression analysis to discriminate between several bi-bi kinetic mechanisms [56]. The data fit equally well to Equation 3 for a steady-state ordered kinetic mechanism and Equation 4 for a ping-pong mechanism; Chi-squared values for the fits to equations 3 and 4 were the same within experimental error. An equilibrium ordered kinetic mechanism would yield one plot of 1/v vs. 1/[substrate] that intersect on the 1/v-axis, clearly inconsistent with the data shown in Figure 2. A pattern of parallel lines for the plot of 1/v vs. 1/[substrate] at fixed concentrations of the second substrate is often indicative of a ping-pong kinetic mechanism; however, a steady-state ordered kinetic mechanism can yield a parallel pattern if the Km for substrate A is greater than the dissociation constant for substrate A [56]. This is exactly what we found for acetyl-CoA, Km = 90 μM and Kd = 0.7 μM [34].
Figure 2.
Bm-iAANAT3 double-reciprocal analysis of initial velocities for acetyl-CoA and tryptamine, fit to equation 3 in a global non-linear regression analysis. (A) Velocities measured at fixed concentrations of tryptamine: 500 μM (□), 250 μM (✦), 120 μM (◆), 60 μM (◯) and 40 μM (▼). (B) Velocities measured at fixed concentrations of acetyl-CoA: 200 μM (✦), 90 μM (□), 40 μM (◆), 25 μM (◯) and 15 μM (▼).
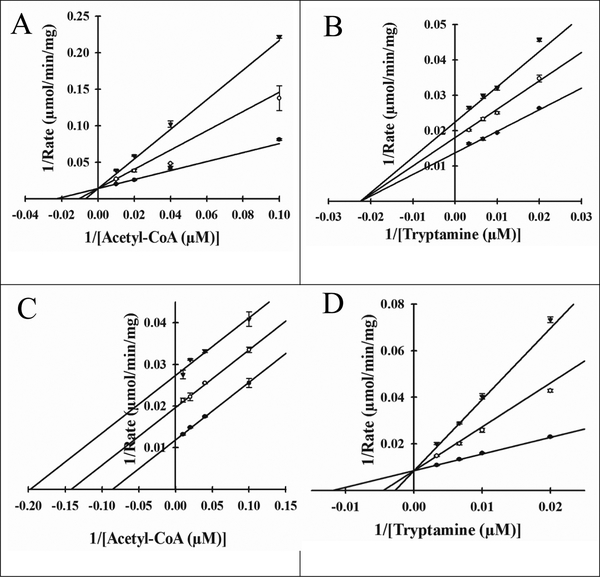
Next, a set of dead-end inhibition studies were performed to provide additional support for an ordered kinetic mechanism. Oleoyl-CoA and tyrosol were exploited as dead-end inhibitors against acetyl-CoA and tryptamine, respectively. Neither analog exhibited a rate of reaction above the baseline rate of hydrolysis, and both have been previously utilized as dead-end inhibitors in studies of the D. melanogaster AANATs [29,31,32] and mammalian serotonin N-acetyltransferase [41]. Oleoyl-CoA was competitive vs. acetyl-CoA (Figure 3A) and noncompetitive vs. tryptamine (Figure 3B) with a Ki,s value of 220 ± 20 nM and Ki,s = Ki,i values of 790 ± 30 nM, respectively. Tyrosol was uncompetitive vs. acetyl-CoA (Figure 3C) and competitive vs. tryptamine (Figure 3D), with a Ki,i value of 230 ± 10 μM and a Ki,s value of 90 ± 10 μM, respectively. Furthermore, these data indicate that enzymatic generation of N-acetyltryptamine from acetyl-CoA and tyramine as catalyzed by Bm-iAANAT3, most likely, occurs via an ordered sequential mechanism with acetyl-CoA binding first followed by the binding of tryptamine to yield the catalytically competent Bm-iAANAT3•acetyl-CoA•tryptamine ternary complex. An ordered sequential kinetic mechanism has been attributed to several D. melanogaster AANATs [29–32] and GNAT enzymes [57–60].
Figure 3.
Dead-end inhibition plots for Bm-iAANAT3. (A) Velocities measured at a fixed concentration of tryptamine (100 μM) with varied concentrations of acetyl-CoA (10 μM, 25 μM, 50 μM, and 100 μM), at varied concentrations of the inhibitor, oleoyl-CoA: 0 nM (●), 250 nM (◯), and 500 nM (▼); Ki = 220 ± 20 nM (B) Velocities measured at a fixed concentration of acetyl-CoA (100 μM) with varied concentrations of tryptamine (50 μM, 100 μM, 150 μM, and 300 μM), at varied concentrations of the inhibitor, oleoyl-CoA: 0 nM (●), 250 nM (◯), and 500 nM (▼), Ki = 790 ± 30 nM (C) Velocities measured at a fixed concentration of tryptamine (100 μM) with varied concentrations of acetyl-CoA (10 μM, 25 μM, 50 μM, and 100 μM), at varied concentrations of the inhibitor, tyrosol: 0 μM (●), 150 μM (◯), and 300 μM (▼); Ki = 230 ± 10 μM (D) Velocities measured at a fixed concentration of acetyl-CoA (100 μM) with varied concentrations of tryptamine (50 μM, 100 μM, 150 μM, and 300 μM), at varied concentrations of the inhibitor, tyrosol: 0 μM (●), 150 μM (◯), and 300 μM (▼); Ki = 90 ± 10 μM
Product inhibition is one method useful in defining the order of product release. N-Acetyltryptamine produced competitive and noncompetitive inhibition plots against acetyl-CoA and tryptamine, respectively (Figure. S2, Supplementary Materials). These data are most consistent with an ordered sequential product release with CoA-SH being released first followed by release of the acetylated product, N-acetyltryptamine. This is similar to the order of product release reported for N-myristoyltransferase [60]. We found that order of product release for D. melanogaster agmatine N-acetyltransferase (AgmNAT) was the N-acetylated product first followed by CoA-SH [32], similar to that reported for human histone acetyltransferase (HAT) [58] and Enterococcus faecium aminoglycoside acetyltransferase (AAC(6’)-I) [59]. The product inhibition patterns for Bm-iAANAT3 (Figure S2) differed from those reported for AgmNAT, HAT, and AAC(6’)-I.
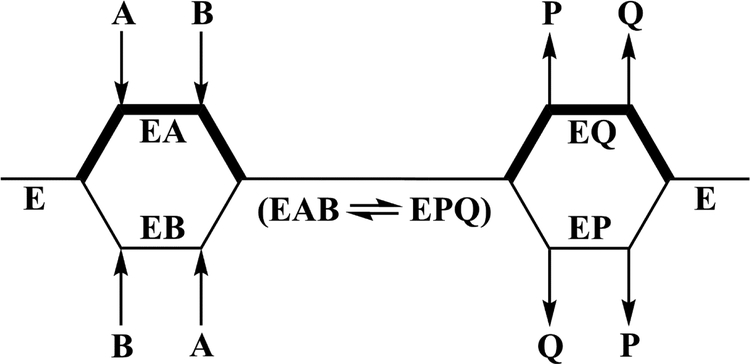
A steady-state ordered bi-bi kinetic mechanism with acetyl-CoA binding first and N-acetyltryptamine being released last (Scheme 1) is consistent with the binding data of Aboalroub et al. [34]. Direct binding measurements by ITC and/or NMR showed that acetyl-CoA and N-acetyltryptamine bound to Bm-iAANAT3 with Kd values of 0.7 ± 0.03 μM and 12 ± 3 μM. The binding of N-acetyltryptamine was eliminated by the addition of acetyl-CoA and was not affected by the addition of CoA-SH. The binding of tryptophol, the non-substrate alchohol analog of tryptamine, was significantly enhanced by the presence of acetyl-CoA, the Kd value decreasing from 230 ± 12 μM to 50 ± 2 μM. Lastly, CoA-SH binds to Bm-iAANAT3 with low affinity, a Kd of 30 ± 1 μM. Relatively weak binding of tryptamine, Kd = 220 ± 8 μM, and CoA SH to free Bm-iAANAT indicates that kinetic mechanism is actually random bi-bi with a strong preference for acetyl-CoA binding first and N-acetyltryptamine being released last (Scheme 1).
Scheme 1.
Proposed kinetic mechanism for the Bm-iAANAT3-catalyzed N-acetylation of tryptamine. The thick lines in the scheme represent a preferred pathway in a random kinetic mechanism with acetyl-CoA binding first and N-acetyltryptamine being released last. A = Acetyl-CoA, B = tryptamine, P = CoA-SH, and Q = N-acetyltryptamine.
3.5. pH dependence of the kinetic constants and identification of a catalytic base
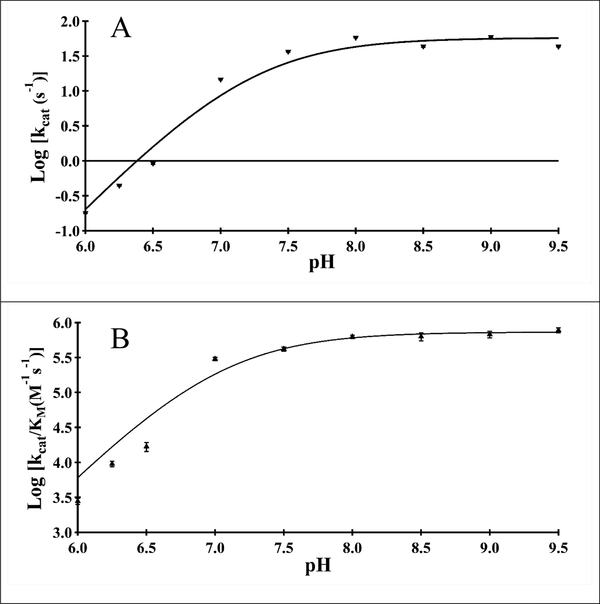
We determined the pH-dependence of the kinetic constants for acetyl-CoA to aid in our identification of amino acids important in catalysis and substrate binding in Bm-iAANAT3. Both the kcat, and (kcat/Km)acetyl-CoA pH-rate profiles exhibited a pH-dependent rise with a slope of 2 from pH 6.25 to 7.5 following by a pH-independent plateau above pH 7.5. These data are best fit to equation 8, pointing towards two unresolvable amino acids that are deprotonated for optimal catalytic activity. For both profiles, a pKa value of 7.2 ± 0.1 provide the fit to the data (Figure 4). These pKa values likely correspond to a general base deprotonating the primary amine of tryptamine; alternatively, it could be attributed to the zwitterionic tetrahedral intermediate that is produced after the nucleophilic attack of the acetyl-CoA thioester by tryptamine. This set of experiments did not uncover a third, higher pKa value; this additional pKa (~8.5 – 10.0) would correspond to a general acid in catalysis, routinely observed for NAT enzymes [9,31,61]. We hypothesize that Bm-iAANAT3 either lacks a general acid in catalysis, or the general acid has a pKa value > 9.5, a number too high for us to ascertain due to limitations of acetyl-CoA hydrolysis and/or DTNB instability at elevated pH ranges.
Figure 4.
pH rate profiles for Bm-iAANAT3. (A) log(kcat) versus pH for acetyl-CoA. (B) log(kcat/Km) versus pH for acetyl-CoA.
3.6. Identification of a catalytic base and a potential chemical mechanism for catalysis
Alignment of the amino acid sequence of Bm-iAANAT3 to those for other insect AANATs shows that Glu-27 of Bm-iAANAT3 corresponds to Glu-47 of AANATA from D. melanogaster (Figure 1). Dempsey et al. [29] have shown that Glu-47 is likely the catalytic base in that enzyme. Thus, we mutated Glu-27 to Ala in Bm-iAANAT3. The E27A mutant enzyme was cultured, purified, and characterized in the same manner as the wild-type Bm-iAANAT3. The kinetic constants we measured for the E27A mutant were: Km,acetyl-CoA = 20 ± 2.2 μM, kcat,acetyl-CoA = 4.0 ± 0.09 × 10−2 s−1, Km, = 9400 ± 990 μM, and k −2 tryptamine , = 8.0 ± 0.3 × 10 −1 cat tryptamine s . Thus, the E27A mutant retained only 0.06% to 0.13% the wild-type kcat while exhibiting 100-fold increase in the Km for tryptamine. The significant decline in catalytic activity as compared to wild-type Bm-iAANAT3, in addition to the large perturbation in the Km,tryptamine, suggests that this residue functions as a general base in catalysis, as well as playing a role in substrate binding. Glu-27 likely represents the pKa value of ~7.2 observed in the pH-rate profile (Figure 4). The ~1000-fold decrease in kcat value for the E27A mutant falls in the range of values measured for general base mutants of other insect N-acyltransferases in GCN5 family (Table 6). A larger kcat effect was measured after the mutation of the catalytic base in mandelate racemase, a ~10,000-fold decrease in the kcat of the D270N mutant [62]. Given the results for the D270N mutant in mandelate racemase, the general base mutants of Bm-iAANAT3 and the other N-acyltransferases in Table 6 have surprisingly high kcat values. Most likely, this results from another chemical species serving as a base in catalysis. We suggest that the second pKa observed in our pH-rate profiles, also with a pKa value of ~7.2, is likely the adjacent aspartate residue (Asp-26), that serves as a “back-up” or redundant role in Bm-iAANAT catalysis. Similarly, Scheibner et al. [61] argue that His-120 is the catalytic base in the mammalian N-acyltransferase, serotonin N-acetyltransferase, and that the nearby His-122 is catalytically redundant, serving as the base in the H120A mutant.
Table 6.
Ratio of kcat,wildtype/kcat,mutant for General Base Mutant Enzymes
| Enzymea | Mutation | (kcat)wildtype/(kcat)mutant | Ref. |
|---|---|---|---|
| Dm-AgmNAT | E34A | ~2100 | 32 |
| Bm-iAANAT3 | E27A | ~1000 | This work |
| Dm-AANATL7 | E26A | ~22 | 31 |
| Dm-AANATA | E47A | ~13–170 | 29,63 |
| Dm-AANATL2b | E29A | ~1.0 | 30 |
Dm = Drosophila melanogaster and Bm = Bombyx mori
Glu-29 is not, most likely, the catalytic base for Dm-AANATL2.
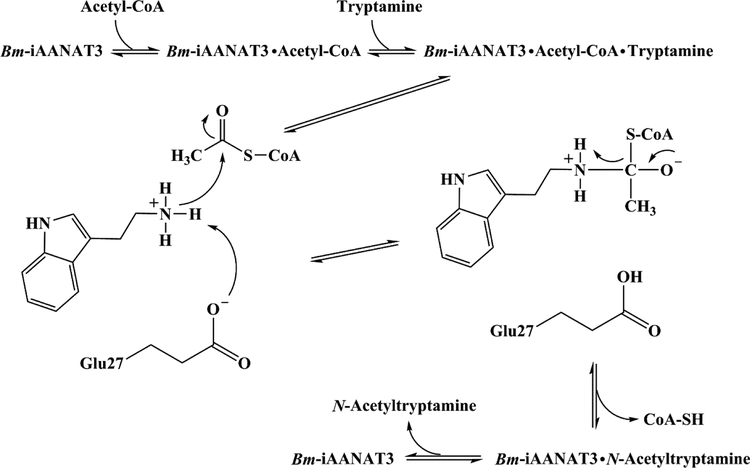
When taken together, our data strongly supports the chemical mechanism depicted in Scheme 2. The double-reciprocal plots and dead-end inhibition analysis indicate a preference for an ordered sequential mechanism: acetyl-CoA binds first, followed by tryptamine, leading to the Bm-iAANAT3•acetyl-CoA•tryptamine ternary complex before catalysis occurs. After the binding of both substrates into the active site, the positively charged amine moiety of tryptamine is deprotonated by Glu-27, the catalytic base. The amine deprotonation step is followed by nucleophilic attack of the deprotonated amine at the carbonyl of the acetyl-CoA thioester to generate a tetrahedral intermediate. The zwitterionic tetrahedral intermediate collapses as the coenzyme A thiolate is protonated by the positively charged amine of the intermediate to yield the two products: CoA-SH and N-acetyltryptamine. Our product inhibition studies show that product release has a preferred order with CoA-SH exiting first, followed by N-acetyltryptamine.
Scheme 2.
Proposed chemical mechanism for the Bm-iAANAT3-catalyzed N-acetylation of tryptamine.
4. Conclusions
We have cloned, over-expressed in E. coli, and characterized a previously unknown iAANAT from B. mori, Bm-iAANAT3. The kinetic mechanism for Bm-iAANAT3 with acetyl-CoA and tryptamine is random bi-bi, but with a strong preference for a steady-state ordered mechanism with acetyl-CoA binding first and N-acetyltryptamine released last. Studies of the chemical mechanism suggest that Glu-27-mediated deprotonation of the amine substrate facilitates nucleophilic attack of the amine at the carbonyl of the acetyl-CoA thioester. Collapse of the resulting tetrahedral intermediate immediately leads to product release, the N-acetylated amine likely leaving first, followed by the CoA-SH by-product.
Substrate specificity data reveal that Bm-iAANAT3 prefers short-chain acyl-CoA thioesters with acetyl-CoA as the best acyl-CoA substrate. While fatty acid amides have been recently identified in B. mori [21], Bm-iAANAT3 is probably not involved in their biosynthesis in vivo. Bm-iAANAT3 accepts a variety of amines as substrates pointing towards a cellular role for this enzyme in amine inactivation and/or excretion. In addition, Bm-iAANAT3 may have role in sclerotization, melanism, and melatonin biosynthesis by catalyzing dopamine and serotonin acetylation. Our identification of malonyl-CoA as a Bm-iAANAT3 substrate suggests that a series of biologically-occurring N-malonylated and N-succinoylated amines await discovery and characterization; a suggestion consistent with the identification of N-succinoylated amines in C. elegans. Why B. mori would express three iAANATs, BmiAANAT, Bm-iAANAT2, and Bm-iAANAT3, with potentially overlapping cellular functions is unknown. Future Bm-iAANAT3 knockdown-experiments in B. mori are planned to address the question of its cellular function.
Supplementary Material
Highlists:
Bm-iAANAT3 exhibits promiscuous substrate specificity for acyl-CoA and amine substrates.
Bm-iAANAT3 catalyzes the formation of N-acetyltryptamine through a bi-bi ordered sequential kinetic mechanism, with acetyl-CoA binding first and the N-acetyltryptamine released last.
pH-activity profiles for Bm-iAANAT3 suggest a chemical mechanism with Glu-27 functioning as a base during catalysis.
5. Acknowledgements
This work has been supported, in part, by grants from the Florida Center for Excellence for Biomolecular and Targeted Therapeutics (FCoE-BITT grant no. GALS020), the University of South Florida (a Creative Scholarship Grant from the College of Arts and Sciences), the Shirley W. and William L. Griffin Foundation, National Institute of Drug Abuse at the National Institutes of Health (R03-DA034323) and National Institute of General Medical Science of the National Institutes of Health (R15-GM107864) to D.J.M. The authors want to thank Dr. Ioannis Gelis and Dr. Daniel R. Dempsey for critical readings of the manuscript.
Abbreviations:
- AANAT
arylalkylamine N-acetyltransferase
- AANATA
arylalkylamine N-acetyltransferase variant A
- AANATL
arylalkylamine N-acyltransferase like
- AgmNAT
agmatine N-acetyltransferase
- CoA
coenzyme A
- DTNB
5,5’-dithiobis(2-nitro-benzoic acid)
- GNAT
GCN5-related N-acetyltransferase
- HAT
histone N-acetyltransferase
- iAANAT
insect arylalkylamine N-acetyltransferase
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SNAT
serotonin N-acetyltransferase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. References
- [1].Tabor H, Mehler AH, Stadtman ER, E.R., The enzymatic acetylation of amines. J. Biol. Chem 204 (1953) 127–138. [PubMed] [Google Scholar]
- [2].Weissbach H, Redfield BG, Axelrod J, The enzymatic acetylation of serotonin and other naturally occurring amines. Biochim. Biophys. Acta 54 (1961) 190–192. [DOI] [PubMed] [Google Scholar]
- [3].Evans PH, Fox PM, Enzymatic N-acetylation of indolealkylamines by brain homogenates of the honeybee, Apis mellifera. J. Insect. Physiol 21 (1975) 343–353. [Google Scholar]
- [4].Puiroux J, Moreau R, Gourdoux L, Simultaneous estimation of dopamine, serotonin and related compounds in the brain of Pieris brassicoe pupae: degradation pathways of biogenic amines in insect nervous tissue. Biogenic Amines 8 (1992) 391–400. [Google Scholar]
- [5].Sloley DB, Metabolism of monoamines in invertebrates: the relative importance of monoamine oxidase in different phyla. NeuroToxicology 25 (2004) 175–183. [DOI] [PubMed] [Google Scholar]
- [6].Zheng WP, Cole PA, Serotonin N-acetyltransferase: mechanism and inhibition. Curr. Med. Chem 9 (2002) 1187–1199. [DOI] [PubMed] [Google Scholar]
- [7].Hiragaki S, Suzuki T, Mohanded AAM, Takeda M, 2015. Structures and functions of insect arylalkylamine N-acetyltransferase (iaaNAT); a key enzyme for physiological and behavioral switch in arthropods. Front. Physiol 6 (2015) 113(DOI: 10.3389/fphys.2015.00113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Dyda F, Klein DC, Hickman AB, GCN5-related N-acetyltransferases: a structural overview. Ann. Rev. Bioph. Biom 29 (2000) 81–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Vetting MW, de Carvalho LPS, Yu M, Hegde SS, Magnet S, Roderick SL, Blanchard JS, Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys 433 (2005) 212–226. [DOI] [PubMed] [Google Scholar]
- [10].Ud-Din AI, Tikhomirova A, Roujeinikova A, Structure and functional diversity of GCN5-related N-acetyltransferases. Int. J. Mol. Sci 17 (2016) 1018 (DOI: 10.3390/ijms17071018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dai F.-y., Qiao L, Tong X-L, Cao C, Chen P, Chen J, Lu C, Xiang Z-H, Mutations of an arylalkylamine-N-acetyltransferase, Bm-iAANAT, are responsible for silkworm melanism mutant. J. Biol. Chem 285 (2010) 19553–19560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhan S, Guo Q, Li M, Li M, Li J, Miao X, Huang Y, Disruption of an N-acetyltransferase gene in the silkworm reveals a novel role in pigmentation. Development 137 (2010) 4083–4090. [DOI] [PubMed] [Google Scholar]
- [13].Hearse DJ, Weber WW, Multiple N-acetyltransferases and drug metabolism. Biochem. J 132 (1973) 519–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Smith TJ, Phylogenetic distribution and function of arylalkylamine N-acetyltransferase. BioEssays 12 (1990) 30–33. [DOI] [PubMed] [Google Scholar]
- [15].Andersen SO, Insect cuticular sclerotization: a review. Insect Biochem. Mol. Biol 40 (2010) 166–178. [DOI] [PubMed] [Google Scholar]
- [16].Jeffries KA, Dempsey DR, Behari AL, Anderson RL, Merkler DJ, Drosophila melanogaster as a model system to study long-chain fatty acid metabolism. FEBS Lett. 588 (2014) 1596–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jeffries KA, Dempsey DR, Farrell EK, Anderson RL, Garbade GJ, Gurina TS, Gruhonjic I, Gunderson CA, Merkler DJ, Glycine N-acyltransferase-like 3 is responsible for long-chain N-acylglycine formation in N18TG2 cells. J. Lipid Res. 57 (2016) 781–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Iannotti FA, Di Marzo V, Petrosino S, Endocannabinoids and endocannabinoid-related mediators: targets, metabolism and role in neurological disorders. Prog. Lipid Res. 62 (2016) 107–128. [DOI] [PubMed] [Google Scholar]
- [19].Waluk DP, Schultz N, Hunt MC, Identification of glycine N-acyltransferase-like 2 (GLYATL2) as a transferase that produces N-acyl glycines in humans. FASEB J. 24 (2010) 2795–2803. [DOI] [PubMed] [Google Scholar]
- [20].Dempsey DR, Jeffries KA, Anderson RL, Carpenter A-M, Rodriguez Opsina S, Merkler DJ, Identification of an arylalkylamine N-acyltransferase from Drosophila melanogaster that catalyzes the formation of long-chain N-acylserotonins. FEBS Lett. 588 (2014) 594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Anderson RL, Battistini MR, Wallis DJ, Shoji C, O’Flynn BG, Dillashaw JE, Merker DJ, Bm-iAANAT and its Potential Role in Fatty Amide Biosynthesis in Bombyx mori. Prostaglandins Leukot. Essent. Fatty Acids 135 (2018) 10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Han Q, Robinson H, Ding H, Christensen BM, Li J, Evolution of insect arylalkylamine N-acetyltransferases: structural evidence from the yellow fever mosquito, Aedes aegypti. Proc. Natl. Acad. Sci. USA 109 (2012) 11669–11674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Witkamp R, Fatty acids, endocannabionoids and inflammation. Eur. J. Pharmacol 785 (2016) 96–107. [DOI] [PubMed] [Google Scholar]
- [24].Tsugehara T, Iwai S, Fujiwara Y, Mita K, Takeda M, Cloning and characterization of insect arylalkylamine N-acetyltransferase from Bombyx mori. Comp. Biochem. Physiol. B Biochem. Mol. Biol 147 (2007) 358–366. [DOI] [PubMed] [Google Scholar]
- [25].Long Y, Li J, Zhao T, Li G, Zhu Y, A new arylalkylamine N-acetyltransferase in silkworm (Bombyx mori) affects integument pigmentation. Appl. Biochem. Biotechnol 175 (2015) 3447–3457. [DOI] [PubMed] [Google Scholar]
- [26].Tsugehara T, Imai S, Takeda M, Characterization of arylalkylamine N-acetyltransferase from silkworm (Antheraea pernyi) and pesticidal drug design based on the baculovirus-expressed enzyme. Comp. Biochem. Physiol. C Toxicol. Pharmacol 157 (2013) 93–102. [DOI] [PubMed] [Google Scholar]
- [27].Lourenço BLA, Silva MVAS, deOliveira EB, de Assis Soares WR, Góes-Neto A, Santos G, Andrade BS, Virtual screening and molecular docking for arylalkylamine-N-acetyltransferase (aaNAT) inhibitors, a key enzyme of Aedes (Stegomyia) aegypti (L.) metabolism. Comput. Mol. Biosci 5 (2015) 35–44. [Google Scholar]
- [28].O’Flynn BG, Hawley AJ, Merkler DJ, Insect arylalkylamine N-acetyltransferases as potential targets for novel insecticide design. Biochem. Mol. Biol. J 4 (2018) 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Dempsey DR, Jeffries KA, Bond JD, Carpenter A-M, Rodriguez-Ospina S, Breydo L, Caswell KK, Merkler DJ, Mechanistic and structural analysis of Drosophila melanogaster arylalkylamine N-acetyltransferases. Biochemistry 53 (2014) 7777–7793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Dempsey DR, Carpenter A-M, Rodriguez Ospina S, Merkler DJ, Probing the chemical mechanism and critical regulatory amino acid residues of Drosophila melanogaster arylalkylamine N-acyltransferase like 2. Insect Biochem. Mol. Biol 66 (2015) 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Dempsey DR, Jeffries KA, Handa S, Carpenter A-M, Rodriguez-Ospina S, Breydo L, Merkler DJ, D.J., Mechanistic and structual analysis of a Drosophila melanogaster enzyme, arylalkylamine N-acetyltransferase like 7, an enzyme that catalyzes the formation of N-acetylarylalkylamides and N-acetylhistamine. Biochemistry 54 (2015) 2644–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Dempsey DR, Nichols DA, Battistini MR, Pemberton O, Rodriquez Ospina S, Zhang X, Carpenter A-M, O’Flynn BG, Leahy JW, Kanwar A, Lewandowski EM, Chen Y, Merkler DJ, Structural and mechanistic analysis of Drosophila melanogaster agmatine N-acetyltransferase, an enzyme that catalyzes the formation of N-acetylagmatine. Sci. Rep 7 (2017) 13432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Aboalroub AA, Zhang Z, Keramisanou D, Gelis I, Backbone resonance assignment of an insect arylalkylamine N-acetyltransferase from Bombyx mori reveals conformational heterogeneity. Biomol. NMR Assign. 11 (2017) 105–109. [DOI] [PubMed] [Google Scholar]
- [34].Aboalroub AA, Bachman AB, Zhang Z, Keramisanou D, Merkler DJ, Gelis I. Acetyl group coordinated progression through the catalytic cycle of an arylalkylamine N-acetyltransferase. PLoS One 12 (2017) e0177270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bradford MM, A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem 72 (1976) 248–254. [DOI] [PubMed] [Google Scholar]
- [36].Barrante JR, Applied Mathematics for Physical Chemistry, third ed Pearson Prentice Hall, Upper Saddle River, NJ, 2004. [Google Scholar]
- [37].Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR, Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77 (1989) 51–59. [DOI] [PubMed] [Google Scholar]
- [38].Kelley M, Vessey DA, Characterization of the acyl-CoA-amino acid N-acyltransferase from primate liver mitochondria. J. Biochem. Toxicol 9 (1994) 153–158. [DOI] [PubMed] [Google Scholar]
- [39].van der Westhuizen FH, Pretorius PJ, Eramus E, The utilization of alanine, glutamic acid, and serine as amino acid substrates for glycine N-acyltransferase. J. Biochem. Mol. Toxicol 14 (2000) 102–109. [DOI] [PubMed] [Google Scholar]
- [40].Dempsey DR, Bond JD, Carpenter A-M, Rodriguez Ospina S, Merkler DJ, Expression, purification, and characterization of mouse glycine N-acyltransferase in Escherichia coli. Protein Expr. Purif 84 (2014) 9–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ferry G, Loynel A, Kucharczyk N, Bertin S, Rodriguez M, Delagrange P, Galizzi J-P, Jacoby E, Volland J-P, Lesieur D, Renard P, Canet E, Fauchere J-L, Boutin JA, Substrate specificity and inhibition studies of human serotonin N-acetyltransferase. J. Biol. Chem 275 (2000) 8794–8805. [DOI] [PubMed] [Google Scholar]
- [42].Hegde SS, Javid-Majd F, Blanchard JS, Overexpression and mechanistic analysis of chromosomally encoded aminoglycoside 2’-N-acetyltransferase (AAC(2’)-Ic) from Mycobacterium tuberculosis. J. Biol. Chem 276 (2001) 45876–45881. [DOI] [PubMed] [Google Scholar]
- [43].Pozo ÓJ, Ibáñez M, Sancho JV, Lahoz-Beneytez J, Farré M, Papeseit E, de la Torre R, Hernández F, Mass spectrometric evaluation of mephedrone in vivo human metabolism: identification of phase I and phase II metabolites, including a novel succinyl conjugate. Drug Metab. Dispos 43 (2014) 248–257. [DOI] [PubMed] [Google Scholar]
- [44].Linhart I, Himl M, Židková M, Balíková M, Lhotková E, Metabolic profile of mephedrone: identification of nor-mephedrone conjugates with dicarboxylic acids as a new type of xenobiotic phase II metabolites. Toxicol. Lett 240 (2016) 114–121. [DOI] [PubMed] [Google Scholar]
- [45].Artyukhin AB, Yim JJ, Srinivasan J, Izrayelit Y, Bose N, von Reuss SH, Jo Y, Jordan JM, Baugh LR, Cheong M, Sternberg PW, Avery L, Schroeder FC, Succinylated octopamine ascarosides amd a new pathway of biogenic amine metabolism in Caenorhabditis elegans. J. Biol. Chem 288 (2013) 18778–18783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kamano Y, Morita H, Takano R, Kotake A, Nogawa T, Hashima H, Takeya K, Itokawa H, Pettit GR, Bufobutanoic acid and bufopyramide, two new indole alkaloids from the Chinese traditional drug Ch’an Su. Heterocycles 50 (1999) 499–503. [Google Scholar]
- [47].Vetting MW, Errey JC, Blanchard JS, Rv802c from Mycobacterium tuberculosis: the first structure of a succinyltransferase with the GNAT fold. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun 64 (2008) 978–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Iqbal A, Arunlanantham H, Brown T Jr., Chowdhury R, Clifton IJ, Kershaw NJ, Hewitson KS, McDonough MA, Schofield CJ, Crystallographic and mass spectrometric analyses of a tandem GNAT protein from the clavulanic acid biosynthesis pathway. Proteins 78 (2010) 1398–1407. [DOI] [PubMed] [Google Scholar]
- [49].Sugumara M, Barek H, Critical analysis of the melanogenic pathway in insects and higher animals. Int. J. Mol. Sci 17 (2016) 1753 (doi: 10.3390/ijms17101753). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Fuji T, Abe H., Kawamoto M, Katsuma S, Banno Y, Shimada T, Albino (al) is a tetrahydrobiopterin (BH4)-deficient mutant of the silkworm Bombyx mori. Insect Biochem. Mol. Biol 43 (2013) 594–600. [DOI] [PubMed] [Google Scholar]
- [51].Takeda N, Takaoka H, Shimizu T, Yazawa M, Yahi S, Biogenic amine levels in the central nervous system and haemolymph of the silkworm, Bombyx mori. Comp. Biochem. Physiol. C 100 (1991) 677–682. [Google Scholar]
- [52].Shimizu T, Takeda N, Yagi S, Levels of biogenic amines in the brain during pupal and adult development of the silkworm, Bombyx mori. Z. Naturforsch. C 52 (1997) 279–282. [Google Scholar]
- [53].Hirashima A, Hirokado S, Ohta H, Suetsugu E, Sakaguchi M, Kuwano E, Taniguchi E, Eto M, Titres of biogenic amines and ecdysteroids: effect of octopamine on the production of ecdysteroids in the silkworm Bombyx mori. J. Insect Physiol 45 (1999) 843–851. [DOI] [PubMed] [Google Scholar]
- [54].Gatellier L, Nagao T, Kanzaki R, Serotonin modifies the sensitivity of the male silkmoth to pheromone. J. Exp. Biol 207 (2004) 2487–2496. [DOI] [PubMed] [Google Scholar]
- [55].Iwano M, Kanzaki R, Immunocytochemical identification of neuroactive substances in the antennal lobe of the male silkworm moth Bombyx mori. Zool. Sci 22 (2005) 199–211. [DOI] [PubMed] [Google Scholar]
- [56].Cook PF, Cleland WW, Enzyme Kinetics and Mechanism. Garland Science, New York, 2007. [Google Scholar]
- [57].De Angelis J, Gastel J, Klein DC, Cole PA, Kinetic analysis of the catalytic mechanism of serotonin N-acetyltransferase (EC 2.3.1.87). J. Biol. Chem 273 (1998) 3045–3050. [DOI] [PubMed] [Google Scholar]
- [58].Tanner KG, Langer MR, Kim Y, Denu JM, Kinetic mechanism of the histone acetyltransferase GCN5 from yeast. J. Biol. Chem 275 (2000), 22048–22055. [DOI] [PubMed] [Google Scholar]
- [59].Draker K.-a., Northrop DB, Wright GD, Kinetic mechanism of the GCN5-related chromosomal aminoglycoside acetyltransferase AAC(6’)-Ii from Enterococcus faecium: evidence of subunit cooperativity. Biochemistry 42 (2003) 6565–6574. [DOI] [PubMed] [Google Scholar]
- [60].Rocque WJ, McWherter CA, Wood DC, Gordon JI, A comparative analysis of the kinetic mechanism and peptide substrate specificity of human and Saccharomyces cerevisiae myristoyl-CoA:protein N-myrostoyltransferase. J. Biol. Chem 268 (1993) 9964–9971. [PubMed] [Google Scholar]
- [61].Scheibner KA, De Angelis J, Burley SK, Cole PA, Investigation of the roles of catalytic resides in serotonin N-acetyltransferase. J. Biol. Chem 277 (2002) 18118–18126. [DOI] [PubMed] [Google Scholar]
- [62].Schafer SL, Barrett WC, Kallarakal AT, Mitra B, Kozarich JW, Gerlt JA, Clifton JG, Petsko GA, Kenyon GL, Mechanism of the reaction catalyzed by mandelate racemase: structure and mechanistic properties of the D270N mutant. Biochemistry 35 (1996) 5662–5669. [DOI] [PubMed] [Google Scholar]
- [63].Cheng K-C, Liao J-N, Lyu P-C, Crystal structure of the dopamine N-acetyltransferase acetyl-CoA complex provides insight into the catalytic mechanism. Biochem. J 446 (2012) 395–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.