Abstract
Background
Insights into the genetic capacities of species to adapt to future climate change can be gained by using comparative genomic and transcriptomic data to reconstruct the genetic changes associated with such adaptations in the past. Here we investigate the genetic changes associated with adaptation to arid environments, specifically climatic extremes and new cactus hosts, through such an analysis of five repleta group Drosophila species.
Results
We find disproportionately high rates of gene gains in internal branches in the species’ phylogeny where cactus use and subsequently cactus specialisation and high heat and desiccation tolerance evolved. The terminal branch leading to the most heat and desiccation resistant species, Drosophila aldrichi, also shows disproportionately high rates of both gene gains and positive selection. Several Gene Ontology terms related to metabolism were enriched in gene gain events in lineages where cactus use was evolving, while some regulatory and developmental genes were strongly selected in the Drosophila aldrichi branch. Transcriptomic analysis of flies subjected to sublethal heat shocks showed many more downregulation responses to the stress in a heat sensitive versus heat resistant species, confirming the existence of widespread regulatory as well as structural changes in the species’ differing adaptations. Gene Ontology terms related to metabolism were enriched in the differentially expressed genes in the resistant species while terms related to stress response were over-represented in the sensitive one.
Conclusion
Adaptations to new cactus hosts and hot desiccating environments were associated with periods of accelerated evolutionary change in diverse biochemistries. The hundreds of genes involved suggest adaptations of this sort would be difficult to achieve in the timeframes projected for anthropogenic climate change.
Electronic supplementary material
The online version of this article (10.1186/s12864-018-5413-3) contains supplementary material, which is available to authorized users.
Keywords: Comparative genomics, Transcriptomics, Cactophilic Drosophila, Heat stress, Host adaptation
Background
One approach to assessing the ability of species to adapt genetically to future climate change is to reconstruct the way such adaptation has occurred in the past. The best way to do this is to compare the genomes of closely related species that have diverged for the relevant phenotypes, but where genetic changes due to drift or other adaptations irrelevant to those phenotypes are minimal [1]. Drosophila is an ideal genus for such an analysis because many of its species have diverged in their responses to climatic extremes [2–4].
One particularly promising species group to study in this respect is the repleta group (subgenus Drosophila), which originated about 15 million years ago in the Americas [5]. Many species in this group, such as the mulleri subgroup species Drosophila mojavensis, D. buzzatii and D. aldrichi, are desert-adapted and display extremely high heat, cold and desiccation tolerance [2, 3] but other species, such as the hydei and repleta subgroup species D. hydei and D. repleta, are largely found outside the desert and are much less tolerant of these stresses [2, 3, 6]. Notably also, while all the repleta group species are saprophagous (feed on rotting tissue) they vary widely in their host preferences; the desert species are dietary specialists that feed and breed on necrotic cactus tissue, whereas D. hydei and D. repleta are dietary generalists which can utilise a wide range of rotting fruits and vegetables, as well as animal faeces and, in the case of D. hydei, cacti as well [7–10].
Some comparative genomic studies have been conducted on two repleta group species, the cactophilic D. mojavensis and D. buzzatii. Both are relatively tolerant to climate stresses [5, 11, 12] but the former is much more restricted geographically and in the range of cacti it will use [13]. Comparisons between these two species and two other drosophilids outside the repleta species group (D. virilis and D. grimshawi) showed expansions of gene families involved in proteolysis, sensory perception and gene regulation in the cactophilic species [5]. The same study also found the cactophilic species were undergoing rapid positive selection in genes involved in gene regulation and the catabolism of some of the heterocyclic toxins found in the cacti [5]. Transcriptomic comparisons of populations living on different hosts within both D. mojavensis and another repleta species group cactophile, D. mettleri, have also highlighted transcriptional changes in key metabolic and sensory pathways which might contribute to desiccation and/or host adaptation [12, 14–16]. There is thus evidence for both regulatory and structural changes, in the form of gene gains as well as positive selection, associated with the acquisition of cactophilism in the repleta group. However interpretation of the associations is limited by the few species studied and in some cases the substantial phylogenetic distance involved in the comparison.
Several genome-wide association (GWAS) studies have also found quantitative trait loci (QTLs) contributing to polymorphic variation in thermal and desiccation stress traits within D. melanogaster [17–20]. Associations have been recorded with hundreds of different genes, including a number of heat shock proteins, but their relevance to the cactophilic repleta species is questionable because of the ecological differences and phylogenetic distance involved, and the fact that most of the D. melanogaster studies are based on microarray rather than sequencing data.
To follow up the work on the cactophilic species above, the current study investigates gene gains and positive selection in five sequenced repleta group species, and transcriptional differences in two of them with very different thermal tolerances. The five species are D. mojavensis and D. buzzatii, plus one additional highly stress tolerant cactophile, D. aldrichi (specifically its Clade A; [21]), and two less tolerant dietary generalists, D. repleta and D. hydei. We use the D. mojavensis genome (generated from its Catalina Island clade; [22]) as published but we re-annotate the published D. buzzatii genome [5] to improve gene model prediction for that species. We present new genomes for the other three species, acknowledging that another version of the D. hydei genome has also recently been published ([23] and see below). Comparative analyses among these four genomes plus D. mojavensis, and between the five repleta species and previously published genomes from other Drosophila groups, are then used to suggest genetic factors contributing to high temperature tolerance and cactus vs generalist dietary adaptations. These analyses are founded on a robust genome-wide phylogeny for a total of 24 Drosophila species for which good quality genomes were available at the time [24]. Orthologue and duplication predictions and branch site modelling are then used to identify lineage-specific gene expansions and bursts of positive selection in the repleta species group. We also compare transcriptomes across a time course of heat shock response for the heat sensitive D. hydei and heat tolerant D. buzzatii, and test whether gene sets showing divergent transcriptomic responses to heat between these species are related to those showing genomic divergence.
Results
Genome assemblies and annotations
Among the three newly sequenced species, the highly inbred D. hydei and D. repleta lines had better assembly statistics than the D. aldrichi line, which was less inbred than the other two (see Materials and Methods and Additional file 1: Text S2). This is apparent from the larger scaffolds and smaller scaffold L50 s for D. hydei and D. repleta compared to D. aldrichi (Additional file 2: Table S1). The D. hydei assembly also had superior assembly (and annotation) statistics to the other recently published version of this genome [23]; compared to the other version, our assembly had an N50 three times larger and covered 90% of the genome in less than half the number of scaffolds (Additional file 2: Tables S1, S2).
The generalist feeders D. hydei and D. repleta yielded assembled genome sizes of ~ 165 Mb, which is very close to previous estimates generated using DAPI staining (177+/− 22 and 167+/− 13 Mb respectively; [25]). No DAPI estimate has been published for the cactophilic D. aldrichi but our assembled genome size (191 Mb) for this species was larger than the two generalists but similar to that previously published for the cactophilic D. mojavensis (194 Mb; [26]), which itself was corroborated by a DAPI estimate (183+/− 3 Mb; [25]). Notably the previously published assembly of the other cactophilic species in our analysis, D. buzzatii, was only estimated at 161 Mb, most likely due to significant underestimation of both repeat and gene content during genome assembly ([27] and see below).
The repeat contents of the various genomes were highly variable across the five assemblies (Additional file 2: Table S3). At the extremes, D. buzzatii had only 8.4% total repeat content while D. aldrichi had 24.5%. The lower D. buzzatii estimate however may in part reflect the tendency of strict short read deBruijn graph assemblies such as that one to underestimate repeat contents [27].
We identified 15,838, 14,790 and 16,070 genes respectively in D. hydei, D. repleta and D. aldrichi, most of which have annotated UTRs (Additional file 2: Table S3). Our gene numbers for these three species are slightly higher than the 14,680 and 13,919 published for D. mojavensis and D. melanogaster respectively, and higher again than the 13,158 published for D. buzzatii. We think this is because the annotation pipeline used in our study yields superior recovery and quality statistics than many of its predecessors [24]. A reannotation of the published D. buzzatii data resulted in the identification of 1374 additional genes (Additional file 2: Table S3) and brings the total gene numbers up to 14,532, which is at the lower end of the range found for the other repleta group species. The new annotation also increased the number of identifiable orthologous genes in D. buzzatii by 485 (Additional file 2: Table S4). Scans for members of the conserved CEGMA [28] and BUSCO [29] gene sets suggested that our new D. buzzatii annotation was still missing about 10% of genes, compared with less than 3% for the other species (data not shown). Accounting for these missing genes would leave D. buzzatii with similar gene numbers to D. hydei, suggesting no consistent difference in gene number between the cactophilic and non-cactophilic species.
Orthonome [24] was used to identify orthologues, inparalogues (species-specific duplications) and de novo gene births (of which there were very few) in the gene sets of these and 11 other Drosophila species (see Materials and Methods). Orthonome classifies sequences as orthologues if they meet three criteria relating to sequence similarity, synteny and, where duplications have occurred, symmetric sister group phylogenetic relationships (see Materials and Methods); sequences only meeting one or two of these criteria were classified as inparalogues and those meeting none classified as gene births. Orthonome and Interproscan (v5.16–55; [30]) were also used to place 87–91% of the genes in each species in various Gene Ontology (GO) categories [31].
Phylogenomics shows a progression from host generalists to cactophilic specialists
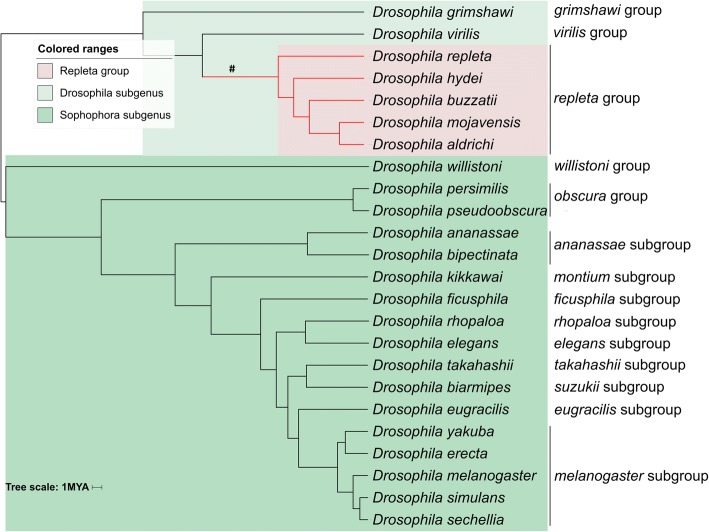
IQ-Tree [32] was used to obtain a species phylogeny from the nucleotide coding sequences of 1802 orthogroups (groups of 1:1 orthologues containing no inparalogues) that had members in all five repleta group species plus the 19 other Drosophila species for which the Drosophila 12 genomes (ftp://ftp.flybase.net/genomes/) and modENCODE (https://www.hgsc.bcm.edu/arthropods/drosophila-modencode-project) projects produced good quality genome sequences. The well-resolved tree obtained (100% bootstrap support for all branches in Fig. 1) generally confirmed earlier phylogenies for the repleta group [5, 13, 33]. Specifically it partitioned the cactophilic species, which are all in the mulleri subgroup, into the mulleri complex species D. aldrichi and D. mojavensis and the buzzatii complex species D. buzzatii, with D. hydei (in the hydei subgroup) the sister clade to that subgroup and D. repleta (in the repleta subgroup) the sister for all four of the other species.
Fig. 1.
Phylogenetic relationships of the five repleta group species and 19 previously sequenced Drosophila species based on concatenated codon alignments of 1802 orthogroups shared by all species. The divergence time was estimated using the RelTime [102] package in MEGA7 [93]. All bootstrap values for nodes from 1000 iterations were equal to 1 and the ancestral repleta group branch is indicated with a hash
Notably in the latter respect our phylogeny agreed with one, but not the other, of the only two previously published phylogenies we know of which included both D. hydei and D. repleta. One of these, by ourselves, which was based on similar methods as here and 257 genes from a partially overlapping set of species, also put the D. repleta split ancestral to that of D. hydei [34]. The other, by Oliviera et al. [13], which applied maximum likelihood methods to four mitochondrial and six nuclear gene fragments, placed D. hydei as the outgroup to the other four species and in so doing suggested D. repleta had lost an ability to use cactus that had itself emerged in the common ancestor of it and D. hydei. In putting D. repleta as an outgroup to the cactus-using species, our current and earlier study do not require D. repleta to have secondarily lost that ability. D. hydei’s ability to eat both fruits/vegetables and faeces as per D. repleta and cacti as per the mulleri subgroup is also consistent with its positioning in our topology between the D. repleta and mulleri subgroup branches. (We note here that the nannoptera species group and its most closely related outgroup D. machalilla, which all also sit in the large virilis-repleta radiation, have independently evolved cactophilism, in their case apparently from flower-feeding ancestors [5, 35].)
We also note that our phylogenetic methods were primarily built upon the supermatrix approach used in the i5K study [36] and only used carefully selected orthologues to run ModelFinder and IQ-Tree ([32, 37] and see Methods below). We obtained the same topology if we used even more stringent filtering of input genes (to remove 22% of the genes which showed compositional heterogeneity [38]) or another concatenated sequence approach using FastTree2 [39] (analyses not shown). On the other hand, we found that a Bayesian consensus tree approach using *BEAST2 [40] could not separate the two divergences and gave lower concordance values [41] and partitioned coalescence support (PCS) [42] across the gene set used (data not shown). We conclude that the two divergences in question are likely very close to one another but the topology with D. repleta as the outgroup which we propose has stronger statistical support.
Our dating analysis does however agree with Oliviera et al. [13] in suggesting an origin for the repleta group around 10–15 MYA (Fig. 1). This was a period of aridification through parts of the Americas where the group is believed to have evolved, together with the flat-leaved Opuntioideae believed to be the hosts for the original cactophiles within the group [13].
Our genome-based tree also clarified some relationships in the Sophophoran subgenus, as detailed in the Additional file 1: Text S1.
Orthogroup generation rate is high when cactophilism evolves but low in the specialist mulleri complex branch
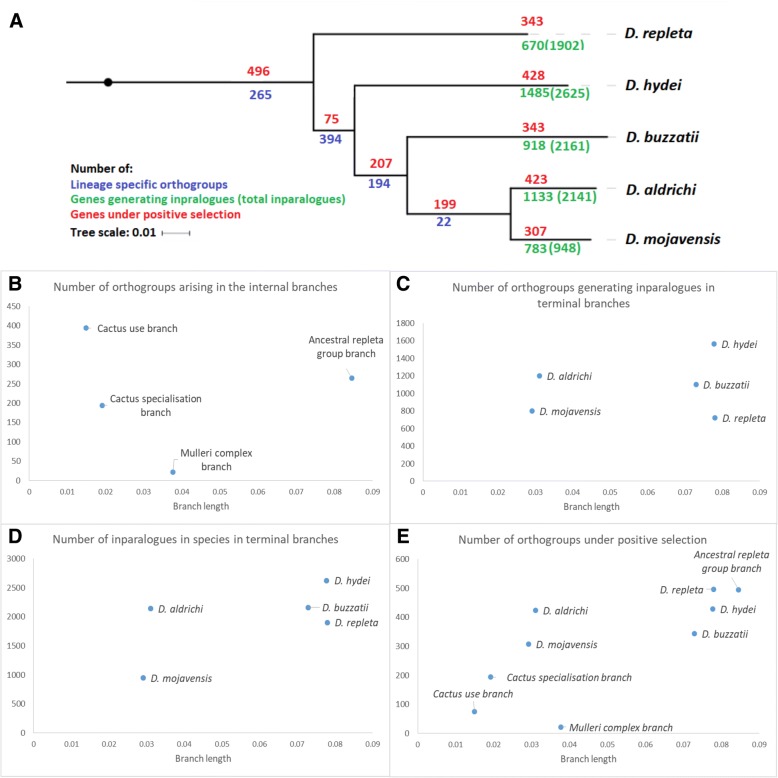
Application of Orthonome to the five repleta species and the 11 other species in the Drosophila 12 genomes project (the somewhat lesser quality modENCODE genomes being excluded from this analysis) identified orthologues for 11,780–13,883 genes in each of the 16 species (Table 1, Additional file 2: Table S4). These orthologues were distributed across 15,907 orthogroups, 971 of which orthogroups arose in the four internal branches within the repleta group phylogeny. Figure 2 shows that 265 of the 971 orthogroups arose in the earliest of these branches (hereafter the ancestral repleta group branch), 394 in the next one (ie after the split of the four cactus using species off from D. repleta, hereafter the cactus use branch), 194 in the next (ie following the split of the three cactus specialists off from D. hydei, hereafter the cactus specialisation branch), and just 22 in the most recent one (ie after the two mulleri complex species split off from D. buzzatii, hereafter the mulleri complex branch). Thus, in proportion to the presumptively neutral silent site branch lengths (calculated from 4-fold degenerate sites in 1802 full length genes as detailed in the Methods section, the rate of orthogroup creation was highest for the branches in which cactus feeding and then, to a lesser extent, cactophilic specialisation evolved, and lowest in the mulleri complex branch within the cactophilic specialists (Fig. 2). The excess of orthogroups arising in the cactus use and cactus specialisation branches in proportion to their silent site branch lengths was of the order of 300 and 150 orthogroups respectively. Relevant here is that the cactus specialisation branch is not only associated with the substantive loss of use of other hosts but also the acquisition of greater heat and desiccation tolerance, all three of the cactophilic species having significantly higher tolerance than D. repleta and D. hydei [2, 3, 43–47].
Table 1.
Summary of Orthonome analysis for D. melanogaster and five repleta group species. Average numbers of one-to-one orthologues (genes identified by Orthonome as having at least one orthologous relationship) are calculated based on pairwise orthologue predictions whereas total genes with orthologues and inparalogues are evaluated using the phylogeny-sensitive Orthonome analysis of all genomes
| Species | Number of genes | Mean number of 1:1 orthologues | Total genes with orthologues | Inparalogues | |
|---|---|---|---|---|---|
| All orthogroups | Repleta group-specific orthogroups | ||||
| D. aldrichi | 16,070 | 11,294 | 12,469 | 2141 | 100 |
| D. mojavensis | 14,680 | 11,669 | 13,147 | 948 | 69 |
| D. buzzatii (improvement compared to original annotation) | 14,532 (1374) | 10,745 (485) | 11,780 | 2161 | 173 |
| D. hydei | 15,838 | 11,096 | 12,523 | 2625 | 514 |
| D. repleta | 14,790 | 11,222 | 12,588 | 1902 | 224 |
| D. melanogaster | 13,919 | 10,894 | 12,992 | 688 | NA |
Fig. 2.
Consensus phylogenetic tree estimated using IQ-Tree with 1802 genes and 1000 bootstraps for the repleta groups species showing the numbers of orthogoups arising in the internal branches ((blue) and generating inparalogues in the terminal branches (green), plus the total numbers of inparalogues in the terminal branches (brackets, green) and the number of positively selected genes in all branches (red), togther with plots of these four metrics against the lengths of the respective branches
In fact the rate estimated for the cactus specialisation branch may have been slightly underestimated, and that for the mulleri complex slightly overestimated, because of the slightly lower rate of gene recovery from our D. buzzatii assembly noted above (which would have meant we assigned a few orthogroups which actually arose in the cactus specialisation branch to the mulleri complex branch).
Inparalogue generation rates are high in D. aldrichi
Orthonome also identified 948 (D. mojavensis) - 2625 (D. hydei) inparalogues arising in the terminal branches of the repleta group phylogeny, with the number of orthogroups giving rise to them varying between 783 (D. mojavensis) and 1485 (D. hydei) (Table 1; Fig. 2). Much of this variation was broadly proportional to the silent site branch lengths. However the most heat and desiccation resistant species, D. aldrichi [2, 3, 43–47], had about twice as many inparalogues and orthogroups giving rise to them (excesses of several hundred in both cases) as might be expected from its silent site branch length. The high number of inparalogues in this species was not obviously explicable in terms of its slightly poorer quality assembly (see above); only 171 out of 2140 of its inparalogues were found at the same locations as their templates in the assembly and were therefore possibly assembly errors (data not shown). On the other hand, inparalogue numbers in D. buzzatii may have been slightly underestimated because of the slightly lower rate of gene recovery from its assembly.
Overall, the terminal branch inparalogue numbers estimated are significantly higher in proportion to the silent site branch lengths than are the numbers of orthogroups which were estimated above to have been generated in the internal branches. This may in part be a statistical artefact; the very rigorous criteria we used for the classification of orthogroups, may mean some real orthologues were assigned as inparalogues (see Materials and Methods). However it likely also has some biological basis; only duplication events that survive long enough as functioning genes to appear in at least two of the five species will be classified as orthogroups, whereas the inparalogues will include some functionless genes, not necessarily transcribed and possibly including disruptions to open reading frames.
There was no obvious correlation between the specific orthogroups generating inparalogues in the different terminal branches (Additional file 2: Figure S1). This would indicate ongoing genetic divergence among the species, even among the cactophiles. As noted, there are considerable differences in the heat and desiccation tolerances of the five species and even the cactophiles differ in host use to a significant degree, specifically in their preferences for various flat-leaved Opuntia species (D. buzzatii) versus specific columnar cacti (the two mulleri complex species, particularly D. mojavensis [33]).
Many positively selected genes in D. aldrichi but few in the internal mulleri complex branch
Adaptive branch-site random effects likelihood (aBSREL; [48]) testing of 7359 orthogroups with 1:1 orthologues in each of the five repleta species and the 11 other FlyBase species found between 75 and 496 orthogroups undergoing positive selection in one or more branches of the repleta group phylogeny (Fig. 2). In proportion to the respective silent site branch lengths, the major outliers were the terminal branch containing D. aldrichi, which showed relatively high numbers of positively selected genes, and the preceding internal branch, the mulleri complex branch, which contained relatively few (of the order of 200 more and 200 less respectively). Drosophila aldrichi, which has the highest heat and desiccation tolerance of the species characterised [2, 3, 43–47], was also associated with relatively high numbers of gene gains (as assessed by inparalogues generated), consistent with its ecological niche having diverged significantly from the others species. Conversely, the mulleri complex branch showed very low relative numbers of both gene gain events and positively selected genes, suggesting a period of relative evolutionary genetic stability. As with the inparalogue analyses, there was little overall correlation between the specific genes showing positive selection in the different branches, even among the terminal cactus specialist branches (Additional file 2: Figure S2).
We note here that this analysis only included the 7359 orthogroups/genes which had orthologues in all 16 species analysed. While this was the most rigorous way to execute the test, it also meant that many orthogroups which were more variable in terms of their presence across species were excluded from the analysis. The analyses below suggest that orthogroups associated with gene gains are more likely to show positive selection than those that do not. These considerations suggest that our estimates of the proportions of genes under positive selection in the various branches above will underestimate the true proportions to some degree.
Genes generating inparalogues are more likely to be positively selected
We used the approach developed by O’Toole et al. [49] to test whether there was an association between the orthogroups/genes generating inparalogues and those that were under positive selection. The structure of this analysis necessarily restricted it to the terminal branches of the phylogeny but a fortunate corollary of this was that we could include more genes than in the aBSREL analysis for positive selection above (see Materials and Methods); specifically we could include all orthogroups which had members in all the repleta group species and at least one other species from the same subgenus. We found that genes generating inparalogues in the terminal branches were more likely to be under positive selection than genes which had not generated inparalogues in those branches (Table 2), although the analysis does not tell us whether the episodes of positive selection involved occurred before or after the generation of the inparalogues.
Table 2.
Proportions of genes with or without inparalogues that are under positive selection. 95% binomial confidence intervals are shown for the percentages in parentheses
| Species | % positively selected genes lacking inparalogues | % positively selected genes generating inparalogues |
|---|---|---|
| D. aldrichi | 429/9615 = 4.47 (0.41, 0.49) | 95/1237 = 7.68 (6.26, 9.31) |
| D. mojavensis | 348/10740 = 3.25 (0.29, 0.36) | 21/431 = 4.88 (3.04, 7.35) |
| D. buzzatii | 327/8951 = 3.66 (0.33, 0.41) | 86/1173 = 7.34 (5.91, 8.98) |
| D. hydei | 496/10070 = 4.93 (0.45, 0.54) | 36/692 = 5.21 (3.67, 7.13) |
| D. repleta | 371/10480 = 3.55 (0.32, 0.39) | 31/456 = 6.80 (4.67, 9.51) |
Metabolic and stress response functions are enriched in the evolutionary changes
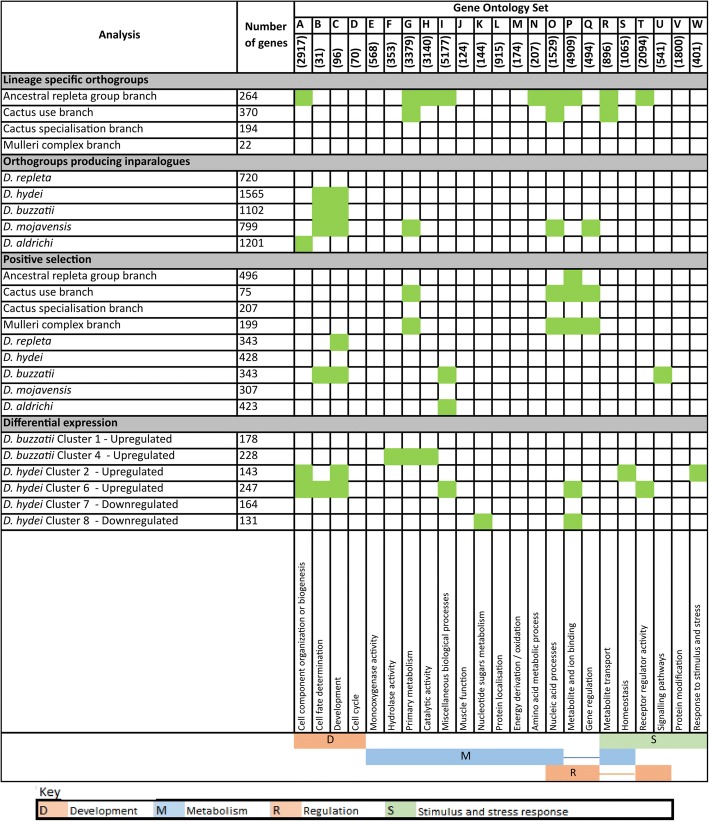
We screened the 18 branch-specific evolutionary analyses above (orthogroups created in the four internal branches, orthogroups generating inparalogues in the five terminal branches and positive selection in each of the nine branches) for the enrichment of 23 mutually exclusive sets of GO terms. These terms were obtained by applying a Louvain clustering approach to all 28,912 terms nested under ‘Biological process’ in the Ontology (see Materials and Methods). We then examined the associations found in more detail by analysing the enrichments of 265 subsets of the 23 sets, again identified by Louvain clustering. The constitutions of the different GO sets and subsets are summarised in Additional file 2: Table S5. Twenty eight significant enrichments of GO sets were found and these 28 were not randomly distributed with respect to either the analyses or the GO sets involved (Fig. 3).
Fig. 3.
Summary of significant enrichments (FDR-corrected P < 0.05) of 23 sets of GO terms for biological processes in the 18 evolutionary analyses in Fig. 2 and in the over-represented species in the six key fuzzy-c means clusters from the heat stress transcriptome analyses (Fig. 5 below)
Nine of the 28 occurred in just one analysis, namely that assessing orthogroup generation in the ancestral repleta group branch. While this branch had not generated disproportionate numbers of orthogroups relative to its silent site branch length, it was nevertheless a relatively long silent site branch. The range of biological functions enriched among the orthogroups generated in this branch may have been prerequisites for the emergence of the very broad host range of D. repleta itself, the flower feeding habits of the ancestors of the nannoptera group, fungus feeding in some fasciola subgroup species, the cactophilic lineage studied here, plus the soil-dwelling D. mettleri that feeds on the exudate of rotting cactus [13, 15]. Another 12 of the 28 significant enrichments occurred in just three other analyses, three in orthogroups arising in the cactus use branch (which had shown a pronounced excess of events relative to the silent site branch length), five in orthogroups generating inparalogues in the D. mojavensis branch (which involved increasing specialisation on particular chemically complex columnar cacti), and four in positive selection events in the D. buzzatii branch (which continues to exploit a wide range of cacti).
Five GO sets were enriched in two or more of the 28 analyses; three (G - Primary metabolism, O - Nucleic acid processes and P - Metabolite and ion transport) in the orthogroups generated in both the ancestral repleta group and cactus use branches above, and two others (B - Cell fate determination and C - Development) each involved in four terminal branch analyses, namely the orthogroups generating inparalogues in the D. hydei, D. buzzatii and D. mojavensis branches and positive selection in the D. buzzatii branch.
We then screened all the GO subsets for enrichments in the 18 analyses, which we did in terms of both absolute number and percentage increases (Fig. 4). Consistent with the set-level analyses, increases in absolute numbers were most evident for the orthogroups generated in the ancestral repleta group and cactus use branches and the orthogroups generating inparalogues in most of the terminal branches, with less marked increases in the positive selection analyses. Notably the four subsets contributing most strongly to these increases were all subsets of the Primary metabolism set (G), and three of these, Organic substance metabolic processes, Nitrogen compound metabolic processes and Primary metabolic process, would have obvious relevance to host use, and in particular the very different carbon and nitrogen contents of cactus over rotting fruit diets [12].
Fig. 4.
GO subsets most represented in the 18 evolutionary analyses, where representation is expressed in terms of either the total number (top) or percentage (below) of genes in the subset involved in the respective gene gain or positive selection events. Only subsets containing more than ten genes were analysed and only those where more than ten genes (top) or more than 5% (below) were involved in the events in any one of the analyses are shown. These values are shaded. However for those subsets included on these criteria, any non-zero involvement in any of the other analyses is also shown, but not shaded. See the Methods section for further details of the methods used to generate this figure
The percentage increase data were more informative for the smaller subsets. While they generally showed patchier distributions across analyses and GO subsets, they nevertheless did identify some relatively large effects for three subsets of Metabolite and ion transport (P) and two subsets of Response to stimulus and stress (W), which again could relate to the acquisition and specialisation of cactophilism.
Evolution in the D. aldrichi branch involves many functions
We were particularly interested to screen for the biochemical functions implicated in the bursts of inparalogue generation and positive selection in the terminal D. aldrichi branch. The GO set-level analysis above showed it was significantly enriched for two relatively ill-defined sets, namely A (Cell component organisation or biogenesis) and I (Miscellaneous biological processes), for inparalogue generation and positive selection respectively. The subset-level analyses also showed this branch was enriched for a wide range of terms, most of which were also involved to some degree in one or more analyses of other branches. These analyses suggest that a broad range of functions were involved in the evolution occurring in this branch, with little differentiation between these genes and those involved in some of the other branches.
To further interrogate the functions involved we then looked at the functional annotations of the 20 genes most strongly implicated (ie having the lowest P values) in this branch in the positive selection screen above (Table 3). Most of these 20 genes had regulatory or developmental functions, suggesting such functions might have been particular targets for the positive selection in this branch. Unfortunately we could not do the equivalent analysis for the inparalogues generated in this branch because the nature of those data prohibited any ranking of genes in terms of strength of the effect.
Table 3.
Top 20 genes undergoing positive selection in D. aldrichi. Orthogroups with Bonferroni corrected p-value < 1.0e− 20 and Likelihood ratio test statistic estimate from aBSREL > 82. Functional information for each gene was summarised based on Flybase descriptions for D. melanogaster orthologues, classification of the protein sequence using gene ontology and/or Pfam family annotations. Asterisks indicate two genes which also have inparalogues in the D. aldrichi branch
| D. aldrichi gene | D. melanogaster orthologue | Gene name | Gene function |
|---|---|---|---|
| DALD015788* | FBgn0001341 | lethal (1) 1Bi (l(1)1Bi) | Transcriptional regulator during larval and embryo stages |
| DALD015521 | FBgn0034583 | CG10527 | DUF3421; Farnesoic acid O-methyl transferase |
| DALD011516 | FBgn0034031 | CG12963 | NA |
| DALD011555 | FBgn0029518 | CG13376 | NA |
| DALD014639 | FBgn0038654 | CG14298 | Serine-type endopeptidase inhibitor activity |
| DALD008141 | FBgn0037244 | CG14647 | Transcriptional regulator of protein homo-oligomerisation |
| DALD001586 | FBgn0029686 | CG2941 | NA |
| DALD015580* | FBgn0050440 | CG30440 | Involved in positive regulation of Rho protein signal transduction |
| DALD010188 | FBgn0266566 | CG45105 | Centrosome organising gene |
| DALD004399 | FBgn0038921 | CG6332 | Testicular haploid expressed repeat |
| DALD000608 | FBgn0036179 | CG7368 | Transcription factor regulating phagocytosis |
| DALD007329 | FBgn0045495 | Gustatory receptor 28b (Gr28b) | Taste receptor involved in perception of chemical stimuli |
| DALD007082 | FBgn0032416 | Gustatory receptor 33a (Gr33a) | Taste receptor involved in perception of chemical stimuli and courtship behaviour |
| DALD009511 | FBgn0031275 | metabotropic GABA-B receptor subtype 3 (GABA-B-R3) | G-protein coupled receptor regulating glucose metabolic processes |
| DALD010061 | FBgn0051025 | Protein phosphatase 1c interacting protein 1 (Ppi1) | DUF4788, involved in protein phosphatase binding |
| DALD012340 | FBgn0003460 | sine oculis (so) | Homeobox gene regulating circadian rhythm, reproduction and development |
| DALD002312 | FBgn0260861 | TRAPP subunit 23 (Trs23) | Trafficking protein involved in vesicle-mediated transport |
| DALD007827 | FBgn0033055 | Tubulin-specific chaperone E (Tbce) | Regulates neurogenesis and neuromuscular synaptic transmission |
| DALD005587 | FBgn0085362 | Vitellin membrane-like (Vml) | vitellin membrane formation and dorsal/ventral axis specification |
| DALD009987 | FBgn0015569 | α-Esterase10 (α-Est10) | Regulator of imaginal disc-derived wing morphogenesis |
Heat stress depresses transcription more in the sensitive D. hydei than tolerant D. buzzatii
Mass bred progeny of wild-caught D. hydei and D. buzzatii females (an appropriate D. aldrichi strain not being available at that time) were subject to a set of pilot heat stresses to determine equivalent sublethal exposures. We found that 37.0 °C and 39.5 °C respectively were the maximum temperatures to which we could expose adults of the two species for 60 min without causing lethality. This difference between the two species’ maximum sublethal heat stress is consistent with previous reports on their relative thermotolerances (Kellermann et al. 2012b). We therefore carried out an experiment in the same generation in which we sampled cohorts of the two strains for transcriptomic analysis at the start, middle and end of their respective 60 min exposures to heat and then at five time points through the first 24 h of a recovery phase at 25.0 °C. The transcriptome analysis yielded 9109 genes from D. hydei for each of which we recovered a total of more than 50 RNA-Seq reads, and 9461 such genes from D. buzzatii. Of these, 1031 and 993 respectively were differentially expressed (DE) (FDR adjusted p-value < 0.05, log2 fold change in expression > 1 in at least two time points) relative to their pre-exposure level of expression.
Our first analyses of these data focused solely on the 10,443 1:1 orthologues in the two species that met the > 50 reads inclusion criterion above in either one of them. We found 123 of these showed DE in both species, 702 did so just in D. hydei and 681 did so just in D. buzzatii. This distribution represents about a 2 fold excess (123 cf. 63. 5; χ2 = 12.4, df = 1, P < 0.001) of shared DE responses compared to a null hypothesis of independent effects in the two species. However the majority of each species’ DE genes were not shared by the other species.
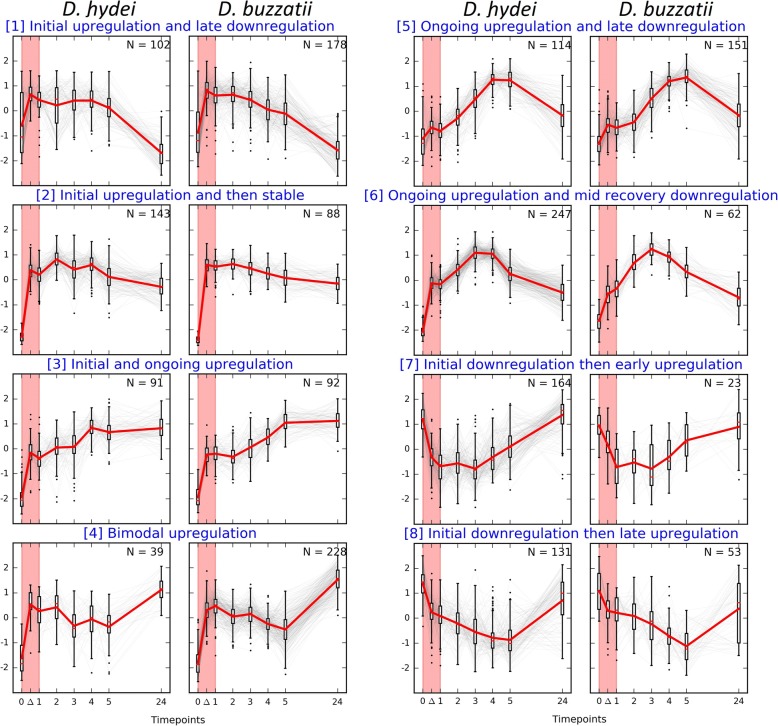
Our second analysis of the DE data used fuzzy c-means clustering of the temporal expression profiles of all 1031 DE genes in D. hydei and 875 of those in D. buzzatii (118 of the D. buzzatii DE genes showed erratic profiles which did not fit within any of the clusters formed) across all eight time points to organize them into eight clusters, representing eight major patterns of response over time (Fig. 5). Both species had genes in each expression trajectory cluster but the number of genes in most clusters varied considerably between the two species.
Fig. 5.
Fuzzy c-means clustering of genes that were differentially expressed (normalized values) in D. hydei and D. buzzatii. The y-axes are standardized expression values with mean = 0 and variance = 1
Two clusters (7 and 8) contained genes that were initially downregulated but subsequently recovered to around pre-exposure transcript abundances. Drosophila hydei genes greatly outnumbered D. buzzatii genes (combined across clusters, 219, or 288%, more genes) in both these clusters, even though D. buzzatii had been exposed to a 2.5 °C higher heat shock.
Genes in the other six clusters all showed initial upregulation, but each cluster showed a distinct trajectory through the recovery phase. Two clusters (3 and 5) containing the most similar numbers of genes from the two species are not considered further. Two others contained a large excess of D. buzzatii genes and were either downregulated relatively late in the recovery phase (cluster 1; 76, or 75%, more D. buzzatii genes) or showed a bimodal response with a second burst of upregulation late in the recovery phase (cluster 4; 189, or 485%, more D. buzzatii genes). Two clusters contained a large excess of D. hydei genes which showed little change through the recovery phase (cluster 2; 65, or 63%, more D. hydei genes) or ongoing upregulation followed by a mid-recovery phase downregulation (cluster 6; 185, or 298%, more D. hydei genes).
Overall there were over 700 genes from the discriminating clusters (ie excluding clusters 3 and 5) in excess in one or other of the species, indicating a profound difference in the transcriptional response of the two species to their respective ~ maximal sublethal high temperature shocks.
Heat stress changes transcription of stress response genes in D. hydei and metabolic genes in D. buzzatii
We investigated the functions of the genes that differentiate the transcriptional responses of D. buzzatii and D. hydei in the heat shock experiment above by screening for enrichments of the GO sets and subsets above among the six discriminating expression trajectory clusters we had identified. Specifically we focussed our attention on the genes in the species in excess in each of these clusters which did not have orthologues in the other species in the same cluster (hereafter the discriminating genes).
The discriminating genes showed significant GO set enrichments (compared to the full gene set for the respective species) in four of the clusters, one with D. buzzatii genes in excess and three with D. hydei genes in excess (Fig. 3). The discriminating D. buzzatii genes in the initially upregulated Cluster 4 were enriched for sets F (Hydrolase activity), G (Primary metabolism) and H (Catalytic activity), suggesting upregulation of many metabolic enzyme activities in response to stress. Of the three clusters with D. hydei genes in excess, the discriminating genes in the initially upregulated cluster 2 were enriched for sets A (Cell component organisation and biogenesis), C (Development), S (Homeostasis) and W (Response to stimulus and stress); those in the initially upregulated cluster 6 were enriched for sets A and C again, plus B (Cell fate determination), I (Miscellaneous biological processes) and T (Receptor regulator activity); and those in the initially downregulated cluster 8 were enriched for sets K (Nucleotide sugars metabolism) and P (Metabolite and ion binding). The enrichments for these latter three clusters suggest impacts on a variety of fundamental cellular processes.
Repeating these analyses at the subset-level (Fig. 6) showed enrichments in terms of absolute numbers and/or percentage increases for various metabolic, stress response and occasionally developmental terms. Of the two initially upregulated clusters with D. buzzatii genes in excess, the discriminating genes in cluster 1 were enriched, albeit weakly, for developmental and metabolic functions, and those in cluster 4 were quite strongly enriched for three metabolic functions and one stress response function. The discriminating genes in the initially upregulated clusters 2 and 6 which had D. hydei genes in excess were also enriched for metabolic and stimulus/stress response functions, but not the same ones as for cluster 4. The discriminating genes in the initially downregulated clusters 7 and 8 with D. hydei genes in excess were weakly enriched for various developmental, metabolic and stimulus/stress response functions.
Fig. 6.
GO subsets most represented among the genes in the species in excess that lack orthologues in the same cluster in the other species in the fuzzy-c means clusters in the differential expression analyses. Representation is expressed in terms of either the total number (top) or percentage (below) of genes from the subset in question in the species in excess (again excluding any that also had orthologues in the other species in the same cluster). Only subsets containing more than 10 genes were analysed and only those where more than ten genes (top) or more than 5% (below) were involved in any one of the six clusters analysed are shown. These values are shaded. However for those subsets included on these criteria, any non-zero involvement in any of the other analyses are also shown, but not shaded. See the Methods section for further details of the methods used to generate this figure
Thus, overall, stress response functions were more often enriched in clusters where D. hydei genes were in excess, whether they were initially up- or downregulated. On the other hand, metabolic functions were more often enriched in the four initially upregulated clusters where D. buzzatii genes were in excess.
We also scrutinised the functions of individual genes contributing most strongly to the discrimination between the species in the six discriminating clusters. Specifically we looked at the ten genes showing the best fit (ie highest membership value) to the profile for each of those clusters in the species in excess which did not have orthologues in the other species in the same cluster (Table 4). The ten discriminating D. buzzatii genes contributing most strongly to each of the two initially upregulated D. buzzatii-dominated clusters principally implicated functions related to non-enzymatic responses to stimuli (cluster 1) and enzymes with potential roles in detoxification (cluster 4). The ten discriminating D. hydei genes contributing most strongly to each of the two initially upregulated D. hydei-dominated clusters principally implicated functions related to stress response (cluster 2) and regulation and stress response (cluster 6). The ten discriminating D. hydei genes contributing most strongly to each of the two initially downregulated D. hydei-dominated clusters principally implicated functions related to regulation (cluster 7) and development (cluster 8), with both clusters also implicating stress response genes. Thus all four D. hydei-dominated clusters involved various stress response genes but such genes were not prominent in either of the D. buzzatii-dominated clusters. This agrees well with the findings from the set and subset enrichment analyses above.
Table 4.
Top 10 genes driving differential expression in six most divergent clusters between D. hydei and D. buzzatii, along with the membership coefficient ranges of the genes in parentheses. Functional information for each gene was summarised based on Flybase descriptions for D. melanogaster orthologues, where available, and otherwise, domain classification of the protein sequence using gene ontology and Pfam family annotations
| Gene ID | Gene name | Gene function |
|---|---|---|
| Cluster 1: Initial upregulation & late downregulation (0.51, 0.59); D. buzzatii in excess, mainly response to stimuli | ||
| DBUZO2014318 | Odorant receptor 10a (Or10a) | Chemoreceptor that mediates response to volatile chemicals |
| DBUZO2013263 | Ucp4C (Ucp4C) | Protein uncouples respiration and energy dissipation |
| DBUZO2011185 | CG17387 | Involved in cilium dependent cell motility |
| DBUZO2010006 | CG11475 (DUF89) | May be involved in protein methylation in response to DNA damage |
| DBUZO2009533 | Ppm1 | Involved in protein serine/threonine phosphatase activity |
| DBUZO2005585 | CG9702 | Transmembrane transporter involved in sulfate transport and regulation of intracellular pH |
| DBUZO2002166 | NA | |
| DBUZO2001878 | Gustatory receptor 97a (Gr97a) | Involved in sensory perception of taste |
| DBUZO2001291 | CG5538 | Voltage gates chloride channel |
| DBUZO2001235 | NA | |
| Cluster 2: Initial upregulation & then stable (0.41, 0.45); D. hydei in excess, mainly stress response | ||
| DHYD012352 | Heat shock protein 83 (Hsp83) | Heat shock protein 90 family, regulates response to abiotic stress and circadian behaviour |
| DHYD011340 | NA | Heat Shock protein 70 family |
| DHYD011253 | CG14516 | Zing ion binding peptidase |
| DHYD011218 | frayed (fray) | Protein serine/threonine kinase involved in several development processes |
| DHYD007159 | Egg-derived tyrosine phosphatase (EDTP) | Protein-tyrosine phosphatase-like gene involved in oogenesis and dephosphorylation |
| DHYD006916 | CG8944 | DNA binding zinc finger domain gene |
| DHYD005532 | NA | |
| DHYD003416 | CG4950 | |
| DHYD001898 | tramtrack (ttk) | DNA binding RNA polymerase promoter involved in response to external stimulus |
| DHYD000023 | Cyclin E (CycE) | Cyclin dependent protein kinase, positively regulates cell cycle and morphogenesis |
| Cluster 4: Bimodal upregulation (0.69, 0.76); D. buzzatii in excess, mainly enzymes involved in detox | ||
| DBUZO2012733 | NA | Cytochrome P450 |
| DBUZO2012084 | NA | S1 peptidase |
| DBUZO2011594 | CG3699 | Short-chain dehydrogenase/reductase, involved in fatty-acid beta-oxidation |
| DBUZO2009194 | NA | Cytochrome P450 |
| DBUZO2005738 | CG31087 (DUF227) | Involved in neurogenesis |
| DBUZO2005447 | alpha-Esterase-5 (alpha-Est5) | Type B carboxylesterase |
| DBUZO2002940 | Maltase A4 (Mal-A4) | Glycosyl hydrolase, involved in carbohydrate metabolism |
| DBUZO2002034 | Tetraspanin 42 Eq (Tsp42Eq) | Cell surface receptor |
| DBUZO2001132 | CG42335 | M1 peptidase involved in proteolysis |
| DBUZO2000745 | CG31198 | M1 peptidase involved in proteolysis |
| Cluster 6: Ongoing upregulation &-mid-recovery downregulation (0.50, 0.53); D. hydei in excess, mainly regulation & stress response | ||
| DHYD014524 | Hira (Hira) | Histone H3-H4 chaperone, involved in various replication-independent nucleosome assembly processes |
| DHYD014213 | ellipsoid body open (ebo) | Complexes with actin, chic, and Ran-GTPase to mediate actin nuclear export |
| DHYD014093 | Ajuba LIM protein (jub) | Protein inhibits activation of the Hippo pathway kinase |
| DHYD011516 | CG7065 | |
| DHYD011485 | maroon-like (mal) | Molybdenum cofactor sulfurase involved in ommochrome biosynthesis |
| DHYD010172 | slow as molasses (slam) | Involved in protein localisation and migration |
| DHYD008182 | withered (whd) | Involved in response to starvation, ethanol, oxidative stress and metal ions |
| DHYD007200 | Enigma (Egm) | Involved in response to endoplasmic reticulum stress; fatty acid beta-oxidation; lipid homeostasis and cellular response to oxidative stress |
| DHYD006980 | CG8611 | Involved in unwinding RNA secondary structure |
| DHYD001526 | Hillarin (Hil) | W180-domain protein, affects cytokinesis, developing nervous system |
| Cluster 7: Initial downregulation & early upregulation (0.42, 0.44); D. hydei in excess, mainly regulation & stress response | ||
| DHYD015984 | Catalytic subunit 3B of the oligosaccharyltransferase complex (Stt3B) | Involved in post-translational protein modification |
| DHYD015297 | Ror | Involved in protein phosphorylation; central nervous system development and transmembrane receptor protein tyrosine kinase signalling pathway |
| DHYD012423 | Myocardin-related transcription factor (Mrtf) | Transcription factor with roles in cell migration during development |
| DHYD009550 | pasilla (ps) | A nuclear RNA binding protein implicated in splicing |
| DHYD008649 | epithelial membrane protein (emp) | Associated with autophagic cell death |
| DHYD006908 | NA | |
| DHYD006727 | NA | Involved in fatty acid elongation |
| DHYD004130 | CG31522 | Involved in fatty acid elongation |
| DHYD004039 | Heterogeneous nuclear ribonucleoprotein at 87F (Hrb87F) | Involved in regulation of alternative mRNA splicing and responses to starvation and heat |
| DHYD001282 | CG16713 | Pancreatic trypsin inhibitor involved in development and signalling |
| Cluster 8: Initial upregulation & late upregulation (0.40, 0.43); D. hydei in excess, mainly development & stress response | ||
| DHYD018935 | short stop (shot) | Cytoskeletal linker molecule in the nervous system and other tissues |
| DHYD016713 | Gp150 (Gp150) | Transmembrane glycoprotein; regulates Notch signalling in development |
| DHYD016696 | acyl-Coenzyme A oxidase at 57D distal (Acox57D-d) | fatty acid beta-oxidation |
| DHYD014956 | rugose (rg) | Involved in olfactory learning and short-term memory |
| DHYD012466 | Laminin A (LanA) | Regulates growth and locomotion of cells |
| DHYD009427 | Glutathione S transferase Z2 (GstZ2) | Involved in aromatic amino acid and glutathione metabolic process |
| DHYD009383 | CG43222 | |
| DHYD001387 | Not1 | Muscular development, morphogenesis, mRNA catabolic processes |
| DHYD001006 | CG42500 | Induced during the immune response of Drosophila |
| DHYD000649 | NA | |
None of the 60 genes contributing most strongly to the discrimination between species in the six discriminating clusters above were among the orthogroups generated in the cactus use branch or those generating inparalogues or under positive selection in the D. aldrichi branch.
Discussion
We have found an unusually high frequency of gene gain events in internal branches of the repleta species group phylogeny during which the topology of our phylogenies suggests the ability to use cactus hosts was acquired. The frequency of gene gains also remained quite high in the subsequent cactus specialisation branch, which was associated with the lost use of other hosts but the gain of relatively high heat and desiccation tolerance. It seems more likely that the gene gains in this branch were associated with the gain of climatic tolerance than the narrowing of host range; a loss of function seems more likely to be linked to gene loss rather than gene gain [50]. The rate of gene gains then slowed considerably in the following mulleri complex branch, before picking up again in certain terminal branches, in particular the D. aldrichi branch, as the species diverged in their use of different cactus hosts and in climatic tolerance and D. aldrichi evolved the highest level of heat and desiccation tolerance.
There is no direct precedent for our analytical approach to assessing rates of accepted duplication events during adaptive evolution. There are many cases where high numbers of duplication events for specific genes or gene families have been associated, in several cases causally, with host shifts [34, 51, 52], or the development of insecticide resistance, in insects [53, 54]. However our data show for the first time a suite of duplications across a range of genes, families and physiologies associated with adaptive changes. Further they do so for both internal and terminal branches of the phylogeny.
We did not find unusually high frequencies of positive selection events in the early branches of the phylogeny with the high frequency of gene gains, but did so in the terminal D. aldrichi branch and overall we found a positive association between the specific genes involved in duplication events and those subject to positive selection in the later branches. Our findings are broadly consistent with the hypothesis that the evolution of new functions is associated with gene duplication and subsequent neofunctionalisation events ([55], but also see [56]).
A broad range of biochemical functions were implicated both in the evolutionary changes above and in the changes in other branches associated with the cactophilic species which did not show such unusually high rates of change. Notably Gene Ontology terms associated with the metabolism of organic and nitrogenous compounds were enriched in a number of branches associated with cactophilism. The nitrogen and secondary compound constitutions of rotting cacti differ markedly from those of rotting fruits and these enrichments are consistent with the idea that some of the evolutionary changes were causally linked to changes in host use. They are also consistent with previous comparative genomic and transcriptomic studies of host races of the cactophilic D. mojavensis and D. mettleri which also associated differences in host use with such biochemical functions [12, 14–16].
We found that gene gains in the relatively long ancestral repleta group branch were enriched for Gene Ontology terms associated with a particularly wide range of metabolic functions. This may relate to the wide array of host uses that evolved in various daughter lineages, ranging from the cactophiles in the hydei subgroup of interest here to the flower feeders in the bromeliae subgroup, the independently evolved cactophilism in the nannoptera subgroup, the fungus feeders in the fasciola subgroup and animal faeces in D. repleta itself.
A range of functions were also enriched among the disproportionately high number of genes generating inparalogues and/or under positive selection in the terminal D. aldrichi branch, but the twenty genes contributing most strongly to the positive selection events in this branch predominantly involved regulatory and developmental processes. These events may contribute to the extreme heat and dessication tolerance of this species.
We also carried out a comparative transcriptome analysis on freshly caught collections of another heat tolerant species, D. buzzatii, and the relatively heat sensitive D. hydei in order to further investigate the potential molecular basis of the thermal tolerance differences in the repleta group. Several hundred genes were found to respond differently to comparable levels of sub-lethal heat stress in the two species. Even though the heat shock administered to D. hydei was less extreme in terms of temperature than that given to D. buzzatii, D. hydei genes more often showed initial downregulation responses than did D. buzzatii genes and more of their responsive genes had orthologues associated with stress responses in other species. However, there was no significant overlap between the heat responsive genes in either species and those implicated in the gene gain and positive selection events in D. aldrichi, suggesting that the genes showing different transcriptional responses to heat shock in the two repleta group species tested are not those on which natural selection has acted to confer extreme heat tolerance in D. aldrichi. Interestingly, there was also no significant overlap (data not shown) between the heat responsive genes in either species and a panel of candidate genes associated with heat stress responses in intraspecific comparisons in D. melanogaster [17, 18]. This could be because of differences in methodologies, the very different ecologies of the different species, or the phylogenetic distance between D. melanogaster and the repleta group species studied here.
Finally we note that hundreds of genes were implicated in the disproportionately high rates of gene gains and positive selection events associated with the phenotypic changes above. This suggests large-scale genomic changes underlay the phenotypic evolution, which is consistent with the findings of the previous comparative analyses involving the cactophilic repleta group species ([5, 14–16] and see Background). The scale of change specifically associated with the cactus use branch is also consistent with the scale of nucleotide differences found in reduced representation genomic analyses of host shifts in Rhagolites flies [57], but it is greater than those so far associated with such analyses of host shifts in Timema stick insects [58] or in full genome analyses of adaptation to the otherwise toxic host Morinda citrifolia in D. sechellia and some D. yakuba [59, 60]. In so much as the rate of adaptation to the anthropogenic climate change now in prospect may be limited by the rate at which new duplication and mutation events are generated, the scale of change in the internal cactus specialisation and terminal D. aldrichi branches associated with increased climatic tolerance also suggests it would be beyond the capacity of many current species to achieve equivalent shifts in climate niches in the timeframes now projected for climate change.
Conclusion
Our phylogenomics analysis of fourteen Drosophila species finds bursts of duplication and selection events associated with adaptation to arid environments. These adaptations include both acquisition of cactophilic host use and greater tolerance of climatic extremes. The bursts of duplication and selection, which support the duplication and neofunctionalisation theory of adaptation, have occurred across both internal and terminal branches of the phylogeny encompassing 6–8 million years. The genes involved cover a wide range of physiological functions and there is little overlap between them and the genes whose transcriptional profiles after heat stress differ between the two species.
Methods
Fly strains
The D. hydei and D. repleta strains used for genome sequencing were collected from Townsville, QLD, Australia and Wandin, VIC, Australia, respectively and then inbred in the laboratory for 17 generations of single pair full-sibling mating to reduce their heterozygosity (expected inbreeding co-efficient > 0.7 [61]). The D. aldrichi strain sequenced was originally caught in Mexico in 2002 and was likely inbred to some degree while being maintained in the University of California San Diego Drosophila Species Stock Center (stock number: 15081–1251.13). It was further inbred for two generations of sibling mating from a single pair of flies prior to sequencing.
Individuals for the mixed life stage transcriptome sequencing that we used to augment the annotations were obtained from the inbred D. hydei and D. repleta lines above and from a wild-caught mass bred line of D. aldrichi from Inglewood, QLD, Australia. RNA was prepared from a mixture of six life stages (embryos, first, second and third instar larvae, pupae and adults, combined in approximately equal weight amounts) for each species.
Recently collected strains of D. hydei and D. buzzatii were used in the thermal stress transcriptomics experiment. The offspring of 50 field females were pooled to establish a mass bred population for each species. The D. hydei females were collected from Pascoe Vale, Melbourne, VIC and the D. buzzatii from Inglewood, QLD, Australia. The mass bred strains were maintained at 25 °C for one generation prior to the experiment.
Genome sequencing and assembly
Sequencing
Adult females from the inbred D. repleta, D. hydei and D. aldrichi strains were harvested and DNA extracted from heads to minimise contamination with DNA from their gut flora. Six libraries were then prepared from the DNA from each species for sequencing on an Illumina HiSeq2000 platform; three paired-end libraries spanning 250 bp, 500 bp and 800 bp using 150 bp or 100 bp paired-end sequencing runs, plus three long insert mate-pair libraries with insert sizes of 2kbp, 5kbp and 10kbp using 49 bp paired-end runs. All library preparation and sequencing were carried out at Beijing Genomics Institute (BGI), Shenzhen. At least 35Gb of raw sequence data were obtained per species, yielding a final read coverage of ~210x of the estimated genome size.
Assembly
The raw paired-end and mate-pair reads were assessed using FastQC [62] and novel contaminant sequences identified using K-mer counting with Jellyfish [63] to provide inputs for trimming and filtering using Trimmomatic [64].The insert size for each library was then re-estimated by aligning the trimmed reads to the D. mojavensis genome [see Additional file 1: Text S2]. Quorum [65] was then used to correct the trimmed reads for D. hydei and D. repleta while BFC [66] was used for D. aldrichi (because higher levels of heterozygosity were expected for this species). The corrected reads were then assembled using the MaSuRCA assembler [67] and contigs extended and scaffolded using the SSPACE 2 scaffolder [68].
The scaffolded genomes were further improved by local realignment and gap-filling carried out using multiple rounds of Pilon [69], with reads aligned using SNAP aligner [70], until no iterative gains were observed. The final assemblies were then benchmarked using the BUSCO pipeline [29] following the authors’ instructions.
Repeat sequence analysis
Tandem repeats and transposable elements (TEs) in D. hydei, D. repleta, D. buzzatii and D. aldrichi were identified following the strategy of Zhang et al. [71]. TRF [72] and RepeatMasker [73] were used to characterise tandem repeats; RepeatMasker was used with the Repbase [74] database of known repeat sequences in D. melanogaster to identify repeats shared with the latter species; and previously undescribed repeats were identified using LTR_FINDER [75], PILER [76] and RepeatScout [77]. Repeat proteins were also identified using RepeatProteinMask (version 3.2.2) as implemented in RepeatMasker. All the repeat sequences in each species identified by the different methods were combined into a final repeat library and categorized in a hierarchical way, as per [71].
Genome annotation
Transcriptome sequencing for gene prediction
We made RNA-Seq transcriptomes from mixed life stage RNA preparations of D. repleta, D. aldrichi and D. hydei on the Illumina HiSeq 2000 platform to assist in gene annotations. The RNAs were prepared using the Zymo Direct-zol™ RNA extraction kits (Zymo Research, Irvine, USA). A standard RNA-Seq library was prepared for D. hydei, while strand-specific libraries were prepared for D. repleta and D. aldrichi according to Borodina et al. [78]. All three libraries were prepared by BGI following standard Illumina protocols. Hundred bp unstranded paired-end RNA-Seq sequencing yielded 4Gb of data for D. hydei and 3Gb each for D. repleta and D. aldrichi. These data were then filtered to remove reads which had more than 10% of bases unknown, more than 40 bases with Phred scores less than 7, or > 20% of the sequence comprising adaptor sequence. This left a high quality read set of ~ 2 Gb for D. hydei and D. repleta and ~ 3 Gb for D. aldrichi.
Annotation of gene models
We identified, evaluated and collated protein coding and putative non-coding genes in D. aldrichi, D. buzzatii, D. hydei and D. repleta using ten sets of RNA-Seq data plus homologue-guided and ab-initio based gene prediction as detailed in Additional file 1: Text S2. The repeat sequences were first used to soft-mask the genome, upon which we mapped 2315 single copy D. melanogaster genes (248 using CEGMA [28] and 2067 from OrthoDB7 [79] using Exonerate [80]) and the D. melanogaster and D. mojavensis proteomes [80] to generate splice-aware alignments.
The resulting splice junctions were used to guide two-pass mapping of RNA-Seq data onto the respective genomes using GMAP/GSNAP (v 2014-08-20; [81] and STAR aligner (v 2.4.0; [82]. The RNA alignments were then used to annotate transcripts using StringTie [83] and genome-guided Trinity [84], while trimmed RNAseq data were used for de novo assembly of the species’ transcripts using Trinity. All three transcript sets were collated using PASA2 [85] to create a comprehensive transcript database and high-quality training dataset (GTrainSet) for training and subsequent annotation of genes using four ab-initio predictors; Genemark-ET [86], Glimmer [87], Augustus (v 3.0.2; [88] and SNAP [89].
All ten sets of gene models created were finally passed to EvidenceModeler [90] to create a comprehensive whole genome annotation. Greatest weight was given to StringTie (15) followed by GTrainSet (12) and homology-based (11) evidence, and least to the ab initio predictions (2–4). The gene set was then updated to annotate alternative splice events, 3′ and 5’ UTRs and to refine gene boundaries using RNA-evidence in PASA2. Finally, nested genes were added using the homology matches in D. melanogaster and D. mojavensis, creating the official gene sets (OGSs).
Comparative genomics
Classification of orthologues
The Orthonome pipeline (http://www.orthonome.com/; [24, 91]) was applied to sets of 1:1 orthologues and orthogroups extracted from the OGSs. The pipeline also identified gene birth events and inparalogues. Only the longest isoform for each gene in each genome was used in this analysis. Note that while Orthonome partitions the output into orthologues, inparalogues and gene births, the number of inparalogues will be inflated to a degree by artefactual inparalogues – which are biologically orthologous genes where one member is missing from the data or where the duplication event occurred so close to a speciation event that it could not be distinguished from two independent post-speciation duplication events.
Three Orthonome analyses were carried out, the first for calculating the phylogeny and the remaining two for comparative genomics. These were (i) one using the twelve Flybase genomes [92] (D. ananassae, D. erecta, D. grimshawi, D. melanogaster, D. mojavensis, D. persimilis, D. pseudoobscura, D. sechellia, D. simulans, D. virilis, D. willistoni and D. yakuba), eight modENCODE genomes (https://www.hgsc.bcm.edu/arthropods/drosophila-modencode-project; D. biarmipes, D. bipectinata, D. elegans, D. eugracilis, D. ficusphila, D. kikkawai, D. rhopaloa and D. takahashii) and our newly annotated D. aldrichi, D. buzzatii, D. hydei and D. repleta genomes, (ii) one using this set minus the eight lesser quality modENCODE genomes, and (iii) one just using the five repleta species and D. melanogaster.
Species phylogeny
The orthologues obtained from analysis (i) above led to the identification of 4935 orthogroups. The orthogroups were then filtered to keep only those that satisfied the following criteria: (1) none of the species had any traceable duplications (i.e. best blast hit was always an orthologue), (2) relatively slowly evolving, with no genes presenting a Ka/Ks > 1 (see below for calculation method), (3) > 200 amino acids in length (unless all members of the orthogroup were within five residues length of each other), and (4) all genes within a 1.5 median absolute deviation from the median length of the orthogroup. The nucleotide coding sequences from the 1802 orthogroups retained were used to construct both a concatenated species phylogeny using FastTree2 [39] with 1000 internal boot-strap replicates and a consensus tree using IQ-Tree [32] allowing a free rate model and with each gene treated as its own partition. The two phylogenies had the same topology but the IQ-Tree tree was chosen for all subsequent analyses. Divergence time was estimated with MEGA7 (Rel-Time) [93] based on a calibration for Drosophila obtained according to Obbard et al. [94].
We also constructed two other trees to interrogate more closely the D. hydei – D. repleta relationship. One was another IQ-Tree based tree and the other was a *BEAST2 Bayesian tree [40]. Both were constructed using the inferred amino acid sequences for the 100 best genes (< 1% gappyness and no compositional heterogeneity; χ2p-value > 0.05). Concordance factors [41] and partitioned coalescence support scores [42] were then used to compare the support for the different phylogenies, finding the *BEAST2 tree performed less well than the others on both measures.
Functional annotations
We first augmented the functional annotations for D. melanogaster genes obtained from FlyBase using InterProScan 5 (January 2016 release) [30]. These updated annotations were transferred to D. aldrichi, D. buzzatii, D. hydei and D. repleta based on orthologous relationships estimated using Orthonome (see above). Functional annotation of the inparalogues remaining was carried out by identifying the phylogenetically closest orthologue or inparalogue from D. melanogaster while de novo annotation was carried out for gene births using InterProScan 5. Gene interaction networks were also transferred from D. melanogaster using orthology [95]. FlyBase Gene Group annotations were transferred to the repleta species in a similar fashion.
Evolutionary rate analyses
Branch-site model tests (adaptive branch-site random effects likelihood; aBSREL) were carried out to identify specific lineages in the phylogeny (both terminal and internal branches) in which orthogroups identified by Orthonome may have been under positive selection. This analysis was carried out using all the FlyBase genomes and the four repleta group genomes annotated in the current study. We assumed fixed branch lengths based on the phylogeny constructed above and calculations used the HYPHY package [48]. Each branch in the repleta lineage was analysed and a False Discovery Rate- (FDR-) corrected χ2P-value cut-off of < 0.05 was used.
As per O’Toole et al. [49], the terminal branches were also analysed to compare the proportions of genes under positive selection in the branches that had versus had not generated inparalogues.
Comparative thermal stress transcriptomics for D. hydei and D. buzzatii
The goal of this experiment was to compare the transcriptional responses to thermal stress of species that differed in high temperature tolerance. Mass-bred populations initiated from recently-collected flies were used for these experiments to minimise issues with changes in gene expression during laboratory adaptation. We also conducted pilot studies on these mass bred populations to identify the most suitable testing temperatures (see Additional file 1: Text S2 for details). These were found to be 37 °C for D. hydei and 39.5 °C for D. buzzatii, consistent with previous findings of their differences in thermal tolerance [3].
For the stress assays, three independent replicates, each of ten virgin adult females aged two days since emergence, were harvested at each time point indicated in Additional file 2: Figure S3 and snap-frozen in liquid nitrogen. One time point was frozen immediately prior to exposure (used as the control for differential expression comparisons), one half way through the exposure, one immediately after the exposure and five at defined intervals over the next 24 h. The snap-frozen material was stored at − 80 °C until RNA extraction (pooling all ten flies in each replicate) using the Zymo Direct-zol™ RNA extraction kits (Zymo Research, Irvine, USA). RNA-Seq libraries were prepared and sequenced with the Illumina HiSeq 2000 platform as above. All 48 libraries (8 time points and 3 replicates for each species) were run on the same instrument in the same flow cell.
The sequence reads for each species were filtered for low quality and adapter sequences and trimmed with Trimmomatic [64] as above. The cleaned reads were then aligned to their respective genomes using a two-pass strategy in STAR aligner (v 2.4.2a) [82], which also produced the read counts matrix for each alignment. These outputs were combined to create an expression value matrix for each gene across all eight time points as input for subsequent analyses.
Counts for all genes with more than 50 reads (threshold for genes with low expression) were normalised across samples using the trimmed mean of M-values method [96] and converted to log2 counts per million values (log2cpm) with associated quality weights using the voom-limma pipeline [97]. Batch effects due to manual handling were corrected with ComBat [98] against an intercept only model. Changes in gene expression over time were then modelled in limma. Genes with a significant difference in expression relative to the first time point, ie the one immediately prior to the treatment, (FDR) adjusted P value less than 0.05 and a log2 fold-change in expression greater than 1) in at least two of the subsequent time points were considered to be heat responsive.
To identify common patterns of expression among differentially expressed genes, the expression of each gene across the different time points was first standardised (independently in each species) to have a mean of 0 and standard deviation of 1. The standardised expression values for both species were then combined and subjected to fuzzy c-means clustering [99]. Trials with from four to 20 clusters yielded the highest fuzzy means partitioning coefficient with eight clusters, so further analysis was based on those eight clusters. Membership exponent values were then calculated for each gene in each cluster to identify the genes best explaining the cluster pattern across time points. The expression patterns for the genes with the highest membership exponent in each cluster were then plotted to visualise trends. The core genes of each cluster (membership value greater than 0.5) were used for the functional enrichment analysis below.
Functional enrichment analyses
Functional enrichment of gene ontologies was carried out after first clustering the 47,676 GO terms ([31]; release: 2016-11-03) into 43 mutually exclusive sets using a Louvain clustering method [100] and allocating semantic similarities as edge weights between connected hierarchical GO terms. We used 23 of these sets that represent biological processes as the framework for our enrichment analyses. Additional file 2: Table S5 summarises the constitution of these sets in terms of the major subsets within them. For each analysis, the respective gene lists were tested for enrichment of each GO term based on frequency with Goatools (https://github.com/tanghaibao/goatools) and the GO terms obtained as significantly enriched were then compressed using REVIGO [101] and assigned to the sets and subsets above. The 23 sets were given names which summarised the dominant highest level GO term(s) within them, while the subsets simply took the names of the dominant highest level GO term (which was always a lower level than in the sets) within them.
Additional files
Text S1. Phylogenetics of sophophoran subgenus. Text S2. Detailed materials and methods for genome assembly and annotation. (DOCX 49 kb)
Figure S1. Overlap between the orthogroups generating inparalogues in the different species. Figure S2. Overlap between orthogroups belonging to genes under positive selection in each repleta group species. Figure S3. Scheme for heat stress and recovery assay. Table S1. Scaffold and contig length based statistics as well as results from BUSCO genome assessment using the dataset for the five assembled repleta group genomes and D. melanogaster. Table S2. Repeat content analysis for the six species analysed in this study, characterising transposable elements as well as tandem repeats. Table S3. Genome annotation statistics for the five assembled repleta group genomes and D. melanogaster. Table S4. Number of pairwise orthologues between D. melanogaster and the five repleta species, plus the previously published annotation of D. buzzatii. Table S5. Sets of functional terms describing hierarchical grouping of gene ontology terms and the number of genes in Drosophila melanogaster within each set. (DOCX 301 kb)
Acknowledgments
We are grateful to Drs Renee Catullo, Kerensa McElroy, Mark Schultz and Alexandre Fournier-level and Mr. Zijun Xiong for their advice and assistance in various parts of the project. We also thank the NeCTAR Research Cloud for their computational facilities.
Funding
This work was supported in part by Australia’s Science and Industry Endowment Fund and the Australian Research Council (under their Laureate Fellowship Scheme). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Availability of data and materials
All DNA and RNA-Seq sequencing data, assembled genomes and alignments used for phylogenomics can be accessed from NCBI, under SRA submission PRJNA476692 and CSIRO Data Access Portal (10.25919/5bdbf09ba0db2).
Authors’ contributions
RVR, CS, GZ, AAH and JGO conceived and designed the study; RVR, CC, MS and JS carried out the empirical work; RVR, SLP, FL, PG, GZ, SFL, AAH and JGO carried out the analyses; RVR, SLP, GZ, AAH and JGO led the writing of the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Rahul V. Rane, Email: Rahul.Rane@csiro.au
Stephen L. Pearce, Email: Stephen.Pearce@csiro.au
Fang Li, Email: lifang1@genomics.cn.
Chris Coppin, Email: Chris.Coppin@csiro.au.
Michele Schiffer, Email: Michele.Schiffer@jcu.edu.au.
Jennifer Shirriffs, Email: jennifer.shirriffs@gmail.com.
Carla M. Sgrò, Email: carla.sgro@monash.edu
Philippa C. Griffin, Email: griffinp@unimelb.edu.au
Goujie Zhang, Email: zhanggj@genomics.cn.
Siu F. Lee, Email: Ronald.Lee@csiro.au
Ary A. Hoffmann, Email: ary@unimelb.edu.au
John G. Oakeshott, Email: John.Oakeshott@csiro.au
References
- 1.Somero G. The physiology of climate change: how potentials for acclimatization and genetic adaptation will determine ‘winners’ and ‘losers. J Exp Biol. 2010;213(6):912–920. doi: 10.1242/jeb.037473. [DOI] [PubMed] [Google Scholar]
- 2.Kellermann V, Loeschcke V, Hoffmann AA, Kristensen TN, Flojgaard C, David JR, et al. Phylogenetic constraints in key functional traits behind species' climate niches: patterns of desiccation and cold resistance across 95 Drosophila species. Evolution. 2012;66(11):3377–3389. doi: 10.1111/j.1558-5646.2012.01685.x. [DOI] [PubMed] [Google Scholar]
- 3.Kellermann V, Overgaard J, Hoffmann AA, Flojgaard C, Svenning JC, Loeschcke V. Upper thermal limits of Drosophila are linked to species distributions and strongly constrained phylogenetically. Proc Natl Acad Sci U S A. 2012;109(40):16228–16233. doi: 10.1073/pnas.1207553109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Overgaard J, Kearney MR, Hoffmann AA. Sensitivity to thermal extremes in Australian Drosophila implies similar impacts of climate change on the distribution of widespread and tropical species. Glob Chang Biol. 2014;20(6):1738–1750. doi: 10.1111/gcb.12521. [DOI] [PubMed] [Google Scholar]
- 5.Guillen Y, Rius N, Delprat A, Williford A, Muyas F, Puig M, et al. Genomics of ecological adaptation in cactophilic Drosophila. Genome Biol Evol. 2015;7(1):349–366. doi: 10.1093/gbe/evu291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kellermann V, van Heerwaarden B, Sgrò CM, Hoffmann AA. Fundamental evolutionary limits in ecological traits drive Drosophila species distributions. Science. 2009;325(5945):1244–1246. doi: 10.1126/science.1175443. [DOI] [PubMed] [Google Scholar]
- 7.Goñi B, Remedios M, González-Vainer P, Martínez M, Vilela CR. Species of Drosophila (Diptera: Drosophilidae) attracted to dung and carrion baited pitfall traps in the Uruguayan eastern Serranías. Zoologia (Curitiba) 2012;29(4):308–317. doi: 10.1590/S1984-46702012000400004. [DOI] [Google Scholar]
- 8.Soto IM, Carreira VP, Corio C, Padro J, Soto EM, Hasson E. Differences in tolerance to host cactus alkaloids in Drosophila koepferae and D. buzzatii. PLoS One. 2014;9(2):e88370. doi: 10.1371/journal.pone.0088370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fogleman JC, Danielson PB. Chemical interactions in the cactus-microorganism-Drosophila model system of the Sonoran Desert. Am Zool. 2001;41(4):877–889. [Google Scholar]
- 10.O’Grady PM, Markow TA. Rapidly evolving genes and genetic systems. New York: Oxford University Press; 2012. Rapid morphological, ecological and behavioral evolution in Drosophila: comparisons between the cactophilic repleta species group and the endemic Hawaiian Drosophila. [Google Scholar]
- 11.Matzkin LM. Ecological Genomics. Dordrecht: Springer; 2014. Ecological genomics of host shifts in Drosophila mojavensis; pp. 233–247. [DOI] [PubMed] [Google Scholar]
- 12.Matzkin LM, Johnson S, Paight C, Bozinovic G, Markow TA. Dietary protein and sugar differentially affect development and metabolic pools in ecologically diverse Drosophila. J Nutr. 2011;141(6):1127–1133. doi: 10.3945/jn.111.138438. [DOI] [PubMed] [Google Scholar]
- 13.Oliveira D, Almeida F, O'Grady P, Armella M, DeSalle R, Etges W. Monophyly, divergence times, and evolution of host plant use inferred from a revised phylogeny of the Drosophila repleta species group. Mol Phylogenet Evol. 2012;64(3):533–544. doi: 10.1016/j.ympev.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 14.Matzkin LM, Markow TA. Transcriptional regulation of metabolism associated with the increased desiccation resistance of the cactophilic Drosophila mojavensis. Genetics. 2009;182(4):1279–1288. doi: 10.1534/genetics.109.104927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoang K, Matzkin LM, Bono JM. Transcriptional variation associated with cactus host plant adaptation in Drosophila mettleri populations. Mol Ecol. 2015;24(20):5186–5199. doi: 10.1111/mec.13388. [DOI] [PubMed] [Google Scholar]
- 16.Rajpurohit S, Oliveira CC, Etges WJ, Gibbs AG. Functional genomic and phenotypic responses to desiccation in natural populations of a desert drosophilid. Mol Ecol. 2013;22(10):2698–2715. doi: 10.1111/mec.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown JB, Boley N, Eisman R, May GE, Stoiber MH, Duff MO, et al. Diversity and dynamics of the Drosophila transcriptome. Nature. 2014;512(7515):393–399. doi: 10.1038/nature12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sørensen JG, Nielsen MM, Loeschcke V. Gene expression profile analysis of Drosophila melanogaster selected for resistance to environmental stressors. J Evol Biol. 2007;20(4):1624–1636. doi: 10.1111/j.1420-9101.2007.01326.x. [DOI] [PubMed] [Google Scholar]
- 19.Telonis-Scott M, Gane M, DeGaris S, Sgrò CM, Hoffmann AA. High resolution mapping of candidate alleles for desiccation resistance in Drosophila melanogaster under selection. Mol Biol Evol. 2012;29(5):1335–1351. doi: 10.1093/molbev/msr294. [DOI] [PubMed] [Google Scholar]
- 20.Telonis-Scott M, van Heerwaarden B, Johnson TK, Hoffmann AA, Sgrò CM. New levels of transcriptome complexity at upper thermal limits in wild Drosophila revealed by exon expression analysis. Genetics. 2013;195(3):809–830. doi: 10.1534/genetics.113.156224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castro Vargas C, Richmond MP, Ramirez Loustalot Laclette M, Markow TA. Early events in speciation: cryptic species of Drosophila aldrichi. Ecol Evol. 2017;7(12):4220–4228. doi: 10.1002/ece3.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reed LK, Nyboer M, Markow TA. Evolutionary relationships of Drosophila mojavensis geographic host races and their sister species Drosophila arizonae. Mol Ecol. 2007;16(5):1007–1022. doi: 10.1111/j.1365-294X.2006.02941.x. [DOI] [PubMed] [Google Scholar]
- 23.Zhao L, Begun DJ. Genomics of parallel adaptation at two timescales in Drosophila. PLoS Genet. 2017;13(10):e1007016. doi: 10.1371/journal.pgen.1007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rane RV, Oakeshott JG, Nguyen T, Hoffmann AA, Lee SF. Orthonome - a new pipeline for predicting high quality orthologue gene sets applicable to complete and draft genomes. BMC Genomics. 2017;18(1):673. doi: 10.1186/s12864-017-4079-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bosco G, Campbell P, Leiva-Neto JT, Markow TA. Analysis of drosophila species genome size and satellite DNA content reveals significant differences among strains as well as between species. Genetics. 2007;177(3):1277–1290. doi: 10.1534/genetics.107.075069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450(7167):203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 27.Rius N, Guillén Y, Delprat A, Kapusta A, Feschotte C, Ruiz A. Exploration of the Drosophila buzzatii transposable element content suggests underestimation of repeats in Drosophila genomes. BMC Genomics. 2016;17(1):344. doi: 10.1186/s12864-016-2648-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23(9):1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 29.Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 30.Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30(9):1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32(1):268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Durando CM, Baker RH, Etges WJ, Heed WB, Wasserman M, DeSalle R. Phylogenetic analysis of the repleta species group of the genus Drosophila using multiple sources of characters. Mol Phylogenet Evol. 2000;16(2):296–307. doi: 10.1006/mpev.2000.0824. [DOI] [PubMed] [Google Scholar]
- 34.Rane RV, Clarke DF, Pearce SL, Zhang G, Hoffmann AA, Oakeshott JG. Detoxification Genes Differ Between Cactus-, Fruit-, and Flower-Feeding Drosophila. J Hered. 2019;110(1):80–91. [DOI] [PubMed]
- 35.Lang M, Polihronakis Richmond M, Acurio AE, Markow TA, Orgogozo V. Radiation of the Drosophila nannoptera species group in Mexico. J Evol Biol. 2014;27(3):575–584. doi: 10.1111/jeb.12325. [DOI] [PubMed] [Google Scholar]
- 36.Misof B, Liu S, Meusemann K, Peters RS, Donath A, Mayer C, et al. Phylogenomics resolves the timing and pattern of insect evolution. Science. 2014;346(6210):763–767. doi: 10.1126/science.1257570. [DOI] [PubMed] [Google Scholar]
- 37.Kalyaanamoorthy S, Minh BQ, Wong TK, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14(6):587. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jermiin LS, Ho SYW, Ababneh F, Robinson J, Larkum AWD. The biasing effect of compositional heterogeneity on phylogenetic estimates may be underestimated. Syst Biol. 2004;53(4):638–643. doi: 10.1080/10635150490468648. [DOI] [PubMed] [Google Scholar]
- 39.Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5(3):e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, et al. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 2014;10(4):e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minh BQ, Hahn M, Lanfear R. New methods to calculate concordance factors for phylogenomic datasets. bioRxiv. 2018:487801. [DOI] [PMC free article] [PubMed]
- 42.Gatesy J, Sloan D, Warren JM, Baker RH, Simmons MP, Springer MS: Partitioned coalescence support reveals biases in species-tree methods and detects gene trees that determine phylogenomic conflicts. bioRxiv 2018:461699. [DOI] [PubMed]
- 43.Stratman R, Markow TA. Resistance to thermal stress in desert Drosophila. Funct Ecol. 1998;12(6):965–970. doi: 10.1046/j.1365-2435.1998.00270.x. [DOI] [Google Scholar]
- 44.Gibbs AG, Matzkin LM. Evolution of water balance in the genus Drosophila. J Exp Biol. 2001;204(Pt 13):2331–2338. doi: 10.1242/jeb.204.13.2331. [DOI] [PubMed] [Google Scholar]
- 45.Matzkin LM, Watts TD, Markow TA. Evolution of stress resistance in Drosophila: interspecific variation in tolerance to desiccation and starvation. Funct Ecol. 2009;23(3):521–527. doi: 10.1111/j.1365-2435.2008.01533.x. [DOI] [Google Scholar]
- 46.Krebs RA, Barker J. Coexistence of ecologically similar colonising species. II. Population differentiation in Drosophila aldrichi and D. buzzatii for competitive effects and responses at different temperatures and allozyme variation in D. aldrichi. J Evol Biol. 1993;6(2):281–298. doi: 10.1046/j.1420-9101.1993.6020281.x. [DOI] [Google Scholar]
- 47.Barker JSF, Krebs RA, Davies HI. Geographical distributions, relative abundance and coexistence of Drosophila aldrichi and Drosophila buzzatii in Australia. Austral Ecol. 2005;30(5):546–557. doi: 10.1111/j.1442-9993.2005.01470.x. [DOI] [Google Scholar]
- 48.Smith MD, Wertheim JO, Weaver S, Murrell B, Scheffler K, Kosakovsky Pond SL. Less is more: an adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Mol Biol Evol. 2015;32(5):1342–1353. doi: 10.1093/molbev/msv022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Toole AN, Hurst LD, McLysaght A. Faster evolving primate genes are more likely to duplicate. Mol Biol Evol. 2018;35(1):107–118. doi: 10.1093/molbev/msx270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Noh MY, Kramer KJ, Muthukrishnan S, Beeman RW, Kanost MR, Arakane Y. Loss of function of the yellow-e gene causes dehydration-induced mortality of adult Tribolium castaneum. Dev Biol. 2015;399(2):315–324. doi: 10.1016/j.ydbio.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 51.Bass C, Zimmer CT, Riveron JM, Wilding CS, Wondji CS, Kaussmann M, et al. Gene amplification and microsatellite polymorphism underlie a recent insect host shift. Proc Natl Acad Sci U S A. 2013;110(48):19460–19465. doi: 10.1073/pnas.1314122110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vieira FG, Sanchez-Gracia A, Rozas J. Comparative genomic analysis of the odorant-binding protein family in 12 Drosophila genomes: purifying selection and birth-and-death evolution. Genome Biol. 2007;8(11):R235. doi: 10.1186/gb-2007-8-11-r235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Devonshire AL, Field LM, Foster SP, Moores GD, Williamson MS, Blackman RL. The evolution of insecticide resistance in the peach-potato aphid, Myzus persicae. Phil Trans Roy Soc (London) Ser B. 1998;353(1376):1677–1684. doi: 10.1098/rstb.1998.0318. [DOI] [Google Scholar]
- 54.Lenormand T, Guillemaud T, Bourguet D, Raymond M. Appearance and sweep of a gene duplication: adaptive response and potential for new functions in the mosquito Culex pipiens. Evolution. 1998;52(6):1705–1712. doi: 10.1111/j.1558-5646.1998.tb02250.x. [DOI] [PubMed] [Google Scholar]
- 55.Rastogi S, Liberles DA. Subfunctionalization of duplicated genes as a transition state to neofunctionalization. BMC Evol Biol. 2005;5(1):28. doi: 10.1186/1471-2148-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Albalat R, Cañestro C. Evolution by gene loss. Nat Rev Genet. 2016;17(7):379–391. doi: 10.1038/nrg.2016.39. [DOI] [PubMed] [Google Scholar]
- 57.Egan SP, Ragland GJ, Assour L, Powell TH, Hood GR, Emrich S, et al. Experimental evidence of genome-wide impact of ecological selection during early stages of speciation-with-gene-flow. Ecol Lett. 2015;18(8):817–825. doi: 10.1111/ele.12460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soria-Carrasco V, Gompert Z, Comeault AA, Farkas TE, Parchman TL, Johnston JS, et al. Stick insect genomes reveal natural selection’s role in parallel speciation. Science. 2014;344(6185):738–742. doi: 10.1126/science.1252136. [DOI] [PubMed] [Google Scholar]
- 59.Dworkin I, Jones CD. Genetic changes accompanying the evolution of host specialization in Drosophila sechellia. Genetics. 2009;181(2):721–736. doi: 10.1534/genetics.108.093419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yassin A, Debat V, Bastide H, Gidaszewski N, David JR, Pool JE. Recurrent specialization on a toxic fruit in an island Drosophila population. Proc Natl Acad Sci U S A. 2016;113(17):4771–4776. doi: 10.1073/pnas.1522559113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rumball W, Franklin IR, Frankham R, Sheldon BL. Decline in heterozygosity under full-sib and double first-cousin inbreeding in Drosophila melanogaster. Genetics. 1994;136(3):1039–1049. doi: 10.1093/genetics/136.3.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.FastQC: A quality control tool for high throughput sequence data http://www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed 17 Apr 2016.
- 63.Marcais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27(6):764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marçais G, Yorke JA, Zimin A. QuorUM: an error corrector for Illumina reads. PLoS One. 2015;10(6):e0130821. doi: 10.1371/journal.pone.0130821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H. BFC: correcting Illumina sequencing errors. Bioinformatics. 2015;31(17):2885–2887. doi: 10.1093/bioinformatics/btv290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zimin AV, Marcais G, Puiu D, Roberts M, Salzberg SL, Yorke JA. The MaSuRCA genome assembler. Bioinformatics. 2013;29(21):2669–2677. doi: 10.1093/bioinformatics/btt476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27(4):578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 69.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9(11):e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zaharia M, Bolosky WJ, Curtis K, Fox A, Patterson D, Shenker S et al: Faster and more accurate sequence alignment with SNAP. arXiv preprint arXiv:11115572 2011.
- 71.Zhang G, Liu X, Quan Z, Cheng S, Xu X, Pan S, et al. Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat Biotechnol. 2012;30(6):549–554. doi: 10.1038/nbt.2195. [DOI] [PubMed] [Google Scholar]
- 72.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27(2):573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tarailo-Graovac M, Chen N: Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protocols Bioinformatics 2009, 25:4.10. 11–14.10. 14. [DOI] [PubMed]
- 74.Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110(1–4):462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 75.Xu Z, Wang H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007;35(Web Server issue):W265–W268. doi: 10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Edgar RC, Myers EW. PILER: identification and classification of genomic repeats. Bioinformatics. 2005;21(Suppl 1):i152–i158. doi: 10.1093/bioinformatics/bti1003. [DOI] [PubMed] [Google Scholar]
- 77.Price AL, Jones NC, Pevzner PA. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21(Suppl 1):i351–i358. doi: 10.1093/bioinformatics/bti1018. [DOI] [PubMed] [Google Scholar]
- 78.Borodina T, Adjaye J, Sultan M. A strand-specific library preparation protocol for RNA sequencing. Methods Enzymol. 2011;500:79–98. doi: 10.1016/B978-0-12-385118-5.00005-0. [DOI] [PubMed] [Google Scholar]
- 79.Waterhouse RM, Zdobnov EM, Tegenfeldt F, Li J, Kriventseva EV. OrthoDB: the hierarchical catalog of eukaryotic orthologs in 2011. Nucleic Acids Res. 2011;39(Database issue):D283–D288. doi: 10.1093/nar/gkq930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Slater GS, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6(1):31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26(7):873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33(3):290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, et al. De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat Protocols. 2013;8(8):1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Haas BJ, Delcher AL, Mount SM, Wortman JR, Smith RK, Hannick LI, et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003;31(19):5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lomsadze A, Burns PD, Borodovsky M. Integration of mapped RNA-Seq reads into automatic training of eukaryotic gene finding algorithm. Nucleic Acids Res. 2014;42(15):e119. doi: 10.1093/nar/gku557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Majoros WH, Pertea M, Salzberg SL. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics. 2004;20(16):2878–2879. doi: 10.1093/bioinformatics/bth315. [DOI] [PubMed] [Google Scholar]
- 88.Stanke M, Tzvetkova A, Morgenstern B. AUGUSTUS at EGASP: using EST, protein and genomic alignments for improved gene prediction in the human genome. Genome Biol. 2006;7(Suppl 1):1–8. doi: 10.1186/gb-2006-7-s1-s11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5(1):59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, Orvis J, et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 2008;9(1):R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rane RV, Walsh TK, Pearce SL, Jermiin LS, Gordon KH, Richards S, et al. Are feeding preferences and insecticide resistance associated with the size of detoxifying enzyme families in insect herbivores? Curr Opin Insect Sci. 2016;13:70–76. doi: 10.1016/j.cois.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 92.McQuilton P, St Pierre SE, Thurmond J, FlyBase C. FlyBase 101--the basics of navigating FlyBase. Nucleic Acids Res. 2012;40(Database issue):D706–D714. doi: 10.1093/nar/gkr1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Obbard DJ, Maclennan J, Kim KW, Rambaut A, O'Grady PM, Jiggins FM. Estimating divergence dates and substitution rates in the Drosophila phylogeny. Mol Biol Evol. 2012;29(11):3459–3473. doi: 10.1093/molbev/mss150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Murali T, Pacifico S, Yu J, Guest S, Roberts GG, Finley RL. DroID 2011: a comprehensive, integrated resource for protein, transcription factor, RNA and gene interactions for Drosophila. Nucleic Acids Res. 2011;39(suppl 1):D736–D743. doi: 10.1093/nar/gkq1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Law CW, Chen Y, Shi W, Smyth GK. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 99.Bezdek JC, Ehrlich R, Full W. FCM: the fuzzy c-means clustering algorithm. Comput Geosci. 1984;10(2):191–203. doi: 10.1016/0098-3004(84)90020-7. [DOI] [Google Scholar]
- 100.Blondel VD, Guillaume J, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. J Stat Mech Theory Exp. 2008;2008(10):P10008. doi: 10.1088/1742-5468/2008/10/P10008. [DOI] [Google Scholar]
- 101.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6(7):e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tamura K, Battistuzzi FU, Billing-Ross P, Murillo O, Filipski A, Kumar S. Estimating divergence times in large molecular phylogenies. Proc Natl Acad Sci U S A. 2012;109(47):19333–19338. doi: 10.1073/pnas.1213199109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Text S1. Phylogenetics of sophophoran subgenus. Text S2. Detailed materials and methods for genome assembly and annotation. (DOCX 49 kb)
Figure S1. Overlap between the orthogroups generating inparalogues in the different species. Figure S2. Overlap between orthogroups belonging to genes under positive selection in each repleta group species. Figure S3. Scheme for heat stress and recovery assay. Table S1. Scaffold and contig length based statistics as well as results from BUSCO genome assessment using the dataset for the five assembled repleta group genomes and D. melanogaster. Table S2. Repeat content analysis for the six species analysed in this study, characterising transposable elements as well as tandem repeats. Table S3. Genome annotation statistics for the five assembled repleta group genomes and D. melanogaster. Table S4. Number of pairwise orthologues between D. melanogaster and the five repleta species, plus the previously published annotation of D. buzzatii. Table S5. Sets of functional terms describing hierarchical grouping of gene ontology terms and the number of genes in Drosophila melanogaster within each set. (DOCX 301 kb)
Data Availability Statement
All DNA and RNA-Seq sequencing data, assembled genomes and alignments used for phylogenomics can be accessed from NCBI, under SRA submission PRJNA476692 and CSIRO Data Access Portal (10.25919/5bdbf09ba0db2).