Abstract
HIV-associated neurocognitive impairment (NCI) is a term established to capture a wide spectrum of HIV related neurocognitive deficits ranging in severity from asymptomatic to dementia. The genetic underpinnings of this complex phenotype are incompletely understood. Mitochondrial function has long been thought to play a role in neurodegeneration, along with iron metabolism and transport. In this work, we aimed to characterize the interplay of mitochondrial DNA (mtDNA) haplogroup and nuclear genetic associations to NCI phenotypes in the CHARTER cohort, encompassing 1025 individuals of European-descent, African-descent, or admixed Hispanic. We first employed a polygenic modeling approach to investigate the global effect of previous marginally associated nuclear SNPs, and to examine how the polygenic effect of these SNPs is influenced by mtDNA haplogroups. We see evidence of a significant interaction between nuclear SNPs en masse and mtDNA haplogroups within European-descent and African-descent individuals. Subsequently, we performed an analysis of each SNP by mtDNA haplogroup, and detected significant interactions between two nuclear SNPs (rs17160128 and rs12460243) and European haplogroups. These findings, which require validation in larger cohorts, indicate a potential new role for nuclear-mitochondrial DNA interactions in susceptibility to NCI and shed light onto the pathophysiology of this neurocognitive phenotype.
Keywords: nuclear mitochondrial interactions, genomics, single nucleotide polymorphism, neurocognitive impairment, HIV, haplogroups
Introduction
The development of combination antiretroviral therapy (CART) has transitioned HIV to a chronic disease for patients able to obtain and adhere to long-term treatment; however, with this change in disease status have come comorbidities and complications that impact quality of life. Neurocognitive Impairment (NCI) is an important neurological complication of HIV infection (McArthur et al., 2010; Schouten et al., 2011), now commonly classified as HIV-Associated Neurocognitive Disorder or HAND. While more severe forms of dementia are now less common in HIV-seropositive (HIV+) individuals in the CART era, up to 50% of unselected HIV+ individuals show some form of NCI (Grant et al., 2014; Heaton et al., 2015). NCI in CART-treated populations can be asymptomatic but can also progress to symptomatic, functional impairment in activities of daily living, affecting quality of life, productivity, and adherence to treatment (Avci et al., 2017). As these populations age, understanding and preventing NCI becomes increasingly important.
Host genetic factors are of great interest in HIV-associated forms of NCI, though few findings have been replicated in subsequent studies. In the pre-CART era, genetic associations of variants within CCR5 (van Rij et al., 1999) and APOE (Corder et al., 1998) were reported for dementia and other severe phenotypes. The APOE-ε4 association has been examined for milder forms of NCI as well, but results are inconsistent and possibly modulated by age (Morgan et al., 2013; Panos et al.). Candidate-gene and genome-wide association studies have reported additional findings which are supported by replication studies, including MCP1, MIP1A/CCL3, DRD2, DRD3, and HLA:DR – see (Kallianpur and Levine, 2014) for a comprehensive review. Genome-wide association studies (GWAS) have also found associations between SNPs within the SLC8A1 and NALCN ion channel genes and processing speed in HIV-infected adults (Levine et al., 2012).
Mitochondrial dysfunction has long been hypothesized to play a role in neurodegenerative diseases through potential defects in energy metabolism and oxidative damage (Beal, 1995). The mitochondrial genome (mtDNA) consists of ~16,000 nucleotides; it is maternally inherited and encodes 13 electron transport chain proteins. Genetic variation in mtDNA is classified into haplogroups that reflect the ancestry of an individual’s maternal lineage, but mtDNA haplogroups also alter mitochondrial function through transcription and replication (Suissa et al., 2009), oxidative phosphorylation (Gómez-Durán et al., 2010), and a variety of other mechanisms (Fang et al., 2016). More recently, cytoplasmic hybrid experiments have shown that the expression of both nuclear and mtDNA-encoded transcripts differs between haplogroups H and J (Kenney et al., 2014). Haplogroups have also been associated to multiple neurological phenotypes, including Alzheimer’s disease (Bi et al., 2015; van der Walt et al., 2004), Parkinson’s disease (Gaweda-Walerych et al., 2008; van der Walt et al., 2003) and schizophrenia (Zhang et al., 2014).
In an observational study of HIV+ individuals within the CHARTER cohort, we previously described a protective association between the B haplogroup and NCI (as defined by the Global Deficit Score) in admixed Hispanic individuals (Hulgan et al., 2015), which was robust to adjustments for comorbidity severity, CART, plasma viral load, an estimate of reading ability at baseline, and the CD4+ T-cell nadir. A recently published GWAS in the CHARTER study also revealed SNPs near the SH3RF3 and TRAα genes with p-values below 1x10−7 (Jia et al., 2017). Based on these prior findings in CHARTER, along with prior literature supporting iron-mitochondrial interplay (Delsite et al., 2002; Dong et al., 2017; Singh et al., 2005) -- some with specific implications for human aging (Tranah, 2011) -- we hypothesized that mtDNA haplogroups may modify the effect of nuclear SNPs on NCI through interactions. In this study, we used mitochondrial haplogroup information, genome-wide genotype data, and clinical data from CHARTER study participants to test the hypothesis that significant interaction effects on NCI exist between mitochondrial haplogroups and either nuclear genomic variants en masse, or key nuclear SNPs that we identified in the prior CHARTER GWAS.
Materials and Methods
Study Design and Participants
CHARTER is a prospective, observational study conducted at six U.S. medical centers: Johns Hopkins University, Baltimore, MD; Mt. Sinai School of Medicine, New York, NY; University of San Diego, San Diego, CA; University of Texas Medical Branch, Galveston, TX; University of Washington, Seattle, WA; and Washington University, St Louis, MO. The Institutional Review Boards at each site approved this research, and each participant provided written informed consent. Data were collected between 2003 and 2007 using a protocol of comprehensive clinical, neuropsychiatric, and laboratory assessments that were standardized across study sites. For this study, we utilized data from 1025 CHARTER participants with available quality-controlled (QC) nuclear and mitochondrial genome-wide genotyping. No other exclusion criteria were used at this stage. The same subjects were used as in the original GWAS (Jia, et al., 2017)
Assessments of Neurocognitive Impairment
Participants were English-speaking and underwent a comprehensive test battery that included seven neurocognitive domains known to be affected by HIV-associated central nervous system (CNS) dysfunction (Heaton et al., 2010). A composite global deficit score (GDS) was derived from standard T-scores using the best available normative standards to correct for learning effects, age, education, sex, and ethnicity in accordance with prior studies. The resulting GDS is a continuous variable reflecting the number and severity of neurocognitive deficits across the test battery, details of which are described elsewhere (Hulgan et al., 2015; Jia et al., 2017). An established cutoff of GDS ≥ 0.50 categorically defines NCI (Blackstone et al., 2012). Detailed review by two senior CHARTER neurologists using published guidelines (Antinori et al., 2007) provided categorization of comorbid conditions for all CHARTER participants as incidental or contributing to NCI, or confounding a diagnosis of NCI. Several conditions (e.g, brain trauma with loss of consciousness, epilepsy or other seizure history, CNS opportunistic diseases) informed this categorization; detailed information on their frequencies are presented elsewhere (Heaton et al., 2010). Individuals with potentially confounding neurocognitive comorbidities (15% of the total CHARTER cohort), which precluded an assessment of the contribution of HIV to their NCI, were excluded from genetic studies.
Mitochondrial DNA Sequencing, Haplogroup Determination, and SNP Selection
Isolation of DNA from whole blood samples was performed using PUREGENE (Gentra Systems Inc, Minneapolis, Minnesota). Full mtDNA sequencing was performed using the GeneChip Human Mitochondrial Resequencing Array v2.0 (Affymetrix, Inc, Santa Clara, California). Array intensity data were processed using the MitoChip Filtering Protocol (Xie et al., 2011), and variants were called relative to the Revised Cambridge Reference Sequence (Andrews et al., 1999), with haplogroups assigned using HaploGrep (http://haplogrep.uibk.ac.at/) (Kloss-Brandstätter et al., 2011), consistent with other published CHARTER studies (Hulgan et al., 2015). European-descent individuals were grouped into H, J, T, UK, and European Other categories; African-descent individuals were grouped into L1, L2, L3, and African Other categories, and Admixed Hispanic individuals were grouped into A, B, C, and American Other categories.
Participants also underwent nuclear DNA genotyping using the Affymetrix Genome-Wide Human SNP Array 6.0 (Affymetrix, Inc, Santa Clara, California). Ancestry-informative markers were analyzed using EIGENSTRAT software (Price et al., 2006) to generate principal components (PC). Model-based clustering on the top 3 PCs, using the mclust R package, was used to assign individuals to genetic ancestry clusters using an ellipsoidal model (Hall et al., 2014). Genetic ancestry clusters showed 97.4% agreement with self-reported race and ethnicity. All analyses used PC-ancestry-based stratifications (European-descent, African-descent, or admixed Hispanic).
Statistical Analyses
Due to the increased statistical power when analyzing continuous traits, our primary outcome of interest was continuous GDS. Primary analyses were stratified by PC-derived genetic ancestry. We performed a hierarchical set of analyses. We first performed a global analysis to test the interaction between all mitochondrial haplogroups (dummy-encoded) and the aggregate effect of all twenty selected SNPs using mixed linear models. Using Genome-wide Complex Trait Analysis (GCTA) (Yang et al., 2011), we generated a genetic relationship matrix (GRM) based on the twenty selected SNPs. We then fit a linear mixed-effects model according to equation 1, containing a random effect from this GRM (the aggregate effect of the twenty SNPs denoted as α), fixed effects for the j dummy-encoded mitochondrial haplogroups (T, J, H, and UK within European-descent, L1, L2, and L3 within African-descent, and A, B, and C within the admixed Hispanics, denoted as β), and interaction terms between each haplogroup and the random effect over each of the k individuals. Using the –gxe option, we estimate the variance explained for each genotype-haplogroup interaction, and test the significance of including the interaction terms by likelihood ratio test (LRT). Each analysis was conducted within ancestry group to account for population stratification, and models were adjusted for the following fixed-effect covariates (covars): comorbidity status (contributing vs. incidental to NCI); current CART use (yes vs. no); plasma viral load; reading ability (by Wide-Range Achievement Test-III [WRAT] Reading subtest score); nadir CD4+ T-cell count; and the first 3 PCs, consistent with prior CHARTER studies (Jia et al., 2017).
Equation 1. Reduced model
Equation 2. Full model
For ancestry groups showing significant global genotype-haplogroup interactions, we then performed linear regression analysis on each of the twenty SNPs. This model contains an additive effect of the minor allele for the selected SNP, a main effect for each haplogroup, and SNP x haplogroup interaction terms (along with the covariate adjustments outlined above). Statistical significance was assessed by performing a likelihood ratio test between this full model and a reduced model containing no interaction terms. In this analysis, a Bonferroni adjustment for multiple comparisons (20) was conducted.
Analyses of gene expression were conducted by accessing two datasets. Harmonized microarray data from lymphoblastoid cell lines were accessed from the GEUVEDIS project (Lappalainen et al., 2013), encompassing 98 European-descent samples (haplo H,J,T,U) with expression and genotype. Haplogroup assignments were obtained from http://www.mitotool.org/1000GmtDNAs.xls. For consistency with our previous analyses of data from CHARTER participants, we called haplogroups using the HaploGrep algorithm, and we performed linear regression analyses on normalized gene expression outcomes with the same modeling strategy outlined above.
Results
Evidence of broad interaction between NCI-Associated SNPs and mitochondrial haplogroups
Using GWAS and MitoChip/HaploGrep-derived mitochondrial haplogroups from 1025 individuals enrolled in the CHARTER study, we performed mixed-model analysis to assess the cumulative significance and variance in GDS explained for 20 SNPs with marginal associations to NCI outcomes in the prior GWAS analysis of this CHARTER dataset. We fit each model separately within each of the three genomically-defined ancestry groups (European-descent, African-descent, and admixed Hispanic). Results from this analysis are shown in Table 1 (model 1). As expected, SNPs nearing significance in the GWAS of NCI in CHARTER showed highly significant effects on continuous GDS across all ethnicities.
Table 1.
SNPs with marginal associations in a prior GWAS of the CHARTER dataset selected for nuclear mitochondrial interaction analysis
| SNP ID | Chr | Position | Str | A1 | A2 | OR | p-value | Trait | MAF | Function | Nearest Gene |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL|EUR|AFR | |||||||||||
| rs829418 | 1 | 21943503 | + | A | G | 0.159 | 2.51×10−6 | GDS cont | 0.106|0.083|0.093 | intronic | RAP1GAP |
| rs11681615 | 2 | 110079980 | + | C | T | 1.659 | 1.25×10−6 | GDS bin | 0.576|0.256|0.43 | intronic | SH3RF3 |
| rs6542826 | 2 | 110049718 | + | A | G | 1.703 | 7.47×10−7 | GDS bin | 0.632|0.278|0.468 | intronic | SH3RF3 |
| rs6723162 | 2 | 71102285 | + | A | T | 0.605 | 1.38×10−6 | GDS bin | 0.411|0.268|0.338 | intergenic | CD207,LINC01143 |
| rs11915964 | 3 | 134348929 | + | G | T | 0.621 | 5.73×10−6 | GDS bin | 0.351|0.387|0.355 | intronic | KY |
| rs17038463 | 3 | 1425167 | + | G | T | 0.173 | 1.11×10−6 | GDS cont | 0.058|0.114|0.085 | intronic | CNTN6 |
| rs9814567 | 3 | 134218556 | + | C | T | 0.592 | 1.57×10−6 | GDS bin | 0.339|0.34|0.327 | intronic | CEP63 |
| rs17154702 | 8 | 8609879 | + | A | G | 0.118 | 2.94×10−6 | GDS cont | 0.26|0.169|0.2 | intergenic | CLDN23,MFHAS1 |
| rs2915495 | 8 | 52279738 | + | A | G | 1.698 | 7.52×10−6 | GDS bin | 0.157|0.354|0.254 | intronic | PXDNL |
| rs7840128 | 8 | 3677986 | + | A | T | 2.17 | 8.15×10−6 | GDS bin | 0.028|0.146|0.084 | intronic | CSMD1 |
| rs876084 | 8 | 121101521 | − | A | G | 0.635 | 4.93×10−6 | GDS bin | 0.611|0.361|0.493 | intergenic | DEPTOR,COL14A1 |
| rs795943 | 12 | 78663466 | + | A | G | 0.589 | 3.30×10−6 | GDS bin | 0.194|0.609|0.398 | intergenic | NAV3,SYT1 |
| rs4772857 | 13 | 107817668 | + | A | G | 1.604 | 2.03×10−6 | GDS bin | 0.45|0.503|0.476 | intergenic | FAM155A |
| rs1076546 | 14 | 22626947 | − | A | T | 0.12 | 7.31×10−7 | GDS cont | 0.257|0.162|0.215 | intergenic | OR4E2,DAD1 |
| rs11157436 | 14 | 22636873 | + | C | T | 0.155 | 1.64×10−7 | GDS cont | 0.212|0.083|0.155 | intergenic | OR4E2,DAD1 |
| rs12437004 | 14 | 22617414 | + | A | C | 0.12 | 4.41×10−7 | GDS cont | 0.254|0.176|0.22 | intergenic | OR4E2,DAD1 |
| rs2293731 | 14 | 22616833 | + | C | G | 0.139 | 2.88×10−6 | GDS cont | 0.21|0.082|0.153 | intergenic | OR4E2,DAD1 |
| rs978490 | 18 | 41924036 | − | C | T | 0.168 | 1.99×10−6 | GDS cont | 0.148|0.044|0.094 | ncRNA | LINC01478 |
| rs12460243 | 19 | 8131239 | + | A | G | 0.166 | 3.73×10−7 | GDS cont | 0.164|0.046|0.105 | intronic | FBN3 |
| rs17160128 | 19 | 8132697 | + | A | G | 0.162 | 1.16×10−6 | GDS cont | 0.162|0.036|0.1 | intronic | FBN3 |
Chromosome (Chr), Strand (Str), Allele1 (A1), Allele2 (A2), Trait association from the prior published GWAS (Trait), either continuous GDS (cont) or GDS >/<0.5 (bin), Minor Allele Frequency (MAF) over all samples (ALL) European-descent (EUR) and African-descent (AFR).
We next examined the potential for statistical interactions between these GWAS-associated SNPs and mitochondrial haplogroups. To conserve statistical power, we first examined the distribution of genotypes across the mitochondrial haplogroup backgrounds within each ethnicity. For each of the 20 SNPs, we ensured that the SNP was polymorphic (MAF > 0.05) across all haplogroups (stratified by ethnicity). We next performed mixed-model analysis of the GDS to explicitly test the interaction between the cumulative effect of GWAS-associated SNPs (a random effect) and mitochondrial haplogroups (which were dummy-encoded fixed effects) within each ancestry. Results from this analysis are also shown in Table 1 (model 2).
From these results, we see no evidence of haplogroup interactions within the admixed Hispanic samples, and either significant (p < 0.05) or marginally significant (p ~ 0.05) interactions in the African and European-descent samples, respectively. We thus excluded admixed Hispanic individuals from the remainder of our analyses.
Specific SNPs interact with mitochondrial haplogroups in European-descent individuals
For European-descent and African-descent individuals, we further analyzed individual SNP-haplogroup interactions on GDS using linear regression. Models were fit using an additive effect of each minor allele of the SNP, an effect for the dummy-encoded mitochondrial haplogroups, and interaction terms between the haplogroup variables and the SNP. A likelihood ratio test was conducted comparing a full model (with interaction terms) to a reduced model (without interaction terms), with a Bonferroni correction for 20 tests (p < 0.0025). Within the African-descent samples, no individual SNP showed a significant interaction with haplogroup. In European-descent samples, however, six SNPs passed Bonferroni correction via the LRT (shown in table 3). We then performed analyses for each haplogroup to further characterize the nature of the haplogroup interaction using a binary (0 or 1) encoding each haplogroup by additive SNP term (shown in table 3). Three SNPs (rs17160128, rs1240243, and rs978490) have one or more haplogroup interactions significant (p < 0.0025) and are thus considered the most interpretable. Of the three remaining SNPs, rs11157436 is driven by weak interactions across the non-H haplogroups, rs17038463 is driven by H and J interactions, and rs2293731 is based largely on an interaction with the “other” haplogroup category.
Table 3.
European-descent SNP-haplogroup regression results for GDS
| SNP | P LRT | P SNPxH | P SNPxJ | P SNPxT | P SNPxUK | P SNPxOther |
|---|---|---|---|---|---|---|
| rs1076546 | 0.0035 | 0.1713 | 0.5772 | 0.2909 | 0.02358 | 0.05014 |
| rs11157436 | 1.84E-16* | 0.4679 | 0.073 | 0.01449 | 0.008973 | 0.005078 |
| rs11681615 | 0.7601 | 0.2464 | 0.5666 | 0.8104 | 0.32 | 0.6593 |
| rs11915964 | 0.7434 | 0.7081 | 0.7225 | 0.6702 | 0.4009 | 0.3716 |
| rs12437004 | 0.0032 | 0.1923 | 0.5959 | 0.3019 | 0.02193 | 0.053 |
| rs12460243 | 0.0003* | 0.002114* | 0.2826 | 0.03088 | 0.8978 | 0.6646 |
| rs17038463 | 0.0011* | 0.01999 | 0.01607 | 0.4157 | 0.9051 | 0.5408 |
| rs17154702 | 0.0858 | 0.4305 | 0.09126 | 0.2986 | 0.0152 | 0.4051 |
| rs17160128 | 0.0003* | 0.00152* | 0.3289 | 0.02849 | 0.6873 | 0.666 |
| rs2293731 | 5.93E-13* | 0.2691 | 0.2293 | 0.09711 | 0.1058 | 0.007946 |
| rs2915495 | 0.2617 | 0.399 | 0.1615 | 0.6733 | 0.5348 | 0.1475 |
| rs4772857 | 0.8515 | 0.4368 | 0.8768 | 0.2947 | 0.7571 | 0.9968 |
| rs6542826 | 0.772 | 0.6197 | 0.7888 | 0.5901 | 0.3838 | 0.7477 |
| rs6723162 | 0.3816 | 0.2266 | 0.5576 | 0.3485 | 0.1358 | 0.4278 |
| rs7840128 | 0.6795 | 0.6417 | 0.9959 | 0.3621 | 0.1044 | 0.8069 |
| rs795943 | 0.5765 | 0.3423 | 0.9994 | 0.08915 | 0.2585 | 0.5672 |
| rs829418 | 0.1982 | 0.7812 | 0.6789 | 0.5565 | 0.2829 | 0.5554 |
| rs876084 | 0.7861 | 0.1068 | 0.3339 | 0.9173 | 0.3165 | 0.6386 |
| rs978490 | 0.0017* | 0.2579 | 0.3472 | 0.001788* | 0.06331 | 0.5532 |
| rs9814567 | 0.7778 | 0.3939 | 0.8298 | 0.9166 | 0.403 | 0.2864 |
indicates p-values meeting Bonferroni significance threshold adjusting for 20 association tests (α = 0.0025)
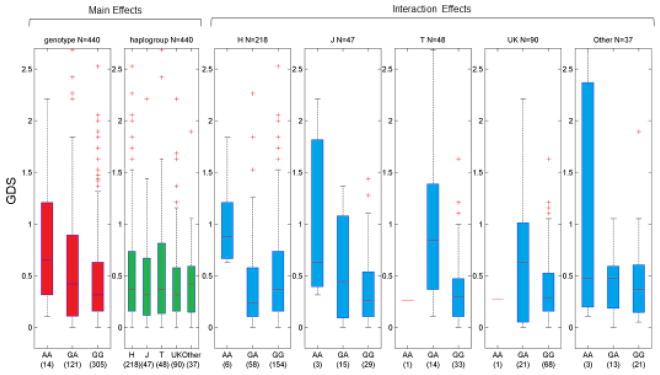
rs17160128 and rs12460243 are two SNPs are in strong linkage disequilibrium within the CHARTER dataset (r-squared =~.9917), and very-likely represent a single association signal within this region of chromosome 19. Box-plots of the GDS by genotype-haplogroup combination are shown in Figure 1. This significant interaction is driven by a difference in effect between haplogroup H and the J, T, and UK haplogroups. Within H haplogroup individuals, heterozygotes for rs17160128 and rs12460243 have significantly lower median GDS values (reduced impairment), but within J, T and UK haplogroup individuals, heterozygotes have increased median GDS values (increased impairment). For T and UK haplogroups, there is only a single individual with the homozygous GG genotype, but the strong increase in GDS values of individuals with the AG genotype makes an additive effect of the G allele the most likely fit (rs12460243).
Figure 1.
Interaction of rs12460243 and Haplogroups. GDS (y-axis) is plotted by genotype/haplogroup combination in European-descent populations. Across all haplogroups, there is a recessive effect of SNP, with increased GDS in the GG genotype. Across all genotypes, all haplogroups have approximately the same median GDS. However, within the H haplogroup, heterozygotes (GA) have lower (better) GDS, compared to J, T, and UK haplogroups where heterozygotes show higher GDS (> 0.5) indicating greater impairment.
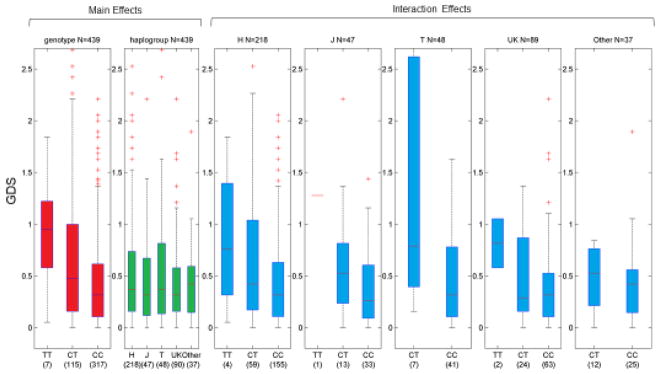
rs978490, located on chromosome 18, shows a significant interaction with haplogroup T, and a more moderate interaction with haplogroup UK. Box-plots of GDS by genotype-haplogroup combination for this SNP are shown in figure 2. An increase in GDS among heterozygotes within the T haplogroup is visible, along with a slight dominant protective effect of the C allele within UK haplogroup individuals.
Figure 2.
Interaction of rs978490 and Haplogroups. GDS (y-axis) is plotted by genotype/haplogroup combination in European-descent populations. There is increased GDS in the CT genotype within T haplogroup individuals relative to other haplogroups.
After observing statistical interactions between SNPs and the H haplogroup, and given evidence of H sub-haplogroup effects in Alzheimer’s disease (Ridge et al., 2012; Santoro et al., 2010), we further evaluated interactions among SNPs and true H, H1, H2, H3, H5, and HV subgroups. No interaction models met our significance criteria of p < 0.0025 (a Bonferroni adjustment for 20 tests), however it is notable that two SNPs approach significance – rs17160128 (p=0.0173) and rs2915495 (p=0.0034).
Effects of SNP-Haplogroup interactions on local gene expression
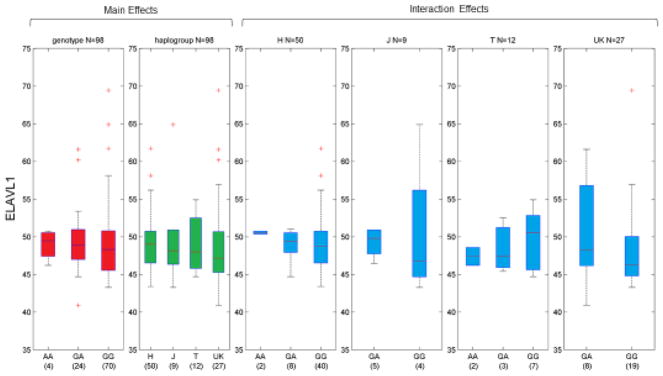
We accessed HaploReg V4.1 (Ward and Kellis, 2012) to explore the regulatory potential of rs12460243, and noted that this SNP falls within predicted enhancer sites for hematopoietic stem cells, neurospheres, and CD14+ monocytes. The SNP also alters a Nuclear transcription factor Y (NF-Y) binding site at position 4 of the motif – two base-pairs upstream of the CCAAT box (Ly et al., 2013) – and thus potentially increases the NF-Y DNA binding affinity. Given the potential regulatory influence of this SNP, we examined the impact of rs12460243 on the expression of nine genes located within the cis-regulatory region (250 Kb upstream and downstream) in 98 lymphoblastoid cell lines from the Geuvadis project, which is based on 1000 Genomes samples and has full mitochondrial sequence available. Of these, one gene showed a significant SNP-haplogroup interaction via likelihood ratio test, ELAVL1 (p = 0.0302), and box-plots of ELAVL1 expression are shown in Figure 3. To further examine the biological mechanisms that may be affected from the rs12460243-Haplogroup interaction, we considered all genes (both within the cis-regulatory and across the genome) that showed nominally significant changes in gene expression (p < 0.05). Using this set of genes, we performed gene-set enrichment analyses to identify gene ontology terms using WebGestalt (Zhang et al., 2005). These analyses revealed multiple mechanisms influenced by this SNP-Haplogroup interaction, notably “mRNA splicing via spliceosome” (GO:0000398) and “regulation of mRNA stability” (GO:0043488) terms, which involve the ELAVL1 gene and thus may be mediated by changes in ELAVL1 expression..
Figure 3.
Interaction Box-Plots for expression of ELAVL1 (Geuvadis). Normalized gene expression (y-axis) is plotted by genotype/haplogroup combination in European-descent populations for rs12460243. We observe a significant interaction between genotype and haplogroup, and similar to GDS in figure 1, we osbserve higher expression in H homozygous (AA) individuals.
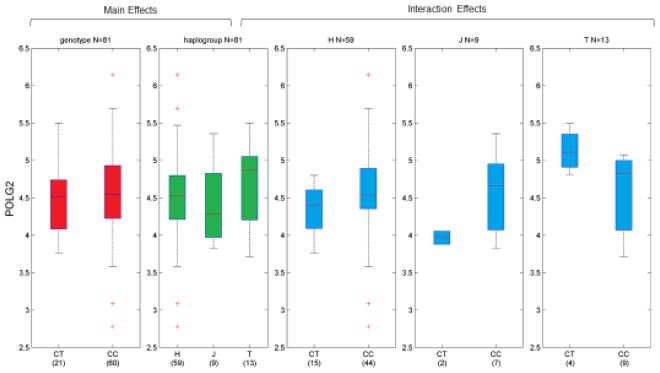
Similarly, rs978490 alters a E26 transformation specific transcription factor binding site, and falls within predicted enhancer sties for astrocytes and primary B-cells, and significantly alters the expression of multiple genes including POLG2 in peripheral blood monoctyes (Zeller et al., 2010). POLG2 is the processivity subunit of the mitochondrial DNA polymerase gamma, which plays a role in mtDNA replication and may mediate changes to mitochondrial DNA accumulation and copy number (Chen et al., 2014; Mahrous et al., 2012). Given the previous relationship between this SNP and POLG2 and the relevance to mitochondrial function, we evaluated SNP-Haplogroup interactions for this gene in the Geuvadis dataset (Figure 4). Via likelihood ratio test, we observe a significant SNP-haplogroup interaction (p=0.038), and an interesting reversal of expression by genotype within the T haplogroup.
Figure 4.
Interaction Box-Plots for expression of POLG2 (Geuvadis). Normalized gene expression (y-axis) is plotted by genotype/haplogroup combination in European-descent populations for rs978490. We observe a significant interaction between genotype and haplogroup, driven by a change in slope in the T haplogroup; for other groups, POLG2 expression is either equal to or slightly increasing with the number of copies of the C allele.
Discussion
Nuclear-mitochondrial interactions are increasingly recognized to play an important role in complex human diseases, but few studies have directly investigated them. This study is the first to address the potential impact of such interaction effects for NCI in HIV-infected individuals. Given the potential influence of mitochondrial genetic variation on aging and neurological phenotypes (Tranah, 2011), we hypothesized that mitochondrial haplogroups interact with selected SNPs to influence NCI, as defined by the GDS. To test our hypothesis, we first employed a polygenic modeling approach to investigate the global effect of previously implicated SNPs and haplogroups, and to explicitly model the interaction between haplogroups and SNPs en masse. Our approach is similar to the random effects models employed by Paliwal et al. used to assess nuclear-“mitotype” interactions in yeast (Paliwal et al., 2014), and could easily be applied to examine other collections of SNPs for haplogroup intereactions, such as biological pathways or other gene groupings. In our models, fixed effects reflecting specific haplogroup associations to GDS are consistent with prior results, most notably the protective effect of the B haplogroup in Hispanics. We estimate that the variance in GDS explained by the 20 selected SNPs was 22.6%, however this is an overestimate as we chose these SNPs specifically based on a previous analysis of this dataset. Our analysis of nuclear-mitochondrial interactions however is unbiased as the haplogroup interactions fit in these models are orthogonal, independent effects. The estimated variance in GDS explained due to SNP-haplogroup interactions was highest for the UK and H haplogroups in European-descent individuals at 2.7% and 4.1% respectively. Haplogroup interactions within the African-descent individuals were more diffuse across the L1, L2, and L3 haplogroups (relative to the Other category), all at approximately 1.2% variance explained. Analyses of the Admixed Hispanic group were non-significant, likely due to limited statistical power is this smaller group (N = 101). Given the significance (p=0.025) and marginal significance (p=0.052) of the African-descent and European-descent groups, we next performed an analysis of each SNP by mtDNA haplogroup within these groups. This analysis detected significant interaction signals between European haplogroups and two nuclear regions, rs12460243 and rs17160128 in strong LD on chromosome 19 and rs978490 on chromosome 18.
These two SNPs (rs12460243 and rs17160128) are generally found at low frequency (0.06 and 0.04) in African-descent individuals but are highly polymorphic within other global populations, with frequencies ranging from 0.15 to 0.49 (Abecasis et al., 2012). The low frequency of these variants in African-descent individuals and significantly smaller sample size of the admixed Hispanic subgroup of the dataset reduced our statistical power to observe an effect outside the European-descent subset.
The SNP-haplogroup interaction effect we detected implicates a binding site of NF-Y, a transcription factor, which is widely expressed in the central nervous system and linked to neurodegeneration of adult neurons (Yamanaka et al., 2014) The NF-Y homolog in yeast, HAP2/3/5, has been shown to be a stress-induced transcriptional activator of the mitochondrial electron transport chain (Benatti et al., 2016), and due to the similarity of electron transport chain gene promoters across organisms, it is hypothesized that NF-Y serves an equivalent regulatory role in humans. Oxidative stress is thought to play an important role in the development of HIV-Associated Neurocognitive Disorders, and anti-oxidant therapies in animal models are neuroprotective (see (Louboutin and Strayer, 2014; Mollace et al., 2001) for an extensive reviews).
The ELAVL1 gene encodes an RNA-binding protein, called HuR, which is thought to play a critical role in post-transcriptional regulation and splicing. HuR has previously been suggested to interact with HIV-1 reverse transcriptase (Lemay et al., 2008), but this interaction was subsequently shown not to influence reverse transcription in vivo, and HuR does not appear to bind to HIV-1 reverse transcriptase directly (Ahn et al., 2010). Interestingly, HIV protease inhibitors, which are frequent components of CART, are known to influence production of the pro-inflammatory cytokine TNF-α and interleUKin-6 in macrophages via a process that is mediated in part by nuclear-cytoplasmic translocation of HuR (Zhou et al., 2007).
Furthermore, rs17160128 (which is in strong linkage disequilibrium with rs12460243) shows moderate association to serum fibrinogen levels (p = 6.324 x 10−5) within the Framingham Heart Study, accessed from PheGenI (Ramos et al., 2014). Fibrinogen levels have been previously implicated in neuronal toxicity and degeneration (Cortes-Canteli et al., 2015; Sonkar et al., 2016), and an interplay between fibrinogen levels and mitochondrial function under inflammatory conditions has been recently proposed (Ueki et al., 2016). Fibrinogen, a marker of a procoagulant state, is elevated in the plasma of HIV+ persons, and these levels decline but do not normalize on CART (Funderburg, 2014). Procoagulant factors, including fibrinogen, have also been inked to poorer neurocognitive functioning (Montoya et al., 2017).
The A allele at rs978490 is at extremely low frequency in African and East Asian populations, but has approximately 10% frequency in other ancestry groups. This SNP influences a differentiating transcription factor (ETS) binding site in both neuronal and immune cell types, and influences expression of POLG2, a subunit of the mitochondrial DNA polymerase. The effect of this SNP on POLG2 trans-acting through an unknown mechanism, as rs978490 is located on chromosome 18 and POLG2 is found on chromosome 17. Interestingly, this nuclear-mitochondrial interaction potentially implicates mtDNA levels, which have been shown to be different between H and UK cybrid lines (Gómez-Durán et al., 2010), and may drive differences in oxidative phosphorylation capacity between these haplogroups.
While these findings are compelling, this study has notable limitations. The CHARTER cohort is a tremendous resource, however the total study sample size (N=1025) and the ethnic stratification of the dataset makes statistical power for interaction analyses a key limitation. Within the European-descent subset (N=440), while some genotype-Haplogroup combinations are at low frequency, our assumption of an additive effect by genotype allows us to estimate differences in slope between haplogroups. Nonetheless, a larger sample of European-descent individuals is ideal to more accurately estimate effects within each haplogroup. We have attempted to limit the scope of our exploratory analysis to nuclear SNPs that already evidence of association from a prior analysis of this dataset, and by first performing an overall test of SNP-Haplogroup interactions within each ethnicity. Our analyses of gene expression within lymphoblastoid cell lines provides additional support for our nuclear-mitochondrial interaction hypotheses, but these do not replace a statistical replication of these effects in an independent sample. Unfortunately, due to the difficulty in ascertainment of this unique phenotype, and the need for both GWAS and full mitochondrial sequence data, we could not identify a suitable replication cohort, which limits our ability to generalize this effect.
Despite its limitations, our work highlights potential new mechanisms through which nuclear and mitochondrial genomic variation may influence cellular pathways of neuroinflammation to influence susceptibility to NCI in HIV+ individuals, and deserve further study.
Table 2.
Mixed-model analysis of GWAS-associated SNPs on continuous GDS
| Ancestry Group | Model Component | Effects | |||||
|---|---|---|---|---|---|---|---|
| Model 1. Selected SNPs and Haplogroups Only (Reduced Model) | |||||||
| European Descent N=440 | (R) 20 SNPs | 0.226 (0.074) | |||||
| (F) Haplogroup | H 0.010 (0.087) | J −0.040 (0.105) | T 0.117 (0.109) | UK −0.041 (0.096) | Other Referent | ||
| Model p-value | 1.731e-20 | ||||||
| Admixed Hispanic N=101 | (R) 20 SNPs | 0.304 (0.124) | |||||
| (F) Haplogroup | A 0.113 (0.105) | B −0.164 (0.121) | C 0.098 (0.132) | Other Referent | |||
| Model p-value | 7.198e-05 | ||||||
| African Descent N=484 | (R) 20 SNPs | 0.12 (0.047) | |||||
| (F) Haplogroup | L1 −0.003 (0.057) | L2 0.012 (0.053) | L3 −0.027 (0.052) | Other Referent | |||
| Model p-value | 1.803e-09 | ||||||
| Model 2. Selected SNPs with Haplogroup Interactions (Full Model) | |||||||
| European Descent N=440 | (R) 20 SNPs | 0.151 (0.096) | |||||
| (F) Haplogroup | H 0.027 (0.087) | J −0.030 (0.105) | T 0.145 (0.105) | UK −0.035 (0.096) | Other Referent | ||
| (R) SNPxHaplogroup | H 0.024 (0.032) | J 0.005 (0.043) | T 0.012 (0.047) | UK 0.041 (0.049) | Other Referent | ||
| variance explained | 0.236 (0.071) | ||||||
| LRT p-value | 0.052 | ||||||
| Admixed Hispanic N=101 | (R) 20 SNPs | 0.013 (0.249) | |||||
| (F) Haplogroup | A 0.131 (0.098) | B −0.141 (0.119) | C 0.095 (0.121) | Other Referent | |||
| (R) SNPxHaplogroup | A 0.191 (0.161) | B 0.245 (0.217) | C 0.008 (0.121) | Other | Referent | ||
| variance explained | 0.364 (0.125) | ||||||
| LRT p-value | 0.50 | ||||||
| African Descent N=484 | (R) 20 SNPs | 0.094 (0.059) | |||||
| (F) Haplogroup | L1 0.004 (0.058) | L2 0.015 (0.053) | L3 −0.023 (0.052) | Other Referent | |||
| (R) SNPxHaplogroup | L1 0.012 (0.037) | L2 0.011 (0.030) | L3 0.015 (0.030) | Other Referent | |||
| variance explained | 0.133 (0.050) | ||||||
| LRT p-value | 0.025 | ||||||
Random effects are denoted by (R) and units are in proportion of GDS variance explained, Fixed effects are denoted by (F) and units are change in GDS relative to Referent. Fixed effects for comorbidity, CART use, plasma viral load, reading ability, nadir CD4+ count and principal components are not shown. LRT: Likelihood Ratio Test between full and reduced model.
Highlights.
Nuclear-mitochondrial interactions are increasingly recognized to play an important role in complex human diseases, but few studies have directly investigated them. This study is the first to address the potential impact of such interaction effects for NCI in HIV-infected individuals.
We employed a polygenic modeling approach to investigate the global effect of previously associated nuclear SNPs, and to examine how the polygenic effect of these SNPs is influenced by mtDNA haplogroups.
We see evidence of a significant interaction between nuclear SNPs en masse and mtDNA haplogroups within European-descent and African-descent individuals.
Our findings indicate a new role for nuclear-mitochondrial DNA interactions in susceptibility to NCI, which requires further study in larger cohorts and potentially sheds light into the pathophysiology of this neurocognitive phenotype.
Our work highlights potential new mechanisms through which nuclear and mitochondrial genomic variation may influence cellular pathways of neuroinflammation to influence susceptibility to NCI in HIV+ individuals
Acknowledgments
This work was supported by National Institutes of Mental Health/National Institutes of Health (NIH) grant number R01 MH095621 to A. R. K. and T. H. The CNS HIV Anti-Retroviral Therapy Effects Research (CHARTER) study was also supported by NIH awards N01 MH22005 and HHSN271201000036C, and HHSN271201000030C. The authors gratefully acknowledge the contributions of all participants in CHARTER. S. L. L. has received support for research projects from Abbott, Merck, Tibotec, Schering-Plough, and GlaxoSmithKline and has received honoraria for speaking from Abbott, GlaxoSmithKline, and Tibotec. R. J. E. gave sponsored talks and received honoraria for serving on the scientific advisory board of GlaxoSmithKline. A. C. C. has received past research support from Schering-Pough, Merck, and Roche Molecular Systems and has served as a data, safety, and monitoring board member for Merck-sponsored studies; she previously owned stock in Abbott Laboratories, Bristol-Myers Squibb, Johnson & Johnson, and Pfizer. C. M. Mreceives royalties from Lippincott Williams and Wilkins and from UptoDate. D. B. C. has received support for consulting/advisory boards from Sanofi, Genentech, Quintiles, Inhibikase, Bristol-Myers Squibb, GlaxoSmithKline, Millennium, Biogen Idec, Amgen, Pfizer, AstraZeneca, Cytheris and Merck and receives research support from Lilly, Roche, Bavarian Nordic, Gilead, the Alzheimer Association and the National Institutes of Health (National Institute of Allergy and Infectious Diseases, National Institute of Mental Health, National Institute of Neurological Disorders and Stroke, National Institute on Aging, and National Institute of Nursing Research). J. A. M. authors chapters on HIV for the Merck Manual. D. M. S. has provided consultancy to Astellas, Acorda, Allergan, Merz, and Ipsen; has received speaking honoraria from Allergan and Acorda; and received research grants from Merz, Allergan, Ipsen, Acorda, and Astellas. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J, Byeon IJL, Dharmasena S, Huber K, Concel J, Gronenborn AM, Sluis-Cremer N. The RNA binding protein HuR does not interact directly with HIV-1 reverse transcriptase and does not affect reverse transcription in vitro. Retrovirology. 2010;7:40. doi: 10.1186/1742-4690-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- Antinori A, Arendt G, Becker JT, Brew BJ, Byrd DA, Cherner M, Clifford DB, Cinque P, Epstein LG, Goodkin K, et al. Updated research nosology for HIV-associated neurocognitive disorders. Neurology. 2007;69:1789–1799. doi: 10.1212/01.WNL.0000287431.88658.8b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avci G, Sheppard DP, Tierney SM, Kordovski VM, Sullivan KL, Woods SP. A systematic review of prospective memory in HIV disease: from the laboratory to daily life. Clin Neuropsychol. 2017:1–33. doi: 10.1080/13854046.2017.1373860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal MF. Aging, energy, and oxidative stress in neurodegenerative diseases. Ann Neurol. 1995;38:357–366. doi: 10.1002/ana.410380304. [DOI] [PubMed] [Google Scholar]
- Benatti P, Chiaramonte ML, Lorenzo M, Hartley JA, Hochhauser D, Gnesutta N, Mantovani R, Imbriano C, Dolfini D. NF-Y activates genes of metabolic pathways altered in cancer cells. Oncotarget. 2016;7:1633–1650. doi: 10.18632/oncotarget.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi R, Zhang W, Yu D, Li X, Wang HZ, Hu QX, Zhang C, Lu W, Ni J, Fang Y, et al. Mitochondrial DNA haplogroup B5 confers genetic susceptibility to Alzheimer’s disease in Han Chinese. Neurobiol Aging. 2015;36:1604e7–16. doi: 10.1016/j.neurobiolaging.2014.10.009. [DOI] [PubMed] [Google Scholar]
- Blackstone K, Moore DJ, Franklin DR, Clifford DB, Collier AC, Marra CM, Gelman BB, McArthur JC, Morgello S, Simpson DM, et al. Defining neurocognitive impairment in HIV: deficit scores versus clinical ratings. Clin Neuropsychol. 2012;26:894–908. doi: 10.1080/13854046.2012.694479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Lin J, Hong J, Han D, Zhang AD, Lan R, Fu L, Wu Z, Lin J, Zhang W, et al. Potential toxicity of quercetin: The repression of mitochondrial copy number via decreased POLG expression and excessive TFAM expression in irradiated murine bone marrow. Toxicol Reports. 2014;1:450–458. doi: 10.1016/j.toxrep.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder EH, Robertson K, Lannfelt L, Bogdanovic N, Eggertsen G, Wilkins J, Hall C. HIV-infected subjects with the E4 allele for APOE have excess dementia and peripheral neuropathy. Nat Med. 1998;4:1182–1184. doi: 10.1038/2677. [DOI] [PubMed] [Google Scholar]
- Cortes-Canteli M, Mattei L, Richards AT, Norris EH, Strickland S. Fibrin deposited in the Alzheimer’s disease brain promotes neuronal degeneration. Neurobiol Aging. 2015;36:608–617. doi: 10.1016/j.neurobiolaging.2014.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsite R, Kachhap S, Anbazhagan R, Gabrielson E, Singh KK. Nuclear genes involved in mitochondria-to-nucleus communication in breast cancer cells. Mol Cancer. 2002;1:6. doi: 10.1186/1476-4598-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Yoshitomi T, Hu JF, Cui J. Long noncoding RNAs coordinate functions between mitochondria and the nucleus. Epigenetics Chromatin. 2017;10:41. doi: 10.1186/s13072-017-0149-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H, Zhang F, Li F, Shi H, Ma L, Du M, You Y, Qiu R, Nie H, Shen L, et al. Mitochondrial DNA haplogroups modify the risk of osteoarthritis by altering mitochondrial function and intracellular mitochondrial signals. Biochim Biophys Acta. 2016;1862:829–836. doi: 10.1016/j.bbadis.2015.12.017. [DOI] [PubMed] [Google Scholar]
- Funderburg NT. Markers of coagulation and inflammation often remain elevated in ART-treated HIV-infected patients. Curr Opin HIV AIDS. 2014;9:80–86. doi: 10.1097/COH.0000000000000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaweda-Walerych K, Maruszak A, Safranow K, Bialecka M, Klodowska-Duda G, Czyzewski K, Slawek J, Rudzinska M, Styczynska M, Opala G, et al. Mitochondrial DNA haplogroups and subhaplogroups are associated with Parkinson’s disease risk in a Polish PD cohort. J Neural Transm. 2008;115:1521–1526. doi: 10.1007/s00702-008-0121-9. [DOI] [PubMed] [Google Scholar]
- Gómez-Durán A, Pacheu-Grau D, López-Gallardo E, Díez-Sánchez C, Montoya J, López-Pérez MJ, Ruiz-Pesini E. Unmasking the causes of multifactorial disorders: OXPHOS differences between mitochondrial haplogroups. Hum Mol Genet. 2010;19:3343–3353. doi: 10.1093/hmg/ddq246. [DOI] [PubMed] [Google Scholar]
- Grant I, Franklin DR, Deutsch R, Woods SP, Vaida F, Ellis RJ, Letendre SL, Marcotte TD, Atkinson JH, Collier AC, et al. Asymptomatic HIV-associated neurocognitive impairment increases risk for symptomatic decline. Neurology. 2014;82:2055–2062. doi: 10.1212/WNL.0000000000000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JB, Dumitrescu L, Dilks HH, Crawford DC, Bush WS. Accuracy of administratively-assigned ancestry for diverse populations in an electronic medical record-linked biobank. PLoS One. 2014;9:e99161. doi: 10.1371/journal.pone.0099161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton RK, Clifford DB, Franklin DR, Woods SP, Ake C, Vaida F, Ellis RJ, Letendre SL, Marcotte TD, Atkinson JH, et al. HIV-associated neurocognitive disorders persist in the era of potent antiretroviral therapy: CHARTER Study. Neurology. 2010;75:2087–2096. doi: 10.1212/WNL.0b013e318200d727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton RK, Franklin DR, Deutsch R, Letendre S, Ellis RJ, Casaletto K, Marquine MJ, Woods SP, Vaida F, Atkinson JH, et al. Neurocognitive change in the era of HIV combination antiretroviral therapy: the longitudinal CHARTER study. Clin Infect Dis. 2015;60:473–480. doi: 10.1093/cid/ciu862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulgan T, Samuels DC, Bush W, Ellis RJ, Letendre SL, Heaton RK, Franklin DR, Straub P, Murdock DG, Clifford DB, et al. Mitochondrial DNA Haplogroups and Neurocognitive Impairment During HIV Infection. Clin Infect Dis. 2015;61:1476–1484. doi: 10.1093/cid/civ527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia P, Zhao Z, Hulgan T, Bush WS, Samuels DC, Bloss CS, Heaton RK, Ellis RJ, Schork N, Marra CM, et al. Genome-wide association study of HIV-associated neurocognitive disorder (HAND): A CHARTER group study. Am J Med Genet Part B Neuropsychiatr Genet. 2017;174:413–426. doi: 10.1002/ajmg.b.32530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallianpur AR, Levine AJ. Host genetic factors predisposing to HIV-associated neurocognitive disorder. Curr HIV/AIDS Rep. 2014;11:336–352. doi: 10.1007/s11904-014-0222-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney MC, Chwa M, Atilano SR, Falatoonzadeh P, Ramirez C, Malik D, Tarek M, Cáceres-del-Carpio J, Nesburn AB, Boyer DS, et al. Inherited mitochondrial DNA variants can affect complement, inflammation and apoptosis pathways: insights into mitochondrial-nuclear interactions. Hum Mol Genet. 2014;23:3537–3551. doi: 10.1093/hmg/ddu065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss-Brandstätter A, Pacher D, Schönherr S, Weissensteiner H, Binna R, Specht G, Kronenberg F. HaploGrep: a fast and reliable algorithm for automatic classification of mitochondrial DNA haplogroups. Hum Mutat. 2011;32:25–32. doi: 10.1002/humu.21382. [DOI] [PubMed] [Google Scholar]
- Lappalainen T, Sammeth M, Friedlander MR, ‘t Hoen PAC, Monlong J, Rivas MA, Gonzalez-Porta M, Kurbatova N, Griebel T, Ferreira PG, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay J, Maidou-Peindara P, Bader T, Ennifar E, Rain JC, Benarous R, Liu LX. HuR interacts with human immunodeficiency virus type 1 reverse transcriptase, and modulates reverse transcription in infected cells. Retrovirology. 2008;5:47. doi: 10.1186/1742-4690-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ, Service S, Miller EN, Reynolds SM, Singer EJ, Shapshak P, Martin EM, Sacktor N, Becker JT, Jacobson LP, et al. Genome-wide association study of neurocognitive impairment and dementia in HIV-infected adults. Am J Med Genet B Neuropsychiatr Genet. 2012;159B:669–683. doi: 10.1002/ajmg.b.32071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louboutin JP, Strayer D. Role of Oxidative Stress in HIV-1-Associated Neurocognitive Disorder and Protection by Gene Delivery of Antioxidant Enzymes. Antioxidants (Basel, Switzerland) 2014;3:770–797. doi: 10.3390/antiox3040770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly LL, Yoshida H, Yamaguchi M. Nuclear transcription factor Y and its roles in cellular processes related to human disease. Am J Cancer Res. 2013;3:339–346. [PMC free article] [PubMed] [Google Scholar]
- Mahrous E, Yang Q, Clarke HJ. Regulation of mitochondrial DNA accumulation during oocyte growth and meiotic maturation in the mouse. Reproduction. 2012;144:177–185. doi: 10.1530/REP-12-0113. [DOI] [PubMed] [Google Scholar]
- McArthur JC, Steiner J, Sacktor N, Nath A. Human immunodeficiency virus-associated neurocognitive disorders: Mind the gap. Ann Neurol. 2010;67:699–714. doi: 10.1002/ana.22053. [DOI] [PubMed] [Google Scholar]
- Mollace V, Nottet HS, Clayette P, Turco MC, Muscoli C, Salvemini D, Perno CF. Oxidative stress and neuroAIDS: triggers, modulators and novel antioxidants. Trends Neurosci. 2001;24:411–416. doi: 10.1016/s0166-2236(00)01819-1. [DOI] [PubMed] [Google Scholar]
- Montoya JL, Iudicello J, Oppenheim HA, Fazeli PL, Potter M, Ma Q, Mills PJ, Ellis RJ, Grant I, Letendre SL, et al. Coagulation imbalance and neurocognitive functioning in older HIV-positive adults on suppressive antiretroviral therapy. AIDS. 2017;31:787–795. doi: 10.1097/QAD.0000000000001404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan EE, Woods SP, Letendre SL, Franklin DR, Bloss C, Goate A, Heaton RK, Collier AC, Marra CM, Gelman BB, et al. Apolipoprotein E4 genotype does not increase risk of HIV-associated neurocognitive disorders. J Neurovirol. 2013;19:150–156. doi: 10.1007/s13365-013-0152-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paliwal S, Fiumera AC, Fiumera HL. Mitochondrial-nuclear epistasis contributes to phenotypic variation and coadaptation in natural isolates of Saccharomyces cerevisiae. Genetics. 2014;198:1251–1265. doi: 10.1534/genetics.114.168575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panos SE, Hinkin CH, Singer EJ, Thames AD, Patel SM, Sinsheimer JS, Del Re AC, Gelman BB, Morgello S, Moore DJ, et al. Apolipoprotein-E genotype and human immunodeficiency virus-associated neurocognitive disorder: the modulating effects of older age and disease severity. Neurobehav HIV Med. 5:11–22. doi: 10.2147/NBHIV.S39573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Ramos EM, Hoffman D, Junkins HA, Maglott D, Phan L, Sherry ST, Feolo M, Hindorff LA. Phenotype-Genotype Integrator (PheGenI): synthesizing genome-wide association study (GWAS) data with existing genomic resources. Eur J Hum Genet. 2014;22:144–147. doi: 10.1038/ejhg.2013.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridge PG, Maxwell TJ, Corcoran CD, Norton MC, Tschanz JT, O’Brien E, Kerber RA, Cawthon RM, Munger RG, Kauwe JSK. Mitochondrial Genomic Analysis of Late Onset Alzheimer’s Disease Reveals Protective Haplogroups H6A1A/H6A1B: The Cache County Study on Memory in Aging. PLoS One. 2012;7:e45134. doi: 10.1371/journal.pone.0045134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rij RP, Portegies P, Hallaby T, Lange JM, Visser J, de Roda Husman AM, van ’t Wout AB, Schuitemaker H. Reduced prevalence of the CCR5 delta32 heterozygous genotype in human immunodeficiency virus-infected individuals with AIDS dementia complex. J Infect Dis. 1999;180:854–857. doi: 10.1086/314940. [DOI] [PubMed] [Google Scholar]
- Santoro A, Balbi V, Balducci E, Pirazzini C, Rosini F, Tavano F, Achilli A, Siviero P, Minicuci N, Bellavista E, et al. Evidence for sub-haplogroup h5 of mitochondrial DNA as a risk factor for late onset Alzheimer’s disease. PLoS One. 2010;5:e12037. doi: 10.1371/journal.pone.0012037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouten J, Cinque P, Gisslen M, Reiss P, Portegies P. HIV-1 infection and cognitive impairment in the cART era: a review. AIDS. 2011;25:561–575. doi: 10.1097/QAD.0b013e3283437f9a. [DOI] [PubMed] [Google Scholar]
- Singh KK, Kulawiec M, Still I, DesoUKi MM, Geradts J, Matsui SI. Inter-genomic cross talk between mitochondria and the nucleus plays an important role in tumorigenesis. Gene. 2005;354:140–146. doi: 10.1016/j.gene.2005.03.027. [DOI] [PubMed] [Google Scholar]
- Sonkar VK, Kulkarni PP, Chaurasia SN, Dash A, Jauhari A, Parmar D, Yadav S, Dash D. Plasma fibrinogen is a natural deterrent to amyloid beta-induced platelet activation and neuronal toxicity. Mol Med. 2016;22 doi: 10.2119/molmed.2016.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suissa S, Wang Z, Poole J, Wittkopp S, Feder J, Shutt TE, Wallace DC, Shadel GS, Mishmar D. Ancient mtDNA genetic variants modulate mtDNA transcription and replication. PLoS Genet. 2009;5:e1000474. doi: 10.1371/journal.pgen.1000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranah GJ. Mitochondrial-nuclear epistasis: implications for human aging and longevity. Ageing Res Rev. 2011;10:238–252. doi: 10.1016/j.arr.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki R, Liu L, Kashiwagi S, Kaneki M, Khan M, Hirose M, Tompkins RG, Martyn JAJ, Yasuhara S. Role of Elevated Fibrinogen in Burn-Induced Mitochondrial Dysfunction: Protective Effects of Glycyrrhizin. Shock. 2016 doi: 10.1097/SHK.0000000000000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Walt JM, Nicodemus KK, Martin ER, Scott WK, Nance MA, Watts RL, Hubble JP, Haines JL, Koller WC, Lyons K, et al. Mitochondrial polymorphisms significantly reduce the risk of Parkinson disease. Am J Hum Genet. 2003;72:804–811. doi: 10.1086/373937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Walt JM, Dementieva YA, Martin ER, Scott WK, Nicodemus KK, Kroner CC, Welsh-Bohmer KA, Saunders AM, Roses AD, Small GW, et al. Analysis of European mitochondrial haplogroups with Alzheimer disease risk. Neurosci Lett. 2004;365:28–32. doi: 10.1016/j.neulet.2004.04.051. [DOI] [PubMed] [Google Scholar]
- Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–4. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie HM, Perin JC, Schurr TG, Dulik MC, Zhadanov SI, Baur JA, King MP, Place E, Clarke C, Grauer M, et al. Mitochondrial genome sequence analysis: a custom bioinformatics pipeline substantially improves Affymetrix MitoChip v2.0 call rate and accuracy. BMC Bioinformatics. 2011;12:402. doi: 10.1186/1471-2105-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka T, Tosaki A, Kurosawa M, Matsumoto G, Koike M, Uchiyama Y, Maity SN, Shimogori T, Hattori N, Nukina N. NF-Y inactivation causes atypical neurodegeneration characterized by ubiquitin and p62 accumulation and endoplasmic reticulum disorganization. Nat Commun. 2014;5:3354. doi: 10.1038/ncomms4354. [DOI] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, Castagne R, Maouche S, Germain M, Lackner K, Rossmann H, et al. Genetics and beyond--the transcriptome of human monocytes and disease susceptibility. PLoS One. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–8. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Tang J, Zhang AM, Peng MS, Xie HB, Tan L, Xu L, Zhang YP, Chen X, Yao YG. A matrilineal genetic legacy from the last glacial maximum confers susceptibility to schizophrenia in Han Chinese. J Genet Genomics. 2014;41:397–407. doi: 10.1016/j.jgg.2014.05.004. [DOI] [PubMed] [Google Scholar]
- Zhou H, Jarujaron S, Gurley EC, Chen L, Ding H, Studer E, Pandak WM, Hu W, Zou T, Wang JY, et al. HIV protease inhibitors increase TNF-α and IL-6 expression in macrophages: Involvement of the RNA-binding protein HuR. Atherosclerosis. 2007;195:e134–e143. doi: 10.1016/j.atherosclerosis.2007.04.008. [DOI] [PubMed] [Google Scholar]