Summary
DACH1 abundance is reduced in human malignancies, including breast cancer. Herein DACH1 was detected among multipotent fetal mammary stem cells in the embryo, among mixed lineage precursors, and in adult basal cells and (ERα+) luminal progenitors. Dach1 gene deletion at 6 weeks in transgenic mice reduced ductal branching, reduced the proportion of mammary basal cells (Lin− CD24med CD29high) and reduced abundance of basal cytokeratin 5, whereas DACH1 overexpression induced ductal branching, increased Gata3 and Notch1, and expanded mammosphere formation in LA-7 breast cells. Mammary gland-transforming growth factor β (TGF-β) activity, known to reduce ductal branching and to reduce the basal cell population, increased upon Dach1 deletion, associated with increased SMAD phosphorylation. Association of the scaffold protein Smad anchor for receptor activation with Smad2/3, which facilitates TGF-β activation, was reduced by endogenous DACH1. DACH1 increases basal cells, enhances ductal formation and restrains TGF-β activity in vivo.
Keywords: DACH, mammary gland, breast cancer, TGF-β, SARA, stem cells
Highlights
-
•
Dach1 is expressed in mammary gland fetal stem cells and adult luminal cells
-
•
Dach1 expands mammary gland basal/myoepithelial cells
-
•
Dach1 induces post-natal mammary gland ductal formation
-
•
Dach1 retrains TGF-β activity in the mammary gland in vivo
DACH1 abundance, is reduced in human malignancies, including breast cancer. In this article, Dr. Pestell and his colleagues showed that DACH1 is expressed in multipotent mammary gland fetal stem cells and both the adult basal and ERα+ luminal cells, promotes mammary gland stem cell and basal/myoepithelial cell expansion, promotes mammary gland ductal branching, and restrains TGF-β action in the pubertal mammary gland.
Introduction
The Drosophila dac gene is a key member of the retinal determination gene network, which also includes eyes absent, eyeless, twin of eyeless, teashirt, and sine oculis. Initially cloned as a dominant inhibitor of a hyperactive egfr allele in Drosophila, dac interacts with the epidermal growth factor receptor, decapentaplegic, and Wingless pathways (Chen et al., 1997, Chen et al., 1999). Dac functions to promote organismal development (Davis and Rebay, 2017), and Dac mutant flies have atretic organs (Davis and Rebay, 2017). Reduced DACH1 (the mammalian ortholog of Dac) expression has been observed in several malignancies including breast, prostate, lung, and endometrial cancer (Chen et al., 2013, Nan et al., 2009, Wu et al., 2006, Wu et al., 2009, Wu et al., 2013, Wu et al., 2014, Wu et al., 2015) (reviewed in Wu et al., 2015). Clinical studies have demonstrated a correlation between poor prognosis and reduced expression of DACH1 in breast cancer (Wu et al., 2006), and DACH1 re-expression was sufficient to inhibit breast cancer tumor metastasis in mice (Wu et al., 2008). The mammalian DACH1 regulates expression of target genes in part through intrinsic DNA sequence-specific binding to Forkhead binding sites and in part through interacting with DNA-binding transcription factors (c-Jun, SMADs, Six, and ERα) (Popov et al., 2009, Wu et al., 2006, Wu et al., 2009, Zhou et al., 2010b).
Dach1 homozygous null mice die at birth, indicating that DACH1 governs essential functions in the organism; however, no morphologic and metabolic alterations have been observed in the analyzed organs (Davis et al., 2001). Given the precedent for Dac promoting organismal development, we sought to define the role for DACH1 function in normal development by examining the role of DACH1 in normal post-natal mammary gland development. Given the importance of mammary stem cells in normal mammary gland development (Visvader and Stingl, 2014), and the prior studies demonstrating that DACH1 restrains breast cancer stem cell expansion (Wu et al., 2011), we conducted careful analysis of the mammary gland developmental hierarchy through generating temporally regulated Dach1fl/fl transgenics.
The current studies were conducted to determine the role of DACH1 in normal mammary gland development. These studies revealed an unexpected role for DACH1 to expand the murine mammary gland progenitor cell pool, and to promote ductal formation. We show that endogenous DACH1 restrains transforming growth factor β (TGF-β) signaling in the murine mammary gland and show that Dach1 governs SARA (also known as the zinc finger FYVE domain-containing protein 9 [ZFYVE9]) abundance and binding to Smad2/3. Given the importance of TGF-β signaling in development and disease, the finding herein that endogenous DACH1 restrains TGF-β signaling in vivo may have broad importance to human disease.
Results
Temporally Regulated Excision of the Dach1 Gene in the Murine Mammary Gland Reduces Cell Proliferation and Ductal Branching
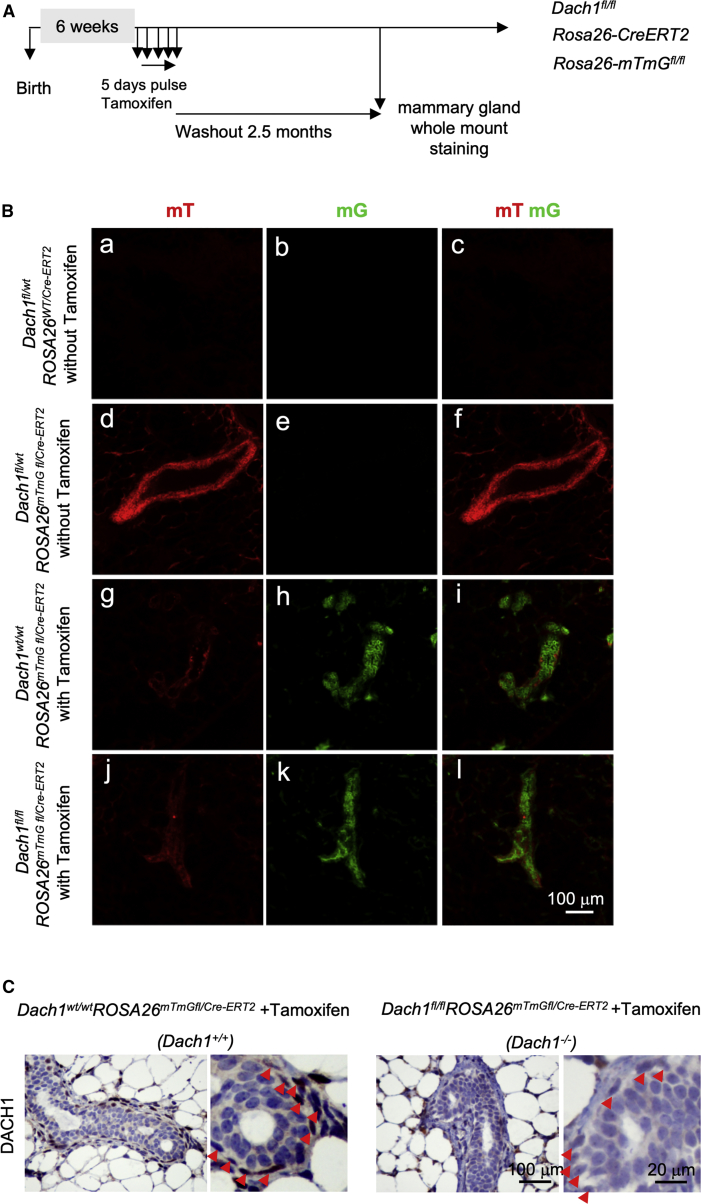
To examine the physiological role of DACH1 in post-natal mammary gland development, transgenic mice were developed in which Dach1fl/fl transgenics (Chen et al., 2015) were intercrossed with the ROSA26CreERT2 transgenics. This mouse strain expresses Cre-ERT2 from the ubiquitously expressed ROSA26 locus. Cre activity utilizes a mutant estrogen hormone-binding domain (ERT) to keep Cre inactive unless the non-steroidal estrogen analog 4-hydroxytamoxifen is present. To follow the efficiency of temporal and spatial regulation of Cre recombination in vivo and in primary cells derived from these mice, bitransgenic mice were intercrossed with double-fluorescent Cre reporter mice (ROSA26-mTmGfl/fl). The ROSA26-mTmGfl/fl transgenics express membrane-targeted tandem dimer Tomato (mT), and after Cre-mediated excision express membrane-targeted GFP (mG) (Muzumdar et al., 2007) (Figure S1A). Tri-transgenic mice at 6 weeks of age were treated with a pulse of tamoxifen and analyzed after a subsequent 2.5 months (Figure 1A). Previous studies had shown that this dose of tamoxifen was without effect on mammary repopulating numbers or on pubertal ductal development at this time of analysis (Shehata et al., 2014). Furthermore, comparison was made herein between tamoxifen-treated transgenics and tamoxifen-treated control transgenics throughout to avoid any potential independent effect of tamoxifen. Genomic DNA analysis of these mice demonstrated the excision of the first exon of Dach1 (Figure S1B). Mammary gland fluorescence without tamoxifen was red throughout the mammary gland and epithelial cells (Figure 1B). Dach1wt/wt-ROSA26CreERT2 mice, which were treated with tamoxifen as a control in the studies, showed efficient excision of the mT transgene and conversion to green fluorescence throughout the mammary gland, without alteration in Dach1 abundance (Figure 1B). Treatment of Dach1fl/fl-ROSA26CreERT2 mice with tamoxifen resulted in the induction of GFP in the mammary gland (Figure 1Bf versus Bi Bl) and DACH1 protein, identified by immunohistochemistry as primarily in the basal cells, was abrogated upon tamoxifen treatment (Figure 1C).
Figure 1.
Inducible Dach1 Deletion in Mouse Mammary Gland
(A) Schematic representation of the tamoxifen treatment schedule for the multigenic Dach1 transgenics (Dach1fl/flROSA26CreERT2/mT-mGfl). (B) Fluorescence microscopy showed the tamoxifen-induced Cre recombinase activity by Tomato-GFP color transformation. (a–c) Dach1fl/wtROSA26wt/CreERT2 mammary gland without tamoxifen treatment (negative control without Cre reporter and Cre induction) showing GFP (mG) and tomato red fluorescence (mT) are both negative. (b–f) Dach1fl/wtROSA26CreERT2/mTmGfl mammary gland without tamoxifen treatment (negative control without Cre induction) showing presence of mT without mG. (g–i) Dach1wt/wtROSA26CreERT2/mTmGfl and (j–l) Dach1fl/flROSA26CreERT2/mTmGfl mammary gland with tamoxifen treatment used for the Dach1 deletion mice analysis shows strong mG and weak mT. The combined images showing mT to mG switch in the mammary ducts after tamoxifen treatment. (C) Immunohistochemical staining for DACH1 protein in the mammary gland of the treated mice (Dach1wt/wtROSA26CreERT2/mTmGfl and Dach1fl/flROSA26CreERT2/mTmGfl).
DACH1 Is Expressed in Multipotent Fetal Mammary Stem Cells and in Both the Adult Basal and ERα+ Luminal Cells
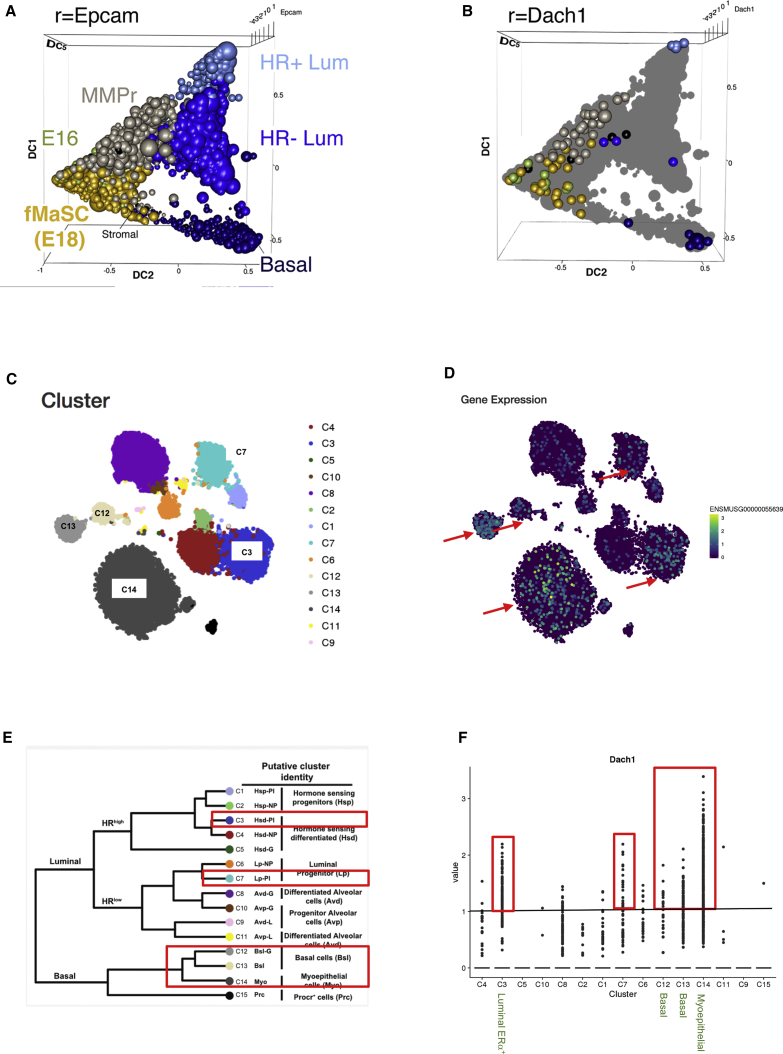
In order to examine the expression of Dach1 by mammary gland cell type, we interrogated two recently published single-cell RNA sequencing (scRNA-seq) studies that had identified mammary gland cellular subtypes (Bach et al., 2017, Giraddi et al., 2018). scRNA-seq transcriptomes annotated by stage of development were generated from Epcam+ mammary epithelial cells (MECs), derived from developing (embryonic day 16 [E16] and E18), post-natal day (P4) and adult mouse mammary tissues (Giraddi et al., 2018) (Figure 2A). The accession number for these data is GEO: GSE106273 and GEO: GSE111113. Relatedness of individual cell transcriptomes was plotted according to diffusion components (DCs) using the webtool (http://uofuhealth.utah.edu/huntsman/labs/spike/d3.php) as previously described (Giraddi et al., 2018). The diffusion map provides a noise-tolerant, nonlinear dimensionality reduction method, revealing a global topology of the data based on local similarities between individual Epcam+ MECs. In this map, DC1 reflects a continuum of relative basal to luminal character. DC2 reflects developmental time from primitive (E16 and E18) at one extreme to P4 to adult mammary cells at the other extreme of the DC2 axis. The abundance of Dach1 transcripts was then assessed in the diffusion map, in order to show the correlation with stage of development and relative abundance in the luminal versus basal subtypes (Figure 2B). Dach1 expression was detected in early developmental stages including among multipotent fetal mammary stem cells (fMaSCs) in the embryo and mixed lineage precursors (MMPr) prior to puberty (Figure 2B). Dach1 expression was also detected in adult basal cells and ERα+ luminal progenitors.
Figure 2.
Dach1 RNA Transcripts in Mammary Epithelial Cells during Development
(A) Diffusion map of single-cell RNA sequencing (scRNA-seq) transcriptomes annotated by stage of development. Epcam+ mammary epithelial cells (MECs) were derived from developing (embryonic day 16 [E16] and E18), post-natal day (P4) and adult MECs (Giraddi et al., 2018), and scRNA-seq was plotted according to diffusion components (DCs) using software (http://uofuhealth.utah.edu/huntsman/labs/spike/d3.php) as described previously (Giraddi et al., 2018). DC1 represents a continuum of relative enrichment from basal to luminal subtype. DC2 represents developmental time from primitive (E16 and E18) to P4 adult mammary cells occupy ends of the DC2 axis.
(B) The abundance of Dach1 transcripts was mapped upon the diffusion map scRNA-seq transcriptomics, in order to show the correlation with stage of development and relative abundance in the luminal versus basal subtypes. Dach1 is expressed among multipotent fetal mammary stem cells in the embryo and among mixed lineage precursors prior to puberty Dach1 expression was detected in adult basal cells (dark blue dots in lower-right corner) and (ERα+) luminal progenitors (light blue dots in upper-right corner).
(C–F) Dach1 RNA transcripts are expressed during post-natal development. (C) scRNA-seq of MECs derived from nulliparous, mid gestation, lactation, and post-involution mammary glands identified 15 clusters of MECs (Bach et al., 2017). (D) Relative expression of Dach1 within each cluster type is shown colorimetrically, with relative expression shown in the bar to the right. (E) Dendrogram of clusters based on the log-transformed mean expression values of the 15 clusters. The red blocks show the cluster subtype in which Dach1 is expressed based on the relative abundance of transcripts shown in (F). Dach1 gene expression was detectable primarily in presumptive basal and myoepithelial cells (clusters C12, C13, and C14), and to a lesser degree also in luminal ERα+ (cluster C3) and luminal progenitor clusters (C6/7).
We next assessed Dach1 transcript abundance in a second distinct transcriptomic analysis. The second study determined the gene expression profile of MECs across four adult developmental stages; nulliparous, mid gestation, lactation, and post involution (Bach et al., 2017). The analysis of 23,184 adult cells identified 15 clusters representing different transcriptional cell states (Figure 2C). Dach1 gene expression was detectable primarily in presumptive basal and myoepithelial cells (clusters C12, C13, and C14), and to a lesser degree also in luminal ERα+ (cluster C3) and luminal progenitor clusters (C6/7) (Figures 2D–2F).
Collectively these studies are consistent between distinct datasets and illustrate that Dach1 is expressed in fMaSCs and in both the adult basal and ERα+ luminal cells.
DACH1 Promotes Mammary Gland Stem Cell and Basal/Myoepithelial Cell Expansion
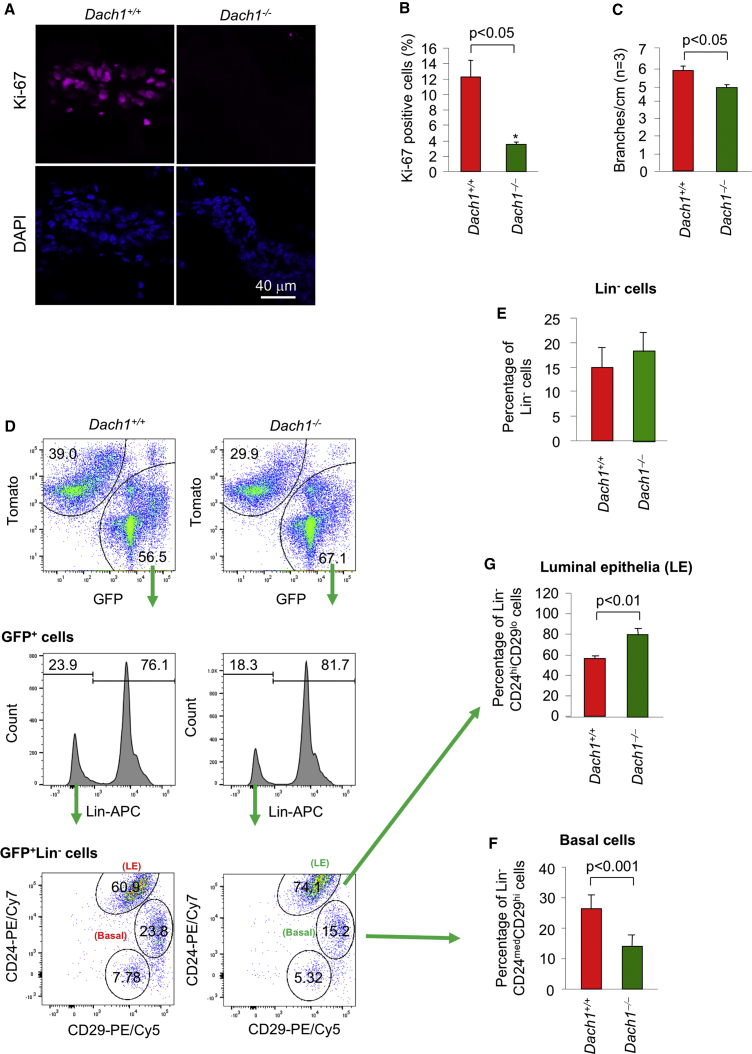
To determine the functional consequence of Dach1 gene deletion, cellular proliferation was assessed using Ki-67 immunohistochemistry. Ki-67 staining was reduced >75% upon excision of the Dach1 gene (Figures 3A and 3B). Apoptosis, as assessed by TUNEL staining, was very low in control mice and was not affected by Dach1 deletion. (Figures S2A and S2C). Treatment of tissues from Dach1 deletion mice (Dach1fl/fl; ROSA26mTmGfl/Cre-ERT2 + tamoxifen) with DNase1, as a form of positive control, induced TUNEL staining in all the epithelial cells (Figures S2B and S2C). Mammary squashes were prepared (Figures S3A and S3B). A careful counting of branches revealed a significant decrease in branching in the Dach1−/− mammary gland (Figure 3C).
Figure 3.
Dach1 Gene Deletion Reduces Mammary Gland Cellular Proliferation, Ductal Branching, and Myoepithelial/Stem Cells
(A) Cellular proliferation assessed by Ki-67 immunostaining with nuclei marked by DAPI staining (A).
(B) As above, but quantified as mean ± SEM for n = 3 separate mice in each group. Data are shown for Dach1wt/wtROSA26CreERT2/mTmGfl versus Dach1fl/flROSA26CreERT2/mTmGfl after tamoxifen treatment as shown in Figure 1A.
(C) Ductal branching analysis (shown as mean ± SEM for n = 3 mice). Representative examples are shown at high magnification in Figure S3.
(D) A representative fluorescence-activated cell sorting (FACS) analysis from the mammary epithelium showing the separation of mT from mG cells, the subsequent separation of Lin− cells and the apportioning of CD24/CD29 status.
(E) The proportion of Lin− cells (cells from mammary gland excluding vascular and hematopoietic cells) was determined by FACS analysis and quantified for n = 4 separate mice. The proportion of Lin− cells was not significantly altered between genotypes.
(F) The percentage of Lin−CD24medCD29hi cells was determined upon FACS analysis for n = 4 separate mice in each group.
(G) Luminal epithelial cells are shown as Lin−CD24hi CD29lo with data as mean ± SEM for n = 4 separate mice.
To determine the role of endogenous DACH1 on the proportion of breast cellular subtypes, fluorescence-activated cell sorting (FACS) analysis was conducted on the mammary gland from transgenic mice (Figure 3D). To enrich for basal cells, Lin− cells were selected (Figures 3D and 3E), with subsequent gating for CD29high and CD24medium (basal) cells from tamoxifen-treated control (Dach1wt/wt-Rosa26mTmGfl/Cre-ERT2) and Dach1fl/fl transgenic (Dach1fl/fl-Rosa26mTmGfl/Cre-ERT2) mice (Figure 3F). Several studies have shown the multilineage potential of cells in this subpopulation (Shackleton et al., 2006, Visvader and Stingl, 2014). This population, herein referred to as basal cells, has previously been shown to be enriched with bipotential mammary stem cells (MaSCs) and gives rise to unipotent stem cells or long-lived progenitors for the basal/myoepithelial and luminal lineages (Visvader and Clevers, 2016). The relative proportion of Lin− CD24med CD29high was reduced >40% in Dach1−/− mammary gland (Figure 3D). Quantitation of the additional cell subtypes, based on epitope markers and FACS analysis revealed an increase in the luminal epithelial pool (CD24high CD29low) in the Dach1−/− mammary gland (Figure 3G).
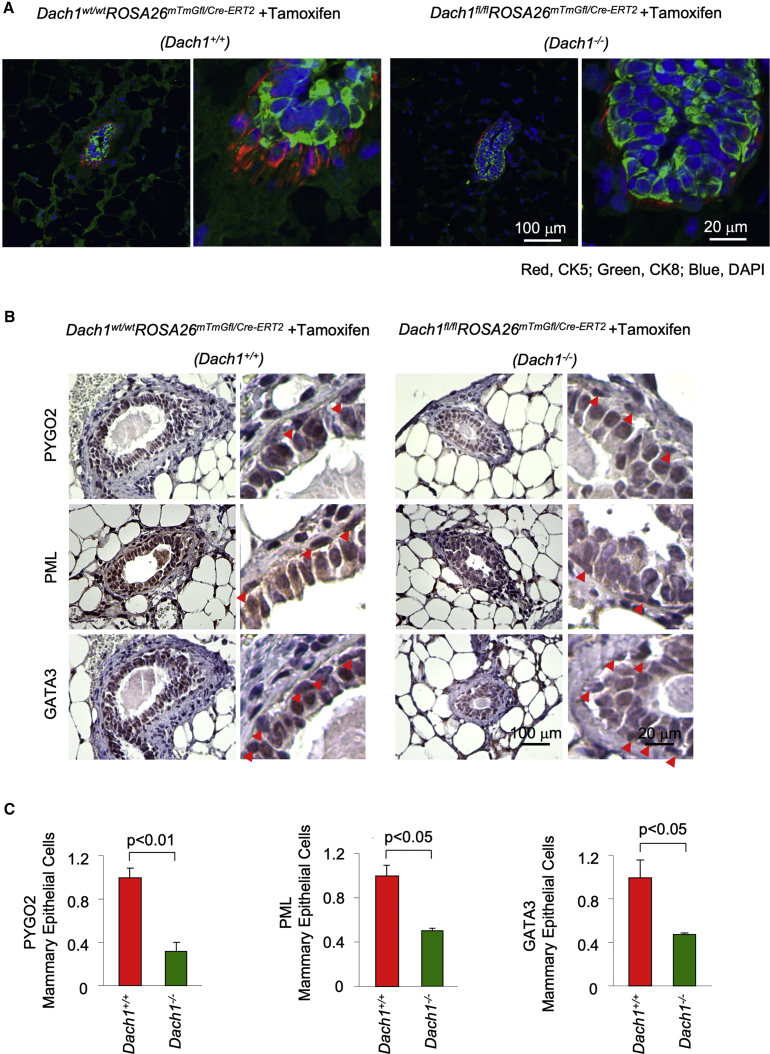
To examine further the impact of endogenous DACH1 on the development of the basal versus the luminal epithelial cells, we conducted immunofluorescence analysis of the mammary glands using traditional cytokeratin lineage markers. Cytokeratin 5 (CK5) was used to identify basal epithelial cells, and CK8 was used for the identification of luminal epithelial cells. In the Dach1−/− mammary gland epithelial cells, the relative abundance of CK5 was reduced and the relative abundance of CK8 was increased (Figures 4A and S4).
Figure 4.
Dach1 Gene Deletion Reduces the Abundance of Mammary Gland Stem Cell and Ductal Cell Lineage Gene Targets
(A) Immunofluorescent double staining for cytokeratin 5 (CK5) and CK8 showed that Dach1 gene deletion decreases the relative amount of CK5-positive cells versus CK8-positive cells.
(B) Immunohistochemical staining for differentiation factors (Gata3, Pml, and Pygo2) in the mammary gland of Dach1+/+ versus Dach1−/− (Cre deletion mice) (Dach1wt/wtROSA26CreERT2/mTmGfl versus Dach1fl/flROSA26CreERT2/mTmGfl). The red arrowheads indicate basal epithelial cells.
(C) Quantitation of immunohistochemical staining shown as mean ± SEM for n = 3 separate mice.
DACH1 Enhances Abundance of Genes Governing Ductal Branching
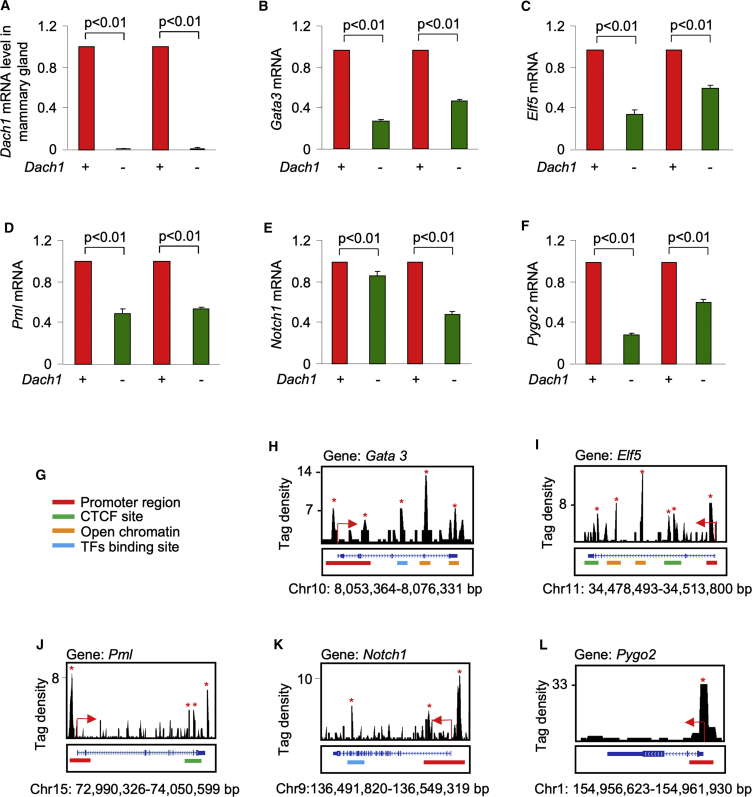
Because DACH1 loss reduced mammary duct branching, we next determined the effect of endogenous DACH1 on the expression of genes known to govern the process. Branching is associated with increased PYGO2 (Gu et al., 2013), ELF5 (Chakrabarti et al., 2012), PML, NOTCH1 (Bouras et al., 2008), and GATA3 (Asselin-Labat et al., 2007). Immunohistochemical analysis of the wild-type mammary gland identified PYGO2 in the luminal cells with occasional basal cells staining, PML in both luminal and epithelial cells, and GATA3 expression in the luminal cells (indicated with the arrows in Figure 4B). In the Dach1−/− mammary gland the abundance of PYGO2, PML, and GATA3 were reduced (Figures 4B and 4C).
qPCR verified the reduction of Dach1 mRNA (Figure 5A) and a reduction in the abundance of the mRNA for Gata3, Elf5, Pml, Notch1, and Pygo2 (Figures 5B–5F) in the Dach1−/− mammary gland. Notch1 mRNA levels were modestly reduced (Figure 5E). Analysis of DACH1 binding to these target genes was assessed using chromatin immunoprecipitation sequencing conducted on a DACH1 stably transfected MDA-MB-231 breast cancer cell line (Wu et al., 2008, Wu et al., 2011). Significant enrichment of DACH1 binding was identified at the target promoters for Gata3, Elf5, Pml, Notch1, and Pygo2 (Figures 5G–5L). DACH1 enrichment was located at the transcriptional start site of each gene, consistent with the known binding of DACH1 to the transcription elongation regulator (TCERG) (TCERG1, also known as CA150 or TAF2S) (Zhou et al., 2010a) and at CTCF sites (Figures 5H–5L).
Figure 5.
DACH1 Occupies the Promoter Regions of Mammary Gland Stem Cell and Ductal Cell Lineage Gene Targets
(A–F) qRT-PCR analysis of mRNA for genes participating in mammary gland stem cell and ductal development, (A) Dach1, (B) Gata3, (C) Elf5, (D)Pml, (E) Notch1, and (F) Pygo2, in the mammary gland of Cre deletion mice (Dach1wt/wtROSA26CreERT2/mTmGfl versus Dach1fl/flROSA26CreERT2/mTmGfl) with quantitation shown as mean ± SEM for n = 3 separate mice. Notch1 mRNA levels were modestly reduced in the Dach1−/− mammary gland.
(G–L) Analysis of DACH1 binding to these target genes was assessed using chromatin immunoprecipitation sequencing assays conducted on DACH1 stable MDA-MB-231 cells (G). (H) Gata3, (I) Elf5, (J) Pml, (K) Notch1, and (L) Pygo2.
DACH1 Enhances Ductal Branching in Epithelial Cultures
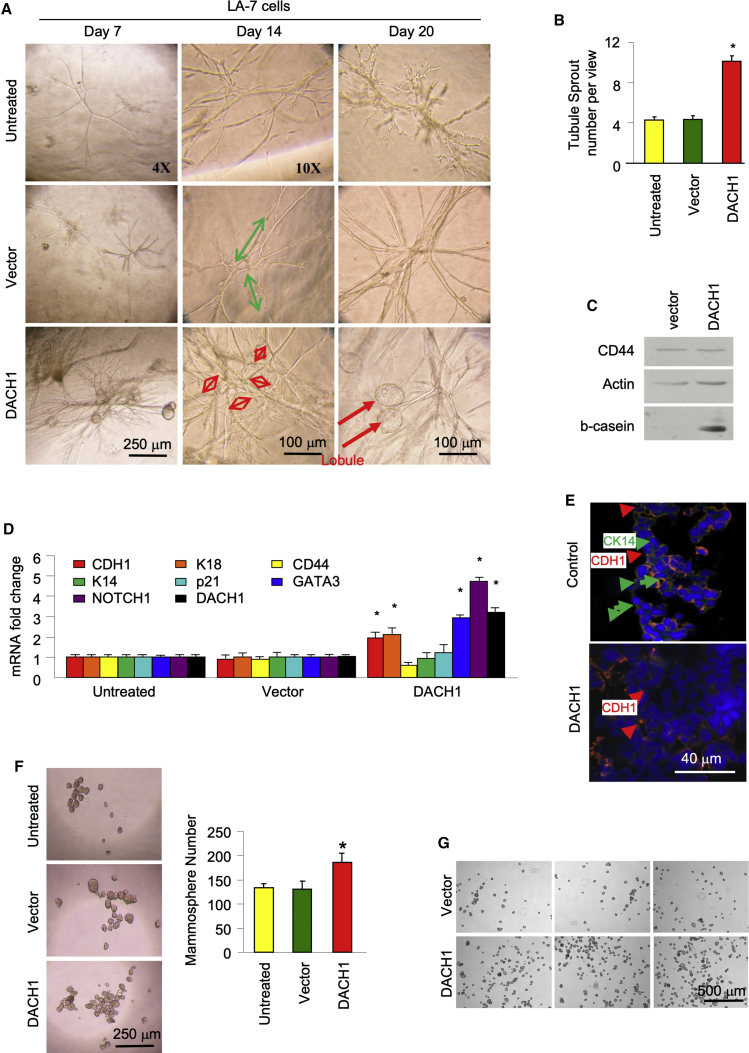
LA-7 cells exhibit the normal mammary gland stem cell properties of self-renewal and multilineage differentiation in vitro and in vivo (Zucchi et al., 2007). LA-7 cells keep the potentials to defferentiate to all the epithelial cell types present in vivo and, when cultured in 3D collagen matrix, develop organoids with 3D tubule-alveolar-like structures that morphologically and functionally recapitulate the 3D architecture of the mammary tree (Zucchi et al., 2007). LA-7 cells expressing DACH1 or control cells seeded onto 3D collagen-generated ductal/acinar structures as assessed by phase contrast microscopy (Figure 6A). Branching occurred by day 7 and increased through day 14 and subsequently (Figure 6A). DACH1-transduced LA-7 cell sprouting was increased 2.3-fold (Figure 6B, n = 3 experiments, p < 0.01, day 20). After day 14 in LA-7-DACH1 cells, the tubule structures were converted into lobular structures when compared with controls (Figure 6A). DACH1 increased β-casein abundance and reduced CD44 at day 4 when normalized to the β-actin loading control (Figure 6C). Transfection of LA-7 cells with the DACH1 expression vector increased DACH1 expression 3-fold, as determined by qPCR (Figure 6D). RT-PCR analysis showed a 2- to 3-fold increase in the luminal markers E-cadherin (CDH1) and CK18, when normalized to hypoxanthine phosphoribosyl (HPR); abundance of the surface glycoprotein CD44 was reduced 40% (Figure 6D, p < 0.05). The ductal morphogenesis target Gata3 was induced 3-fold and Notch1 was induced 4.8-fold (Figure 6D).
Figure 6.
DACH1 Induces Ductal Branching and Mammospheres in LA-7 Cells
(A) Representative phase contrast microscopy images of LA-7 cells cultured on collagen plates (upper panels), LA-7 cells transduced with control vector (middle panel), or vector-expressing DACH1 (lower panels) are shown.
(B and C) Cells were analyzed for ductal formation at day 20 (B). LA-7 tubule sprout number per view shown as mean ± SEM (C). Western blot with antibodies as indicated of either LA-7 cells transduced with control vector or DACH1 expression vector. β-Actin is used as a protein loading control.
(D) RT-PCR analysis of mammary stem cell regulatory genes was conducted. Results are shown as mean ± SEM for n = 3 separate experiments. ∗p < 0.05.
(E) Immunofluorescence of LA-7 cells transfected with DACH1 or with empty vector, grown in 3D organoids structures. DACH1 reduced myoepithelial markers (CK14, green), whereas luminal markers (CDH1) were upregulated.
(F) LA-7 cells, or LA-7 cells transduced with control vector or vector expressing DACH1, were analyzed for mammosphere formation. Cells were plated in limiting dilutions of 500 cells/1.4 mL of media and mammosphere (150–200 cells) were counted at day 8. Phase contrast image of a representative field is shown and the number of mammospheres (mean ± SEM is shown for n = 3 separate experiments, ∗p < 0.01).
(G) A separate clone of LA-7 cells transduced with DACH1 or vector control were grown as mammospheres in 24-well plates and representative images are shown at 2×.
We next examined the impact of DACH1 on the abundance of luminal versus myoepithelial cell lineage markers during LA-7 cell organoid formation using immunofluorescence. LA-7 cells transfected with DACH1 or with empty vector as a control, were cultured in collagen for 10 days and then digested with collagenase. The 3D structures were then embedded in OCT and sectioned with a cryotome. Organoids generated by LA-7 in which DACH1 was transfected express epithelial luminal markers (CDH1) but less of the myoepithelial markers (CK14) (Figures 6E and S5).
Because endogenous DACH1 enhanced the proportion of Lin− CD24med CD29high, which we herein referred to as basal cells, we considered the possibility that DACH1 may drive the expansion of MaSCs and therefore deployed a surrogate assay of mammosphere formation using LA-7 cells. DACH1 transduction of LA-7 cells increased mammosphere number by 30% (Figures 6F and 6G, n = 3 separate experiments, p < 0.01). Transduction efficiency of the LA-7 cells was 70%–80%. Thus, DACH1 promotes formation of mammary gland ducts and mammospheres in LA-7 cells.
DACH1 Restrains TGF-β Action in the Pubertal Mammary Gland In Vivo
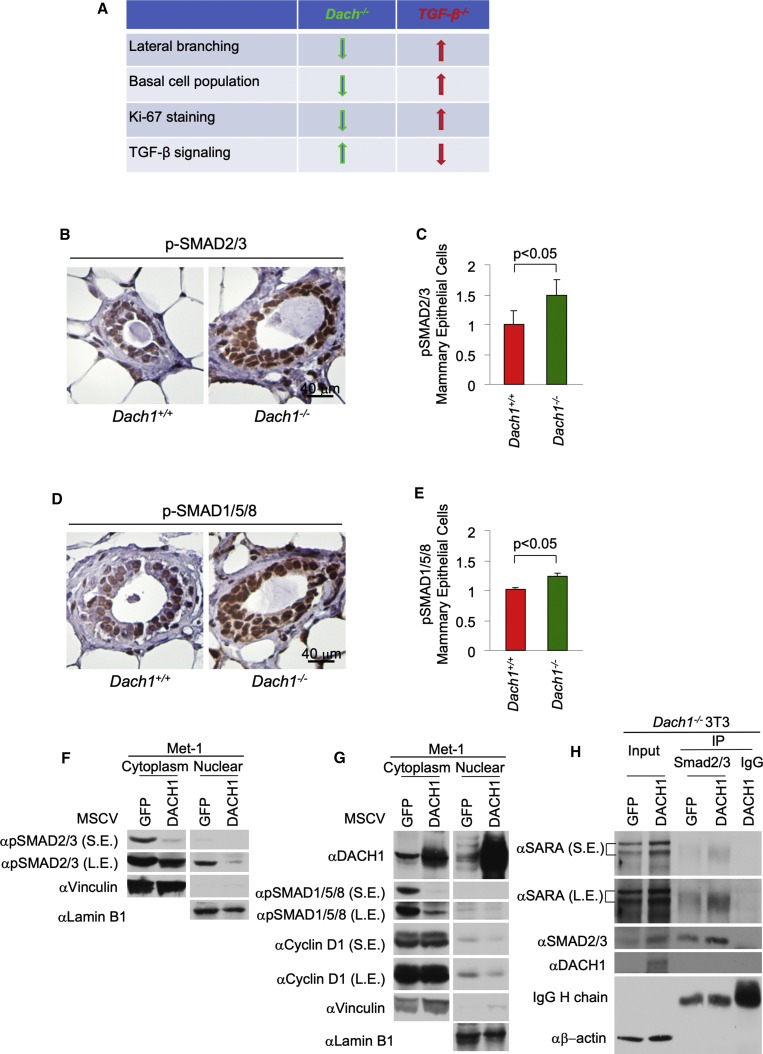
The TGF-β signaling pathway restrains mammary progenitor self-renewal (Martinez-Ruiz et al., 2016) and lineage commitment decisions, decreasing myoepithelial cell progenitors (Lindley and Briegel, 2010, Martinez-Ruiz et al., 2016). Furthermore, TGF-β is known to restrain mammary ductal branching (Bottinger et al., 1997, Daniel and Robinson, 1992, Moses and Barcellos-Hoff, 2011, Nelson et al., 2006, Pierce et al., 1993) (Figure 7A). We therefore determined the possible role of endogenous Dach1 in restraining mammary gland TGF-β activity by first assessing the phosphorylation of SMADs. Immunohistochemical staining demonstrated increased phosphorylation of SMAD 2/3 and SMAD 1/5/8 in both the basal and luminal MECs (Figures 7B–7E).
Figure 7.
Dach1 Gene Deletion Activates TGF-β Signaling in the Mammary Gland
(A) A summary of the mammary gland phenotype identified herein upon Dach1 gene deletion, with comparison to the effect of TGF-β gene deletion summarized from the literature (Daniel and Robinson, 1992, Moses and Barcellos-Hoff, 2011, Bottinger et al., 1997, Nelson et al., 2006, Pierce et al., 1993).
(B and C) Representative example (B) and quantitative analysis (C) of immunohistochemical staining for pSMAD2/3 in the mammary gland from mice shown as Dach1+/+ (Dach1wt/wtROSA26CreERT2/mTmGfl) and DACH1–/– (Dach1fl/flROSA26CreERT2/mTmGfl).
(D and E) Representative example of immunostaining for pSMAD 1/5/8 (D), with quantification (E) shown as mean ± SEM for n = 3 separate animals.
(F and G) Western blots of Met-1 breast cancer cells transduced with a DACH1 expression vector using antibodies to the proteins as indicated (F) for p-SMAD2/3 and (G) for DACH1, p-SMAD1/5/8, and Cyclin D1.
(H) Western blot of Dach1wt versus Dach1−/− 3T3 cells for SARA association after immune precipitation for Smad 2/3. S.E., short exposure; L.E., longer exposure. β-Actin is used as a protein loading control.
DACH1 Inhibits SMAD Association with Smad Anchor for Receptor Activation
Reintroduction of DACH1 via a retroviral expression vector into the metastatic Met-1 breast cancer cell line, increased DACH1 abundance and reduced Smad anchor for receptor activation (SMAD) 2/3 phosphorylation (Figure 7F) and SMAD 1/5/8 phosphorylation (Figure 7G) in both the nuclear and cytoplasmic fractions. The scaffold protein SARA, also known as the zinc finger FYVE domain-containing protein 9 (ZFYVE9), brings SMAD 2/3 to TGF-β receptors, facilitating SMAD 2/3 activation and thereby augmenting TGF-β signaling (Tsukazaki et al., 1998). The relative abundance of SARA was increased 4-fold upon Dach1 deletion (Figure 7H). Immunoprecipitation (IP) with an antibody directed to SMAD 2/3 co-precipitated SARA; however, the relative abundance of SARA in the SMAD 2 IP was dramatically increased in the Dach1−/− cells, consistent with a role for DACH1 to inhibit the SARA-SMAD 2/3 association and thereby reduce basal SMAD activity (Figures 7H, S6B, and S6C).
Discussion
DACH1 Skews Mammary Gland Lineage Development
In this current study, we show that endogenous DACH1 increases the proportion of basal cells and reduces the proportion of luminal cells in the post-natal mammary gland. Interrogation of Dach1 transcripts from scRNA-seq (Giraddi et al., 2018, Latha and Saddala, 2017) demonstrated that Dach1 gene expression was detectable in multiple lineages and stages, including multipotent fMaSCs in the embryo and among MMPr prior to puberty. DACH1 expression was detected in presumptive basal and myoepithelial cells, adult basal cells and (ERα+), luminal progenitor, and in ERα+ luminal cells. The mechanism by which DACH1 alters the relative proportion of basal versus luminal cells may involve lineage skewing during differentiation, intrinsic effects in different lineage progenitors, or transitive effects from myoepithelial cells that normally restrain the luminal cell number in a juxtacrine/paracrine fashion and may involve both cell intrinsic or heterotypic signaling.
What might be the consequence of DACH1 increasing the proportion of basal cells? Myoepithelial cells express a variety of recognized tumor-suppressor proteins (p63, p73, 14-3-3-s, and maspin) and provide the interface between tumor cells of ductal carcinoma in situ and the microenvironment (Pandey et al., 2010b), conveying antiangiogenic, antiproliferative, and anti-invasive properties. Previous studies have shown that DACH1 reduced growth of breast tumor cells in tissue culture and in immune-deficient mice (Popov et al., 2009, Popov et al., 2010, Wu et al., 2006, Wu et al., 2011). Collectively, these studies suggest that DACH1 is required for normal myoepithelial cell number and function and may thereby play a regulatory role as “natural tumor suppressors” in the mammary gland (reviewed in Adriance et al., 2005, Bissell and Labarge, 2005, Lakhani and O'Hare, 2001, Pandey et al., 2010a, Sternlicht and Barsky, 1997, Sternlicht et al., 1997).
DACH1 Promotes Mammary Gland Ductal Branching
The current studies demonstrate that Dach1 deletion reduced mammary gland ductal branching and DACH1 expression in LA-7 cells enhanced ductal branching. Prior studies demonstrated a role for DACH1 in promoting migration in several different cell types including fibroblasts (Wu et al., 2008), MECs (Wu et al., 2008), prostate epithelial cells (Chen et al., 2015), and vascular endothelial cells (Chang et al., 2017). TGF-β1 is a major negative regulator of mammary ductal branching, restricting end bud bifurcation and branch formation (Ewan et al., 2002, Ingman and Robertson, 2008, Silberstein and Daniel, 1987). Epithelial cell or stromal expression of a dominant-negative TGF-β allele (dnIIR) increased mammary ductal branching (Bottinger et al., 1997). Dach1 deletion phenocopies TGF-β overactivity. The finding that Dach1 gene deletion phenocopies TGF-β over activity, and that Dach1 deletion results in TGF-β hyperactivity demonstrated by increased SMAD phosphorylation in the mammary gland, suggests that Dach1-mediated TGF-β restraint may govern the mammary gland ductal branching phenotype. SMAD phosphorylation was increased in both the basal and luminal cells. DACH1 was expressed in both basal and luminal cells during development; however, in the adult mammary gland, immunohistochemistry showed that DACH1 was predominantly in the basal cells, raising the possibility that Dach1 exerts both cell-autonomous and non-autonomous effects on TGF-β signaling.
Several other genes are essential for normal mammary duct development including Esr1, Srib, and Ovol2, and pubertal branching development is disrupted in mice lacking GH, insulin-like growth factor 1 (Igf1), or estrogen receptor α (Esr1). DACH1 can inhibit IGF1 and ERα signaling to restrain EMT (DeAngelis et al., 2011, Popov et al., 2009, Wu et al., 2011), which may contribute to the mammary gland developmental changes of Dach1−/− mice.
DACH1 Expands the Mammary Gland Stem Cell Population
Herein DACH1 enhanced LA-7 cell mammosphere formation and endogenous Dach1 enhanced the proportion of basal cells, characterized by CD24medCD29hiLin− cells, in vivo. The expansion of the basal cell population may be secondary to the restraint of TGF-β activity, as TGF-β decreases MEC repopulating activity (Martinez-Ruiz et al., 2016). Alternatively the expansion may be secondary to transcriptional effects by Dach1 on several target genes known to participate in the expansion of MaSCs, including PYGO2, NOTCH1, and BRCA1 (Gu et al., 2013, Martinez-Ruiz et al., 2016, Visvader and Stingl, 2014) (Figures S6B and S6C). PML serves to enhance the transition of luminal progenitors to alveolar progenitors, which may in turn increase the myoepithelial progenitor pool. The histone methylation reader PYGO2, a coactivator of the WNT pathway, is necessary for suppressing luminal and alveolar differentiation of the MaSC-enriched population by coordinating the activity of the Wnt and Notch pathways (Gu et al., 2013). GATA3 is a crucial regulator of the luminal lineage as GATA3 restrains luminal progenitors (Asselin-Labat et al., 2007). Herein DACH1 enhanced basal cells and restrained TGF-β activity, and TGF-β is known to restrain basal cell expansion. The frequency of basal cells and the development of breast cancer subtypes suggests that inactivation of lineage determinant proteins are mechanistically linked (Visvader and Stingl, 2014). The current studies contrast with findings that Dach1 restrains breast cancer (Wu et al., 2011) and brain cancer (Watanabe et al., 2011) stem cells. These findings may reflect the dual effects of TGF-β early in the mammary gland as a tumor suppressor and at a later stage to evade immune surveillance and promote metastasis (Massague, 2008, Massague, 2012).
Experimental Procedures
Transgenic Mice
All the animal studies were approved by the Institutional Animal Care and Use Committee of Thomas Jefferson University. The Dach1fl/fl mice (Chen et al., 2015, Pierce et al., 1993) (which remove the same section of the Dach1 locus that is deleted in the knockout mice strain [Davis et al., 2001]), were intercrossed with the ROSA26CreERT2 mice (expressing CRE-ERT2 fusion protein under the control of the ubiquitous ROSA26 promoter; the mice were kindly provided by Dr. Thomas Ludwig at Columbia University, New York, NY). Dach1fl/fl mice were mated with either ROSA26CreERT2/CreERT2 or ROSA26mTmG/mTmG mice to generate Dach1fl/wtROSA26CreERT2/CreERT2 (Cre mice) and Dach1fl/wtROSA26mTmG/mTmG (Cre reporter mice). Dach1fl/wtROSA26CreERT2/+ mice were then mated with Dach1fl/wtROSA26mTmG/+ mice to generate Dach1fl/flROSA26CreERT2/mTmG mice and Dach1wt/wtROSA26CreERT2/mTmG littermates.
Immunohistochemistry, Immunofluorescence, and TUNEL Staining
Immunohistochemical analysis of paraffin-embedded murine mammary gland tissues was conducted using a polyclonal DACH1 antibody (Proteintech, cat. no. 10,914-1-AP) (Wu et al., 2006), anti-PYGO2 (Thermo Fisher Scientific, cat no. PA5-34878), anti-PML1 (Abcam, cat. no ab67761), anti-GATA3 (Santa Cruz Biotechnology, cat. no, sc-9009), anti-vWF (A0082, Dako), anti-phosphorylated SMAD2/3 (Santa Cruz Biotechnology, cat. no. sc-11769-R), or anti-phosphorylated SMAD1/5/8 (Ser 423/425) (Santa Cruz Biotechnology, cat. no. sc-12353-R). Immunofluorescent analysis of frozen mouse mammary gland was conducted using anti-Ki-67 clone SP6 rabbit monoclonal antibody (Abcam, cat. no. ab16667), anti-CK5 antibody (PRB-160P, Covance), rabbit polyclonal and/or anti-CK8 mouse monoclonal antibody (MMS-162P, Covance). DAPI was used as counter staining. Alexa Fluor 633- or Alexa Fluor 565-conjugated goat anti-rabbit antibody and Alexa Fluor 488-conjugated goat anti-mouse antibody were used as secondary antibodies (Jiao et al., 2008, Ju et al., 2013). Frozen sections of murine mammary gland tissues were used in immunofluorescence TUNEL staining. E-cadherin-FITC conjugated raised in mouse (BD Bioscience, cat no. 612131), cytokeratin 14 raised in rabbit (Sigma cat no. HAPA023040), and the secondary antibody raised in goat against rabbit IGG Alexa Fluor 488 (Life Technologies, cat no. A11034) were used in LA-7 cell immunofluorescence.
Cell Culture, Reagents, Mammosphere Assays, Expression Vectors, DNA Transfection, and Statistical Analyses
The generation and culture of 3T3 and mouse embryonic fibroblasts derived from Dach1−/− mice, the Met-1 breast cancer cell line (Wu et al., 2008, Wu et al., 2011), and LA-7 rat mammary gland stem cells (Zucchi et al., 2007) were described previously (Wu et al., 2008, Wu et al., 2011). TGF-β was obtained from Calbiochem. Met-1 cells were infected with either MSCV-IRES-GFP (murine stem cell virus) or MSCV-DACH1-IRES-GFP (Wu et al., 2006). LA7 cells were transiently transfected with Amaxa system (Amaxa Nucleofector Kit, Lonza) according to the manufacturer’s protocol. 1 × 106 LA7 cells were tansfected with 2 μg of each plasmid (GFP-empty, pKW-empty, and pKW-DACH1). GFP-plasmid was used as endogenous control of transfection. Data were expressed as the means ± SEM. Statistical analyses were performed using the Student's t test, and p values were <0.05.
Tubule and Organoid Formation in 3D Culture
For tubule/organoid formation, cells were suspended on ice in rat tail-derived collagen prepared as described previously (Zucchi et al., 2007). Organoid cultures were examined daily with light microscopy to assess tubule and organoid formation and photographed at days 7, 14, and 20. Evaluation of branching was assessed 3 days after seeding by examining 10 tubules for each condition in 3 independent experiments.
IP and Western Blot
IP and western blot assays were performed in cells as indicated. For each IP, 1 mL lysate (1 mg protein) and 2 μg anti-SMAD2/3 (BD, cat. no. 610843) or anti-SARA (Proteintech, cat. no. 14821-1-AP) were incubated overnight at 4°C. Immunoprecipitates were washed 5 times in IP buffer, and 30 μL of 2× sample buffer was added to the bead pellet. Laminin B1 antibody (Abcam, cat. no. ab16048) was used as an internal control for nuclear protein abundance and vinculin (Sigma, cat. no. V9131) was used as a control for cytoplasmic fraction enrichment. Polyclonal anti-Beta casein (Santa Cruz, cat no. SC-30042), monoclonal anti CD44 (Santa Cruz cat no. SC-53068), and polyclonal anti-Beta actin (Santa Cruz, cat no. SC-1615) were used in LA-7 cell Western-blot.
FACS Analysis of Stem Cells
FACS analysis for mammary epithelial stem cells was conducted as outlined in prior publications (dos Santos et al., 2013, Jiao et al., 2016, Shackleton et al., 2006). In brief, mammary glands were removed from 12-week-old Dach1fl/flROSA26CreERT2/mTmG and Dach1wt/wtROSA26CreERT2/mTmG mice. Both strains of mice were treated with a 5-day pulse of tamoxifen when mice reached 6 weeks of age and were analyzed after a subsequent 2.5 months. The data were analyzed with FlowJo single-cell analysis software (Tree Star, Ashland, OR).
Real-Time qPCR
PCR analysis for luminal markers (Cdh1 [E-cadherin]), CK18, alveolar marker (β-casein), and CD44 was conducted of LA-7 mRNA as described previously (Zucchi et al., 2007). Primers for all the genes/gene transcripts including 18s rRNA and HPRT (hypoxanthine-guanine phosphoribosyltransferase) transferase are listed in Table S1, except Cdh1, CK18, and CD44 which described previously (Zucchi et al., 2007).
Author Contributions
R.G.P. initiated and supervised the experiments and wrote the paper. X.J. revised the manuscripts. X.J. and Z.L. conducted transgenic experiments and analysis, including immunohistochemistry and FACS. C.C., D.C., and R.R. performed all the experiments with LA-7 cells grown as mammospheres or as organoids, all the RT-PCR and western blot experiments, and analyzed the data. R.R. and I.Z. supervised all the LA-7 experiments, analyzed the data, and wrote the LA-7 section of the paper. K.W., I.T. and B.T.S. participated in discussing and interpreting the data. Z.L., M.F., and A.P.S. conducted transgenic generation. Z.L., M.W., and X.J. conducted analysis of the transgenic mice.
Acknowledgments
This work was supported in part by R01CA132115, (to R.G.P.), a generous grant from the Dr. Ralph and Marian Falk Medical Research Trust (to R.G.P.), a grant from the Breast Cancer Research Foundation. The Department disclaims responsibility for any analysis, interpretations, or conclusions. R.G.P. holds ownership interests in, and serves as CSO/Founder of the biopharmaceutical companies ProstaGene and LightSeed, and is CMO of CytoDyn. R.G.P. holds ownership interests (value unknown) in several issued and submitted patent applications. This work was also supported by the CNR-Flag Projects Epigen and Interomics (to I.Z.). M.F. is supported by the Sigrid Jusélius Foundation, Orion Research Foundation, and the Finnish Society of Dermatology. K.W. is supported by the National Natural Science Foundation of China no. 81572608 and no. 81874120.
Published: December 13, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, six figures, and one table and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.11.010.
Supplemental Information
References
- Adriance M.C., Inman J.L., Petersen O.W., Bissell M.J. Myoepithelial cells: good fences make good neighbors. Breast Cancer Res. 2005;7:190–197. doi: 10.1186/bcr1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselin-Labat M.L., Sutherland K.D., Barker H., Thomas R., Shackleton M., Forrest N.C., Hartley L., Robb L., Grosveld F.G., van der Wees J. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat. Cell Biol. 2007;9:201–209. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- Bach K., Pensa S., Grzelak M., Hadfield J., Adams D.J., Marioni J.C., Khaled W.T. Differentiation dynamics of mammary epithelial cells revealed by single-cell RNA sequencing. Nat. Commun. 2017;8:2128. doi: 10.1038/s41467-017-02001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell M.J., Labarge M.A. Context, tissue plasticity, and cancer: are tumor stem cells also regulated by the microenvironment? Cancer Cell. 2005;7:17–23. doi: 10.1016/j.ccr.2004.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottinger E.P., Jakubczak J.L., Haines D.C., Bagnall K., Wakefield L.M. Transgenic mice overexpressing a dominant-negative mutant type II transforming growth factor beta receptor show enhanced tumorigenesis in the mammary gland and lung in response to the carcinogen 7,12-dimethylbenz-[a]-anthracene. Cancer Res. 1997;57:5564–5570. [PubMed] [Google Scholar]
- Bouras T., Pal B., Vaillant F., Harburg G., Asselin-Labat M.L., Oakes S.R., Lindeman G.J., Visvader J.E. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell. 2008;3:429–441. doi: 10.1016/j.stem.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Chakrabarti R., Wei Y., Romano R.A., DeCoste C., Kang Y., Sinha S. Elf5 regulates mammary gland stem/progenitor cell fate by influencing notch signaling. Stem Cells. 2012;30:1496–1508. doi: 10.1002/stem.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A.H., Raftrey B.C., D'Amato G., Surya V.N., Poduri A., Chen H.I., Goldstone A.B., Woo J., Fuller G.G., Dunn A.R. DACH1 stimulates shear stress-guided endothelial cell migration and coronary artery growth through the CXCL12-CXCR4 signaling axis. Genes Dev. 2017;31:1308–1324. doi: 10.1101/gad.301549.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Wu K., Cai S., Zhang W., Zhou J., Wang J., Ertel A., Li Z., Rui H., Quong A. Dachshund binds p53 to block the growth of lung adenocarcinoma cells. Cancer Res. 2013;73:3262–3274. doi: 10.1158/0008-5472.CAN-12-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Wu K., Jiao X., Wang L., Ju X., Wang M., Di Sante G., Xu S., Wang Q., Li K. The endogenous cell-fate factor dachshund restrains prostate epithelial cell migration via repression of cytokine secretion via a cxcl signaling module. Cancer Res. 2015;75:1992–2004. doi: 10.1158/0008-5472.CAN-14-0611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Amoui M., Zhang Z., Mardon G. Dachshund and eyes absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell. 1997;91:893–903. doi: 10.1016/s0092-8674(00)80481-x. [DOI] [PubMed] [Google Scholar]
- Chen R., Halder G., Zhang Z., Mardon G. Signaling by the TGF-beta homolog decapentaplegic functions reiteratively within the network of genes controlling retinal cell fate determination in Drosophila. Development. 1999;126:935–943. doi: 10.1242/dev.126.5.935. [DOI] [PubMed] [Google Scholar]
- Daniel C.W., Robinson S.D. Regulation of mammary growth and function by TGF-beta. Mol. Reprod. Dev. 1992;32:145–151. doi: 10.1002/mrd.1080320210. [DOI] [PubMed] [Google Scholar]
- Davis R.J., Shen W., Sandler Y.I., Amoui M., Purcell P., Maas R., Ou C.N., Vogel H., Beaudet A.L., Mardon G. Dach1 mutant mice bear no gross abnormalities in eye, limb, and brain development and exhibit postnatal lethality. Mol. Cell. Biol. 2001;21:1484–1490. doi: 10.1128/MCB.21.5.1484-1490.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis T.L., Rebay I. Master regulators in development: views from the Drosophila retinal determination and mammalian pluripotency gene networks. Dev. Biol. 2017;421:93–107. doi: 10.1016/j.ydbio.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAngelis T., Wu K., Pestell R., Baserga R. The type 1 insulin-like growth factor receptor and resistance to DACH1. Cell Cycle. 2011;10:1956–1959. doi: 10.4161/cc.10.12.15800. [DOI] [PubMed] [Google Scholar]
- dos Santos C.O., Rebbeck C., Rozhkova E., Valentine A., Samuels A., Kadiri L.R., Osten P., Harris E.Y., Uren P.J., Smith A.D. Molecular hierarchy of mammary differentiation yields refined markers of mammary stem cells. Proc. Natl. Acad. Sci. U S A. 2013;110:7123–7130. doi: 10.1073/pnas.1303919110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewan K.B., Shyamala G., Ravani S.A., Tang Y., Akhurst R., Wakefield L., Barcellos-Hoff M.H. Latent transforming growth factor-beta activation in mammary gland: regulation by ovarian hormones affects ductal and alveolar proliferation. Am. J. Pathol. 2002;160:2081–2093. doi: 10.1016/s0002-9440(10)61158-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraddi R.R., Chung C.Y., Heinz R.E., Balcioglu O., Novotny M., Trejo C.L., Dravis C., Hagos B.M., Mehrabad E.M., Rodewald L.W. Single-cell transcriptomes distinguish stem cell state changes and lineage specification programs in early mammary gland development. Cell Rep. 2018;24:1653–1666.e7. doi: 10.1016/j.celrep.2018.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu B., Watanabe K., Sun P., Fallahi M., Dai X. Chromatin effector Pygo2 mediates Wnt-notch crosstalk to suppress luminal/alveolar potential of mammary stem and basal cells. Cell Stem Cell. 2013;13:48–61. doi: 10.1016/j.stem.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingman W.V., Robertson S.A. Mammary gland development in transforming growth factor beta1 null mutant mice: systemic and epithelial effects. Biol. Reprod. 2008;79:711–717. doi: 10.1095/biolreprod.107.067272. [DOI] [PubMed] [Google Scholar]
- Jiao X., Katiyar S., Liu M., Mueller S.C., Lisanti M.P., Li A., Pestell T.G., Wu K., Ju X., Li Z. Disruption of c-Jun reduces cellular migration and invasion through inhibition of c-Src and hyperactivation of ROCK II kinase. Mol. Biol. Cell. 2008;19:1378–1390. doi: 10.1091/mbc.E07-08-0753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X., Rizvanov A.A., Cristofanilli M., Miftakhova R.R., Pestell R.G. Breast cancer stem cell isolation. Methods Mol. Biol. 2016;1406:121–135. doi: 10.1007/978-1-4939-3444-7_10. [DOI] [PubMed] [Google Scholar]
- Ju X., Ertel A., Casimiro M.C., Yu Z., Meng H., McCue P.A., Walters R., Fortina P., Lisanti M.P., Pestell R.G. Novel oncogene-induced metastatic prostate cancer cell lines define human prostate cancer progression signatures. Cancer Res. 2013;73:978–989. doi: 10.1158/0008-5472.CAN-12-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakhani S.R., O'Hare M.J. The mammary myoepithelial cell – Cinderella or ugly sister? Breast Cancer Res. 2001;3:1–4. doi: 10.1186/bcr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latha M.S., Saddala M.S. Molecular docking based screening of a simulated HIF-1 protein model for potential inhibitors. Bioinformation. 2017;13:388–393. doi: 10.6026/97320630013388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindley L.E., Briegel K.J. Molecular characterization of TGFbeta-induced epithelial-mesenchymal transition in normal finite lifespan human mammary epithelial cells. Biochem. Biophys. Res. Commun. 2010;399:659–664. doi: 10.1016/j.bbrc.2010.07.138. [DOI] [PubMed] [Google Scholar]
- Martinez-Ruiz H., Illa-Bochaca I., Omene C., Hanniford D., Liu Q., Hernando E., Barcellos-Hoff M.H. A TGFbeta-miR-182-BRCA1 axis controls the mammary differentiation hierarchy. Sci. Signal. 2016;9:ra118. doi: 10.1126/scisignal.aaf5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J. TGFbeta in cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J. TGFbeta signalling in context. Nat. Rev. Mol. Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses H., Barcellos-Hoff M.H. TGF-beta biology in mammary development and breast cancer. Cold Spring Harb. Perspect. Biol. 2011;3:a003277. doi: 10.1101/cshperspect.a003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar M.D., Tasic B., Miyamichi K., Li L., Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Nan F., Lu Q., Zhou J., Cheng L., Popov V.M., Wei S., Kong B., Pestell R.G., Lisanti M.P., Jiang J. Altered expression of DACH1 and cyclin D1 in endometrial cancer. Cancer Biol. Ther. 2009;8:1534–1539. doi: 10.4161/cbt.8.16.8963. [DOI] [PubMed] [Google Scholar]
- Nelson C.M., Vanduijn M.M., Inman J.L., Fletcher D.A., Bissell M.J. Tissue geometry determines sites of mammary branching morphogenesis in organotypic cultures. Science. 2006;314:298–300. doi: 10.1126/science.1131000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey G., Zhang B., Chang A.N., Myers C.L., Zhu J., Kumar V., Schadt E.E. An integrative multi-network and multi-classifier approach to predict genetic interactions. PLoS Comput Biol. 2010;6 doi: 10.1371/journal.pcbi.1000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey P.R., Saidou J., Watabe K. Role of myoepithelial cells in breast tumor progression. Front. Biosci. 2010;15:226–236. doi: 10.2741/3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce D.F., Jr., Johnson M.D., Matsui Y., Robinson S.D., Gold L.I., Purchio A.F., Daniel C.W., Hogan B.L., Moses H.L. Inhibition of mammary duct development but not alveolar outgrowth during pregnancy in transgenic mice expressing active TGF-beta 1. Genes Dev. 1993;7:2308–2317. doi: 10.1101/gad.7.12a.2308. [DOI] [PubMed] [Google Scholar]
- Popov V.M., Wu K., Zhou J., Powell M.J., Mardon G., Wang C., Pestell R.G. The Dachshund gene in development and hormone-responsive tumorigenesis. Trends Endocrinol. Metab. 2010;21:41–49. doi: 10.1016/j.tem.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov V.M., Zhou J., Shirley L.A., Quong J., Yeow W.S., Wright J.A., Wu K., Rui H., Vadlamudi R.K., Jiang J. The cell fate determination factor DACH1 is expressed in estrogen receptor-alpha-positive breast cancer and represses estrogen receptor-alpha signaling. Cancer Res. 2009;69:5752–5760. doi: 10.1158/0008-5472.CAN-08-3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleton M., Vaillant F., Simpson K.J., Stingl J., Smyth G.K., Asselin-Labat M.L., Wu L., Lindeman G.J., Visvader J.E. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- Shehata M., van Amerongen R., Zeeman A.L., Giraddi R.R., Stingl J. The influence of tamoxifen on normal mouse mammary gland homeostasis. Breast Cancer Res. 2014;16:411. doi: 10.1186/s13058-014-0411-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberstein G.B., Daniel C.W. Reversible inhibition of mammary gland growth by transforming growth factor-beta. Science. 1987;237:291–293. doi: 10.1126/science.3474783. [DOI] [PubMed] [Google Scholar]
- Sternlicht M.D., Barsky S.H. The myoepithelial defense: a host defense against cancer. Med. Hypotheses. 1997;48:37–46. doi: 10.1016/s0306-9877(97)90022-0. [DOI] [PubMed] [Google Scholar]
- Sternlicht M.D., Kedeshian P., Shao Z.M., Safarians S., Barsky S.H. The human myoepithelial cell is a natural tumor suppressor. Clin. Cancer Res. 1997;3:1949–1958. [PubMed] [Google Scholar]
- Tsukazaki T., Chiang T.A., Davison A.F., Attisano L., Wrana J.L. SARA, a FYVE domain protein that recruits Smad2 to the TGFbeta receptor. Cell. 1998;95:779–791. doi: 10.1016/s0092-8674(00)81701-8. [DOI] [PubMed] [Google Scholar]
- Visvader J.E., Clevers H. Tissue-specific designs of stem cell hierarchies. Nat. Cell Biol. 2016;18:349–355. doi: 10.1038/ncb3332. [DOI] [PubMed] [Google Scholar]
- Visvader J.E., Stingl J. Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes Dev. 2014;28:1143–1158. doi: 10.1101/gad.242511.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A., Ogiwara H., Ehata S., Mukasa A., Ishikawa S., Maeda D., Ueki K., Ino Y., Todo T., Yamada Y. Homozygously deleted gene DACH1 regulates tumor-initiating activity of glioma cells. Proc. Natl. Acad. Sci. U S A. 2011;108:12384–12389. doi: 10.1073/pnas.0906930108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Chen K., Wang C., Jiao X., Wang L., Zhou J., Wang J., Li Z., Addya S., Sorensen P.H. Cell fate factor DACH1 represses YB-1-mediated oncogenic transcription and translation. Cancer Res. 2014;74:829–839. doi: 10.1158/0008-5472.CAN-13-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Jiao X., Li Z., Katiyar S., Casimiro M.C., Yang W., Zhang Q., Willmarth N.E., Chepelev I., Crosariol M. Cell fate determination factor Dachshund reprograms breast cancer stem cell function. J. Biol. Chem. 2011;286:2132–2142. doi: 10.1074/jbc.M110.148395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Katiyar S., Li A., Liu M., Ju X., Popov V.M., Jiao X., Lisanti M.P., Casola A., Pestell R.G. Dachshund inhibits oncogene-induced breast cancer cellular migration and invasion through suppression of interleukin-8. Proc. Natl. Acad. Sci. U S A. 2008;105:6924–6929. doi: 10.1073/pnas.0802085105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Katiyar S., Witkiewicz A., Li A., McCue P., Song L.N., Tian L., Jin M., Pestell R.G. The cell fate determination factor dachshund inhibits androgen receptor signaling and prostate cancer cellular growth. Cancer Res. 2009;69:3347–3355. doi: 10.1158/0008-5472.CAN-08-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Li A., Rao M., Liu M., Dailey V., Yang Y., Di Vizio D., Wang C., Lisanti M.P., Sauter G. DACH1 is a cell fate determination factor that inhibits cyclin D1 and breast tumor growth. Mol. Cell. Biol. 2006;26:7116–7129. doi: 10.1128/MCB.00268-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Li Z., Cai S., Tian L., Chen K., Wang J., Hu J., Sun Y., Li X., Ertel A. EYA1 phosphatase function is essential to drive breast cancer cell proliferation through cyclin D1. Cancer Res. 2013;73:4488–4499. doi: 10.1158/0008-5472.CAN-12-4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Yuan X., Pestell R. Endogenous Dach1 in cancer. Oncoscience. 2015;2:803–804. doi: 10.18632/oncoscience.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Liu Y., Zhang W., Popov V.M., Wang M., Pattabiraman N., Sune C., Cvekl A., Wu K., Jiang J. Transcription elongation regulator 1 is a co-integrator of the cell fate determination factor Dachshund homolog 1. J. Biol. Chem. 2010;285:40342–40350. doi: 10.1074/jbc.M110.156141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wang C., Wang Z., Dampier W., Wu K., Casimiro M.C., Chepelev I., Popov V.M., Quong A., Tozeren A. Attenuation of Forkhead signaling by the retinal determination factor DACH1. Proc. Natl. Acad. Sci. U S A. 2010;107:6864–6869. doi: 10.1073/pnas.1002746107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucchi I., Sanzone S., Astigiano S., Pelucchi P., Scotti M., Valsecchi V., Barbieri O., Bertoli G., Albertini A., Reinbold R.A. The properties of a mammary gland cancer stem cell. Proc. Natl. Acad. Sci. U S A. 2007;104:10476–10481. doi: 10.1073/pnas.0703071104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.